Abstract
The SARS-CoV-2 infection rate, as well as mortality rate, is high. There is an urgent need for a high-throughput, accurate and reliable method of diagnosing COVID-19 pneumonia. We included references from databases, such as PubMed, Cochrane Library, Web of Science, and Embase, and extracted data. Then, MetaDisc and STATA were used to establish forest plots and funnel plots for meta-analysis. We collected 14 articles and performed a systematic review. The following results were obtained: sensitivity and specificity were 0.97 (0.96 to 0.98) and 0.97 (0.96 to 0.98) respectively; PLR and NLR were 24.51 (16.63–36.12) and 0.03 (0.01 to 0.10) respectively, DOR was 975.15 (430.11–2210.88), and AUC was 0.9926. When Xpress detects SARS-CoV-2 in different samples, the heterogeneity is small and the specificity and sensitivity are extremely high. We recommend the employment of Xpert Xpress analysis in rapid screening.
Keywords: Xpert Xpress, COVID-19 pneumonia, SARS-CoV-2, Diagnostic accuracy, Systemic review
1. Introduction
A new coronavirus, severe acute respiratory syndrome-related coronavirus-2 (SARS-CoV-2), was identified as the pathogen causing a new severe acute respiratory disease named coronavirus disease 2019 (COVID-19) (Anon, 2022). Coronaviruses are a group of enveloped viruses with non-segmented, single-stranded positive-sense RNA genomes (Huang et al., 2020; Lu and Liu, 2012). SARS-CoV-2 is the seventh coronavirus that is known to cause human diseases (Wang et al., 2020).
A classic feature of patients with SARS-CoV-2 is pneumonia (COVID-19 pneumonia). The main manifestations of COVID-19 pneumonia are fever, fatigue and dry cough. A few patients also present nasal congestion, nasal discharge, sore throat and diarrhea symptoms (Wiersinga et al., 2020). In the early stages of the infection, most patients show symptoms of acute respiratory infection, such as fever and cough. Severely affected patients develop dyspnea and/or hypoxemia one week after SARS-CoV-2 infection and may develop rapidly into acute respiratory distress syndrome, septic shock, refractory metabolic acidosis and coagulopathy, and multiple organ failure. Impaired function of the heart, brain, lung, liver, kidney and coagulation system is a complication of COVID-19 pneumonia (Zheng, 2020). Respiratory droplets, respiratory secretions, and direct contact are generally thought to be the most common routes of transmission of the coronaviruses. Later, it was reported that viral nucleotide could be isolated from feces and blood, which indicates that novel coronavirus may spread through other ways (Rabi et al., 2020). As of November 18, 2021, SARS-CoV-2 had a total of 254,847,065 confirmed cases worldwide, including 5,120,712 deaths (WHO, 2022). In view of the above, a rapid, accurate and reliable method for COVID-19 pneumonia diagnosis is needed urgently.
The GeneXpert® molecular diagnostic system is a fully-automated and integrated PCR-based nucleic acid detection system (Gotham et al., 2021a). It can integrate sample preparations and quantitative PCR processes into a closed kit and complete sample preparation, nucleic acid purification and concentration, quantitative PCR amplification and detection, data analysis, and output the results automatically (Loeffelholz et al., 2020). The Cepheid Xpert Xpress SARS-CoV-2 Test, for SARS-CoV-2 rapid detection, is a fully automated in vitro diagnostic assay that targets the envelope gene (E gene) and nucleocapsid gene (N2 gene) on the GeneXpert® platform (Wong et al., 2020a).
With the global outbreak of SARS-CoV-2, rapid diagnosis is essential for the current pandemic. The routine use of real-time reverse transcription polymerase chain reaction (RT-qPCR) can process large quantities of samples at the same time, good sensitivity, and specificity. However, it also has shortcomings such as a long turnover time. In the case of high infection rate and high mortality rate of SARS-CoV-2, rapid access to test results can effectively control transmission and assist in isolation of patients and contacts (Loeffelholz et al., 2020). In this larger context, molecular point-of-care test can be an option to providing diagnostic tests since this quick testing technology for SARS-CoV-2 plays a vital role in early diagnosis and timely control of infection (Wong et al., 2020a). The Cepheid Xpert Xpress SARS-CoV-2 Test, compared with the gold standard RT-qPCR, is convenient, fast and less demanding in laboratory technology (Gotham et al., 2021b).
