RAD51AP2 is required specifically for efficient meiotic recombination to form crossover between X and Y chromosomes.
Abstract
Faithful segregation of X and Y chromosomes requires meiotic recombination to form a crossover between them in the pseudoautosomal region (PAR). Unlike autosomes that have approximately 10-fold more double-strand breaks (DSBs) than crossovers, one crossover must be formed from the one or two DSBs in PARs, implying the existence of a sex chromosome–specific recombination mechanism. Here, we found that RAD51AP2, a meiosis-specific partner of RAD51, is specifically required for the crossover formation on the XY chromosomes, but not autosomes. The decreased crossover formation between X and Y chromosomes in Rad51ap2 mutant mice results from compromised DSB repair in PARs due to destabilization of recombination intermediates rather than defects in DSB generation or synapsis. Our findings provide direct experimental evidence that XY recombination may use a PAR-specific DSB repair mechanism mediated by factors that are not essential for recombination on autosomes.
INTRODUCTION
Meiosis, a cell division process compulsory for the production of haploid gametes, involves an ordered series of events, including pairing, synapsis, recombination, and separation, occurring between homologous chromosomes (1, 2). Meiotic recombination results in crossovers that hold homologous chromosomes together and ensure correct alignment of chromosomes at metaphase I and accurate segregation during meiotic division (2–4). Failure in crossover formation impedes the alignment of chromosomes at the metaphase plate, causes meiosis arrest, and ultimately leads to infertility (5–10).
Meiotic recombination is initiated with programmed DNA double-strand breaks (DSBs). In most mammals, these DSBs occur at discrete hotspots defined by the PR domain-containing 9 (PRDM9)-deposited histone modifications and are catalyzed by a topoisomerase-like enzyme complex consisting of SPO11 and type 2 DNA topisomerase 6 subunit B-like protein (TOP6BL) (11–15). The DSB ends are resected to generate 3′ single-stranded tails for strand invasion, leading to the formation of recombination intermediates (3, 16–18). These recombination intermediates can either be stabilized, mature into a four-way branched DNA structure to yield crossovers by resolution, or otherwise, they are disassembled to generate noncrossovers (3, 19–22). A far greater number of meiotic DSBs form and then mature into crossovers. In mice, 200 to 300 meiotic DSBs are generated but produce only about 25 crossovers per spermatocyte, with each pair of homologs obtaining at least one crossover (23–25). Therefore, crossover formation is controlled not only by the formation of programmed DSBs but also by the process of DSB repair.
Unlike autosomes that are homologous throughout the whole chromosome region [estimated 61 to 195 megabases (Mb) in length, National Center for Biotechnology Information build GRCm39], the XY chromosomes are paired, synapsed at a very short homologous region known as the pseudoautosomal region (PAR), and thus face a greater challenge in crossover formation. The sizes of the PARs are limited, with a length of about 0.7 Mb in mice (26) and about 2.6 and 0.4 Mb for PAR1 and PAR2, respectively, in humans (27). Compared with autosomes, PAR has an extremely high density of crossovers (~570-fold higher than the genome average in mice), postulating a specific mechanism for ensuring XY crossover formation (3, 7, 23). Studies in the past decade have uncovered that the DSB formation is differently regulated at the PAR than on autosomes in mice. For example, PAR DSB formation is activated by a PRDM9-independent pathway (15); most PAR DSBs require SPO11α for formation and are generated after late zygonema, while the autosome DSBs are generated by SPO11β mostly from leptonema to mid-zygonema (7). These specific mechanisms ensure the formation of one or two DSBs at the PAR, which are subsequently repaired to produce one crossover, yielding a ~5-fold higher rate of crossovers per DSB than the genome average (23, 25). Thus, both the PAR-specific mechanism for DSB formation and the disproportionally high yield of crossover per DSB imply that spermatocytes may also use a PAR-specific DSB repair process to ensure XY recombination. However, so far, little experimental evidence has been reported to support this deduction.
Here, we demonstrate that RAD51-associated protein 2 (RAD51AP2), a meiosis-specific and functionally uncharacterized protein, is required for recombination and crossover formation on sex chromosomes, but not autosomes. This gene was found to be mutated in two men with unaligned chromosomes in meiotic metaphase I (MMI). Mouse models mimicking the patients’ mutation failed to form crossover between XY chromosomes due to DSB repair defects in PAR recombination. Further analysis showed that the DSB repair defects were caused by destabilized recombination intermediates. These findings thus provide direct experimental evidence that XY recombination may use a PAR-specific DSB repair mechanism mediated by factors that are not essential for the recombination on autosomes.
RESULTS
Biallelic RAD51AP2 frameshift mutations identified in patients with unaligned chromosomes at MMI
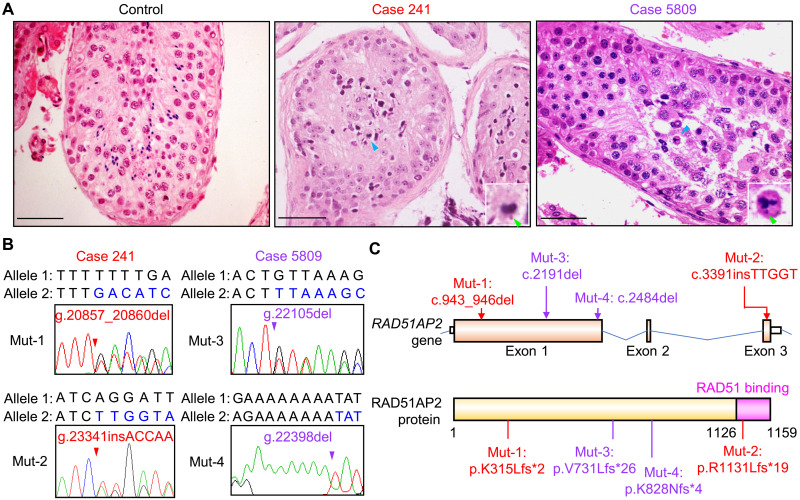
We recruited 60 patients with sporadic nonobstructive azoospermia to investigate genetic causes of male infertility. All the patients displayed abnormal spermatocytes maturation that was identified as an obvious reduction in the number of post-meiotic germ cells in testicular histology. They had normal karyotypes and no Y chromosome microdeletion. Through whole-exome sequencing (WES), we identified four frameshift mutations in RAD51AP2 (NM_001099218): c.943_946del (p.K315Lfs*2) and c.3391insTTGGT (p.R1131Lfs*19) mutations in one patient (registered number at the Human Reproductive Disease Resource Bank, case 241) and c.2191del (p.V731Lfs*26) and c. 2484del (p.K828Nfs*4) mutations in another patient (case 5809) (Fig. 1 and table S1). Subsequent Sanger sequencing of genomic DNA extracted from blood samples validated the presence of these mutations in cases 241 and 5809 (Fig. 1B) and confirmed that these mutations are localized on two different homologous chromosomes in each patient (fig. S1). Hence, these two patients harbor compound heterozygous RAD51AP2 frameshift mutations that are predicted to yield truncated protein products resulting from premature termination of translation.
Fig. 1. Biallelic RAD51AP2 frameshift mutations identified in two patients with unaligned chromosomes in metaphase spermatocytes.
(A) Representative images of testicular sections after hematoxylin and eosin staining. A man who was diagnosed with obstructive azoospermia served as the control. Blue arrowheads indicate metaphase cells with unaligned chromosomes. Green arrowheads indicate the unaligned chromosomes. Scale bars, 50 μm. (B) Sanger sequencing chromatograms of RAD51AP2 frameshift mutations in genomic DNA from the patients. Arrowheads indicate the mutation sites. The nucleotides altered after the site of mutation are written in blue in the mutant allele. (C) Schematic representation of the positions of the mutations identified from patients in the RAD51AP2 gene and protein. The domain interacting with RAD51 is indicated by the pink box. The mutations in case 241 are indicated by red arrows, and the mutations in case 5809 are indicated by purple arrows.
Both patients had normal sex hormone levels and secondary sexual characteristic but small bilateral testicular sizes and no spermatozoa in their semen samples (table S1). Comparative analysis with a control man who was diagnosed with obstructive azoospermia revealed an interesting distinction (Fig. 1A): Cross sections of seminiferous tubules from the control man had abundant spermatogonia, spermatocytes, spermatids, and spermatozoa, whereas there were many spermatogonia and spermatocytes but few spermatids or spermatozoa in the seminiferous tubules of case 5809. Moreover, some metaphase spermatocytes were seen lying close to the lumen, displaying condensed nuclei and obviously unaligned chromosomes (Fig. 1A). In the testicular biopsy of case 241, metaphase spermatocytes with unaligned chromosomes could also be identified (Fig. 1A).
RAD51AP2 is a meiosis-specific protein that localizes to chromosome axes
RAD51AP2 is known as an RAD51-associated protein that interacts with RAD51 through its C terminus by yeast two-hybrid assay (28), but the expression, localization, and biological functions of this protein remain uninvestigated. Here, using the customized antibodies against mouse RAD51AP2, we found that the RAD51AP2 protein was abundant in the testis but was not detected in somatic tissues or in adult ovaries (in which germ cells are arrested at the diplotene stage) (fig. S2A), which is consistent with the previous report showing the detection of RAD51AP2 mRNA in human testis and fetal ovaries, but not in somatic tissues (28). Immunofluorescence staining performed on the testicular cross sections of wild-type (WT) mice revealed the specific presence of RAD51AP2 in the nuclei of spermatocytes (fig. S2B). RAD51AP2 foci were detected on chromosome axes starting from leptonema up until early/mid-pachynema (fig. S2, C and D), with the average number of foci per nucleus peaking in early/mid-zygonema (152.0 ± 43.9), before decreasing to 29.9 ± 10.8 in early/mid-pachynema and totally disappearing before late pachynema. At least one RAD51AP2 focus was always observed in the PARs of early/mid-pachytene spermatocytes. To determine whether the recruitment of RAD51AP2 is dependent on meiotic programmed DSBs, we examined RAD51AP2 foci in spermatocytes from Spo11−/− mice [known as Spo11tm1Mjn, originally described in (29); ID in the Mouse Genome Informatics (MGI) resource, MGI: 2178805], in which meiotic programmed DSBs are not generated. No RAD51AP2 foci were observed in any of the Spo11−/− spread spermatocytes (fig. S2E), indicating that the recruitment of RAD51AP2 to the chromosome axes depends on programmed meiotic DSBs.
