ABSTRACT
Zinc-finger protein 36, CCCH type-like 1 (ZFP36L1), containing tandem CCCH-type zinc-finger motifs with an RNA-binding property, plays an important role in cellular RNA metabolism mainly by RNA decay pathways. Recently, we demonstrated that human ZFP36L1 has potent antiviral activity against influenza A virus infection. However, its role in the host defense response against flaviviruses has not been addressed. Here, we demonstrate that ZFP36L1 functions as a host innate defender against flaviviruses, including Japanese encephalitis virus (JEV) and dengue virus (DENV). Overexpression of ZFP36L1 reduced JEV and DENV infection, and ZFP36L1 knockdown enhanced viral replication. ZFP36L1 destabilized the JEV genome by targeting and degrading viral RNA mediated by both 5′-3′ XRN1 and 3′-5′ RNA-exosome RNA decay pathways. Mutation in both zinc-finger motifs of ZFP36L1 disrupted RNA-binding and antiviral activity. Furthermore, the viral RNA sequences specifically recognized by ZFP36L1 were mapped to the 3′-untranslated region of the JEV genome with the AU-rich element (AUUUA) motif. We extend the function of ZFP36L1 to host antiviral defense by directly binding and destabilizing the viral genome via recruiting cellular mRNA decay machineries.
IMPORTANCE Cellular RNA-binding proteins are among the first lines of defense against various viruses, particularly RNA viruses. ZFP36L1 belongs to the CCCH-type zinc-finger protein family and has RNA-binding activity; it has been reported to bind directly to the AU-rich elements (AREs) of a subset of cellular mRNAs and then lead to mRNA decay by recruiting mRNA-degrading enzymes. However, the antiviral potential of ZFP36L1 against flaviviruses has not yet been fully demonstrated. Here, we reveal the antiviral potential of human ZFP36L1 against Japanese encephalitis virus (JEV) and dengue virus (DENV). ZFP36L1 specifically targeted the ARE motif within viral RNA and triggered the degradation of viral RNA transcripts via cellular degrading enzymes 5′-3′ XRN1 and 3′-5′ RNA exosome. These findings provide mechanistic insights into how human ZFP36L1 serves as a host antiviral factor to restrict flavivirus replication.
KEYWORDS: ZFP36L1, zinc-finger RNA-binding protein, flavivirus, antiviral mechanism
INTRODUCTION
Cellular RNA decay mediated by CCCH-type zinc-finger (ZF) proteins plays a pivotal role in cellular mRNA metabolism as well as in regulating immune responses and in antiviral innate immunity (1–4). The CCCH-type ZF proteins are generally considered RNA-binding proteins, and some are host defense factors against virus infection, such as tristetraprolin (5), monocyte chemoattractant protein 1 (MCP-1)-induced protein 1 (MCPIP1) (6–8), ZF antiviral protein (ZAP) (9–15), tetrachlorodibenzo-p-dioxin-inducible poly(ADP ribose) polymerase (TIPARP) (16), target of Egr1 (17), and the PARP family proteins (18). MCPIP1, ZAP, and TIPARP bind directly to and degrade viral RNA by itself or recruit the cellular mRNA decay machineries. MCPIP1 acts as a host innate defender against virus infection by directly binding and degrading viral RNA transcripts (6–8). ZAP binds to viral RNA via its ZF motifs and then recruits cellular mRNA decay enzymes—5′-3′ exoribonuclease XRN1 and 3′-5′ RNA processing exosome—to degrade RNA molecules from the 5′ end and 3′ end, respectively (11, 12, 15). TIPARP, whose functions are similar to the antiviral activity of ZAP, inhibits Sindbis virus replication by binding viral RNA via its ZF motifs and recruits the 3′-5′ exosome complex for RNA degradation (16).
Human ZF protein 36, CCCH type-like 1 (ZFP36L1; also named TIS11b/BRF-1/ERF-1) belongs to the ZFP36 (also known as tristetraprolin) ZF protein family, which contains two other paralog members, namely, ZFP36 and ZFP36L2 (19). ZFP36 family members are characterized by two CCCH-type ZF motifs that are responsible for binding to the AU-rich element (ARE) in the 3′-untranslated region (UTR) of target cellular mRNA (20, 21). ZFP36L1 triggers ARE-dependent mRNA decay by recruiting the CCR4-NOT deadenylase complex via shortening the 3′-poly(A) tail of the target mRNA and further association with the 5′-3′ XRN1- and 3′-5′ RNA exosome-mediated RNA decay machineries for degrading target mRNAs (22). ZFP36L1 serves as an RNA-binding protein to destabilize target mRNAs. It specifically targets and destabilizes tumor necrosis factor α (TNF-α) (23), interleukin 3 (IL-3) (24), granulocyte-macrophage colony-stimulating factor (25), vascular endothelial growth factor (26, 27), low-density lipoprotein receptor (28), B cell lymphoma 2 (29), cholesterol 7α-hydroxylase (30), transcription factors interferon regulatory factor 8 and Krüppel-like factor 2 (31), Notch1 (32), and heat shock protein 70 family members (33).
Japanese encephalitis virus (JEV) and dengue virus (DENV), members of the Flavivirus genus of the Flaviviridae family, are the major mosquito-borne pathogens that cause hemorrhagic, febrile, and severe encephalitic illnesses in individuals worldwide (34, 35). Flaviviruses are enveloped viruses with a single-stranded, positive-sense RNA genome with a 5′ cap but no 3′ poly(A) tail. Flavivirus RNA flanked with 5′ and 3′ UTRs encodes a polyprotein that is proteolytically processed by cellular and viral proteases into three structural proteins and seven nonstructural proteins for the whole viral life cycle (36). The 5′ and 3′ UTRs of the flaviviral genome contain conserved complementary sequences and form highly conserved secondary and tertiary structures required for viral genome replication and translation (37, 38). We previously showed that the CCCH-type ZF proteins, including ZAP and MCPIP1, provided an innate host defense against flavivirus by diverse mechanisms (6, 39). Nevertheless, the antiviral potential and possible mechanisms of ZFP36L1 against flaviviruses have not been studied. Here, we examined the antiviral potential of human ZFP36L1 and found that ZFP36L1 has potent antiviral activity against JEV and DENV infection by binding directly to the viral RNA genome and recruiting both 5′-3′ XRN1- and 3′-5′ RNA exosome-mediated RNA decay machineries for degradation. Finally, we further demonstrated that the possible binding site of ZFP36L1 is located in the ARE region of the JEV 3′ UTR.
RESULTS
Human ZFP36L1 inhibits JEV and DENV infection.
