Trio-based whole-genome sequencing identified the role of chromatin modification in OCD pathology.
Abstract
Obsessive-compulsive disorder (OCD) is a chronic anxiety disorder with a substantial genetic basis and a broadly undiscovered etiology. Recent studies of de novo mutation (DNM) exome-sequencing studies for OCD have reinforced the hypothesis that rare variation contributes to the risk. We performed, to our knowledge, the first whole-genome sequencing on 53 parent-offspring families with offspring affected with OCD to investigate all rare de novo variants and insertions/deletions. We observed higher mutation rates in promoter-anchored chromatin loops (empirical P = 0.0015) and regions with high frequencies of histone marks (empirical P = 0.0001). Mutations affecting coding regions were significantly enriched within coexpression modules of genes involved in chromatin modification during human brain development. Four genes—SETD5, KDM3B, ASXL3, and FBL—had strong aggregated evidence and functionally converged on transcription’s epigenetic regulation, suggesting an important OCD risk mechanism. Our data characterized different genome-wide DNMs and highlighted the contribution of chromatin modification in the etiology of OCD.
INTRODUCTION
Obsessive-compulsive disorder (OCD) is a severe, disabling neuropsychiatric disorder characterized by repetitive, compulsive thoughts, and/or compulsive movements or rituals, with an estimated lifetime prevalence of 1 to 3% (1–3). OCD is typically diagnosed on the basis of observed behaviors, duration of symptoms, and impairment of function, rather than based on a biological understanding of the disease etiology. This has notably hindered progress in developing more precise diagnoses and better therapeutics to improve patient outcomes. Epidemiological studies have demonstrated the important contributions of genetic factors to the etiology of OCD. On the basis of concordance rates in monozygotic and dizygotic twin studies, the heritability of OCD is approximately 50% (4). Although some putative chromosomal regions have been highlighted by traditional genetic study (5), no causative genes for OCD have been confidently identified (6).
More recently, genome-wide association studies (GWASs) (7, 8) have identified single-nucleotide polymorphisms (SNPs) associated with OCD with roles in glutamate signaling and synaptic functions. However, the SNP-based heritability of OCD is estimated to be 0.22 (7), suggesting that the remaining contribution to the disease risk cannot be explained by weakly associated common SNPs (9). The apparent contradiction between the profound heritability of OCD based on epidemiological studies and the limited polygenic risks estimated by GWAS, the so-called “missing heritability,” has given rise to the hypothesis of a “de novo paradigm,” which suggests the potential role of rare variants in the development of OCD. More recently, two whole-exome sequencing (WES) studies of parent-offspring OCD trios by Cappi et al. (10, 11) have shown compelling evidence for the role of de novo mutations (DNMs) in OCD, which adds to the accumulating proof for the hypothesis that de novo variants have a significant role in the genetic architecture of psychiatric disorders (12). The two OCD exome studies (10, 11) (the number of trio = 20 and 184, respectively) estimated that about 22% of patients with OCD carried a damaging coding DNM that could estimate the occurrence of the disease. In the meantime, they highlighted genes, such as CHD8 and SCUBE1, which have a potential role in the pathology of OCD.
However, most of the large-scale investigations on OCD susceptibility have been focused on attributing disease risk to the protein-coding regions of the genome by targeted-sequencing (13) or exome-sequencing approaches (10, 11). Apparent highly penetrant pathogenic variants in intergenic, noncoding RNA (ncRNA), and large structural variant (SV) regions (14, 15) are also known, suggesting that whole-genome sequencing (WGS), which detects more classes and sizes of mutations than WES, could be considered as the preferred genomic platform to study OCD. Furthermore, the application of WGS in other neuropsychiatric disorder studies, such as autism disorder spectrum (ASD) (14–16) and intellectual disability (ID) (17), has already demonstrated its unparalleled power for understanding the overall burden of DNMs at the genome scale and their contributions to diseases.
Thus, following the previous exome studies in OCD, we applied WGS and bioinformatics analysis to investigate a cohort of OCD parent-child trios to identify genome-scale de novo variants, including single-nucleotide variants (SNVs), small insertions and deletions (indels), and SVs. In 53 parent-offspring families, we found strong evidence for a high occurrence of mutation at promoter-interacting loops. We also observed that DNMs preferentially occurred on intolerant genes and affected genes regulating chromatin modification. Furthermore, we identified three high-confidence chromatin modifiers—SETD5 (SET-containing-domain 5), KDM3B (lysine demethylase 3B), and ASXL3 (transcriptional regulator 3)—as OCD candidate risk genes based on multiple lines of evidence. Last, our integrated analysis found that genes affected by OCD and Tourette disorder (TD) DNMs (18–20) had similar biological functions but affected distinct brain regions and cell-type expression patterns, which, in part, explains the complex comorbidity relationship between the two neurodevelopmental disorders.
RESULTS
Global identification of de novo variants and their distributions in OCD
We performed WGS of 160 DNA samples from a cohort of 53 families with 54 OCD probands and their unaffected parents without major psychoses (Materials and Methods and table S1). After the quality control, 53 probands from 51 trios and one quad, as well as their unaffected parents, remained for subsequent analysis with one trio family removed (the total number of participants is 157). On average, 89.1% of genomic regions were covered by 20 or more reads at the individual level. The average sequencing depth was 29.92× (fig. S1). Among 53 parent-offspring families, we identified 4143 de novo SNVs and short indels initially. After the Sanger-sequencing validation with ~96% positive rate (see Materials and Methods and table S2), we retained a list of 4062 DNMs (table S2) with a mutation rate of 1.34 × 10−8 (95% confidence interval: 1.26 × 10−8 to 1.41 × 10−8) per base pair. The number of DNMs in each proband ranged from 51 to 117 in the genome (zero to four in the exome). Thirty-five probands carried one or more exonic DNMs. The per-individual number of DNMs was 0.93 for SNVs and 0.13 for indels across coding regions, 62.5 for SNVs, and 14.7 for indels across the genome, similar to those reported in other family-based WES and WGS studies of psychiatric diseases (14–16).
Next, we calculated the relative mutation rates (number of DNMs of a particular type divided by the total number of DNMs in the study) globally for five general mutation types: exonic, intronic, untranslated region (UTR), ncRNA, and intergenic. Similar to the results reported previously in a large WGS study in ASD (14), differences between OCD probands and Simons Simplex Collection (SSC) controls (15) in most types did not reach statistical significance after the multiple testing correction except for ncRNA (P = 0.02). Furthermore, we defined a damaging DNM category as one causing de novo loss-of-function (LoF; frameshift indel, stop-gain, or splice-site mutation) or pathogenic missense [predicted by SIFT (21) and PolyPhen-2 (22) with a damaging effect, and the PHRED-scaled score of combined annotation dependent depletion (CADD) (23) ≥ 20]. We then evaluated the rate of damaging mutations in OCD against SSC controls (15) by calculating the relative exonic mutation rates against the synonymous mutation rates, since the usage of synonymous mutations as an internal control should be resistant to potential artifacts caused by the comparison of data from different studies. We observed a significantly higher mutation rate resulting in damaging DNMs in OCD [P = 0.02, odds ratio (OR) = 2.32]. Moreover, no significant difference was observed in guanine-cytosine content (GC content) (Wilcoxon’s test P = 0.1) or gene length (Wilcoxon’s test P = 0.9) between DNM genes in cases and those in controls, indicating that the observed significance in the rate of OCD damaging mutation was not introduced by biases, such as GC content or gene length.
DNMs in OCD are enriched in promoter-interacting loops and regions with high frequencies of histone marks
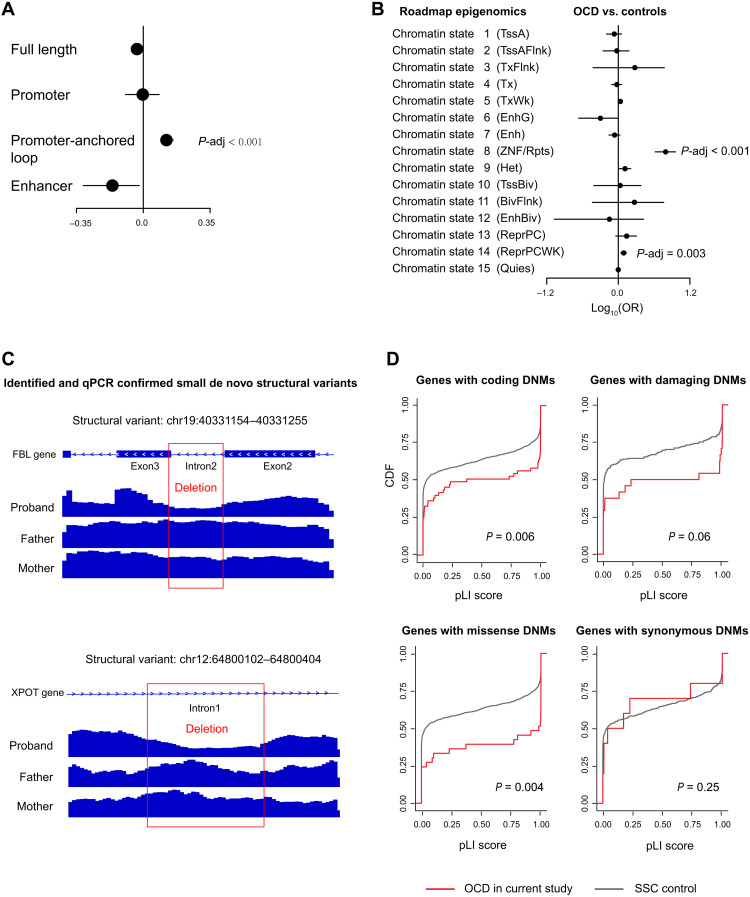
Next, we investigated whether OCD mutations occurred more frequently in functional genomic regions by integrating genomic annotations from multiple different data sources, including PsychEncode (24), ENSEMBL (25), and Roadmap epigenomics (26). We observed no global enrichment of DNMs directly hitting promoters, enhancers, or full-length genes with noncoding regions after correcting for multiple testing, which was also similar to a previous large WGS study of schizophrenia (SCZ) trios (14). However, we observed that our OCD mutations were significantly enriched in the promoter-anchored chromatin loops (empirical Pcorrected < 0.001 and OR = 1.23; Fig. 1A), which are promoter-interacting distal regulatory elements. To rule out the potential bias by study design and sequencing technical issues, we collected two additional control datasets (27, 28) and found that the control DNM distribution in promoter-anchored chromatin loops was similar in different studies (fig. S2; Cochran test P = 0.91). We compared our OCD data to the three control data, estimated SE by subsampling test and leave-one-chromosome-out (LOCO) (Materials and Methods), and obtained consistent enrichment with no cross-study heterogeneity (subsampling: meta-analysis OR = 1.25, P < 2.2 × 10−16, Cochran test P = 0.94; LOCO: meta-analysis OR = 1.25, P < 2.2 × 10−16, Cochran test P = 0.15).
Fig. 1. Rate of DNMs across genomic regions.
(A and B) Distribution of the per-individual rate of mutations within (A) different functional genomic regions and (B) different chromatin states. The P values represent the significance of differences in controls or patients with other diseases and were corrected for multiple testing using a false discovery rate. The X axes corresponded to ORs estimated by the Fisher’s tests, and error bars represent 95% confidence intervals. (C) Schematic and Integrative Genomics Viewer plots are showing two dSVs validated by quantitative polymerase chain reaction (qPCR). (D) Comparison of intolerance of genes affected by the same four types of DNMs. SSC control, Simons Simplex Collection siblings; pLI, the probability that a gene is intolerant to a LoF mutation; dSV, de novo SV.
Chromatin states, which are the combination of multiple epigenomic marks in a spatial context, can capture information describing different classes of genomic elements such as promoters, enhancers, transcribed, repressed, and repetitive regions. Thus, we extended our analysis of DNM burden to investigate their occurrence at epigenomic marks in cases versus controls by mapping mutations to the 15 core chromatin states (26). We observed a significantly higher mutation occurrence within regions of zinc finger genes and repeats (chromatin state 8; empirical Pcorrected < 0.0001 and OR = 9.65; Fig. 1B), which is characterized by high frequencies of histone marks H3K9me3, H4K20me3, and H3K36me3 and relatively low frequencies of other marks. Similar to above, the DNM distributions among chromatin states were highly concordant between different control studies [Pearson correlation coefficient (R) > 0.96; fig. S2], and the enrichment result was consistent across these studies (subsampling: meta-analysis OR = 7.24, P = 1.23 × 10−10, Cochran test P = 0.87; LOCO: meta-analysis OR = 7.46, P < 2.2 × 10−16, Cochran test P = 0.07).
