Auxin concentration–dependent cell expansion coordinates hypocotyl elongation and apical hook development for soil emergence.
Abstract
Seedling emergence is critical for food security. It requires rapid hypocotyl elongation and apical hook formation, both of which are mediated by regulated cell expansion. How these events are coordinated in etiolated seedlings is unclear. Here, we show that biphasic control of cell expansion by the phytohormone auxin underlies this process. Shortly after germination, high auxin levels restrain elongation. This provides a temporal window for apical hook formation, involving a gravity-induced auxin maximum on the eventual concave side of the hook. This auxin maximum induces PP2C.D1 expression, leading to asymmetrical H+-ATPase activity across the hypocotyl that contributes to the differential cell elongation underlying hook development. Subsequently, auxin concentrations decline acropetally and switch from restraining to promoting elongation, thereby driving hypocotyl elongation. Our findings demonstrate how differential auxin concentrations throughout the hypocotyl coordinate etiolated development, leading to successful soil emergence.
INTRODUCTION
Successful seedling emergence enables plants to produce photosynthates to fuel growth and development. In most dicots, rapid hypocotyl elongation drives seedling emergence from the soil (1). Simultaneously, the apical hook structure develops at the hypocotyl apex, which protects the shoot apical meristem from damage (2, 3). This strategy of seedling emergence was described 140 years ago in Darwin’s pioneering work (2). The phytohormone auxin plays crucial roles in regulating both hypocotyl elongation and apical hook development (4–9) through its ability to mediate changes in cell expansion in a tissue- and concentration-dependent manner (10). However, key questions related to these processes remain. First, hypocotyl elongation proceeds slowly initially and accelerates markedly at around 24 hours post germination (HPG) with cells elongating as an acropetal (from bottom to top) wave (1, 11). How these temporal and spatial patterns of cell expansion are controlled is unclear. Second, while auxin can promote cell elongation in the growing parts of the hypocotyl (4), it inhibits elongation at the apical hook region (9). How auxin exerts these differential effects throughout the hypocotyl is unknown. Third, auxin asymmetrically accumulates at the concave side of the hook to inhibit cell elongation and drive hook formation (5, 9), but both the underlying mechanism for this inhibition and the signal that initiates the asymmetric auxin distribution remain uncertain (12, 13). Finally, fourth, auxin-mediated promotion of hypocotyl cell elongation can be explained by the acid growth theory (14). This requires auxin-induced small auxin-up RNA (SAUR)–mediated repression of PP2C.D (D clade type 2C protein phosphatase) phosphatases to activate plasma membrane (PM) H+–ATPases (adenosine triphosphatases) (4, 15–17). These downstream effectors of auxin may also participate in apical hook development (16, 17), but the mechanism of inhibition of cell elongation on the concave side of the hook has yet to be identified.
RESULTS AND DISCUSSION
Auxin regulates hypocotyl cell elongation in a biphasic manner
Auxin regulates cell expansion in a concentration- and tissue-dependent manner (4). In general, auxin promotes cell expansion in shoots (16) while inhibiting it in roots (18, 19). A notable exception exists at the concave side of the apical hook, where auxin inhibits cell expansion (9). To obtain insights into how auxin coordinates hypocotyl elongation and apical hook development, we first analyzed the spatial distribution of auxin signaling in the epidermis of etiolated Arabidopsis hypocotyls, in which cell elongation proceeds acropetally from the base to the tip over time (1, 11). We used the R2D2 reporter, which consists of auxin-degradable (DII) and auxin-insensitive (mDII) fluorescent proteins (20). The mDII/DII signal ratio may serve as a proxy for cellular auxin levels (20). Treatment with the natural auxin, indole-3-acetic acid (IAA), increased the mDII/DII ratio, whereas the IAA biosynthesis inhibitor KOK2153 (21) led to a decrease in the ratio (fig. S1, A to C), validating the use of this reporter as a proxy for auxin distribution in these cells. Visual inspection of R2D2 signals suggested an auxin gradient down the hypocotyl, with the highest auxin signal in the upper cells (fig. S1A). This was confirmed by quantifying the mDII/DII ratio at the cellular level in epidermal cells along the length of young hypocotyls (Fig. 1A) and at the organ level by IAA quantification in the upper and lower halves of 36HPG hypocotyls (fig. S1D).
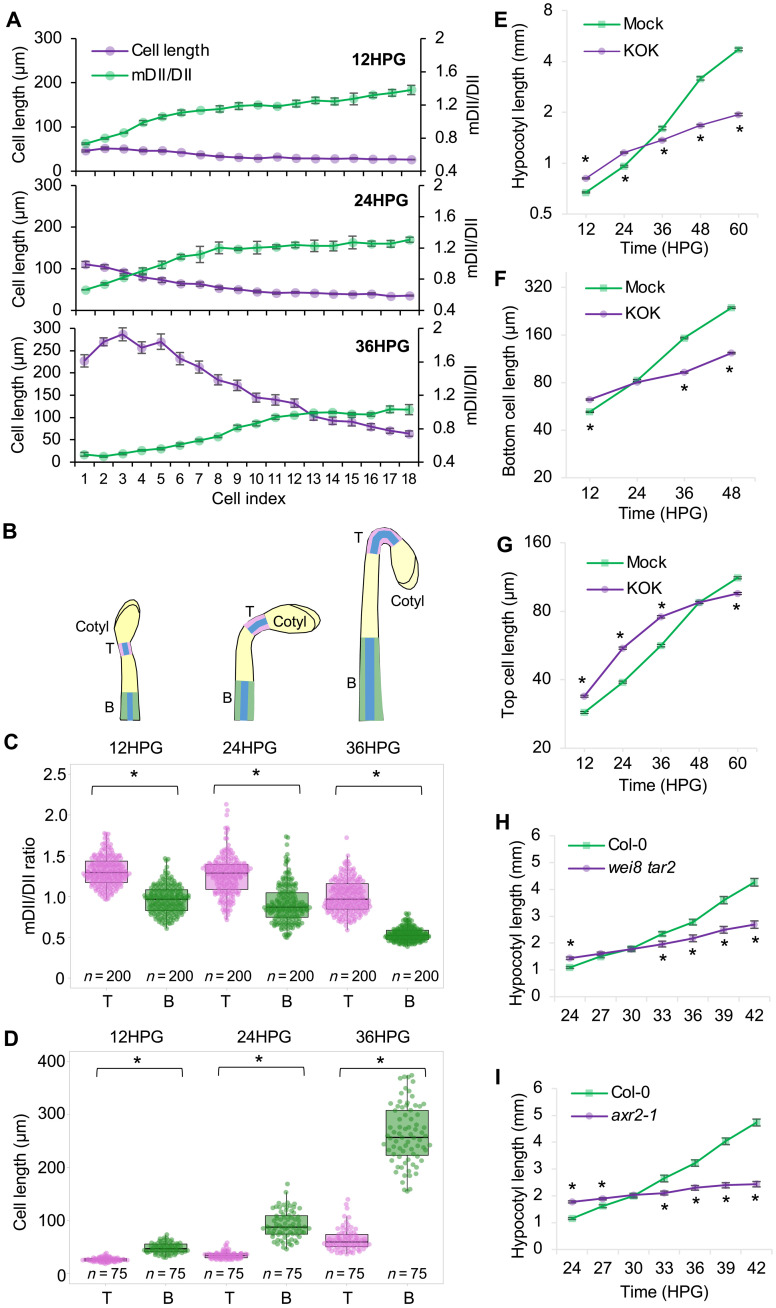
Fig. 1. Auxin regulates hypocotyl cell elongation in a biphasic manner.
(A) Cell length (magenta) and corresponding auxin signaling (green) from bottom (cell index 1) to top (cell index 18) of the hypocotyl epidermis during early hypocotyl development. Bars represent the SEM. (B) Schematic diagrams showing the positions of the top cells (T) and bottom cells (B) analyzed in etiolated hypocotyls. The epidermal cells where the R2D2 signals and cell lengths were measured are highlighted in blue. (C and D) Quantification of the mDII/DII ratio (C) and cell lengths (D) in the top (T) and bottom (B) cells of R2D2 hypocotyls for the indicated time. Five bottom cells (B) and five top cells (T) were used for quantification from each hypocotyl. (E) Quantification of hypocotyl length during etiolated seedling development, n = 60 hypocotyls. (F and G) Quantification of epidermal cell length of the bottom (F) and top (G) cells during etiolated seedling development, n = 300 cells. (E to G) Germinated seeds were transferred to ½ Murashige and Skoog (MS) medium containing 1% sucrose supplemented with dimethyl sulfoxide (DMSO) (mock) or KOK2153 (KOK). Five bottom cells and five top cells from each hypocotyl were used for quantification. Values represent sample means ± SEM from three replicates. (H) Quantification of hypocotyl length during etiolated seedling development in Col-0 and the wei8 tar2 mutant. (I) Quantification of hypocotyl length during etiolated seedling development in Col-0 and the axr2-1 mutant. *P < 0.01.