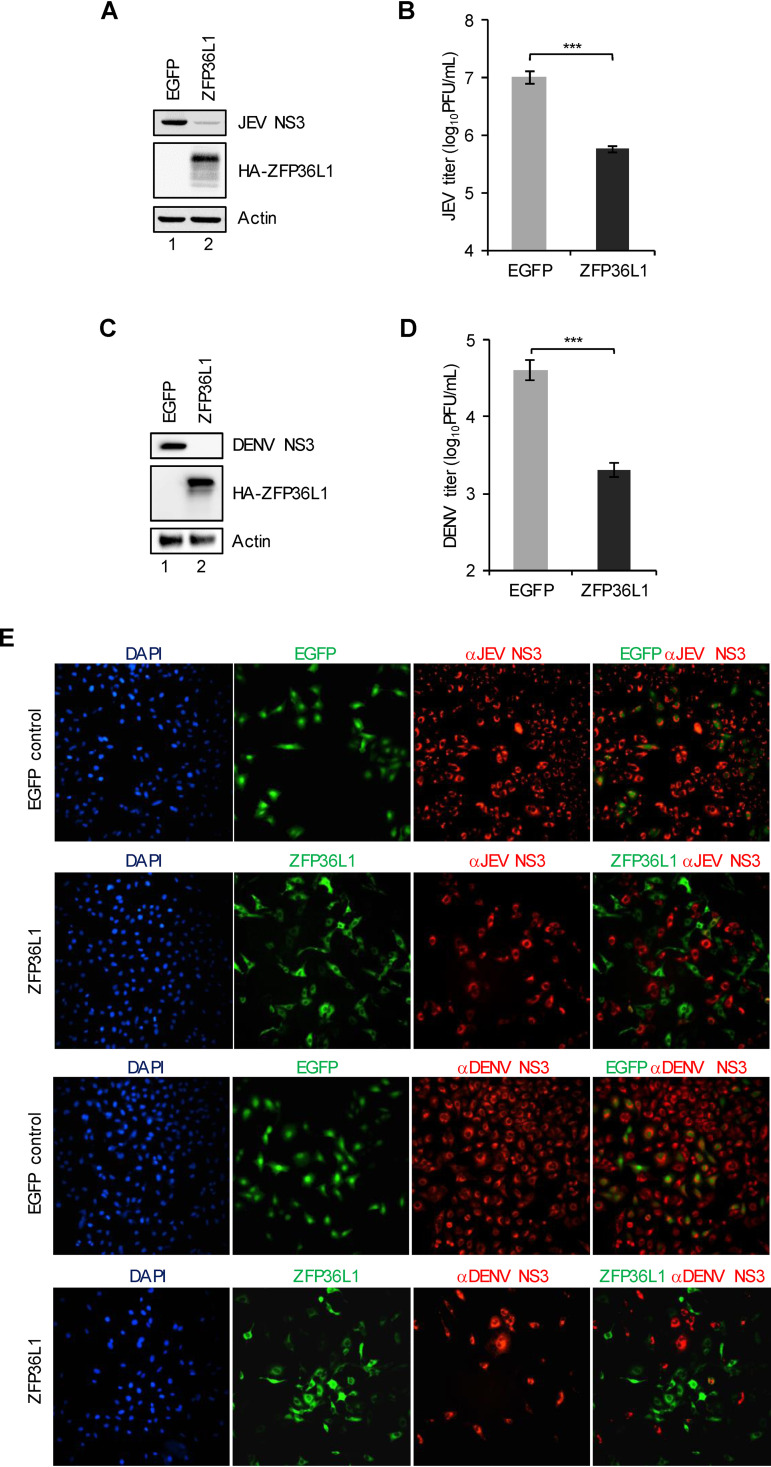
To evaluate the antiviral potential of the human ZFP36L1 protein, A549 cells were transduced with lentiviral vectors to ectopically express HA-tagged ZFP36L1 (HA-ZFP36L1) and control enhanced green fluorescent protein (EGFP). Cells with ZFP36L1 or EGFP overexpression were infected with a high multiplicity of infection (MOI) of JEV and DENV (MOI, 5); virus replication was then analyzed for JEV and DENV NS3 protein expression by conducting a Western blot analysis of the viral NS3 protein (Fig. 1A and C), determining viral titers by plaque-forming assay (Fig. 1B and D), and performing an immunofluorescence assay of the viral NS3 protein (Fig. 1E). At 24 h postinfection, ZFP36L1-overexpressing cells showed a greatly reduced viral NS3 protein level and viral propagation upon JEV and DENV infection (Fig. 1A to D). On immunofluorescence assay, overexpression of ZFP36L1 but not the EGFP control rendered the cells resistant to JEV and DENV infection (Fig. 1E). Thus, the human ZFP36L1 protein has potent antiviral activity against JEV and DENV infection.
FIG 1.
Antiviral activity of human ZFP36L1 against flavivirus infection. A549 cells transduced with lentiviruses expressing EGFP or HA-tagged ZFP36L1 (HA-ZFP36L1) for 72 h were infected with JEV (A and B) and DENV (C and D) (MOI, 5). Cell lysates and culture supernatants were harvested at 24 hpi. (A and C) Cell lysates were used to determine protein levels of viral JEV or DENV NS3 protein, HA-ZFP36L1, and actin by Western blot analysis. (B and D) Culture supernatants were used to determine viral titers by plaque assay. Representative data are shown as mean ± SD (n = 3), and statistical significance was analyzed by the two-tailed Student’s t test. **, P ≤ 0.01. (E) Cells were fixed and permeabilized for an immunofluorescent assay. HA-tagged ZFP36L1 (green)-, viral protein NS3 (red)-, and 4′,6-diamidino-2-phenylindole (DAPI; blue)-stained cells were photographed using a fluorescence microscope.
The ZF motifs of ZFP36L1 are essential for its antiviral activity.
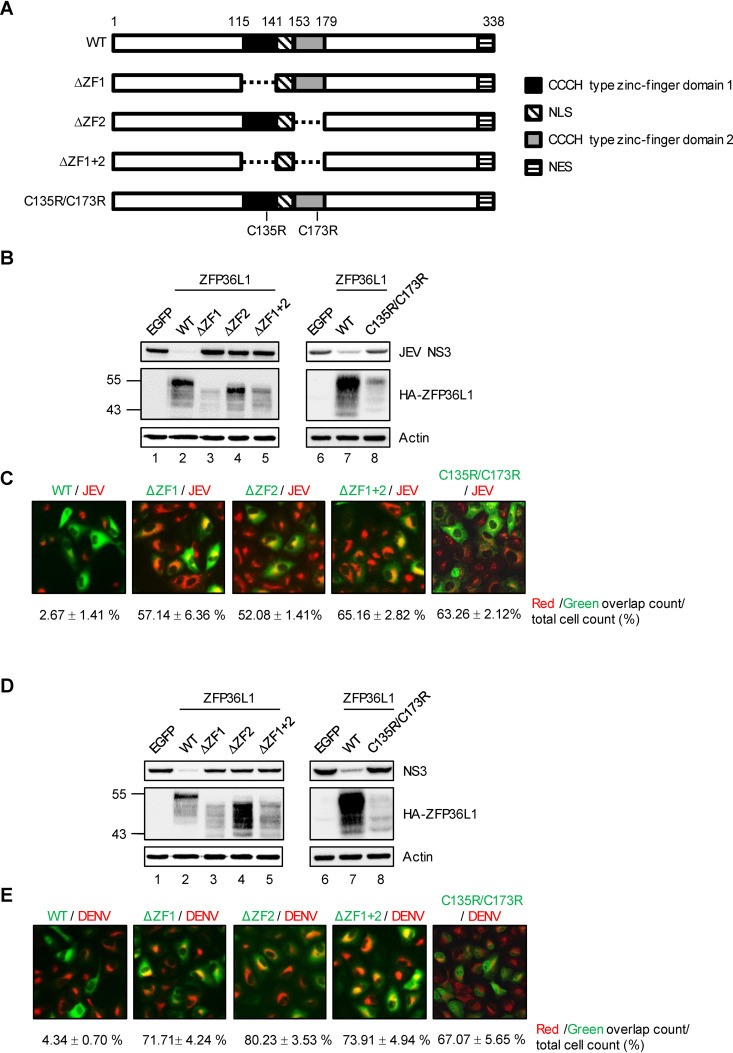
ZFP36L1 has two CCCH-type ZF motifs that are conserved and shared by members of the ZFP36 family and are pivotal for their RNA-binding abilities and biological functions (32, 40, 41). To assess whether the ZF motifs of ZFP36L1 are essential for its anti-flavivirus effects, we generated three ZF-deleted (ΔZF1, ΔZF2, and ΔZF1 + 2) constructs and one ZF mutant C135R/C173R (32) construct (Fig. 2A). We first examined the antiviral activities of these constructs against JEV and DENV infection. By measuring the level of the viral NS3 protein, ZFP36L1 with deleted or mutant ZF greatly reduced antiviral activity compared with wild-type (WT) ZFP36L1 (Fig. 2B and D). This finding was further supported by results of the immunofluorescence assay. Cells with ectopic expression (green signal) of ZFP36L1 with deleted or mutant ZF but not WT ZFP36L1 showed markedly higher viral NS3 protein expression (red signals) (Fig. 2C and E), which indicates the role of ZF motifs of ZFP36L1 against flavivirus infection.
FIG 2.