Small de novo structural variation affecting FBL gene in patient with OCD
Aside from identifying point mutations and indel, we also explored whether de novo SVs (dSVs) had a role in OCD. After machine learning detection, manual inspection, and population frequency filtration (Materials and Methods), we identified seven potential dSVs not presented in the GnomAD population. Two of them were quantitative polymerase chain reaction (qPCR) verified as true de novo variants on the basis of the sample availability. One of them was identified as a heterozygous de novo deletion (chr19:40331154–40331255) of the second intron of gene FBL on a male patient (ID: WOC3_114_1_PT) with early-onset OCD (table S3). This dSV skipped most of the second intron and two bases of the 5′ of exon3 and inserted one base to the 3′ of exon2, leading to a frameshift-like alteration on FBL (Fig. 1C). FBL encodes Fibrillarin, a nuclear protein that methylates glutamine-104 of histone H2A (H2AQ104me) and modulates chromatin structure and activity (29). The other confirmed dSV was on another male patient (ID: WOC4_34), also with early-onset OCD. The dSV was a heterozygous deletion of a 703–base pair (bp) fragment (chr12:64800102–64800404), which affected the part of the first intron of XPOT (Fig. 1C and table S3) and was predicted to have little effect on the gene-based function annotations from ANNOVAR (30).
De novo coding mutations preferentially hit intolerant genes and affected genes regulating chromatin modification
Next, we explored the functional impact of the identified DNMs. Since genes differ in their tolerance to mutations, as measured by the probability of being LoF intolerant (pLI) (31), we then investigated whether this intolerance differed across genes affected by different mutation classes (Fig. 1D). We observed that OCD-associated genes were significantly more mutation-intolerant than SSC controls with regard to coding DNMs (P = 0.003), damaging DNMs (P = 0.03), or missense DNMs (P = 0.002), but not for synonymous DNMs (P = 0.25).
Moreover, we reasoned that transcriptomic data from the developmental human brain would improve our understanding of OCD pathophysiology, as the cerebral cortex has been consistently implicated in OCD (32). We examined the expression data from the Brainspan (33) and constructed prenatal and postnatal coexpression networks by weighted gene coexpression network analysis (WGCNA) (34) to access three sets of gene lists: set 1 contained protein-altering DNMs (Table 1), set 2 contained genes with promoters hit by DNMs, and set 3 was a combination of set 1 and set 2.
Table 1. List of all protein-altering DNMs identified in the present study.
VUS, the variant with uncertain significance. NA, not available.
| Proband ID | Position | Mutation |
Gene
symbol |
Mutation type |
Protein
change |
SIFT |
Polyphen2
HDIV |
CADD | Classification |
| WOC5_160_1 | 18:31324188 | GC > G | ASXL3 | Frameshift deletion |
p.F1460Lfs*5 | NA | NA | 35 | Damaging |
| WOC3_114_1_PT | 6:29912028 | AGG > AG | HLA-A | Frameshift deletion |
p.D251Tfs*45 | NA | NA | 32 | Damaging |
| WOC5_137_1 | 1:109242401 | CAAATAAA>CAAA | PRPF38B | Frameshift deletion |
p.K324Efs*59 | NA | NA | 34 | Damaging |
| WOC5_26 | 2:114482963 | CA > C | SLC35F5 | Frameshift deletion |
p.L408Rfs*11 | NA | NA | 35 | Damaging |
| WOC5_160_1 | 19:16611793 | CT > CTT | C19orf44 | Frameshift insertion |
p.K65Efs*15 | NA | NA | 14.66 | Damaging |
| WOC5_6 | 19:2939254 | AGTGAGGGAATGACA CCACCCTTACCCAAG GAGGCA>A |
ZNF77 | Splice-site mutation (frameshift deletion) |
p.A41Lfs*5 | NA | NA | 16.62 | Damaging |
| WOC5_160_1 | 3:49713641 | C > T | APEH | Nonsense | p.Q199X | NA | NA | 35 | Damaging |
| WOC4_28 | 13:32937534 | T > G | BRCA2 | Nonsense | p.L2732X | NA | NA | 39 | Damaging |
| WOC5_102_1 | 14:20779873 | G > A | CCNB1IP1 | Nonsense | p.R224X | NA | NA | 37 | Damaging |
| WOC4_34 | 4:47427852 | C > G | GABRB1 | Nonsense | p.Y414X | NA | NA | 27 | Damaging |
| WOC4_165_1 | 6:39286844 | C > T | KCNK16 | Nonsense | p.W93X | NA | NA | 43 | Damaging |
| WOC4_170_1 | 22:25320164 | C > T | SGSM1 | Nonsense | p.R1008X | NA | NA | 46 | Damaging |
| WOC84 | 1:118616533 | G > A | SPAG17 | Missense | p.R777C | D | D | 29.5 | Damaging |
| WOC4_170_1 | X:51640699 | C > T | MAGED1 | Missense | p.R515C | D | D | 28.9 | Damaging |
| WOC5_157_1 | 5:137722015 | A > G | KDM3B | Missense | p.E362G | D | D | 28.6 | Damaging |
| WOC5_096_1 | 17:19284412 | A > G | MAPK7 | Missense | p.Y158C | D | D | 27.5 | Damaging |
| WOC5_62 | 1:36479519 | C > T | AGO3 | Missense | p.R192W | D | D | 27.4 | Damaging |
| WOC5_178_1 | 3:142741802 | C > G | U2SURP | Missense | p.P376A | D | D | 25.9 | Damaging |
| WOC4_130_1_WY | 3:9476069 | C > T | SETD5 | Missense | p.R77C | D | D | 24.6 | Damaging |
| WOC5_093_1 | 17:18195985 | G > C | TOP3A | Missense | p.P324A | D | D | 25.2 | Damaging |
| WOL3_046_1_ZXY | 19:58438675 | C > T | ZNF418 | Missense | p.G207R | D | D | 21.7 | Damaging |
| WOC5_093_1 | 1:206648328 | C > T | IKBKE | Missense | p.R32C | D | D | 28.8 | Damaging |
| WOC5_099_1 | 4:142949945 | G > T | INPP4B | Missense | p.A922D | D | D | 25.7 | Damaging |
| WOC5_149_1 | 8:131072874 | G > A | ASAP1 | Missense | p.T1048M | D | D | 24.5 | Damaging |
| WOC5_17 | 2:71817395 | C > T | DYSF | Missense | p.A1152V | D | P | 27.9 | VUS |
| WOC5_164_1 | 12:96617430 | A > T | ELK3 | Missense | p.N29I | D | P | 25 | VUS |
| WOC5_4F | 13:76381720 | G > T | LMO7 | Missense | p.R201L | D | P | 23.2 | VUS |
| WOC5_097_1 | 3:38595773 | C > T | SCN5A | Missense | p.V1586M | D | P | 23.1 | VUS |
| WOC5_130_1 | 20:33594249 | T > C | TRPC4AP | Missense | p.T606A | T | P | 25.9 | VUS |
| WOC5_161_1 | X:123519751 | G > A | TENM1 | Missense | p.T1944I | D | B | 23.6 | VUS |
| WOC5_155_1 | 11:62416110 | A > G | INTS5 | Missense | p.V481A | D | B | 23.5 | VUS |
| WOC5_078_1_X | 15:41810235 | C > T | RPAP1 | Missense | p.S1314N | T | B | 22 | VUS |
| WOC5_60 | 16:68225447 | A > G | NFATC3 | Missense | p.T959A | D | B | 21.7 | VUS |
| WOC5_091_1 | 6:32123547 | G > A | PPT2 | Missense | p.M140I | D | B | 20.8 | VUS |
| WOC5_076_1_WSH | 14:91928489 | T > C | PPP4R3A | Missense | p.M451V | T | B | 20.4 | VUS |
| WOC5_15 | 17:40149144 | T > C | DNAJC7 | Missense | p.S38G | T | B | 19.65 | VUS |
| WOC5_4F | 3:102157378 | C > T | ZPLD1 | Missense | p.P32L | T | B | 18.89 | VUS |
| WOC5_094_1 | 19:47207624 | G > A | PRKD2 | Missense | p.P74S | T | B | 16.23 | VUS |
| WOC5_138_1 | 19:36223863 | C > T | KMT2B | Missense | p.P2138L | D | P | 15.46 | VUS |
| WOC5_116_1 | 19:57036758 | G > A | ZNF471 | Missense | p.S367N | D | B | 15.06 | VUS |
| WOC5_152_1 | 12:113759149 | C > T | SLC8B1 | Missense | p.R54H | T | B | 14.72 | VUS |
| WOC5_39 | 2:217012900 | C > T | XRCC5 | Missense | p.T524I | T | B | 14.16 | VUS |
| WOC5_39 | 9:125551863 | A > C | OR5C1 | Missense | p.I218L | T | B | 12.96 | VUS |
| WOC5_084_1_PS | 6:151671185 | T > A | AKAP12 | Missense | p.D455E | T | B | 6.24 | VUS |
| WOC5_150_1 | 18:76753591 | G > C | SALL3 | Missense | p.V534L | T | B | 0.008 | VUS |
| WOC5_091_1 | X:114468473 | CCCGCCGCCGCCGCCGCC> CCCGCCGCCGCCGCC |
LRCH2 | Nonframeshift deletion |
p.G44del | NA | NA | 16.51 | VUS |
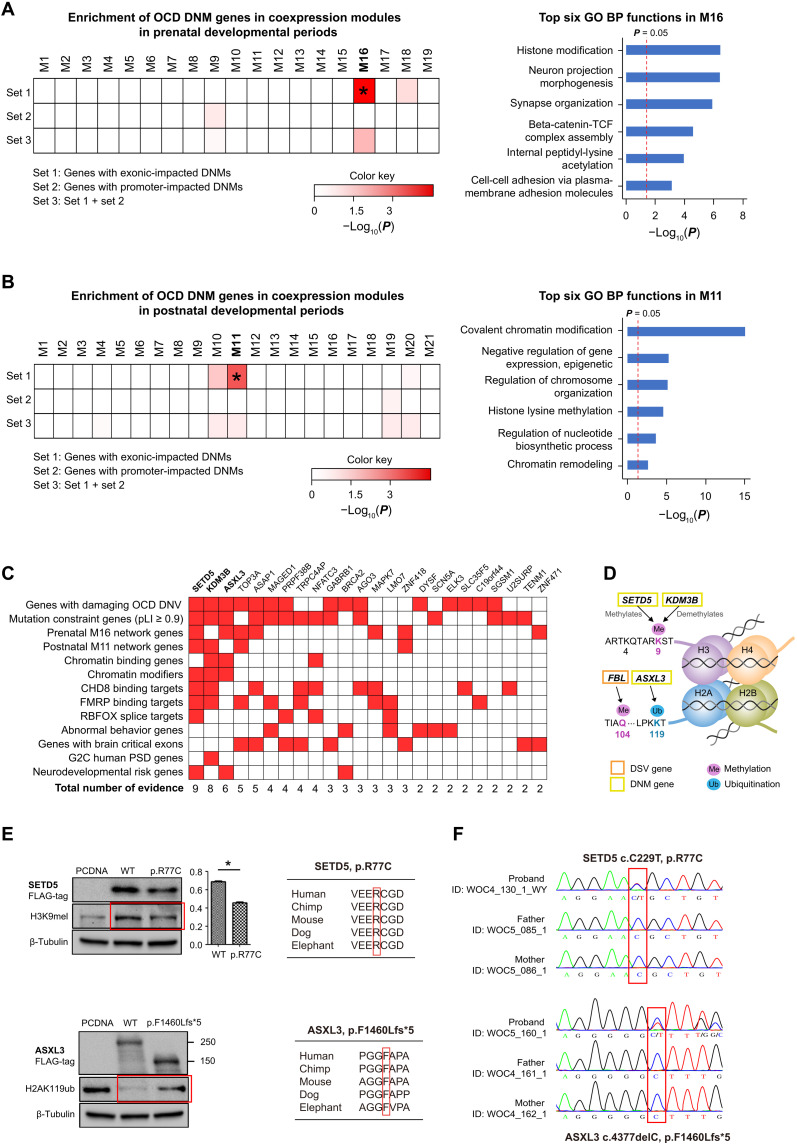
We first observed that module M16 in the prenatal network contained significantly more set1 genes than expected (two-sided Fisher’s exact test P = 0.00012). M16 was enriched in functions including histone modification, neuron projection morphogenesis, and synapse organization, according to Metascape (Fig. 2A and table S4). When exploring the connectivity of DNM genes in M16, we observed three DNM genes acting as hubs, i.e., ASXL3 [degree = 161, kME (a measure of intramodular connectivity) = 0.78], SETD5 (degree = 45, kME = 0.75), and KDM3B (degree = 246, kME = 0.87), in M16 at an average degree of 41. Next, we observed that module M11 in the postnatal network contained significantly more set1 genes than expected (P = 0.007). M11 was mainly enriched in epigenetic regulation–related functions, such as covalent chromatin modification, histone lysine methylation, and chromatin remodeling (Fig. 2B and table S4). When investigating the connectivity of OCD DNM genes within M11, we also found two OCD genes, SETD5 (degree = 26, kME = 0.92) and KDM3B (degree = 27, kME = 0.84), that act as hubs in M11 with an average degree of 9. However, we did not observe similar results for set2 or set3 genes in both networks.
