ABSTRACT
Background
The microbiome of the digestive tract exerts fundamental roles in host physiology. Extrinsic factors including lifestyle and diet are widely recognized as key drivers of gut and oral microbiome compositions. Although drinking water is among the food items consumed in the largest amount, little is known about its potential impact on the microbiome.
Objectives
We explored the associations of plain drinking water source and intake with gut and oral microbiota compositions in a population-based cohort.
Methods
Microbiota, health, lifestyle, and food intake data were extracted from the American Gut Project public database. Associations of drinking water source (bottled, tap, filtered, or well water) and intake with global microbiota composition were evaluated using linear and logistic models adjusted for anthropometric, diet, and lifestyle factors in 3413 and 3794 individuals, respectively (fecal samples; 56% female, median [IQR] age: 48 [36–59] y; median [IQR] BMI: 23.3 [20.9–26.3] kg/m2), and in 283 and 309 individuals, respectively (oral samples).
Results
Drinking water source ranked among the key contributing factors explaining the gut microbiota variation, accounting for 13% [Faith's phylogenetic diversity (Faith's PD)] and 47% (Bray–Curtis dissimilarity) of the age effect size. Drinking water source was associated with differences in gut microbiota signatures, as revealed by β diversity analyses (P < 0.05; Bray–Curtis dissimilarity, weighted UniFrac distance). Subjects drinking mostly well water had higher fecal α diversity (P < 0.05; Faith's PD, observed amplicon sequence variants), higher Dorea, and lower Bacteroides, Odoribacter, and Streptococcus than the other groups. Low water drinkers also exhibited gut microbiota differences compared with high water drinkers (P < 0.05; Bray–Curtis dissimilarity, unweighted UniFrac distance) and a higher abundance of Campylobacter. No associations were found between oral microbiota composition and drinking water consumption.
Conclusions
Our results indicate that drinking water may be an important factor in shaping the human gut microbiome and that integrating drinking water source and intake as covariates in future microbiome analyses is warranted.
Keywords: drinking water, water source, water intake, human microbiome, gut microbiota diversity, oral microbiota, American Gut Project
Introduction
Environmental exposure and lifestyle factors strongly influence the composition of the human microbiome. Both short- and long-term dietary patterns are among the main modifiable drivers shaping the digestive tract microbiome structure (1, 2). From the oral cavity to the large intestine, microbial communities are in close interaction with the external environment and the epithelial barriers that delineate the inner self (3). These communities exert a marked influence on the host during homeostasis and disease episodes through a range of physiological functions (4).
Whereas the role of dietary patterns on oral and gut microbial communities has been extensively explored, drinking water, a vital source of fluids to replace daily body water losses and maintain homeostasis, has rarely been a consideration in human microbiome research. The US Institute of Medicine has set a daily Adequate Intake (AI) for total water for the adult population of 2.7 L for women and 3.7 L for men, of which 70%–80% comes from plain drinking water and other beverages (5). In Europe, the daily AI for total water for the adult population set by the European Food Safety Authority is 2.0 L for women and 2.5 L for men, of which 80% is estimated to come from fluids (6).
Drinking water may come from various origins and be subjected to different treatments before its consumption. Whereas natural mineral water, spring water, and well water solely come from groundwater, tap water originates either from surface water or groundwater, depending on community water systems’ location and setting. Drinking water may be subjected to various treatments, depending on the quality of raw water and applicable regulations. Common treatments consist of flocculation, sedimentation, and filtration to remove debris and control the chemical and mineral composition. Some drinking waters may further be disinfected to remove or control disease-causing microorganisms (7, 8). The combination of environmental conditions underground and at the source, and along the processing, distribution, and storage, results in multiple and distinct physicochemical (9), mineral (10), and microbial signatures (11–14) of drinking waters. Consumed in large amounts on a daily basis, drinking water may be considered a potential source of microbial gut diversity (15).
The effect of drinking water on the gut microbiome is poorly understood. A limited number of studies have explored microbiota composition after the ingestion of different types of drinking water and observed that water type led to differences in gut microbiota compositions (16). Consumption of tap water resulted in an increased amount of bacteria associated with antibiotic resistance in the mouse gut microbiota compared with sterilized water (16). Today, the effect of drinking water on human gut and oral microbiota is even less explored. During infant microbiome development, tap water and boiled water intake correlated with gut microbiota signatures, indicating that drinking water may be a determinant of microbiome acquisition (17). In adults, limited evidence suggests that the source of drinking water consumed is associated with oral (9) or gut microbiota compositions (18, 19). The mechanisms by which drinking water may interact with microbial communities, if any, remain to be elucidated but could involve water pH (20, 21), solute and mineral composition (9, 16, 18, 19), natural intrinsic microbial communities (16, 18), or residual chlorine and disinfection byproducts that remain in most tap waters (22, 23).
To our knowledge, most population-based gut microbiota cohorts do not consider plain drinking water as part of the diet (24, 25). Despite its primary role for health (26), water is under-researched and often referred to as the forgotten nutrient (27). A recent citizen-science microbiota-based cohort, the American Gut Project (AGP), provided a unique opportunity to analyze multiple environmental and lifestyle factors, including diet and the consumption of plain drinking water. In this exploratory analysis, we investigated whether plain drinking water source and intake were associated with fecal and oral microbiota compositions as measured by 16S ribosomal RNA (rRNA) gene sequencing in multivariate-adjusted models.
Methods
Participant recruitment and sample processing
Data for this research originate from the AGP database, a self-selected citizen-scientist cohort initiated by the University of California San Diego (28). It contains analyses of fecal, oral, and skin samples sent by >24,000 volunteers worldwide. After contributing $99, participants receive a kit to collect their biological samples and mail them for 16S rRNA gene sequencing to establish their microbial composition. Self-reported metadata are collected through a web portal (https://microsetta.ucsd.edu/). Surveys, methodology, sampling, and laboratory test procedures have been described elsewhere (28).
Ethical committee approval for the collection of AGP data was obtained either from the University of Colorado Boulder Review Board (protocol no. 12-0582; December 2012–March 2015) or from the University of California Review Board, San Diego (protocol no. 141853; February 2015–present) in accordance with the Declaration of Helsinki and all participants provided informed consent. Return of results to participants, public deposition of de-identified data, and subsequent analyses are allowed by the IRB-approved protocol (28). To investigate the associations between drinking water consumption and gut and oral microbiota composition, Redbiom (29) was used to fetch in Qiita (30) the 100-nt-long sequences associated with 20,454 stool sample identifiers available in the database on 5 December, 2019 using the Deblur-Illumina-16S-V4-100nt-fbc5b2 context (28).
Participant and sample selection
Participants who completed both the standard AGP questionnaire (general health status, disease history, and lifestyle data) and a validated picture-based FFQ (VioScreen™; http://www.viocare.com/vioscreen.html) (n = 3022 participants out of 15,254) were screened to exclude participants with aberrant total water intake, based on the variable “Water in g” available in Vioscreen™ (see Supplemental Methods for more details). When available, coherence of fluid variables between the general questionnaire and Vioscreen™ was evaluated. Fecal samples were then screened for unusual dominant microbiota (see Supplemental Methods). This first set of exclusion criteria resulted in the exclusion of 2045 and 4564 fecal samples for the drinking water source and the drinking water intake analysis, respectively (Figure 1, Supplemental Figure 1). Individuals with missing information on confounding factors were filtered out (see Supplemental Methods). Participants were further filtered based on their age, geographical location, and antibiotics intake to reduce variability (see Supplemental Methods). The resulting study population consisted of 3413 subjects for the drinking water source analysis and 3794 subjects for the drinking water intake analysis.