The CCCH-type zinc-finger motifs of ZFP36L1 are required for its antiviral activity. (A) Schematic representation of wild-type (WT) and zinc-finger motif deletion/mutation constructs of ZFP36L1. The two zinc-finger motifs within ZFP36L1 are shown as solid black and gray boxes. Deletion of the zinc-finger motifs is represented by a dashed line. Point mutations from cysteine (C) to arginine (R) at amino acid (aa) 135 and 173 of ZFP36L1. (B to E) A549 cells overexpressing EGFP, WT ZFP36L1, ZF-deleted (ΔZF1, ΔZF2, and ΔZF1 + 2), ZFP36L1, and ZF-mutated (C135R/C173R) ZFP36L1 by lentiviral transduction were infected with JEV and DENV (MOI, 5) for 24 h. (B and D) Western blot analysis of the expression of viral NS3 protein, HA-ZFP36L1, and actin. (C and E) Immunofluorescence assay of HA-ZFP36L1 and viral NS3 protein stained with anti-HA and anti-JEV/DENV NS3 antibodies. Overlapping red and green signals were quantified by using ImageJ. Data are presented as mean ± SD from two independent experiments.
ZF motifs of ZFP36L1 are required for viral RNA binding.
Next, we assessed the requirement for ZF motifs of ZFP36L1 in viral RNA binding. JEV and DENV-infected 293T/17 cells transfected with HA-tagged WT or C135R/C173R ZFP36L1 and control HA-tagged LacZ-expressing plasmids were processed for RNA immunoprecipitation assay. WT ZFP36L1 immunoprecipitated by anti-HA antibody pulled down JEV and DENV RNA; however, C135R/C173R ZFP36L1 lost its viral RNA binding activity (Fig. 3A and B), which indicates the requirement for ZF motifs of ZFP36L1 in viral RNA binding.
FIG 3.
The CCCH-type zinc-finger motifs of human ZFP36L1 are required for its RNA-binding property. 293T/17 cells were infected with JEV (MOI, 5) for 16 h or DENV (MOI, 5) for 36 h and then transfected with the indicated plasmids expressing HA-ZFP36L1 (WT or C135R/C173R) or HA-LacZ for 16 h. The viral RNA bound to HA-ZFP36L1 and HA-LacZ was pulled down with anti-HA beads and amplified by RT-PCR with JEV or DENV 3′ UTR-specific primers (middle panel). Input RT-PCR shown for RNA levels of JEV or DENV 3′ UTR in virus-infected cells (bottom panel). Western blot analysis of HA-ZFP36L1 (WT and C135R/C173R) and HA-LacZ expression in cell lysates is shown (top panel).
ZFP36L1 restricts JEV replication and destabilizes viral RNA.
To evaluate whether viral replication was affected by ZFP36L1, we used JEV as a model and monitored the viral RNA level in ZFP36L1- and EGFP-overexpressing cells simultaneously infected with JEV. Before 3 h postinfection (hpi), viral RNA was not significantly affected in cells with or without ZFP36L1 overexpression (Fig. 4A), implying that ZFP36L1 might not interfere with viral entry. However, the reduced viral RNA in cells with ZFP36L1 overexpression was noted after 6 hpi and was proportional to the decrease in viral titers (Fig. 4A), so the viral replication was affected by ZFP36L1. To further clarify whether ZFP36L1 influenced JEV RNA stability, we used a replication-dead JEV replicon RNA transcript transfected into 293T/17-ZFP36L1- and -EGFP-overexpressing cells. The replicon RNA level was decreased in cells with ZFP36L1 versus EGFP overexpression (Fig. 4B), which suggests that ZFP36L1 promoted the decrease of JEV viral RNA. We also used a cytomegalovirus (CMV) promoter-driven, replication-dead JEV infectious clone to transfect 293T/17 cells with or without ZFP36L1 overexpression. ZFP36L1 overexpression reduced both viral protein level and viral RNA at 24 hpi (Fig. 4C). Thus, ZFP36L1 can interfere with viral replication by destabilizing viral RNA.
FIG 4.
ZFP36L1 represses JEV replication by impairing viral RNA stability. (A) A549 cells overexpressing EGFP and HA-ZFP36L1 were absorbed with JEV (MOI, 10) on ice for 2 h and then incubated at 37°C. Total RNA was harvested at the indicated times for RT-qPCR. Relative JEV RNA levels were normalized to that of GAPDH and shown as mean ± SD (n = 3). (B) A549 cells were cotransfected with 5′-capped RdRP-dead JEV replicon RNA and control Firefly luciferase RNA for the indicated hours, and total RNA was harvested for RT-qPCR. The relative RNA levels of replicon were normalized to that of Firefly luciferase and shown as mean ± SD (n = 3). Statistical significance was analyzed by the two-tailed Student’s t test. **, P ≤ 0.01; ***, P ≤ 0.001. (C) 293T/17 cells were cotransfected with a CMV promoter-driven, RdRP-dead JEV infectious clone plus HA-ZFP36L1/pcDNA5/TO plasmid or control pcDNA5/TO vector. After 24 h of transfection, cell lysates and total RNA were assayed for viral NS3 protein level and viral genome level by Western blot and RT-PCR analysis, respectively. The relative quantification of JEV NS3 proteins normalized by actin was quantified by using ImageJ.
Both 5′-3′ and 3′-5′ cellular RNA decay pathways are involved in the antiviral activity of ZFP36L1.
ZFP36 family members, including ZFP36, ZFP36L1, and ZFP36L2, are reported to be recruiters to attract the cellular RNA decay machineries for degrading target RNAs, such as the 5′-3′ exonuclease XRN1 and the 3′-5′ RNA exosome complex (22). To determine whether the 5′-3′ and 3′-5′ RNA decay machineries were involved in the antiviral activity of ZP36L1, we depleted the 5′-3′ exoribonuclease XRN1 and 3′-5′ exosome component EXOSC5 in ZFP36L1- and EGFP-overexpressing cells by lentiviral transduction. Compared with shLacZ cells, cells with XRN1 or EXOSC5 knockdown (shXRN1 or shEXOSC5) did not show the anti-JEV effects of ZFP36L1 on viral RNA and viral titers (Fig. 5A to F). However, with ZFP36L1 overexpression, the antiviral activity of ZFP36L1 was lost in XRN1 and EXOSC5 double-knockdown cells (Fig. 5G to I). Therefore, both XRN1- and exosome-mediated RNA decay were involved in the antiviral activity of ZFP36L1.
FIG 5.
The cellular RNA decay pathways are involved in the antiviral activity of ZFP36L1. A549 cells with stable knockdown of LacZ, XRN1, and EXOSC5 by specific shRNA were transduced with lentiviruses expressing EGFP or HA-ZFP36L1. After 24 h of JEV infection (MOI, 5), cell lysates were examined by Western blot analysis for the indicated proteins (A, D, and G). Total RNA and culture supernatants were collected for measuring viral RNA by RT-qPCR (B, E, and H) and determining viral titers by plaque assay (C, F, and I). Relative JEV RNA levels were normalized to that of GAPDH and shown as mean ± SD (n = 3). Representative data analyzed by two-tailed Student’s t test are shown as mean ± SD (n = 3). **, P ≤ 0.01; ***, P ≤ 0.001; ns, not significant.
ZFP36L1 targets the AU-rich elements (AREs) in JEV 3′ UTR.