Fig. 2. The biological significance of coding DNMs.
(A and B) Enrichment of DNM-affected genes within coexpression modules in (A) prenatal and (B) postnatal human brains (left) and Gene Ontology (GO) biological process (BP) enrichment results according to Metascape for corresponding modules (right). Heatmaps indicate the P values of the degree of enrichment. (C) Integrative analysis of the potential pathogenicity of coding DNM-affected genes. Thirteen independent sources of evidence were used to rank DNM-affected genes. (D) Schematic of the chromatin-regulating activities of SETD5, KDM3B, and ASXL3. (E) Western blot result (left) and cross-species conservation at the discovered mutation site (right). (F) qPCR validation of mutation of (E). CHD8, chromodomain helicase DNA binding protein 8; FMRP, fragile X mental retardation protein; RBFOX, RNA binding fox-1 homolog 1; G2C, the genes to cognition program.
Chromatin modifiers as potential contributors to OCD pathology
As noted, our analysis of mutation enrichment, dSV location, and coding DNM coexpression consistently implicated chromatin regions and chromatin modification–related pathways in OCD. Thus, we hypothesized that a subset of DNM genes related to the highlighted pathways might be more relevant to OCD. Thus, we prioritized all 45 DNM genes (Table 1) according to 13 independent sources of evidence (median Jaccard similarity index = 0.0244) (Fig. 2C; Supplementary Materials and Methods for details). We found three genes—i.e., SETD5, KDM3B, and ASXL3—with the most supporting evidence (≥6) and were all mutation-intolerant (pLI ≥ 0.9) chromatin modifiers that acted as network hubs. SETD5 functions as a histone methyltransferase and monomethylates Lys9 of histone H3, whereas KDM3B is a histone demethylase that explicitly demethylates Lys9 of histone H3. Also, ASXL3 is a part of the polycomb repressive deubiquitinase (PR-DUB) complex that deubiquitinates histone H2A lysine119 (Fig. 2D). Furthermore, as for FBL, a gene affected by de novo structure variant of patient WOC3_114_1_PT, a previous study has already confirmed its ability to methylated histone H2A glutamine-104 and regulated the nucleolar activity (Fig. 2D).
To confirm whether these mutations affected genes’ functions in chromatin modifications as expected, we selected SETD5 and ASXL3 genes to perform wild-type (WT) versus mutant experiments in human embryonic kidney (HEK) 293T cells for the hypothesis verification (Materials and Methods). We first constructed expression plasmids with WT/mutant SETD5 and ASXL3, fused with a FLAG-tag for overexpression experiments in HEK293T cells. Next, we transfected HEK293T cells with plasmids expressing the empty pcDNA3.1 vector (as a control), WT SETD5 or ASXL3, and p.R77C SETD5 or p.F1460Lfs*5 ASXL3. We collected protein lysates from each cell group and examined histone H3K9me1 or H2AK119ub levels via Western blotting after 48 hours. We found that the p.R77C SETD5 mutant’s transfection could not retain the monomethylation level of the histone mark H3K9 compared to the transfection of empty pcDNA3.1 vector or WT SETD5, indicating that p.R77C mutation impaired the enzymatic activity of SETD5 (Fig. 2E). Meanwhile, we observed that transfection of WT ASXL3 decreased the H2AK119 ubiquitination level compared with control with pcDNA, confirming the histone deubiquitination function of WT ASXL3. In contrast, the H2AK119 ubiquitination level of cells with ASXL3 mutant of p.F1460Lfs*5 resembled the pcDNA control, suggesting that this mutant lacked the normal histone deubiquitination function (Fig. 2E).
Chromatin modifiers might involve in OCD through neurotransmitters
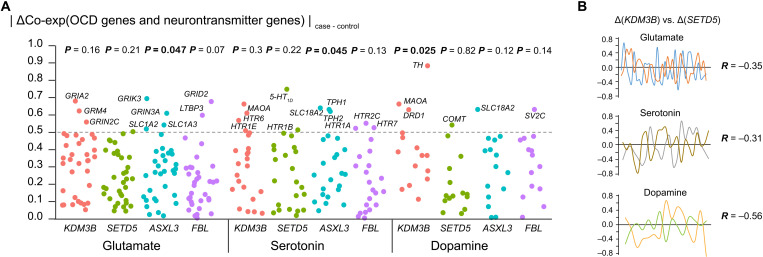
OCD has been hypothesized to be caused by alterations in neurotransmitter pathways, such as the glutamate system, dopamine system, and serotonin system. Therefore, we hypothesized that chromatin modifiers identified in the present study—such as SETD5, KDM3B, ASXL3, and FBL—may regulate neurotransmitters’ expression by epigenetic modification. To investigate the expression regulation between chromatin modifiers and neurotransmitters in OCD, we used brain gene expression data from a study conducted by Jaffe et al. (35), which contains the expression profile of the dorsolateral prefrontal cortex of postmortem brains isolated from OCD cases and nonpsychiatric controls. We first calculated the Pearson’s Rs between three chromatin modifiers and neurotransmitters in OCD cases and in controls. We then measured the coexpression regulation (ΔCo-exp) by calculating the absolute coexpression difference between patients with OCD and controls (|ΔCo-exp| = |ROCD – Rcontrol|) (Fig. 3A and table S5). We found that the overall coexpression between KDM3B and dopamine genes was significantly altered between OCD cases and controls (Wilcoxon’s test P = 0.025). In contrast, the coexpression between ASXL3 and glutamate and between ASXL3 (Wilcoxon’s test P = 0.047) and serotonin were marginally disrupted (Wilcoxon’s test P = 0.045; Fig. 3A).
Fig. 3. Disruption of coexpression between key de novo genes.
(A) Coexpression between FBL, SETD5, KDM3B, and ASXL3 and neurotransmitter (glutamate, serotonin, and dopamine)–related genes in the prefrontal cortex of patients with OCD. Each dot represents a gene involved in one neurotransmitter system, and the Y axis shows the absolute differences in the coexpression (ΔCo-exp) between patients and controls. P values were calculated using Fisher’s tests and indicate whether ΔCo-exp was significantly larger than expected. (B) Disruptions caused by KDM3B and SETD5 had opposite trends. For each neurotransmitter system, the ΔR values of SETD5 and KDM3B were plotted against each other, and their correlations were calculated.
By examining the neurotransmitters with the most altered coexpression between controls and OCD cases, we observed that the tyrosine hydroxylase (TH) gene, which is involved in the conversion of tyrosine to dopamine, had the highest disruption by KDM3B (ΔKDM3B = 0.89 from R = 0.22 in control to R = −0.66 in OCD, P = 0.0065; Fig. 3A and table S5). The MAOA gene, which is involved in serotonin and dopamine metabolism, was also found to be disrupted in patients with OCD by KDM3B (ΔKDM3B = 0.66 from R = −0.07 in control to R = 0.59 in OCD, P = 0.06). The coexpression between SETD5 and the serotonin gene HTR1D, also known as 5-HT1D, was significantly changed (ΔSETD5 = 0.75 from R = 0.35 in control to R = −0.4 in OCD, P = 0.002). The coexpression between FBL and dopamine gene SV2C, which regulated secretion in neural and endocrine cells, was significantly changed (ΔFBL = 0.76 from R = −0.23 in control to R = 0.53 in OCD).
Since SETD5 methylates the histone mark H3K9, and KDM3B demethylates the histone mark H3K9 (Fig. 2D), we hypothesized that SETD5 and KDM3B might regulate the expression of the same neurotransmitters differently. As expected, we observed negative Rs between ΔCo-expSETD5 and ΔCo-expKDM3 in the glutamate, serotonin, and dopamine systems with R = −0.35, −0.31, and − 0.56, respectively (Fig. 3B). In contrast, ASXL3, which promotes deubiquitination of the histone mark H2A119, a different histone mark, was not correlated with either SETD5 or KDM3B in any of the neurotransmitter regulatory systems.
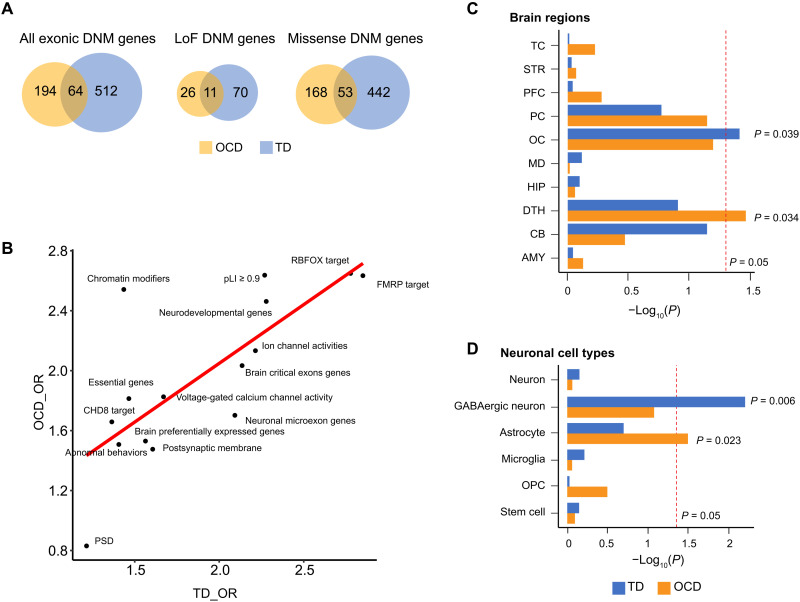
Integrated analysis of DNMs identified convergent and divergent patterns between OCD and TD
OCD and TD exhibit many similarities, including genetics, phenotypes, and epidemiology (36). Thus, to further understand the complex genetics of OCD, we compared and contrasted OCD and TD through the analysis of rare DNMs (Supplementary Materials and Methods). By directly comparing the overlapping genes between OCD and TD, combined from the current study and published exome data (10, 11, 18–20), we observed more gene overlaps between these disorders than expected (all exonic DNMs: P = 1.21 × 10−38; LoF DNMs: P = 9.19 × 10−18; missense DNMs: P = 1.01 × 10−34) (Fig. 4A). Next, we investigated whether the enrichment of OCD and TD genes across 15 disease-related gene sets (Supplementary Materials and Methods) were also similar. We observed a strong positive correlation (Pearson R = 0.54, P = 0.01) between enrichments of the two disorders by comparing the log10-transformed enrichment ORs (Fig. 4B), indicating that a number of functional biological pathways are shared between the two disorders.
Fig. 4. Comparison of genes affected by coding mutations in OCD and TDA.
(A) Venn plots showing the relationships between TD DNM–affected genes (curated from three studies) and OCD DNM–affected genes [both DNMs from Cappi et al. (10, 11) and the current study]. (B) Correlations between gene-set enrichments of TD DNM and OCD DNM–affected genes, which were not introduced by similarity or dependency between functional gene sets (average Jaccard similarity index = 0.025). Each dot represents one of the 15 manually curated gene sets (Supplementary Materials and Methods). The X axis represents the OR of TD DNM gene enrichment within this gene set, and the Y axis represents that of the OCD DNM–affected genes. Rs and P values were calculated using Pearson’s correlation analysis. (C and D) Distinct expression patterns of TD and OCD genes among (C) brain regions and (D) brain cell types. Red lines indicate the nominal P value threshold (0.05). TC, temporal cortex; STR, striatum; PFC, prefrontal cortex; PC, parietal cortex; OC, occipital cerebral wall; MD, mediodorsal nucleus of the thalamus; HIP, hippocampus; DTH, dorsal thalamus; CB, cerebellar cortex; AMY, amygdala.
Since a large proportion (75%) of DNM genes in OCD are not shared by TD, we hypothesized that this difference might elucidate specific developmental timing, brain regions, or neuronal cells likely to be involved in OCD pathogenesis. Using BrainSpan data (33) for developmental periods and brain regions, we observed that the OCD mutation genes were enriched in the dorsal thalamus [EWCE (37) P = 0.034], whereas the TD mutation genes were enriched in the occipital cortex (P = 0.039; Fig. 4C). On the basis of a study conducted by Dronc (38) for adult brain cell–type data, the OCD DNM genes were found to be enriched in astrocytes (P = 0.023), whereas the TD mutation genes were found to be enriched in GABAergic neurons (P = 0.006; Fig. 3D). No enrichment differences were observed according to brain developmental periods. These results indicate that there are clear, distinct developmental patterns that separate these two disorders.
DISCUSSION
Motivated by the growing interest in identifying ultra-rare, potentially highly penetrant genetic variants underlying the pathogenesis of neuropsychiatric disorders (14, 15, 17), in the present study, we described the first genome-wide DNM profiling of families of trios with OCD by WGS. We identified 4147 genome-wide DNMs, 57 of which affected exonic regions in probands. By exploring the global burden of DNMs in OCD probands, we observed a higher-than-expected proportion of mutations occurring at promoter-anchored chromatin loops and regions with a high frequency of histone marks, such as H3K9me3, H4K20me3, and H3K36me3 (chromatin state 8). When examining the properties of genes directly affected by mutations in OCD, we observed that DNM genes were significantly more mutation-intolerant than SSC controls, consistent with the hypothesis that DNMs might be a significant risk factor for OCD (10, 11).