An inverse correlation between auxin level and cell length along the hypocotyl (Fig. 1A) suggested a critical role for the auxin gradient in regulating hypocotyl elongation. A shift in the auxin signal along the hypocotyl appeared to move upward over time, which coincided with the front of the cellular elongation wave (Fig. 1A). To further characterize the relationship between auxin and cell length during hypocotyl elongation, we selected the bottom five cells (low auxin signal) and the top five cells (high auxin signal) of the hypocotyl for further detailed analysis (Fig. 1B). A significant difference in the auxin signal between the upper and lower cells was apparent at all time points examined (Fig. 1C). As previously described, etiolated hypocotyls showed a transition from slow to rapid elongation beginning at around 24HPG (1, 11). From 24 to 36HPG, the bottom cells more than doubled their length (Fig. 1D), and a strong reduction in the auxin signal in these cells was evident (Fig. 1C). To further address the relationship between auxin and this growth wave, we analyzed the kinematics of hypocotyl elongation using pharmacologic and genetic tools. Inclusion of KOK2153 in the medium significantly promoted hypocotyl elongation at the early stages (before 24HPG) but inhibited elongation thereafter (Fig. 1E). Consistently, epidermal cell lengths in the presence of the inhibitor were significantly increased at the early stages but decreased at the later stages (Fig. 1, F and G), compared with untreated controls. The inhibitor’s effects during the early stages were largely reversed by cotreatment with 100 nM IAA, indicating that these effects were caused by inhibition of auxin biosynthesis (fig. S2, A and B). These observations suggest a biphasic role for auxin in hypocotyl cell elongation: Auxin inhibits individual cell elongation at the early stages but promotes it at the later stages. Consistent with this notion, treatment with auxinole, an antagonist of the TIR1/AFB (transport inhibitor response 1/auxin signaling F-box protein) auxin receptors (22), also promoted hypocotyl cell elongation at 12HPG (fig. S2, C to E). To genetically challenge this hypothesis, we analyzed the Arabidopsis thaliana mutants wei8-3 tar2-1 and tir1 afb2 that display decreased auxin biosynthesis (23, 24) and defective auxin perception (25), respectively. Both mutants exhibited longer hypocotyl cells at 12HPG when compared with the wild type (WT) (fig. S2, F to H). By contrast, the yuc1-D mutant, which overproduces IAA (26), displayed shorter hypocotyl cells (fig. S2, F to H). Furthermore, kinematic analysis of etiolated hypocotyl lengths in both moderate (tir1 afb2) and strong (axr2-1) (27) auxin signaling mutants, as well as the wei8-3 tar2-1 biosynthesis mutant, further demonstrated the biphasic role for auxin in hypocotyl cell elongation, as these mutants exhibited longer hypocotyls than WT controls during early development, but shorter hypocotyls at later stages (Fig. 1, H and I, and fig. S2I). Although auxin is typically thought to promote cell elongation in shoots, here we demonstrate, through fine temporal and spatial analyses, an inhibitory effect on hypocotyl cell elongation during the early stages of etiolated seedling development.
Auxin concentrations correlate with the biphasic hypocotyl elongation
The biphasic role of auxin in hypocotyl cell elongation indicates that auxin both restrains and promotes hypocotyl elongation in precise developmental contexts. This transition from auxin inhibiting to promoting elongation occurred earlier in the bottom cells (at ~24HPG) than in the top cells (after 36HPG) (Fig. 1, F and G). The cell length at which the transition occurred was very similar between the bottom and top cells (~80 μm; Fig. 1, F and G), suggesting that cell size might be a key factor affecting the transition. Because the R2D2 reporter suggested that bottom cells contained less auxin (Fig. 1C and fig. S1D), we hypothesized that the transition is driven by the decreased IAA concentration in cells to levels that promote cell elongation (10). As hypocotyls elongated, the cellular mDII/DII ratio gradually dropped, and the ratio at which the transition occurred was very similar between the bottom (24HPG) and top (36HPG) cells (~0.8; Fig. 2, A and B). These findings support the idea that the transition is the result of reduced cellular IAA concentration. Conceivably, this reduction in IAA concentration could be caused by the slow increase in cell volume that occurs during the time at which auxin is restraining elongation. To explore this possibility, we quantified the IAA concentration in elongating hypocotyls over time and found a significant decrease in auxin concentration (as expressed per volume of hypocotyl) between 12HPG, 24HPG, and 36HPG (Fig. 2, C and D). While cell volume increases are generally thought to be the result of increased vacuole size, nuclei containing the TIR1/AFB receptors also increase in size as a result of endoreduplication (1), which could contribute to the dilution of the active IAA pool.
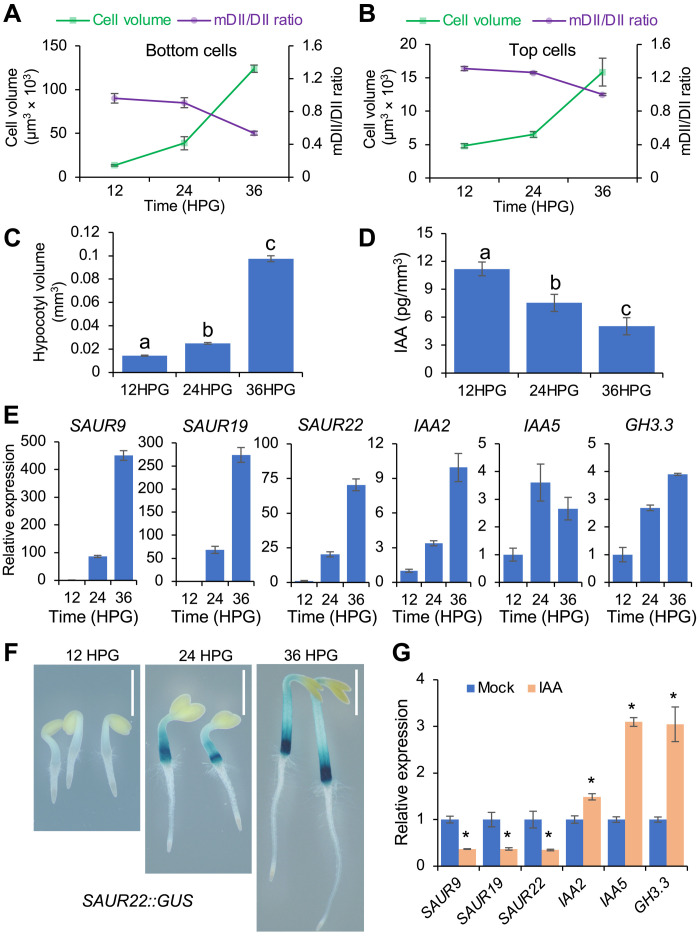
Fig. 2. Decreased auxin levels correlate with increased cell volume and SAUR expression during hypocotyl elongation.
(A and B) Decreased auxin levels correlate with increased cell volume of bottom (A) and top (B) cells of R2D2 hypocotyls for the indicated time. Auxin levels were proxied by the inverse n3 × Venus/ntdTomato signal ratio (mDII/DII). Cells were assumed to have a cylindrical shape, and cell volume was estimated by the formula for the volume of a cylinder [π * (width/2)2 * length]. Five cells each from the bottom and the top of the hypocotyl were used for analysis. Values represent sample means ± SEM from three replicates. (C) Quantification of hypocotyl volumes at indicated times. (D) Quantification of endogenous IAA in the elongating hypocotyls at indicated times. Auxin concentrations were expressed in picograms of IAA per volume of hypocotyl (mm3). Different letters indicate statistical differences between groups with P < 0.01 using analysis of variance (ANOVA) test. (E) Expression of auxin-responsive genes in etiolated hypocotyls at the indicated times. (F) SAUR22::GUS expression in etiolated hypocotyls at the indicated times. Scale bars, 1 mm. (G) Expression of auxin-responsive genes in etiolated hypocotyls grown on different media. Germinated seeds were transferred to ½ MS medium supplemented with 1% sucrose and with DMSO (mock) or 1 μM IAA (IAA) for growth. Expression analysis was performed at 36HPG. (E and G) Expression levels of indicated genes were normalized against ACTIN7 expression and presented as the fold change to the expression at 12HPG (E) or mock (G). Data are means ± SEM from three biological replicates. *P < 0.01.
We next examined how this decrease in auxin concentrations is reflected on the expression of SAUR genes, which promote auxin-mediated cell elongation. The expression of SAUR genes increased markedly (>60-fold) as hypocotyls grew (Fig. 2, E and F), suggesting that high IAA levels inhibit SAUR expression at the early stages of hypocotyl elongation. Consistent with this possibility, 1 μM IAA treatment repressed the expression of SAUR genes but induced the expression of other auxin-responsive genes in hypocotyls (Fig. 2G). These results suggested that auxin-mediated repression of SAUR expression might contribute to its inhibitory effect on hypocotyl cell elongation at the early stages. This notion is supported by the observation that constitutive expression of SAUR19 from the 35S promoter resulted in increased hypocotyl cell length (fig. S2, J to L).
Together, these findings suggest that high auxin levels inhibit SAUR expression and cell elongation during the early stages of etiolated seedling development. As the hypocotyl cells elongate following an acropetal wave, the auxin concentration drops, perhaps as a result of the increased cell volume, to reach levels that promote cell elongation. This concentration-dependent inhibition suggests parallels with apical hook formation, where high IAA levels in cells of the presumptive concave side of the developing hook are believed to inhibit cell elongation (6). We therefore undertook an investigation to determine whether and how this inhibition is involved in apical hook formation during early seedling development.
