ABSTRACT
Background
Although diets rich in carotenoids are associated with reduced risks of cardiovascular disease, age-related macular degeneration, disability, and other adverse aging outcomes, the underlying biological mechanisms are not fully elucidated.
Objectives
To characterize the plasma proteome fingerprint associated with circulating carotenoid and retinol concentrations in older adults.
Methods
In 728 adults ≥65 y participating in the Invecchiare in Chianti (InCHIANTI) Study, plasma α-carotene, β-carotene, β-cryptoxanthin, lutein, zeaxanthin, and lycopene were measured using HPLC. The SOMAscan assay was used to measure 1301 plasma proteins. Multivariable linear regression models were used to examine the relationship of individual carotenoids and retinol with plasma proteins. A false discovery rate approach was used to deal with multiple comparisons using a q-value < 0.05.
Results
Plasma β-carotene, β-cryptoxanthin, lutein, zeaxanthin, and lycopene were associated with 85, 39, 4, 2, and 5 plasma proteins, respectively, in multivariable linear regression models adjusting for potential confounders (q < 0.05). No proteins were associated with α-carotene or retinol. Two or more carotenoids were positively associated with ferritin, 6-phosphogluconate dehydrogenase (decarboxylating), hepcidin, thrombospondin-2, and choline/ethanolamine kinase. The proteins associated with circulating carotenoids were related to energy metabolism, sirtuin signaling, inflammation and oxidative stress, iron metabolism, proteostasis, innate immunity, and longevity.
Conclusions
The plasma proteomic fingerprint associated with elevated circulating carotenoids in older adults provides insight into the mechanisms underlying the protective role of carotenoids on health.
Keywords: carotene, carotenoid, cryptoxanthin, lutein, lycopene, protein, proteomics, zeaxanthin, retinol
Introduction
Carotenoids are natural pigments found in most fruits and vegetables, algae, and photosynthetic bacteria. Humans cannot synthesize carotenoids and must obtain them through diet or supplementation. Carotenoids have a conjugated polyene structure that allows these molecules to serve as efficient quenchers of singlet oxygen. Carotenoids can accept electrons from reactive oxygen species and neutralize free radicals. While carotenoids show potent antioxidant effects in in vitro and ex vivo studies, provitamin A carotenoids can also serve as precursors to retinol, which plays a role in growth factor signaling, cell growth and differentiation, gap junctional communication, and immunoregulation (1). Lutein and zeaxanthin are found in the macula in high concentrations, where they are thought to protect photoreceptors from light-induced oxidative damage (2). A high dietary intake or high concentrations of circulating carotenoids have been associated with reduced risks of cardiovascular disease (3, 4), diabetes (5), age-related macular degeneration (6), disability (7), some cancers (8–10), and all-cause mortality (11).
The most abundant carotenoids in human plasma are α-carotene, β-carotene, β-cryptoxanthin, lutein, zeaxanthin, and lycopene. These can be further divided into carotenes (α-carotene, β-carotene, lycopene), which contain a parent hydrocarbon chain without any functional units, and xanthophylls (β-cryptoxanthin, lutein, and zeaxanthin), which are oxidized carotenoids. Since they can serve as precursors to vitamin A, α-carotene, β-carotene, and β-cryptoxanthin are considered provitamin A carotenoids. Plasma carotenoids are considered biomarkers of diets rich in green leafy vegetables and fruits (7).
Recent advances in proteomics allow the identification and quantification of hundreds to thousands of proteins in biological fluids and tissues. Although diets rich in carotenoids are considered to be healthy, the biological mechanisms by which high circulating carotenoids may protect against age-related conditions and diseases are not well understood. One approach to further understand the mechanisms underlying the protective effect of carotenoids is to identify a “fingerprint” of circulating proteins that are associated with high circulating carotenoid and retinol concentrations. In order to provide further insight into the biological pathways that may be associated with carotenoids, we conducted an agnostic discovery analysis to identify a proteomic fingerprint associated with circulating carotenoids and retinol in older adults.
Methods
The study participants consisted of men and women, aged ≥65 y, who participated in the Invecchiare in Chianti, “Aging in the Chianti Area” (InCHIANTI) Study, conducted in 2 small towns in Tuscany, Italy. The rationale, design, and data collection have been described elsewhere (12). The original screening and enrollment in the InCHIANTI Study are summarized as follows: in August 1998, 1299 subjects ≥65 y and 431 subjects from age strata 20–29, 30–39, 40–49, 50–59, and 60–64 y were randomly selected from a population registry of 2 sites in the region of Florence, from Greve, in Chianti (population 11,709), and Bagno a Ripoli (population 4704). Of 1749 eligible subjects, 1453 (86.1%) agreed to participate. The baseline round of data collection was conducted in 1998–2000. In this cross-sectional analysis, there were 728 participants at baseline ≥65 y who had both plasma carotenoid measurements and plasma protein measurements (Supplemental Figure 1). All participants received an extensive description of the study and participated after written, informed consent. The study protocol complied with the Declaration of Helsinki and was approved by the Italian National Institute of Research and Care on Aging Ethical Committee.
Information on demographics, including the total number years of formal schooling and smoking, alcohol, and medication use, were collected using standardized questionnaires. All participants were examined by a project geriatrician, and diseases were ascertained according to standard, preestablished criteria and algorithms based upon those used in the Women's Health and Aging Study for coronary heart disease, diabetes mellitus, stroke, and cancer (13). Weight was measured using a high-precision mechanical scale. Standing height was measured to the nearest 0.1 cm. BMI was calculated as weight/height2 (kg/m2). The Mini-Mental Status Examination (MMSE) was administered at enrollment (14). Commercial enzymatic tests (Roche Diagnostics) were used for measuring serum total cholesterol, triglycerides, and HDL cholesterol concentrations. LDL cholesterol was calculated by the Friedewald formula (15). High-sensitivity serum C-reactive protein (CRP) was measured by ELISA, using purified protein and polyclonal anti-CRP antibodies (Calbiochem).
Laboratory studies
Blood collection and storage
Blood samples were collected in the morning after a 12-h fast and after a 15-min rest. Blood samples were immediately stored at 4°C, centrifuged within 4 h, then immediately aliquoted and frozen at −80°C. Plasma samples were shipped on dry ice to RDS’ laboratory (the Semba laboratory) for measurements of plasma carotenoids (α-carotene, β-carotene, β-cryptoxanthin, lutein, zeaxanthin, and lycopene) and retinol. Plasma samples were stored at -80°C in the Semba laboratory and not thawed until analysis in 2007.
Plasma carotenoid and retinol analyses
Carotenoids and retinol were measured using a method validated by the US CDC and used for the National Eye Institute Age-Related Eye Disease Study 2 (16). Briefly, we used a chromatographic system consisting of a Waters 2695 separations module and a Waters 996 photodiode array detector with Maxima software (all Waters Corporation). All sample preparation was conducted at 27°C under gold fluorescent lights to avoid photooxidation of the analytes. Fat-soluble compounds were extracted from human plasma by combining 200 μL of plasma with 200 μL of ethanol containing nonapreno-β-carotene and retinyl butyrate as internal standards in a glass 12 × 75 mm culture tube. Samples were vortex mixed for 10 seconds, after which 1000 μL of hexane were added. Samples were vortex mixed for another 30 seconds. The tubes were centrifugated for 5 min at 1500 g to separate the phases. The hexane layer (900 μL) was transferred to a 12 × 75 mm tube and dried under a stream of nitrogen gas to a waxy or glassy consistency. The residue was dissolved by adding 100 μL of ethanol. Acetonitrile (100 μL) was added to the tube, followed by vortex mixing and filtering through a 0.45-μm pore-size filter (Millipore) into an injection vial. The following standards were used: retinol, retinyl palmitate, α-carotene, β-carotene, lycopene (Sigma Chemical Corp.), lutein, zeaxanthin, β-cryptoxanthin (Hoffmann-LaRoche), and nonapreno-β-carotene, 2’3’-anhydrolutein (courtesy of Frederick Khachik, Nutrient Composition Laboratory, US Department of Agriculture), and apo-8’-carotenal (Fluka Chemie AG). Columns, solvents, and gradients were used and run as described on pages 96–103 of the Age-Related Eye Disease Study protocol (16).
