ABSTRACT
Over the past 20 years, the severe acute respiratory syndrome coronavirus (SARS-CoV), Middle East respiratory syndrome CoV (MERS-CoV), and SARS-CoV-2 emerged, causing severe human respiratory diseases throughout the globe. Developing broad-spectrum drugs would be invaluable in responding to new, emerging coronaviruses and to address unmet urgent clinical needs. Main protease (Mpro; also known as 3CLpro) has a major role in the coronavirus life cycle and is one of the most important targets for anti-coronavirus agents. We show that a natural product, noncovalent inhibitor, shikonin, is a pan-main protease inhibitor of SARS-CoV-2, SARS-CoV, MERS-CoV, human coronavirus (HCoV)-HKU1, HCoV-NL63, and HCoV-229E with micromolar half maximal inhibitory concentration (IC50) values. Structures of the main protease of different coronavirus genus, SARS-CoV from the betacoronavirus genus and HCoV-NL63 from the alphacoronavirus genus, were determined by X-ray crystallography and revealed that the inhibitor interacts with key active site residues in a unique mode. The structure of the main protease inhibitor complex presents an opportunity to discover a novel series of broad-spectrum inhibitors. These data provide substantial evidence that shikonin and its derivatives may be effective against most coronaviruses as well as emerging coronaviruses of the future. Given the importance of the main protease for coronavirus therapeutic indication, insights from these studies should accelerate the development and design of safer and more effective antiviral agents.
IMPORTANCE The current pandemic has created an urgent need for broad-spectrum inhibitors of SARS-CoV-2. The main protease is relatively conservative compared to the spike protein and, thus, is one of the most promising targets in developing anti-coronavirus agents. We solved the crystal structures of the main protease of SARS-CoV and HCoV-NL63 that bound to shikonin. The structures provide important insights, have broad implications for understanding the structural basis underlying enzyme activity, and can facilitate rational design of broad-spectrum anti-coronavirus ligands as new therapeutic agents.
KEYWORDS: broad-spectrum, coronavirus, inhibitor, main protease, nature product
INTRODUCTION
To date, there are seven coronaviruses in nature that infect humans, of which the severe acute respiratory syndrome coronavirus (SARS-CoV) in 2002, the Middle East respiratory syndrome coronavirus (MERS-CoV) in 2012, and the SARS-CoV-2 in 2019 can cause severe human respiratory diseases (1–4). The potential threat to global public health as well as the lack of drugs and therapies has driven drug development and optimization campaigns for safer and more effective antiviral agents since the global pandemic caused by coronavirus (5–7). More alarmingly, SARS-like CoVs currently circulating in animal reservoirs can replicate in human airway cells and cause in vivo pathogenesis (8). The outbreak of COVID-19 foreshadows a potential risk of coronavirus reemergence. More efficient and broad-spectrum antiviral drugs are urgently needed for future emerging coronaviruses outbreaks in humans.
The main protease (Mpro; also known as 3CLpro) and the RNA-dependent RNA polymerase (RdRp) are the most attractive small molecule targets in the viral life cycle. At present, only one small molecule inhibitor, remdesivir, that targets the RdRp polymerase and showed broad-spectrum antiviral activity against human coronaviruses has been approved with emergency use authorization by the Food and Drug Administration (FDA) in the United States (9). The main protease, Mpro, has a major role in the replication of coronavirus life cycle and is one of the most important targets for anti-coronavirus agents (10, 11). Despite the introduction of potent, broad-spectrum, peptidomimetic inhibitors like α-ketoamides and N3, developing antiviral drugs without the drawbacks of classic covalent inhibitors has remained an elusive goal.
Natural products act as important sources of therapeutic agents and are regarded as the cornerstone of modern drug discovery and development. To date, few broad-spectrum, natural products are available for the inhibition of human coronaviruses. Given that Mpro is of great potential for the treatment of SARS-CoV-2, SARS-CoV, and MERS-CoV, Mpro inhibitors have therapeutic promise. Several peptidomimetic, covalent inhibitor structures of Mpro have been reported (12–15). Crystal structures of SARS-CoV-2 Mpro with three noncanonical Mpro inhibitors have also been solved (carmofur, baicailen, shikonin) (16–18). In this study, we aim to find broad-spectrum, natural product inhibitors that target the Mpro of coronavirus using a structure-based approach. We found that shikonin has broad-spectrum Mpro inhibition for six human CoVs that infect humans. To better understand the molecular basis for shikonin interactions with the different coronaviruses, we solved the structures of the Mpro of SARS-CoV from the betacoronavirus genus and HCoV-NL63 from the alphacoronavirus genus. We elucidated subtype-selectivity by clarifying the binding mode of the noncovalent inhibitor shikonin with the main proteases of SARS-CoV and HCoV-NL63. These insights should facilitate the design of antivirals for the treatment of coronavirus-associated diseases.
RESULTS
Structure-based docking and discovery of broad-spectrum compounds.