Gravity triggers asymmetric auxin distribution for apical hook formation
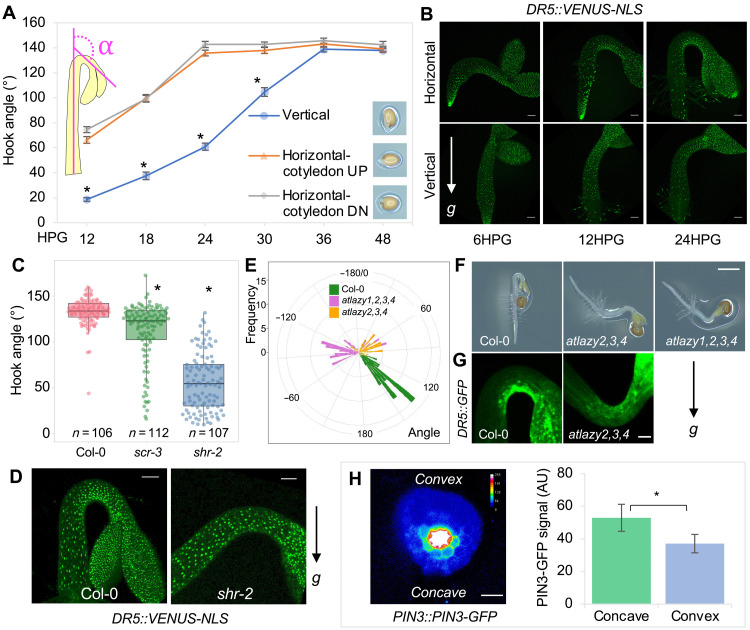
Apical hook formation is driven by asymmetric auxin distribution across the hypocotyl (3, 5, 28). The signal that initiates the asymmetric auxin distribution remains unclear (12, 13). Gravity has long been suggested to play a role in apical hook formation (29–31). Early studies using clinostats identified a major role for gravity in hypocotyl apical hook formation in cress (29), although other studies using pea seedlings that form an epicotyl hook yielded conflicting results (32). Recently, a signal originating from gravity-induced root bending, but not from gravity per se, was proposed to direct apical hook formation (12). In that study, however, apical hooks still formed after removal of the root, suggesting the signal may not originate from the root. We therefore hypothesized that apical hook formation is a hypocotyl gravitropic response. We first noticed that, upon rotating etiolated seedlings to a horizontal plane, the hypocotyl curved against gravity, while apical hooks actively reoriented positively with the new gravity vector (movie S1). Because shoot gravitropic response was reported to be sensitive to the inclination angle of the organ from the direction of gravity (33, 34), we tested whether the inclination angle of the hypocotyl would affect hook formation. We placed the germinated seeds at a vertical orientation (inclination angle θ = 0, vertical group) or at a horizontal orientation (inclination angle θ = 90°, horizontal group) (Fig. 3A and fig. S3A) and monitored the development of the hook. Apical hooks formed significantly earlier in the horizontal group than in the vertical group, regardless of cotyledon orientation (Fig. 3A, fig. S3A, and movie S2). In the horizontal group, hook bending was observed as early as 6HPG, and the hook angle reached its maximum around 24HPG. By contrast, in the vertical group, hypocotyls started to bend at about 12HPG and fully formed the hook after 30HPG. The asymmetric distribution of auxin signal, at the organ level, as shown by the DR5::VENUS-NLS reporter (35), was also established earlier in the horizontal group than in the vertical group (Fig. 3B). Furthermore, hook formation was affected by periodically rotating the seedlings to counteract the effect of gravity (fig. S3, B and C). Removal of the root delayed hook formation, potentially as a result of wounding or impaired water/nutrient uptake (12). However, periodic rotation of rootless seedlings conferred a nearly identical reduction in hook angle as was observed for intact seedlings, suggesting that gravity’s effect on hook formation was independent of the root (fig. S3D). These data indicate that hook formation might be a gravity-dependent and root-independent response. To genetically support this notion, we analyzed mutants that are defective in shoot gravitropism. Two radial pattern mutants, scarecrow (scr-3/shoot gravitropism1) and short-root (shr-2/shoot gravitropism7), exhibit impaired shoot gravitropism, because of the absence of the gravity-sensing endodermal cell layer (36). Both mutants were defective in apical hook formation, with the more severe shr-2 mutant exhibiting a stronger defect (Fig. 3C, fig. S3E, and movie S3). Consistently, the normally asymmetric distribution of the DR5::VENUS-NLS signal at the hook region was largely abolished in the shr-2 mutant (Fig. 3D). Recently, the LAZY family of genes have been implicated in the control of gravity-induced directional auxin transport (37). In the atlazy2,3,4 triple mutant and the atlazy1,2,3,4 quadruple mutant, auxin accumulated at the nongravistimulated side, and accordingly, the mutants exhibited reversed gravitropic responses (38). Hence, we used these atlazy mutants to examine gravity’s role in apical hook formation. When placed at a horizontal orientation, 14% of the atlazy2,3,4 hooks (8 of 57) and 92% of the atlazy1,2,3,4 hooks (56 of 60) were formed with the concave side facing upward instead of downward (Fig. 3, E and F, and movie S4), and DR5::GFP (green fluorescent protein) (39) signal accumulated at the nongravistimulated side (Fig. 3G). Furthermore, consistent with a previous study showing that gravity-induced polarization of the auxin efflux carrier PIN-FORMED3 (PIN3) mediated auxin flow toward the lower side of the hypocotyl (40), we found that, when the hook is forming, PIN3-GFP (8) exhibited stronger accumulation in the endodermis of the concave side compared with the convex side (Fig. 3H), supporting the idea that gravity-induced PIN3 asymmetric distribution is also involved in auxin mobilization for apical hook formation. These results strongly imply that gravity is an important signal that triggers asymmetric auxin distribution for apical hook formation.
Fig. 3. Gravity triggers asymmetric auxin distribution during apical hook formation.
(A) Quantification of the kinematics of apical hook development in differentially oriented seedlings. Germinated seeds were initially placed at a vertical or horizontal orientation, for which cotyledons were oriented either above or below the hypocotyl. Hook curvature angles were measured over 48 hours; n = 36 hooks. Values represent sample means ± SEM from three replicates. The inset depicts how the angle of hook curvature was determined. (B) DR5::VENUS-NLS expression during apical hook development in differentially oriented seedlings. Scale bars, 100 μm. (C) Quantification of the hook angle of Col-0 and scr-3 and shr-2 mutants. (D) DR5::VENUS-NLS expression in the apical hook of Col-0 and shr-2. Scale bars, 100 μm. (E) Quantification of the hook angle and direction of Col-0 and atlazy2,3,4 and atlazy1,2,3,4 mutants. Hook curvature angles of concave-side-up seedlings have negative values. (F) Representative pictures showing the apical hook of Col-0, atlazy2,3,4, and atlazy1,2,3,4. Scale bar, 1 mm. (G) DR5::GFP expression in the apical hook of Col-0 and an atlazy2,3,4 seedling that developed a concave-side-up hook. Scale bar, 100 μm. (C to G) Germinated seeds were initially placed at a horizontal orientation, and the hook pictures were acquired at 24HPG. (H) Localization and signal quantification of PIN-FORMED3 green fluorescent protein (PIN3-GFP) in a transverse section of the hook at 6HPG. Scale bar, 50 μm. Description of the quantification of the PIN3-GFP signals in the endodermis of the concave and convex sides can be found in Materials and Methods. For (A), (C), and (H), *P < 0.01.
During plant gravitropic responses, shoots bend upward (negative gravitropism) while roots bend downward (positive gravitropism) (41). The basis of this opposite gravitropism is the regulation by auxin, which generally promotes cell elongation in shoots, while inhibiting it in roots. That is, gravity-induced auxin accumulation on the lower side will strengthen the growth inhibition on that side of the root to cause the downward bending. By contrast, in the shoot, gravity-induced auxin accumulation on the lower side will increase the growth promotion on that side, leading to an upward curvature (42). The differential regulation of gravitropism by auxin is likely concentration dependent, since the application of high auxin concentration induced a switch in tomato hypocotyl gravitropic response from negative to positive (43). We verified this finding in tomato seedlings and Arabidopsis shoots (fig. S4A and movies S5 and S6). Our finding that high auxin concentrations also inhibit cell elongation in hypocotyls during early seedling development (Fig. 1 and fig. S2) suggested the possibility that the auxin maximum at the presumptive concave side of developing hooks promotes downward bending in response to gravity. Consistent with this notion, hypocotyls bent upward when the gravity-directed auxin flow was reversed in the atlazy mutants (Fig. 3, F and G). The downward bending of the hypocotyl is the beginning of the hook formation. Subsequently, as auxin concentrations decline to levels that promote cell elongation in the bottom cells (Figs. 1 and 2), the bottom part starts to bend upward, while the top part is still in a downward direction, gradually causing the hook to be fully formed. Therefore, hook curvature is the consequence of the opposite directional bending of the bottom and top parts of the young hypocotyl. This notion was further supported by the observation that the top segments of tomato hypocotyls displayed positive gravitropism, while the bottom segments exhibited negative gravitropism (fig. S4B and movies S7 and S8).
Hook-expressed PP2C.D1 is specifically activated by auxin via ARF7
We next explored the molecular mechanism by which the gravity-induced auxin maximum at the concave side of the hook inhibits cell elongation. We paid particular attention to the PP2C.D1 protein phosphatase because it is a negative regulator of cell elongation, exhibits a specific expression pattern in epidermal cells at the concave side of the hook (17) (Fig. 4A), and its expression is induced by IAA (17). In addition, pp2c.d1 mutants exhibit impaired apical hook development (17, 44) (Fig. 4, B and C) because of the release of growth inhibition at the concave side (fig. S5A). The highly specific expression of PP2C.D1 coincides with the auxin maximum at the concave side of the hook (Fig. 4A). We found that the expression of PP2C.D1::EGFP-GUS (17) in the apical hook was auxin regulated as shown in auxin-related mutants pin3-3 (28), arf7-1 (45), and yuc1-D (fig. S5B) (26). Furthermore, treatments with 10 μM IAA or 1 μM auxin efflux inhibitor N-(1-naphthyl)phthalamic acid (NPA), abolished the asymmetric expression of PP2C.D1 (fig. S5C), indicating that asymmetric auxin distribution determines the pattern of PP2C.D1 expression in the apical hook. Using reverse transcription quantitative polymerase chain reaction (RT-qPCR) and Western blot analyses, we further confirmed that PP2C.D1 expression was induced by auxin in a dose-dependent manner (fig. S5, D and E).