FIGURE 1.
American Gut Project participant selection and data filtering process for gut microbiota analysis according to their drinking water source. 1Based on age- and sex-specific Adequate Intake as defined by the European Food Safety Authority and TWI expressed as mL/kg. Participants with either TWI < 25% AI or aged < 14 y with TWI > 300% AI, or TWI > 200 mL/kg of body weight were excluded. 2Participants were excluded if Alcohol_frequency = “Never” AND Alcohol_consumption = “Yes.” 3The top 10 genera list in adults and the top 10 genera list in nonadults were merged, resulting in 12 genera. Samples were flagged as outliers when their respective read mass had <25% of those 12 genera. SSB, sugar-sweetened beverage; TWI, total water intake.
For oral microbiota analyses, data from 1383 oral samples coming from 1140 participants were extracted. Supplemental Figure 2 details exclusion criteria. Complete data were available for n = 283 participants for the drinking water source analysis and n = 309 participants for the water intake analysis.
Numerical ecology and statistical analyses
All statistical analyses were performed using R Statistical Software version 3.6 (R Core Team). Characteristics of participants were compared between groups of drinking water source and groups of low and high water drinkers using ANOVA with post hoc tests for continuous variables and chi-square tests for categorical variables (31). All P values reported were adjusted for multiplicity with Benjamini–Hochberg's correction procedure.
Microbiota α diversity
Observed amplicon sequence variants (ASVs), Chao1 (32), Shannon index (33), and Faith's phylogenetic diversity (Faith's PD) (34) were used as response variables in multivariate-adjusted linear models (see Supplemental Methods). For all analyses performed in this study, α = 0.05 was considered the significance threshold for adjusted P values. In the water intake analysis, participants were separated into different age categories to reduce α-diversity variability induced by age (35). Adjusted means of α diversity were computed, then compared between all pairs of groups using pairwise comparisons of estimated marginal means. P values were corrected with Benjamini–Hochberg's procedure (31) to account for multiple testing. To measure variables’ effect sizes, the proportion of variance (R2) captured by a given variable was calculated as the ratio of the sum of squares explained by the variable to the total sum of squares in type II ANOVAs when applicable (see Supplemental Methods).
Microbiota β diversity
β Diversity was assessed using unweighted and weighted UniFrac distances as well as Bray–Curtis dissimilarity in QIIME at ASV level, with a rarefaction at 1000 sequences (see Supplemental Methods). To investigate interindividual differences in microbiota β diversity in relation to consumption of drinking water, permutational multivariate analysis of variance (PERMANOVA) was performed using the adonis function in the vegan R package with full adjustment. Only confounding factors with equivalent dispersion for all β diversity metrics were retained, i.e., age, BMI, and diet type.
A permutation test was applied to assess whether dispersion of groups of interest was equivalent (see Supplemental Methods). To measure variables’ effect sizes, the proportion of variance (R2) captured by a given variable was calculated as the ratio of the sum of squares explained by the variable to the total sum of squares in PERMANOVAs when applicable.
Microbiota taxonomy
Differences in the abundance of bacterial genera between drinking water groups were assessed in adjusted models using the DESeq2 R package (36) (see Supplemental Methods). DESeq2 is a differential abundance detection method for 16S gene sequencing but with limited performance on low-abundant ASVs with relatively high variance. Therefore, ASVs were trimmed to those with a mean relative abundance ≥ 0.01% and prevalence ≥ 10%.
Results
Participants’ characteristics
Participants were classified into groups of drinking water source or intake based on their answers to the AGP standard questionnaire. Participants’ characteristics according to their drinking water source and intake are shown in Table 1 and Supplemental Table 1, respectively. “Drinking water source” means the source of drinking water usually consumed at home by participants, i.e., bottled, city (tap water), filtered, or well. A total of 3413 participants were included in this analysis. In a separate analysis, 3794 participants were classified as either high water drinkers (if they reported “daily” or “regularly”) or low water drinkers (if they reported “never,” “rarely,” “occasionally”) based on the variable “One liter of water a day frequency.”
TABLE 1.
Characteristics of the American Gut Project participants according to their drinking water source1
| All | Bottled | City | Filtered | Well | P value | |
|---|---|---|---|---|---|---|
| (n = 3413) 2 | (n = 284) | (n = 1784) | (n = 1147) | (n = 198) | ||
| Age, y | 48 [36–59] | 49 [39–62] | 47 [35–59] | 47 [36–58] | 52 [42–61] | 0.001 |
| Sex | 0.014 | |||||
| Female | 1909 (56) | 164 (58) | 951 (53) | 678 (59) | 116 (59) | |
| Male | 1504 (44) | 120 (42) | 833 (47) | 469 (41) | 82 (41) | |
| BMI | 0.070 | |||||
| Normal (BMI = 18.5–24.9) | 2073 (61) | 154 (54) | 1079 (61) | 723 (63) | 117 (59) | |
| Underweight (BMI ≤ 18.4) | 351 (10) | 32 (11) | 200 (11) | 97 (9) | 22 (11) | |
| Overweight (BMI = 25.0–29.9) | 824 (24) | 79 (28) | 432 (24) | 265 (23) | 48 (24) | |
| Obese (BMI ≥ 30.0) | 165 (5) | 19 (7) | 73 (4) | 62 (5) | 11 (6) | |
| Diabetes3, 4 | 88 (3) | 8 (3) | 44 (3) | 28 (2) | 8 (4) | 0.392 |
| Cardiovascular disease3, 5 | 85 (3) | 7 (3) | 46 (3) | 30 (3) | 2 (1) | 0.786 |