Using expression data from multiple regions of the developing human brain, we discovered that the DNM genes identified in our OCD whole-genome screening showed highly correlated gene expression patterns in prenatal and postnatal development separately. The prenatal phase coexpression network included one single enriched module, M16, related to histone modifications and neuronal and synapse organization, whereas the postnatal phase included one single enriched module, M11, highlighting mainly chromatin modifications. These enriched modules from two sequential phases both identified chromatin regulation as a related top pathway, emphasizing its importance in OCD. These results also suggest that additional functional activities are involved in the development of OCD during prenatal development because M16 has other complex neuronal functions, such as neuron projection morphogenesis, synapse organization, and cell-cell adhesion, in addition to epigenetic regulation.
Chromatin-modifier genes are associated with the risk of various neurodevelopmental disorders (39, 40). In the present study, we observed three genes with top evidence-based rankings in OCD (SETD5, KDM3B, and ASXL3) and a gene disrupted by a damaging dSV (FBL). These genes are mainly chromatin modifiers and are affected by de novo damaging mutations despite their extremely high intolerance to protein-altering variants. SETD5 and KDM3B both regulate H3K9 histone methylation, which is critical in heterochromatin maintenance (41). This is consistent with our observation that patients with OCD harbor more mutations in chromatin regions (chromatin state 8) packed with a high frequency of H3K9 histone marks (42). LoF mutations in SETD5 are already implicated heavily in ASD (39) and ID (40). Mouse models of SETD5 haploinsufficiency have demonstrated impaired cognition and memory as well as inflexibility in behaviors (43), suggesting possible similarities to OCD clinical phenotypes. KDM3B is part of an important group of histone lysine methylases (KMTs) and histone lysine demethylases (KDMs), which are involved in gene regulation and expression. De novo and inherited pathogenic variants in KDM3B can cause a syndrome characterized by ID, short stature, and facial dysmorphism (44). The ASXL3 gene (OMIM® number: 615485) is associated with the Bainbridge-Ropers syndrome, a developmental disorder characterized by delayed psychomotor development and severe ID (45). The gene encodes a polycomb group protein that is part of the PR-DUB complex, functioning to deubiquitinate H2AUb1 (45). Thus, previous genetic studies have established their important connections to various other mental disorders through different epigenetic regulatory mechanisms. FBL acts as a protein methyltransferase by mediating methylation of “Gln105” of histone H2A (H2AQ104me), a modification that impairs the binding of the FACT complex (29). It is expressed in both mRNA and proteins in the human brain (46), with a pLI mutation intolerance score of >0.95. Although there is no direct link between FBL and psychiatric disorders, previous study has linked SNPs of FBL to phenotypes, such as nicotine metabolite ratio (47), suggesting that it might be a potential candidate for studying disorders with addictive behaviors.
An exome study by Cappi et al. (11) identified a chromatin remodeler gene, CHD8 (encoding chromodomain helicase DNA binding protein 8), as a risk gene for OCD and multiple other psychiatric diseases (48, 49). Although we did not find DNMs in CHD8 in our study because of the polygenic nature of complex diseases and our sample size, both SETD5 and KDM3B have been found to be binding targets of CHD8 (49). This, therefore, confirms the concept of the involvement of chromatin regulation in OCD pathogenesis.
The pathophysiology of OCD is associated with abnormalities in the cortico-striatal-thalamic-cortical (CSTC) circuitry and dysregulation of glutamate, serotonin, and dopamine systems within this network. Recently, an integrative OCD model that incorporates circuitry, neurochemistry, and genetic/epigenetic elements has been proposed by Pauls et al. (4), suggesting that individuals with OCD carrying genetic risks may be vulnerable to the impact of environmental factors that may trigger expression modifications of glutamate, serotonin, and dopamine system–related genes through epigenetic mechanisms. Our analysis showed that the coexpression patterns between three system genes and chromatin modifiers were significantly altered in the prefrontal cortex in patients with OCD. These results suggest that chromatin modifications involving SETD5, KDM3B, ASXL3, and FBL may be upstream regulators of neurotransmitter system expression, which controls necessary neurocognitive functions. Disruption of any part of this cascade may lead to abnormal obsessive phenotypes.
Despite only having 53 parent-offspring families in the current study, one of the most consistent observations in our findings is the enrichment of chromatin modification function, indicating highly significant, robust evidence for epigenetic regulation in OCD and suggests potential epigenomic vulnerability in OCD. Consequently, although synaptic proteins are commonly believed to be involved in OCD pathogenesis (8), our findings highlight the existence of other contributing mechanisms extending beyond the synapse. Although further large-scale studies are required to prove the pathogenic role of epigenetic regulation by DNMs in OCD, given the moderate sample size and statistical significance in the present study, our observations are credible, considering the fact that similar findings have been reported for ASD and SCZ, along with the fact that CHD8 was identified as a top candidate risk in a recent OCD exome study and is also a chromatin remodeler gene.
Last, our findings provide evidence that OCD shares a strong genetic etiology with TD, although the transcriptional patterns differ by brain region and neuronal cell types in these two neuropsychiatric disorders. Similar enrichment in neurodevelopmental genes and prenatal periods was observed for TD and OCD genes, consistent with similar abnormalities in neurotransmitters’ metabolism (50) and symptoms. Our findings further support that both diseases are developmental disorders (11). However, OCD genes show a uniquely high expression in the dorsal thalamus, consistent with the CSTC circuit in the OCD pathology model. This convergence and divergence between TS and OCD may be interpreted from the aspect of polygenic risk. Given the extensive genetic and phenotypic heterogeneity underlying both diseases and that only DNMs have been examined in the present study, our findings likely represent only a small fraction of the convergent and/or divergent aspects of the two disorders. Nonetheless, our analytic approach provides an important step in understanding the underlying comorbidities between OCD and TD.
MATERIALS AND METHODS
Subjects
We collected 53 unrelated parent-offspring families consisting of 160 individuals based on the availability of genomic DNA from whole-blood and the completeness of phenotype information. The parent-offspring families were composed of 52 trios (each family with one OCD-affected offspring, 156 samples in total) and one family with two OCD-affected offspring (four samples in total). Detailed information for each participant is provided in table S1. All the OCD affected patients met the following conditions: (i) The patients were diagnosed as having OCD according to Diagnostic and Statistical Manual of Mental Disorders, Fourth Edition (DSM-IV) (51) criteria. (ii) between the age of 18 and 65 years, and (iii)with a Yale-Brown Obsessive-Compulsive Scale total score cutoff of 16. The patients were excluded if they (i) included DSM-IV criteria for other disorders other than OCD, (ii) had moderate to severe suicidal ideation, and (iii) were pregnant or lactating females. Patients diagnosed with SCZ, schizoaffective disorder, ASD, pervasive developmental disorder not otherwise specified, or ID were excluded from the present study. All of the parents were screened for mental disorders by a structured interview using Mini International Neuropsychiatric Interview (52), and the families with the parents of any DSM-IV Axis I psychiatric diagnosis were excluded. All participants’ informed consent was obtained, as approved by the Institutional Review Boards of Shanghai Mental Health Center.
WGS, data processing, and DNM identification
Genomic DNA extracted from whole-blood– or lymphoblast-derived cell lines were assessed for quality by PicoGreen and gel electrophoresis and then sequenced by Novogene (Novogene Biosciences Inc., Beijing, China). DNA quantity was measured by Qubit 3.0, and at least 1 μg of nondegraded genomic DNA was used for genomic library preparation and WGS. We sequenced all trio samples, which have never been previously sequenced, on an Illumina HiSeq 4000 sequencing platform (150-bp paired-end reads). Using the processing pipeline, all de novo variants were identified as high quality and then were manually inspected by visualization of aligned reads using the Integrative Genomics Viewer (IGV) (53). Variants were annotated by using wANNOVAR (30). The average depth of coverage of our WGS data was 29.9×, with an average of 99.87% median alignment rate. After one trio was excluded for quality control, the remaining 53 trios were submitted for subsequent analyses.
Using the Sentieon DNAseq (54) processing pipeline, reads were aligned to GRCh37.63 human genome reference build using BWA-mem (55) v0.7.15-r1140 and sorted by Sentieon Dedup. Then, duplicate reads were marked and removed by Sentieon LocusCollector. The indel realignment and base quality score recalibration were performed using Sentieon Realigner and QualCal. Next, we performed the quality check on the BAM files using SAMtools (version 1.3.1) flagstat (56) and WGA metric by Picard (57) (version 2.5.0).
De novo SNVs and indels were called using a combination of two algorithms, SAMtools bcftools (version 1.3.1) (56) and the Bayesian framework TrioDeNovo (58) with the default settings on a per-family basis. Putative de novo variants were filtered on the basis of the following thresholds: (i) genotype quality ≥ 30; (ii) minimum sequencing depth is 15 read points in the proband and both parents; (iii) mapping quality ≥ 30; (iv) homozygous in father and mother with allele balance (AB) < 0.05; (v) heterozygous in a proband with AB between 0.3 and 0.7; (vi) de novo quality score from TrioDeNovo ≥7; (vii) no overlap with known regions annotated as segmental duplication; (viii) minor allele frequency ≤ 5.0 × 10−3 in 1000 Genomes project (59), ExAC (31), EVS (60), and gnomAD (61) databases; and (ix) variant clustered within a distance of 50 bp has been removed. Last, all de novo variants were detected with aligned reads from families in silico BLAT search (62) and then were manually inspected by visualization of aligned reads using IGV (53).
SNV and indel annotation
Variants were annotated by four different annotation groups as follows: (1) Variant type: Each variant was classified by mutation types using wANNOVAR (30), including SNV and indel (<50 bp), such as frameshift, stop gain (nonsense), nonsynonymous SNV (missense), and synonymous SNV, etc. (2) Genomic location annotation: Genomic locations of variants were annotated by wANNOVAR (30) and assigned in the following priority: coding, intron, UTR, upstream, downstream, ncRNA, and intergenic. (3) Gene sets: Gene lists associated with brain development or neuropsychiatric disorders were selected as follows: (3.1) CHD8 target genes were defined as the union of lists from two chromatin immunoprecipitation sequencing studies (49, 63). (3.2) FMRP target genes were defined as the union of lists from Darnell et al. (64) and Ascano et al. (65). (3.3) RBFOX splice targets were selected from Weyn-Vanhentenryck et al. (66). (3.4) Human postsynaptic density proteins were extracted from the Genes2Cognition database (67). (3.5) Constrained genes were defined as having a pLI score ≥ 0.9 in the ExAC database (31). (3.6) Genes encoding chromatin modifiers were downloaded from Chen et al. (68). (3.7) Neurodevelopmental disorder risk genes were obtained from Stessman et al. (69).
Identification of dSV
Four types of methods have been used to detect SVs: Manta (70), cn.MOPS (71), DELLY (72), and LUMPY (73). We applied SV2 (74) to merge SVs within 50 bp to 10 Mb into a VCF file for each family, and SVs with “PASS” in both FILTER tag and DENOVO_FILTER tag are selected as the candidate dSVs. In addition, we perform different filter conditions for final dSVs as follows: For male patients, we screen for the homozygous SVs that are absent in unaffected parent on autosome and X chromosome or absent in unaffected father on Y chromosome. For a female patient, we screen for the heterozygous/homozygous SVs, which are absent in unaffected parents on autosome and X chromosomes. The potential dSV detected by SV2 was further filtered by two procedures. First, we inspected the IGV of each dSV in the corresponding trios with at least two experts and removed those without corresponding dosage change. Second, we browsed GnomAD database and removed all dSV that had ever appeared (i.e., more than 50% length of dSV was covered by a SV of same type from GnomAD) in the nondisease Han Chinese population. Last, we validated the existence of candidate dSV by qPCR.
Estimation of relative mutation rates and definition of subtypes of coding mutations
To calculate the genome- and exome-wide mutation rate, we first downloaded the ranges of all exonic and UTR regions of hg19 from the UCSC Genome Browser and calculated the total number of base pairs in coding regions with more than 20× coverage across the whole genome. Then, we estimated the average mutation rates in a 95% confidence interval for all 53 patients with OCD using the “t.test” function in R. To determine the differences in DNM distribution between patients with OCD and healthy individuals, we included DNMs from SSC siblings (15) used as controls. Since the detected number of DNM slightly differed between studies, we applied Fisher exact test to evaluate the ORs, which measure the relative rate of DNMs in healthy controls compared to those in patients with OCD. We then partitioned all coding mutations according to Annovar (30) annotations: We defined frameshift indel or start site/stop site/splice site mutations as LoF mutations and nonsynonymous SNVs as missense mutations.