Unaligned chromosomes at MMI in mice carrying Rad51ap2 frameshift mutations similar to that in our patient
To assess the potential impacts of the RAD51AP2 mutations identified in patients on spermatogenesis, we generated Rad51ap2 mutant mice carrying mutations similar to the Mut-1 variant identified in case 241. Two mutant mouse lines were obtained: ΔC741 (p.K236Hfs*2) and ΔC689 (p.S288Pfs*4) (fig. S3, A to C). Western blotting using an antibody recognizing an N-terminal RAD51AP2 epitope confirmed the absence of full-length RAD51AP2 proteins and revealed the presence of truncated proteins of the predicted sizes in the mutant mice (fig. S3D). Notably, no RAD51AP2 foci were detected on axes of the sex chromosomes or autosomes in spread spermatocytes from mutant mice of both lines (fig. S3E).
Given that RAD51AP2 localization on chromosome axes was completely abolished in both mouse lines, the following experiments focused on one line (ΔC741). Rad51ap2ΔC741/ΔC741 male mice were viable and displayed no overt developmental anomalies but reduced testicular weight and sperm number (fig. S4, A and B). Histological analyses of testes revealed many MMI spermatocytes with obvious unaligned chromosomes and a reduction in the number of post-meiotic germ cells in the Rad51ap2ΔC741/ΔC741 mutant testes (Fig. 2A). TUNEL (terminal deoxynucleotidyl transferase–mediated deoxyuridine triphosphate nick end labeling) assays detected more apoptotic metaphase I spermatocytes from the mutant mice than in controls (fig. S4, C to E).
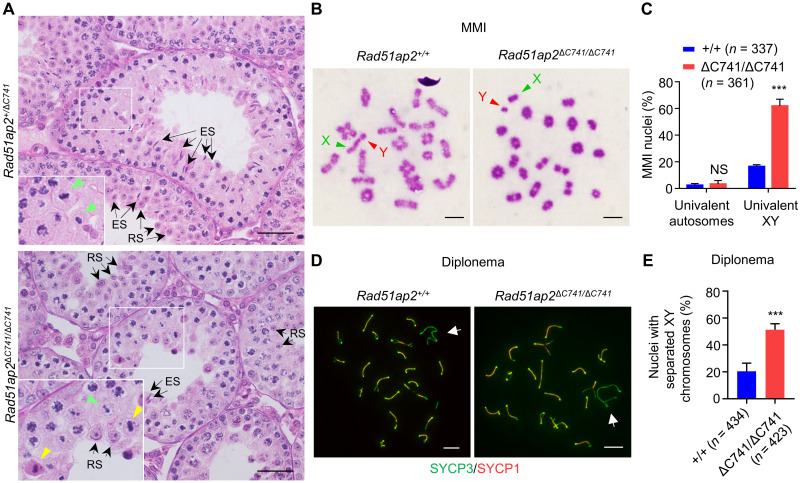
Fig. 2. Precocious separation of XY chromosomes in Rad51ap2 mutant spermatocytes.
(A) Testicular histology from 6-week-old WT and Rad51ap2 mutant mice. Green arrowheads indicate normal metaphase cells, yellow arrowheads indicate metaphase cells with unaligned chromosomes, and arrows indicate representative round spermatids (RSs) or elongating/elongated spermatids (ESs). Scale bars, 50 μm. (B) MMI spermatocytes stained with Giemsa. Chromosomes X and Y are indicated. Scale bars, 10 μm. (C) Frequencies of nuclei with XY chromosome or autosome dissociation in MMI spermatocytes. n, the number of MMI cells scored from four mice per genotype. NS, not significant. ***P < 0.001, two-way analysis of variance. (D) Representative spread spermatocytes in diplonema, stained for SYCP3 (green) and SYCP1 (red). Arrows indicate the X and Y chromosomes. Scale bars, 10 μm. (E) Frequencies of nuclei with XY separation in diplotene spermatocytes. n, the number of cells scored from at least two mice per genotype. ***P < 0.001, two-tailed Student’s t test.
These trends were confirmed by histological analysis of the first wave of spermatogenesis. At the age of 21 days postnatal (dpp), in contrast to the metaphase cells in control mice for which all of the chromosomes aligned at metaphase plate, tubules from Rad51ap2ΔC741/ΔC741 mice contained many MMI spermatocytes with unaligned chromosomes and/or a condensed (indicative of apoptosis) nucleus (fig. S4F). At 28 dpp, elongated spermatids were evident in control mice, but in Rad51ap2ΔC741/ΔC741 tubules, there were many metaphase spermatocytes with unaligned chromosomes and/or condensed nuclei but very few post-meiotic cells and almost no elongating/elongated spermatids were observed, collectively suggesting an incomplete metaphase arrest (fig. S4F) (30). To quantify the frequencies of MMI spermatocytes with unaligned chromosomes, we prepared testicular cell smears and counted MMI cells with and without unaligned chromosomes after Giemsa staining. In the Rad51ap2 mutants, 65.6% of MMI cells showed chromosome(s) that did not line up at the metaphase plate (either unilaterally or bilaterally), which was much higher than in WT mice (11.4%) (fig. S4, G and H). Collectively, these data demonstrate that, in congruent with the observations in patients’ testicular histology (Fig. 1A), RAD51AP2 mutant mice display a high rate of MMI spermatocytes with unaligned chromosomes.
Increased frequency of XY dissociation in Rad51ap2 mutant MMI spermatocytes
The unaligned chromosomes at MMI can be caused by the absence of physical links (or chiasmata) between homologous chromosomes, which usually present as univalent chromosomes (5, 6, 10). We thus prepared metaphase chromosome spreads from control and Rad51ap2ΔC741/ΔC741 testes to detect the univalent chromosomes at MMI. More than 300 MMI spermatocytes were scored for each genotype, with univalent autosomes observed in 2.5 ± 0.7% and 3.3 ± 1.5% of cells from WT and Rad51ap2ΔC741/ΔC741 mice, respectively [P = 0.9513, two-way analysis of variance (ANOVA)] (Fig. 2, B and C). By contrast, XY univalents were observed in 61.9 ± 2.5% of Rad51ap2ΔC741/ΔC741 MMI spermatocytes, compared to 16.5 ± 0.7% of WT MMI spermatocyte (P < 0.001, two-way ANOVA) (Fig. 2, B and C). Consistently, a significantly increased proportion of diplotene cells with univalent XY was observed in spermatocyte spreads from mutant mice (Fig. 2, D and E). Hence, the RAD51AP2 loss-of-function truncating mutation renders XY chromosomes, but not autosomes, more susceptible to precocious separation in meiotic prophase.
Rad51ap2 mutant spermatocytes exhibit decreased XY crossover
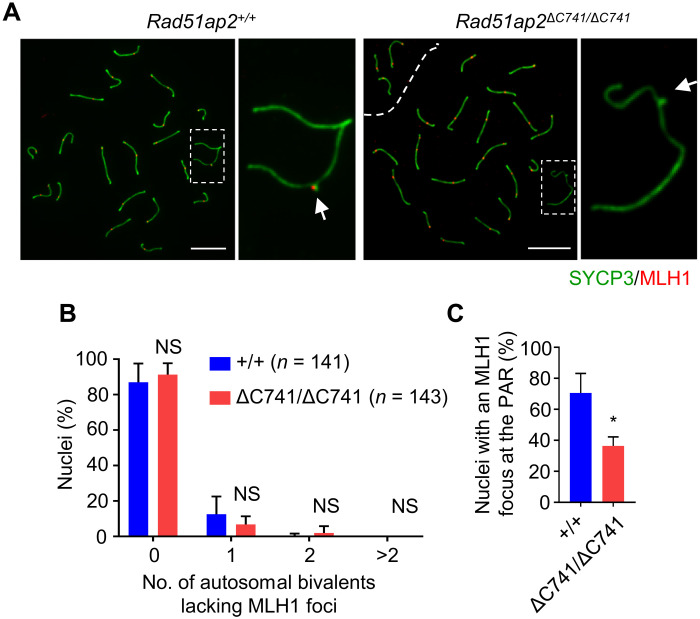
The observed increase in precocious XY dissociation suggested that crossover formation in PARs is likely disrupted. We thus stained spermatocyte spreads for MLH1, a marker of crossovers, and synaptonemal complex protein 3 (SYCP3), the lateral element component, and counted MLH1 foci on each bivalent, including the XY bivalent. The average number of MLH1 foci per nucleus was 23.9 ± 2.3 in WT pachytene spermatocytes, which was decreased to 22.3 ± 1.8 in the Rad51ap2 mutants (P < 0.01, two-way ANOVA) (Fig. 3A and fig. S5). More specifically, for autosomes, no significant difference was found in the frequency of autosomal bivalents lacking MLH1 foci between Rad51ap2 mutant and WT mice (Fig. 3B); however, for XY chromosomes, the frequency of nuclei lacking an MLH1 focus was significantly higher in Rad51ap2 mutant mice than in WT mice (Fig. 3C). These observations are consistent with the precocious XY separation in MMI cells, which together indicate that crossover formation is decreased on XY chromosomes, but not on autosomes, in Rad51ap2 mutant mice.
Fig. 3. RAD51AP2 disruption decreases XY crossover formation but does not affect autosome crossover formation.
(A) Representative spread spermatocytes stained for SYCP3 (green) and MLH1 (red) in mid- to late pachynema. Magnified views of the X and Y chromosomes are shown on the right, and PAR is indicated by an arrow. Scale bars, 10 μm. (B) Fractions of nuclei in which zero, one, two, or more autosomal bivalents lacking an MLH1 focus. Two-way ANOVA. (C) Frequencies of nuclei with an MLH1 focus detected at the PAR. A total of 141 WT and 143 mutant spermatocytes from three mice per genotype were scored. *P < 0.05, two-tailed Student’s t test.