The Semba laboratory regularly participated in the biannual round robin proficiency testing program for retinol, retinyl palmitate, α- and γ-tocopherol, lutein, zeaxanthin, β-cryptoxanthin, lycopene, and α- and β-carotene sponsored by the National Institute of Standards and Technology, Gaithersburg, MD. Our laboratory had highly consistent and accurate laboratory performance for plasma retinol and carotenoid analyses. Within-run and between-run CVs were, respectively, 7.3 and 9.6% for α-carotene, 4.5 and 5.4% for β-carotene, 2.7 and 3.5% for β-cryptoxanthin, 2.6 and 7.1% for lutein, 6.2 and 6.8% for zeaxanthin, 7.5 and 7.8% for lycopene, and 2.8% and 3.3% for retinol.
Plasma protein analyses
The collection of EDTA plasma in the InCHIANTI Study was consistent with guidelines for protein biomarker work (17). Preanalytical studies show that plasma proteins are stable for 14–17 y in storage at −80°C and for up to 25 freeze-thaw cycles (18, 19). Plasma proteins were measured using the 1.3k HTS SOMAscan assay (SOMALogic) at the Trans‐NIH Center for Human Immunology, Autoimmunity, and Inflammation (CHI), National Institute of Allergy and Infectious Disease, National Institutes of Health. The plasma protein concentrations were expressed in relative fluorescence units. The protein abundance data were normalized as follows: hybridization control normalization removed individual sample variance on the basis of signaling differences between the microarray or Agilent scanner; median signal normalization removed inter-sample differences within a plate due to technical differences such as pipetting variation; calibration normalization removed variance across assay runs; and interplate normalization procedures using CHI site-specific calibrators from pooled healthy donors were performed to allow quality control of the normalization across all experiments conducted at the CHI (20). An interactive Shiny web tool was used during the CHI quality control process (20). The overall technical variability of the assay is low, with a median intraplate CV in the ∼3% to 4% range. The SOMAscan assay platform includes 1305 SOMAmer reagents, and 4 are nonspecifically targeted SOMAmers. A final analysis was conducted with 1301 human proteins in this study. The reference source for protein names was the UniProt designation. The reference sources for protein function were UniProt and String (String Consortium 2020: Swiss Institute of Bioinformatics, Novo Nordisk Foundation Center Protein Research, and European Molecular Biology Laboratory).
Statistical analysis
Differences in baseline characteristics across total plasma carotenoids were tested using the bivariate linear regression model. The distributions of the individual plasma carotenoids were evaluated using the Kolmogorov-Smirnov test using the normal distribution for reference. Since all plasma carotenoid concentrations were positively skewed, they were log transformed. Multivariate linear regression models were used to examine relationships between log plasma carotenoids or retinol and plasma proteins, adjusting for age, sex, alcohol use, smoking status, BMI, CRP, total cholesterol, MMSE score, heart failure, stroke, and diabetes. All plasma proteins were used in the same linear multivariable model for each respective carotenoid or retinol. Data analyses were performed using SPSS version 25.0 for Windows (IBM Corp.) and R Studio version 1.2.1335 (2009–2019 RStudio, Inc.). A Benjamini and Hochberg (21) false discovery rate approach was used to adjust for multiple testing (22). A q-value cutoff of <0.05 was used to define statistically significant associations (22). The q-value represents correction for 1301 comparisons.
Results
The demographic and health characteristics of the study population are shown in Table 1. Out of 728 adults, the mean age was 73.6 y, and 55.8% were female. Bivariate associations between demographic and disease variables and total plasma carotenoids are shown in Table 2. Sex (female compared with males), a lower frequency of alcohol intake, being a current smoker, and having a lower BMI, a lower CRP, higher total cholesterol, higher HDL cholesterol, higher LDL cholesterol, an MMSE score <22, heart failure, stroke, and diabetes were significantly associated with total plasma carotenoids.
TABLE 1.
Baseline characteristics of 728 adults in the InCHIANTI Study
| Characteristics | Mean ± SD or % |
|---|---|
| Age, y | 73.6 ± 6.4 |
| Sex, female, % | 55.8 |
| Education, y | 5.5 ± 3.3 |
| Alcohol intake, g/d | 15.1 ± 21.0 |
| Current smoker, % | 14.2 |
| BMI, kg/m2 | 27.5 ± 4.1 |
| Physical activity, % | |
| Inactive | 71.8 |
| Low | 6.9 |
| Moderate to high | 21.3 |
| Serum C-reactive protein, μg/mL | 0.91 ± 0.18 |
| Serum total cholesterol, mg/dL | 219.5 ± 38.6 |
| Serum HDL cholesterol, mg/dL | 56.1 ± 14.9 |
| Serum LDL cholesterol, mg/dL | 137.8 ± 34.4 |
| Serum triglycerides, mg/dL | 128.3 ± 68.9 |
| Mini-Mental State Exam score <22, % | 12.5 |
| Hypertension, % | 47.3 |
| Coronary artery disease, % | 1.1 |
| Heart failure, % | 3.3 |
| Peripheral artery disease, % | 8.9 |
| Stroke, % | 4.3 |
| Diabetes mellitus, % | 7.4 |
| Cancer, % | 6.2 |
TABLE 2.
Bivariate relationship between demographic and disease variables with total plasma carotenoids among 728 adults in the InCHIANTI Study
| Characteristics | Beta | SE | P value |
|---|---|---|---|
| Age, y | −0.040 | 0.004 | 0.28 |
| Sex, female, % | 0.126 | 0.049 | 0.001 |
| Education, y | 0.028 | 0.007 | 0.45 |
| Alcohol intake, g/d | −0.118 | 0.001 | 0.001 |
| Current smoker, % | −0.094 | 0.070 | 0.01 |
| BMI, kg/m2 | −0.092 | 0.006 | 0.01 |
| Physical activity, % | −0.008 | 0.030 | 0.84 |
| Log serum C-reactive protein, μg/mL | −0.115 | 0.285 | 0.002 |
| Serum total cholesterol, mg/dL | 0.305 | 0.001 | <0.0001 |
| Serum HDL cholesterol, mg/dL | 0.106 | 0.002 | 0.004 |
| Serum LDL cholesterol, mg/dL | 0.299 | 0.001 | <0.0001 |
| Serum triglycerides, mg/dL | −0.008 | 0.0004 | 0.83 |
| Mini-Mental State Exam score <22, % | −0.102 | 0.074 | 0.006 |
| Hypertension, % | 0.010 | 0.049 | 0.79 |
| Coronary artery disease, % | 0.058 | 0.236 | 0.12 |
| Heart failure, % | −0.082 | 0.138 | 0.03 |
| Peripheral artery disease, % | 0.016 | 0.086 | 0.66 |
| Stroke, % | −0.084 | 0.122 | 0.02 |
| Diabetes mellitus, % | −0.097 | 0.094 | 0.009 |
| Cancer, % | −0.018 | 0.102 | 0.62 |
Abbreviation: InCHIANTI, Invecchiare in Chianti.
The mean (± SD) plasma concentrations of α-carotene, β-carotene, β-cryptoxanthin, lutein, zeaxanthin, lycopene, and retinol were 0.04 ± 0.04, 0.37 ± 0.29, 0.17 ± 0.15, 0.38 ± 0.18, 0.06 ± 0.03, 0.64 ± 0.42, and 2.51 ± 0.39 μmol/L, respectively. The proportion of each of the 6 carotenoids as part of total carotenoid concentrations is shown in Figure 1. Lycopene was the most abundant carotenoid in plasma, accounting for 38.5% of total carotenoids. Alpha-carotene was the least abundant of the 6 carotenoids, accounting for 2.7% of total carotenoids.
FIGURE 1.