Comparison of mutation severity, gene intolerance, and mutation distribution
We compared CADD-phred scores between patients with OCD and SSC siblings from An et al. (15) for both exonic and genomic mutations. For the evaluation of gene intolerance, we collected pLI (31) scores for genes affected by different kinds of coding DNMs (LoF, missense, damaging, and synonymous) in patients with OCD. P values were calculated by a two-sided Wilcoxon rank-sum test. Since CADD-phred corresponds to the percentile rank of mutation severity and does not follow a normal distribution, we applied the cumulative distribution curve to visualize the distribution of CADD-phred score and Wilcoxon rank-sum test to compare the predicted severity. To evaluate the influence of GC bias in different genome regions, we compared the GC content and gene length between DNMs in our OCD data and SSC controls. Data for GC content and gene length were collected from GenBank. P values were calculated by a two-sided Wilcoxon rank-sum test.
We compared the results for the OCD patients with those of other neuropsychiatric diseases (developmental delay, ID, SCZ, and ASD) obtained from the PsyMuKB database (46). Since detailed information for single individuals in some of these studies was unavailable, we applied a straightforward Fisher’s test on mutation counts adjusted by synonymous mutation.
To investigate the functional genomic distribution of mutations, we first obtained four definitions of functional regions of the human genome from ENSEMBL (25) and PsychENCODE (24): (i) Full-length gene: We obtained ranges of all coding genes from biomaRt with entries “start_position” and “end_position.” (ii) Promoter: For all genes obtained in (i), we found their longest transcripts and the corresponding transcription start sites (TSSs). We defined the promoter region as 2-kb upstream and 1-kb downstream of TSSs. (iii) Promoter Interaction Region: We downloaded from PsychENCODE resource page the promoter anchored loop file, which was obtained from HI-C data of the human prefrontal cortex. (iv) Enhancer: From PsychENCODE resource page, we downloaded the enhancer-gene interaction file generated using HI-C data of the prefrontal cortex and extracted all enhancer regions. We also downloaded the core 15 chromatin state annotations from the Roadmap project (26), which was calculated by chromHMM (75).
Permutation test
To verify that the mutation distribution enrichment was not driven by random effects, we applied permutation tests to the positive results (promoter loop and chromatin state 8). First, we randomly chose 53 controls and repeated the Fisher’s test. This procedure was repeated 10,000 times, and the distribution of OR was used to generate a P value for OR = 1. Last, we removed all mutations on one chromosome and repeated Fisher’s test on the remaining mutations. This procedure was repeated for each of the 22 autosomal chromosomes.
WGCNA and analysis of coexpression patterns of SETD5, KDM3B, FBL, and ASXL3
BrainSpan RNA sequencing data (33) were applied to the following coexpression network analysis. We split the expression profiles into two different period sets by prenatal and postnatal samples, including all brain regions. For each subset, only genes with Reads Per Kilobase per Million mapped reads (RPKM) of >0 in at least half of the samples and coefficient of variance of >0.3 were retained for WGCNA (34) analysis. The remaining data were log10-transformed. The soft threshold was set at 12 for the prenatal subset and 16 for a postnatal subset. The minimum module size was set at 20. We constructed the signed networks by blockwiseModules function in the WGCNA R package based on Pearson R and partitioned genes into modules with a tree cut height of 0.3. After the detection of coexpression modules in both subsets, we tested whether genes carrying OCD coding mutations were enriched in some of the modules by the Fisher’s exact test. The enriched modules were then used for the Gene Ontology enrichment and network analysis.
To explore the potential dysregulation between four key genes (SETD5, KDM3B, FBL, and ASXL3) and three neurotransmitter systems (glutamate, serotonin, and dopamine), we first calculated the Pearson’s R between the key genes and all other genes in the prefrontal cortex expression data from Jaffe et al. (35). We used the two-sided Wilcoxon rank-sum test to check whether the overall coexpression between the chromatin modifiers and neurotransmitters was significant between patients with OCD and controls. To see whether any single neurotransmitter gene was significantly dysregulated, we compared the |ΔCo-exp| of each gene to the whole distribution of |ΔCo-exp| of corresponding key genes and generated a nominal P value.
Plasmids
The SETD5-FLAG and ASXL3-FLAG expression plasmids were purchased from Youze Biotechnology Co. Ltd. (Hunan, China), and the SETD5 R77C and ASXL3 F1460Lfs*5 mutated plasmids were introduced using the Q5 Site-Directed Mutagenesis Kit (New England BioLabs). All plasmids were confirmed via Sanger sequencing.
Western blot analysis and antibody
HEK293T cells were lysed in 1× lysis buffer on ice for 30 min and centrifuged at 12,000 rpm for 15 min. Cell lysates were separated by electrophoresis on 4 to 20% SDS–polyacrylamide gel electrophoresis gels and then transferred onto nitrocellulose membranes. The membranes were blocked with 5% milk in TBST (Tris-buffered saline, 0.1% Tween® 20) for 1 hour and incubated overnight at 4°C with the corresponding primary antibodies: rabbit anti-FLAG and anti–β-tubulin (Sigma-Aldrich), rabbit anti-H3K9me1 (Abcam), and rabbit anti-H2AK119ub (Cell Signaling Technology). Next, the experiment was followed by incubation with the horseradish peroxidase–labeled Goat Anti-Rabbit immunoglobulin G (Beyotime) for 1 hour at room temperature 20° to 25°C. Quantitation of immunoblots was then performed via densitometric analysis using the ImageJ software.
Comparison of OCD and TD DNM genes
We downloaded publicly available DNM data for OCD and TD from the psyMuKB database (46). A total of two OCD studies (10, 11) and three TD studies (18–20) were included in the present dataset. All genes with at least one missense or LoF mutation were recorded. We combined our nonsynonymous mutation genes with published OCD DNM genes and compared them to all previous TD DNM genes.
To analyze the overlap of TD and OCD genes, we performed hypergeometric tests with a background gene list as all protein-coding genes. P values were calculated for two-tailed tests. To analyze the functional characteristics of TD and OCD genes, we first obtained a list of 15 gene sets, mainly about the central nervous system, from psyMuKB (46). We tested enrichment TD/OCD genes in all these gene sets by Fisher’s exact test. Then, we performed a Pearson correlation analysis on −log10 (OR) of genes from two diseases. To analyze the spatiotemporal- and cell type–specific expression patterns of TD and OCD genes, we applied EWCE R package (37), a bootstrap enrichment tool based on gene cell-type specificity matrix, to conduct enrichment analysis on two datasets: (i) Brainspan (33) for period and region analysis and (ii) Dronc data (38) for adult brain cell–type analysis. Specificity was defined in the corresponding paper (37). We first calculated the specificity matrix of two expression datasets by “generate.celltype.data” function. Next, the enrichment of all TD/OCD genes was tested by “bootstrap.enrichment.test” function. We defined the background gene list as all genes annotated in the tested expression dataset.
Acknowledgments
Funding: This work was supported by grants from National Natural Science Foundation of China (grant nos. 81971292 and 82071518), Natural Science Foundation of Shanghai (no. 21ZR1428600), Program for Professor of Special Appointment (Eastern Scholar) at Shanghai Institutions of Higher Learning (no. 1610000043), the Innovation Research Plan by Shanghai Municipal Education Commission (no. ZXWF082101), Shanghai Municipal Health Commission (no. 2019ZB0201), and Shanghai Key Laboratory of Psychotic Disorders grant (no. 13dz2260500).
Author contributions: G.N.L. and Z.W. conceived and coordinated the project. G.N.L. coordinated the whole-genome sequencing experiments. P.W. and Z.W. coordinated, recruited, managed, diagnosed, and examined the recruited participants. G.N.L., W.W., W.C., X.J. W.H., H.Y., W.Q., Y.C., and M.C. processed the whole-genome sequencing data, variant calling, and validation. G.N.L and W.S. designed and performed experiments for variant characterization and different components of analysis. S.Y., T.X., Y.J., Q.L., C.Z., Z.Y., Q.F., and J.C. helped in participant recruitments and diagnoses and sample collections. G.N.L. and W.S. wrote the manuscript. All authors read and approved the manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Supplementary Materials and Methods
Figs. S1 to S3
Table S1
Legends for tables S2 to S5
Other Supplementary Material for this manuscript includes the following:
Tables S2 to S5
REFERENCES AND NOTES
- 1.Ruscio A. M., Stein D. J., Chiu W. T., Kessler R. C., The epidemiology of obsessive-compulsive disorder in the National Comorbidity Survey Replication. Mol. Psychiatry 15, 53–63 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fontenelle L. F., Mendlowicz M. V., Versiani M., The descriptive epidemiology of obsessive-compulsive disorder. Prog. Neuro Psychopharmacol. Biol. Psychiatry 30, 327–337 (2006). [DOI] [PubMed] [Google Scholar]
- 3.Huang Y., Wang Y., Wang H., Liu Z., Yu X., Yan J., Yu Y., Kou C., Xu X., Lu J., Wang Z., He S., Xu Y., He Y., Li T., Guo W., Tian H., Xu G., Xu X., Ma Y., Wang L., Wang L., Yan Y., Wang B., Xiao S., Zhou L., Li L., Tan L., Zhang T., Ma C., Li Q., Ding H., Geng H., Jia F., Shi J., Wang S., Zhang N., Du X., Du X., Wu Y., Prevalence of mental disorders in China: A cross-sectional epidemiological study. Lancet Psychiatry 6, 211–224 (2019). [DOI] [PubMed] [Google Scholar]
- 4.D. L. Pauls, A. Abramovitch, S. L. Rauch, D. A. Geller, Obsessive-compulsive Disorder: An Integrative Genetic and Neurobiological Perspective (Nature Publishing Group, 2014), vol. 15. [DOI] [PubMed] [Google Scholar]
- 5.Kinoshita M., Numata S., Tajima A., Shimodera S., Imoto I., Ohmori T., Plasma total homocysteine is associated with DNA methylation in patients with schizophrenia. Epigenetics 8, 584–590 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shugart Y. Y., Samuels J., Willour V. L., Grados M. A., Greenberg B. D., Knowles J. A., McCracken J. T., Rauch S. L., Murphy D. L., Wang Y., Pinto A., Fyer A. J., Piacentini J., Pauls D. L., Cullen B., Page J., Rasmussen S. A., Bienvenu O. J., Hoehn-Saric R., Valle D., Liang K. Y., Riddle M. A., Nestadt G., Genomewide linkage scan for obsessive-compulsive disorder: Evidence for susceptibility loci on chromosomes 3q, 7p, 1q, 15q, and 6q. Mol. Psychiatry 11, 763–770 (2006). [DOI] [PubMed] [Google Scholar]