ZFP36 family proteins recognize the AREs containing overlapping sequences (5′-UUAUUUAUU-3′) within the target transcripts (21, 40, 42, 43). By searching for the minimal ARE pentameric unit AUUUA, the JEV genome is predicted to have two AUUUA sequences located in NS5 (nucleotide [nt.] position 9319 to 9323) and the 3′ UTR (nt. position 10399 to 10403) (Fig. 6A). To examine the potential of the two possible AREs targeted by ZFP36L1, we constructed the reporter plasmid pRL-TK/JNS5 expressing renilla luciferase (Rluc) flanked by the JEV NS5 fragment at the 3′ end. The in vitro-transcribed 5′-capped Rluc/JNS5 plus control Firefly luciferase (Fluc) RNA or 5′-capped Fluc/J3′-UTR RNA (39) plus control Rluc RNA were cotransfected into 293T/17 cells with ZFP36L1 and EGFP overexpression for the dual-luciferase reporter assay. Compared with EGFP-overexpressing cells, ZFP36L1-overexpressing cells did not show reduced luciferase activity of the Rluc reporter RNA with NS5 (Fig. 6B), whereas ZFP36L1-overexpressing cells showed significantly decreased luciferase activity of Fluc reporter RNA with the 3′ UTR (Fig. 6C). These results imply that the ARE recognized by ZFP36L1 was located in the 3′ UTR of the JEV genome. To validate the AUUUA sequence in the JEV 3′ UTR responsible for ZFP36L1 targeting, we generated an ARE mutant Fluc/J3′-UTR reporter (AUUUA→AGGGA). Wild-type ZFP36L1 but not the deleted/mutant (ΔZF2 and C135R/C173R) decreased the luciferase activity of the reporter RNA-containing (AUUUA) ARE in ZFP36L1-overexpressing cells (Fig. 6D). Conversely, wild-type ZFP36L1 had no significant effect on the luciferase activity of the reporter RNA-containing mutant (AGGGA) ARE (Fig. 6D). These results suggest that the ARE in the JEV 3′ UTR is recognized by ZFP36L1 via its ZF motifs.
FIG 6.
Mapping of the ZFP36L1-targeting regions/sequences in the JEV genome. (A) Basic ARE units (AUUUA) in the JEV genome. (B and C) Schematic diagram of the reporter RNAs (top panel). 293T/17 cells overexpressing EGFP and HA-ZFP36L1 were cotransfected with 5′-capped renilla luciferase (Rluc) flanked by JEV NS5 RNA and control Firefly luciferase (Fluc) RNA (bottom panel) (B) or 5′-capped Fluc fused with JEV 3′-UTR RNA and control Rluc RNA bottom panel) (C) for 24 h. Relative luciferase activity was measured by a dual-luciferase reporter assay. (D) The reporter RNAs containing WT or mutant AREs are shown. A solid box indicates wild-type ARE (AUUUA) and open box mutant ARE (AGGGA) of JEV 3′ UTR. 293T/17 cells expressing EGFP, WT, and different zinc-finger-defective forms of ZFP36L1 were cotransfected with 5′-capped Fluc JEV 3′ UTR RNA (WT and mutant) and control Rluc RNA. Relative luciferase activity was measured by a dual-luciferase reporter assay. Data are from three repeated experiments and shown as mean ± SD. Statistical significance was analyzed by a two-tailed Student’s t test. **, P ≤ 0.01; ***, P ≤ 0.001.
Antiviral potential of endogenous ZFP36L1 against JEV and DENV infection.
To address the role of endogenous ZFP36L1 in JEV and DENV infection, A549 cells were infected with JEV and DENV (MOI, 5) at the indicated times, and the protein expression of endogenous ZFP36L1 was determined. The expression of endogenous ZFP36L1 was mainly downregulated in JEV- and DENV-infected cells (Fig. 7A). To further investigate the physiological role of ZFP36L1 upon virus infection, we infected control and ZFP36L1-knockdown A549 cells established by transduction with lentiviral vectors expressing LacZ or ZFP36L1 shRNA (Fig. 7B) with JEV or DENV (MOI, 5) for 24 h. Compared with control shLacZ cells, shZFP36L1 cells with JEV (Fig. 7C and D) and DENV (Fig. 7E and F) infection showed increased viral RNA and viral progeny level, respectively. Furthermore, rescue of the ZFP36L1 knockdown by transduction with a lentiviral vector overexpressing ZFP36L1 significantly decreased the infectious JEV and DENV production (Fig. 7G). These results reveal the intrinsic antiviral potential of human ZFP36L1 against flaviviruses.
FIG 7.
Antiviral potential of endogenous ZFP36L1 against flavivirus infection. (A) A549 cells were mock infected or infected with JEV or DENV (MOI, 5) for the indicated times. Western blot analysis of protein levels of ZFP36L1, viral NS3, and actin as a loading control is shown. The relative quantification of the ZFP36L1 protein level normalized to actin level was quantified by using ImageJ. (B) A549 cells were transduced by lentiviruses containing shRNA targeting LacZ or ZFP36L1. Western blot analysis of protein levels of ZFP36L1 and actin. (C to F) A549-shLacZ and -shZFP36L1 cells were infected with JEV or DENV (MOI, 5) for 24 h and then total RNA and culture supernatants were harvested. (C and E) Relative viral RNA levels normalized to that of GAPDH determined by RT-qPCR. (D and F) Viral titers determined by plaque assay. (G) A549-shLacZ, -shZFP36L1, or -shZFP36L1 cells transduced with a lentiviral vector expressing HA-tagged ZFP36L1 (wobble mutant) were infected with JEV or DENV-2 (MOI, 5) for 24 h and then cell lysates and culture supernatants were harvested. Cell lysates were examined for ZFP36L1 and actin protein levels by Western blot analysis. Viral titers from culture supernatants were determined by plaque assay. Data from three repeated experiments are shown as mean ± SD. Statistical significance was analyzed by two-tailed Student’s t test. *, P ≤ 0.05; **, P ≤ 0.01; ***, P < 0.001.
DISCUSSION
ZFP36L1 is a multifunctional protein involved in a broad range of physiological processes, such as the regulation of inflammatory responses (23, 24, 44), angiogenesis (45), senescence (46), quiescence during B cell development (47), maintenance of marginal zone B cells (31), chorioallantoic fusion (48), skeletal muscle myogenesis (49), osteoarthritis pathogenesis (33), perturbation of thymic development (32), and tumor suppression (29, 32). Recently, we demonstrated that human ZFP36L1 had potent antiviral activity against influenza A virus infection by blocking the translational process of influenza A virus mRNA (50). In this report, we further extend the functions of ZFP36L1 to host defense against flavivirus infection. Overexpression of human ZFP36L1 inhibited JEV and DENV infection, and depletion of endogenous ZFP36L1 enhanced JEV and DENV replication, which shows the intrinsic antiviral potential of ZFP36L1. Different from the related family member ZFP36, which acts as a restriction factor against HIV-1 infection by directly binding and dysregulating HIV-1 viral RNA splicing (5), ZFP36L1 serves as a recruiter and uses cellular mRNA decay machineries and enzymes, either the 5′-3′ exoribonuclease XRN1 or the 3′-5′ RNA exosome complex, to impair the accumulation of target viral RNA.
After binding to target mRNA, mRNA decay mediated by ZFP36 family proteins involves recruiting cellular proteins or complexes, including the deadenylase CCR4-NOT complex, 3′-5′ RNA exosome complex, decapping enzyme Dcp1/2, and 5′-3′ exoribonuclease XRN1 (22, 51). Depletion of XRN1 or the component of RNA exosome (EXOSC5) did not hamper the antiviral activity of ZFP36L1 against flaviviruses (Fig. 5A to F), but knockdown of both XRN1 and EXOSC5 rescued JEV replication in cells with ZFP36L1 overexpression (Fig. 5G to I). These results indicate that ZFP36L1-mediated viral RNA decay can occur in 5′-3′ or 3′-5′ ways, which suggests the complementary cooperation between 5′-3′ XRN1-mediated and 3′-5′ RNA exosome-mediated RNA degradation in the antiviral activity of ZFP36L1.
Posttranscriptional regulation of cellular mRNA by ZFP36L1 plays a pivotal role in controlling gene expression. ZFP36L1 regulates vascular endothelial growth factor expression via mRNA degradation and also translation inhibition (45, 52). Recently, we demonstrated that ZAP specifically targets JEV viral RNA and restricts JEV infection by both translation inhibition and RNA degradation (39). Nevertheless, ZFP36L1 suppresses JEV infection mainly via RNA decay but not translation inhibition. We found that ZFP36L1 exhibits potent antiviral activity against influenza A virus by translational repression of HA, M, and NS mRNA transcripts (50), which suggests that ZFP36L1 can exert antiviral activity by diverse antiviral mechanisms.