XY synapsis occurs in Rad51ap2 mutant spermatocytes
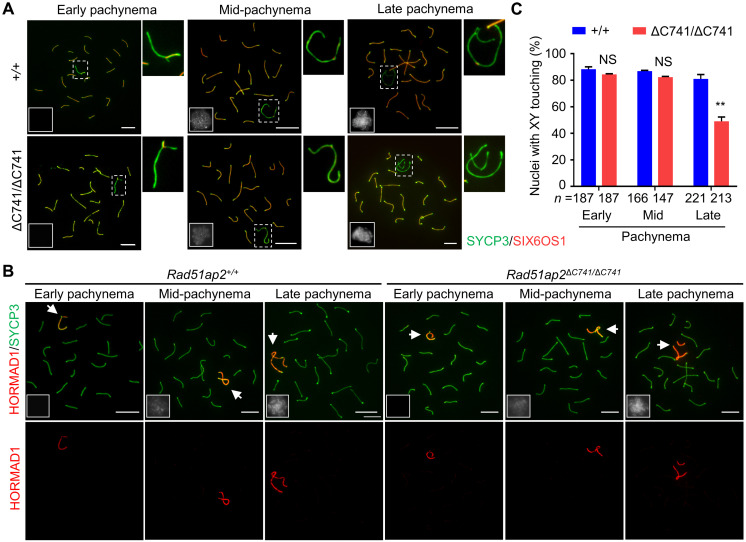
As homologous recombination and synapsis are interdependent, recombination promotes synapsis formation, and synapsis is the basis for crossover formation (17), we next tracked synapsis by immunofluorescence staining for SYCP3 and SIX6OS1, a central element component of the synaptonemal complex (31, 32), together with HORMA domain-containing protein 1 (HORMAD1) or pSer1083-SMC3, on spread spermatocytes, followed by cautious analyses of the early, mid-, and late pachytene cells. In pachynema, HORMAD1 is removed from synapsed chromosome axes and only retained on unsynapsed regions of the sex chromosomes (33), while pSer1083-SMC3 localizes only on unsynapsed sex chromosomal regions, but not synapsed or desynapsed regions (34). Same as the observation in WT mice, all the autosomes were fully synapsed, with SIX6OS1 signals detected consecutively throughout the whole axis length and no HORMAD1 or pSer1083-SMC3 detected, in all the pachytene cells of the pachytene cells of the Rad51ap2ΔC741/ΔC741 mice (Fig. 4, A and B, and fig. S6A) nor were any aberrant γH2AX signals on autosomes observed in the mutant pachytene spermatocytes (fig. S6B). In early and mid-pachynema, the XY chromosomes were synapsed at the PAR in 84.4 ± 0.5% and 82.3 ± 0.6% of the mutant spermatocytes, respectively, which were similar to those in WT mice (early pachynema, 88.3 ± 1.8%, P = 0.7326, two-way ANOVA; mid-pachynema, 86.88 ± 0.6%, P = 0.8513, two-way ANOVA) (Fig. 4C). In late pachytene, XY desynapsis was observed in both WT and mutant spermatocytes (Fig. 4A). However, the XY remained touching in 176 of 221 (81.0 ± 3.3%) WT cells analyzed, owing to crossovers, but in only 110 of 213 (49.1 ± 3.3%) Rad51ap2ΔC741/ΔC741 spermatocytes analyzed (P < 0.01, two-way ANOVA) (Fig. 4C). In addition, meiotic prophase progression and the sex bodies in each substages of prophase were comparable between WT and Rad51ap2ΔC741/ΔC741 mice (fig. S7). These findings showed that in Rad51ap2 mutant spermatocytes, the XY chromosomes are synapsed normally but precociously separated when desynapsis occurs during late pachynema due to the lack of crossovers.
Fig. 4. XY chromosomes are synapsed in early/midpachynema but undergo precocious separation from late pachynema in Rad51ap2 mutant spermatocytes.
(A) Representative spread spermatocytes stained for SYCP3 (green) and the central element (SIX6OS1, red) in pachynema. X and Y chromosomes are magnified and shown on the right. (B) Representative spread spermatocytes stained for SYCP3 (green) and HORMAD1 (red, localizing only on unsynapsed chromosome axes). Arrows indicate the sex chromosomes. (C) Quantification of nuclei with XY touching in early, mid-, and late pachynema. n, the number of cells scored from at least two mice for each genotype. NS, P > 0.05 and **P < 0.01, two-way ANOVA. For (A) and (B), miniaturized H1t staining (white) images are shown in the left lower corner of the overlay images. Scale bars, 10 μm.
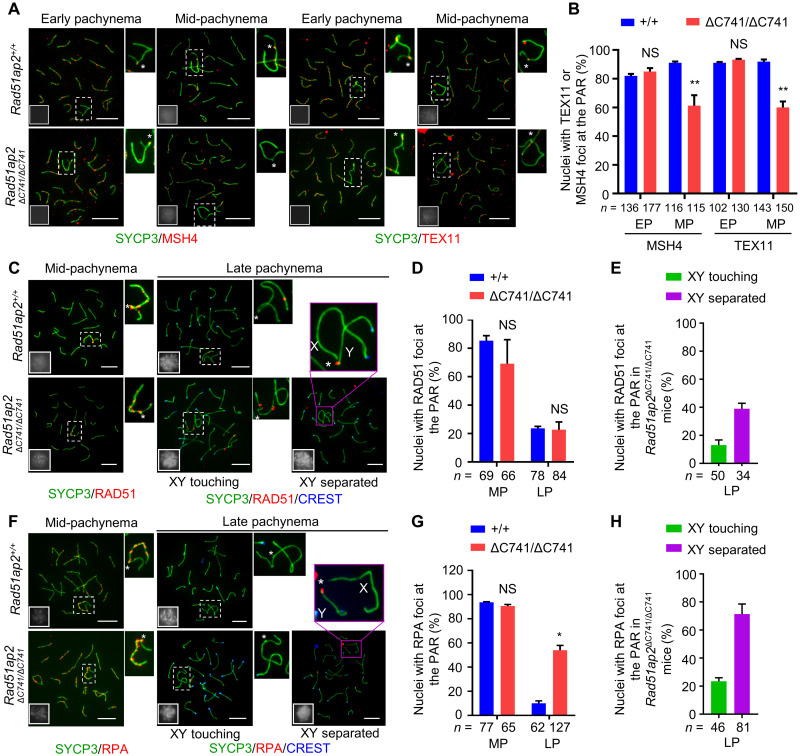
Compromised PAR DSB repair due to destabilized recombination intermediates in Rad51ap2 mutant spermatocytes
Normal synapsis of XY chromosomes indicates that DSBs are formed, and strand invasion, the early event of recombination DSB repair, occurs, leading to the formation of recombination intermediates. The recombination intermediates are then stabilized by MutS homolog 4 (MSH4) and Testis-expressed protein 11 (TEX11), and destabilized intermediates, marked by precocious dissociation of MSH4 and TEX11 foci from chromosome axes, would lead to reduced crossover formation (22, 35). To further check the stabilization of recombination intermediates formed after strand invasion, we analyzed the kinetics of MSH4 and TEX11 foci in pachytene spermatocytes of the mutant mice. Quantification of MSH4 and TEX11 foci in early pachynema indicated that the frequencies of cells with MSH4 and TEX11 foci at the PAR in the Rad51ap2ΔC741/ΔC741 mice were comparable to those in the control mice, in accordance with the normal synapsis of XY chromosomes (Fig. 5, A and B). However, in mid-pachynema, the mutant spermatocytes showed significant reductions in the frequencies of nuclei with PAR-resident foci for MSH4 and TEX11 (for MSH4: 61.1 ± 10.4% in mutant versus 91.1 ± 1.2% in WT, P < 0.01, two-way ANOVA; for TEX11: 60.0 ± 5.9% in mutant versus 91.9 ± 2.1% in WT, P < 0.01, two-way ANOVA) (Fig. 5, A and B), indicating that the recombination intermediates in the mutant spermatocytes are susceptible to be precociously dismantled.
Fig. 5. RAD51AP2 is required for stabilizing recombination intermediates for efficient DSB repair in PARs.
(A) Representative spread early and mid-pachytene spermatocytes stained for SYCP3 (green) and MSH4 or TEX11 (red). (B) Frequencies of nuclei with MSH4 or TEX11 foci detected at the PAR in spread early and mid-pachytene spermatocytes with XY touching. NS, P > 0.05 and **P < 0.01, two-way ANOVA. (C to H) Representative spread spermatocytes stained for SYCP3 (green) and RAD51 (C) or RPA (F) (red), frequencies of nuclei with RAD51 (D) and RPA (G) foci detected at the PAR in spread mid- and late pachytene spermatocytes, and frequencies of nuclei with RAD51 (E) or RPA (H) foci detected at the PAR in spread Rad51ap2ΔC741/ΔC741 spermatocytes with XY touching or XY separated in late pachynema are shown. NS, P > 0.05 and *P < 0.05, two-way ANOVA. For (A), (C), and (F), magnified views of the X and Y chromosomes are shown on the right or above, with an asterisk indicating the PAR. Miniaturized H1t staining (white) images are shown in the left-lower corner of the overlay images. Scale bars, 10 μm. The boundaries of PARs were determined on the basis of the average ratios of PAR axial length to the total axial length of X or Y chromosomes (fig. S16). n, the number of spermatocytes scored from at least two mice per genotype. EP, early pachynema; MP, mid-pachynema; LP, late pachynema.
We theorized that this precocious dismantling of recombination intermediates could result from either completion of DSB repair to produce noncrossovers or result from destabilization of the recombination intermediate structure before DSB repair is completed. We thus detected DSBs in mid- and late pachytene cells from WT and Rad51ap2ΔC741/ΔC741 mice (Fig. 5, C to H). In WT mice, RAD51 and replication protein A (RPA) foci were detected at the PAR in 85.4 and 93.7% of mid-pachytene cells and gradually disappeared during late pachynema. In Rad51ap2ΔC741/ΔC741 mice, the frequencies of nuclei with RAD51 foci at the PAR were comparable to those in control mice in mid- and late pachynema (Fig. 5, C and D). The frequency of nuclei with PAR-resident RPA foci in mid-pachytene mutant spermatocytes was similar to those in controls; however, in late pachynema, the frequency of nuclei with RPA foci at the PAR was significantly higher than in control mice (54.0 ± 5.6% in mutant versus 10.1 ± 2.8% in WT, P < 0.05, two-way ANOVA) (Fig. 5, F and G). Since approximately half of the spermatocytes displayed XY separation in late pachynema, we determined the frequency of PAR-resident RAD51 and RPA foci in spermatocytes with XY separated or XY touching, respectively. In the mutant late pachytene spermatocytes, 71.3 ± 10.2% and 38.9 ± 0.1% of cells with separated XY and 23.5 ± 3.5% and 13.0 ± 0.1% of cells with XY touching retained RPA foci and RAD51 foci at the PAR, respectively, although these foci have vanished before entering diplonema, indicating that the DSBs in PARs are not repaired efficiently (Fig. 5, E and H). These findings suggest that the recombination intermediates in PARs were dismantled before completion of DSB repair, thus leading to failed XY crossover formation in the Rad51ap2 mutant spermatocytes.