- 7.Arnold P. D., Askland K. D., Barlassina C., Bellodi L., Bienvenu O. J., Black D., Bloch M., Brentani H., Burton C. L., Camarena B., Cappi C., Cath D., Cavallini M., Conti D., Cook E., Coric V., Cullen B. A., Cusi D., Davis L. K., Delorme R., Denys D., Derks E., Eapen V., Edlund C., Erdman L., Falkai P., Figee M., Fyer A. J., Geller D. A., Goes F. S., Grabe H., Grados M. A., Greenberg B. D., Grünblatt E., Guo W., Hanna G. L., Hemmings S., Hounie A. G., Jenicke M., Keenan C., Kennedy J., Khramtsova E. A., Konkashbaev A., Knowles J. A., Krasnow J., Lange C., Lanzagorta N., Leboyer M., Lennertz L., Li B., Liang K.-Y. Y., Lochner C., Macciardi F., Maher B., Maier W., Marconi M., Mathews C. A., Matthesien M., McCracken J. T., McLaughlin N. C., Miguel E. C., Moessner R., Murphy D. L., Neale B., Nestadt G., Nestadt P., Nicolini H., Nurmi E., Osiecki L., Pauls D. L., Piacentini J., Posthuma D., Pulver A. E., Qin H.-D. D., Rasmussen S. A., Rauch S., Richter M. A., Riddle M. A., Ripke S., Ruhrmann S., Sampaio A. S., Samuels J. F., Scharf J. M., Shugart Y. Y., Smit J., Stein D., Stewart S. E., Turiel M., Vallada H., Veenstra-VanderWeele J., Wagner M., Walitza S., Wang Y., Wendland J., Vulink N., Yu D., Zai G.; P. D. International Obsessive Compulsive Disorder Foundation Genetics Collaborative (IOCDF-GC); OCD Collaborative Genetics Association Studies (OCGAS), Askland K. D., Barlassina C., Bellodi L., Bienvenu O. J., Black D., Bloch M., Brentani H., Burton C. L., Camarena B., Cappi C., Cath D., Cavallini M., Conti D., Cook E., Coric V., Cullen B. A., Cusi D., Davis L. K., Delorme R., Denys D., Derks E., Eapen V., Edlund C., Erdman L., Falkai P., Figee M., Fyer A. J., Geller D. A., Goes F. S., Grabe H., Grados M. A., Greenberg B. D., Grünblatt E., Guo W., Hanna G. L., Hemmings S., Hounie A. G., Jenicke M., Keenan C., Kennedy J., Khramtsova E. A., Konkashbaev A., Knowles J. A., Krasnow J., Lange C., Lanzagorta N., Leboyer M., Lennertz L., Li B., Liang K.-Y. Y., Lochner C., Macciardi F., Maher B., Maier W., Marconi M., Mathews C. A., Matthesien M., McCracken J. T., McLaughlin N. C., Miguel E. C., Moessner R., Murphy D. L., Neale B., Nestadt G., Nestadt P., Nicolini H., Nurmi E., Osiecki L., Pauls D. L., Piacentini J., Posthuma D., Pulver A. E., Qin H.-D. D., Rasmussen S. A., Rauch S., Richter M. A., Riddle M. A., Ripke S., Ruhrmann S., Sampaio A. S., Samuels J. F., Scharf J. M., Shugart Y. Y., Smit J., Stein D., Stewart S. E., Turiel M., Vallada H., Veenstra-VanderWeele J., Wagner M., Walitza S., Wang Y., Wendland J., Vulink N., Yu D., Zai G., Revealing the complex genetic architecture of obsessive-compulsive disorder using meta-analysis. Mol. Psychiatry 23, 1181–1188 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mattheisen M., Samuels J. F., Wang Y., Greenberg B. D., Fyer A. J., McCracken J. T., Geller D. A., Murphy D. L., Knowles J. A., Grados M. A., Riddle M. A., Rasmussen S. A., McLaughlin N. C., Nurmi E. L., Askland K. D., Qin H.-D., Cullen B. A., Piacentini J., Pauls D. L., Bienvenu O. J., Stewart S. E., Liang K.-Y., Goes F. S., Maher B., Pulver A. E., Shugart Y. Y., Valle D., Lange C., Nestadt G., Genome-wide association study in obsessive-compulsive disorder: Results from the OCGAS. Mol. Psychiatry 20, 337–344 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee S. H., Wray N. R., Goddard M. E., Visscher P. M., Estimating missing heritability for disease from genome-wide association studies. Am. J. Hum. Genet. 88, 294–305 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cappi C., Brentani H., Lima L., Sanders S. J., Zai G., Diniz B. J., Reis V. N. S., Hounie A. G., do Rosário M. C., Mariani D., Requena G. L., Puga R., Souza-Duran F. L., Shavitt R. G., Pauls D. L., Miguel E. C., Fernandez T. V., Whole-exome sequencing in obsessive-compulsive disorder identifies rare mutations in immunological and neurodevelopmental pathways. Transl. Psychiatry 6, e764 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cappi C., Oliphant M. E., Péter Z., Zai G., do Rosário M. C., Sullivan C. A. W., Gupta A. R., Hoffman E. J., Virdee M., Olfson E., Abdallah S. B., Willsey A. J., Shavitt R. G., Miguel E. C., Kennedy J. L., Richter M. A., Fernandez T. V., De novo damaging DNA coding mutations are associated with obsessive-compulsive disorder and overlap with Tourette’s disorder and autism. Biol. Psychiatry 87, 1035–1044 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang W., Corominas R., Lin G. N., De novo mutations from whole exome sequencing in neurodevelopmental and psychiatric disorders: From discovery to application. Front. Genet. 10, 258 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Noh H. J., Tang R., Flannick J., O’dushlaine C., Swofford R., Howrigan D., Genereux D. P., Johnson J., Van Grootheest G., Grünblatt E., Andersson E., Djurfeldt D. R., Patel P. D., Koltookian M., Hultman C. M., Pato M. T., Pato C. N., Rasmussen S. A., Jenike M. A., Hanna G. L., Stewart S. E., Knowles J. A., Ruhrmann S., Grabe H. J., Wagner M., Rück C., Mathews C. A., Walitza S., Cath D. C., Feng G., Karlsson E. K., Lindblad-Toh K., Integrating evolutionary and regulatory information with a multispecies approach implicates genes and pathways in obsessive-compulsive disorder. Nat. Commun. 8, 774 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Werling D. M., Brand H., An J.-Y., Stone M. R., Zhu L., Glessner J. T., Collins R. L., Dong S., Layer R. M., Markenscoff-Papadimitriou E., Farrell A., Schwartz G. B., Wang H. Z., Currall B. B., Zhao X., Dea J., Duhn C., Erdman C. A., Gilson M. C., Yadav R., Handsaker R. E., Kashin S., Klei L., Mandell J. D., Nowakowski T. J., Liu Y., Pochareddy S., Smith L., Walker M. F., Waterman M. J., He X., Kriegstein A. R., Rubenstein J. L., Sestan N., McCarroll S. A., Neale B. M., Coon H., Willsey A. J., Buxbaum J. D., Daly M. J., State M. W., Quinlan A. R., Marth G. T., Roeder K., Devlin B., Talkowski M. E., Sanders S. J., An analytical framework for whole-genome sequence association studies and its implications for autism spectrum disorder. Nat. Genet. 50, 727–736 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.An J. Y., Lin K., Zhu L., Werling D. M., Dong S., Brand H., Wang H. Z., Zhao X., Schwartz G. B., Collins R. L., Currall B. B., Dastmalchi C., Dea J., Duhn C., Gilson M. C., Klei L., Liang L., Markenscoff-Papadimitriou E., Pochareddy S., Ahituv N., Buxbaum J. D., Coon H., Daly M. J., Kim Y. S., Marth G. T., Neale B. M., Quinlan A. R., Rubenstein J. L., Sestan N., State M. W., Willsey A. J., Talkowski M. E., Devlin B., Roeder K., Sanders S. J., Genome-wide de novo risk score implicates promoter variation in autism spectrum disorder. Science 362, eaat6576 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yuen R. K. C., Merico D., Bookman M., Howe J. L., Thiruvahindrapuram B., Patel R. V., Whitney J., Deflaux N., Bingham J., Wang Z., Pellecchia G., Buchanan J. A., Walker S., Marshall C. R., Uddin M., Zarrei M., Deneault E., D’Abate L., Chan A. J. S., Koyanagi S., Paton T., Pereira S. L., Hoang N., Engchuan W., Higginbotham E. J., Ho K., Lamoureux S., Li W., MacDonald J. R., Nalpathamkalam T., Sung W. W. L., Tsoi F. J., Wei J., Xu L., Tasse A.-M., Kirby E., Van Etten W., Twigger S., Roberts W., Drmic I., Jilderda S., Modi B. M., Kellam B., Szego M., Cytrynbaum C., Weksberg R., Zwaigenbaum L., Woodbury-Smith M., Brian J., Senman L., Iaboni A., Doyle-Thomas K., Thompson A., Chrysler C., Leef J., Savion-Lemieux T., Smith I. M., Liu X., Nicolson R., Seifer V., Fedele A., Cook E. H., Dager S., Estes A., Gallagher L., Malow B. A., Parr J. R., Spence S. J., Vorstman J., Frey B. J., Robinson J. T., Strug L. J., Fernandez B. A., Elsabbagh M., Carter M. T., Hallmayer J., Knoppers B. M., Anagnostou E., Szatmari P., Ring R. H., Glazer D., Pletcher M. T., Scherer S. W., Whole genome sequencing resource identifies 18 new candidate genes for autism spectrum disorder. Nat. Neurosci. 20, 602–611 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gilissen C., Hehir-Kwa J. Y., Thung D. T., Van De Vorst M., Van Bon B. W. M., Willemsen M. H., Kwint M., Janssen I. M., Hoischen A., Schenck A., Leach R., Klein R., Tearle R., Bo T., Pfundt R., Yntema H. G., De Vries B. B. A., Kleefstra T., Brunner H. G., Vissers L. E. L. M., Veltman J. A., Genome sequencing identifies major causes of severe intellectual disability. Nature 511, 344–347 (2014). [DOI] [PubMed] [Google Scholar]
- 18.Wang S., Mandell J. D., Kumar Y., Sun N., Morris M. T., Arbelaez J., Nasello C., Dong S., Duhn C., Zhao X., Yang Z., Padmanabhuni S. S., Yu D., King R. A., Dietrich A., Khalifa N., Dahl N., Huang A. Y., Neale B. M., Coppola G., Mathews C. A., Scharf J. M., Fernandez T. V., Buxbaum J. D., De Rubeis S., Grice D. E., Xing J., Heiman G. A., Tischfield J. A., Paschou P., Willsey A. J., State M. W., De novo sequence and copy number variants are strongly associated with tourette disorder and implicate cell polarity in pathogenesis. Cell Rep. 24, 3441–3454.e12 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Willsey A. J., Fernandez T. V., Yu D., King R. A., Dietrich A., Xing J., Sanders S. J., Mandell J. D., Huang A. Y., Richer P., Smith L., Dong S., Samocha K. E.; B. M. Tourette International Collaborative Genetics (TIC Genetics); G. Tourette Syndrome Association International Consortium for Genetics (TSAICG), Neale B. M., Coppola G., Mathews C. A., Tischfield J. A., Scharf J. M., State M. W., Heiman G. A., Bromberg Y., Brown L. W., Cheon K.-A., Coffey B. J., Deng L., Dietrich A., Dong S., Elzerman L., Fernandez T. V., Fründt O., Garcia-Delgar B., Gedvilaite E., Gilbert D. L., Grice D. E., Hagstrøm J., Hedderly T., Heiman G. A., Heyman I., Hoekstra P. J., Hong H. J., Huyser C., Ibanez-Gomez L., Kim Y. K., Kim Y.-S., King R. A., Koh Y.-J., Kook S., Kuperman S., Lamerz A., Leventhal B., Ludolph A. G., da Silva C. L., Madruga-Garrido M., Mandell J. D., Maras A., Mir P., Morer A., Münchau A., Murphy T. L., Nasello C., Openneer T. J. C., Plessen K. J., Richer P., Roessner V., Sanders S., Shin E.-Y., Sival D. A., Smith L., Song D.-H., Song J., State M. W., Stolte A. M., Sun N., Tischfield J. A., Tübing J., Visscher F., Walker M. F., Wanderer S., Wang S., Willsey A. J., Woods M., Xing J., Zhang Y., Zhou A., Zinner S. H., Barr C. L., Batterson J. R., Berlin C., Bruun R. D., Budman C. L., Cath D. C., Chouinard S., Coppola G., Cox N. J., Darrow S., Davis L. K., Dion Y., Freimer N. B., Grados M. A., Hirschtritt M. E., Huang A. Y., Illmann C., King R. A., Kurlan R., Leckman J. F., Lyon G. J., Malaty I. A., Mathews C. A., MaMahon W. M., Neale B. M., Okun M. S., Osiecki L., Pauls D. L., Posthuma D., Ramensky V., Robertson M. M., Rouleau G. A., Sandor P., Scharf J. M., Singer H. S., Smit J., Sul J.-H., Yu D., De novo coding variants are strongly associated with tourette disorder. Neuron 94, 486–499.e9 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eriguchi Y., Kuwabara H., Inai A., Kawakubo Y., Nishimura F., Kakiuchi C., Tochigi M., Ohashi J., Aoki N., Kato K., Ishiura H., Mitsui J., Tsuji S., Doi K., Yoshimura J., Morishita S., Shimada T., Furukawa M., Umekage T., Sasaki T., Kasai K., Kano Y., Identification of candidate genes involved in the etiology of sporadic Tourette syndrome by exome sequencing. Am. J. Med. Genet. B Neuropsychiatr. Genet. 174, 712–723 (2017). [DOI] [PubMed] [Google Scholar]