| Kidney disease3, 6 | 44 (1) | 8 (3) | 21 (1) | 15 (1) | 0 (0) | 0.020 |
| IBS7 | 0.001 | |||||
| Diagnosed by a medical professional | 406 (12) | 25 (9) | 224 (13) | 132 (12) | 25 (13) | |
| Diagnosed by an alternative medicine practitioner | 33 (1) | 3 (1) | 12 (1) | 16 (1) | 2 (1) | |
| Self-diagnosed | 213 (6) | 13 (5) | 119 (7) | 73 (6) | 8 (4) | |
| IBD3, 8 | 105 (3) | 8 (3) | 54 (3) | 39 (3) | 4 (2) | 0.161 |
| Smoking frequency9 | 0.112 | |||||
| Never | 3190 (94) | 260 (92) | 1656 (93) | 1089 (95) | 185 (93) | |
| Rarely | 111 (3) | 8 (3) | 62 (4) | 35 (3) | 6 (3) | |
| Occasionally | 37 (1) | 3 (1) | 25 (1) | 7 (1) | 2 (1) | |
| Regularly | 20 (1) | 2 (1) | 13 (1) | 4 (0.4) | 1 (1) | |
| Daily | 50 (1) | 10 (4) | 27 (2) | 9 (1) | 4 (2) | |
| Exercise frequency | <0.001 | |||||
| Never | 96 (3) | 17 (6) | 52 (3) | 24 (2) | 3 (2) | |
| Rarely | 377 (11) | 44 (16) | 201 (11) | 115 (10) | 17 (9) | |
| Occasionally | 832 (24) | 77 (27) | 434 (24) | 270 (24) | 51 (26) | |
| Regularly | 1393 (41) | 104 (37) | 749 (42) | 467 (41) | 73 (37) | |
| Daily | 715 (21) | 42 (15) | 348 (20) | 271 (24) | 54 (27) | |
| Country | <0.001 | |||||
| USA | 1887 (55) | 195 (69) | 792 (44) | 740 (65) | 160 (81) | |
| United Kingdom | 1526 (45) | 89 (31) | 992 (56) | 407 (36) | 38 (19) | |
| Collection season | 0.008 | |||||
| Spring | 887 (26) | 59 (21) | 454 (25) | 320 (28) | 54 (27) | |
| Summer | 699 (20) | 49 (17) | 377 (21) | 228 (20) | 45 (23) | |
| Fall | 803 (24) | 73 (26) | 451 (25) | 244 (21) | 35 (18) | |
| Winter | 1024 (30) | 103 (36) | 502 (28) | 355 (31) | 64 (32) | |
| Level of education | <0.001 | |||||
| Did not complete high school | 96 (2) | 10 (4) | 44 (3) | 35 (3) | 7 (4) | |
| High school or GED equivalent | 126 (4) | 14 (5) | 65 (4) | 43 (4) | 4 (2) | |
| Some college or technical school | 335 (10) | 44 (16) | 150 (8) | 120 (11) | 21 (11) | |
| Associate's degree | 78 (2) | 12 (4) | 21 (1) | 36 (3) | 9 (5) | |
| Bachelor's degree | 906 (27) | 74 (26) | 455 (26) | 317 (28) | 60 (30) | |
| Some graduate school or professional | 248 (7) | 20 (7) | 112 (6) | 98 (9) | 18 (9) | |
| Graduate or Professional degree | 1624 (48) | 110 (39) | 937 (53) | 498 (43) | 79 (40) | |
| Fed as infant | <0.001 | |||||
| Primarily breast milk | 1873 (55) | 143 (50) | 1029 (58) | 601 (52) | 100 (51) | |
| Primarily infant formula | 930 (27) | 90 (32) | 429 (24) | 339 (30) | 72 (36) | |
| Both | 610 (18) | 51 (18) | 326 (18) | 207 (18) | 26 (13) | |
| Diet type | 0.007 | |||||
| Omnivore | 2681 (79) | 231 (81) | 1425 (80) | 866 (76) | 159 (80) | |
| Omnivore but no red meat | 238 (7) | 20 (7) | 114 (6) | 93 (8) | 11 (6) | |
| Vegetarian | 174 (5) | 15 (5) | 101 (6) | 52 (5) | 6 (3) | |
| Vegetarian but eat seafood | 227 (7) | 14 (5) | 108 (6) | 89 (8) | 16 (8) | |
| Vegan | 93 (3) | 4 (1) | 36 (2) | 47 (4) | 6 (3) | |
| Types of plants per week, n | <0.001 | |||||
| <5 | 205 (6) | 43 (15) | 97 (5) | 53 (5) | 12 (6) | |
| 6–10 | 792 (23) | 87 (31) | 416 (23) | 252 (22) | 37 (19) | |
| 11–20 | 1243 (36) | 90 (32) | 672 (38) | 410 (36) | 71 (36) | |
| 21–30 | 752 (22) | 35 (12) | 397 (22) | 270 (24) | 50 (25) | |
| >30 | 421 (12) | 29 (10) | 202 (11) | 162 (14) | 28 (14) | |
| Sugar-sweetened beverage frequency | <0.001 | |||||
| Never | 2503 (73) | 200 (70) | 1266 (71) | 886 (77) | 151 (76) | |
| Rarely | 663 (19) | 45 (16) | 401 (23) | 182 (16) | 35 (18) | |
| Occasionally | 143 (4) | 19 (7) | 72 (4) | 43 (4) | 9 (5) | |
| Regularly | 60 (2) | 10 (4) | 21 (1) | 27 (2) | 2 (1) | |
| Daily | 44 (1) | 10 (4) | 24 (1) | 9 (1) | 1 (0.5) | |
| Alcohol frequency | <0.001 | |||||
| Never | 667 (20) | 79 (28) | 293 (16) | 249 (22) | 46 (23) | |
| Rarely | 937 (27) | 89 (31) | 460 (26) | 332 (29) | 56 (28) | |
| Occasionally | 765 (22) | 56 (20) | 428 (24) | 254 (22) | 27 (14) | |
| Regularly | 757 (22) | 42 (15) | 449 (25) | 223 (19) | 43 (22) | |
| Daily | 287 (8) | 18 (6) | 154 (9) | 89 (8) | 26 (13) | |
| One liter of water a day frequency10 | <0.001 | |||||
| Never | 71 (2) | 11 (4) | 42 (2) | 16 (1) | 2 (1) | |
| Rarely | 239 (7) | 17 (6) | 152 (9) | 57 (5) | 13 (7) | |
| Occasionally | 379 (11) | 28 (10) | 220 (12) | 114 (10) | 17 (9) | |
| Regularly | 852 (25) | 68 (24) | 476 (27) | 263 (23) | 45 (23) | |
| Daily | 1865 (55) | 160 (56) | 887 (50) | 697 (61) | 121 (61) |
1Values are medians [IQRs] for continuous variables and n (%) for categorical variables. Adjusted P values are calculated from global chi-square test. Occasionally means 1–2 times/wk; rarely means a few times per month); regularly means 3–5 times/wk. GED, General Educational Development; IBD, Inflammatory Bowel Disease; IBS, Irritable Bowel Syndrome.
2Unless stated otherwise.
3Diagnosed by a medical professional (doctor, physician assistant).
4 n = 3391, missing data from n = 9 (city), n = 11 (filtered), n = 2 (well).
5 n = 3385, missing data from n = 1 (bottled), n = 15 (city), n = 9 (filtered), n = 3 (well).
6 n = 3389, missing data from n = 1 (bottled), n = 10 (city), n = 10 (filtered), n = 3 (well).
7 n = 3380, missing data from n = 2 (bottled), n = 18 (city), n = 10 (filtered), n = 3 (well).
8 n = 3354, missing data from n = 3 (bottled), n = 27 (city), n = 27 (filtered), n = 2 (well).
9 n = 3408, missing data from n = 1 (bottled), n = 1 (city), n = 3 (filtered).
10 n = 3406, missing data from n = 7 (city), n = 3 (filtered).
Participants included in these analyses were 56% women, the median [IQR] age was 48 [36–59] y, and the majority of participants had a normal BMI (61%) or were overweight (24%). The vast majority of participants were nonsmokers (94%), and only a small proportion reported having a chronic disease (3% diabetes, 3% cardiovascular disease, 1% chronic kidney disease, 3% inflammatory bowel disease).