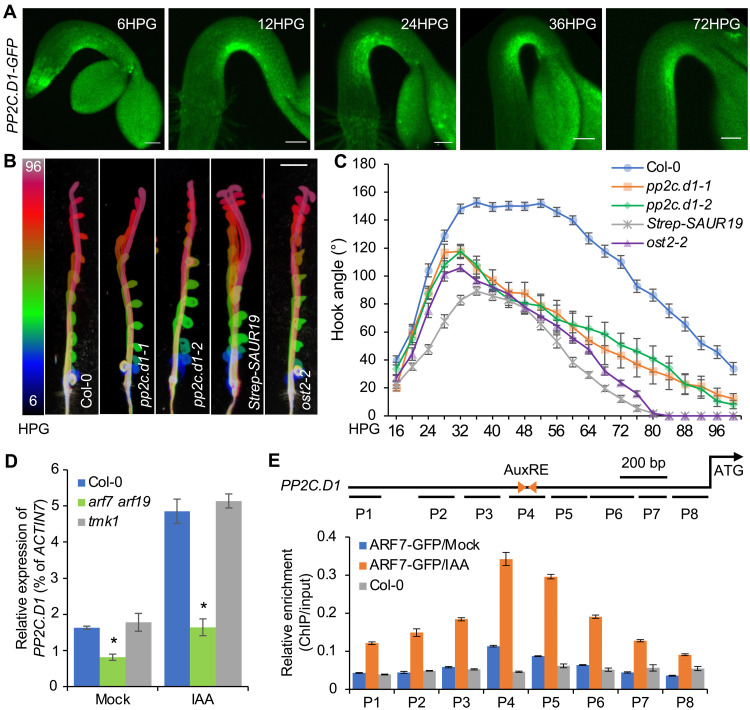
Fig. 4. Auxin induces PP2C.D1 expression via ARF7.
(A) PP2C.D1::PP2C.D1-GFP expression during apical hook development. Germinated seeds were initially placed at a horizontal orientation at 0HPG. Scale bars, 100 μm. (B) Apical hook development in Col-0, pp2c.d1-1, pp2c.d1-2, 35S::Strep-SAUR19, and ost2-2. The color scale indicates the time points (6HPG to 96HPG) with 6-hour intervals. Scale bar, 2 mm. (C) Kinematics of apical hook development in Col-0 (n = 39), pp2c.d1-1 (n = 13), pp2c.d1-2 (n = 16), 35S::Strep-SAUR19 (n = 22), and ost2-2 (n = 25). Values represent sample means ± SEM from three replicates. (D) Quantitative reverse transcription polymerase chain reaction (RT-qPCR) analysis of PP2C.D1 expression in the whole seedlings of WT, tmk1-1, and arf7 arf719 double mutant. Three-day-old dark-grown seedlings were transferred to ½ MS liquid medium containing 1% sucrose with or without 10 μM IAA for 2 hours. PP2C.D1 transcript levels were normalized against ACTIN7 expression. Data are means ± SEM from three biological replicates. *P < 0.01. (E) Schematic representation of the PP2C.D1 gene and chromatin immunoprecipitation (ChIP)–qPCR assays showing the binding of ARF7-GFP to the PP2C.D1 promoter upstream of the ATG start codon. Triangles represent the inverted auxin response elements (AuxREs), and short lines represent the DNA probes followed in the ChIP-qPCR assays. Each ChIP value was normalized to its respective input DNA value, and enrichment is shown as the percentage of input. Error bars represent the SEM from three biological replicates.
The transmembrane kinase 1 (TMK1)–mediated noncanonical auxin pathway was recently reported to regulate apical hook development through transcriptional regulation (9). We asked whether PP2C.D1 expression is induced through the canonical TIR1/AFB auxin pathway (25) or noncanonical TMK1-mediated signaling. PP2C.D1 expression in the hook region was reduced upon inhibiting the SCFTIR1 auxin receptor with auxinole (fig. S5F). In contrast, RT-qPCR analysis showed that auxin-induced PP2C.D1 expression was comparable in WT and the tmk1 mutant (Fig. 4D). These results suggest that PP2C.D1 expression is regulated by the canonical TIR1/AFB pathway, rather than TMK1-mediated signaling. ARF7 (auxin response factor 7) is an important transcriptional activator downstream of the TIR1/AFB receptors and is critical for apical hook development (fig. S5G) (45). The markedly decreased PP2C.D1 expression in the arf7-1 arf19-1 mutant (Fig. 4D) (46) led us to reason that PP2C.D1 may be directly regulated by ARF7 and/or ARF19. We performed chromatin immunoprecipitation (ChIP) experiments followed by qPCR using arf7 arf19 pARF7::ARF7-GFP seedlings (47). Strong ARF7-GFP enrichment was detected within a PP2C.D1 promoter region (P4) containing two inverted auxin response elements (AuxRE) (Fig. 4E) (48). Together, these observations indicate that auxin induces PP2C.D1 expression at the concave side of the hook through ARF7. ARF19 may also contribute to this regulation, as the arf19 mutation confers a modest apical hook phenotype and enhances the strong hook development defect of arf7 mutants (8).
PP2C.D1 controls asymmetric acid growth for apical hook development
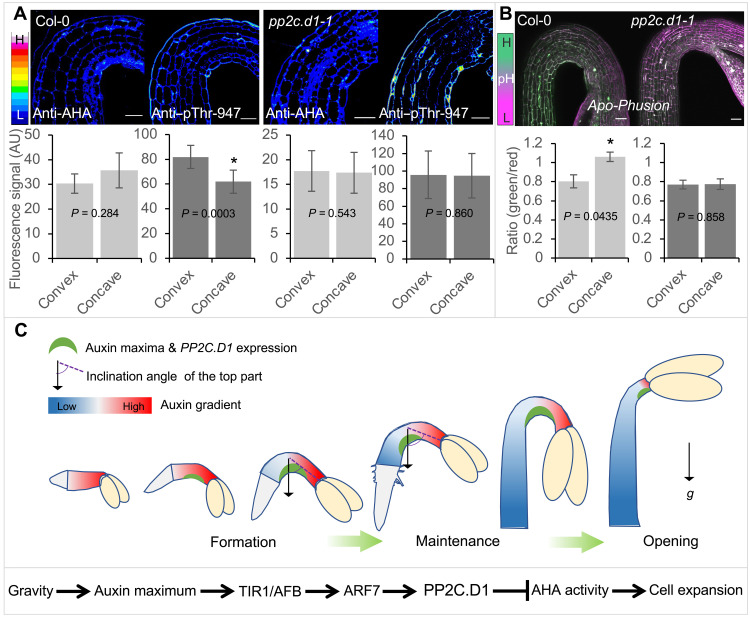
PP2C.D1 overexpression inhibits the activity of PM H+-ATPases (16) by dephosphorylating the penultimate threonine residue [Thr947 in AHA2 (autoinhibited H+-ATPase 2)] (15). While other PP2C.D family members have been implicated in regulating PM H+-ATPase activity to control hypocotyl elongation, the lack of hypocotyl elongation defects in pp2c.d1 mutants (17) indicates that PP2C.D1 does not play a major role in this process. Instead, the specific PP2C.D1 expression at the concave side of the hook suggests that PP2C.D1 predominantly inhibits PM H+-ATPase activity in these cells, thereby causing differential efflux of protons and altered apoplastic pH across the hook. We analyzed the in vivo distribution pattern of PM H+-ATPase at the hook region by immunolabeling with H+-ATPase and pThr947 antibodies (49). In WT, while PM H+-ATPase was nearly equally distributed across the hook epidermis, we observed a higher accumulation of phosphorylated H+-ATPase at the convex side (Fig. 5A and fig. S6A). Consistently, using the genetically encoded apoplastic pH sensor Apo-pHusion (50), the pH at the convex side was lower than that at the concave side (Fig. 5B), suggesting that differential activation of proton pumps and the consequent asymmetric acid growth between the concave and convex sides drive hook curvature. Consistent with this possibility and the fact that SAUR19 promotes H+-ATPase activation (16), 35S:Strep-SAUR19 expression abolished the asymmetric distribution of phosphorylated proton pumps (fig. S6B) and conferred severe defects in hook development (Fig. 4, B and C). The hook defects in these seedlings correlated with increased epidermal cell length at both sides of the hook (fig. S6, C and D). Similar defects were also seen in the open stomata2 (ost2-2) mutant (Fig. 4, B and C, and fig. S6, C and D), which contains a constitutively active allele of the PM H+-ATPase encoded by AHA1 (51). To chemically abolish the differential call wall acidification between the concave and convex sides of the hook, we used the fungal toxin fusicoccin that is known to constitutively activate PM H+-ATPases (16). Fusicoccin treatment induced a strong and significant reduction in the hook angle compared with the mock treatment (fig. S6E), highlighting the importance of asymmetric apoplastic pH in hook development. Asymmetries in PM H+-ATPase phosphorylation and apoplastic pH were also largely abolished in the pp2c.d1-1 mutant (Fig. 5, A and B). These results demonstrate that differential acid growth across the hook is established through PP2C.D1-mediated inhibition of PM H+-ATPase activity at the concave side.
Fig. 5. PP2C.D1 is required for asymmetric acid growth during apical hook development.