The RNA sequence and structure of the mRNA target are important for specific recognition by CCCH-type ZF proteins. For example, ZAP preferentially binds to mRNA targets containing a high frequency of CG dinucleotide plus predicted stem-loop structures (53–55). MCPIP1 preferentially recognizes and cleaves the unpaired regions of microRNAs (miRNAs) around the structure of the terminal loop in a stem-loop (56, 57). ZFP36 family proteins are considered to preferentially recognize the RNA target sequence, a 5′-UUAUUUAUU-3′ nonamer, located in AREs (42, 43). In this study, we screened the RNA sequences of the JEV genome and found two core sequences of AUUUA located in NS5 and 3′ UTR regions. However, ZFP36L1 appeared to target the AUUUA sequence in the 3′ UTR but not the NS5 region of the JEV genome (Fig. 7). Thus, besides a core sequence of AUUUA, whether a specific RNA structure is required for recognition of ZFP36L1 remains to be elucidated.
ZFP36 family proteins are well known to control inflammatory responses. ZFP36, ZFP36L1, and ZFP36L2 are negative regulators of granulocyte-macrophage colony-stimulating factor, TNF-α, IL-2, IL-3, and IL-6 (58). Patients with severe dengue disease and those with acute Japanese encephalitis showed an excessive level of proinflammatory cytokines (i.e., “cytokine storm”), such as TNF-α, IL-6, IL-1β, and MCP-1 (59–61). Therefore, the benefits of host defense by ZFP36L1 are counteracting flavivirus infection but also controlling virus-induced inflammation. Taken together, we first showed ZFP36L1 as an anti-flavivirus cellular factor and provide insight into the molecular antiviral mechanism of ZFP36L1 via its authentic features in RNA binding and decay.
MATERIALS AND METHODS
Viruses, cell lines, and chemicals.
JEV PR-9 strain (GenBank accession AF014161) (62) and DENV serotype 2 (DENV-2) PL046 strain (GenBank accession KJ734727) (63) were propagated in mosquito C6/36 cells (ATCC; CRL-1660) maintained in RPMI 1640 medium supplemented with 5% fetal bovine serum (FBS). JEV and DENV titers was determined by plaque assay in baby hamster kidney (BHK-21) cells (ATCC; CCL-10) grown in RPMI 1640 medium supplemented with 5% FBS, 2 mM l-glutamine, and 1% penicillin-streptomycin (P/S). Human lung carcinoma A549 cells (ATCC; CCL-185) were cultured in F-12 medium supplemented with 10% FBS, 2 mM l-glutamine, and 1% P/S. Human embryonic kidney 293T/17 cells (ATCC; CRL-11268) were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% FBS, 2 mM l-glutamine, and 1% P/S. Lipofectamine 2000 (Invitrogen) was used for DNA and RNA transfection and puromycin (InvivoGen) for stable cell line establishment.
Plasmid constructs, lentivirus generation, and establishment of shRNA-expressing stable cell lines.
The cDNA of human ZFP36L1 (GenBank accession NM_004926) was amplified from A549 cells with the primers 5′-ATGACCACCACCCTCGTGTC-3′ and 5′-TTAGTCATCTGAGATGGAAAGTCTGC-3′ and cloned into the self-inactivating lentiviral vector (pSIN) with an N-terminal hemagglutinin (HA) tag, with the expression of an inserted gene under the control of a constitutive spleen focus-forming virus (SFFV) promoter (64). The lentiviral vector (pSIN) overexpressing ZFP36L1 with wobble position are shown below: GGC AAT AAA ATG CTC AAT TAC at its cDNA (nt. position 58 to 78) (mutation underlined), that encodes an altered mRNA and is resistant to short hairpin RNA (shRNA) silencing in knockdown cells. The CMV promoter-driven, replication-dead JEV infectious clone containing NS5 mutations (GDD→AAG) was generated from a replication-competent JEV infectious clone (65) by single-primer mutagenesis (66) with primers described previously (67). The ZF-deleted ZFP36L1 (ΔZF1, ΔZF2, and ΔZF1 + 2) was generated by single-primer mutagenesis with the ΔZF1 primer (5′-CAGGTCAACTCCAGCCGC/CACGAGCTCCGCAGCC-3′) and ΔZF2 primer (5′-ACCCGCCACCCCAAG/GAAGAGCGCCGTGCCC-3′). The ZF mutant ZFP36L1 (C135R/C173R) was generated by single-primer mutagenesis with the C135R primer (5′-CTACGGGCCCCGCCGCCACTTCATCCACAACGC-3′) and C173R primer (5′-GGAAGCCTGTAAGTACGGGGACAAGCGCCAGTTCGCACACGGC-3′) (mutation underlined). The JEV 5′ + 3′ UTR/pGL3-promoter plasmid for the RNA-based reporter assay was described previously (39). The JEV NS5/pRL-TK plasmid for the RNA-based reporter assay consisted of a renilla luciferase gene flanked by the NS5 sequence (nt. position 7677 to 10391) at its 3′ end downstream of a herpes simplex virus thymidine kinase (HSV TK) promoter. From the National RNAi Core Facility, Taiwan, we obtained lentiviral vectors containing short hairpin RNA (shRNA) targeting specific genes, human ZFP36L1 (TRCN0000013620; 5′-GTAACAAGATGCTCAACTATA-3′), human XRN1 (TRCN0000296739; 5′-GTTACTCACAGGTCGTAAATA-3′), human EXOSC5 (TRCN0000050621; 5′-GTGAAGGTCAGCAAAGAGATT-3′), and LacZ (TRCN0000072223; 5′-CGCGATCGTAATCACCCGAGT-3′). The lentivirus preparation for the expression of the protein or shRNA was as described (68). The knockdown cells were established by transduction with lentiviruses expressing specific shRNAs and were selected with puromycin (10 μg/ml) (68).
Antibodies.
Mouse monoclonal anti-JEV NS3 and anti-DENV NS3 antibodies were described previously (63, 69). The following commercial antibodies were used in this study: anti-actin (catalog no. NB600-501; 1:5,000) and anti-XRN1 (NB500-191; 1:1,000) (Novus Biologicals); anti-HA (C29F4; 1:1,000) and anti-BRF1/2 (2119; 1:1,000) for ZFP36L1 detection (Cell Signaling Technology); and anti-EXOSC5 (ab168804; 1:1,000) (Abcam).
Immunofluorescence assay (IFA).
Cells were fixed by 4% paraformaldehyde in phosphate-buffered saline (PBS) for 20 min, permeabilized with 0.5% Triton X-100 in PBS for 10 min, and blocked with 5% skim milk in PBS for 30 min at room temperature. Cells were incubated with the antibody anti-JEV NS3 (1:100) or anti-DENV NS3 (1:1,000) plus anti-HA antibody (catalog no. C29F4; 1:1,000) overnight at 4°C. After being washed with PBS for three times, cells were incubated with Alexa Fluor 568 goat anti-mouse IgG (A11031; Molecular Probes) for JEV/DENV NS3 staining and Alexa Fluor 488 goat anti-rabbit IgG (A11008; Molecular Probes) for HA-ZFP36L1 (wild-type and deleted/mutant forms) staining for 1 h at room temperature. Next, 4′,6′-diamidino-2-phenylindole (DAPI) (Molecular Probes) was used for nuclei staining. The signal was photographed by fluorescence microscopy (IX71; Olympus).
RNA immunoprecipitation assay.
RNA immunoprecipitation was used to detect the association between ZFP36L1 and viral RNA. Briefly, cells were lysed by radioimmunoprecipitation assay (RIPA) lysis buffer (10 mM Tris-HCl [pH 7.5], 150 mM NaCl, 5 mM EDTA, 0.1% SDS, 1% Triton X-100, and 1% sodium deoxycholate) containing a protease inhibitor cocktail (Roche) and RNasin (Promega) and incubated with EZview red anti-HA affinity gel (E6779; Sigma-Aldrich) for 16 h at 4°C. The antibody-protein-RNA complexes were washed three times with PBS, and the pulldown RNA was extracted by an RNeasy minikit (Qiagen). RT-PCR involved using JEV and DENV 3′ UTR-specific primers described previously (39), 2% agarose gel electrophoresis, and ethidium bromide staining.