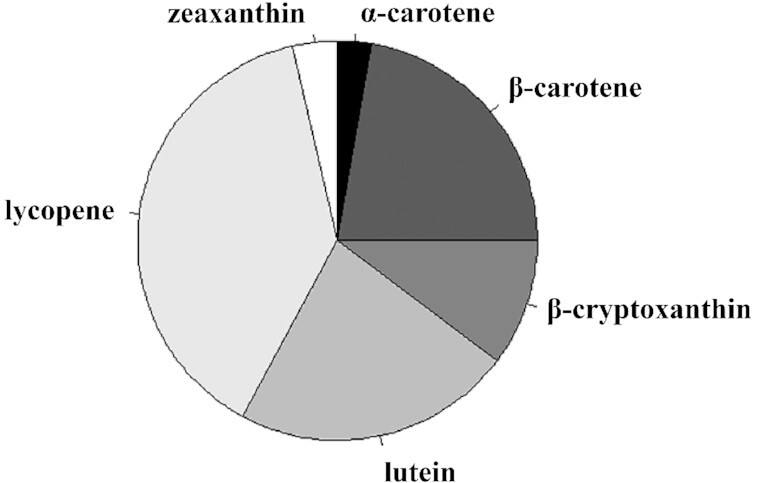
Pie diagram showing the relative abundance of plasma carotenoids in 728 adults, ≥65 y, living in rural Tuscany, Italy.
The relationships of individual plasma carotenoids and retinol with plasma proteins were assessed using multivariable linear regression models adjusted for age, sex, alcohol use, smoking status, BMI, CRP, total cholesterol, MMSE score, heart failure, stroke, and diabetes (Supplemental Table 1). There were no proteins associated with plasma α-carotene or retinol at a q-value < 0.05. Plasma β-carotene was associated with 85 plasma proteins (Table 3). There were 60 proteins positively associated and 25 plasma proteins negatively associated with plasma β-carotene (Supplemental Tables 2and3). Sixteen plasma proteins were positively associated and 23 proteins were negatively associated with plasma β-cryptoxanthin (Table 3; Supplemental Tables 2 and 3). Four proteins were negatively associated with plasma lutein (Table 3; Supplemental Table 3). There were 2 proteins negatively associated with zeaxanthin (Table 3; Supplemental Table 3). Three proteins were positively associated and 1 protein was negatively associated with plasma lycopene (Table 3; Supplemental Tables 2 and 3). We ran an alternative multivariable model with all the variables included in the final multivariable model, but excluded total cholesterol. In the alternative model, the results were markedly similar, with no changes in the proteins that were significant between the final multivariable model and the alternative model (data not shown).
TABLE 3.
Significant plasma proteins associated with carotenoids in older adults in the InCHIANTI Study
| Protein name | Gene | β | SE | P value | q-value |
|---|---|---|---|---|---|
| Beta-carotene | |||||
| Neuronal growth regulator 1 | NEGR1 | 0.507 | 0.105 | <0.0001 | 0.011 |
| Matrilin-2 | MATN2 | 0.463 | 0.106 | <0.0001 | 0.011 |
| Neurogenic locus notch homolog protein 1 | NOTCH1 | 0.421 | 0.133 | 0.002 | 0.034 |
| Limbic system–associated membrane protein | LSAMP | 0.404 | 0.107 | 0.0002 | 0.017 |
| Epidermal growth factor receptor | EGFR | 0.388 | 0.107 | 0.0003 | 0.017 |
| Hepatocyte growth factor receptor | MET | 0.379 | 0.100 | 0.0002 | 0.017 |
| Kin of IRRE-like protein 3 | KIRREL3 | 0.362 | 0.095 | 0.0002 | 0.017 |
| Repulsive guidance molecule domain family member B | RGMB | 0.348 | 0.109 | 0.001 | 0.022 |
| CD59 glycoprotein | CD59 | 0.339 | 0.108 | 0.002 | 0.034 |
| Tumor necrosis factor ligand superfamily member 8 | TNFRSF8 | 0.338 | 0.094 | 0.0004 | 0.017 |
| Neural cell adhesion molecule 1 | NCAM1 | 0.336 | 0.095 | 0.0004 | 0.017 |
| Vascular endothelial growth factor receptors 2 | KDR | 0.329 | 0.105 | 0.002 | 0.034 |
| Tyrosine-protein kinase receptor Tie-1 | TIE1 | 0.314 | 0.099 | 0.002 | 0.034 |
| Complement decay-accelerating factor | CD55 | 0.310 | 0.102 | 0.003 | 0.046 |
| Cadherin-5 | CDH5 | 0.309 | 0.086 | 0.0003 | 0.017 |
| OX-2 membrane glycoprotein | CD200 | 0.305 | 0.088 | 0.001 | 0.022 |
| Angiopoietin-1 receptor | TEK | 0.300 | 0.089 | 0.001 | 0.022 |
| Dermatopontin | DPT | 0.289 | 0.085 | 0.001 | 0.022 |
| Brain derived neurotrophic factor /Neurotrophin-3 growth factors receptor | NTRK2 | 0.286 | 0.095 | 0.003 | 0.046 |
| Delta-like protein 4 | DLL4 | 0.286 | 0.068 | <0.0001 | 0.011 |
| Melanoma-derived growth regulatory protein | MIA | 0.282 | 0.082 | 0.001 | 0.022 |
| Insulin-like growth factor–binding protein 6 | IGFBP6 | 0.280 | 0.089 | 0.002 | 0.034 |
| Superoxide dismutase [Mn], mitochondrial | SOD2 | 0.278 | 0.090 | 0.002 | 0.034 |
| Ficolin-2 | FCN2 | 0.269 | 0.083 | 0.001 | 0.022 |
| Interleukin-6 receptor subunit alpha | IL6R | 0.265 | 0.067 | 0.0001 | 0.011 |
| Neural cell adhesion molecule L1 | L1CAM | 0.256 | 0.075 | 0.001 | 0.022 |
| Insulin-like growth factor–binding protein 3 | IGFBP3 | 0.253 | 0.065 | 0.0001 | 0.011 |
| Interleukin-34 | IL34 | 0.250 | 0.074 | 0.001 | 0.022 |
| Collagen alpha-1(XVIII) chain | COL18A1 | 0.247 | 0.084 | 0.003 | 0.046 |
| Ectodysplasin-A | EDA | 0.234 | 0.059 | 0.0001 | 0.011 |
| Ciliary neurotrophic factor receptor subunit alpha | CNTFR | 0.232 | 0.064 | 0.0003 | 0.017 |
| NT-3 growth factor receptor | NTRK3 | 0.231 | 0.062 | 0.0002 | 0.017 |
| Ephrin-A5 | EFNA5 | 0.227 | 0.066 | 0.001 | 0.022 |
| Oncostatin-M | OSM | 0.221 | 0.069 | 0.001 | 0.022 |
| CD109 antigen | CD109 | 0.220 | 0.054 | 0.0001 | 0.011 |
| Immunoglobulin superfamily containing leucine-rich repeat protein 2 | ISLR2 | 0.219 | 0.068 | 0.001 | 0.022 |
| Carbonic anhydrase 4 | CA4 | 0.213 | 0.061 | 0.001 | 0.022 |
| Tropomyosin beta chain | TPM2 | 0.209 | 0.070 | 0.003 | 0.046 |
| Cell adhesion molecule 1 | CADM1 | 0.201 | 0.061 | 0.001 | 0.022 |
| Nicotinamide phosphoribosyltransferase | NAMPT | 0.199 | 0.051 | 0.0001 | 0.011 |
| Endoglin | ENG | 0.199 | 0.059 | 0.001 | 0.022 |
| Tumor necrosis factor ligand superfamily member 15 | TNFSF15 | 0.196 | 0.061 | 0.001 | 0.022 |
| Interleukin-1 receptor accessory protein | IL1RAP | 0.195 | 0.060 | 0.001 | 0.022 |
| Dual specificity mitogen-activated protein kinase kinase 4 | MAP2K4 | 0.191 | 0.061 | 0.002 | 0.034 |
| Iduronate 2-sulfatase | IDS | 0.190 | 0.062 | 0.002 | 0.034 |
| Myoglobin | MB | 0.189 | 0.060 | 0.002 | 0.034 |
| Mast/stem cell growth factor receptor kit | KIT | 0.188 | 0.057 | 0.001 | 0.022 |
| Netrin receptor UNC5D | UNC5D | 0.187 | 0.060 | 0.002 | 0.034 |
| Sialoadhesin | SIGLEC1 | 0.183 | 0.053 | 0.001 | 0.022 |
| Growth/differentiation factor 8 | MSTN | 0.180 | 0.054 | 0.001 | 0.022 |
| Contactin-2 | CNTN2 | 0.180 | 0.054 | 0.001 | 0.022 |
| Interleukin-1 receptor type 2 | IL1R1 | 0.178 | 0.059 | 0.003 | 0.046 |
| PILR alpha-associated neural protein | PIANP | 0.178 | 0.057 | 0.002 | 0.034 |
| Ephrin-B2 | EFNB2 | 0.177 | 0.056 | 0.002 | 0.034 |
| Interleukin-17 receptor C | IL17RC | 0.175 | 0.059 | 0.003 | 0.046 |
| G2/mitotic-specific cyclin-B1 | CCNB1 | 0.166 | 0.047 | 0.0004 | 0.017 |