In addition, we also examined the nucleus-wide MSH4 and TEX11 foci counts in early and mid-pachytene cells and found that their numbers were slightly reduced in Rad51ap2ΔC741/ΔC741 spermatocytes compared with those in control cells (fig. S8). To be noted, no obvious differences were observed in the nucleus-wide kinetics of RPA foci in prophase between WT and the mutant spermatocytes, except that the number of RPA foci was significantly increased at the early pachynema at the axes of X-specific region of the X chromosome (fig. S9), nor were any unrepaired DSBs, marked by aberrant γH2AX signals, observed in the mutant pachytene spermatocytes (fig. S6B). These findings indicate that, although MSH4 and TEX11 tend to vanish faster on the autosomes, the overall DSB repair capacity seems undisturbed in the Rad51ap2 mutant mice.
RAD51AP2 is dispensable for crossover formation and meiosis in oocytes
RAD51AP2 mRNA was previously detected in fetal ovaries (28), suggesting that it may play a role in oogenesis. Nevertheless, we did not notice any obvious impairment in follicle development for the Rad51ap2 mutant female mice. Analysis of the ovaries from mice at 4 dpp and 2 months of age revealed that the ovary size and morphology of Rad51ap2ΔC741/ΔC741 females were similar to those of WT mice (fig. S10). Ovarian histological analyses indicated no notable difference in the numbers of total oocytes, primordial, primary, secondary, and/or antral follicles between WT mice and the mutants, suggesting that the follicle development was not affected by RAD51AP2 disruption (fig. S10). To check whether RAD51AP2 disruption would lead to defects in meiosis of females, oocyte spreads from embryos at 17.5 days postcoitus (dpc) were prepared for analyzing synapsis and recombination. Synapsis and the number of MLH1 foci showed no obvious difference between Rad51ap2ΔC741/ΔC741 and WT oocytes (fig. S11, A to D). To analyze the meiotic maturation process, we next cultured oocytes retrieved from 3-week-old control and Rad51ap2ΔC741/ΔC741 mice after superovulation treatment and found that the rates of germinal vesicle breakdown at 8 hours of culture and the rates of first polar body extrusion at 16 hours of culture were comparable to those observed for WT mice, indicating that the mutant oocytes are competent to complete meiosis I (fig. S11, E and F). Chromosome spreading analysis further showed that all the examined Rad51ap2ΔC741/ΔC741 oocytes exhibited 20 typical bivalents at metaphase I and 20 chromosomes, each with two chromatids, at metaphase II, the manifestation of normal meiotic recombination in the mutant oocytes (fig. S11G). Together, these findings indicate that the oogenesis is not affected after RAD51AP2 functional disruption.
Interaction with RAD51 is essential for RAD51AP2 to ensure XY recombination
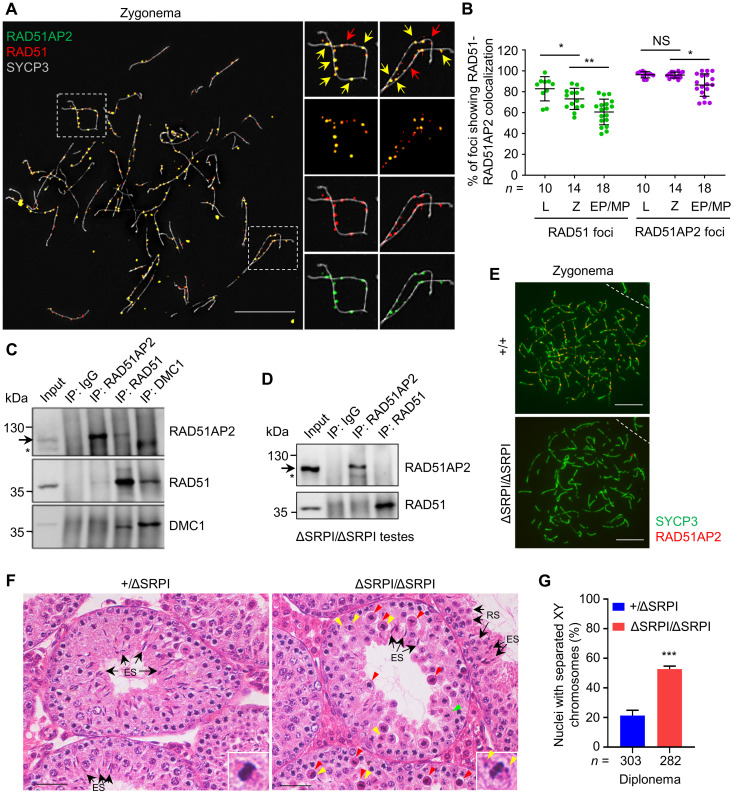
Since RAD51AP2 has been shown to interact with RAD51 in yeast two-hybrid assay (28), we wondered whether they are also associated in vivo in meiosis. Immunofluorescence staining revealed that RAD51AP2 and RAD51 foci showed a high degree of colocalization on chromosome axes, with almost a complete overlay of foci signals at the colocalization site, under superresolution structured illumination microscopy (SIM) (Fig. 6A and fig. S12). The average fraction of RAD51 foci colocalizing with RAD51AP2 was 83% in leptonema, 73% in zygonema, and 62% in early/mid-pachynema, while the average fraction of RAD51AP2 colocalizing with RAD51 was 96% in both leptonema and zygonema and 86% in early/mid-pachynema (Fig. 6B). RAD51AP2 foci completely vanished before late pachynema, while RAD51 foci could still be detected in a subset of cells at this stage (Fig. 5C and fig. S2, C and D). Thus, we infer that RAD51AP2 may be recruited to chromosome axes relying on RAD51 and largely disappear earlier than RAD51. RAD51AP2 also colocalized with DMC1 under SIM, mostly in a side-by-side manner, and the colocalization frequency is lower than with RAD51 (fig. S13, A and B). Similar to the situation with RAD51—which is known to locate closer to axis than DMC1 (16, 36)—our data showed that RAD51AP2 foci tend to be closer to the axis than DMC1 foci (fig. S13A). We found that RAD51AP2 foci remained on chromosomal axes in Dmc1−/− mice, which is similar to what has been found for RAD51 as well (37) (fig. S13C). Coimmunoprecipitation (Co-IP) assays using mouse testicular lysates revealed that RAD51AP2 interacted with RAD51, but not with DMC1 (Fig. 6C). Together, the protein-protein interaction and colocalization patterns allowed us to deduce that the interaction with RAD51 may be required for the localization and function of RAD51AP2.
Fig. 6. The RAD51AP2 C terminus mediates the interaction with RAD51 and is essential for RAD51AP2 loading onto chromosome axes.
(A) Representative WT zygotene spread spermatocytes stained for RAD51AP2 (green), RAD51 (red), and SYCP3 (gray) under SIM. Magnified views of the boxed areas are shown on the right. Yellow arrows indicate representative RAD51AP2 foci colocalizing with RAD51. Red arrows indicate representative RAD51 foci existing alone. Scale bars, 10 μm. (B) Ratios of RAD51-RAD51AP2 colocalizing foci to the RAD51 or RAD51AP2 total foci assessed under SIM. n, the number of nuclei scored for each substage. L, leptonema; Z, zygonema. NS, P > 0.05; *P < 0.05; and **P < 0.01, Mann-Whitney U test. (C and D) Co-IP assay with whole-testis extracts of WT (C) and Rad51ap2ΔSRPI/ΔSRPI (D) mice, followed by Western blotting. Arrows indicate the bands corresponding to WT RAD51AP2 proteins (predicted molecular weight, 112.2 kDa) or RAD51AP2 ΔSRPI (predicted molecular weight, 111.7 kDa). Asterisks, a nonspecific band. (E) Representative spread spermatocytes stained for RAD51AP2 (red) and SYCP3 (green). Scale bars, 10 μm. (F) Testicular histology of 6-week-old mice. Green arrowheads, metaphase cells that appear normal. Red arrowheads, metaphase cells with a condensed nucleus indicative of apoptosis. Yellow arrowheads indicate unaligned chromosomes. Arrows indicate representative round spermatids or elongating/elongated spermatids. Scale bars, 50 μm. (G) Frequency of nuclei with XY separation in diplonema. n, the number of cells scored from two mice per genotype. ***P < 0.001, two-tailed Student’s t test.
As it has been reported that the interaction with RAD51 is mediated via the C-terminal 33 amino acids of human RAD51AP2 (corresponding to C-terminal 22 amino acids in mouse protein) (28), we thus generated mutant mouse models with the RAD51AP2 C terminus disrupted. Rad51ap2ΔC741/ΔC741 mice, carrying a frameshift mutation that is predicted to result in a truncated protein lacking the C-terminal 22 amino acids, and Rad51ap2ΔSRPI/ΔSRPI mice, with a deletion of 4 amino acid residues (955_958SRPI) that are highly conserved in mammals (fig. S14), were obtained. Western blotting confirmed the presence of mutant RAD51AP2 proteins in testes (Fig. 6D and fig. S15A), but RAD51AP2 focus localization on chromosome axes was completely abolished, in both mutant mouse models (Fig. 6E and fig. S15B), indicating that the C terminus is essential for the loading of RAD51AP2 onto chromosomal axes. Further analyses of the Rad51ap2ΔSRPI/ΔSRPI mice revealed that RAD51AP2 ΔSRPI mutant proteins could not interact with RAD51 (Fig. 6D). Precocious separation of XY chromosomes was seen in 52.8 ± 2.0% of diplotene spermatocytes, and unaligned chromosomes in MMI cells were frequently observed in the mutant mice (Fig. 6, F and G), resembling the phenotypes of Rad51ap2ΔC741/ΔC741 mice. These results collectively show that the interaction between RAD51 and RAD51AP2’s C terminus is essential for the loading of RAD51AP2 on chromosome axes.