Real-time quantitative RT-PCR (RT-qPCR).
Total RNA was extracted by using the RNeasy minikit (Qiagen) and reverse transcribed with a random hexamer primer by using the SuperScript III first-strand synthesis system (Invitrogen). Real-time PCR (qPCR) involved the TaqMan assay and ABI-Prism 7500 system. The relative RNA levels were normalized to that of GAPDH or Firefly luciferase by the comparative threshold cycle (ΔΔCT) method. The TaqMan master mix with UNG (Applied Biosystems), commercial probes for human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Hs02758991) and firefly luciferase (Mr03987587) (Applied Biosystems), and JEV and DENV viral RNA primers previously described (70, 71) were used in qPCRs.
In vitro transcription.
The RdRP-dead JEV replicon DNA with a SP6 promoter was amplified from the SP6-JR2A NS5mt/pBR22 plasmid (67) with primers described previously (39). DNA templates of Firefly luciferase (Fluc), renilla luciferase (Rluc), and Fluc/3′ UTR were acquired from the pGL3-promoter vector, pRL-TK vector, and JEV 5′+3′-UTR/pGL3-promoter plasmid described previously (39). The Rluc/NS5 reporter DNA containing an SP6 promoter was amplified from the JEV NS5/pRL-TK plasmid with the primers 5′-ATTTAGGTGACACTATAGATGACTTCGAAAGTTTATGATCCAG-3′ and 5′-GATGACCCTGTCTTCCTGGATCAAG-3′ (SP6 promoter sequence underlined). The 5′-capped RNA transcripts were in vitro-synthesized by using the cap analog [m7G(5′)ppp(5′)G] (Ambion) and RiboMAX large-scale RNA production system-SP6 (Promega) according to the manufacturer’s instructions.
Luciferase assay.
After transfection with reporter plasmids, cells were lysed by passive lysis buffer (E1941; Promega) then underwent a dual-luciferase reporter assay (E1910; Promega). The results are represented by relative luciferase activity.
Statistical analysis.
The two-tailed Student’s t test was used to estimate the statistical significance between two groups. Representative data from repeated independent experiments are shown as mean ± standard deviation (SD) from triplicate experiments (n = 3). A P value of ≤0.05 was considered statistically significant, as follows: *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; and NS, not significant.
Contributor Information
Yi-Ling Lin, Email: yll@ibms.sinica.edu.tw.
Ren-Jye Lin, Email: linrenjye@nhri.edu.tw.
Stacey Schultz-Cherry, St. Jude Children's Research Hospital.
REFERENCES
- 1.Fu M, Blackshear PJ. 2017. RNA-binding proteins in immune regulation: a focus on CCCH zinc finger proteins. Nat Rev Immunol 17:130–143. 10.1038/nri.2016.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maeda K, Akira S. 2017. Regulation of mRNA stability by CCCH-type zinc-finger proteins in immune cells. Int Immunol 29:149–155. 10.1093/intimm/dxx015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mino T, Takeuchi O. 2018. Post-transcriptional regulation of immune responses by RNA binding proteins. Proc Jpn Acad Ser B Phys Biol Sci 94:248–258. 10.2183/pjab.94.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Turner M, Diaz-Munoz MD. 2018. RNA-binding proteins control gene expression and cell fate in the immune system. Nat Immunol 19:120–129. 10.1038/s41590-017-0028-4. [DOI] [PubMed] [Google Scholar]
- 5.Maeda M, Sawa H, Tobiume M, Tokunaga K, Hasegawa H, Ichinohe T, Sata T, Moriyama M, Hall WW, Kurata T, Takahashi H. 2006. Tristetraprolin inhibits HIV-1 production by binding to genomic RNA. Microbes Infect 8:2647–2656. 10.1016/j.micinf.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 6.Lin RJ, Chien HL, Lin SY, Chang BL, Yu HP, Tang WC, Lin YL. 2013. MCPIP1 ribonuclease exhibits broad-spectrum antiviral effects through viral RNA binding and degradation. Nucleic Acids Res 41:3314–3326. 10.1093/nar/gkt019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin RJ, Chu JS, Chien HL, Tseng CH, Ko PC, Mei YY, Tang WC, Kao YT, Cheng HY, Liang YC, Lin SY. 2014. MCPIP1 suppresses hepatitis C virus replication and negatively regulates virus-induced proinflammatory cytokine responses. J Immunol 193:4159–4168. 10.4049/jimmunol.1400337. [DOI] [PubMed] [Google Scholar]
- 8.Liu S, Qiu C, Miao R, Zhou J, Lee A, Liu B, Lester SN, Fu W, Zhu L, Zhang L, Xu J, Fan D, Li K, Fu M, Wang T. 2013. MCPIP1 restricts HIV infection and is rapidly degraded in activated CD4+ T cells. Proc Natl Acad Sci USA 110:19083–19088. 10.1073/pnas.1316208110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bick MJ, Carroll JW, Gao G, Goff SP, Rice CM, MacDonald MR. 2003. Expression of the zinc-finger antiviral protein inhibits alphavirus replication. J Virol 77:11555–11562. 10.1128/jvi.77.21.11555-11562.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gao G, Guo X, Goff SP. 2002. Inhibition of retroviral RNA production by ZAP, a CCCH-type zinc finger protein. Science 297:1703–1706. 10.1126/science.1074276. [DOI] [PubMed] [Google Scholar]
- 11.Guo X, Carroll JW, Macdonald MR, Goff SP, Gao G. 2004. The zinc finger antiviral protein directly binds to specific viral mRNAs through the CCCH zinc finger motifs. J Virol 78:12781–12787. 10.1128/JVI.78.23.12781-12787.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guo X, Ma J, Sun J, Gao G. 2007. The zinc-finger antiviral protein recruits the RNA processing exosome to degrade the target mRNA. Proc Natl Acad Sci USA 104:151–156. 10.1073/pnas.0607063104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Muller S, Moller P, Bick MJ, Wurr S, Becker S, Gunther S, Kummerer BM. 2007. Inhibition of filovirus replication by the zinc finger antiviral protein. J Virol 81:2391–2400. 10.1128/JVI.01601-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang X, Tu F, Zhu Y, Gao G. 2012. Zinc-finger antiviral protein inhibits XMRV infection. PLoS One 7:e39159. 10.1371/journal.pone.0039159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu Y, Chen G, Lv F, Wang X, Ji X, Xu Y, Sun J, Wu L, Zheng YT, Gao G. 2011. Zinc-finger antiviral protein inhibits HIV-1 infection by selectively targeting multiply spliced viral mRNAs for degradation. Proc Natl Acad Sci USA 108:15834–15839. 10.1073/pnas.1101676108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kozaki T, Komano J, Kanbayashi D, Takahama M, Misawa T, Satoh T, Takeuchi O, Kawai T, Shimizu S, Matsuura Y, Akira S, Saitoh T. 2017. Mitochondrial damage elicits a TCDD-inducible poly(ADP-ribose) polymerase-mediated antiviral response. Proc Natl Acad Sci USA 114:2681–2686. 10.1073/pnas.1621508114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sperandio S, Barat C, Cabrita MA, Gargaun A, Berezovski MV, Tremblay MJ, de Belle I. 2015. TOE1 is an inhibitor of HIV-1 replication with cell-penetrating capability. Proc Natl Acad Sci USA 112:E3392–3401. 10.1073/pnas.1500857112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Atasheva S, Frolova EI, Frolov I. 2014. Interferon-stimulated poly(ADP-ribose) polymerases are potent inhibitors of cellular translation and virus replication. J Virol 88:2116–2130. 10.1128/JVI.03443-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sanduja S, Blanco FF, Dixon DA. 2011. The roles of TTP and BRF proteins in regulated mRNA decay. Wiley Interdiscip Rev RNA 2:42–57. 10.1002/wrna.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Amann BT, Worthington MT, Berg JM. 2003. A Cys3His zinc-binding domain from Nup475/tristetraprolin: a novel fold with a disklike structure. Biochemistry 42:217–221. 10.1021/bi026988m. [DOI] [PubMed] [Google Scholar]