We conducted a systematic review and meta-analysis on the performance of commercial Cepheid Xpert Xpress SARS-CoV-2 test for the identification of SARS-CoV-2. We applied various samples to the Xpert Xpress diagnostic technique and evaluated the detection efficacy and clinical auxiliary advantages of the Xpert Xpress in many aspects.
2. Materials and methods
2.1. Research identification and selection
The reviewer (Xun-Jie Cao) searched four online electronic databases until September 21, 2020. Databases that have been searched consist of Embase, PubMed, Cochrane Library and Web of Science. We included articles that met these requirements: (1) The data were provided as two-by-two tables (True positives (TP), true negatives (TN), false positives (FP) and false negatives (FN)), (2) full-text publications, and (3) The reference standard was viral culture or RT-PCR. The exclusion criteria consisted of (1) Meta-analysis, comments, letters and (2) Animal research.
2.2. Quality assessment and data extraction
For each article that met the requirements, four investigators (Xun-Jie Cao, Ya-Ping Li, Jie Zhou, and Ke-Ying Fang) grouped in pairs, and then we independently extracted the following information: year of publication, the first author, country, sample type, reference standard, gender, sample size and data for two-by two tables. The results of data extraction were compared between the two groups. If the results are different, they will be solved through discussion.
Because QUADAS-2 is a relatively subjective standard, and different people’s evaluations may cause differences. We use QUADAS-2 as the standard to discuss and resolve differences in four researchers’ understanding of the standard. Four researchers were divided into two groups to reviewed the quality of eligible articles respectively in terms of the inclusion and exclusion of cases and samples, the selection of diagnostic gold standards, and the setting of thresholds based on the quality assessment of the Diagnostic Accuracy Research Tool 2 (QUADAS-2) recommended by the Cochrane Collaborative Organization (Whiting et al., 2011). During the evaluation process, differences were resolved through negotiation. The quality assessment results are drawn using Revman (version 5.3) software.
2.3. Measurement of diagnostic parameters
For the specific evaluation of the diagnostic accuracy of Xpert Xpress to SARS-Cov-2, we established two by two tables. True positives (TP), true negatives (TN), false positives (FP) and false negatives (FN) were directly extracted from the original research or obtained through calculation. We calculate TP, FP, FN, TN by sensitivity, specificity, the number of positives, the number of negatives, positive predict value (PPV) and negative predict value (NPV). For example, sensitivity is the true positive rate, sensitivity = a/(a + c), TP = a = sensitivity×(a + c). Specificity is the true negative rate, specificity = d/(b + d), TN = specificity×(b + d). Then, the forest plot was set up to evaluate the specificity and sensitivity of each included study, with a confidence interval of 95 % (95 % CIs). The Summary Receiver Operating Characteristic (SROC) curve was established to summarize the combined distribution of sensitivity and specificity. The area under the SROC curve (AUC) was used to evaluate the accuracy of the overall test. In addition, the combined SPE and SEN were used to calculate the negative likelihood ratio (NLR) and positive likelihood ratio (PLR). Fagan plot was set up to show the relationship between pre-probability, likelihood ratio, and post-probability. A funnel plot was constructed to visually verify any potential publication bias. Furthermore, a bivariate boxplot was established to perform heterogeneity testing. These analyses were performed by using Stata statistical software package, version 12.0 (Stata Corp LP, College Station, U.S.A.), Review Manager 5.3, and Meta-DiSc 1.4. Fagan plot, bivariate boxplot and funnel plot was constructed by Stata statistical software. The forest plots were constructed by Meta-DiSc 1.4.
3. Results
3.1. Search result
A total of 140 papers were retrieved, 47 duplicate articles were eliminated. According to the inclusion/exclusion criteria, 67 unrelated articles were excluded by screening the abstracts. After full-text reading, full-text articles excluded, with reasons including 1 meta-analysis, 1 comment, 3 letters, 2 notes and 5 reviews. A total of 11 articles were included. The exclusion reasons are shown in Fig. S1. Finally, we included 14 qualified articles which included a total of 1999 patients and performed the meta-analysis.
3.2. Characteristics of eligible studies
A total of 14 studies and 1647 samples were included. The characteristics of eligible studies are presented in Table 1 . The sample types contain nasopharyngeal swabs, saliva specimens, sputum specimens, tracheal aspirate and bronchoalveolar lavage specimens, remnant stool specimens, and mixed specimens.