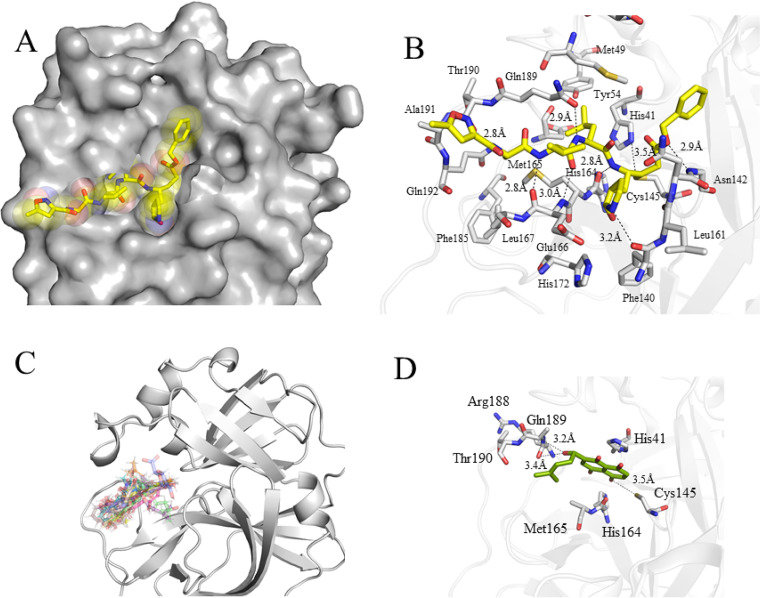
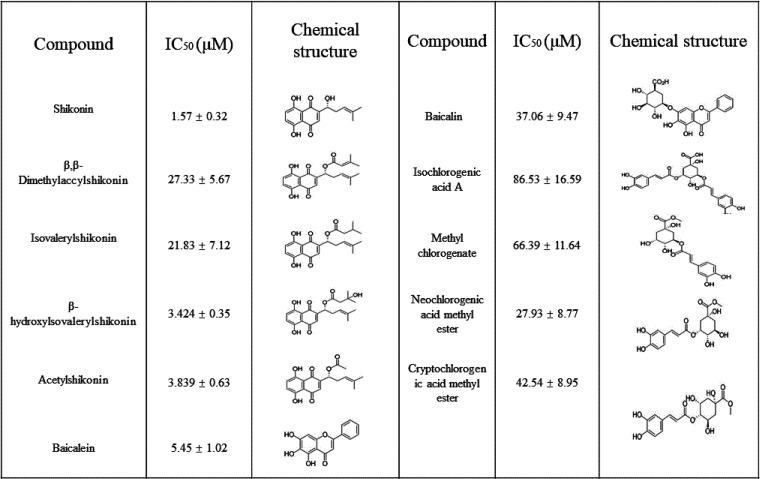
To combat COVID-19, Glide SP mode and Glide XP mode were used to screen an in-house library consisting of a database of available small molecules, including natural products, traditional Chinese medicine monomers, and a synthetic compound library. The determination of the crystal structure of the main protease of SARS-CoV-2, SARS-CoV, and MERS-CoV provides an opportunity to seek a new scaffold of small molecules by structure-based approaches (13, 19–21). We targeted the Mpro of SARS-CoV-2 that was bound to N3 for docking to seek broad-spectrum ligands and new chemotypes (13) (Fig. 1A and B). N3 is a covalent and broad-spectrum inhibitor that has been demonstrated to have inhibitory activity against the main protease of SARS-CoV-2, SARS-CoV, MERS-CoV, HCoV-HKU1, HCoV-NL63, and HCoV-229E. The chemical structure of N3 is similar to that of the substrate of the main protease. Given that N3 shows broad-spectrum inhibition against the main protease of coronaviruses, we chose the N3 binding site as the docking model for structure-based drug screening. This structure was used as a template to screen a combined, in-house natural products database of 216,580 using a molecular docking calculation. To identify a broad-spectrum small molecule, we screened a panel of nature products for inhibitory activity against the binding pocket of Mpro of inactive SARS-CoV-2, beginning with the consensus structure of different inhibitor-bound structures (Fig. 1B). We ordered and tested 220 top-scoring compounds for their inhibitory activity against Mpro using a fluorescence resonance energy transfer (FRET) assay. Naphthoquinone, chlorogenic acid, and baicalin derivatives exhibited better enzyme inhibition activity than other compounds (Table 1). The inhibitory effects of naphthoquinone analogues were moderate with IC50 values ranging from 1.57 μM to 86.53 μM (Table 1). Although some compounds, such as tubuloside A and maltopentaose, showed a high score in the docking analysis, none of them showed potent inhibition against Mpro.
FIG 1.
Structure-based compound discovery for the main protease of SARS-CoV-2. (A) N3 binding site of SARS-CoV-2-Mpro. Structure of SARS-CoV-2 Mpro is shown as gray sphere. N3 is presented as yellow sticks. (B) Close-up view of the N3 binding pocket. N3 recognition region is conserved between SARS-CoV Mpro and SARS-CoV-2 Mpro. Hydrogen bonds are shown as dashed lines. (C) Overlaid docking poses of 17 compounds selected for experimental testing. (D) Shikonin binding site of crystal structure of SARS-CoV-2 Mpro. Shikonin is shown as green sticks. Hydrogen bonds are shown as dashed lines.
TABLE 1.