| Trefoil factor 2 | TFF2 | 0.147 | 0.047 | 0.002 | 0.034 |
| Complement C3b | C3 | 0.145 | 0.041 | 0.0004 | 0.017 |
| Leukotriene A-4 hydrolase | LTA4H | 0.136 | 0.046 | 0.003 | 0.046 |
| Myeloid cell surface antigen CD33 | CD33 | 0.130 | 0.044 | 0.003 | 0.046 |
| 6-phosphogluconate dehydrogenase, decarboxylating | PGD | −0.078 | 0.023 | 0.001 | 0.022 |
| Signal transducer and activator of transcription 1-alpha/beta | STAT1 | −0.081 | 0.027 | 0.003 | 0.046 |
| Proto-oncogene vav | VAV1 | −0.084 | 0.028 | 0.003 | 0.046 |
| Platelet glycoprotein VI | GP6 | −0.093 | 0.032 | 0.003 | 0.046 |
| Interstitial collagenase | MMP1 | −0.098 | 0.028 | 0.0004 | 0.017 |
| Methionine aminopeptidase 2 | METAP2 | −0.102 | 0.032 | 0.001 | 0.022 |
| N-6 adenine-specific DNA methyltransferase 1 | N6AMT1 | −0.110 | 0.036 | 0.003 | 0.046 |
| Inosine-5'-monophosphate dehydrogenase 2 | IMPDH2 | −0.118 | 0.059 | 0.001 | 0.022 |
| Protein kinase C theta type | PRKCQ | −0.136 | 0.039 | 0.0004 | 0.017 |
| Coiled-coils motif chemokine 28 | CCL28 | −0.137 | 0.044 | 0.002 | 0.034 |
| Midkine | MDK | −0.144 | 0.042 | 0.001 | 0.022 |
| Choline/ethanolamine kinase | CHKB | −0.151 | 0.050 | 0.003 | 0.046 |
| Lysosomal protective protein | CTSA | −0.162 | 0.052 | 0.002 | 0.034 |
| Nucleoside diphosphate kinase A | NME1 | −0.175 | 0.055 | 0.002 | 0.034 |
| Ras GTPase-activating protein 1 | RASA1 | −0.176 | 0.055 | 0.002 | 0.034 |
| Non-receptor tyrosine-protein kinase tyrosine kinase 2 | TYK2 | −0.179 | 0.059 | 0.003 | 0.046 |
| Dual specificity mitogen-activated protein kinase kinase 3 | MAP2K3 | −0.181 | 0.055 | 0.001 | 0.022 |
| C-reactive protein | CRP | −0.197 | 0.034 | 0.001 | 0.022 |
| Aminoacylase-1 | ACY1 | −0.199 | 0.044 | <0.0001 | 0.011 |
| AH receptor-interacting protein | AIP | −0.200 | 0.054 | 0.0002 | 0.017 |
| N-acetyl-D-glucosamine kinase | NAGK | −0.210 | 0.053 | 0.0001 | 0.011 |
| Vascular endothelial growth factor C | VEGFC | −0.219 | 0.067 | 0.001 | 0.022 |
| Laminin subunit alpha-1/laminin subunit beta-1 | LAMA1 LAMB1 LAMC1 | −0.316 | 0.082 | 0.0001 | 0.011 |
| Metalloproteinase inhibitor 1 | TIMP1 | −0.321 | 0.091 | 0.0005 | 0.020 |
| Complement C3 | C3 | −0.372 | 0.097 | 0.0001 | 0.011 |
| Beta-cryptoxanthin | |||||
| Vesicular integral-membrane protein VIP36 | LMAN2 | 0.516 | 0.100 | <0.0001 | 0.003 |
| Opioid-binding protein/cell adhesion molecule | OPCML | 0.512 | 0.112 | <0.0001 | 0.003 |
| Dickkopf-like protein 1 | DKKL1 | 0.478 | 0.097 | <0.0001 | 0.003 |
| Calcineurin subunit B type 1 | PPP3R1 | 0.386 | 0.105 | 0.0003 | 0.016 |
| Clusterin | CLU | 0.375 | 0.115 | 0.001 | 0.036 |
| Ephrin type-B receptor 4 | EPHB4 | 0.373 | 0.095 | 0.0001 | 0.009 |
| Calcium-/calmodulin-dependent protein kinase type 1 | CAMK1 | 0.350 | 0.091 | 0.0001 | 0.009 |
| Cyclic guanosine monophosphate-dependent 3',5'-cyclic phosphodiesterase | PDE2A | 0.306 | 0.077 | 0.0001 | 0.005 |
| CCN family member 4 | WISP1 | 0.305 | 0.086 | 0.0004 | 0.024 |
| Choline/ethanolamine kinase | CHKB | 0.260 | 0.062 | <0.0001 | 0.005 |
| 14-3-3 protein theta | YWHAQ | 0.227 | 0.071 | 0.001 | 0.036 |
| ATP synthase subunit beta, mitochondrial | ATP5B | 0.225 | 0.067 | 0.001 | 0.036 |
| Transforming growth factor beta-2 proprotein | TGFB2 | 0.189 | 0.058 | 0.001 | 0.036 |
| Beta-Ala-His dipeptidase | CNDP1 | 0.146 | 0.041 | 0.0004 | 0.024 |
| 6-phosphogluconate dehydrogenase, decarboxylating | PGD | 0.094 | 0.028 | 0.008 | 0.036 |
| SRC kinase signaling inhibitor 1 | SRC | 0.078 | 0.022 | 0.0005 | 0.024 |
| Glyceraldehyde-3-phosphate dehydrogenase | GAPDH | −0.110 | 0.026 | <0.0001 | 0.005 |
| Creatine kinase B-type / Creatine kinase M-type | CKB CKM | −0.137 | 0.041 | 0.008 | 0.036 |
| SLAM family member 7 | SLAMF7 | −0.144 | 0.043 | 0.001 | 0.036 |
| Oxidized LDL receptor 1 | OLR1 | −0.200 | 0.060 | 0.001 | 0.036 |
| Thrombospondin-2 | THBS2 | −0.216 | 0.061 | 0.0005 | 0.024 |
| Vesicle transport protein glutamic-oxaloacetic transaminase 1 | GOT1 | −0.218 | 0.061 | 0.0004 | 0.024 |
| Bone morphogenetic protein 6 | BMP6 | −0.234 | 0.071 | 0.001 | 0.036 |
| Neuroligin-4, X-linked | NLGN4X | −0.237 | 0.073 | 0.001 | 0.036 |
| Semaphorin-6B | SEMA6B | −0.253 | 0.077 | 0.001 | 0.036 |
| Kallikrein-11 | KLK11 | −0.255 | 0.073 | 0.001 | 0.036 |
| Serotransferrin | F3 | −0.258 | 0.061 | <0.0001 | 0.005 |
| Periostin | POSTN | −0.270 | 0.072 | 0.0002 | 0.016 |
| Cathepsin L2 | CTSV | −0.283 | 0.066 | <0.0001 | 0.005 |
| Ephrin type-A receptor 2 | EPHA2 | −0.292 | 0.076 | 0.0001 | 0.009 |
| Eotaxin | CCL11 | −0.302 | 0.072 | <0.0001 | 0.005 |
| Growth/differentiation factor 15 | GDF15 | −0.307 | 0.079 | 0.0001 | 0.009 |
| Trefoil factor 3 | TFF3 | −0.320 | 0.085 | 0.0002 | 0.016 |
| Ephrin-A4 | EFNA4 | −0.336 | 0.091 | 0.0002 | 0.016 |
| Spondin-1 | SPON1 | −0.342 | 0.106 | 0.001 | 0.036 |
| Tumor necrosis factor receptor superfamily member 17 | TNFRSF17 | −0.343 | 0.097 | 0.0005 | 0.024 |
| Decorin | DCN | −0.436 | 0.122 | 0.0004 | 0.024 |
| Follistatin-related protein 3 | FSTL3 | −0.472 | 0.124 | 0.0002 | 0.016 |
| Cystatin-C | CST3 | −0.534 | 0.139 | 0.0001 | 0.009 |
| Lutein | |||||
| Hepcidin | HAMP | −0.046 | 0.013 | 0.0003 | 0.002 |
| Ferritin heavy chain/ferritin light chain | FTH1 FTL | −0.081 | 0.017 | <0.0001 | 0.002 |
| Ephrin type-A receptor 10 | EPHA10 | −0.129 | 0.033 | 0.0001 | 0.002 |
| Thrombospondin-2 | THBS2 | −0.145 | 0.035 | 0.0001 | 0.002 |
| Zeaxanthin | |||||
| Hepcidin | HAMP | −0.047 | 0.012 | 0.0001 | 0.049 |
| Ferritin heavy chain/ferritin light chain | FTH1 FTL | −0.065 | 0.016 | 0.0001 | 0.049 |
| Lycopene | |||||
| Quinone oxidoreductase-like protein 1 | CRYZL1 | 0.291 | 0.056 | <0.0001 | 0.010 |
| Stromelysin-1 | MMP3 | 0.266 | 0.068 | 0.0002 | 0.036 |
| Apolipoprotein E | apoE | 0.173 | 0.044 | 0.0001 | 0.025 |
| Ferritin heavy chain/ferritin light chain | FTH1 FTL | −0.095 | 0.023 | <0.0001 | 0.022 |
Abbreviation: InCHIANTI, Invecchiare in Chianti.