Table 1.
Characteristics of 11 Studies Included in the Meta-analysis.
| Author | Year | Study Design | Sample type | Country | Reference Test | Gender | Sample size |
|---|---|---|---|---|---|---|---|
| (Lieberman et al., 2020) | 2020 | Retrospective | Nasopharyngeal (NP) swabs | America | CDC-based LDT | all genders | 26 |
| (Zhen et al., 2020) | 2020 | Retrospective and Prospective | nasopharyngeal swabs | China | the Hologic Panther Fusion® SARS-CoV-2 assay | all genders | 108 |
| (Wong et al., 2020b) | 2020 | Retrospective and Prospective | posterior oropharyngeal saliva(DTS specimens) | China | the TIB-Molbiol LightMix® SarbecoV E-gene assay | all genders | 120 |
| (Wong et al., 2020b) | 2020 | Retrospective and Prospective | sputum, tracheal aspirate and bronchoalveolar lavagespecimens(LRT specimens) | China | the TIB-Molbiol LightMix® SarbecoV E-gene assay | all genders | 42 |
| (Wong et al., 2020b) | 2020 | Retrospective and Prospective | posterior oropharyngeal saliva(DTS specimens), sputum, tracheal aspirate and bronchoalveolar lavagespecimens(LRT specimens)(total) | China | the TIB-Molbiol LightMix® SarbecoV E-gene assay | all genders | 162 |
| (Szymczak et al., 2020) | 2020 | Retrospective and Prospective | remnant stool specimens | USA | Hologic Panther Fusion SARS-CoV-2 assay | all genders | 79 |
| (Stevens et al., 2020) | 2020 | Retrospective | nasopharyngeal swabs | USA | Panther Fusion | all genders | 104 |
| (Whiting et al., 2011) | 2020 | Prospective | nasopharyngeal swabs | USA | Cobas Ct Category | all genders | 113 |
| (Loeffelholz et al., 2020) | 2020 | Retrospective | swab specimens | USA | nucleic acid amplification tests (NAATs) | all genders | 481 |
| (Jokela et al., 2020) | 2020 | Cross Sectional Study | respiratory | Finland | LDT | all genders | 39 |
| (Jokela et al., 2020) | 2020 | Cross Sectional Study | respiratory | Finland | cobas® SARS-CoV2 | all genders | 30 |
| (Jokela et al., 2020) | 2020 | Cross Sectional Study | respiratory | Finland | Amplidiag® COVID-19 | all genders | 21 |