(A) Immunolabeling and signal quantification of PM H+-ATPase and Thr947-phosphorylated PM H+-ATPase in WT and pp2c.d1-1 mutant. Scale bars, 50 μm. (B) Apoplast pH and signal quantification at the hook region using the Apo-pHusion marker line in WT and pp2c.d1-1 backgrounds. Scale bars, 50 μm. (A and B) Details of the signal quantification can be found in Materials and Methods. (C) Proposed model for hypocotyl elongation and apical hook development during etiolated seedling development. A molecular signaling pathway underlying auxin-mediated inhibition of cell elongation at the concave side of the hook is shown at the bottom.
Together, these results indicate that PP2C.D1 acts downstream of TIR1/AFB-based canonical auxin signaling to inhibit cell elongation at the concave side of the hook. At lower concentrations (e.g., in the elongating part of the hypocotyl), auxin induces SAUR expression and therefore inhibits the PP2C.D2/5/6 phosphatases to promote cell elongation (fig. S7) (16, 17). By contrast, high auxin concentrations (e.g., at the concave side of the hook) induce PP2C.D1 expression, allowing the system to bypass SAUR regulation and inhibit cell elongation through PP2C.D1 dephosphorylation of PM H+-ATPases (fig. S7). A recent study found that SAUR17 has a high affinity for PP2C.D1 without inhibiting its activity (52). This allows SAUR17 to shield PP2C.D1 from inhibition caused by other SAUR proteins and could serve to reinforce the PP2C.D1-mediated H+-ATPase inhibition resulting from localized phosphatase expression. Although we demonstrate that auxin-regulated differential acid growth is a major contributing component to hook development, it is almost certainly not the only driver of this process. In addition to canonical auxin signaling regulating PP2C.D1 and SAUR expression, TMK1-mediated auxin signaling (9), as well as contributions from other hormones (13), also plays important roles in hook development.
Our findings suggest a mechanistic framework for auxin-regulated hypocotyl elongation and apical hook development during seedling emergence (Fig. 5C). During early etiolated seedling development, high levels of auxin restrain cell elongation in the hypocotyl. This delay in growth coincides with the beginning of apical hook formation. Simultaneously, gravity generates an auxin maximum on one side of the hypocotyl, which induces high PP2C.D1 expression. Consequently, growth inhibition is strengthened at the lower side through PP2C.D1-dependent inhibition of PM H+-ATPase activity, and released at the opposite side, causing the hypocotyl to bend toward the gravity vector and form the hook. As the bottom cells of the hypocotyl slowly elongate and cell volumes increase, auxin levels fall below the threshold for inhibition and begin to promote hypocotyl elongation against the gravity vector. This in turn increases the inclination angle of the top of the hypocotyl to the direction of gravity (Fig. 5C). With the bottom part of the hypocotyl having a more vertical orientation, the gravity-induced auxin maximum is shifted upward, moving the hook on its way. This is supported by the observations that the PP2C.D1-GFP signal moved upward (Fig. 4A) and the number of cells above the center of the hook dropped during hook development (fig. S8). As the hypocotyl cells elongate acropetally, the proportion of the lower part of the hypocotyl that is negatively responding to gravity increases, while the number of cells above the hook that are positively oriented with the gravity vector decreases (fig. S8). Last, when the acropetal wave of hypocotyl elongation reaches the very top cells, the hook starts to open.
This study provides critical insights into the coordination of hypocotyl elongation and apical hook development for successful seedling emergence. Auxin’s biphasic effect on cell elongation underlies this process through a concentration-dependent manner. Early inhibition of hypocotyl elongation may facilitate emergence, as elongation before hook formation would increase kinetic friction, hindering soil penetration and potentially increasing mechanical damage to the cotyledons and shoot meristem. Consistent with this notion, SAUR19 overexpression seedlings and pp2c.d1 mutants displayed substantial reductions in soil emergence compared with WT controls (fig. S9). Coordination of elongation and hook development enables the seedling to safely and efficiently emerge from the soil and initiate phototropic development.
MATERIALS AND METHODS
Plant materials
All Arabidopsis (A. thaliana) lines used in this study were in the Col-0 background except the R2D2 reporter (20), which is in the Columbia-Utrecht ecotype. The transgenic and mutant lines used have been described previously: 35S::StrepII-SAUR19 (53), PP2C.D1::PP2C.D1-GFP (17), PP2C.D1::EGFP-GUS (17), DR5::VENUS-NLS (35), MYR-YFP (54), PIN3::PIN3-GFP (8), Apo-pHusion (50), tmk1 (9), yuc1-D (26), wei8-1 tar2-1 (23) (24), tir1 afb2 (25), axr2-1 (27), atlazy2,3,4 (38), atlazy1,2,3,4 (38), scr-3 (36), shr-2 (36), pp2c.d1-1 (44), pp2c.d1-2 (44), ost2-2 (51), pin3-3 (28), arf7-1 (45), arf7-1 arf19-1 (46), and arf7 arf19 ARF7::ARF7-GFP (47).
Growth conditions
All Arabidopsis materials were grown in continuous darkness at 22°C, unless otherwise indicated. For ChIP-qPCR analysis, seeds were sterilized with 30% bleach for 15 min, washed three times with sterile water, and sown on ½ Murashige and Skoog (MS; PhytoTech Labs, M524) plates containing 1% sucrose (Macron, 8360-06) and 0.6% agargel (Sigma-Aldrich, A3301). Media pH was adjusted to 5.7 using KOH (Fisher Chemical, P250-1). After sowing, the seeds were left at 4°C for 48 hours in darkness, then transferred to white light for 6 hours to stimulate germination, and subsequently to darkness and left at 22°C for the desired time. For other experiments, seeds were sterilized with 70% ethanol for 5 min and germinated on ½ MS plates containing 0.6% agargel (pH 5.7). Germination was defined as the time when the radicle broke through the endosperm (0HPG). At this time, seedlings were selected and aligned horizontally or vertically on ½ MS plates containing 1% sucrose and 0.6% agargel (pH 5.7). The plates were wrapped with two layers of foil to simulate constant darkness.
Quantification of angles of hook curvature
The angle of hook curvature is defined as 180° minus the angle between the tangential of the apical part with the axis of the lower part of the hypocotyl (8). For quantifying the hook angles of scr-3, shr-2, atlazy2,3,4, atlazy1,2,3,4, and arf7 mutants, germinated seeds were initially placed at a horizontal orientation on ½ MS plates containing 1% sucrose and 0.6% agargel (pH 5.7) at 0HPG, which were wrapped with two layers of foil and kept vertically in the growth chamber. Pictures of the seedlings were taken at 24HPG or 36HPG (arf7) for measuring the hook angles using ImageJ (National Institutes of Health, Bethesda, MD, USA). For measuring the hook angles in the rotation experiment, germinated seeds were initially placed horizontally. The wrapped plates were rotated 180° once per hour (rotations were done at 1, 2, 3, …11HPG). Pictures of the seedlings were taken at 12HPG for measuring the hook angles. To investigate the gravity effect on the hook angle in the absence of roots, T0 seedlings were placed vertically and left to grow for 6 hours in the dark. Roots were then decapitated under green light and seedlings were placed horizontally on fresh plates and rotated clockwise in the dark every hour for an extra 18 hours. Controls consisted of nonrotated rootless seedlings and intact seedlings with and without hourly rotations. For fusicoccin treatment, seedlings were transferred at T0 and placed horizontally in cell culture plates containing ½ MS media with 1% sucrose and 5 μM fusicoccin (ENZO, BML-EI334-0001) and imaged at 36HPG. For analyzing the kinematics of apical hook development, germinated seeds were initially placed at a vertical orientation on ½ MS plates with 1% sucrose and 0.6% agargel (pH 5.7) at 0HPG, which were kept vertically in darkness at 22°C. Development of seedlings was recorded at 15-min intervals with an infrared light source (880-nm light-emitting diode; Advanced Illumination) by a Stingray F146B CCD camera (Allied Vision Technologies) equipped with a 18- to 108-mm macro video zoom lens with an infrared long-pass filter. Images for selected time points were extracted, and angles of hook curvature were measured using ImageJ. Frames with 6-hour intervals were imported in ImageJ and color coded using the (Rainbow RGB-LUT projection) in the temporal-color coding tool. For the kinematic analysis of hypocotyl length of Col-0, axr2-1, wei8 tar2, and tir1 afb2 grown under the infrared setup, time-lapse images with 3- or 6-hour intervals were collected, and hypocotyl length was measured using ImageJ.
Plant gravitropic responses
Two-week-old tomato and 4-week-old Arabidopsis seedlings were uniformly sprayed with either 0.5 mM IAA (tomato) or 1 mM IAA (Arabidopsis) using a liquid mister. Mock treatments with dimethyl sulfoxide (DMSO) were used as controls. Plants were left upright for 20 min before placing them horizontally and time-lapse imaging with 2-minute intervals. Four-day-old etiolated tomato seedlings were cut either above the hook (top cut; fig. S4) or near the hypocotyl base (bottom cut; fig. S4). Segments were placed horizontally on ½ MS media supplemented with 1% sucrose and imaged over time in the dark using the infrared setup. Images were analyzed using ImageJ.
Measurement of epidermal cell length
For measuring the cell length of hypocotyl epidermal cells during etiolated seedling development, Arabidopsis hypocotyls were synchronized by selecting seeds at germination (0HPG; radical emergence from endosperm). Germinated seeds were placed at a vertical orientation and then used for measurements at different time points. Seedlings were stained with propidium iodide (0.1 mg/mL) (Molecular Probes, P3566) for 10 min before imaging, and the outline of epidermal cells was imaged using a Leica DM5000B fluorescence microscope equipped with a 490- to 510-nm excitation and 520- to 550-nm band-pass emission filter cube and a 20× air objective lens. Epidermal cell length was measured using ImageJ software. Analyses were performed on middle epidermal files with no cell division.