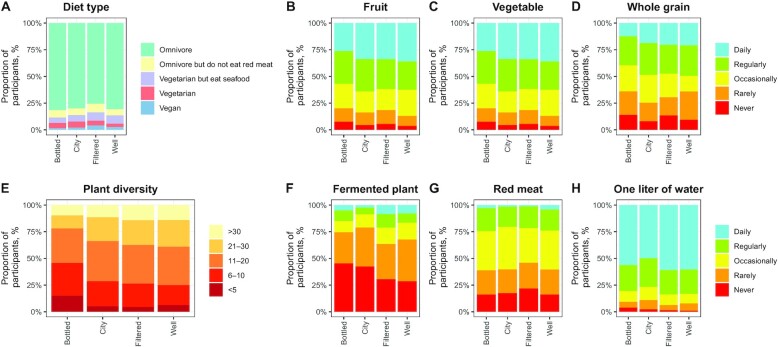
In the drinking water source subsample, 55% of participants were American, whereas 45% were from the United Kingdom (Table 1). The proportion of subjects coming from the United Kingdom and the average education level were higher in the city group than in other groups. Subjects drinking bottled water tended to consume more sugar-sweetened beverages, less alcohol, and fewer fiber-containing foods (fruits, vegetables, and plants) (Table 1, Figure 2). Fewer omnivores and red meat eaters were in the filtered water group.
FIGURE 2.
Distribution of dietary profiles of American Gut Project participants according to the drinking water source (n = 3413). (A) Diet type, (B) fruit intake, (C) vegetable intake, (D) whole grain intake, (E) plant diversity of intake, (F) fermented plant intake, (G) red meat intake, (H) intake of 1 L water/d. Occasionally meant 1–2 times/wk; rarely meant a few times per month; regularly meant 3–5 times/wk.
In the water intake analysis, the proportion of subjects exercising regularly or daily was higher among the high water drinkers than among the low water drinkers (Supplemental Table 1). High water drinkers were younger than low water drinkers. High water drinkers also had healthier dietary habits because they drank less sugar-sweetened beverages and alcohol and consumed more plants than low water drinkers.
Subjects’ demographic, dietary, and lifestyle variables associated with drinking water source and intake and that are known to be associated with gut microbiota composition were regarded as confounding factors and included in the statistical models (Supplemental Figure 3, Supplemental Methods).
Plain drinking water and microbiota α diversity
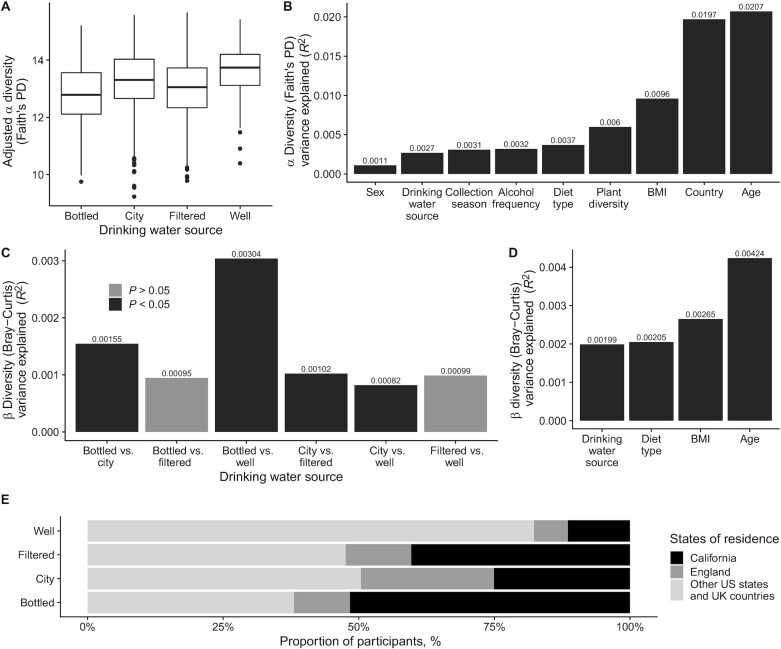
α Diversity, as evaluated by Faith's PD and observed ASVs, was associated with drinking water source in fully adjusted models (P < 0.05, type II ANOVA) (Figure 3A, Supplemental Figure 4A). Post hoc pairwise comparisons of the 4 groups of drinking water source revealed that the α diversity in the participants drinking mostly well water was higher than in the other groups (bottled compared with well, P < 0.05; city compared with well, P = 0.05; filtered compared with well, P < 0.05; pairwise comparisons of estimated marginal means; Faith's PD).
FIGURE 3.
Diversity analysis of fecal microbiota in American Gut Project participants according to their drinking water source (n = 3413). (A) α Diversity measured by Faith's PD (type II ANOVA, P < 0.05). Data are presented as adjusted means and 95% CIs. Adjusted for age, sex, BMI, infant feeding, level of education, country, collection season, exercise frequency, diet type, plant diversity (number of types of plants consumed per week), alcohol, and sugar-sweetened beverage consumption. Groups are bottled (n = 284), city (n = 1784), filtered (n = 1147), and well (n = 198). (B) Faith's PD effect sizes. Proportions of variance captured by significant variables in fully adjusted models. (C) β Diversity measured by Bray–Curtis dissimilarity. Proportions of variance captured by pairwise comparisons of drinking water source groups; adjusted for age, BMI, and diet type. (D) Bray–Curtis dissimilarity effect sizes. (E) State or country of residence of participants according to their drinking water source. Faith's PD, Faith's phylogenetic diversity.
Results obtained for Shannon and Chao1 also showed a trend for higher α diversity in participants drinking well water, although not reaching statistical significance (Shannon, P = 0.14; Chao1, P = 0.38; type II ANOVA) (Supplemental Figure 4C, D). Whereas the majority of bottled water drinkers came from California, well water drinkers were more evenly distributed across the United States and United Kingdom (Figure 3E). Figure 3B presents effect sizes, calculated as the proportions of α-diversity variability explained by each variable. The amounts of variance (R2) explained by age, country, and BMI were 2.1 × 10−2, 2.0 × 10−2, and 9.6 × 10−3, respectively. Among all diet-related variables, plant diversity (number of types of plants consumed per week) and diet type were those explaining the most variance in α diversity with R2 of 6.0 × 10−3 and 3.7 × 10−3, respectively. The influence of drinking water source was 13% of that of age, 73% of that of diet type, and of the same magnitude as those of collection season and alcohol frequency (Faith's PD) (Figure 3B). In addition, drinking water source variance was of the same magnitude as those of alcohol frequency and exercise frequency for observed ASVs (Supplemental Figure 4E). No associations between drinking water intake and any of the α diversity indexes for gut microbiota were observed (data not shown). There were no differences in oral microbiota α diversity between drinking water sources or between high and low water drinkers (Supplemental Figure 5A, Supplemental Figure 6A).
Plain drinking water and microbiota β diversity
Analysis of microbiota variability revealed differences in β diversity between groups of drinking water source, as measured by Bray–Curtis dissimilarity and weighted UniFrac distance (P < 0.05; PERMANOVAs) in models adjusted for age, BMI, and diet type (Figure 3C, Supplemental Figure 5B). All pairs comparing city water and other water sources were significant ( P < 0.05) (Figure 3C), with R2 ranging between 8.2 × 10−4 and 1.6 × 10−3. The highest variance explained was observed when comparing bottled with well water sources (R2 = 3.0 × 10−3). Several pairwise comparisons were significant across the 3 diversity metrics, with differences between bottled and city consistently detected (Supplemental Figure 5B). The amounts of variance ( R2) explained by age and BMI were 4.2 × 10−3 and 2.7 × 10−3, respectively. The amounts of variance captured by drinking water source were 47% and 50% of those captured by age for Bray–Curtis dissimilarity and weighted UniFrac distance, respectively (Figure 3D, Supplemental Figure 5C). In addition, low and high water drinkers were detected to have different global fecal microbiota compositions as measured by unweighted UniFrac distance and Bray–Curtis dissimilarity in adjusted models (P < 0.05, unweighted UniFrac distance; P = 0.05, Bray–Curtis dissimilarity; PERMANOVAs) (Supplemental Figure 6B). “One liter of water a day frequency” captured 5.6% and 10.6% of the amounts of variance captured by age for unweighted UniFrac distance (Supplemental Figure 6C) and Bray–Curtis dissimilarity (data not shown), respectively. Differences in β diversity of oral microbiota samples were detected between low and high water drinkers for Bray–Curtis dissimilarity and weighted UniFrac distance only when the model was not adjusted. Adjustments for age, BMI, diet type, collection season, and alcohol frequency attenuated these differences (Supplemental Figure 6D). Analysis of the effect sizes revealed that collection season was the factor explaining the most oral microbiota diversity (Supplemental Figure 6E).