Compounds with SARS-CoV-2 Mpro activity identified in the initial screen
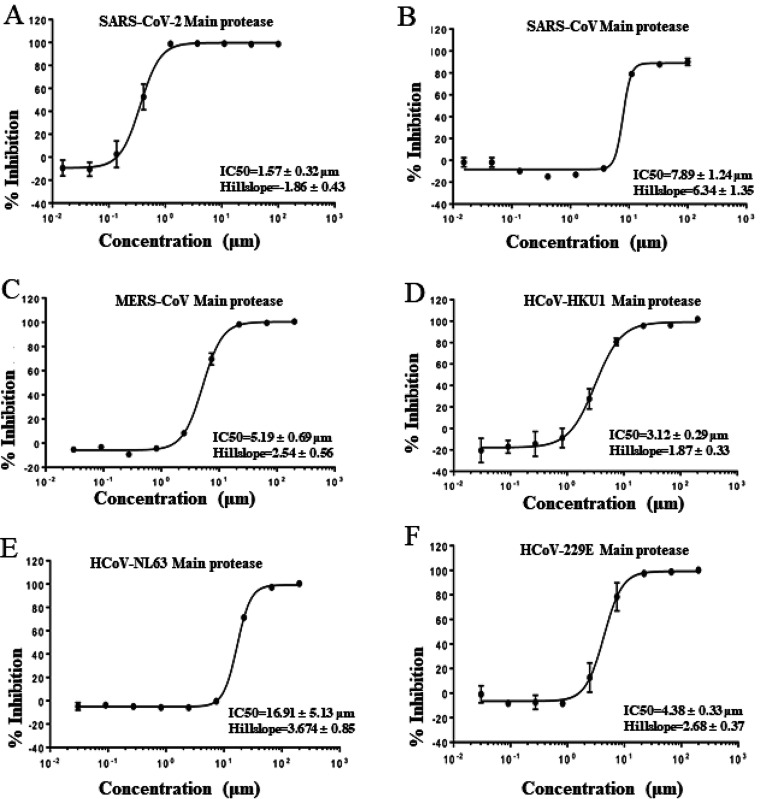
There is high sequence conservation in the main proteases across seven human CoVs (3, 22) (Fig. S1). For example, the main protease of SARS-CoV-2 has 98% similarity and 95% amino acid (aa) identity to that of SARS-CoV (Fig. S1). To test the broad-spectrum activity of shikonin against coronaviruses, we purified six coronavirus main proteases except for HCoV-OC43. The shikonin of naphthoquinone derivatives was effective against the main proteases of SARS-CoV-2, SARS-CoV, MERS-CoV, HCoV-HKU1, HCoV-NL63, and HCoV-229E with IC50s ranging from 1.57 μM to 16.91 μM under our FRET assay conditions. Shikonin showed similar activity against SARS-CoV-2, SARS-CoV, MERS, HCoV-HKU1, and HCoV-229E proteases but was substantially less effective against the HCoV-NL63 protease (Fig. 2).
FIG 2.
Inhibition of shikonin on the recombinant Mpro of SARS-CoV-2 (A), SARS-CoV (B), MERS-CoV (C), HCoV-HKU1 (D), HCoV-NL63 (E), and HCoV-229E (F) against the inhibitor shikonin. IC50 is given as mean ± standard deviation. The values are the mean ±standard deviation from three replicates. The main proteases from different coronaviruses were preincubated in the reaction buffer (50 mM Tris 7.3,150mM NaCl, 1 mM EDTA) with various concentrations of shikonin at room temperature for 30 min. The enzymatic reaction was initiated by adding the FRET substrate. The IC50 of shikonin was evaluated using the a dose-response curve in GraphPad Prism as described in Materials and Methods.
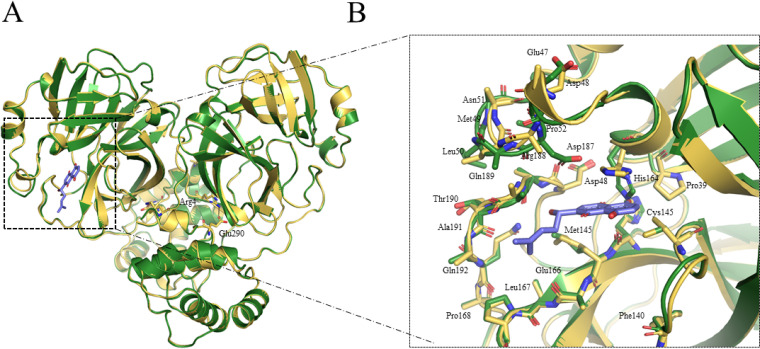
Structure of apo and shikonin with the Mpro of SARS-CoV.
The SARS-CoV Mpro with shikonin forms a dimer in the crystal and has three distinct dimer interfaces that were similar to that observed for the apo crystal form (Fig. 3). These two Mpro protomers had similar configurations, and the root mean square (rms) deviation was ∼0.02 Å for all equivalent Cα atoms. Three key residues (Arg4, Glu290, and Arg298) were involved in the monomer-monomer interface. A conserved Arg4-Glu290 salt bridge was found in the apo state of SARS-CoV Mpro (Fig. 3A). Mutational studies of SARS have identified that these residues lead to the dissociation of the dimer (23). Dimerization and oligomerization modulated various protease functions, such as enzymatic activity, cooperativity, and activation. Although the structure revealed here shares similar overall structure with other published structures, there are several key differences that highlight potential features that could be exploited (Fig. 3A). The catalytic dyad His41-Cys145 underwent dramatic conformational changes, and the structure revealed an unusual arrangement of oxyanion loop stabilized by the substrate. The most intriguing difference was observed in the catalytic dyad His41, which underwent a conformational change to accommodate π-π stacking interaction with the naphthoquinone ring of shikonin (Fig. 3B). Another large difference was found in a flexible loop of the protease, including Cys44 to Tyr54, Asp187 to Ala191, and Leu141 to Ser144, which were not located in the dimerization region and were irrelevant to crystal packing (Fig. 3B). These loops are relative flexible and involved in the substrate interaction and protease acidity (24).
FIG 3.