| (Jokela et al., 2020) | 2020 | Cross Sectional Study | respiratory | Finland | total | all genders | 90 |
| (Goldenberger et al., 2020) | 2020 | Prospective | nasopharyngeal swabs | Switzerland | cobas® SARSCoV-2 assay (Roche) on the COBAS 6800 system (Roche) | all genders | 19 |
| (Dust et al., 2020) | 2020 | Cross Sectional Study | nasopharyngeal swabs | Canada | LDT-1 | all genders | 38 |
| (Wolters et al., 2020) | 2020 | Prospective | nasopharyngeal, mid-turbinate and oropharyngeal swabs | Netherlands | RT-PCR | all genders | 88 |
| (Hou et al., 2020) | 2020 | Retrospective | oropharyngeal swab | China | RT-PCR | all genders | 285 |
| (Lowe et al., 2020) | 2020 | Retrospective | nasopharyngeal swabs | Canada | cobas®or the Lightmix®assay | NA | 37 |
| (Falasca et al., 2020) | 2020 | Retrospective | nasopharyngeal swabs | Italy | RT-PCR | NA | 17 |
3.3. Quality assessment
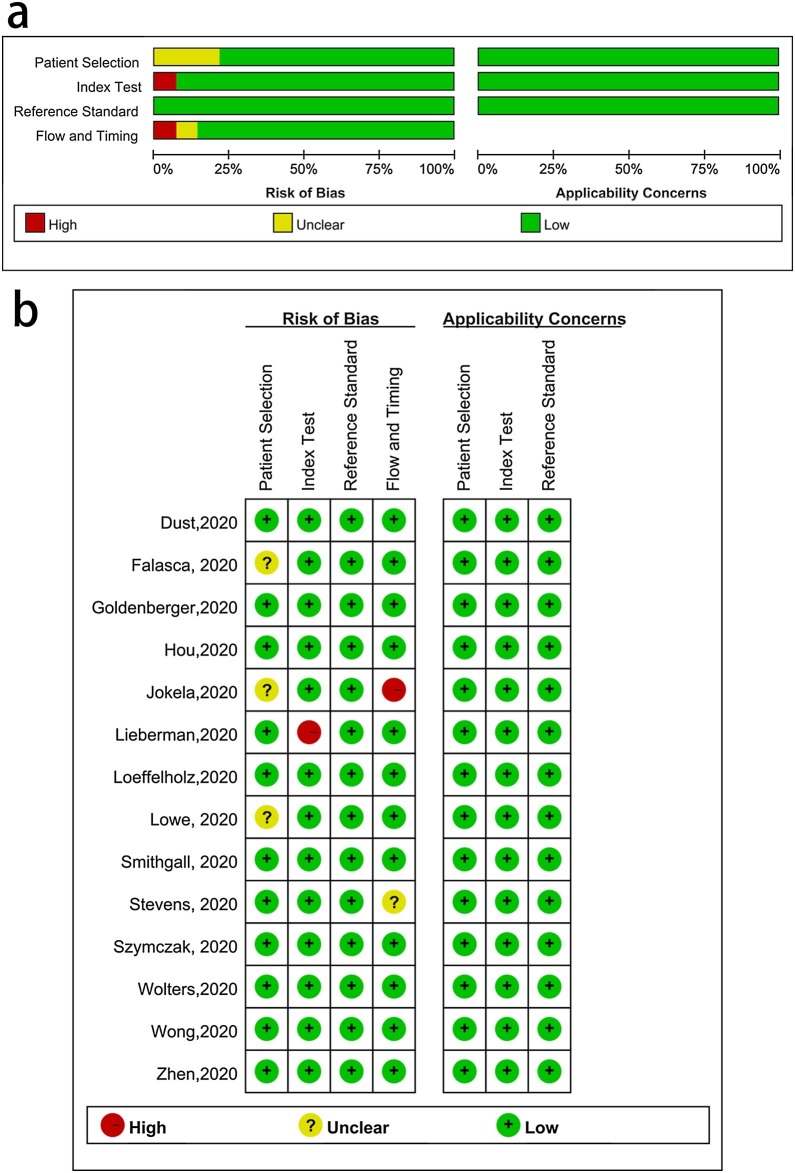
The overall quality of the 14 included studies was excellent, and the results are shown in Fig. 1 .
Fig. 1.
a. Overall quality assessment of the included studies. b. Quality assessment of the individual studies.
3.4. Overall accuracy of Cepheid Xpert Xpress assay
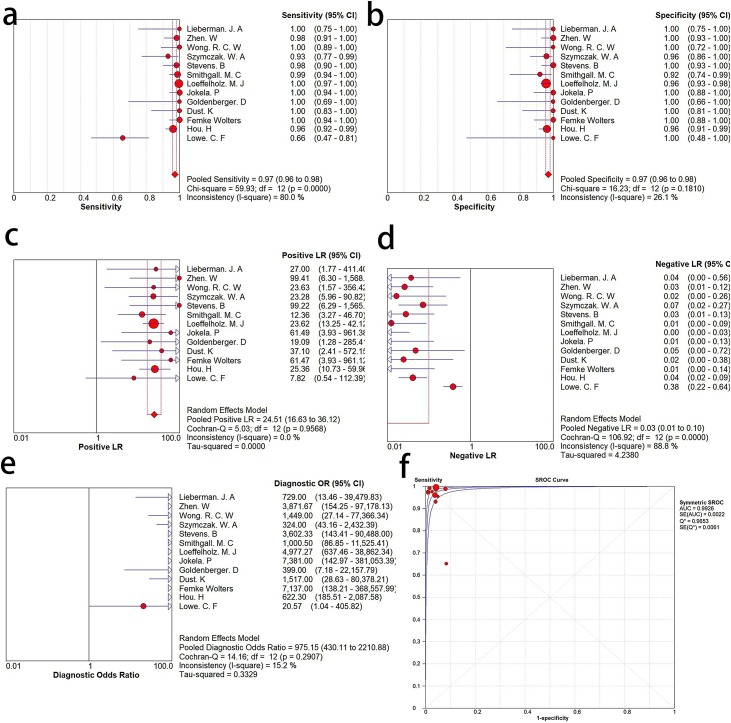
For the determination of the diagnostic accuracy of the Xpert Xpress assay, a random effects model was employed in our research. Xpert Xpress assay showed excellent diagnostic performance, and there was no obvious heterogeneity among the 11 studies. Overall, the sensitivity was 0.97 (95% CI :0.96−0.98), I2 = 80 %; Fig. 2 a), the specificity was 0.97 (95 % CI: 0.96−0.98, I2 = 21.6 %; Fig. 2b), the positive likelihood ratio (PLR) was 24.51 (95 % CI: 24.51 16.63–36.12, I2 = 0.0 %; Fig. 2c), the negative likelihood ratio (NLR) was 0.03 (95 % CI: 0.01 to 0.10, and I2 = 88.8 %; Fig. 2d), the diagnostic odds ratio (DOR) was 975.15 (95 % CI: 430.11–2210.88, I2 = 15.2 %; Fig. 2e), and the area under the SROC curve (AUC) was 0.9953 (Fig. 2f).