Each carotenoid had a unique protein fingerprint. However, there were 5 proteins that were associated with more than 1 carotenoid (Table 3). Ferritin [ferritin heavy chain (FTH1)/ferritin light chain (FTL)] was associated with 3 carotenoids, and 4 proteins [6-phosphogluconate dehydrogenase, decarboxylating; hepcidin (HAMP); thrombospondin-2 (THBS2); and choline/ethanolamine kinase] were associated with 2 carotenoids.
Discussion
In the present study, the proteomic fingerprint associated with the 6 major plasma carotenoids and retinol were characterized. To our knowledge, this is the first investigation of the plasma proteome in relation to circulating carotenoids in older adults. Eighty plasma proteins were positively associated and 59 plasma proteins were negatively associated with plasma carotenoids. Overall, the plasma protein fingerprint was unique for individual carotenoids. There were only 5 proteins associated with multiple carotenoids. No circulating proteins were associated with either plasma α-carotene or retinol. The contrasting fingerprints may reflect differences in the chemistry, metabolism, and function of individual carotenoids.
Plasma β-carotene was positively associated with ficolin-2, interleukin-1 receptor accessory protein (IL1RAP), G2/mitotic-specific cyclin-B1 (CCNB1), manganese superoxide dismutase (SOD2), and nicotinamide phosphoribosyltransferase (NAMPT). Plasma β-carotene was negatively associated with aminoacylase-1 (ACY1), a hydrolase that degrades amino acids. Ficolin-2 plays an important role in immune surveillance and innate immunity by binding molecules present on microorganisms and dying host cells to initiate activation of the lectin pathway of complement (23, 24). Lower circulating ficolin-2 concentrations are associated with bronchiectasis (25, 26) and pulmonary tuberculosis (27). Circulating IL1RAP may reduce inflammation by acting as a decoy receptor and binding the proinflammatory cytokine IL-1β (28). Plasma IL1RAP concentrations were negatively associated with obesity in adults (28).
CCNB1 coordinates mitochondrial respiration in the cell cycle at the G2/M (mitosis) transition (29). CCNB1 and cyclin-dependent kinase 1 (CDK1) bind to form cyclin B1/CDK1, a complex that regulates mitochondrial bioenergetics (30). Cyclin B1/CDK1 plays a role in the genotoxic stress response by phosphorylating SOD2 (31). SOD2 clears mitochondrial reactive oxygen species and protects against premature cell death (32). Cyclin B1/CDK1 enhances Sirtuin3 (SIRT3) activity in response to genotoxic stress (32). SIRT3 influences nutrient oxidation, ATP generation, detoxification of reactive oxygen species, and the mitochondrial unfolded protein response (32). SIRT3 has been strongly implicated in aging-related diseases (31).
NAMPT is a rate-limiting enzyme in nicotinamide adenine dinucleotide (NAD+) synthesis. NAD+ plays a vital role as a coenzyme for ATP synthesis, a co-factor in DNA repair mechanisms, and as a cosubstrate for various enzymes, including the sirtuins (33). The age-related systematic decline of NAD+ is thought to play a critical role in the health span and longevity (34). NAD+ is synthesized by the de novo pathway from dietary tryptophan in the kynurenine pathway or recycled from nicotinic acid via NAMPT in the salvage pathway. The salvage pathway and NAMPT provide the main source of NAD+. SIRT1 mediates the deacetylation of NAMPT and promotes its extracellular secretion (35). Higher circulating NAMPT levels have been shown to delay aging in mice (36). NAMPT/NAD+/SIRT1 has been hypothesized as the master time regulator that determines the biological age of an organism: the biological age of organs and tissues is regulated and synchronized through NAMPT secretion in the blood (37).
Plasma β-cryptoxanthin was positively associated with transforming growth factor beta-2 proprotein (TGFB2) and clusterin (CLU), and negatively associated with cathepsin L2 (CTSV) and Growth/differentiation factor 15 (GDF15). TGFB2 maintains TGF-beta in a latent state and is negatively associated with proteins involved in immune activation, such as Signaling lymphocytic activation molecule family member 7 eotaxin-1, which induces eosinophil chemotaxis, and tumor necrosis factor receptor superfamily member 17, which plays a role in humoral immunity. CLU influences the protein folding and is discussed further below. CTSV is a protease that degrades albumin, collagen, and other proteins. GDF15 plays a role in the regulation of food intake, energy expenditure, and body weight (38). GDF15 expression increases in tissues in response to cellular stress, is elevated in conditions such as cancer and cardiovascular disease (39), and independently predicts anemia (40), a decline in physical performance (41), and mortality (39).
Plasma lutein, zeaxanthin, and lycopene were negatively associated with plasma ferritin. Ferritin, the major iron storage protein of the body, is mostly located in the cytoplasm but is also secreted into the circulation, where it functions as an iron carrier (42). Iron is an essential nutrient that is required as a cofactor and enzymes for oxidative phosphorylation, oxygen transport, and metabolite oxidation (42). Excess iron can contribute to the formation of free radicals that can damage other biomolecules; thus, iron homeostasis is tightly regulated. The nuclear factor erythroid 2–related factor 2 (NRF2) regulates the labile iron pool and controls the transcription of FTH1 and FTL genes (43). The expression of ferritin is increased by oxidative stress through NRF2, and greater ferritin availability may protect cells against oxidative damage by reducing the labile iron pool (43). High levels of circulating carotenoids are associated with reduced oxidative stress and may play a role in modulating the low expression of circulating ferritin, but further evidence to support this hypothesis is needed from intervention studies and laboratory models.