The interaction with RAD51 was abolished by the RAD51AP2 frameshift mutations identified in the patients
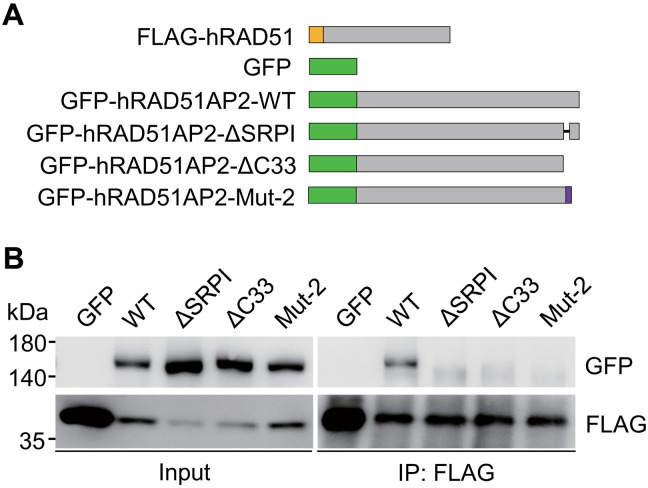
For the four RAD51AP2 mutations identified in our patients, the C-terminal residues needed for interacting with RAD51 are completely lost in Mut-1, Mut-3, and Mut-4 and are partially lost in Mut-2 lacking the C-terminal 29 amino acids. To assess potential impacts of the RAD51AP2 mutations identified in patients on RAD51AP2-RAD51 interaction, we first coexpressed green fluorescent protein (GFP)–tagged WT human RAD51AP2, mutant human RAD51AP2 lacking its 33 C-terminal amino acids (ΔC33) or SRPI (ΔSRPI), or human RAD51AP2 Mut-2 with the FLAG-tagged human RAD51 in human embryonic kidney (HEK) 293T cells (Fig. 7A). Then, Co-IP assays were performed and revealed that GFP-tagged WT RAD51AP2 proteins were coimmunoprecipitated with FLAG-RAD51, while RAD51AP2 ΔC33, ΔSRPI, or Mut-2 proteins are not (Fig. 7B), indicating that these RAD51AP2 mutants fail to interact with RAD51. We therefore conclude that all the RAD51AP2 mutations identified in the patients abolish the interaction with RAD51.
Fig. 7. Disruptions of human RAD51AP2 C terminus abolish the interaction with RAD51.
(A) Schematic of the tagged human proteins. The FLAG tag is indicated by the yellow box, and GFP tags are indicated by the green boxes. The amino acids altered after the Mut-2 frameshift mutation are indicated by the purple box. (B) Co-IP assays with transfected HEK293T cells, showing that the interaction with RAD51 is disrupted for the human RAD51AP2 variants lacking its C-terminal 33 amino acids or SRPI and for an RAD51AP2 variant, Mut-2 (p.R1131Lfs*19, identified in case 241)”.
All these findings from our mouse models and cultured cells demonstrate an important role of RAD51AP2, as a meiosis-specific partner of RAD51, in XY crossover formation by stabilizing recombination intermediates in PARs. Mutations in RAD51AP2, which disrupt the interaction with RAD51, lead to unaligned chromosomes in MMI and ultimately impaired spermatogenesis in humans and mice.
DISCUSSION
Given the heterologous nature of the X and Y chromosomes, faithful segregation of XY chromosomes is much more challenging than autosomes because the homology-dependent processes of pairing, synapsis, and recombination are restricted to the PAR. Nevertheless, XY nondisjunction is rare, which suggests that there are distinct mechanisms that ensure a successful and high rate of XY crossing-over in this tiny region. Studies have provided extensive evidence for the PAR-specific mechanisms underlying meiotic DSB formation (7–9, 15, 38), which promotes the pairing and synapsis between X and Y. Nonetheless, to ensure that the XY chromosomes remain held together after the late pachytene stage, DSBs at the PAR have to be repaired to yield a crossover. It remains unclear how the generation of XY crossover from one or two DSBs at the tiny PAR is guaranteed. A plausible explanation is that DSB repair at the PAR is regulated by the mechanism that ensures the formation of at least one obligate crossover for each pair of homologs (39, 40). However, our results did not favor this explanation, as we did not observe failed obligate crossover formation between autosomal homologs in Rad51ap2 mutant spermatocytes. Nonetheless, our results provide direct experimental evidence that XY recombination at the DSB repair mechanism at the PAR may be mediated by proteins that are not essential for that on autosomes, such as RAD51AP2.
The temporal dynamics of RAD51AP2-RAD51 colocalization pattern and the phenotype of Rad51ap2 mutant mice suggested that RAD51AP2 is likely recruited by RAD51 and functions subsequent to the ssDNA-RAD51 nucleofilament formation, which is similar to another well-known RAD51-interacting protein, RAD51AP1. RAD51AP1 is expressed in various tissues, including testis and fetal ovaries, and is found to stimulate the RAD51-mediated D-loop formation to enhance homologous recombination repair in somatic cells (28, 41–43). RAD51AP1 contains an RAD51-binding region showing 81% homology to that in RAD51AP2, implying that they may interact with the same region of RAD51 (28). In meiosis, RAD51AP1 colocalizes with RAD51 and DMC1 with a similar intimacy, but their colocalization levels are much lower than those of RAD51AP2 with RAD51 and DMC1 (44). Given their similarity in localization and potential activities, RAD51AP1 and RAD51AP2 may exhibit similar functions in meiotic recombination by directly associating with and promoting the activities of RAD51. Noticeably, no reproductive phenotype was annotated for Rad51ap1 knockout mice in the MGI resource (MGI: 5635282). Thus, it is inferred that RAD51AP2 may play a more predominant role in recombination than RAD51AP1, although we cannot exclude the possibility that RAD51AP1 may compensate for the loss of RAD51AP2 in meiosis. Hence, it would be interesting to investigate the meiosis, particularly recombination at the PAR, in mice with Rad51ap1 and Rad51ap2 both mutated.
To our knowledge, RAD51AP2 is the first protein that specifically participates in PAR DSB repair for ensuing XY crossover formation but is dispensable for autosomal crossover formation. Several genes have been reported to increase precocious XY separation during meiosis due to defects in DSB repair after disruption in mice (19, 22, 45–49). It should be noted that all these genes, when mutated in mice, not only increased the frequency of XY separation but also impaired autosomal chromosome behaviors. For example, Hfm1-deficient mice not only showed precocious XY separation in 43.5% of pachytene cells but also displayed the absence of class I crossovers on autosomes in all the spermatocytes examined (45). Palb2 mutant mice carrying a mutation that impairs the interaction with BRCA1 not only exhibited precocious XY separation in ~32% of pachytene cells but also showed nonhomologous synapsis of autosomes (46). However, in the current study, disruption of RAD51AP2 compromises the recombination repair only on XY chromosomes, but not on autosomal homologs. Furthermore, we also found that, although RAD51AP2 expression were detected in fetal ovaries (28), no defects in synapsis, the counts of MLH1 foci, and meiotic maturation were observed after RAD51AP2 disruption in mouse oocytes. Looking from another aspect, these observations in female mice, which represent a non-PAR situation in another physiological context, also corroborate the notion that RAD51AP2 is not essential for normal crossover formation on autosomes in spermatocytes.
Our results indicated that RAD51AP2 promotes XY recombination repair by stabilizing recombination intermediates during male meiosis. In Rad51ap2 mutant mice, normal loading of MSH4 and TEX11 at the PAR was observed in early pachynema, in congruent with normal XY synapsis. However, the frequency of cells with PAR-resident MSH4 and TEX11 foci was decreased by ~30% when compared with that in controls in mid-pachynema with XY touching, which was in accordance with the increase in the frequency of cells with XY separation from mid-pachynema (17.7%) to late pachynema (50.1%). Thus, these findings indicate that recombination intermediates at the PAR are formed but precociously dismantled in the absence of RAD51AP2, leading to failure in crossover formation and consequently precocious separation of XY chromosomes. This indication was further confirmed by the observation of the persistence of RPA foci, at the PAR of late pachytene spermatocytes with separated XY in Rad51ap2 mutant mice. It should be noted that two types of RPA foci have been identified, RPA binding the single-stranded DNA (ssDNA) overhangs that are subsequently replaced by RAD51/DMC1 to form the nucleoprotein filaments and RPA binding to the recombination intermediates (such as D-loops) at a later stage (16, 50). The normal synapsis of XY chromosomes in Rad51ap2 mutant spermatocytes indicates that the loading of ssDNA-bound RPA and the subsequent replacement by RAD51/DMC1 are not largely affected. The RPA foci detected at the PAR in late pachytene mutant spermatocytes are likely the second type, marking the persisting intermediates, and thus signify that the DSBs failed to be repaired efficiently at the PAR with XY separated.
Given that RAD51AP2 is detected on chromosome axes of both autosomes and sex chromosomes, it is speculated that it may have a broader role than just at the PAR. Consistent with this view, we did observe a mild decrease of MSH4 and TEX11 foci in Rad51ap2 mutant spermatocytes at early/mid-pachytene stage, suggesting a genome-wide destabilization of intermediates. This raises the possibility that the impaired XY crossover formation caused by RAD51AP2 disruption could result from a common defect in recombination with a higher susceptibility for the sex chromosomes than autosomes. Nonetheless, the reduction ratios of the PAR-associated MSH4 and TEX11 foci were approximately three times the reduction ratios observed for nucleus-wide foci numbers, and the possibility that RAD51AP2 may have a PAR-specific role, in addition to its role on the autosomes, could not be entirely ruled out. Future studies are warrant to further dissect the molecular functions of RAD51AP2, which would provide intriguing clues for understanding of the recombination regulation, particularly at the PAR.
MATERIALS AND METHODS
Participants and genetic studies
This study was approved by Ethical Committee of University of Science and Technology China (USTC) and conducted conforming to the committee guidelines. Written informed consent was obtained from all the participants. Patients, who were diagnosed with non-obstructive azoozpermia (NOA) and had testicular biopsy, were recruited. Semen analyses were performed according to the guidelines of World Health Organization (51). The testicular biopsies were fixed in 4% paraformaldehyde (PFA) and embedded into paraffin blocks for sectioning and histological analyses. Those patients with abnormal spermatocyte maturation in histology were included in this study for WES.
Peripheral blood samples were obtained from patients and stored in EDTA tubes or TRIzol reagent (Thermo Fisher Scientific) for genomic DNA and RNA extraction, respectively. For WES, exons were captured from genomic DNAs of peripheral blood cells using the Agilent SureSelect XT 50 Mb Exon Capture Kit, followed by high-throughput sequencing performed on an Illumina HiSeq 2000 platform. Variants were called using Genome Analysis Toolkit Unified Genotyper and annotated on the basis of Ensembl (www.ensembl.org). Variants within exons or exon-intron boundaries were retained and filtered through the criteria as we previously described (32). Among the variants after the filtration, we identified four candidate pathogenic RAD51AP2 variants in two unrelated patients and validated these four variants by Sanger sequencing. The information of primers is shown in table S2.