Fig. 2.
Forest plots of a. sensitivity, b. specificity, c. positive LR, d. negative LR, e. the diagnostic odds ratio (DOR) of Xpert Xpress for diagnosis of SARS-CoV-2 infection, f. SROC curve.
3.5. Publication bias and heterogeneity
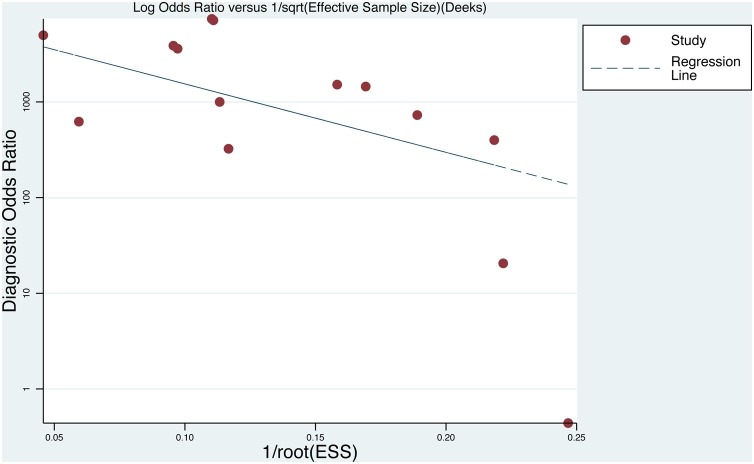
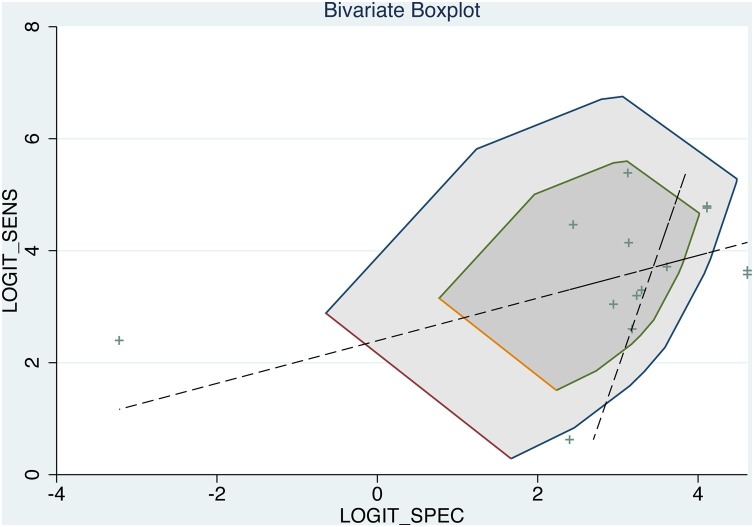
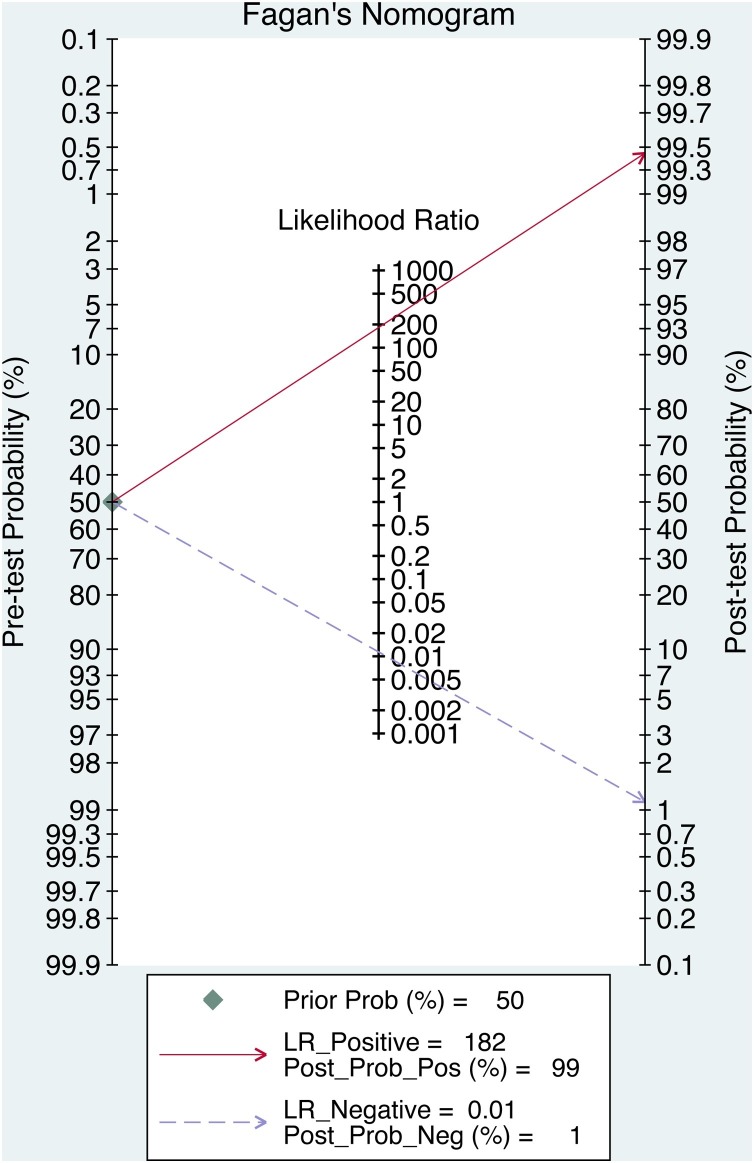
Deek’s funnel plot, with a p value of 0.036 (Fig. 3 ), showed that there was no significant publication bias in the included studies. Bivariate boxplot (Fig. 4 ) showed that there were 3 articles out of the circles which indicated there was heterogeneity between articles we included. Fagan’s Nomogram indicates that when the prediction probability of the sample is 50 %, the probability of a positive result after the test was 98 %, while the probability of a negative result was 1% (Fig. 5 ).
Fig. 3.
Deeks’ funnel plot asymmetry test to assess publication bias for Cepheid Xpert Xpress detection of SARS-CoV-2.
Studies are almost symmetrically distributed near the regression line which means there is no publication bias.
Fig. 4.
Bivariate boxplot showing the relationship between sensitivity and specificity.
Fig. 5.
Fagan nomogram of disease probabilities based on Bayes’ theorem.
4. Discussion
The SARS-CoV-2 infection rate, as well as mortality rate, is high. There is an urgent need for a fast and convenient method of diagnosing COVID-19 pneumonia (WHO, 2022). Cepheid Xpert Xpress assay has been FDA emergency authorized for detection of SARS-CoV-2. To understand the test performance of the Xpert Xpress assays, we compared the clinical efficacy of Xpert Xpress with other SARS-CoV-2 diagnostic techniques and found the high accuracy of Xpert Xpress test and its diagnostic advantages over other methods.
At present, there are many studies that are compared with Xpert Xpress. For example, the study by Daniel Goldenberger et al. Goldenberger et al. (2020) compared a commercial SARS-CoV-2 specific nucleic acid testing, which proved that Xpert Xpress has good diagnostic performance. The research of Marie C. Smithgall et al. Smithgall et al. (2020) also reached a similar conclusion. In addition to its good diagnostic accuracy, Xpert Xpress has an advantage in running time. Compared to the commonly used Roche Cobas SARS-CoV-2, Cepheid Xpert Xpress has a shorter run time. Compared to the new Abbott ID Now method, Cepheid Xpert Xpress SARS-CoV-2 has a diagnostic accuracy which is more specific despite its longer run time (the run time of Abbott ID Now is 40 min).