- 21.Hudson BP, Martinez-Yamout MA, Dyson HJ, Wright PE. 2004. Recognition of the mRNA AU-rich element by the zinc finger domain of TIS11d. Nat Struct Mol Biol 11:257–264. 10.1038/nsmb738. [DOI] [PubMed] [Google Scholar]
- 22.Lykke-Andersen J, Wagner E. 2005. Recruitment and activation of mRNA decay enzymes by two ARE-mediated decay activation domains in the proteins TTP and BRF-1. Genes Dev 19:351–361. 10.1101/gad.1282305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lai WS, Carballo E, Thorn JM, Kennington EA, Blackshear PJ. 2000. Interactions of CCCH zinc finger proteins with mRNA. Binding of tristetraprolin-related zinc finger proteins to Au-rich elements and destabilization of mRNA. J Biol Chem 275:17827–17837. 10.1074/jbc.M001696200. [DOI] [PubMed] [Google Scholar]
- 24.Stoecklin G, Colombi M, Raineri I, Leuenberger S, Mallaun M, Schmidlin M, Gross B, Lu M, Kitamura T, Moroni C. 2002. Functional cloning of BRF1, a regulator of ARE-dependent mRNA turnover. EMBO J 21:4709–4718. 10.1093/emboj/cdf444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carballo E, Lai WS, Blackshear PJ. 2000. Evidence that tristetraprolin is a physiological regulator of granulocyte-macrophage colony-stimulating factor messenger RNA deadenylation and stability. Blood 95:1891–1899. 10.1182/blood.V95.6.1891. [DOI] [PubMed] [Google Scholar]
- 26.Hacker C, Valchanova R, Adams S, Munz B. 2010. ZFP36L1 is regulated by growth factors and cytokines in keratinocytes and influences their VEGF production. Growth Factors 28:178–190. 10.3109/08977190903578660. [DOI] [PubMed] [Google Scholar]
- 27.Prenzler F, Fragasso A, Schmitt A, Munz B. 2016. Functional analysis of ZFP36 proteins in keratinocytes. Eur J Cell Biol 95:277–284. 10.1016/j.ejcb.2016.04.007. [DOI] [PubMed] [Google Scholar]
- 28.Adachi S, Homoto M, Tanaka R, Hioki Y, Murakami H, Suga H, Matsumoto M, Nakayama KI, Hatta T, Iemura S, Natsume T. 2014. ZFP36L1 and ZFP36L2 control LDLR mRNA stability via the ERK-RSK pathway. Nucleic Acids Res 42:10037–10049. 10.1093/nar/gku652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zekavati A, Nasir A, Alcaraz A, Aldrovandi M, Marsh P, Norton JD, Murphy JJ. 2014. Post-transcriptional regulation of BCL2 mRNA by the RNA-binding protein ZFP36L1 in malignant B cells. PLoS One 9:e102625. 10.1371/journal.pone.0102625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tarling EJ, Clifford BL, Cheng J, Morand P, Cheng A, Lester E, Sallam T, Turner M, de Aguiar Vallim TQ. 2017. RNA-binding protein ZFP36L1 maintains posttranscriptional regulation of bile acid metabolism. J Clin Invest 127:3741–3754. 10.1172/JCI94029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Newman R, Ahlfors H, Saveliev A, Galloway A, Hodson DJ, Williams R, Besra GS, Cook CN, Cunningham AF, Bell SE, Turner M. 2017. Maintenance of the marginal-zone B cell compartment specifically requires the RNA-binding protein ZFP36L1. Nat Immunol 18:683–693. 10.1038/ni.3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hodson DJ, Janas ML, Galloway A, Bell SE, Andrews S, Li CM, Pannell R, Siebel CW, MacDonald HR, De Keersmaecker K, Ferrando AA, Grutz G, Turner M. 2010. Deletion of the RNA-binding proteins ZFP36L1 and ZFP36L2 leads to perturbed thymic development and T lymphoblastic leukemia. Nat Immunol 11:717–724. 10.1038/ni.1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Son YO, Kim HE, Choi WS, Chun CH, Chun JS. 2019. RNA-binding protein ZFP36L1 regulates osteoarthritis by modulating members of the heat shock protein 70 family. Nat Commun 10:77. 10.1038/s41467-018-08035-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gubler DJ. 2002. Epidemic dengue/dengue hemorrhagic fever as a public health, social and economic problem in the 21st century. Trends Microbiol 10:100–103. 10.1016/s0966-842x(01)02288-0. [DOI] [PubMed] [Google Scholar]
- 35.Solomon T. 2003. Recent advances in Japanese encephalitis. J Neurovirol 9:274–283. 10.1080/13550280390194037. [DOI] [PubMed] [Google Scholar]
- 36.Fernandez-Garcia MD, Mazzon M, Jacobs M, Amara A. 2009. Pathogenesis of flavivirus infections: using and abusing the host cell. Cell Host Microbe 5:318–328. 10.1016/j.chom.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 37.Selisko B, Wang C, Harris E, Canard B. 2014. Regulation of Flavivirus RNA synthesis and replication. Curr Opin Virol 9:74–83. 10.1016/j.coviro.2014.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brinton MA, Basu M. 2015. Functions of the 3′ and 5′ genome RNA regions of members of the genus Flavivirus. Virus Res 206:108–119. 10.1016/j.virusres.2015.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chiu HP, Chiu H, Yang CF, Lee YL, Chiu FL, Kuo HC, Lin RJ, Lin YL. 2018. Inhibition of Japanese encephalitis virus infection by the host zinc-finger antiviral protein. PLoS Pathog 14:e1007166. 10.1371/journal.ppat.1007166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lai WS, Kennington EA, Blackshear PJ. 2002. Interactions of CCCH zinc finger proteins with mRNA: non-binding tristetraprolin mutants exert an inhibitory effect on degradation of AU-rich element-containing mRNAs. J Biol Chem 277:9606–9613. 10.1074/jbc.M110395200. [DOI] [PubMed] [Google Scholar]
- 41.Morgan BR, Massi F. 2010. A computational study of RNA binding and specificity in the tandem zinc finger domain of TIS11d. Protein Sci 19:1222–1234. 10.1002/pro.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zubiaga AM, Belasco JG, Greenberg ME. 1995. The nonamer UUAUUUAUU is the key AU-rich sequence motif that mediates mRNA degradation. Mol Cell Biol 15:2219–2230. 10.1128/MCB.15.4.2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Worthington MT, Pelo JW, Sachedina MA, Applegate JL, Arseneau KO, Pizarro TT. 2002. RNA binding properties of the AU-rich element-binding recombinant Nup475/TIS11/tristetraprolin protein. J Biol Chem 277:48558–48564. 10.1074/jbc.M206505200. [DOI] [PubMed] [Google Scholar]
- 44.Wang KT, Wang HH, Wu YY, Su YL, Chiang PY, Lin NY, Wang SC, Chang GD, Chang CJ. 2015. Functional regulation of Zfp36l1 and Zfp36l2 in response to lipopolysaccharide in mouse RAW264.7 macrophages. J Inflamm (Lond) 12:42. 10.1186/s12950-015-0088-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ciais D, Cherradi N, Bailly S, Grenier E, Berra E, Pouyssegur J, Lamarre J, Feige JJ. 2004. Destabilization of vascular endothelial growth factor mRNA by the zinc-finger protein TIS11b. Oncogene 23:8673–8680. 10.1038/sj.onc.1207939. [DOI] [PubMed] [Google Scholar]