Crystal structures in apo form of SARS-CoV Mpro and SARS-CoV-2 Mpro in complex with shikonin. (A) Comparison of overall structures in apo form of SARS-CoV Mpro- and SARS-CoV-2 Mpro-shikonin. Structure in apo form of SARS-CoV Mpro is shown in green. Structure of Mpro with shikonin is shown in yellow. The shikonin is shown in blue. Carbon atoms of shikonin are blue, and oxygen atoms are red. (B) A zoomed in view of shikonin binding pocket for SARS-CoV Mpro.
Comparison of structures of SARS-CoV-2 Mpro, SARS-CoV Mpro, and HCoV-NL63 Mpro in complex with shikonin.
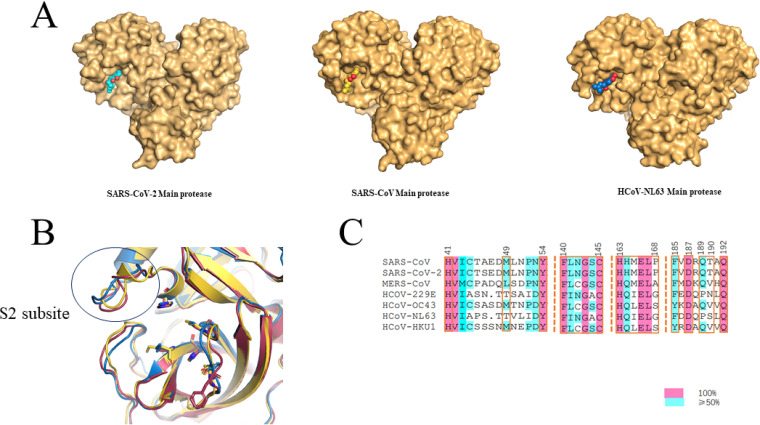
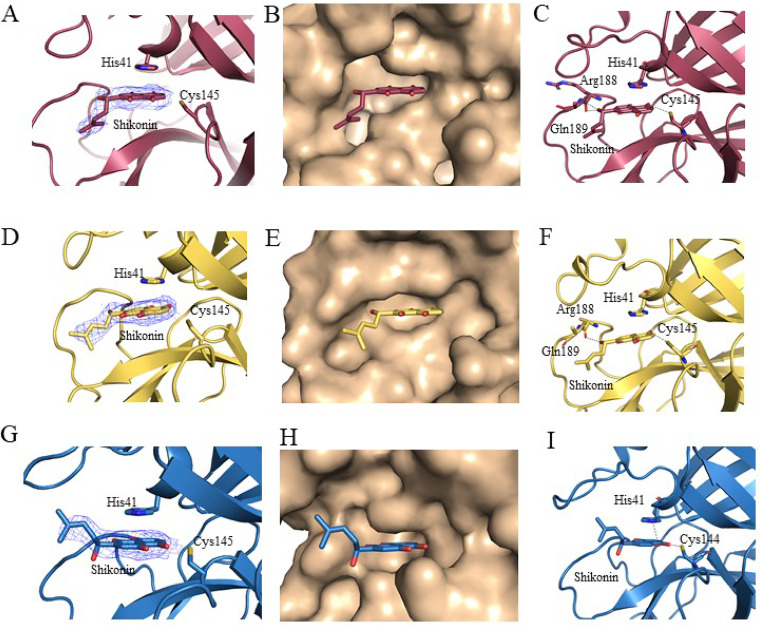
The Mpro of SARS-CoV and HCoV-NL63 were selected to study the binding mode with shikonin in comparison to the binding mode of shikonin with the SARS-CoV-2 Mpro. To identify residues critical for shikonin binding, we obtained the crystal structures of the SARS-CoV Mpro-shikonin complex at a resolution of 2.28 Å and of the HCoV-NL63 Mpro-shikonin complex at a resolution of 2.25 Å (Fig. S2 and Table S1). Examining the substrate binding site revealed a striking difference in electron density in each subunit that was consistent with shikonin (Fig. 4 and 5). Like the case with SARS-CoV-2 Mpro, the 1,4-naphthoquinone (1,4-NQ) of shikonin of both the SARS-CoV Mpro-shikonin and HCoV-NL63 Mpro-shikonin structures fit into the S1 and S2 subsites (Fig. 4) as expected. The key residues interacting with shikonin were highly conserved (Fig. 4B and C). Hydrogen bonding interactions between SARS-CoV Mpro or HCoV-NL63 Mpro and shikonin are shown in Fig. 5. For the SARS-CoV Mpro and SARS-CoV-2 Mpro complexes, the hydrogen bond interactions between amino acid residues Cys145, His164, Arg188, and Gln189 in the protease and the compound are shown in Fig. 5. The hydroxyl group of the naphthalene formed a hydrogen bond with Cys145 in the presumed orthosteric site, and the shikonin ring system formed a face-to-face π-π stack with His41 in the dyad catalytic site, which is commonly conserved between coronaviruses. The binding mode of shikonin in the HCoV-NL63 Mpro-shikonin structure as compared to those of shikonin bound to SARS-CoV or SARS-CoV-2 revealed a subtly different positioning of the ligand tail (chiral six-carbon side chain with the hydroxy group at C-1) toward the outside with the 1,4-naphthoquinone being the deepest between Cys145 and His41 in the orthosteric site (Fig. 5). This differential positioning is likely caused by the flexible loop (aa 44 to 54) on S2 subsite and loop (aa 185 to 192) on S4 subsite, which is less conserved in coronaviruses (Fig. 5 and Fig. S3).
FIG 4.