Plasma lutein and lycopene were negatively associated with plasma hepcidin, a major regulator of iron metabolism. Hepcidin is secreted by the liver and inhibits the entry of iron into plasma from 3 main sources: the absorption of dietary iron in the duodenum, the release of recycled iron from macrophages, and the release of iron from hepatocytes (44). Hepcidin is regulated at the transcriptional level by intracellular and extracellular iron, inflammation, and increased erythropoietic activity (44).
Higher plasma carotenoids were associated with lower plasma levels of 3 proteins that have been identified as independent predictors of mortality: CRP (45), THBS2 (46), and GDF15 (38). THBS2 mediates cell-to-cell and cell-to-matrix interactions and has antiangiogenic effects mediated by CD36 (47). THBS2 is highly expressed during acute inflammation (48). Elevated circulating TBP2 has been associated with diabetes mellitus, atrial fibrillation, cardiovascular disease, cerebrovascular disease (48, 49), and cardiovascular mortality (48). The association of circulating carotenoids with mortality could also be due to the fact that people who do not eat fruits and vegetables have a shorter lifespan that is not causally related to carotenoids.
Of the 1301 proteins in the SOMAscan platform, 9 proteins (CD36, apoA1, apoE, apoB, LDLR, GSTP1, SOD2, CXCL8, and RBP4) are known to be specific to carotenoid uptake, transport, metabolism, and cleavage to vitamin A, or have been identified in genome-wide association studies to have single nucleotide polymorphisms associated with circulating retinol or carotenoid concentrations (50–54). Of the 9 proteins, only 2 were significantly associated with carotenoids (β-carotene and SOD2; lycopene and apoE). There are other important proteins involved in carotenoid metabolism—for example, Beta-Carotene Oxygenase 1, which cleaves provitamin A carotenoids to vitamin A, Beta-Carotene Oxygenase 2, and Niemann-Pick C1-Like 1—that were not among the circulating proteins measured by the SOMAscan platform. Moreover, some of the proteins involved with carotenoid and retinol metabolism are intracellular or membrane proteins that may not be found in the circulation.
The strengths of this study include a population-based sample of older adults, highly standardized data collection and laboratory assessments, use of a targeted proteomics platform, and a robust statistical approach that accounted for multiple comparisons to reduce the risk of false-positive results. The cross-sectional nature of the present study can only show associations and limits any inferences that can be made regarding causality. However, this discovery phase study is important for generating hypotheses that could be evaluated in interventional studies with dietary carotenoids.
The results of this study cannot be extrapolated to other study populations and age groups. The relationship between plasma carotenoids and plasma proteins was characterized in malnourished children, aged 6–8 y, in rural Nepal (55). There were 982 plasma proteins measured by mass spectrometry, of which 66 proteins were significantly associated with plasma carotenoids (β-carotene, β-cryptoxanthin, or lutein/zeaxanthin). Of the 66 significant proteins associated with circulating carotenoids in children (55), only 2 proteins had a similar association in older adults in the present study [plasma β-cryptoxanthin with beta Ala-His dipeptidase (CNDP1) and CLU]. CNDP1 is the rate-limiting enzyme in hydrolysis of carnosine, an antioxidant and free-radical scavenger, to β-alanine and L-histidine (56). CLU is a molecular chaperone that prevents aggregation of nonnative proteins and plays important roles in protein homeostasis/proteostasis, inhibition of cell death pathways, and modulation of prosurvival signaling and transcriptional networks (57). There were large differences in the methodology of protein assessments, participant ages, diets, nutritional statuses, and health between older adults in the present study and the study of children in Nepal. Further work could be done to address the hypothesis that carotenoids affect health through CNDP1 and CLU in intervention studies or laboratory models. Further studies are needed to fully characterize the proteomic profile of plasma carotenoids in humans in different populations.
Ideal diets that have been associated with optimal health and longevity, such as the Mediterranean diet and Japanese washoku diet, are rich in dietary carotenoids. A basic principle of washoku is that each meal should contain 5 colors: red, yellow, green, black, and white (58). The first 3 colors generally reflect carotenoid-rich foods. The reference diet proposed by the EAT-Lancet Commission, based upon the best nutritional evidence available for optimizing health, emphasizes high intakes of fruit, dark green vegetables, and red and orange vegetables (59).
In conclusion, the present study provides new insights into the potential biomarkers and biological mechanisms by which plasma carotenoids could contribute to health in older adults. The plasma proteomic fingerprint associated with elevated circulating carotenoids in older adults is characterized by proteins that are known to be related to sirtuins (NAMPT, CCNB1), inflammation and oxidative stress (CCNB1, SOD2, IL1RAP, CNDP1), iron metabolism (HAMP, ferritin), proteostasis (clusterin, CTSV, ACY1, CLU), innate immunity (ficolin-2), and longevity (CRP, GDF15, THBS2). Future studies are needed to examine the response of the plasma proteome to dietary interventions with carotenoids.
Supplementary Material
Acknowledgments
The authors’ contributions were as follows—RDS, LF: designed the study; YY, TT: created the data set; YY, MZ, RDS: analyzed the data; YY, RDS: drafted the manuscript; RDS: had primary responsibility for the final content; and all authors: read and approved the final manuscript.
Notes
The Invecchiare in Chianti study baseline (1998–2000) was supported as a “targeted project” (ICS110.1/RF97.71) by the Italian Ministry of Health and in part by the US National Institute on Aging (contracts 263 MD 9164 and 263 MD 821336) and was supported in part by the Intramural Research Program of the National Institute on Aging, National Institutes of Health, Baltimore, MD. This study was also supported by the National Institutes of Health R01 AG057723.
Author disclosure: The authors report no conflicts of interest.
Supplemental Figure 1 and Supplemental Tables 1–3 are available from the “Supplementary data” link in the online posting of the article and from the same link in the online table of contents at https://academic.oup.com/jn/.
Abbreviations used: ACY1, aminoacylase-1; CCNB1, G2/mitotic-specific cyclin-B1; CDK1, cyclin-dependent kinase 1; CHI, Center for Human Immunology, Autoimmunity, and Inflammation; CLU, clusterin; CNDP1, beta Ala-His dipeptidase; CRP, C-reactive protein; CTSV, cathepsin L2; FTH1, ferritin heavy chain; FTL, ferritin light chain; HAMP, hepcidin; IL1RAP, interleukin-1 receptor accessory protein; InCHIANTI, Invecchiare in Chianti; MMSE, Mini-Mental State Examination; NAD+, nicotinamide adenine dinucleotide; NAMPT, nicotinamide phosphoribosyltransferase; NRF2, nuclear factor erythroid 2–related factor 2; SIRT3, Sirtuin3; SOD2, superoxide dismutase [Mn], mitochondrial; TGFB2, transforming growth factor beta-2 proprotein; THBS2, thrombospondin-2.
Contributor Information
Yuko Yamaguchi, Wilmer Eye Institute, Johns Hopkins University School of Medicine, Baltimore, MD, USA; Graduate School of Health Sciences, Kobe University, Kobe, Hyogo, Japan.
Marta Zampino, National Institutes on Aging, National Institutes of Health, Baltimore, MD, USA.
Toshiko Tanaka, National Institutes on Aging, National Institutes of Health, Baltimore, MD, USA.
Stefania Bandinelli, Azienda USL TOSCANA CENTRO, Geriatric Unit, Firenze, Italy.
Ruin Moaddel, National Institutes on Aging, National Institutes of Health, Baltimore, MD, USA.
Giovanna Fantoni, National Institutes on Aging, National Institutes of Health, Baltimore, MD, USA.
Julián Candia, National Institutes on Aging, National Institutes of Health, Baltimore, MD, USA.
Luigi Ferrucci, National Institutes on Aging, National Institutes of Health, Baltimore, MD, USA.