Plain drinking water and microbiota taxonomy
Differential abundance analyses revealed consistent differences in participant fecal microbiota between drinking water source groups after adjustment for confounding factors (see Supplemental Methods). Bottled and city water drinkers were notably enriched in Bacteroides, Odoribacter, and Streptococcus genera compared with well water drinkers (Figure 4). The same trend was observed for other genera, including Veillonella and Fusobacterium. Well water drinkers had higher Dorea genus than city and filtered water drinkers. Bottled water drinkers’ fecal microbiota was enriched in different genera from the Lachnospiraceae family compared with the city water drinkers group. Comparing low and high water drinkers revealed a differential pattern, with Campylobacter abundance enriched in the low drinkers group. No differences were detected in participants’ oral microbiota (Supplemental Figure 7).
FIGURE 4.
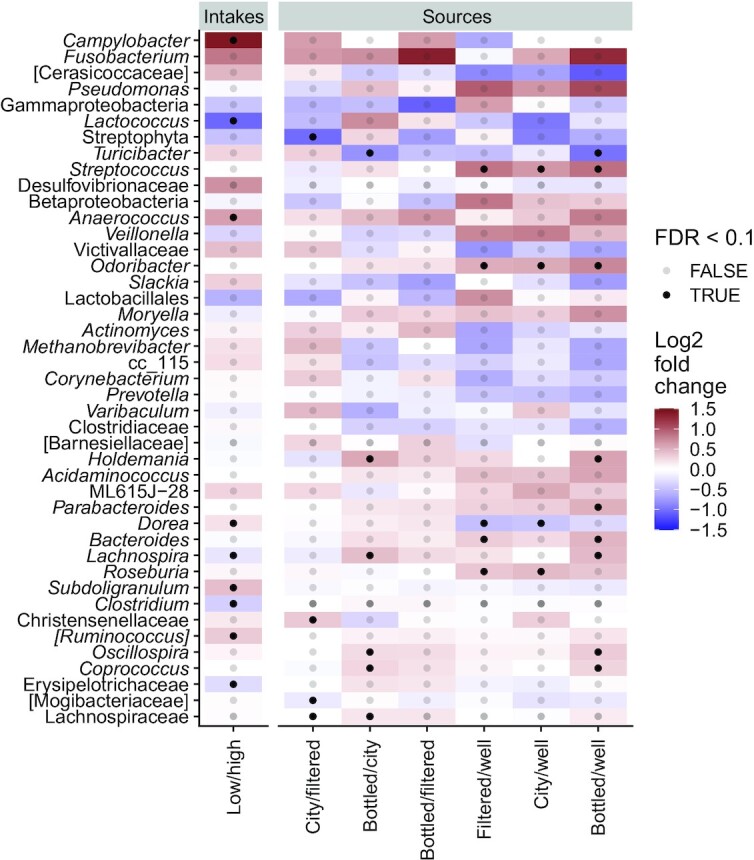
Taxonomic analysis of fecal microbiota samples of American Gut Project participants according to their drinking water source (n = 3413) or intake (n = 3794). Heatmap of log2 fold changes of pairs. Water source analysis adjusted for age, sex, BMI, infant feeding, level of education, country, collection season, exercise frequency, diet type, plant diversity (number of types of plants consumed per week), alcohol, and sugar-sweetened beverage consumption. Water intake analysis adjusted for age, sex, BMI, infant feeding, level of education, continent, collection season, exercise frequency, diet type, plant diversity, alcohol, and sugar-sweetened beverage consumption. All genera that had a log2 fold change >0.5 or an FDR <0.1 were selected. Colors account for log2 fold change between groups and dots account for FDR significance. P values for Wald's test. FDR, false discovery rate.
Discussion
In this study, we explored the relations of plain drinking water source and intake with the composition of the gut and oral microbiota in a large cohort of self-selected participants. This work is, to the best of our knowledge, the first to specifically explore the links between drinking water consumption, in terms of both origin and amount consumed, and the microbiota of the digestive tract independently of a wide range of confounding factors (demographics, lifestyle, and diet) in a large sample of the general population. We observed that drinking water source was among the major contributing factors to the variation in gut microbiota composition. We showed that drinking water source was associated with differences in gut microbiota as assessed by Bray–Curtis dissimilarity and weighted Unifrac distance. Subjects drinking mostly well water had higher fecal microbiota α diversity as measured by Faith's PD and observed ASVs than the other groups drinking either bottled, tap, or filtered water. We further showed that drinking water intake was associated with differences in gut microbiota composition between participants who consumed low and high amounts of drinking water. Finally, we failed to find any links between drinking water consumption and oral microbiota composition in this cohort.
Lower gut bacterial diversity, as well as gene diversity, has consistently been associated with impaired gut health and diseases (37–39), indicating that diverse gut microbial communities and functions may be more resilient and robust against environmental influences (40, 41). Hence, higher diversity appears a hallmark of healthy gut microbiomes. Although the link between drinking water source and gut microbiota composition deserves further investigation to establish whether it is causal, it could be hypothesized that either the physicochemical, mineral, or microbial composition of water, or a combination of the 3, may influence the gut microbiome. Results from a UK cohort suggest an association between the composition of fecal microbiota and the mineral composition of tap water (19). Specifically, the intraindividual fecal microbiota diversity was associated with the average daily dose of sodium, i.e., a higher dose of sodium was associated with a lower microbiota diversity. In addition, preclinical studies have reported differences in the gut microbiota composition of mice drinking acidic and neutral water, thereby suggesting that water pH may affect gut microbial communities (20, 21). High intakes of mineral substances, including magnesium and sodium sulfates, may alleviate constipation and improve stool consistency in functionally constipated individuals (42, 43). These functional effects are thought to derive either from a change in the intraluminal osmotic pressure, the stimulation of NO synthase, or stimulation of intestinal water channels regulating the fecal water content, which all affect intestinal motility (43). Although there is a link between intestinal motility, stool consistency, and fecal microbiota composition (44–46), it is unlikely that a difference in gut motility may explain the observed differences in α diversity in this study. Our study population neither consisted of functionally constipated participants nor revealed any differences in bowel movement quality and frequency between groups (Supplemental Figure 8). Although we could not investigate the mineral content of the diet of participants nor the mineral composition of drinking waters because of the absence of such data, we can hypothesize that it is unlikely that the mineral content of the drinking water may explain the differences observed in gut microbiota composition. Indeed, the variety of geographical locations of the participants likely introduced various water sources and mineral and physicochemical signatures within each group of drinking water sources. Alternatively, the microbial community in the drinking water, which varies between different water sources, may also explain the differences observed in gut microbiota composition. Drinking water contains an intrinsic community of microorganisms that may differ between natural sources, along the processing and distribution system, and under different storage conditions (11–14). Whereas tap and processed waters are subjected to systematic treatment to control microbial communities, some others, like natural mineral waters, are not subjected to any disinfection treatment (47, 48). In addition, it has been recently described that the drinking water microbiome is structurally and functionally less diverse and variable across disinfected than across nondisinfected systems (8). Therefore, the higher α diversity observed in the well water group could be due to more diverse natural microbial communities in drinking water, because water coming from wells, especially private wells, may not be systematically subjected to disinfection treatment either (44, 45). However, we cannot fully ascertain the correctness of this hypothesis owing to the absence of information about the operation of wells in this database. Previous research reported that exposure to different types of water with distinct endogenous microbial communities affects the gut microbiota composition of mice. In particular, mice drinking autoclaved tap water exhibited the smallest intragroup variation in gut microbial diversity, and the largest distance from other groups receiving either tap water, disinfected water, or natural mineral water (16).