Comparison of the binding pocket of Mpro from different coronaviruses. (A) Surface of SARS-CoV-2 Mpro, SARS-CoV Mpro, and HCoV-NL63 Mpro. Shikonin is shown as cyan, yellow, and blue sticks in SARS-CoV-2 Mpro, SARS-CoV Mpro and HCoV-NL63 Mpro, respectively. Oxygen atoms are red. (B) Superposition of the binding pocket of SARS-CoV-2 Mpro, SARS-CoV Mpro, and HCoV-NL63 Mpro. (C) Sequence alignment of the binding pocket of seven human CoVs.
FIG 5.
Cocrystal structures of SARS-CoV-2 Mpro, SARS-CoV Mpro, and HCoV-NL63 Mpro in complex with shikonin. (A to C) SARS-CoV-2 Mpro-shikonin complex. (A) Fo_Fc omit map contoured at 0.5 σ for shikonin. (B) Shikonin (red) in the S1, S2, and S3 positions of the active site of SARS-CoV-2 Mpro. (C) Hydrogen bonding (dashed lines) interactions between SARS-CoV-2 Mpro and shikonin. (D to F) SARS-CoV Mpr o-shikonin complex. (D) Fo_Fc omit map contoured at 0.5 σ for shikonin. (E) Shikonin (yellow) in the S1, S2, and S3 positions of the active site of SARS-CoV Mpro. (F) Hydrogen bonding (dashed lines) interactions between SARS-CoV Mpro and shikonin. (G to I) HCoV-NL63 Mpro-shikonin complex. (G) Fo_Fc omit map contoured at 0.5 σ for shikonin. (H) Shikonin (blue) in the S1, S2, and S3 positions of the active site of HCoV-NL63 Mpro. (I) Hydrogen bonding (dashed lines) interactions between HCoV-NL63 Mpro and shikonin.
DISCUSSION
Three novel human CoVs (SARS-CoV-2, SARS-CoV, and MERS-CoV) from the betacoronavirus genus have emerged in the past 20 years and threatened global health due to infectivity, virulence, and pathogenicity (1, 2, 4, 25, 26). Currently there are different types of COVID-19 vaccines, including mRNA vaccines, live-attenuated vaccines, recombinant protein vaccines, and inactivated vaccines, that elicit antibody responses. However, the effective period of the vaccine may be shortened to 3 to 6 months due to the high variability and mutation speed of the virus (27). The main protease represents a promising target for the development of antiviral drugs because the main proteases of human CoVs share sequence and structural similarities.
Here, we presented a combination of structural, computational, and biochemical studies that illuminated the structure and function of a natural product, shikonin, for coronavirus main proteases. Using a structure-based approach latterly supported by an activity assay and X-ray structure determination, we can reliably identify new scaffolds and chemotypes for COVID-19 ligand discovery. A series of optimal natural ligands that displayed micromolar affinities was discovered. We further showed that shikonin has broad-spectrum inhibition activity for the main protease of six human coronaviruses. The viruses selected for structural evaluation in our study are important human pathogens, including HCoV-NL63 from the alphacoronavirus genus and SARS-CoV and SARS-CoV-2 from the betacoronavirus genus. The determination of the crystal structures of shikonin bound to SARS-CoV and SARS-CoV-2 provided an opportunity to seek new scaffold and broad-spectrum chemotypes by structure-based approaches. Thus, to better understand the molecular basis for inhibitor interactions with SARS-CoV and SARS-CoV-2, we sought to obtain the crystal structures of both subtypes. In addition to our previously solved structure of the main protease bound to shikonin in SARS-CoV-2 (18), we reported the apo- and shikonin-bound main protease structures for SARS-CoV and HCoV-NL63.
We then established a binding model based on the X-ray crystal structures of SARS-CoV and HCoV-NL63 complexes to broad-spectrum inhibitor shikonin. The binding of shikonin to SARS-CoV resembled that of SARS-CoV-2 with a π-π stacking interaction with His41 and hydrogen bonding with Cys145. The cocrystal structure of the HCoV-NL63-shikonin complex also demonstrated similar binding interactions between the protease and the ligand (Fig. 5). These results confirmed the conserved key sites of the main protease as a potential target for the broad-spectrum antiviral design.
In conclusion, the Mpro inhibitor shikonin has broad-spectrum antiviral activity. Whether shikonin and its derivatives will be the basis of a new class of therapeutics, however, remains to be determined. All of the broad-spectrum antiviral agents have relatively low affinities for their targets, and the range of antiviral activity of shikonin highlights the potential for the development of broad-spectrum antiviral compounds for multiple viruses. Such a potent, low molecular mass (288 Da) weight compound made an excellent starting point for optimization. It is worth noting that shikonin has strong cytotoxicity. The optimized shikonin-derived compounds with low toxicity may be suitable candidates for further development as antivirals against one or more viruses.
MATERIALS AND METHODS
Virtual screening.
The crystal structure of the SARS-CoV-2 Mpro in complex with N3 (PDB accession 6LU7) (13) was used to construct the molecular docking-based virtual screening model. Glide program (Schrödinger, LLC, New York, NY, 2015) was chosen to perform the molecular docking. The coordinates of the SARS-CoV-2 Mpro were first prepared by the Protein Preparation Wizard panel with default settings. The docking grid was then created by defining residues located within 15 Å around baicalein. Finally, molecules in our natural product and synthetic compound libraries were prepared to dock to the grid using the extra precision (XP) docking mode. The top natural products and 220 synthetic compounds ranked by the XP Gscore were selected for further biological evaluation.
Expression and purification of human CoVs.