- 21.Sim N.-L., Kumar P., Hu J., Henikoff S., Schneider G., Ng P. C., SIFT web server: Predicting effects of amino acid substitutions on proteins. Nucleic Acids Res. 40, W452–W457 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Adzhubei I. A., Schmidt S., Peshkin L., Ramensky V. E., Gerasimova A., Bork P., Kondrashov A. S., Sunyaev S. R., A method and server for predicting damaging missense mutations. Nat. Methods 7, 248–249 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kircher M., Witten D. M., Jain P., O’Roak B. J., Cooper G. M., Shendure J., A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 46, 310–315 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang D., Liu S., Warrell J., Won H., Shi X., Navarro F. C. P., Clarke D., Gu M., Emani P., Yang Y. T., Xu M., Gandal M. J., Lou S., Zhang J., Park J. J., Yan C., Rhie S. K., Manakongtreecheep K., Zhou H., Nathan A., Peters M., Mattei E., Fitzgerald D., Brunetti T., Moore J., Jiang Y., Girdhar K., Hoffman G. E., Kalayci S., Gümüş Z. H., Crawford G. E., Consortium P. P. E. N. C. O. D. E., Roussos P., Akbarian S., Jaffe A. E., White K. P., Weng Z., Sestan N., Geschwind D. H., Knowles J. A., Gerstein M. B., Comprehensive functional genomic resource and integrative model for the human brain. Science 362, eaat8464 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zerbino D. R., Achuthan P., Akanni W., Amode M. R., Barrell D., Bhai J., Billis K., Cummins C., Gall A., Girón C. G., Gil L., Gordon L., Haggerty L., Haskell E., Hourlier T., Izuogu O. G., Janacek S. H., Juettemann T., To J. K., Laird M. R., Lavidas I., Liu Z., Loveland J. E., Maurel T., McLaren W., Moore B., Mudge J., Murphy D. N., Newman V., Nuhn M., Ogeh D., Ong C. K., Parker A., Patricio M., Riat H. S., Schuilenburg H., Sheppard D., Sparrow H., Taylor K., Thormann A., Vullo A., Walts B., Zadissa A., Frankish A., Hunt S. E., Kostadima M., Langridge N., Martin F. J., Muffato M., Perry E., Ruffier M., Staines D. M., Trevanion S. J., Aken B. L., Cunningham F., Yates A., Flicek P., Ensembl 2018. Nucleic Acids Res. 46, D754–D761 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kundaje A., Meuleman W., Ernst J., Bilenky M., Yen A., Heravi-Moussavi A., Kheradpour P., Zhang Z., Wang J., Ziller M. J., Amin V., Whitaker J. W., Schultz M. D., Ward L. D., Sarkar A., Quon G., Sandstrom R. S., Eaton M. L., Wu Y.-C., Pfenning A. R., Wang X., Claussnitzer M., Liu Y., Coarfa C., Harris R. A., Shoresh N., Epstein C. B., Gjoneska E., Leung D., Xie W., Hawkins R. D., Lister R., Hong C., Gascard P., Mungall A. J., Moore R., Chuah E., Tam A., Canfield T. K., Hansen R. S., Kaul R., Sabo P. J., Bansal M. S., Carles A., Dixon J. R., Farh K.-H., Feizi S., Karlic R., Kim A.-R., Kulkarni A., Li D., Lowdon R., Elliott G., Mercer T. R., Neph S. J., Onuchic V., Polak P., Rajagopal N., Ray P., Sallari R. C., Siebenthall K. T., Sinnott-Armstrong N. A., Stevens M., Thurman R. E., Wu J., Zhang B., Zhou X., Beaudet A. E., Boyer L. A., De Jager P. L., Farnham P. J., Fisher S. J., Haussler D., Jones S. J. M., Li W., Marra M. A., McManus M. T., Sunyaev S., Thomson J. A., Tlsty T. D., Tsai L.-H., Wang W., Waterland R. A., Zhang M. Q., Chadwick L. H., Bernstein B. E., Costello J. F., Ecker J. R., Hirst M., Meissner A., Milosavljevic A., Ren B., Stamatoyannopoulos J. A., Wang T., Kellis M., Kellis M., Integrative analysis of 111 reference human epigenomes. Nature 518, 317–330 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Halldorsson B. V., Palsson G., Stefansson O. A., Jonsson H., Hardarson M. T., Eggertsson H. P., Gunnarsson B., Oddsson A., Halldorsson G. H., Zink F., Gudjonsson S. A., Frigge M. L., Thorleifsson G., Sigurdsson A., Stacey S. N., Sulem P., Masson G., Helgason A., Gudbjartsson D. F., Thorsteinsdottir U., Stefansson K., Human genetics: Characterizing mutagenic effects of recombination through a sequence-level genetic map. Science 363, eaau1043 (2019). [DOI] [PubMed] [Google Scholar]
- 28.Jónsson H., Sulem P., Kehr B., Kristmundsdottir S., Zink F., Hjartarson E., Hardarson M. T., Hjorleifsson K. E., Eggertsson H. P., Gudjonsson S. A., Ward L. D., Arnadottir G. A., Helgason E. A., Helgason H., Gylfason A., Jonasdottir A., Jonasdottir A., Rafnar T., Frigge M., Stacey S. N., Magnusson O. T., Thorsteinsdottir U., Masson G., Kong A., Halldorsson B. V., Helgason A., Gudbjartsson D. F., Stefansson K., Parental influence on human germline de novo mutations in 1,548 trios from Iceland. Nature 549, 519–522 (2017). [DOI] [PubMed] [Google Scholar]
- 29.Iyer-Bierhoff A., Krogh N., Tessarz P., Ruppert T., Nielsen H., Grummt I., SIRT7-dependent deacetylation of fibrillarin controls histone H2A methylation and rRNA synthesis during the cell cycle. Cell Rep. 25, 2946–2954.e5 (2018). [DOI] [PubMed] [Google Scholar]
- 30.Yang H., Wang K., Genomic variant annotation and prioritization with ANNOVAR and wANNOVAR. Nat. Protoc. 10, 1556–1566 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lek M., Karczewski K. J., Minikel E. V., Samocha K. E., Banks E., Fennell T., O’Donnell-Luria A. H., Ware J. S., Hill A. J., Cummings B. B., Tukiainen T., Birnbaum D. P., Kosmicki J. A., Duncan L. E., Estrada K., Zhao F., Zou J., Pierce-Hoffman E., Berghout J., Cooper D. N., Deflaux N., DePristo M., Do R., Flannick J., Fromer M., Gauthier L., Goldstein J., Gupta N., Howrigan D., Kiezun A., Kurki M. I., Moonshine A. L., Natarajan P., Orozco L., Peloso G. M., Poplin R., Rivas M. A., Ruano-Rubio V., Rose S. A., Ruderfer D. M., Shakir K., Stenson P. D., Stevens C., Thomas B. P., Tiao G., Tusie-Luna M. T., Weisburd B., Won H.-H., Yu D., Altshuler D. M., Ardissino D., Boehnke M., Danesh J., Donnelly S., Elosua R., Florez J. C., Gabriel S. B., Getz G., Glatt S. J., Hultman C. M., Kathiresan S., Laakso M., McCarroll S., McCarthy M. I., McGovern D., McPherson R., Neale B. M., Palotie A., Purcell S. M., Saleheen D., Scharf J. M., Sklar P., Sullivan P. F., Tuomilehto J., Tsuang M. T., Watkins H. C., Wilson J. G., Daly M. J., MacArthur D. G.; Exome Aggregation Consortium , Analysis of protein-coding genetic variation in 60,706 humans. Nature 536, 285–291 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Norman L. J., Carlisi C., Lukito S., Hart H., Mataix-Cols D., Radua J., Rubia K., Structural and functional brain abnormalities in attention-deficit/hyperactivity disorder and obsessive-compulsive disorder: A comparative meta-analysis. JAMA Psychiat. 73, 815–825 (2016). [DOI] [PubMed] [Google Scholar]
- 33.Kang H. J., Kawasawa Y. I., Cheng F., Zhu Y., Xu X., Li M., Sousa A. M. M., Pletikos M., Meyer K. A., Sedmak G., Guennel T., Shin Y., Johnson M. B., Krsnik Ž., Mayer S., Fertuzinhos S., Umlauf S., Lisgo S. N., Vortmeyer A., Weinberger D. R., Mane S., Hyde T. M., Huttner A., Reimers M., Kleinman J. E., Šestan N., Spatio-temporal transcriptome of the human brain. Nature 478, 483–489 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Langfelder P., Horvath S., WGCNA: An R package for weighted correlation network analysis. BMC Bioinformatics 9, 559 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jaffe A. E., Deep-Soboslay A., Tao R., Hauptman D. T., Kaye W. H., Arango V., Weinberger D. R., Hyde T. M., Kleinman J. E., Genetic neuropathology of obsessive psychiatric syndromes. Transl. Psychiatry 4, e432 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hirschtritt M. E., Lee P. C., Pauls D. L., Dion Y., Grados M. A., Illmann C., King R. A., Sandor P., McMahon W. M., Lyon G. J., Cath D. C., Kurlan R., Robertson M. M., Osiecki L., Scharf J. M., Mathews C. A., Tourette Syndrome Association International Consortium for genetics, lifetime prevalence, age of risk, and genetic relationships of comorbid psychiatric disorders in tourette syndrome. JAMA Psychiat. 72, 325–333 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Skene N. G., Grant S. G. N., Identification of vulnerable cell types in major brain disorders using single cell transcriptomes and expression weighted cell type enrichment. Front. Neurosci. 10, 16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lake B. B., Chen S., Sos B. C., Fan J., Kaeser G. E., Yung Y. C., Duong T. E., Gao D., Chun J., Kharchenko P. V., Zhang K., Integrative single-cell analysis of transcriptional and epigenetic states in the human adult brain. Nat. Biotechnol. 36, 70–80 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Geisheker M. R., Heymann G., Wang T., Coe B. P., Turner T. N., Stessman H. A. F., Hoekzema K., Kvarnung M., Shaw M., Friend K., Liebelt J., Barnett C., Thompson E. M., Haan E., Guo H., Anderlid B. M., Nordgren A., Lindstrand A., Vandeweyer G., Alberti A., Avola E., Vinci M., Giusto S., Pramparo T., Pierce K., Nalabolu S., Michaelson J. J., Sedlacek Z., Santen G. W. E., Peeters H., Hakonarson H., Courchesne E., Romano C., Kooy R. F., Bernier R. A., Nordenskjöld M., Gecz J., Xia K., Zweifel L. S., Eichler E. E., Hotspots of missense mutation identify neurodevelopmental disorder genes and functional domains. Nat. Neurosci. 20, 1043–1051 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McRae J. F., Clayton S., Fitzgerald T. W., Kaplanis J., Prigmore E., Rajan D., Sifrim A., Aitken S., Akawi N., Alvi M., Ambridge K., Barrett D. M., Bayzetinova T., Jones P., Jones W. D., King D., Krishnappa N., Mason L. E., Singh T., Tivey A. R., Ahmed M., Anjum U., Archer H., Armstrong R., Awada J., Balasubramanian M., Banka S., Baralle D., Barnicoat A., Batstone P., Baty D., Bennett C., Berg J., Bernhard B., Bevan A. P., Bitner-Glindzicz M., Blair E., Blyth M., Bohanna D., Bourdon L., Bourn D., Bradley L., Brady A., Brent S., Brewer C., Brunstrom K., Bunyan D. J., Burn J., Canham N., Castle B., Chandler K., Chatzimichali E., Cilliers D., Clarke A., Clasper S., Clayton-Smith J., Clowes V., Coates A., Cole T., Colgiu I., Collins A., Collinson M. N., Connell F., Cooper N., Cox H., Cresswell L., Cross G., Crow Y., D’Alessandro M., Dabir T., Davidson R., Davies S., De Vries D., Dean J., Deshpande C., Devlin G., Dixit A., Dobbie A., Donaldson A., Donnai D., Donnelly D., Donnelly C., Douglas A., Douzgou S., Duncan A., Eason J., Ellard S., Ellis I., Elmslie F., Evans K., Everest S., Fendick T., Fisher R., Flinter F., Foulds N., Fry A., Fryer A., Gardiner C., Gaunt L., Ghali N., Gibbons R., Gill H., Goodship J., Goudie D., Gray E., Green A., Greene P., Greenhalgh L., Gribble S., Harrison R., Harrison L., Harrison V., Hawkins R., He L., Hellens S., Henderson A., Hewitt S., Hildyard L., Hobson E., Holden S., Holder M., Holder S., Hollingsworth G., Homfray T., Humphreys M., Hurst J., Hutton B., Ingram S., Irving M., Islam L., Jackson A., Jarvis J., Jenkins L., Johnson D., Jones E., Josifova D., Joss S., Kaemba B., Kazembe S., Kelsell R., Kerr B., Kingston H., Kini U., Kinning E., Kirby G., Kirk C., Kivuva E., Kraus A., Kumar D., Kumar V. K. A., Lachlan K., Lam W., Lampe A., Langman C., Lees M., Lim D., Longman C., Lowther G., Lynch S. A., Magee A., Maher E., Male A., Mansour S., Marks K., Martin K., Maye U., McCann E., McConnell V., McEntagart M., McGowan R., McKay K., McKee S., McMullan D. J., McNerlan S., McWilliam C., Mehta S., Metcalfe K., Middleton A., Miedzybrodzka Z., Miles E., Mohammed S., Montgomery T., Moore D., Morgan S., Morton J., Mugalaasi H., Murday V., Murphy H., Naik S., Nemeth A., Nevitt L., Newbury-Ecob R., Norman A., O’Shea R., Ogilvie C., Ong K. R., Park S. M., Parker M. J., Patel C., Paterson J., Payne S., Perrett D., Phipps J., Pilz D. T., Pollard M., Pottinger C., Poulton J., Pratt N., Prescott K., Price S., Pridham A., Procter A., Purnell H., Quarrell O., Ragge N., Rahbari R., Randall J., Rankin J., Raymond L., Rice D., Robert L., Roberts E., Roberts J., Roberts P., Roberts G., Ross A., Rosser E., Saggar A., Samant S., Sampson J., Sandford R., Sarkar A., Schweiger S., Scott R., Scurr I., Selby A., Seller A., Sequeira C., Shannon N., Sharif S., Shaw-Smith C., Shearing E., Shears D., Sheridan E., Simonic I., Singzon R., Skitt Z., Smith A., Smith K., Smithson S., Sneddon L., Splitt M., Squires M., Stewart F., Stewart H., Straub V., Suri M., Sutton V., Swaminathan G. J., Sweeney E., Tatton-Brown K., Taylor C., Taylor R., Tein M., Temple I. K., Thomson J., Tischkowitz M., Tomkins S., Torokwa A., Treacy B., Turner C., Turnpenny P., Tysoe C., Vandersteen A., Varghese V., Vasudevan P., Vijayarangakannan P., Vogt J., Wakeling E., Wallwark S., Waters J., Weber A., Wellesley D., Whiteford M., Widaa S., Wilcox S., Wilkinson E., Williams D., Williams N., Wilson L., Woods G., Wragg C., Wright M., Yates L., Yau M., Nellåker C., Firth H. V., Wright C. F., FitzPatrick D. R., Barrett J. C., Hurles M. E., Prevalence and architecture of de novo mutations in developmental disorders. Nature 542, 433–438 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pinheiro I., Margueron R., Shukeir N., Eisold M., Fritzsch C., Richter F. M., Mittler G., Genoud C., Goyama S., Kurokawa M., Son J., Reinberg D., Lachner M., Jenuwein T., Prdm3 and Prdm16 are H3K9me1 methyltransferases required for mammalian heterochromatin integrity. Cell 150, 948–960 (2012). [DOI] [PubMed] [Google Scholar]