In this study, we observed fecal microbiota taxonomic differences between individuals drinking mostly well water and those drinking other sources of water. The Odoribacter genus, which is positively associated with symptoms of functional gastrointestinal disorders (46) and negatively correlated with stool consistency (49), was depleted in well water drinkers’ fecal microbiota. Bacteroides genus relative abundance, which is negatively associated with diversity in the human gut microbiome (50), was also lower in well water drinkers than in the other groups. This is in line with our findings showing that well water drinkers have higher α diversity. Interestingly, Streptoccocus, together with Veillonella and Fusobacterium, known to be dominant in the upper digestive tract (51, 52), was found in higher abundance in fecal samples of bottled, tap, and filtered water drinkers than in the well water drinkers group. Recent studies report an enrichment of oral and small intestinal species in the lower digestive tract of patients with colorectal cancer (53) and liver cirrhosis (54). These results might indicate some ecological shifts in bottled, tap, and filtered water drinkers, with upper digestive tract–related species found in higher abundances in the lower gut, although the differences observed are small. In our study, well water was the only source of water coming solely from groundwater, i.e., water that exists underground beneath the land surface after percolation through earth layers (44, 45), a distinction that may contribute to explaining the taxonomic differences observed. Although we found that the increased diversity in the gut microbiota of well water drinkers was independent of the country or state of residence, the geographical locations of well water drinkers were evenly distributed across each country, which may have confounded the results. In addition, it may be hypothesized that well water drinkers may more frequently live in rural than in urban areas and that their exposure to environmental microbial diversity could be increased, resulting in increased gut microbiota diversity (55, 56).
In our study, drinking water source ranked among the key contributing factors explaining gut microbiota variation in the α- and β-diversity analyses, with effect sizes comparable with those of alcohol or diet type, thereby adding water as a novel variable to report in gut microbiota studies. This finding is consistent with the general observation that environmental factors, including diet, influence the gut microbiome (1, 2). The identification of alcohol as a contributor to gut microbiota composition is also consistent with a recent investigation (57). In our exploration, the amount of variance of β diversity captured by drinking water sources was half of the variance captured by age, a factor highly associated with microbiota compositions (35, 58). The ranking of effect sizes reported in our study between age and BMI is similar to that previously reported in a different cohort (24). Our findings are also in line with recent evidence showing differences in gut microbiota composition in native Himalayan populations associated with drinking water source (stream compared with underground well water) (18). The source of drinking water was identified as the factor most strongly associated with the gut microbiota composition of these populations, as assessed by Bray–Curtis dissimilarity, unweighted UniFrac, and weighted UniFrac distances. This finding was replicated in another native East-African population of hunter-gatherers (18).
Surprisingly, we found that drinking water intake was associated with β diversity, as measured by unweighted UniFrac distance and Bray–Curtis dissimilarity, suggesting that water intake is associated with global fecal microbiota compositions. Because this observational study cannot prove causality, we cannot assess whether these drinking water variables directly affect fecal microbiota compositions. Still, our findings emphasize the need to use them as proxies for lifestyle habits to control in future fecal microbiota analyses. These differences in gut microbiota compositions were also reflected in taxonomic analyses where we found that high water drinkers had a lower abundance of the Campylobacter genus known to cause gastrointestinal infection (59). Although it is known that increased water intake can alleviate functional constipation (43, 60), little is known about whether the volume of water consumed could alter gut microbiota composition in the general population.
When considering the oral microbiota composition, drinking water source was not associated with differences in α diversity, β diversity, or taxonomy. Previous research reported that the chemical composition of tap water was the environmental factor with the highest impact on the composition of the oral microbiota in a Spanish cohort (9), with some microbial abundance following a geographical pattern similar to that of public water quality parameters, e.g., water alkalinity and water hardness. Our study is likely underpowered to detect any changes in oral microbiota composition owing to the small sample size and the wide geographical diversity of samples. The association observed between drinking water intake and β diversity in unadjusted models may reflect differences in lifestyle habits that may confound the association. Further research with a larger sample size is warranted to confirm these conclusions.
The present study comes with limitations. Although a wide range of known confounders was either considered in the exclusion criteria or adjusted for in the statistical models, we neither excluded nor adjusted for pregnancy, cesarean deliveries, or some chronic diseases, which accounted for 1%, 9%, and 1%–3% of the study population, respectively. The reasons for this include that we wanted to limit the collinearity deriving from multiple adjustments and maintain a fair representation of the subjects’ population. It is recalled that AGP is a self-selected cohort that is not representative of the US or the UK population. In addition, we excluded participants with missing data on all confounding factors, which may have introduced an additional selection bias. A key limitation of any citizen-based approach is that it relies on self-reported, subjective measures as well as variables with low granularity to reduce participants’ burden, which may have introduced some variability in our analyses. For example, whereas drinking water source referred to the main source of water available at home, subjects may have diverse drinking water sources outside of their home. In addition, bottled water could be either natural mineral water, spring, or treated water. Finally, in this study, we used 16S rRNA gene data to explore the overall gut microbiota composition, which limits taxonomy resolution. Further studies using shotgun metagenomics sequencing would allow functional characterization and improve analysis resolution at species level. Beyond compositional and functional diversity, gut microbiome biomarkers such as SCFAs should be profiled to increase knowledge of host–microbe interactions. Nonetheless, such a citizen-science-based approach enables the collection of a large number and a diversity of samples. This allowed us to analyze data from both oral and fecal samples, although AGP sampling guidelines for the collection of oral samples should be standardized to gain precision on the sample location (e.g., saliva, tongue).
This study showed that drinking water source and intake were associated with fecal microbiota composition and found limited associations with oral microbiota composition. The work performed here contributed to formulating some hypotheses about the links between water consumption and fecal and oral microbiota compositions. It may guide future microbiota analyses, because it pointed out the importance of reporting the source and intake of drinking water consumed by participants in studies or population-based cohorts investigating the human microbiome.
Supplementary Material
Acknowledgments
We are indebted to Aurélie Cotillard for helpful discussion on statistical analysis, and to Erica T Perrier, Muriel Derrien, Hana Koutnikova, Lodovico Di Gioia, Matthieu Pichaud, and Patrick Veiga (all of Danone Research) for the critical reading of the manuscript. The authors’ responsibilities were as follows—TV: designed the research; TV and JT: wrote the paper and had primary responsibility for the final content; and all authors: conducted the research, analyzed the data, performed the statistical analyses, and read and approved the final manuscript.