The codon-optimized cDNAs for the Mpro of SARS-CoV-2, SARS-CoV, MERS-CoV, HCoV-HKU1, HCoV-NL63, and HCoV-229E were synthesized fused with 6× His at the N terminus. Synthesized genes were subcloned into the pET-28a vector. The expression and purification of each main protease was performed by a standard method previously described (18).
Enzymatic assays.
Fluorogenic substrates as a donor and quencher pair were synthesized (28). The IC50 values of the screening compounds against SARS-CoV-2, SARS-CoV, MERS-CoV, HCoV-HKU1, HCoV-NL63, and HCoV-229E main proteases were measured with a common protocol as the following: First, one μl of six human CoVs proteases (200 nM) was incubated with various concentrations of testing inhibitors at room temperature for 30 min in its reaction buffer (50 mM Tris 7.3,150mM NaCl, 1 mM EDTA) in a 384-well plate, and then FRET substrate was added to the reaction system. The reaction was monitored for 20 min, and the data were calculated at 10 min intervals using a linear regression. The IC50s of the compounds were determined by plotting the initial velocity against various concentrations of the test inhibitor by using the dose-response curve in GraphPad Prism software.
X-ray crystallography.
Details of the crystallization, data collection, structure solution, and refinement are provided in Table S1. Briefly, all crystallization trials were conducted using a sitting-drop vapor diffusion method at 20°C. Shikonin was soaked with crystals of SARS-CoV-apo or HCoV-NL63-apo within 24 h, and the X-ray diffraction data were collected at beamline17U1 (BL17U1) at the Shanghai Synchrotron Radiation Facility (SSRF, Shanghai, China). The structure solution was conducted by molecular replacement using SARS-CoV-apo PDB (21) and HCoV-NL63 PDB (29) as an initial model. Refinement and model building were carried out using Phenix (30) and Coot (31), respectively.
Data availability.
Coordinates and structure factors for SARS-CoV-apo, SARS-CoV-shikonin, and HCoV-NL63-shikonin complexes have been deposited in the Protein Data Bank (PDB) under accession numbers 7DQZ, 7EO8, and 7EO7, respectively.
ACKNOWLEDGMENTS
J.L. was supported by the Open Project of Key Laboratory of Prevention and Treatment of Cardiovascular and Cerebrovascular Diseases, Ministry of Education (grant no. XN201904), Gannan Medical University (grant no. QD201910), Jiangxi key research and development program (grant no. 20203BBG73063) and Jiangxi “Double Thousand Plan”. J.Z. was supported by the Thousand Young Talents Program of China, the National Natural Science Foundation of China (grant no. 31770795; grant no. 81974514), and the Jiangxi Province Natural Science Foundation (grant no. 20181ACB20014). P.J.M. was supported by the Foreign Talent project of Jiangxi Province. F.J. was supported by the National Natural Science Foundation of China (grant no. 21961003), Natural Science Foundation of Jiangxi Province (grant no. 20192BAB205114), Talent project of Jiangxi Province. J.D. was supported by the Natural Science Foundation of China (grant no. 31971043). This work was also supported by Ganzhou COVID-19 Emergency Research Project (grant no. 2020.17), Major science and technology programs of Ganzhou City (grant no. 2020.67) and Ganzhou Zhanggong District COVID-19 prevention and control key research projects (grant no. 2020.67), Shenzhen Science and Technology Program (grant no. JCYJ20210324115611032), China Postdoctoral Science Foundation COVID-19 Prevention Special Project (grant no. 2020T130151ZX), China Postdoctoral Science Foundation COVID-19 Prevention Special Project (grant no. 2020T130151ZX).
J.L. and J.Z. initiated and supervised the project. Y.Z., H.G. and X.H. crystallized the protein complexes and performed the soaking experiments. J.L., Q.W., F.Z., X.Z., C.L., H.Z. and J.Z. collected X-ray data and solved and refined structures. H.Z., Y.Y., J.W., W.D., H.H., W.H. J.F., B.Q. and D.W. performed docking and identified compounds to be tested in the initial screens. H.Z., Y.Z., H.J., Q.L., Y.Z., L.L. L.C., Z.Z. and J.L. synthesized compounds and performed enzymatic assays by FRET. P.J.M., Q.C., Y.F., J.D., T.Z. assisted with the design of experiments, project management, and interpretation of results.
Footnotes
Supplemental material is available online only.
Contributor Information
Jin Zhang, Email: zhangxiaokong@hotmail.com.
Jian Li, Email: rmsl_2040@163.com.
Tom Gallagher, Loyola University Chicago.