For measuring the epidermal cell length at the concave and convex sides of the hook, germinated seeds were initially placed at a horizontal orientation at 0HPG. At 36HPG, hooks were dissected using custom-made blades, placed in longitudinal slits on 1.5% agar pads, and imaged using transmitted light on a Leica DM5000B microscope. Measurements were done using ImageJ.
Immunolabeling
Seedlings were fixed in a phosphate-buffered saline (PBS) solution containing 2% formaldehyde (Acros, 11969) and 2.5% glutaraldehyde (Sigma-Aldrich, G6257) under vacuum for 1 hour and left at 4°C overnight. Samples were washed twice in PBS and then dehydrated in a series of increasing ethanol concentrations. Samples were then embedded in medium-grade LR White (London Resin Company), which was left to polymerize at 60°C overnight. Thin sections (0.5 μm) were generated using glass knives on a Leica UltraCut microtome. Sections were blocked in PBS solution containing 2% bovine serum albumin (BSA) (Sigma-Aldrich, 2153) for 1 hour. The primary antibodies (Anti-AHA and anti-pThr947) were diluted 500 times in PBS-BSA and left on the samples overnight at 4°C. Samples were then washed four times for 5 min each in PBS-BSA. Samples were then incubated for 3 hours in DyLight 488 donkey–anti-rabbit secondary antibody (BioLegend, 406404), which was diluted 400 times in PBS-BSA. Samples were then washed six times for 5 min each in PBS. Sections were mounted in citifluor CFM-3 (Electron Microscopy Sciences), and slides were sealed with nail polish and imaged within 2 days from mounting them.
Confocal imaging
For R2D2 imaging, germinated seeds were placed at a vertical orientation at 0HPG, and the seedlings were examined at indicated times. For IAA treatment, 12HPG old seedlings were incubated with DMSO (mock) or 100 nM IAA for 30 min before imaging. For KOK2153 treatment, germinated seeds were grown on the plates supplemented with DMSO (mock) or 10 μM KOK2153 (KOK; custom synthesis by LabSeeker, Wujiang City, China) for 12 hours before imaging. For PIN3::PIN3-GFP imaging, germinated seeds were placed at a horizontal orientation at 0HPG, and the seedlings were examined at 6HPG. For PIN3::PIN3-GFP sample preparation, seedlings were dissected out of the seed coat and cut perpendicular to the hypocotyl axis using custom-designed pins and blades. They were then placed in an agar pad with the cut side facing the outside. A drop of water was applied, and the samples were covered with a coverslip and imaged using a confocal microscope. For Apo-pHusion imaging, germinated seeds were placed at a horizontal orientation at 0HPG, and the seedlings were examined at 36HPG. For MYR-YFP imaging, germinated seeds were placed at a horizontal orientation at 0HPG, and the seedlings were examined at different time points. The tip of the hook was determined using the kappa plugin in FIJI. Briefly, the points with highest curvature on the concave and convex sides were determined, and a line joining them indicates the middle of the hook.
Confocal imaging was conducted using a Nikon A1si confocal microscope. For DR5::GFP, PIN3::PIN3-GFP, and DyLight 488 imaging, a 20×, 0.75 numerical aperture objective was used. Samples were excited with a 480-nm laser, and the emitted laser was collected between 500 and 550 nm. For MYR-YFP and DR5::VENUS-NLS, a 10×, 0.45 numerical aperture objective was used. A 514.5-nm excitation laser and a 510- to 570-mm emission filter were used. For R2D2 and Apo-pHusion, images were acquired using a 10×, 0.45 numerical aperture objective. A 488-nm excitation laser and 500- to 550-mn emission filter were used for the first channel, and 562-nm excitation and 570- to 620-nm emission filter were used for the second channel.
Image processing and analysis
All confocal images were processed using ImageJ. Maximum projection images were generated, and background noise was subtracted using a 50-pixel rolling ball radius. For immunolabeling images, a 16-color look-up table was applied. A 3-pixel-wide line was used in ImageJ to plot the fluorescence intensity profile on the outer periclinal walls of the epidermal cells on the concave and the convex sides. For processing R2D2 images, after background removal, a Gaussian blur was applied with a 2.0 sigma (radius). The red channel was then divided by the yellow channel using the image calculator function to generate an image that was used to quantify the signal ratio in the nucleus. The signal was measured using the average intensity per nucleus, and data were exported to excel. For the Apo-pHusion images, a 20-pixel-wide line was used in ImageJ to plot the fluorescence intensity profile of the green and red channels on the epidermal cells both at the concave and the convex sides of the hook. For PIN3::PIN3-GFP, a 16-color look-up table was applied. A 3-pixel-wide line was used in ImageJ to plot the fluorescence intensity profile on the outer periclinal walls of the endodermis. All data were then exported to and analyzed in Microsoft Excel.
Quantification of endogenous IAA
To quantify the IAA levels in the elongating hypocotyls at different times, seedlings were grown on ½ MS plates containing 1% sucrose and 1.5% agargel (pH 5.7). The shoot meristem, cotyledons, and root were removed before harvesting the hypocotyls for analysis. For measuring the IAA levels in the bottoms and tops of hypocotyls, the hypocotyls of 36HPG seedlings were cut into two equal halves and analyzed separately.
Free IAA was measured by isotope dilution essentially as previously described (55). The samples were analyzed by liquid chromatography with tandem mass spectrometry (LC-MS/MS) at high resolution. [13C6]IAA internal standard was added with 2-propanol/buffer (56) to plant tissue samples weighing 7 to 21 mg, which were then homogenized and incubated for 1 hour on ice. Samples were then diluted with water, centrifuged, and IAA was extracted from the supernatant using amino and polymethylmethacrylate epoxide (PMME) solid phase extraction resins in Top Tips spin tips (Glygen, Columbia, MD, USA). After elution from PMME tips with methanol, sample volumes were reduced to approximately 20 μl and transferred to autosampler vials for LC-MS/MS analysis with a Dionex Ultimate 3000 RSLC HPLC coupled to a hybrid quadrupole Orbitrap Q Exactive mass spectrometer (Thermo Fisher Scientific). Extract (8 to 10 μl) was injected onto a 50 × 2.1 mm Force C18 column with 1.8-μm particle size (Restek, Bellefonte, PA, USA) and run with a solvent gradient of 0.1% formic acid in water (solvent A) and 0.1% formic acid in acetonitrile (solvent B) at a flow rate of 0.4 ml min−1. Gradient parameters were as follows: −1 to 0 min, 5% B; 0 to 3 min, 5 to 20% B; 3 to 6 min, 20 to 80% B; and 6 to 6.5 min, 80% B. Mass spectrometry data were collected in the parallel reaction monitoring (PRM) scan mode with the [M + 1] for IAA and [13C6]IAA at 176.08 and 182.1 mass/charge ratio (m/z), respectively, in the inclusion list. Extracted ion chromatogram peaks for 130.0641 to 130.0661 m/z (corresponding to unlabeled quinolinium ion) and 136.0843 to 136.0863 m/z ([13C6] quinolinium produced from [13C6]IAA internal standard) were selected at 4.4 to 4.8 min. Retention time and peak areas were used to calculate endogenous IAA levels by isotope dilution (57, 58).
For calculating the IAA concentration, the IAA amount was divided by the fresh weight or the total volume of the hypocotyls, which was estimated by hypocotyl number times hypocotyl volume. We considered the hypocotyl as a truncate cone, whose volume was calculated using the formula: volume = (1/3) * π * L * ((D/2)2 + (d/2)2 + (D/2) * (d/2)), where L is the hypocotyl length, D is the diameter of the base of the hypocotyl, and d is the diameter of the top of the hypocotyl.
GUS staining
SAUR22::GUS and PP2C.D1::EGFP-GUS seedlings at indicated times were used for β-glucuronidase (GUS) staining. Seedlings were incubated in the staining buffer containing 0.1 M sodium phosphate buffer (pH 7), 1 mM K4Fe(CN)6, 1 mM K3Fe(CN)6, 0.1% Triton X-100, and X-Gluc (1 mg/ml) for 2 hours at 37°C. GUS expression patterns were imaged with an Olympus SZX12 dissecting microscope using SPOT Advanced imaging software.
Phytohormone and chemical treatment
Germinated seeds were grown on ½ MS plates containing 1% sucrose and 0.6% agargel (pH 5.7) supplemented with various chemicals: 100 nM IAA (Chem-Implex International, 00188) or 1 μM NPA (Duchefa Biochemie), 10 μM KOK2153 (also called Pyruvamine 2153) (21), or 10 μM auxinole. For gene expression, Western blot, and ChIP assays, we transferred 3-day-old dark-grown seedlings to ½ MS liquid medium containing 1% sucrose and different concentrations of IAA for 2 hours.
RNA extraction and RT-qPCR analysis
Dark-grown seedlings were treated with different concentrations of IAA in ½ MS liquid medium for 2 hours. Total RNA was extracted from harvested seedlings using the NucleoSpin RNA Plant kit (Macherey-Nagel), and the quality of the total RNA was determined using an Implen NanoPhotometer P330. Two micrograms of total RNA was used to synthesize the first-strand cDNA with the M-MLV reverse transcriptase kit (Promega, M1701). RT-qPCR was performed on a StepOnePlus Real-Time PCR System (Applied Biosystems) with the Brilliant III Ultra-Fast SYBR Green QPCR Master Mix (Agilent Technologies, 600882). The expression levels of genes were normalized to that of the ACTIN7 gene. Primer information is given in table S1.