Richard D Semba, Wilmer Eye Institute, Johns Hopkins University School of Medicine, Baltimore, MD, USA; Center for a Livable Future, Johns Hopkins Bloomberg School of Public Health, Baltimore, MD, USA.
References
- 1. Milani A, Basirnejad M, Shahbazi S, Bolhassani A. Carotenoids: Biochemistry, pharmacology and treatment. Br J Pharmacol. 2017;174(11):1290– 324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bernstein PS, Li B, Vachali PP, Gorusupudi A, Shyam R, Henriksen BS, Nolan JM. Lutein, zeaxanthin, and meso-zeaxanthin: The basic and clinical science underlying carotenoid-based nutritional interventions against ocular disease. Prog Retin Eye Res. 2016;50:34–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Aune D, Keum N, Giovannucci E, Fadnes LT, Boffetta P, Greenwood DC, Tonstad S, Vatten LJ, Riboli E, Norat T. Dietary intake and blood concentrations of antioxidants and the risk of cardiovascular disease, total cancer, and all-cause mortality: A systematic review and dose-response meta-analysis of prospective studies. Am J Clin Nutr. 2018;108(5):1069–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wang Y, Chung SJ, McCullough ML, Song WO, Fernandez ML, Koo SI, Chun OK. Dietary carotenoids are associated with cardiovascular disease risk biomarkers mediated by serum carotenoid concentrations. J Nutr. 2014;144(7):1067–74. [DOI] [PubMed] [Google Scholar]
- 5. Sluijs I, Cadier E, Beulens JW, van der A DL, Spijkerman AM, van der Schouw YT. Dietary intake of carotenoids and risk of type 2 diabetes. Nutr Metab Cardiovasc Dis. 2015;25(4):376–81. [DOI] [PubMed] [Google Scholar]
- 6. Ma L, Dou HL, Wu YQ, Huang YM, Huang YB, Xu XR, Zou ZY, Lin XM. Lutein and zeaxanthin intake and the risk of age-related macular degeneration: A systematic review and meta-analysis. Br J Nutr. 2012;107(3):350–9. [DOI] [PubMed] [Google Scholar]
- 7. Lauretani F, Semba RD, Bandinelli S, Dayhoff-Brannigan M, Lauretani F, Corsi AM, Guralnik JM, Ferrucci L. Carotenoids as protection against disability in older persons. Rejuvenation Res. 2008;11(3):557–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Slattery ML, Benson J, Curtin K, Ma KN, Schaeffer D, Potter JD. Carotenoids and colon cancer. Am J Clin Nutr. 2000;71(2):575–82. [DOI] [PubMed] [Google Scholar]
- 9. Aune D, Chan DS, Vieira AR, Navarro Rosenblatt DA, Vieira R, Greenwood DC, Norat T. Dietary compared with blood concentrations of carotenoids and breast cancer risk: A systematic review and meta-analysis of prospective studies. Am J Clin Nutr. 2012;96(2):356–73. [DOI] [PubMed] [Google Scholar]
- 10. Chen P, Zhang W, Wang X, Zhao K, Negi DS, Zhuo L, Qi M, Wang X, Zhang X. Lycopene and risk of prostate cancer: A systematic review and meta-analysis. Medicine (Baltimore). 2015;94(33):e1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shardell MD, Alley DE, Hicks GE, El-Kamary SS, Miller RR, Semba RD, Ferrucci L. Low-serum carotenoid concentrations and carotenoid interactions predict mortality in US adults: The Third National Health and Nutrition Examination Survey. Nutr Res. 2011;31(3):178–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ferrucci L, Bandinelli S, Benvenuti E, Di Iorio A, Macchi C, Harris TB, Guralnik JM. Subsystems contributing to the decline in ability to walk: Bridging the gap between epidemiology and geriatric practice in the InCHIANTI study. J Am Geriatr Soc. 2000;48(12):1618–25. [DOI] [PubMed] [Google Scholar]
- 13. Guralnik JM, Fried LP, Simonsick EM, Kasper JD, Lafferty ME. The Women's Health and Aging Study: Health and social characteristics of older women with disability. Bethesda, MD: National Institute on Aging; 1995. [Google Scholar]
- 14. Folstein MF, Folstein SE, McHugh PR. “Mini-mental state.” A practical method for grading the cognitive state of patients for the clinician. J Psychiatr Res. 1975;12(3):189–98. [DOI] [PubMed] [Google Scholar]
- 15. Friedewald WT, Levy RI, Frederickson DS. Estimation of the concentration of low-density lipoprotein cholesterol in plasma, without use of the preparation ultracentrifuge. Clin Chem. 1972;18(6):499–502. [PubMed] [Google Scholar]
- 16. National Eye Institute. Age-related eye disease study 2. Manual of procedures. Vitamin A/E/carotenoid profile. Exhibit 14-4 [Internet]. Available from: https://documents.pub/document/age-related-eye-disease-study-2-manual-of-procedures-90-august-23-2012-age-related [Google Scholar]
- 17. Tuck MK, Chan DW, Chia D, Godwin AK, Grizzle WE, Krueger KE, Rom W, Sanda M, Sorbara L, Stass S et al. Standard operating procedures for serum and plasma collection: Early detection research network consensus statement standard operating procedure integration working group. J Proteome Res. 2009;8(1):113–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hassis ME, Niles RK, Braten MN, Albertolle ME, Ewa Witkowska H, Hubel CA, Fisher SJ, Williams KE. Evaluating the effects of preanalytical variables on the stability of the human plasma proteome. Anal Biochem. 2015;478:14–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zimmerman LJ, Li M, Yarbrough WG, Slebos RJ, Liebler DC. Global stability of plasma proteomes for mass spectrometry-based analyses. Mol Cell Proteomics. 2012;11(6):M111.014340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Candia J, Cheung F, Kotliarov Y, Fantoni G, Sellers B, Griesman T, Huang J, Stuccio S, Zingone A, Ryan BM et al. Assessment of variability in the SOMAscan assay. Sci Rep. 2017;7(1):14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol. 1995;57:298–300. [Google Scholar]
- 22. Boca SM, Leek JT. A direct approach to estimating false discovery rates conditional on covariates. PeerJ. 2018;6:e6035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Garred P, Honoré C, Ma YJ, Munthe-Fog L, Hummelshøj T. MBL2, FCN1, FCN2 and FCN3–44. The genes behind the initiation of the lectin pathway of complement. Mol Immunol. 2009;46(14):2737–44. [DOI] [PubMed] [Google Scholar]
- 24. Kilpatrick DC, Chalmers JD. Human L-ficolin (ficolin-2) and its clinical significance. J Biomed Biotechnol. 2012;2012:138797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kilpatrick DC, Chalmers JD, MacDonald SL, Murray M, Mohammed A, Hart SP, Matsushita M, Hill A. Stable bronchiectasis is associated with low serum L-ficolin concentrations. Clin Respir J. 2009;3:29–33. [DOI] [PubMed] [Google Scholar]