REFERENCES
- 1.Chan JF, Yuan S, Kok KH, To KK, Chu H, Yang J, Xing F, Liu J, Yip CC, Poon RW, Tsoi HW, Lo SK, Chan KH, Poon VK, Chan WM, Ip JD, Cai JP, Cheng VC, Chen H, Hui CK, Yuen KY. 2020. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet 395:514–523. 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li Q, Guan X, Wu P, Wang X, Zhou L, Tong Y, Ren R, Leung KSM, Lau EHY, Wong JY, Xing X, Xiang N, Wu Y, Li C, Chen Q, Li D, Liu T, Zhao J, Liu M, Tu W, Chen C, Jin L, Yang R, Wang Q, Zhou S, Wang R, Liu H, Luo Y, Liu Y, Shao G, Li H, Tao Z, Yang Y, Deng Z, Liu B, Ma Z, Zhang Y, Shi G, Lam TTY, Wu JT, Gao GF, Cowling BJ, Yang B, Leung GM, Feng Z. 2020. Early Transmission Dynamics in Wuhan, China, of Novel Coronavirus-Infected Pneumonia. N Engl J Med 382:1199–1207. 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, Wang W, Song H, Huang B, Zhu N, Bi Y, Ma X, Zhan F, Wang L, Hu T, Zhou H, Hu Z, Zhou W, Zhao L, Chen J, Meng Y, Wang J, Lin Y, Yuan J, Xie Z, Ma J, Liu WJ, Wang D, Xu W, Holmes EC, Gao GF, Wu G, Chen W, Shi W, Tan W. 2020. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet 395:565–574. 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao GF, Tan W, China Novel Coronavirus Investigating and Research Team. 2020. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 382:727–733. 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huynh J, Li S, Yount B, Smith A, Sturges L, Olsen JC, Nagel J, Johnson JB, Agnihothram S, Gates JE, Frieman MB, Baric RS, Donaldson EF. 2012. Evidence supporting a zoonotic origin of human coronavirus strain NL63. J Virol 86:12816–12825. 10.1128/JVI.00906-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu B, Ge X, Wang LF, Shi Z. 2015. Bat origin of human coronaviruses. Virol J 12:221. 10.1186/s12985-015-0422-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anthony SJ, Gilardi K, Menachery VD, Goldstein T, Ssebide B, Mbabazi R, Navarrete-Macias I, Liang E, Wells H, Hicks A, Petrosov A, Byarugaba DK, Debbink K, Dinnon KH, Scobey T, Randell SH, Yount BL, Cranfield M, Johnson CK, Baric RS, Lipkin WI, Mazet JA. 2017. Further evidence for bats as the evolutionary source of Middle East respiratory syndrome coronavirus. mBio 8:e00373-17. 10.1128/mBio.00373-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Menachery VD, Yount BL, Jr., Debbink K, Agnihothram S, Gralinski LE, Plante JA, Graham RL, Scobey T, Ge XY, Donaldson EF, Randell SH, Lanzavecchia A, Marasco WA, Shi ZL, Baric RS. 2015. A SARS-like cluster of circulating bat coronaviruses shows potential for human emergence. Nat Med 21:1508–1513. 10.1038/nm.3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sheahan TP, Sims AC, Graham RL, Menachery VD, Gralinski LE, Case JB, Leist SR, Pyrc K, Feng JY, Trantcheva I, Bannister R, Park Y, Babusis D, Clarke MO, Mackman RL, Spahn JE, Palmiotti CA, Siegel D, Ray AS, Cihlar T, Jordan R, Denison MR, Baric RS. 2017. Broad-spectrum antiviral GS-5734 inhibits both epidemic and zoonotic coronaviruses. Sci Transl Med 9:eaal3653. 10.1126/scitranslmed.aal3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zumla A, Chan JF, Azhar EI, Hui DS, Yuen KY. 2016. Coronaviruses - drug discovery and therapeutic options. Nat Rev Drug Discov 15:327–347. 10.1038/nrd.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anand K, Ziebuhr J, Wadhwani P, Mesters JR, Hilgenfeld R. 2003. Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science 300:1763–1767. 10.1126/science.1085658. [DOI] [PubMed] [Google Scholar]
- 12.Dai W, Zhang B, Jiang X-M, Su H, Li J, Zhao Y, Xie X, Jin Z, Peng J, Liu F, Li C, Li Y, Bai F, Wang H, Cheng X, Cen X, Hu S, Yang X, Wang J, Liu X, Xiao G, Jiang H, Rao Z, Zhang L-K, Xu Y, Yang H, Liu H. 2020. Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Science 368:1331–1335. 10.1126/science.abb4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jin Z, Du X, Xu Y, Deng Y, Liu M, Zhao Y, Zhang B, Li X, Zhang L, Peng C, Duan Y, Yu J, Wang L, Yang K, Liu F, Jiang R, Yang X, You T, Liu X, Yang X, Bai F, Liu H, Liu X, Guddat LW, Xu W, Xiao G, Qin C, Shi Z, Jiang H, Rao Z, Yang H. 2020. Structure of M(pro) from COVID-19 virus and discovery of its inhibitors. Nature 582:289–293. 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 14.Zhang L, Lin D, Kusov Y, Nian Y, Ma Q, Wang J, von Brunn A, Leyssen P, Lanko K, Neyts J, de Wilde A, Snijder EJ, Liu H, Hilgenfeld R. 2020. α-Ketoamides as broad-spectrum inhibitors of coronavirus and enterovirus replication: structure-based design, synthesis, and activity assessment. J Med Chem 63:4562–4578. 10.1021/acs.jmedchem.9b01828. [DOI] [PubMed] [Google Scholar]
- 15.Zhang L, Lin D, Sun X, Curth U, Drosten C, Sauerhering L, Becker S, Rox K, Hilgenfeld R. 2020. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science 368:409–412. 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jin Z, Zhao Y, Sun Y, Zhang B, Wang H, Wu Y, Zhu Y, Zhu C, Hu T, Du X, Duan Y, Yu J, Yang X, Yang X, Yang K, Liu X, Guddat LW, Xiao G, Zhang L, Yang H, Rao Z. 2020. Structural basis for the inhibition of SARS-CoV-2 main protease by antineoplastic drug carmofur. Nat Struct Mol Biol 27:529–532. 10.1038/s41594-020-0440-6. [DOI] [PubMed] [Google Scholar]