Notes
The authors reported no funding received for this study.
Author disclosures: TV and JT are full-time employees of Danone Research. OB was an intern at Danone Research at the time of analysis. MP is a consultant at IT&M Innovation and performs statistical analyses for Danone Research.
Supplemental Methods, Supplemental Figures 1–8, and Supplemental Table 1 are available from the “Supplementary data” link in the online posting of the article and from the same link in the online table of contents at https://academic.oup.com/jn/.
Abbreviations used: AGP, American Gut Project; AI, Adequate Intake; ASV, amplicon sequence variant; Faith's PD, Faith's phylogenetic diversity; PERMANOVA, permutational multivariate analysis of variance; rRNA, ribosomal RNA.
Contributor Information
Tiphaine Vanhaecke, Danone Research, Palaiseau, France.
Oriane Bretin, Danone Research, Palaiseau, France.
Marion Poirel, IT&M Innovation.
Julien Tap, Danone Research, Palaiseau, France.
References
- 1. Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, Bewtra M, Knights D, Walters WA, Knight R et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334(6052):105–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ding T, Schloss PD. Dynamics and associations of microbial community types across the human body. Nature. 2014;509(7500):357–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kundu P, Blacher E, Elinav E, Pettersson S. Our gut microbiome: the evolving inner self. Cell. 2017;171(7):1481–93. [DOI] [PubMed] [Google Scholar]
- 4. Relman DA. The human microbiome: ecosystem resilience and health. Nutr Rev. 2012;70(Suppl 1):S2–S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Institute of Medicine . Dietary Reference Intakes for water, potassium, sodium, chloride, and sulfate. Washington (DC): National Academies Press; 2005. [Google Scholar]
- 6. EFSA Panel on Dietetic Products, Nutrition, and Allergies (NDA) . Scientific opinion on dietary reference values for water. EFSA J. 2010;8(3):1459. [Google Scholar]
- 7. CDC . Community water treatment. [Internet]. Atlanta, GA: Centers for Disease Control and Prevention; 2015; [cited May 2021]. Available from: https://www.cdc.gov/healthywater/drinking/public/water_treatment.html. [Google Scholar]
- 8. Dai Z, Sevillano-Rivera MC, Calus ST, Bautista-de los Santos QM, Eren AM, van der Wielen PWJJ, Ijaz UZ, Pinto AJ. Disinfection exhibits systematic impacts on the drinking water microbiome. Microbiome. 2020;8(1):42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Willis JR, González-Torres P, Pittis AA, Bejarano LA, Cozzuto L, Andreu-Somavilla N, Alloza-Trabado M, Valentín A, Ksiezopolska E, Company C et al. Citizen science charts two major “stomatotypes” in the oral microbiome of adolescents and reveals links with habits and drinking water composition. Microbiome. 2018;6(1):218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Patterson KY, Pehrsson PR, Perry CR. The mineral content of tap water in United States households. J Food Compos Anal. 2013;31(1):46–50. [Google Scholar]
- 11. Leclerc H, Moreau A. Microbiological safety of natural mineral water. FEMS Microbiol Rev. 2002;26(2):207–22. [DOI] [PubMed] [Google Scholar]
- 12. De Roy K, Clement L, Thas O, Wang Y, Boon N. Flow cytometry for fast microbial community fingerprinting. Water Res. 2012;46(3):907–19. [DOI] [PubMed] [Google Scholar]
- 13. Zhang Y, Oh S, Liu W-T. Impact of drinking water treatment and distribution on the microbiome continuum: an ecological disturbance's perspective. Environ Microbiol. 2017;19(8):3163–74. [DOI] [PubMed] [Google Scholar]
- 14. Sacchetti R, De Luca G, Dormi A, Guberti E, Zanetti F. Microbial quality of drinking water from microfiltered water dispensers. Int J Hyg Environ Health. 2014;217(2–3):255–9. [DOI] [PubMed] [Google Scholar]
- 15. Rechenburg A. WHO CC Newsletter. Microbiomes: an undervalued impact of water treatment on human gut health?. 2019. December 29. https://www.ukbonn.de/site/assets/files/16453/water_and_risk_vol29_low.pdf [Google Scholar]
- 16. Dias MF, Reis MP, Acurcio LB, Carmo AO, Diamantino CF, Motta AM, Kalapothakis E, Nicoli JR, Nascimento AMA. Changes in mouse gut bacterial community in response to different types of drinking water. Water Res. 2018;132:79–89. [DOI] [PubMed] [Google Scholar]
- 17. Baumann-Dudenhoeffer AM, D'Souza AW, Tarr PI, Warner BB, Dantas G. Infant diet and maternal gestational weight gain predict early metabolic maturation of gut microbiomes. Nat Med. 2018;24(12):1822–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jha AR, Davenport ER, Gautam Y, Bhandari D, Tandukar S, Ng KM, Fragiadakis GK, Holmes S, Gautam GP, Leach J et al. Gut microbiome transition across a lifestyle gradient in Himalaya. PLoS Biol. 2018;16(11):e2005396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bowyer RCE, Schillereff DN, Jackson MA, Le Roy C, Wells PM, Spector TD, Steves CJ. Associations between UK tap water and gut microbiota composition suggest the gut microbiome as a potential mediator of health differences linked to water quality. Sci Total Environ. 2020;739:139697. [DOI] [PubMed] [Google Scholar]
- 20. Sofi MH, Gudi R, Karumuthil-Melethil S, Perez N, Johnson BM, Vasu C. pH of drinking water influences the composition of gut microbiome and type 1 diabetes incidence. Diabetes. 2014;63(2):632–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wolf KJ, Daft JG, Tanner SM, Hartmann R, Khafipour E, Lorenz RG. Consumption of acidic water alters the gut microbiome and decreases the risk of diabetes in NOD mice. J Histochem Cytochem. 2014;62(4):237–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Boorman GA. Drinking water disinfection byproducts: review and approach to toxicity evaluation. Environ Health Perspect. 1999;107(suppl 1):207–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sasada T, Hinoi T, Saito Y, Adachi T, Takakura Y, Kawaguchi Y, Sotomaru Y, Sentani K, Oue N, Yasui W et al. Chlorinated water modulates the development of colorectal tumors with chromosomal instability and gut microbiota in Apc-deficient mice. PLoS One. 2015;10(7):e0132435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Falony G, Joossens M, Vieira-Silva S, Wang J, Darzi Y, Faust K, Kurilshikov A, Bonder MJ, Valles-Colomer M, Vandeputte D et al. Population-level analysis of gut microbiome variation. Science. 2016;352(6285):560–4. [DOI] [PubMed] [Google Scholar]
- 25. Zhernakova A, Kurilshikov A, Bonder MJ, Tigchelaar EF, Schirmer M, Vatanen T, Mujagic Z, Vila AV, Falony G, Vieira-Silva S et al. Population-based metagenomics analysis reveals markers for gut microbiome composition and diversity. Science. 2016;352(6285):565–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Perrier ET, Armstrong LE, Bottin JH, Clark WF, Dolci A, Guelinckx I, Iroz A, Kavouras SA, Lang F, Lieberman HR et al. Hydration for health hypothesis: a narrative review of supporting evidence. Eur J Nutr. 2021;60(3):1167–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Benton D, Young HA. Water: the Cinderella nutrient. J Nutr. 2019;149(12):2081–2. [DOI] [PubMed] [Google Scholar]