Antibody generation
Polyclonal antibodies against mouse RAD51AP2 were raised in rabbits and rats against peptide antigen (the epitope region of mouse RAD51AP2 from 87 to 326 amino acids and 131 to 320 amino acids, respectively) and purified from serum using Protein A/G spin columns (GE Healthcare) by ABclonal Biotechnology Co. Ltd. The specificities of antibodies were determined and verified by Western blotting and immunofluorescence staining in WT and Rad51ap2 mutant mice.
Immunostaining of testis sections
Immunostaining of testicular sections was performed as described previously, with the following modifications (52). Testes were fixed in 4% PFA for 5 hours, dehydrated in 30% sucrose for at least 8 hours, and embedded in optimal cutting temperature compound (Sakura Finetek, CA). Seven-micrometer-thick sections were fixed for 30 min in precold 4% PFA (w/v) at room temperature, then washed twice in phosphate-buffered saline (PBS) for 5 min each time, and permeabilized in PBST (0.2% Triton X-100 in PBS) for 20 min. Sections were blocked in antibody dilution buffer [10% normal donkey serum, 3% bovine serum albumin (BSA), 0.05% Triton X-100 in PBS] for 45 min, followed by overnight incubation at 4°C with primary antibodies. Four washes with PBST were performed before secondary antibody incubation at room temperature for 1.5 hours. Last, sections were mounted in VECTASHIELD mounting medium (Vector Laboratories, H-1000) containing Hoechst 33342 (Invitrogen). Images were captured using a Nikon ECLIPSE 80i microscope (Nikon) equipped with a charge-coupled device camera (Hamamatsu) and analyzed using NIS-Element Microscope imaging software (Nikon). The primary and secondary antibodies used and their dilutions are shown in table S3.
Western blotting
Protein lysates were prepared as we previously described (53). The lysates were denatured for 10 min and separated by SDS–polyacrylamide gel electrophoresis, followed by transferring the proteins to 0.45-μm pore size immobilon-P membranes (Millipore, IPVH00010). Membranes were blocked in Tris-buffered saline with Tween 20 [50 mM tris (pH 7.4), 150 mM NaCl, and 0.1% Tween-20] containing 5% nonfat milk for 1 hour and incubated with primary antibodies at 4°C overnight. Following incubation with horseradish peroxidase–conjugated secondary antibodies for 1 hour, the membranes were developed with chemiluminescence substrate by an ImageQuant LAS 4000 imaging system (GE Healthcare). The primary and secondary antibodies used and their dilutions are shown in table S3.
Spermatocyte spreading and immunofluorescence staining
Mouse testicular cells were prepared for surface spreading and subsequent immunofluorescence staining as we previously described (52, 54) with the following modifications. Seminiferous tubules were incubated in hypotonic extraction buffer [30 mM tris (pH 8.2), 50 mM sucrose, 17 mM trisodium citrate dihydrate, 5 mM EDTA, 2.5 mM dithiothreitol (DTT), and 1 mM phenylmethylsulfonyl fluoride (PMSF)] for 25 min at room temperature. Subsequently, cell suspension was made in 100 mM sucrose and spread on slides with 1% PFA (pH 9.2) containing 0.15% Triton X-100. Slides were then placed in a humidified chamber for at least 3 hours. Last, slides were washed twice for 3 min in 0.4% Photoflo (Kodak) and air-dried at room temperature. Slides were either used for immunofluorescence staining immediately or stored at −80°C. Immunofluorescence staining was conducted as we previously described (32, 52). The primary and secondary antibodies used and their dilutions are shown in table S3. Spermatocytes were staged according to previously published papers (8, 9, 55). Briefly, in leptonema, there are short stretches of axis (axis stretches shorter than 15× the widths of axes) without any evidence of synapsis; in early/mid zygonema, there are relatively long and incomplete stretches of axes with some synapsis; and in late zygonema, there are complete axes with substantial (>70%) synapsis. Staging of early, mid-, and late pachynema was based on the staining of H1t, a testis-specific linker histone that is transcribed from the mid-pachynema (56) and axis morphology assessed by SYCP3. In the early pachynema, thickness of synapsed axis pairs is largely uniform along their entire length and H1t signal is absent or very weak; in mid-pachynema, besides the uniform thickness of synapsed axis pairs, H1t signal is weak to intermediate in strength; in the late pachynema, synapsed axes of autosomes are clearly thicker at ends than at interstitial sites, and H1t signal intensity is intermediate to strong.
Mice
All animal experiments were approved by the Institutional Animal Care and Use Committee of the University of Science and Technology of China and conducted according to the committee guidelines. Spo11−/− (29) and Dmc1−/− (37) mice were maintained on a C57BL/6 background. Rad51ap2 mutant mice were generated using CRISPR-Cas9 genome editing as previously described (52). Briefly, we coinjected Cas9 mRNAs and two guide RNAs targeting exon 1 or one guide RNA targeting exon 3 into B6D2F1 (C57BL/6 × DBA/2) zygotes, followed by embryo transfer into pseudopregnant ICR females. Genomic DNA was extracted from tail biopsies from founder mice, and genotyping was performed by polymerase chain reaction and Sanger sequencing. The founder mice that carried the mutant Rad51ap2 allele(s) were backcrossed onto the C57BL/6 background. Homozygous mutants obtained from the F2 generation were used for subsequent experiments. The C57BL/6, DBA/2, and ICR mice were purchased from Beijing Vital River Laboratory Animal Technology Co. Ltd. The information of primers is shown in table S2.
Histology
Testes were collected and fixed in Bouin’s fixative solution. After dehydration through a graded series of ethanol, tissues were embedded in paraffin and serially sectioned, followed by hematoxylin and eosin or periodic acid–Schiff staining. Slides were examined, and images were captured under a light microscope (Nikon) equipped with a charge-coupled device camera (Nikon). Ovaries were harvested from mice at the indicated ages, fixed in modified Davidson’s fluid overnight, and embedded in paraffin. Paraffin-embedded ovaries were serially sectioned at a thickness of 5 μm and mounted on slides, followed by hematoxylin staining. The numbers of follicles were counted as we previously described (57).
TUNEL assay
Testicular sections were deparaffinized with xylene, rehydrated through gradient ethanol, and permeabilized with proteinase K (20 μg/ml) in 10 mM tris-HCl (pH 7.5) for 15 min at room temperature. After two washes with PBS, sections were blocked with blocking buffer [3% BSA and 10% normal donkey serum in 10 mM tris-HCl (pH 7.5)]. TUNEL assay was then performed using the In Situ Cell Death Detection Kit, Fluorescein (Roche, 11684795910) according to the manufacturer’s protocol. Images were captured using a Nikon ECLIPSE 80i microscope (Nikon) equipped with a charge-coupled device camera (Hamamatsu) and analyzed using the NIS-Element Microscope imaging software (Nikon).
Diakinesis/metaphase I chromosome spreading
Chromosome spreads of meiotic diakinesis/metaphase cells were prepared as previously described (52, 58) and stained with Giemsa. Images were captured under a light microscopy (Nikon) equipped with a charge-coupled device camera (Nikon).
Testicular cell smears
Diakinesis/metaphase I chromosome spreading assay was modified to obtain appropriate metaphase cells with chromosomes aligning on the equatorial plate maintained. Briefly, testicular cell suspensions were harvested in PBS and filtered through a PBS-prewetted 75-μm mesh. After centrifugation at 800 rpm for 10 min at room temperature, the cells were resuspended with fixative and mixed well in 7 ml of fixative (7:1 ratio of methanol and acetic acid glacial) at room temperature for 1 hour. After centrifugation again, cells were resuspended in 500 μl of fixative and dropped on room temperature or prewarmed (37°C) slides. Air-dried slides were used for subsequent Giemsa staining.
Immuno-FISH
To determine the relative axial length of PAR, immuno-FISH (fluorescence in situ hybridization) for PAR boundary (PARb) probes, which recognizes PAR sequences that border the heterologous parts of X and Y chromosomes, was performed as previously described with minor modifications (7). To generate PARb probes, plasmids BAC RP24-500I4 (Children’s Hospital Oakland Research Institute, Oakland, CA) was labeled with SpectumRed dUTP (Vysis 30-803400) using the BioPrime DNA Labeling System (Invitrogen, 18094-011) following the manufacturer’s instructions. Following immunofluorescence staining, slides of spermatocyte spreads were dehydrated through a serial of gradient ethanol, aged at 65°C for 1 hour, denatured in 70% formamide at 72°C for 7 min, immersed immediately in precooled 50% ethanol, and redehydrated through gradient ethanol. The slides were dried, then incubated with 2× SSC incubation buffer containing PARb probes and fish sperm DNA (Roche, 11467140001), and denatured at 80°C for 10 min, followed by overnight incubation in a humid chamber at 37°C for 24 to 48 hours. After three washes with 4× Saline-sodium citrate (SSC)/0.2% Tween-20 at 42°C, the slides were mounted in VECTASHIELD mounting medium (Vector Laboratories, H-1000) supplemented with Hoechst 33342. The average ratios of PAR axial length to the total axial lengths of X and Y chromosomes in early, mid-, and late pachynema were calculated (fig. S16).
Preparation of oocyte spreads
Fetal oocytes were collected from embryos at 17.5 dpc and spread as previously described (59). Briefly, ovaries were placed in hypoextraction buffer for 15 min, transferred into 100 mM sucrose drop, and teased apart with two needles to release cells. Twenty microliters of the cell suspension was added onto slides mounted with 200 μl of 1% PFA containing 0.15% Triton X-100, evenly spread on the slides, and then incubated in a closed humid chamber for 2 hours at room temperature. The slides were then air-dried, and subsequent immunofluorescence staining was performed as described above for spermatocyte spreads.
Oocyte collection and in vitro culture
Mice at 21 days of age were injected with 10 IU of pregnant mare serum gonadotropin (PMSG) (Ningbo Sansheng Pharmaceutical Co. Ltd.) and euthanized 46 to 48 hours later. Fully grown germinal vesicle-stage oocytes were collected in M2 medium (Sigma-Aldrich, M7167) and cultured in minidrops of M16 medium (Sigma-Aldrich, M7292) covered with mineral oil at 37°C in a 5% CO2 atmosphere.