For the selection of sample types, WHO recommends the nasopharyngeal specimens. However, other sample types, such as posterior oropharyngeal saliva, sputum, tracheal aspirate, bronchoalveolar lavage specimens, and so on, still have important diagnostic value. In the included studies, in addition to the above-mentioned specimens, there are samples of non-respiratory stool specimens (remnant stool specimens). However, the analysis results still show the good diagnostic performance of Xpert Xpress. This proves that Xpert Xpress may be used in the detection of samples from different sources and types. Xpert Xpress SARS-CoV-2 analysis is a completely automated in vitro diagnostic test which can target for envelope genes and nucleocapsid genes on the GeneXpert platform. Nevertheless, only one study showed that the detection rate of N2 in nasopharyngeal swabs samples was slightly higher than the detection rate of E gene target genes. Therefore, the detection rate of N2 gene and E gene in different samples requires further study.
However, in the current practical applications the majority of commercial tests (including the Xpert Xpress test) mostly perform verification analysis of nasopharyngeal samples and rarely include other types of samples. In our study, although the sample types are diverse, there is still the limitation of small sample size and the mucus and high viscosity of posterior oropharyngeal saliva and tracheal aspirate and bronchoalveolar lavage specimens make it difficult to process the automated sample-to-answer platform. Although the detection time of Xpert Xpress is only 45 min and the price is low, the throughput of the ink cartridge detection is small compared to that of the high-throughput sequencing, and it may not become the main popular detection method in the world environment of the new coronavirus pandemic, which also indicates that Xpert Xpress can be used as the detection method in the large environment with low sample size and relatively backward economy and can also be applied to small laboratories for accurate evaluation.
However, there were some limitations of the review processes used. We only included the articles published in the English language, which may contribute to language bias. This review only included peer-reviewed studies with primary data. Despite the systematic search, there is still the possibility of missing literature. We didn’t perform a subgroup analysis based on sample type due to the limitation of the amount of data.
5. Conclusion
In this article, we concluded that the heterogeneity of the Cepheid Xpress SARS-CoV-2 assay in different samples is small and the detection in different samples does not reflect the limitations of the technology itself. Due to its extremely high specificity and sensitivity, the Cepheid Xpress SARS-CoV-2 assay can be used in rapid screening. It will become the mainstream method of SARS-CoV-2 detection and is superior to the existing mainstream methods in run time.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Author contributions
Xu-Guang Guo conceived and designed the experiments. Xun-Jie Cao, Ke-Ying Fang, Ya-Ping Li and Jie Zhou analyzed the data and made the tables. Xun-Jie Cao, Ke-Ying Fang, Ya-Ping Li and Jie Zhou participated in the writing, reading, and revising of the manuscript and approved the final version of the manuscript.
Declaration of Competing Interest
The authors report no declarations of interest.
Acknowledgment
Not applicable.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.jviromet.2022.114460.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- Coronavirus disease (COVID-19) pandemic at https://www.who.int/emergencies/diseases/novel-coronavirus-2019.
- Dust K., et al. Comparison of commercial assays and laboratory developed tests for detection of SARS-CoV-2. J. Virol. Methods. 2020;285 doi: 10.1016/j.jviromet.2020.113970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falasca F., Sciandra I., Di Carlo D., Gentile M., Deales A., Antonelli G., Turriziani O. Detection of SARS-COV N2 gene: very low amounts of viral RNA or false positive? J. Clin. Virol. 2020;133(December) doi: 10.1016/j.jcv.2020.104660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldenberger D., et al. Brief validation of the novel GeneXpert xpress SARS-CoV-2 PCR assay. J. Virol. Methods. 2020;284 doi: 10.1016/j.jviromet.2020.113925. 