- 46.Herranz N, Gallage S, Mellone M, Wuestefeld T, Klotz S, Hanley CJ, Raguz S, Acosta JC, Innes AJ, Banito A, Georgilis A, Montoya A, Wolter K, Dharmalingam G, Faull P, Carroll T, Martinez-Barbera JP, Cutillas P, Reisinger F, Heikenwalder M, Miller RA, Withers D, Zender L, Thomas GJ, Gil J. 2015. mTOR regulates MAPKAPK2 translation to control the senescence-associated secretory phenotype. Nat Cell Biol 17:1205–1217. 10.1038/ncb3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Galloway A, Saveliev A, Łukasiak S, Hodson DJ, Bolland D, Balmanno K, Ahlfors H, Monzón-Casanova E, Mannurita SC, Bell LS, Andrews S, Díaz-Muñoz MD, Cook SJ, Corcoran A, Turner M. 2016. RNA-binding proteins ZFP36L1 and ZFP36L2 promote cell quiescence. Science 352:453–459. 10.1126/science.aad5978. [DOI] [PubMed] [Google Scholar]
- 48.Stumpo DJ, Byrd NA, Phillips RS, Ghosh S, Maronpot RR, Castranio T, Meyers EN, Mishina Y, Blackshear PJ. 2004. Chorioallantoic fusion defects and embryonic lethality resulting from disruption of Zfp36L1, a gene encoding a CCCH tandem zinc finger protein of the Tristetraprolin family. Mol Cell Biol 24:6445–6455. 10.1128/MCB.24.14.6445-6455.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bye AJH, Pugazhendhi D, Woodhouse S, Brien P, Watson R, Turner M, Pell J. 2018. The RNA-binding proteins Zfp36l1 and Zfp36l2 act redundantly in myogenesis. Skelet Muscle 8:37. 10.1186/s13395-018-0183-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lin RJ, Huang CH, Liu PC, Lin IC, Huang YL, Chen AY, Chiu HP, Shih SR, Lin LH, Lien SP, Yen LC, Liao CL. 2020. Zinc finger protein ZFP36L1 inhibits influenza A virus through translational repression by targeting HA, M and NS RNA transcripts. Nucleic Acids Res 48:7371–7384. 10.1093/nar/gkaa458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hau HH, Walsh RJ, Ogilvie RL, Williams DA, Reilly CS, Bohjanen PR. 2007. Tristetraprolin recruits functional mRNA decay complexes to ARE sequences. J Cell Biochem 100:1477–1492. 10.1002/jcb.21130. [DOI] [PubMed] [Google Scholar]
- 52.Bell SE, Sanchez MJ, Spasic-Boskovic O, Santalucia T, Gambardella L, Burton GJ, Murphy JJ, Norton JD, Clark AR, Turner M. 2006. The RNA binding protein Zfp36l1 is required for normal vascularisation and post-transcriptionally regulates VEGF expression. Dev Dyn 235:3144–3155. 10.1002/dvdy.20949. [DOI] [PubMed] [Google Scholar]
- 53.Meagher JL, Takata M, Goncalves-Carneiro D, Keane SC, Rebendenne A, Ong H, Orr VK, MacDonald MR, Stuckey JA, Bieniasz PD, Smith JL. 2019. Structure of the zinc-finger antiviral protein in complex with RNA reveals a mechanism for selective targeting of CG-rich viral sequences. Proc Natl Acad Sci USA 116:24303–24309. 10.1073/pnas.1913232116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Takata MA, Goncalves-Carneiro D, Zang TM, Soll SJ, York A, Blanco-Melo D, Bieniasz PD. 2017. CG dinucleotide suppression enables antiviral defence targeting non-self RNA. Nature 550:124–127. 10.1038/nature24039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huang Z, Wang X, Gao G. 2010. Analyses of SELEX-derived ZAP-binding RNA aptamers suggest that the binding specificity is determined by both structure and sequence of the RNA. Protein Cell 1:752–759. 10.1007/s13238-010-0096-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wilamowski M, Gorecki A, Dziedzicka-Wasylewska M, Jura J. 2018. Substrate specificity of human MCPIP1 endoribonuclease. Sci Rep 8:7381. 10.1038/s41598-018-25765-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li M, Cao W, Liu H, Zhang W, Liu X, Cai Z, Guo J, Wang X, Hui Z, Zhang H, Wang J, Wang L. 2012. MCPIP1 down-regulates IL-2 expression through an ARE-independent pathway. PLoS One 7:e49841. 10.1371/journal.pone.0049841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Baou M, Jewell A, Murphy JJ. 2009. TIS11 family proteins and their roles in posttranscriptional gene regulation. BioMed Res Int 2009:1–11. 10.1155/2009/634520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ravi V, Parida S, Desai A, Chandramuki A, Gourie-Devi M, Grau GE. 1997. Correlation of tumor necrosis factor levels in the serum and cerebrospinal fluid with clinical outcome in Japanese encephalitis patients. J Med Virol 51:132–136. . [DOI] [PubMed] [Google Scholar]
- 60.Winter PM, Dung NM, Loan HT, Kneen R, Wills B, Thu l.T, House D, White NJ, Farrar JJ, Hart CA, Solomon T. 2004. Proinflammatory cytokines and chemokines in humans with Japanese encephalitis. J Infect Dis 190:1618–1626. 10.1086/423328. [DOI] [PubMed] [Google Scholar]
- 61.Malavige GN, Huang LC, Salimi M, Gomes L, Jayaratne SD, Ogg GS. 2012. Cellular and cytokine correlates of severe dengue infection. PLoS One 7:e50387. 10.1371/journal.pone.0050387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen LK, Lin YL, Liao CL, Lin CG, Huang YL, Yeh CT, Lai SC, Jan JT, Chin C. 1996. Generation and characterization of organ-tropism mutants of Japanese encephalitis virus in vivo and in vitro. Virology 223:79–88. 10.1006/viro.1996.0457. [DOI] [PubMed] [Google Scholar]
- 63.Lin YL, Liao CL, Chen LK, Yeh CT, Liu CI, Ma SH, Huang YY, Huang YL, Kao CL, King CC. 1998. Study of Dengue virus infection in SCID mice engrafted with human K562 cells. J Virol 72:9729–9737. 10.1128/JVI.72.12.9729-9737.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Clements MO, Godfrey A, Crossley J, Wilson SJ, Takeuchi Y, Boshoff C. 2006. Lentiviral manipulation of gene expression in human adult and embryonic stem cells. Tissue Eng 12:1741–1751. 10.1089/ten.2006.12.1741. [DOI] [PubMed] [Google Scholar]
- 65.Liang JJ, Liao CL, Liao JT, Lee YL, Lin YL. 2009. A Japanese encephalitis virus vaccine candidate strain is attenuated by decreasing its interferon antagonistic ability. Vaccine 27:2746–2754. 10.1016/j.vaccine.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 66.Makarova O, Kamberov E, Margolis B. 2000. Generation of deletion and point mutations with one primer in a single cloning step. Biotechniques 29:970–972. 10.2144/00295bm08. [DOI] [PubMed] [Google Scholar]
- 67.Chien HL, Liao CL, Lin YL. 2011. FUSE binding protein 1 interacts with untranslated regions of Japanese encephalitis virus RNA and negatively regulates viral replication. J Virol 85:4698–4706. 10.1128/JVI.01950-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lin RJ, Yu HP, Chang BL, Tang WC, Liao CL, Lin YL. 2009. Distinct antiviral roles for human 2′,5′-oligoadenylate synthetase family members against dengue virus infection. J Immunol 183:8035–8043. 10.4049/jimmunol.0902728. [DOI] [PubMed] [Google Scholar]
- 69.Chen LK, Liao CL, Lin CG, Lai SC, Liu CI, Ma SH, Huang YY, Lin YL. 1996. Persistence of Japanese encephalitis virus is associated with abnormal expression of the nonstructural protein NS1 in host cells. Virology 217:220–229. 10.1006/viro.1996.0109. [DOI] [PubMed] [Google Scholar]
- 70.Kao YT, Chang BL, Liang JJ, Tsai HJ, Lee YL, Lin RJ, Lin YL. 2015. Japanese encephalitis virus nonstructural protein NS5 interacts with mitochondrial trifunctional protein and impairs fatty acid beta-oxidation. PLoS Pathog 11:e1004750. 10.1371/journal.ppat.1004750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tang WC, Lin RJ, Liao CL, Lin YL. 2014. Rab18 facilitates dengue virus infection by targeting fatty acid synthase to sites of viral replication. J Virol 88:6793–6804. 10.1128/JVI.00045-14. [DOI] [PMC free article] [PubMed] [Google Scholar]