Oocyte chromosome spreading
Oocytes were collected at 8 and 16 hours of in vitro culture for chromosome spreading analysis. After removing the zona pellucida, oocytes were fixed in a solution containing 1% PFA, 0.15% Triton X-100, and 3 mM DTT on glass slides for 30 min and air dried. The slides were washed with PBS for three times and then mounted with VECTASHIELD mounting medium (Vector Laboratories, H-1000) supplemented with Hoechst 33342 (Invitrogen, H21492).
Coimmunoprecipitation
Testes were lysed in lysis buffer [50 mM tris (pH 7.5), 150 mM NaCl, 5 mM EDTA, and 0.1% NP-40] supplemented with PMSF, Protease Inhibitor Cocktail (Abcam), and MG132 (Selleckchem) on ice using a glass homogenizer, followed by rotation at 4°C for 30 min. After centrifugation, 100 μl of the supernatant was taken out as the input. Protein A/G beads (Santa Cruz Biotechnology) were washed with precooled lysis buffer three times and split into two equal aliquots, one aliquot was incubated with remaining supernatant, and the other aliquot was incubated with 2 μg of primary antibodies at 4°C for 2 to 3 hours. After washing with lysis buffer, the antibody-coated beads were incubated with precleared protein lysates at 4°C overnight. After five washes, the beads were boiled in 2× SDS sample loading buffer for 10 min. The samples were either analyzed by Western blotting immediately or stored at −80°C.
For Co-IP in cultured cells, HEK293T cells were plated in six-well plates, followed by transfection using Lipofectamine 3000 reagents (Thermo Fisher Scientific). MG132 (5 μM) was added into the cultured cells 6 hours before cell harvesting. Cells were collected and lyzed in the same lysis buffer used for Co-IP from testicular samples 48-hours after transfection. The cell lysates were then incubated with prewashed FLAG M2 Magnetic Beads (Sigma-Aldrich) at 4°C overnight, followed by washing and boiling as described above for the Co-IP from testicular samples.
Statistical analysis
Statistical analyses were conducted using GraphPad Prism software version 7.0 (GraphPad Software Inc.). All tests and P values are described in the corresponding figure legends and/or results. Data are presented as means ± standard error in figures and main text.
Acknowledgments
We thank all the participants for cooperation. We thank S. Keeney for Spo11 mice; P. J. Wang and M. Luo for antibodies; X. Huang for plasmids of the CRISPR-Cas9 system; L. Zhong, X. Zhang, and H. Cheng for the generation and care of genetically modified mice; J. Gao, J. Xie, and Y. Xue for experimental assistance; and C. Wei and Y. Xu for critical reading of the manuscript.
Funding: This work was supported by State Key Program of National Natural Science Foundation of China (31630050 to Q.S.), Major Program of National Natural Science Foundation of China (31890780 to Q.S.), Natural Science Foundation of China (3200060244 to B.S.), Strategic Priority Research Program of the Chinese Academy of Sciences (XDB19000000 to Q.S.), and National Key Research and Developmental Program of China (2016YFC1000600 to Q.S. and 2018YFC1003400 to H.J.).
Author contributions: Q.S. and Y.Z. conceived the initial study. H.M. and H.Y. designed and supervised subsequent investigations. T.L., X.X., L.J., H.Y., J.Y., C.G., B.S., B.Z., X.J., Y.L., X.Z., J.X., and X.H. carried out the experiments. Y.Z., H.Z., and J.Z. conducted the WES and analysis. H.M., T.L., H.Y., and X.X. analyzed and interpreted the data. H.M. wrote the manuscript with the input from Q.S., H.J., H.Y., T.L., and S.F.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S16
Tables S1 to S3
REFERENCES AND NOTES
- 1.Kleckner N., Meiosis: How could it work? Proc. Natl. Acad. Sci. U.S.A. 93, 8167–8174 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Page S. L., Hawley R. S., Chromosome choreography: The meiotic ballet. Science 301, 785–789 (2003). [DOI] [PubMed] [Google Scholar]
- 3.Baudat F., Imai Y., de Massy B., Meiotic recombination in mammals: Localization and regulation. Nat. Rev. Genet. 14, 794–806 (2013). [DOI] [PubMed] [Google Scholar]
- 4.Shi Q., Spriggs E., Field L. L., Ko E., Barclay L., Martin R. H., Single sperm typing demonstrates that reduced recombination is associated with the production of aneuploid 24,XY human sperm. Am. J. Med. Genet. 99, 34–38 (2001). [DOI] [PubMed] [Google Scholar]
- 5.Edelmann W., Cohen P. E., Kane M., Lau K., Morrow B., Bennett S., Umar A., Kunkel T., Cattoretti G., Chaganti R., Pollard J. W., Kolodner R. D., Kucherlapati R., Meiotic pachytene arrest in MLH1-deficient mice. Cell 85, 1125–1134 (1996). [DOI] [PubMed] [Google Scholar]
- 6.Lipkin S. M., Moens P. B., Wang V., Lenzi M., Shanmugarajah D., Gilgeous A., Thomas J., Cheng J., Touchman J. W., Green E. D., Schwartzberg P., Collins F. S., Cohen P. E., Meiotic arrest and aneuploidy in MLH3-deficient mice. Nat. Genet. 31, 385–390 (2002). [DOI] [PubMed] [Google Scholar]
- 7.Kauppi L., Barchi M., Baudat F., Romanienko P. J., Keeney S., Jasin M., Distinct properties of the XY pseudoautosomal region crucial for male meiosis. Science 331, 916–920 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Papanikos F., Clément J. A. J., Testa E., Ravindranathan R., Grey C., Dereli I., Bondarieva A., Valerio-Cabrera S., Stanzione M., Schleiffer A., Jansa P., Lustyk D., Fei J. F., Adams I. R., Forejt J., Barchi M., de Massy B., Toth A., Mouse ANKRD31 regulates spatiotemporal patterning of meiotic recombination initiation and ensures recombination between X and Y sex chromosomes. Mol Cell 74, 1069–1085.e11 (2019). [DOI] [PubMed] [Google Scholar]
- 9.Boekhout M., Karasu M. E., Wang J., Acquaviva L., Pratto F., Brick K., Eng D. Y., Xu J., Camerini-Otero R. D., Patel D. J., Keeney S., REC114 partner ANKRD31 controls number, timing, and location of meiotic DNA breaks. Mol. Cell. 74, 1053–1068.e8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eaker S., Cobb J., Pyle A., Handel M. A., Meiotic prophase abnormalities and metaphase cell death in MLH1-deficient mouse spermatocytes: Insights into regulation of spermatogenic progress. Dev. Biol. 249, 85–95 (2002). [DOI] [PubMed] [Google Scholar]
- 11.Robert T., Nore A., Brun C., Maffre C., Crimi B., Guichard V., Bourbon H.-M., de Massy B., The TopoVIB-Like protein family is required for meiotic DNA double-strand break formation. Science 351, 943–949 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Vrielynck N., Chambon A., Vezon D., Pereira L., Chelysheva L., De Muyt A., Mézard C., Mayer C., Grelon M., A DNA topoisomerase VI–like complex initiates meiotic recombination. Science 351, 939–943 (2016). [DOI] [PubMed] [Google Scholar]
- 13.Keeney S., Giroux C. N., Kleckner N., Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88, 375–384 (1997). [DOI] [PubMed] [Google Scholar]
- 14.Bergerat A., de Massy B., Gadelle D., Varoutas P.-C., Nicolas A., Forterre P., An atypical topoisomerase II from archaea with implications for meiotic recombination. Nature 386, 414–417 (1997). [DOI] [PubMed] [Google Scholar]
- 15.Brick K., Smagulova F., Khil P., Camerini-Otero R. D., Petukhova G. V., Genetic recombination is directed away from functional genomic elements in mice. Nature 485, 642–645 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hinch A. G., Becker P. W., Li T., Moralli D., Zhang G., Bycroft C., Green C., Keeney S., Shi Q., Davies B., Donnelly P., The configuration of RPA, RAD51, and DMC1 binding in meiosis reveals the nature of critical recombination intermediates. Mol Cell 79, 689–701.e10 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hunter N., Meiotic recombination: The essence of heredity. Cold Spring Harb. Perspect. Biol. 7, a016618 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brown M. S., Bishop D. K., DNA strand exchange and RecA homologs in meiosis. Cold Spring Harb. Perspect. Biol. 7, a016659 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang F., Gell K., van der Heijden G. W., Eckardt S., Leu N. A., Page D. C., Benavente R., Her C., Höög C., McLaughlin K. J., Wang P. J., Meiotic failure in male mice lacking an X-linked factor. Genes Dev. 22, 682–691 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Snowden T., Acharya S., Butz C., Berardini M., Fishel R., hMSH4-hMSH5 recognizes Holliday junctions and forms a meiosis-specific sliding clamp that embraces homologous chromosomes. Mol. Cell 15, 437–451 (2004). [DOI] [PubMed] [Google Scholar]
- 21.Cannavo E., Sanchez A., Anand R., Ranjha L., Hugener J., Adam C., Acharya A., Weyland N., Aran-Guiu X., Charbonnier J.-B., Hoffmann E. R., Borde V., Matos J., Cejka P., Regulation of the MLH1–MLH3 endonuclease in meiosis. Nature 586, 618–622 (2020). [DOI] [PubMed] [Google Scholar]
- 22.Reynolds A., Qiao H., Yang Y., Chen J. K., Jackson N., Biswas K., Holloway J. K., Baudat F., de Massy B., Wang J., Höög C., Cohen P. E., Hunter N., RNF212 is a dosage-sensitive regulator of crossing-over during mammalian meiosis. Nat. Genet. 45, 269–278 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lange J., Yamada S., Tischfield S. E., Pan J., Kim S., Zhu X., Socci N. D., Jasin M., Keeney S., The landscape of mouse meiotic double-dtrand break formation, processing, and repair. Cell 167, 695–708.e16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moens P. B., Marcon E., Shore J. S., Kochakpour N., Spyropoulos B., Initiation and resolution of interhomolog connections: Crossover and non-crossover sites along mouse synaptonemal complexes. J. Cell Sci. 120, 1017–1027 (2007). [DOI] [PubMed] [Google Scholar]