- 26. Metzger ML, Michelfelder I, Goldacker S, Melkaoui K, Litzman J, Guzman D, Grimbacher B, Salzer U. Low ficolin-2 levels in common variable immunodeficiency patients with bronchiectasis. Clin Exp Immunol. 2015;179:256–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Luo F, Sun X, Wang Y, Wang Q, Wu Y, Pan Q, Fang C, Zhang XL. Ficolin-2 defends against virulent Mycobacteria tuberculosis infection in vivo, and its insufficiency is associated with infection in humans. PLoS One. 2013;8:e73859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bozaoglu K, Attard C, Kulkarni H, Cummings N, Diego VP, Carless MA, Shields KA, Johnson MP, Kowlessur S, Dyer TD et al. Plasma levels of soluble interleukin 1 receptor accessory protein are reduced in obesity. J Clin Endocrinol Metab. 2014;99:3435–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Xie B, Wang S, Jiang N, Li JJ. Cyclin B1/CDK1-regulated mitochondrial bioenergetics in cell cycle progression and tumor resistance. Cancer Lett. 2019;443:56–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Candas D, Fan M, Nantajit D, Vaughan AT, Murley JS, Woloschak GE, Grdina DJ, Li JJ. CyclinB1/Cdk1 phosphorylates mitochondrial antioxidant MnSOD in cell adaptive response to radiation stress. J Mol Cell Biol. 2013;5(3):166–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. McDonnell E, Peterson BS, Bomze HM, Hirschey MD. SIRT3 regulates progression and development of diseases of aging. Trends Endocrinol Metab. 2015;26:486–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liu R, Fan M, Candas D, Qin L, Zhang X, Eldridge A, Zou JX, Zhang T, Juma S, Jin C et al. CDK1-mediated SIRT3 activation enhances mitochondrial function and tumor radioresistance. Mol Cancer Ther. 2015;14(9):2090–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Verdin E. NAD⁺ in aging, metabolism, and neurodegeneration. Science. 2015;350(6265):1208–13. [DOI] [PubMed] [Google Scholar]
- 34. Yoon MJ, Yoshida M, Johnson S, Takikawa A, Usui I, Tobe K, Nakagawa T, Yoshino J, Imai S. SIRT1-mediated eNAMPT secretion from adipose tissue regulates hypothalamic NAD+ and function in mice. Cell Metab. 2015;21(5):706–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yoshida M, Satoh A, Lin JB, Mills KF, Sasaki Y, Rensing N, Wong M, Apte RS, Imai SI. Extracellular vesicle-contained eNAMPT delays aging and extends lifespan in mice. Cell Metab. 2019;30(2):329–42.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Poljsak B. NAMPT-mediated NAD biosynthesis as the internal timing mechanism: In NAD+ world, time is running in its own way. Rejuvenation Res. 2018;21(3):210–24. [DOI] [PubMed] [Google Scholar]
- 37. Wang Z, Fan M, Candas D, Zhang TQ, Qin L, Eldridge A, Wachsmann-Hogiu S, Ahmed KM, Chromy BA, Nantajit D et al. Cyclin B1/Cdk1 coordinates mitochondrial respiration for cell-cycle G2/M progression. Dev Cell. 2014;29(2):217–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wollert KC, Kempf T, Wallentin L. Growth differentiation factor 15 as a biomarker in cardiovascular disease. Clin Chem. 2017;63(1):140–51. [DOI] [PubMed] [Google Scholar]
- 39. Breit SN, Johnen H, Cook AD, Tsai VW, Mohammad MG, Kuffner T, Zhang HP, Marquis CP, Jiang L, Lockwood G et al. The TGF-β superfamily cytokine, MIC-1/GDF15: A pleotrophic cytokine with roles in inflammation, cancer and metabolism. Growth Factors. 2011;29(5):187–95. [DOI] [PubMed] [Google Scholar]
- 40. Yamaguchi Y, Zampino M, Tanaka T, Bandinelli S, Osawa Y, Ferrucci L, Semba RD. Elevated plasma growth and differentiation factor 15 predicts incident anemia in older adults aged 60 years and older. J Gerontol A Biol Sci Med Sci. 2021;76(7):1192–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Semba RD, Gonzalez-Freire M, Tanaka T, Biancotto A, Zhang P, Shardell M, Moaddel R, Consortium CHI, Ferrucci L. Elevated plasma growth and differentiation factor 15 is associated with slower gait speed and lower physical performance in healthy community-dwelling adults. J Gerontol A Biol Sci Med Sci. 2020;75:175–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Arosio P, Elia L, Poli M. Ferritin, cellular iron storage and regulation. IUBMB Life. 2017;69:414–22. [DOI] [PubMed] [Google Scholar]
- 43. Kerins MJ, Ooi A. The roles of NRF2 in modulating cellular iron homeostasis. Antioxid Redox Signal. 2018;29:1756–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ganz T, Nemeth E. Hepcidin and iron homeostasis. Biochim Biophys Acta. 2012;1823:1434–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Li Y, Zhong X, Cheng G, Zhao C, Zhang L, Hong Y, Wan Q, He R, Wang Z. Hs-CRP and all-cause, cardiovascular, and cancer mortality risk: A meta-analysis. Atherosclerosis. 2017;259:75–82. [DOI] [PubMed] [Google Scholar]
- 46. Kimura Y, Izumiya Y, Hanatani S, Yamamoto E, Kusaka H, Tokitsu T, Takashio S, Sakamoto K, Tsujita K, Tanaka T et al. High serum levels of thrombospondin-2 correlate with poor prognosis of patients with heart failure with preserved ejection fraction. Heart Vessels. 2016;31(1):52–9. [DOI] [PubMed] [Google Scholar]
- 47. Simantov R, Febbraio M, Silverstein RL. The antiangiogenic effect of thrombospondin-2 is mediated by CD36 and modulated by histidine-rich glycoprotein. Matrix Biol. 2005;24(1):27–34. [DOI] [PubMed] [Google Scholar]
- 48. Morikawa N, Adachi H, Enomoto M, Fukami A, Kumagai E, Nakamura S, Nohara Y, Nakao E, Kono S, Tsuru T et al. Thrombospondin-2 as a potential risk factor in a general population. Int Heart J. 2019;60:310–17. [DOI] [PubMed] [Google Scholar]
- 49. Egerstedt A, Berntsson J, Smith ML, Gidlöf O, Nilsson R, Benson M, Wells QS, Celik S, Lejonberg C, Farrell L et al. Profiling of the plasma proteome across different stages of human heart failure. Nat Commun. 2019;10(1):5830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mondul AM, Yu K, Wheeler W, Zhang H, Weinstein SJ, Major JM, Cornelis MC, Männistö S, Hazra A, Hsing AW et al. Genome-wide association study of circulating retinol levels. Hum Mol Genet. 2011;20(23):4724–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Bohn T, Desmarchelier C, Dragsted LO, Nielsen CS, Stahl W, Rühl R, Keijer J, Borel P. Host-related factors explaining interindividual variability of carotenoid bioavailability and tissue concentrations in humans. Mol Nutr Food Res. 2017;61(6):1600685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bohn T, Bonet ML, Borel P, Keijer J, Landrier JF, Milisav I, Ribot J, Riso P, Winklhofer-Roob B, Sharoni Y et al. Mechanistic aspects of carotenoid health benefits–Where are we now?. Nutr Res Rev. [accessed 2021 September 29]. doi: 10.1017/S0954422421000147. [DOI] [PubMed] [Google Scholar]
- 53. Reboul E. Mechanisms of carotenoid intestinal absorption: where do we stand?. Nutrients. 2019;11(4):838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Moran NE, Thomas-Ahner JM, Fleming JL, McElroy JP, Mehl R, Grainger EM, Riedl KM, Toland AE, Schwartz SJ, Clinton SK. Single nucleotide polymorphisms in β-carotene oxygenase 1 are associated with plasma lycopene responses to a tomato-soy juice intervention in men with prostate cancer. J Nutr. 2019;149(3):381–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Eroglu A, Schulze KJ, Yager J, Cole RN, Christian P, Nonyane BAS, Lee SE, Wu LSF, Khatry S, Groopman J et al. Plasma proteins associated with circulating carotenoids in Nepalese school-aged children. Arch Biochem Biophys. 2018;646:153–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Peters V, Yard B, Schmitt CP. Carnosine and diabetic nephropathy. Curr Med Chem. 2020;27(11):1801–12. [DOI] [PubMed] [Google Scholar]
- 57. Wittwer J, Bradley D. Clusterin and its role in insulin resistance and the cardiometabolic syndrome. Front Immunol. 2021;12:612496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Andoh E. Washoku: Recipes from the Japanese Home Kitchen. Berkeley, CA: Ten Speed Press; 2006. [Google Scholar]
- 59. Willett W, Rockström J, Loken B, Springmann M, Lang T, Vermeulen S, Garnett T, Tilman D, DeClerck F, Wood A et al. Food in the anthropocene: The EAT-Lancet Commission on healthy diets from sustainable food systems. Lancet North Am Ed. 2019;393(10170):447–92. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