- 28. McDonald D, Hyde E, Debelius JW, Morton JT, Gonzalez A, Ackermann G, Aksenov AA, Behsaz B, Brennan C, Chen Y et al. American gut: an open platform for citizen science microbiome research. mSystems. 2018;3(3):e00031–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. McDonald D, Kaehler B, Gonzalez A, DeReus J, Ackermann G, Marotz C, Huttley G, Knight R. redbiom: a rapid sample discovery and feature characterization system. mSystems. 2019;4(4):e00215–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gonzalez A, Navas-Molina JA, Kosciolek T, McDonald D, Vázquez-Baeza Y, Ackermann G, DeReus J, Janssen S, Swafford AD, Orchanian SB et al. Qiita: rapid, web-enabled microbiome meta-analysis. Nat Methods. 2018;15(10):796–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Statist Soc B. 1995;57(1):289–300. [Google Scholar]
- 32. Chao A. Nonparametric estimation of the number of classes in a population. Scand J Statist. 1984;11(4):265–70. [Google Scholar]
- 33. Shannon CE. A mathematical theory of communication. Bell Syst Tech J. 1948;27(3):379–423. [Google Scholar]
- 34. Faith DP. Conservation evaluation and phylogenetic diversity. Biol Conserv. 1992;61(1):1–10. [Google Scholar]
- 35. Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP et al. Human gut microbiome viewed across age and geography. Nature. 2012;486(7402):222–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489(7415):220–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sommer F, Anderson JM, Bharti R, Raes J, Rosenstiel P. The resilience of the intestinal microbiota influences health and disease. Nat Rev Microbiol. 2017;15(10):630–8. [DOI] [PubMed] [Google Scholar]
- 39. Le Chatelier E, Nielsen T, Qin J, Prifti E, Hildebrand F, Falony G, Almeida M, Arumugam M, Batto J-M, Kennedy S et al. Richness of human gut microbiome correlates with metabolic markers. Nature. 2013;500(7464):541–6. [DOI] [PubMed] [Google Scholar]
- 40. Valdes AM, Walter J, Segal E, Spector TD. Role of the gut microbiota in nutrition and health. BMJ. 2018;361:k2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cotillard A, Kennedy SP, Kong LC, Prifti E, Pons N, Le Chatelier E, Almeida M, Quinquis B, Levenez F, Galleron N et al. Dietary intervention impact on gut microbial gene richness. Nature. 2013;500(7464):585–8. [DOI] [PubMed] [Google Scholar]
- 42. Dupont C, Campagne A, Constant F. Efficacy and safety of a magnesium sulfate–rich natural mineral water for patients with functional constipation. Clin Gastroenterol Hepatol. 2014;12(8):1280–7. [DOI] [PubMed] [Google Scholar]
- 43. Bothe G, Coh A, Auinger A. Efficacy and safety of a natural mineral water rich in magnesium and sulphate for bowel function: a double-blind, randomized, placebo-controlled study. Eur J Nutr. 2017;56(2):491–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. US Environmental Protection Agency (EPA) . Learn about private well water. [Internet]. Washington (DC): US EPA; [cited May 2021]. Available from: https://www.epa.gov/privatewells/learn-about-private-water-wells. [Google Scholar]
- 45. CDC . Private ground water wells. [Internet]. Atlanta, GA: Centers for Disease Control and Prevention; 2014; [cited May 2021]. Available from: https://www.cdc.gov/healthywater/drinking/private/wells/. [Google Scholar]
- 46. Jeffery IB, O'Toole PW, Öhman L, Claesson MJ, Deane J, Quigley EM, Simrén M. An irritable bowel syndrome subtype defined by species-specific alterations in faecal microbiota. Gut. 2012;61(7):997–1006. [DOI] [PubMed] [Google Scholar]
- 47. European Commission Directorate-General for Health and Food Safety . Natural mineral waters and spring water. 2020; [Internet]. Brussels, Belgium: European Commission; [cited May 2021]. Available from: https://ec.europa.eu/food/safety/labelling_nutrition/mineral_waters_en. [Google Scholar]
- 48. European Parliament . Directive 2009/54/EC of the European Parliament and of the Council of 18 June 2009 on the exploitation and marketing of natural mineral waters. Brussels, Belgium: European Parliament; 2009. [Google Scholar]
- 49. Vandeputte D, Falony G, Vieira-Silva S, Tito RY, Joossens M, Raes J. Stool consistency is strongly associated with gut microbiota richness and composition, enterotypes and bacterial growth rates. Gut. 2016;65(1):57–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tap J, Derrien M, Törnblom H, Brazeilles R, Cools-Portier S, Doré J, Störsrud S, Le Nevé B, Öhman L, Simrén M. Identification of an intestinal microbiota signature associated with severity of irritable bowel syndrome. Gastroenterology. 2017;152(1):111–23.e8. [DOI] [PubMed] [Google Scholar]
- 51. van den Bogert B, Erkus O, Boekhorst J, de Goffau M, Smid EJ, Zoetendal EG, Kleerebezem M. Diversity of human small intestinal Streptococcus and Veillonella populations. FEMS Microbiol Ecol. 2013;85(2):376–88. [DOI] [PubMed] [Google Scholar]
- 52. The Human Microbiome Project Consortium . Structure, function and diversity of the healthy human microbiome. Nature. 2012;486(7402):207–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zeller G, Tap J, Voigt AY, Sunagawa S, Kultima JR, Costea PI, Amiot A, Böhm J, Brunetti F, Habermann N et al. Potential of fecal microbiota for early-stage detection of colorectal cancer. Mol Syst Biol. 2014;10(11):766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Qin N, Yang F, Li A, Prifti E, Chen Y, Shao L, Guo J, Le Chatelier E, Yao J, Wu L et al. Alterations of the human gut microbiome in liver cirrhosis. Nature. 2014;513(7516):59–64. [DOI] [PubMed] [Google Scholar]
- 55. Ayeni FA, Biagi E, Rampelli S, Fiori J, Soverini M, Audu HJ, Cristino S, Caporali L, Schnorr SL, Carelli V et al. Infant and adult gut microbiome and metabolome in rural Bassa and urban settlers from Nigeria. Cell Rep. 2018;23(10):3056–67. [DOI] [PubMed] [Google Scholar]
- 56. Tyakht AV, Kostryukova ES, Popenko AS, Belenikin MS, Pavlenko AV, Larin AK, Karpova IY, Selezneva OV, Semashko TA, Ospanova EA et al. Human gut microbiota community structures in urban and rural populations in Russia. Nat Commun. 2013;4(1):2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Vujkovic-Cvijin I, Sklar J, Jiang L, Natarajan L, Knight R, Belkaid Y. Host variables confound gut microbiota studies of human disease. Nature. 2020;587(7834):448–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Derrien M, Alvarez AS, de Vos WM. The gut microbiota in the first decade of life. Trends Microbiol. 2019;27(12):997–1010. [DOI] [PubMed] [Google Scholar]
- 59. Igwaran A, Okoh AI. Human campylobacteriosis: a public health concern of global importance. Heliyon. 2019;5(11):e02814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Anti M, Pignataro G, Armuzzi A, Valenti A, Iascone E, Marmo R, Lamazza A, Pretaroli AR, Pace V, Leo P et al. Water supplementation enhances the effect of high-fiber diet on stool frequency and laxative consumption in adult patients with functional constipation. Hepatogastroenterology. 1998;45(21):727–32. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.