- 25.Kauppi L., Barchi M., Lange J., Baudat F., Jasin M., Keeney S., Numerical constraints and feedback control of double-strand breaks in mouse meiosis. Genes Dev. 27, 873–886 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perry J., Palmer S., Gabriel A., Ashworth A., A short pseudoautosomal region in laboratory mice. Genome Res. 11, 1826–1832 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rappold G. A., The pseudoautosomal regions of the human sex chromosomes. Hum. Genet. 92, 315–324 (1993). [DOI] [PubMed] [Google Scholar]
- 28.Kovalenko O. V., Wiese C., Schild D., RAD51AP2, a novel vertebrate- and meiotic-specific protein, shares a conserved RAD51-interacting C-terminal domain with RAD51AP1/PIR51. Nucleic Acids Res. 34, 5081–5092 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baudat F., Manova K., Yuen J. P., Jasin M., Keeney S., Chromosome synapsis defects and sexually dimorphic meiotic progression in mice lacking Spo11. Mol. Cell 6, 989–998 (2000). [DOI] [PubMed] [Google Scholar]
- 30.Lara-Gonzalez P., Westhorpe F. G., Taylor S. S., The spindle assembly checkpoint. Curr. Biol. 22, R966–R980 (2012). [DOI] [PubMed] [Google Scholar]
- 31.Gomez H. L., Felipe-Medina N., Sánchez-Martín M., Davies O. R., Ramos I., García-Tuñón I., de Rooij D. G., Dereli I., Toth A., Barbero J. L., Benavente R., Llano E., Pendas A. M., C14ORF39/SIX6OS1 is a constituent of the synaptonemal complex and is essential for mouse fertility. Nat. Commun. 7, 13298 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fan S., Jiao Y., Khan R., Jiang X., Javed A. R., Ali A., Zhang H., Zhou J., Naeem M., Murtaza G., Li Y., Yang G., Zaman Q., Zubair M., Guan H., Zhang X., Ma H., Jiang H., Ali H., Dil S., Shah W., Ahmad N., Zhang Y., Shi Q., Homozygous mutations in C14orf39/SIX6OS1 cause non-obstructive azoospermia and premature ovarian insufficiency in humans. Am. J. Hum. Genet. 108, 324–336 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wojtasz L., Daniel K., Roig I., Bolcun-Filas E., Xu H., Boonsanay V., Eckmann C. R., Cooke H. J., Jasin M., Keeney S., McKay M. J., Toth A., Mouse HORMAD1 and HORMAD2, two conserved meiotic chromosomal proteins, are depleted from synapsed chromosome axes with the help of TRIP13 AAA-ATPase. PLOS Genet. 5, e1000702 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fukuda T., Pratto F., Schimenti J. C., Turner J. M., Camerini-Otero R. D., Höög C., Phosphorylation of chromosome core components may serve as axis marks for the status of chromosomal events during mammalian meiosis. PLOS Genet. 8, e1002485 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guo R., Xu Y., Leu N. A., Zhang L., Fuchs S. Y., Ye L., Wang P. J., The ssDNA-binding protein MEIOB acts as a dosage-sensitive regulator of meiotic recombination. Nucleic Acids Res. 48, 12219–12233 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Slotman J. A., Paul M. W., Carofiglio F., de Gruiter H. M., Vergroesen T., Koornneef L., van Cappellen W. A., Houtsmuller A. B., Baarends W. M., Super-resolution imaging of RAD51 and DMC1 in DNA repair foci reveals dynamic distribution patterns in meiotic prophase. PLOS Genet. 16, e1008595 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pittman D. L., Cobb J., Schimenti K. J., Wilson L. A., Cooper D. M., Brignull E., Handel M. A., Schimenti J. C., Meiotic prophase arrest with failure of chromosome synapsis in mice deficient for Dmc1, a germline-specific RecA homolog. Mol. Cell 1, 697–705 (1998). [DOI] [PubMed] [Google Scholar]
- 38.Acquaviva L., Boekhout M., Karasu M. E., Brick K., Pratto F., Li T., van Overbeek M., Kauppi L., Camerini-Otero R. D., Jasin M., Keeney S., Ensuring meiotic DNA break formation in the mouse pseudoautosomal region. Nature 582, 426–431 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martini E., Diaz R. L., Hunter N., Keeney S., Crossover homeostasis in yeast meiosis. Cell 126, 285–295 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jones G. H., Franklin F. C. H., Meiotic crossing-over: Obligation and interference. Cell 126, 246–248 (2006). [DOI] [PubMed] [Google Scholar]
- 41.Modesti M., Budzowska M., Baldeyron C., Demmers J. A. A., Ghirlando R., Kanaar R., RAD51AP1 is a structure-specific DNA binding protein that stimulates joint molecule formation during RAD51-mediated homologous recombination. Mol. Cell 28, 468–481 (2007). [DOI] [PubMed] [Google Scholar]
- 42.Wiese C., Dray E., Groesser T., San Filippo J., Shi I., Collins D. W., Tsai M.-S., Williams G. J., Rydberg B., Sung P., Schild D., Promotion of homologous recombination and genomic stability by RAD51AP1 via RAD51 recombinase enhancement. Mol. Cell 28, 482–490 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ouyang J., Yadav T., Zhang J.-M., Yang H., Rheinbay E., Guo H., Haber D. A., Lan L., Zou L., RNA transcripts stimulate homologous recombination by forming DR-loops. Nature 594, 283–288 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dray E., Dunlop M. H., Kauppi L., Filippo J. S., Wiese C., Tsai M.-S., Begovic S., Schild D., Jasin M., Keeney S., Sung P., Molecular basis for enhancement of the meiotic DMC1 recombinase by RAD51 associated protein 1 (RAD51AP1). Proc. Natl. Acad. Sci. U.S.A. 108, 3560–3565 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guiraldelli M. F., Eyster C., Wilkerson J. L., Dresser M. E., Pezza R. J., Mouse HFM1/Mer3 is required for crossover formation and complete synapsis of homologous chromosomes during meiosis. PLOS Genet. 9, e1003383 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Simhadri S., Peterson S., Patel D. S., Huo Y., Cai H., Bowman-Colin C., Miller S., Ludwig T., Ganesan S., Bhaumik M., Bunting S. F., Jasin M., Xia B., Male fertility defect associated with disrupted BRCA1-PALB2 interaction in mice. J. Biol. Chem. 289, 24617–24629 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li H., Watford W., Li C., Parmelee A., Bryant M. A., Deng C., O’Shea J., Lee S. B., Ewing sarcoma gene EWS is essential for meiosis and B lymphocyte development. J. Clin. Invest. 117, 1314–1323 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Inagaki A., Sleddens-Linkels E., Wassenaar E., Ooms M., van Cappellen W. A., Hoeijmakers J. H. J., Seibler J., Vogt T. F., Shin M. K., Grootegoed J. A., Baarends W. M., Meiotic functions of RAD18. J. Cell Sci. 124, 2837–2850 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dai X. X., Jiang Y., Gu J. H., Jiang Z. Y., Wu Y. W., Yu C., Yin H., Zhang J., Shi Q. H., Shen L., Sha Q. Q., Fan H. Y., The CNOT4 subunit of the CCR4-NOT complex is involved in mRNA degradation, efficient DNA damage repair, and XY chromosome crossover during male germ cell meiosis. Adv. Sci. 8, 2003636 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.de Boer E., Stam P., Dietrich A. J., Pastink A., Heyting C., Two levels of interference in mouse meiotic recombination. Proc. Natl. Acad. Sci. U.S.A. 103, 9607–9612 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.World Health Organization, WHO Laboratory Manual for the Examination and Processing of Human Semen (WHO Team, ed. 5, 2010). [Google Scholar]
- 52.Jiang L., Li T., Zhang X., Zhang B., Yu C., Li Y., Fan S., Jiang X., Khan T., Hao Q., Xu P., Nadano D., Huleihel M., Lunenfeld E., Wang P. J., Zhang Y., Shi Q., RPL10L is required for male meiotic division by compensating for RPL10 during meiotic sex chromosome inactivation in mice. Curr Biol 27, 1498–1505.e6 (2017). [DOI] [PubMed] [Google Scholar]
- 53.Zhang B., Ma H., Khan T., Ma A., Li T., Zhang H., Gao J., Zhou J., Li Y., Yu C., Bao J., Ali A., Murtaza G., Yin H., Gao Q., Jiang X., Zhang F., Liu C., Khan I., Zubair M., Hussain H. M. J., Khan R., Yousaf A., Yuan L., Lu Y., Xu X., Wang Y., Tao Q., Hao Q., Fang H., Cheng H., Zhang Y., Shi Q., A DNAH17 missense variant causes flagella destabilization and asthenozoospermia. J. Exp. Med. 217, e20182365 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Peters A. H., Plug A. W., van Vugt M. J., de Boer P., A drying-down technique for the spreading of mammalian meiocytes from the male and female germline. Chromosome Res. 5, 66–68 (1997). [DOI] [PubMed] [Google Scholar]
- 55.Stanzione M., Baumann M., Papanikos F., Dereli I., Lange J., Ramlal A., Trankner D., Shibuya H., de Massy B., Watanabe Y., Jasin M., Keeney S., Toth A., Meiotic DNA break formation requires the unsynapsed chromosome axis-binding protein IHO1 (CCDC36) in mice. Nat. Cell Biol. 18, 1208–1220 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Drabent B., Bode C., Bramlage B., Doenecke D., Expression of the mouse testicular histone gene H1t during spermatogenesis. Histochem. Cell Biol. 106, 247–251 (1996). [DOI] [PubMed] [Google Scholar]
- 57.Jiao Y., Fan S., Jabeen N., Zhang H., Khan R., Murtaza G., Jiang H., Ali A., Li Y., Bao J., Zhang B., Xu J., Xu B., Hussain H. M. J., Zaman Q., Khan I., Bukhari I., Iqbal F., Yousaf A., Dil S., Khan M., Ahmad N., Ma H., Jiang X., Zhang Y., Shi Q., A TOP6BL mutation abolishes meiotic DNA double-strand break formation and causes human infertility. Sci. Bull. 65, 2120–2129 (2020). [DOI] [PubMed] [Google Scholar]
- 58.Evans E. P., Breckon G., Ford C. E., An air-drying method for meiotic preparations from mammalian testes. Cytogenetics 3, 289–294 (1964). [DOI] [PubMed] [Google Scholar]
- 59.Hwang G. H., Hopkins J. L., Jordan P. W., Chromatin spread preparations for the analysis of mouse oocyte progression from prophase to metaphase II. J. Vis. Exp. 2018, 56736 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S16
Tables S1 to S3