- 42.Fellous A., Earley R. L., Silvestre F., The Kdm/Kmt gene families in the self-fertilizing mangrove rivulus fish, Kryptolebias marmoratus, suggest involvement of histone methylation machinery in development and reproduction. Gene 687, 173–187 (2019). [DOI] [PubMed] [Google Scholar]
- 43.Deliu E., Arecco N., Morandell J., Dotter C. P., Contreras X., Girardot C., Käsper E. L., Kozlova A., Kishi K., Chiaradia I., Noh K. M., Novarino G., Haploinsufficiency of the intellectual disability gene SETD5 disturbs developmental gene expression and cognition. Nat. Neurosci. 21, 1717–1727 (2018). [DOI] [PubMed] [Google Scholar]
- 44.Diets I. J., van der Donk R., Baltrunaite K., Waanders E., Reijnders M. R. F., Dingemans A. J. M., Pfundt R., Vulto-van Silfhout A. T., Wiel L., Gilissen C., Thevenon J., Perrin L., Afenjar A., Nava C., Keren B., Bartz S., Peri B., Beunders G., Verbeek N., van Gassen K., Thiffault I., Cadieux-Dion M., Huerta-Saenz L., Wagner M., Konstantopoulou V., Vodopiutz J., Griese M., Boel A., Callewaert B., Brunner H. G., Kleefstra T., Hoogerbrugge N., de Vries B. B. A., Hwa V., Dauber A., Hehir-Kwa J. Y., Kuiper R. P., Jongmans M. C. J., De novo and inherited pathogenic variants in KDM3B cause intellectual disability, short stature, and facial dysmorphism. Am. J. Hum. Genet. 104, 758–766 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Srivastava A., Ritesh K. C., Tsan Y. C., Liao R., Su F., Cao X., Hannibal M. C., Keegan C. E., Chinnaiyan A. M., Martin D. M., Bielas S. L., De novo dominant ASXL3 mutations alter H2A deubiquitination and transcription in Bainbridge-Ropers syndrome. Hum. Mol. Genet. 25, 597–608 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lin G. N., Guo S., Tan X., Wang W., Qian W., Song W., Wang J., Yu S., Wang Z., Cui D., Wang H., PsyMuKB: An integrative de novo variant knowledge base for developmental disorders. Genomics Proteomics Bioinformatics 17, 453–464 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chenoweth M. J., Ware J. J., Zhu A. Z. X., Cole C. B., Cox L. S., Nollen N., Ahluwalia J. S., Benowitz N. L., Schnoll R. A., Hawk L. W., Cinciripini P. M., George T. P., Lerman C., Knight J., Tyndale R. F., Genome-wide association study of a nicotine metabolism biomarker in African American smokers: Impact of chromosome 19 genetic influences. Addiction 113, 509–523 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McCarthy S. E., Gillis J., Kramer M., Lihm J., Yoon S., Berstein Y., Mistry M., Pavlidis P., Solomon R., Ghiban E., Antoniou E., Kelleher E., O’Brien C., Donohoe G., Gill M., Morris D. W., McCombie W. R., Corvin A., De novo mutations in schizophrenia implicate chromatin remodeling and support a genetic overlap with autism and intellectual disability. Mol. Psychiatry 19, 652–658 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cotney J., Muhle R. A., Sanders S. J., Liu L., Willsey A. J., Niu W., Liu W., Klei L., Lei J., Yin J., Reilly S. K., Tebbenkamp A. T., Bichsel C., Pletikos M., Sestan N., Roeder K., State M. W., Devlin B., Noonan J. P., The autism-associated chromatin modifier CHD8 regulates other autism risk genes during human neurodevelopment. Nat. Commun. 6, 6404 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fan S., Cath D. C., van den Heuvel O. A., van der Werf Y. D., Schöls C., Veltman D. J., Pouwels P. J. W., Abnormalities in metabolite concentrations in tourette’s disorder and obsessive-compulsive disorder—A proton magnetic resonance spectroscopy study. Psychoneuroendocrinology 77, 211–217 (2017). [DOI] [PubMed] [Google Scholar]
- 51.American Psychiatric Association, Diagnostic and statistical manual of mental disorders: DSM-IV (ed. 4, 1994).
- 52.Sheehan D. V, Lecrubier Y., Sheehan K. H., Amorim P., Janavs J., Weiller E., Hergueta T., Baker R., Dunbar G. C., The Mini-International Neuropsychiatric Interview (M.I.N.I.): The development and validation of a structured diagnostic psychiatric interview for DSM-IV and ICD-10. J. Clin. Psychiatry 59( suppl. 2), 22–33; quiz 34-57 (1998). [PubMed] [Google Scholar]
- 53.Robinson J. T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E. S., Getz G., Mesirov J. P., Integrative Genomics Viewer. Nat. Biotechnol. 29, 24–26 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kendig K. I., Baheti S., Bockol M. A., Drucker T. M., Hart S. N., Heldenbrand J. R., Hernaez M., Hudson M. E., Kalmbach M. T., Klee E. W., Mattson N. R., Ross C. A., Taschuk M., Wieben E. D., Wiepert M., Wildman D. E., Mainzer L. S., Sentieon DNASeq variant calling workflow demonstrates strong computational performance and accuracy. Front. Genet. 10, 736 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li H., Durbin R., Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26, 589–595 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R.; 1000 Genome Project Data Processing Subgroup , The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Broad Institute, Picard Toolkit (2019). GitHub Repository. http://broadinstitute.github.io/picard Broad Institute.
- 58.Wei Q., Zhan X., Zhong X., Liu Y., Han Y., Chen W., Li B., A Bayesian framework for de novo mutation calling in parents-offspring trios. Bioinformatics 31, 1375–1381 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.1000 Genomes Project Consortium , A global reference for human genetic variation. Nature 526, 68–74 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.NHLBI GO Exome Sequencing Project (ESP), Exome Variant Server; https://evs.gs.washington.edu/EVS/.
- 61.Karczewski K. J., Francioli L. C., Tiao G., Cummings B. B., Alföldi J., Wang Q., Collins R. L., Laricchia K. M., Ganna A., Birnbaum D. P., Gauthier L. D., Brand H., Solomonson M., Watts N. A., Rhodes D., Singer-Berk M., Seaby E. G., Kosmicki J. A., Walters R. K., Tashman K., Farjoun Y., Banks E., Poterba T., Wang A., Seed C., Whiffin N., Chong J. X., Samocha K. E., Pierce-Hoffman E., Zappala Z., O’Donnell-Luria A. H., Minikel E. V., Weisburd B., Lek M., Ware J. S., Vittal C., Armean I. M., Bergelson L., Cibulskis K., Connolly K. M., Covarrubias M., Donnelly S., Ferriera S., Gabriel S., Gentry J., Gupta N., Jeandet T., Kaplan D., Llanwarne C., Munshi R., Novod S., Petrillo N., Roazen D., Ruano-Rubio V., Saltzman A., Schleicher M., Soto J., Tibbetts K., Tolonen C., Wade G., Talkowski M. E.; Genome Aggregation Database (gnomAD) Consortium, Neale B. M., Daly M. J., MacArthur D. G., Variation across 141,456 human exomes and genomes reveals the spectrum of loss-of-function intolerance across human protein-coding genes. bioRxiv, 531210 (2019). [Google Scholar]
- 62.Kent W. J., BLAT—The BLAST-like alignment tool. Genome Res. 12, 656–664 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sugathan A., Biagioli M., Golzio C., Erdin S., Blumenthal I., Manavalan P., Ragavendran A., Brand H., Lucente D., Miles J., Sheridan S. D., Stortchevoi A., Kellis M., Haggarty S. J., Katsanis N., Gusella J. F., Talkowski M. E., CHD8 regulates neurodevelopmental pathways associated with autism spectrum disorder in neural progenitors. Proc. Natl. Acad. Sci. U.S.A. 111, E4468–E4477 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Darnell J. C., Van Driesche S. J., Zhang C., Hung K. Y. S., Mele A., Fraser C. E., Stone E. F., Chen C., Fak J. J., Chi S. W., Licatalosi D. D., Richter J. D., Darnell R. B., FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell 146, 247–261 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ascano M., Mukherjee N., Bandaru P., Miller J. B., Nusbaum J. D., Corcoran D. L., Langlois C., Munschauer M., Dewell S., Hafner M., Williams Z., Ohler U., Tuschl T., FMRP targets distinct mRNA sequence elements to regulate protein expression. Nature 492, 382–386 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Weyn-Vanhentenryck S. M., Mele A., Yan Q., Sun S., Farny N., Zhang Z., Xue C., Herre M., Silver P. A., Zhang M. Q., Krainer A. R., Darnell R. B., Zhang C., HITS-CLIP and integrative modeling define the Rbfox splicing-regulatory network linked to brain development and autism. Cell Rep. 6, 1139–1152 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bayés A., van de Lagemaat L. N., Collins M. O., Croning M. D. R., Whittle I. R., Choudhary J. S., Grant S. G. N., Characterization of the proteome, diseases and evolution of the human postsynaptic density. Nat. Neurosci. 14, 19–21 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen S., Fragoza R., Klei L., Liu Y., Wang J., Roeder K., Devlin B., Yu H., An interactome perturbation framework prioritizes damaging missense mutations for developmental disorders. Nat. Genet. 50, 1032–1040 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stessman H. A. F., Xiong B., Coe B. P., Wang T., Hoekzema K., Fenckova M., Kvarnung M., Gerdts J., Trinh S., Cosemans N., Vives L., Lin J., Turner T. N., Santen G., Ruivenkamp C., Kriek M., van Haeringen A., Aten E., Friend K., Liebelt J., Barnett C., Haan E., Shaw M., Gecz J., Anderlid B.-M., Nordgren A., Lindstrand A., Schwartz C., Kooy R. F., Vandeweyer G., Helsmoortel C., Romano C., Alberti A., Vinci M., Avola E., Giusto S., Courchesne E., Pramparo T., Pierce K., Nalabolu S., Amaral D. G., Scheffer I. E., Delatycki M. B., Lockhart P. J., Hormozdiari F., Harich B., Castells-Nobau A., Xia K., Peeters H., Nordenskjöld M., Schenck A., Bernier R. A., Eichler E. E., Targeted sequencing identifies 91 neurodevelopmental-disorder risk genes with autism and developmental-disability biases. Nat. Genet. 49, 515–526 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen X., Schulz-Trieglaff O., Shaw R., Barnes B., Schlesinger F., Källberg M., Cox A. J., Kruglyak S., Saunders C. T., Manta: Rapid detection of structural variants and indels for germline and cancer sequencing applications. Bioinformatics 32, 1220–1222 (2016). [DOI] [PubMed] [Google Scholar]
- 71.Klambauer G., Schwarzbauer K., Mayr A., Clevert D. A., Mitterecker A., Bodenhofer U., Hochreiter S., Cn.MOPS: Mixture of Poissons for discovering copy number variations in next-generation sequencing data with a low false discovery rate. Nucleic Acids Res. 40, e69 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rausch T., Zichner T., Schlattl A., Stütz A. M., Benes V., Korbel J. O., DELLY: Structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics 28, i333–i339 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Layer R. M., Chiang C., Quinlan A. R., Hall I. M., LUMPY: A probabilistic framework for structural variant discovery. Genome Biol. 15, R84 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Antaki D., Brandler W. M., Sebat J., SV2: Accurate structural variation genotyping and de novo mutation detection from whole genomes. Bioinformatics 34, 1774–1777 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ernst J., Kellis M., ChromHMM: Automating chromatin-state discovery and characterization. Nat. Methods 9, 215–216 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Materials and Methods
Figs. S1 to S3
Table S1
Legends for tables S2 to S5
Tables S2 to S5