Protein extraction and Western blot
Three-day-old dark-grown PP2C.D1::PP2C.D1-GFP seedlings were treated with different concentrations of IAA in ½ MS liquid medium containing 1% sucrose for 2 hours. Microsomal fractions were prepared by two-phase partitioning as previously described (16). Twenty micrograms of microsomal proteins was mixed with SDS–polyacrylamide gel electrophoresis (PAGE) sample buffer, separated by SDS-PAGE, and blotted to nitrocellulose. The proteins were detected by Western blot with anti-GFP primary antibody (Covance, MMS-118R). A nonspecific band detected by the antibody was served as a loading control.
ChIP-qPCR assay
Three-day-old dark-grown arf7 arf19 ARF7::ARF7-GFP seedlings treated with or without 10 μM IAA were used for ChIP-qPCR analysis. The ChIP-qPCR assay was conducted as described previously (59). The ChIP signal was quantified by qPCR. The primers used in qPCR were designed to amplify various regions of the PP2C.D1 promoter. Each ChIP value was normalized to its respective input DNA value, and enrichment of DNA is shown as the percentage of input.
Statistical analysis
All box plots were generated using the PlotsOfData (60) or ggplot2 in RStudio. In the boxplots, the top, bottom, and middle lines represent the 75th percentile, the 25th percentile, and the median, respectively. Bar graphs and connecting lines were generated using Microsoft Excel. All the experiments were performed at least three times. The values were collected from three biological replicates. Student’s t test and analysis of variance (ANOVA) were used for statistical analysis, using Microsoft Excel.
Acknowledgments
We thank E. Spalding for providing the seeds of atlazy mutants, J. Reed for the gift of axr2-1 seeds, H. Fukaki and M. Furutani for providing the arf7 arf19 pARF7::ARF7-GFP seeds, T. Kinoshita for providing α-AHA and α-pThr947 antisera, K.-i. Hayashi for providing auxinole, and C. Carter for providing his real-time PCR system for our qPCR experiments. We also thank the University Imaging Center for assistance with confocal microscopy.
Funding: This work was supported by grants from the NIH (GM067203 to W.M.G.), the USDA National Institute of Food and Agriculture (2018-67013-27503 to J.D.C.), and the K.C. Wong Education Foundation (to C.L.).
Author contributions: Conceptualization: M.D., F.B., and W.M.G. Methodology: M.D., F.B., A.S., M.T., and C.L. Investigation: M.D., F.B., A.S., M.T., Y.L., X.Z., J.H.W., and H.R. Visualization: M.D. and F.B. Funding acquisition: W.M.G., J.D.C., and C.L. Project administration: W.M.G. and M.D. Supervision: W.M.G. Writing—original draft: M.D. Writing—review and editing: M.D., W.M.G., F.B. Research strategy: M.D., F.B., and W.M.G. Experimentation: M.D., F.B., Y.L., X.Z., J.H.W., and H.R. IAA quantification: A.S. and M.T., with assistance from J.C. C.L. assisted with ChIP experiments and hosted M.D. to perform collaborative study in his laboratory. Data analysis: M.D., F.B., J.D.C., and W.M.G. Writing: M.D., W.M.G., and F.B.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Raw image and measurement data have been deposited in the University Digital Conservancy and are available at https://doi.org/10.13020/spr0-2r11. Correspondence and requests for materials should be addressed to W.M.G.
Supplementary Materials
This PDF file includes:
Legends for movies S1 to S8
Figs. S1 to S9
Table S1
Other Supplementary Material for this manuscript includes the following:
Movies S1 to S8
REFERENCES AND NOTES
- 1.Gendreau E., Traas J., Desnos T., Grandjean O., Caboche M., Hofte H., Cellular basis of hypocotyl growth in Arabidopsis thaliana. Plant Physiol. 114, 295–305 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.C. Darwin, F. Darwin, The power of movement in plants. New York (Republished 1892), (1881).
- 3.Raz V., Ecker J. R., Regulation of differential growth in the apical hook of Arabidopsis. Development 126, 3661–3668 (1999). [DOI] [PubMed] [Google Scholar]
- 4.Du M., Spalding E. P., Gray W. M., Rapid auxin-mediated cell expansion. Annu. Rev. Plant Biol. 71, 379–402 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li H., Johnson P., Stepanova A., Alonso J. M., Ecker J. R., Convergence of signaling pathways in the control of differential cell growth in Arabidopsis. Dev. Cell 7, 193–204 (2004). [DOI] [PubMed] [Google Scholar]
- 6.Beziat C., Kleine-Vehn J., The road to auxin-dependent growth repression and promotion in apical hooks. Curr. Biol. 28, R519–R525 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Vandenbussche F., Petrášek J., Žádníková P., Hoyerová K., Pešek B., Raz V., Swarup R., Bennett M., Zažímalová E., Benková E., van der Straeten D., The auxin influx carriers AUX1 and LAX3 are involved in auxin-ethylene interactions during apical hook development in Arabidopsis thaliana seedlings. Development 137, 597–606 (2010). [DOI] [PubMed] [Google Scholar]
- 8.Zadnikova P., Petrásek J., Marhavy P., Raz V., Vandenbussche F., Ding Z., Schwarzerová K., Morita M. T., Tasaka M., Hejátko J., Van Der Straeten D., Friml J., Benková E., Role of PIN-mediated auxin efflux in apical hook development of Arabidopsis thaliana. Development 137, 607–617 (2010). [DOI] [PubMed] [Google Scholar]
- 9.Cao M., Chen R., Li P., Yu Y., Zheng R., Ge D., Zheng W., Wang X., Gu Y., Gelová Z., Friml J., Zhang H., Liu R., He J., Xu T., TMK1-mediated auxin signalling regulates differential growth of the apical hook. Nature 568, 240–243 (2019). [DOI] [PubMed] [Google Scholar]
- 10.Thimann K. V., Auxins and the inhibition of plant growth. Biol. Rev. 14, 314–337 (1939). [Google Scholar]
- 11.Bou Daher F., Chen Y., Bozorg B., Clough J., Jönsson H., Braybrook S. A., Anisotropic growth is achieved through the additive mechanical effect of material anisotropy and elastic asymmetry. eLife 7, e38161 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu Q., Gallemí M., Pospíšil J., Žádníková P., Strnad M., Benková E., Root gravity response module guides differential growth determining both root bending and apical hook formation in Arabidopsis. Development 146, dev175919 (2019). [DOI] [PubMed] [Google Scholar]
- 13.Wang Y., Guo H., On hormonal regulation of the dynamic apical hook development. New Phytol. 222, 1230–1234 (2019). [DOI] [PubMed] [Google Scholar]
- 14.Rayle D. L., Cleland R. E., The acid growth theory of auxin-induced cell elongation is alive and well. Plant Physiol. 99, 1271–1274 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takahashi K., Hayashi K., Kinoshita T., Auxin activates the plasma membrane H+-ATPase by phosphorylation during hypocotyl elongation in Arabidopsis. Plant Physiol. 159, 632–641 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Spartz A. K., Ren H., Park M. Y., Grandt K. N., Lee S. H., Murphy A. S., Sussman M. R., Overvoorde P. J., Gray W. M., SAUR inhibition of PP2C-D phosphatases activates plasma membrane H+-ATPases to promote cell expansion in Arabidopsis. Plant Cell 26, 2129–2142 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ren H., Park M. Y., Spartz A. K., Wong J. H., Gray W. M., A subset of plasma membrane-localized PP2C.D phosphatases negatively regulate SAUR-mediated cell expansion in Arabidopsis. PLOS Genet. 14, e1007455 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fendrych M., Akhmanova M., Merrin J., Glanc M., Hagihara S., Takahashi K., Uchida N., Torii K. U., Friml J., Rapid and reversible root growth inhibition by TIR1 auxin signalling. Nat. Plants 4, 453–459 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barbez E., Dünser K., Gaidora A., Lendl T., Busch W., Auxin steers root cell expansion via apoplastic pH regulation in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 114, E4884–E4893 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liao C.-Y., Smet W., Brunoud G., Yoshida S., Vernoux T., Weijers D., Reporters for sensitive and quantitative measurement of auxin response. Nat. Methods 12, 207–210 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Narukawa-Nara M., Nakamura A., Kikuzato K., Kakei Y., Sato A., Mitani Y., Yamasaki-Kokudo Y., Ishii T., Hayashi K. I., Asami T., Ogura T., Yoshida S., Fujioka S., Kamakura T., Kawatsu T., Tachikawa M., Soeno K., Shimada Y., Aminooxy-naphthylpropionic acid and its derivatives are inhibitors of auxin biosynthesis targeting l-tryptophan aminotransferase: Structure-activity relationships. Plant J. 87, 245–257 (2016). [DOI] [PubMed] [Google Scholar]
- 22.Hayashi K., Neve J., Hirose M., Kuboki A., Shimada Y., Kepinski S., Nozaki H., Rational design of an auxin antagonist of the SCFTIR1 auxin receptor complex. ACS Chem. Biol. 7, 590–598 (2012). [DOI] [PubMed] [Google Scholar]
- 23.Stepanova A. N., Robertson-Hoyt J., Yun J., Benavente L. M., Xie D. Y., Doležal K., Schlereth A., Jürgens G., Alonso J. M., TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell 133, 177–191 (2008). [DOI] [PubMed] [Google Scholar]