2020.113925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotham D., McKenna L., Deborggraeve S., Madoori S., Branigan D. Public investments in the development of GeneXpert molecular diagnostic technology. PLoS One. 2021;16(8) doi: 10.1371/journal.pone.0256883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotham D., McKenna L., Deborggraeve S., Madoori S., Branigan D. Public investments in the development of GeneXpert molecular diagnostic technology. PLoS One. 2021;16(8) doi: 10.1371/journal.pone.0256883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou H., Chen J., Wang Y., Lu Y., Zhu Y., Zhang B., Wang F., Mao L., Tang Y.W., Hu B., Ren Y., Sun Z. Multicenter evaluation of the Cepheid Xpert Xpress SARS-CoV-2 assay for the detection of SARS-CoV-2 in oropharyngeal swab specimens. J. Clin. Microbiol. 2020;58(July (8)):e01288–20. doi: 10.1128/JCM.01288-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jokela P., et al. SARS-CoV-2 sample-to-answer nucleic acid testing in a tertiary care emergency department: evaluation and utility. J. Clin. Virol. 2020;131(13) doi: 10.1016/j.jcv.2020.104614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman J.A., et al. Comparison of commercially available and laboratory-developed assays for in vitro detection of SARS-CoV-2 in clinical laboratories. J. Clin. Microbiol. 2020;58(8) doi: 10.1128/JCM.00821-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeffelholz M.J., et al. Multicenter evaluation of the Cepheid Xpert Xpress SARS-CoV-2 test. J. Clin. Microbiol. 2020;58 doi: 10.1128/JCM.00926-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe C.F., Matic N., Ritchie G., Lawson T., Stefanovic A., Champagne S., Leung V., Romney M.G. Detection of low levels of SARS-CoV-2 RNA from nasopharyngeal swabs using three commercial molecular assays. J. Clin. Virol. 2020;128(Jul) doi: 10.1016/j.jcv.2020.104387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu G., Liu D. SARS-like virus in the Middle East: a truly bat-related coronavirus causing human diseases. Protein Cell. 2012;3(11):803–805. doi: 10.1007/s13238-012-2811-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabi F.A., Al Zoubi M.S., Kasasbeh G.A., Salameh D.M., Al-Nasser A.D. SARS-CoV-2 and coronavirus disease 2019: what we know so far. Pathogens. 2020;9 doi: 10.3390/pathogens9030231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smithgall M.C., Scherberkova I., Whittier S., Green D.A. Comparison of Cepheid Xpert Xpress and Abbott ID now to Roche cobas for the rapid detection of SARS-CoV-2. J. Clin. Virol. 2020;128 doi: 10.1016/j.jcv.2020.104428. Epub 2020 May 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens B., et al. Comparison of a point-of-care assay and a high-complexity assay for detection of SARS-CoV-2 RNA. J. Appl. Lab. Med. 2020 doi: 10.1093/jalm/jfaa135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymczak W.A., et al. Utility of stool PCR for the diagnosis of COVID-19: comparison of two commercial platforms. J. Clin. Microbiol. 2020;58 doi: 10.1128/jcm.01369-20. 10.1128/JCM.01369-20. Print 2020 Aug 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., et al. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020;181:894–904. doi: 10.1016/j.cell.2020.03.045. e899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiting P.F., et al. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann. Intern. Med. 2011;155(8):529–536. doi: 10.7326/0003-4819-155-8-201110180-00009. [DOI] [PubMed] [Google Scholar]
- WHO Coronavirus (COVID-19) Dashboard at https://covid19.who.int/.
- Wiersinga W.J., Rhodes A., Cheng A.C., Peacock S.J., Prescott H.C. Pathophysiology, transmission, diagnosis, and treatment of coronavirus disease 2019 (COVID-19): a review. JAMA. 2020;324:782–793. doi: 10.1001/jama.2020.12839. [DOI] [PubMed] [Google Scholar]
- Wolters F., et al. Multi-center evaluation of cepheid Xpert(R) Xpress SARS-CoV-2 point-of-care test during the SARS-CoV-2 pandemic. J. Clin. Virol. 2020;128:104426. doi: 10.1016/j.jcv.2020.104426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong R.C., Wong A.H., Ho Y.I., Leung E.C., Lai R.W. Evaluation on testing of deep throat saliva and lower respiratory tract specimens with Xpert Xpress SARS-CoV-2 assay. J. Clin. Virol. 2020;131 doi: 10.1016/j.jcv.2020.104593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong R.C.W., Wong A.H., Ho Y.I.I., Leung E.C.M., Lai R.W.M. Evaluation on testing of deep throat saliva and lower respiratory tract specimens with Xpert Xpress SARS-CoV-2 assay. J. Clin. Virol. 2020;131 doi: 10.1016/j.jcv.2020.104593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhen W., et al. Clinical evaluation of three sample-to-answer platforms for detection of SARS-CoV-2. J. Clin. Microbiol. 2020;58(8) doi: 10.1128/JCM.00783-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J. SARS-CoV-2: an emerging coronavirus that causes a global threat. Int. J. Biol. Sci. 2020;16:1678–1685. doi: 10.7150/ijbs.45053. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.