- 17.Su H, Yao S, Zhao W, Li M, Liu J, Shang W, Xie H, Ke C, Gao M, Yu K, Liu H, Shen J, Tang W, Zhang L, Zuo J, Jiang H, Bai F, Wu Y, Ye Y, Xu Y. 2020. Discovery of baicalin and baicalein as novel, natural product inhibitors of SARS-CoV-2 3CL protease in vitro. bioRxiv 10.1101/2020.04.13.038687:2020.04.13.038687. [DOI]
- 18.Li J, Zhou X, Zhang Y, Zhong F, Lin C, McCormick PJ, Jiang F, Luo J, Zhou H, Wang Q, Fu Y, Duan J, Zhang J. 2021. Crystal structure of SARS-CoV-2 main protease in complex with the natural product inhibitor shikonin illuminates a unique binding mode. Sci Bull (Beijing) 66:661–663. 10.1016/j.scib.2020.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tomar S, Johnston ML, St John SE, Osswald HL, Nyalapatla PR, Paul LN, Ghosh AK, Denison MR, Mesecar AD. 2015. Ligand-induced dimerization of Middle East respiratory syndrome (MERS) coronavirus nsp5 protease (3CLpro): implications for nsp5 regulation and the development of antivirals. J Biol Chem 290:19403–19422. 10.1074/jbc.M115.651463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Galasiti Kankanamalage AC, Kim Y, Damalanka VC, Rathnayake AD, Fehr AR, Mehzabeen N, Battaile KP, Lovell S, Lushington GH, Perlman S, Chang KO, Groutas WC. 2018. Structure-guided design of potent and permeable inhibitors of MERS coronavirus 3CL protease that utilize a piperidine moiety as a novel design element. Eur J Med Chem 150:334–346. 10.1016/j.ejmech.2018.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang H, Yang M, Ding Y, Liu Y, Lou Z, Zhou Z, Sun L, Mo L, Ye S, Pang H, Gao GF, Anand K, Bartlam M, Hilgenfeld R, Rao Z. 2003. The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor. Proc Natl Acad Sci USA 100:13190–13195. 10.1073/pnas.1835675100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu A, Peng Y, Huang B, Ding X, Wang X, Niu P, Meng J, Zhu Z, Zhang Z, Wang J, Sheng J, Quan L, Xia Z, Tan W, Cheng G, Jiang T. 2020. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe 27:325–328. 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen S, Jonas F, Shen C, Hilgenfeld R, Higenfeld R. 2010. Liberation of SARS-CoV main protease from the viral polyprotein: N-terminal autocleavage does not depend on the mature dimerization mode. Protein Cell 1:59–74. 10.1007/s13238-010-0011-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hussey RJ, Coates L, Gill RS, Erskine PT, Coker SF, Mitchell E, Cooper JB, Wood S, Broadbridge R, Clarke IN, Lambden PR, Shoolingin-Jordan PM. 2011. A structural study of norovirus 3C protease specificity: binding of a designed active site-directed peptide inhibitor. Biochemistry 50:240–249. 10.1021/bi1008497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alexander E, Gorbalenya SCB, Baric RS, de Groot RJ, Drosten C, Gulyaeva AA, Haagmans BL, Lauber C, Leontovich AM, Neuman BW, Penzar D, Perlman S, Poon LLM, Samborskiy DV, Sidorov IA, Isabel S, Ziebuhr J. 2020. The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol 5:536–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, Hu Y, Tao ZW, Tian JH, Pei YY, Yuan ML, Zhang YL, Dai FH, Liu Y, Wang QM, Zheng JJ, Xu L, Holmes EC, Zhang YZ. 2020. A new coronavirus associated with human respiratory disease in China. Nature 579:265–269. 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Buss LF, Prete CA, Jr., Abrahim CMM, Mendrone A, Jr., Salomon T, de Almeida-Neto C, França RFO, Belotti MC, Carvalho M, Costa AG, Crispim MAE, Ferreira SC, Fraiji NA, Gurzenda S, Whittaker C, Kamaura LT, Takecian PL, da Silva Peixoto P, Oikawa MK, Nishiya AS, Rocha V, Salles NA, de Souza Santos AA, da Silva MA, Custer B, Parag KV, Barral-Netto M, Kraemer MUG, Pereira RHM, Pybus OG, Busch MP, Castro MC, Dye C, Nascimento VH, Faria NR, Sabino EC. 2021. Three-quarters attack rate of SARS-CoV-2 in the Brazilian Amazon during a largely unmitigated epidemic. Science 371:288–292. 10.1126/science.abe9728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ma C, Sacco MD, Hurst B, Townsend JA, Hu Y, Szeto T, Zhang X, Tarbet B, Marty MT, Chen Y, Wang J. 2020. Boceprevir, GC-376, and calpain inhibitors II, XII inhibit SARS-CoV-2 viral replication by targeting the viral main protease. Cell Res 30:678–692. 10.1038/s41422-020-0356-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang F, Chen C, Tan W, Yang K, Yang H. 2016. Structure of main protease from human coronavirus NL63: insights for wide spectrum anti-coronavirus drug design. Sci Rep 6:22677. 10.1038/srep22677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. 2002. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr 58:1948–1954. 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 31.Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60:2126–2132. 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 to S3; Table S1<br>. Download jvi.01253-21-s0001.pdf, PDF file, 0.8 MB (850.3KB, pdf)
Data Availability Statement
Coordinates and structure factors for SARS-CoV-apo, SARS-CoV-shikonin, and HCoV-NL63-shikonin complexes have been deposited in the Protein Data Bank (PDB) under accession numbers 7DQZ, 7EO8, and 7EO7, respectively.