- 24.Tao Y., Ferrer J. L., Ljung K., Pojer F., Hong F., Long J. A., Li L., Moreno J. E., Bowman M. E., Ivans L. J., Cheng Y., Lim J., Zhao Y., Ballaré C. L., Sandberg G., Noel J. P., Chory J., Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell 133, 164–176 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dharmasiri N., Dharmasiri S., Weijers D., Lechner E., Yamada M., Hobbie L., Ehrismann J. S., Jürgens G., Estelle M., Plant development is regulated by a family of auxin receptor F box proteins. Dev. Cell 9, 109–119 (2005). [DOI] [PubMed] [Google Scholar]
- 26.Zhao Y., Christensen S. K., Fankhauser C., Cashman J. R., Cohen J. D., Weigel D., Chory J., A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science 291, 306–309 (2001). [DOI] [PubMed] [Google Scholar]
- 27.Timpte C., Wilson A. K., Estelle M., The axr2-1 mutation of Arabidopsis thaliana is a gain-of-function mutation that disrupts an early step in auxin response. Genetics 138, 1239–1249 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Friml J., Wisniewska J., Benkova E., Mendgen K., Palme K., Lateral relocation of auxin efflux regulator PIN3 mediates tropism in Arabidopsis. Nature 415, 806–809 (2002). [DOI] [PubMed] [Google Scholar]
- 29.MacDonald I. R., Gordon D. C., Hart J. W., Maher E. P., The positive hook:The role of gravity in the formation and opening of the apical hook. Planta 158, 76–81 (1983). [DOI] [PubMed] [Google Scholar]
- 30.Myers A. B., Firn R. D., Digby J., Gravitropic sign reversal—a fundamental feature of the gravitropic perception or response mechanisms in some plant organs. J. Exp. Bot. 45, 77–83 (1994). [Google Scholar]
- 31.Migliaccio F., Micciulla O., Ferrari S., Hook formation in sunflower seedlings is directed by both positive gravitropism and a form of circumnutation. Plant Biosyst. 132, 11–16 (1998). [Google Scholar]
- 32.Miyamoto K., Yamasaki T., Uheda E., Ueda J., Analysis of apical hook formation in Alaska pea with a 3-D clinostat and agravitropic mutant ageotropum. Front. Plant Sci. 5, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chauvet H., Pouliquen O., Forterre Y., Legué V., Moulia B., Inclination not force is sensed by plants during shoot gravitropism. Sci. Rep. 6, 35431 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pouliquen O., Forterre Y., Bérut A., Chauvet H., Bizet F., Legué V., Moulia B., A new scenario for gravity detection in plants: The position sensor hypothesis. Phys. Biol. 14, 035005 (2017). [DOI] [PubMed] [Google Scholar]
- 35.Heisler M. G., Ohno C., Das P., Sieber P., Reddy G. V., Long J. A., Meyerowitz E. M., Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Curr. Biol. 15, 1899–1911 (2005). [DOI] [PubMed] [Google Scholar]
- 36.Fukaki H., Wysocka-Diller J., Kato T., Fujisawa H., Benfey P. N., Tasaka M., Genetic evidence that the endodermis is essential for shoot gravitropism in Arabidopsis thaliana. Plant J. 14, 425–430 (1998). [DOI] [PubMed] [Google Scholar]
- 37.Nakamura M., Nishimura T., Morita M. T., Bridging the gap between amyloplasts and directional auxin transport in plant gravitropism. Curr. Opin. Plant Biol. 52, 54–60 (2019). [DOI] [PubMed] [Google Scholar]
- 38.Yoshihara T., Spalding E. P., LAZY genes mediate the effects of gravity on auxin gradients and plant architecture. Plant Physiol. 175, 959–969 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Friml J., Vieten A., Sauer M., Weijers D., Schwarz H., Hamann T., Offringa R., Jürgens G., Efflux-dependent auxin gradients establish the apical–basal axis of Arabidopsis. Nature 426, 147–153 (2003). [DOI] [PubMed] [Google Scholar]
- 40.Rakusová H., Gallego-Bartolomé J., Vanstraelen M., Robert H. S., Alabadí D., Blázquez M. A., Benková E., Friml J., Polarization of PIN3-dependent auxin transport for hypocotyl gravitropic response in Arabidopsis thaliana. Plant J. 67, 817–826 (2011). [DOI] [PubMed] [Google Scholar]
- 41.Masson P. H., Tasaka M., Morita M. T., Guan C., Chen R., Boonsirichai K., Arabidopsis thaliana: A model for the study of root and shoot gravitropism. The Arabidopsis Book 1, e0043 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lomax T. L., Molecular genetic analysis of plant gravitropism. Gravit. Space Biol. Bull. 10, 75–82 (1997). [PubMed] [Google Scholar]
- 43.Harrison M. A., Pickard B. G., Auxin asymmetry during gravitropism by tomato hypocotyls. Plant Physiol. 89, 652–657 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sentandreu M., Martín G., González-Schain N., Leivar P., Soy J., Tepperman J. M., Quail P. H., Monte E., Functional profiling identifies genes involved in organ-specific branches of the PIF3 regulatory network in Arabidopsis. Plant Cell 23, 3974–3991 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Harper R. M., Stowe-Evans E. L., Luesse D. R., Muto H., Tatematsu K., Watahiki M. K., Yamamoto K., Liscum E., The NPH4 locus encodes the auxin response factor ARF7, a conditional regulator of differential growth in aerial Arabidopsis tissue. Plant Cell 12, 757–770 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Okushima Y., Overvoorde P. J., Arima K., Alonso J. M., Chan A., Chang C., Ecker J. R., Hughes B., Lui A., Nguyen D., Onodera C., Quach H., Smith A., Yu G., Theologis A., Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: Unique and overlapping functions of ARF7 and ARF19. Plant Cell 17, 444–463 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ito J., Fukaki H., Onoda M., Li L., Li C., Tasaka M., Furutani M., Auxin-dependent compositional change in Mediator in ARF7- and ARF19-mediated transcription. Proc. Natl. Acad. Sci. U.S.A. 113, 6562–6567 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Freire-Rios A., Tanaka K., Crespo I., van der Wijk E., Sizentsova Y., Levitsky V., Lindhoud S., Fontana M., Hohlbein J., Boer D. R., Mironova V., Weijers D., Architecture of DNA elements mediating ARF transcription factor binding and auxin-responsive gene expression in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 117, 24557–24566 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hayashi Y., Nakamura S., Takemiya A., Takahashi Y., Shimazaki K. I., Kinoshita T., Biochemical characterization of in vitro phosphorylation and dephosphorylation of the plasma membrane H+-ATPase. Plant Cell Physiol. 51, 1186–1196 (2010). [DOI] [PubMed] [Google Scholar]
- 50.Gjetting K. S., Ytting C. K., Schulz A., Fuglsang A. T., Live imaging of intra- and extracellular pH in plants using pHusion, a novel genetically encoded biosensor. J. Exp. Bot. 63, 3207–3218 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Merlot S., Leonhardt N., Fenzi F., Valon C., Costa M., Piette L., Vavasseur A., Genty B., Boivin K., Müller A., Giraudat J., Leung J., Constitutive activation of a plasma membrane H+-ATPase prevents abscisic acid-mediated stomatal closure. EMBO J. 26, 3216–3226 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang J., Sun N., Zhang F., Yu R., Chen H., Deng X. W., Wei N., SAUR17 and SAUR50 differentially regulate PP2C-D1 during apical hook development and cotyledon opening in Arabidopsis. Plant Cell 32, 3792–3811 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Spartz A. K., Lee S. H., Wenger J. P., Gonzalez N., Itoh H., Inzé D., Peer W. A., Murphy A. S., Overvoorde P. J., Gray W. M., The SAUR19 subfamily of SMALL AUXIN UP RNA genes promote cell expansion. Plant J. 70, 978–990 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Willis L., Refahi Y., Wightman R., Landrein B., Teles J., Huang K. C., Meyerowitz E. M., Jönsson H., Cell size and growth regulation in the Arabidopsis thaliana apical stem cell niche. Proc. Natl. Acad. Sci. U.S.A. 113, E8238–E8246 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tang Q., Yu P., Tillmann M., Cohen J. D., Slovin J. P., Indole-3-acetylaspartate and indole-3-acetylglutamate, the IAA-amide conjugates in the diploid strawberry achene, are hydrolyzed in growing seedlings. Planta 249, 1073–1085 (2019). [DOI] [PubMed] [Google Scholar]
- 56.Liu X., Hegeman A. D., Gardner G., Cohen J. D., Protocol: High-throughput and quantitative assays of auxin and auxin precursors from minute tissue samples. Plant Methods 8, 31 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cohen J. D., Baldi B. G., Slovin J. P., 13C6-[benzene ring]-indole-3-acetic acid. Plant Physiol. 80, 14–19 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Barkawi L. S., Tam Y. Y., Tillman J. A., Normanly J., Cohen J. D., A high-throughput method for the quantitative analysis of auxins. Nat. Protoc. 5, 1609–1618 (2010). [DOI] [PubMed] [Google Scholar]
- 59.Du M., Zhao J., Tzeng D. T. W., Liu Y., Deng L., Yang T., Zhai Q., Wu F., Huang Z., Zhou M., Wang Q., Chen Q., Zhong S., Li C.-B., Li C., MYC2 orchestrates a hierarchical transcriptional cascade that regulates jasmonate-mediated plant immunity in tomato. Plant Cell 29, 1883–1906 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Postma M., Goedhart J., PlotsOfData—A web app for visualizing data together with their summaries. PLOS Biol. 17, e3000202 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Legends for movies S1 to S8
Figs. S1 to S9
Table S1
Movies S1 to S8