Abstract
RNA G-quadruplexes (rG4s) have functional roles in many cellular processes in diverse organisms. While a number of rG4 examples have been reported in coding messenger RNAs (mRNA), so far only limited works have studied rG4s in non-coding RNAs (ncRNAs), especially in long non-coding RNAs (lncRNAs) that are of emerging interest and significance in biology. Herein, we report that MALAT1 lncRNA contains conserved rG4 motifs, forming thermostable rG4 structures with parallel topology. We also show that rG4s in MALAT1 lncRNA can interact with NONO protein with high specificity and affinity in vitro and in nuclear cell lysate, and we provide cellular data to support that NONO protein recognizes MALAT1 lncRNA via rG4 motifs. Notably, we demonstrate that rG4s in MALAT1 lncRNA can be targeted by the rG4-specific small molecule, peptide, and L-aptamer, leading to the dissociation of MALAT1 rG4-NONO protein interaction. Altogether, this study uncovers new and important rG4s in MALAT1 lncRNAs, reveals their specific interactions with NONO protein, offers multiple strategies for targeting MALAT1 and its RNA–protein complex via its rG4 structure and illustrates the prevalence and significance of rG4s in ncRNAs.
INTRODUCTION
G-quadruplexes (G4s) are four-stranded nucleic acid secondary structures formed by G-rich DNA or RNA sequences, and the G-quartet layers in G4s are stabilized by monovalent ions such as potassium ion (K+) and sodium ion (Na+) (1,2). According to the classical definition, canonical G4 sequences contain four runs of guanine tracks separated by three loops (GGGN1–7GGGN1–7GGGN1–7GGG) (3). Recently, non-canonical G4s such as two-quartet G4s, bulged G4s and long-loop G4s were found to be prevalent in the human genome and transcriptome (4,5), highlighting the structural diversity and complexity of G4s (6,7). Studies have shown that the intervening nucleotides and the loop length can affect the stability and properties of G4 structures (8,9). Over the past decades, the structure information and the functional roles of DNA G4s (dG4s) have been studied extensively, and recent evidence has shown that they were involved in various cellular processes, including but not limited to replication, transcription, genomic instability and epigenetic regulation (10–12).
Our understanding and study of RNA G4s (rG4s) are still in their infancy (13,14). RG4s were reported to take part in numerous biological functions (14) such as microRNA (miRNAs) targeting (15,16), RNA localization (17,18), translational regulation (19,20), and alternative polyadenylation (21,22). Currently, most rG4s identified were localized in the open reading frame (ORF) (23–25) and untranslated (UTR) regions (21,26,27) of protein-coding messenger RNAs (mRNAs), which is a small fraction (<5%) of the entire human transcriptome (28). The advent of next-generation sequencing has enabled the robust identification of non-coding RNAs (ncRNAs), which significantly facilitated our understanding and investigation of their structures and functions in cells. Interestingly, latest studies have begun to explore the existence and roles of rG4s in ncRNAs (13,14) such as long non-coding RNAs (lncRNAs) (29–33), miRNAs (34–36), Piwi-interacting RNAs (piRNAs) (37,38), ribosomal RNAs (rRNAs) (39,40), transfer RNAs (tRNAs) (41,42). The initial findings were exciting and showed ncRNA rG4s to have key regulatory roles in gene expression, RNA metabolism, and protein recognition and function (13,14,43). Nevertheless, compared with mRNA G4s, which have been studied more extensively, there is still a lot of work to be done for ncRNA G4s.
RNA binding proteins (RBPs) play significant roles in regulating the RNA structure, which in turn control diverse biological processes (44,45). And rG4 structure and rG4 binding protein is no exception. Since the first rG4 binding protein, fragile X mental retardation protein (FMRP), was identified in 2001 (46,47), many rG4-binding proteins have been reported to play regulatory roles by folding or unfolding the G4 structures or convening other binding motifs (25,48,49). For instance, the binding of hnRNP F to an rG4 located in the intron of CD44 promotes the inclusion of CD44 variable exon v8 and regulates alternative splicing (25). The binding of DHX36 to rG4 located at the 5′ end of the lncRNA human telomerase RNA (hTR) can unwind the RNA structure and facilitate hTR maturation and telomerase function (50,51). Moreover, the binding of nucleolin (NCL) to the rG4 in lncRNA LUCAT1 regulates the expression of MYC, thus modulating the CRC cell proliferation (49). Therefore, targeting and interfering with the rG4-protein interaction can serve as a potential strategy for manipulating gene function and regulation and may have impact and implications in treating and preventing rG4-associated diseases.
In this study, we first identified hundreds of rG4-containing ncRNAs and then focused on investigating rG4s on a biologically important lncRNA, MALAT1. We demonstrated the formation of the lncRNA MALAT1 rG4 structures by multiple spectroscopic assays and found MALAT1 rG4s to be highly conserved using comparative sequence analysis. Besides, we reported for the first time the specific binding between MALAT1 rG4s and NONO protein in vitro and in nuclear lysate and provided substantial evidence of their interaction through rG4 motifs in cells. Furthermore, to explore the targeting approach and potential application, we showed that the MALAT1 rG4-NONO protein interaction could be targeted by rG4-specific small molecule pyridostatin (PDS), peptide RHAU53, and our recently developed rG4-targeting L-RNA aptamer (L-Apt.4–1c), leading to the dissociation of MALAT1 rG4-NONO protein interaction.
MATERIALS AND METHODS
Oligonucleotide, protein, peptide and small molecule preparation
The 5′ FAM-labeled and 5′ Biotin-labelled oligonucleotides (oligos) used in this study were synthesized by Integrated DNA Technologies (IDT, USA). The primers used for qPCR were synthesized by Genewiz Biotechnology Co., Ltd. (China). The L-Apt.4–1c aptamer used in this study was synthesized by ChemGenes Corporation (USA). They were dissolved to a concentration of 100 μM (according to the supplier's instruction) with ultra-pure nuclease-free distilled water (Thermo, USA). All the dissolved oligos were stored at –20°C before the experiment. The oligonucleotide sequences are listed in Table 1 and Supplementary Table S2. Thioflavin T (ThT) was ordered from Solarbio Life Science (China). Recombinant protein NONO (53–312) was synthesized by MerryBio Co., Ltd. (China). Recombinant protein DHX36 was purchased from OriGene Technologies Inc (USA). RHAU53 peptide was synthesized by SynPeptide Co., Ltd. (China).
Table 1.
Oligonucleotide sequences of MALAT1 rG4s used in this study.
| Name | Sequence (5′–3′) | Length (nt) | rG4 type | Tm (°C) |
|---|---|---|---|---|
| MALAT1 rG4_02 | UGGGAUGGUCUUAACAGGGAAGAGAGAGGGUGGGGGA | 37 | Long-loop | 70°C |
| MALAT1 rG4_04 | AGGGUGGGCUUUUGUUGAUGAGGGAGGGGA | 30 | Long-loop | >85°C |
| MALAT1 rG4_05 | AGGGAAGGGAGGGGGUGCCUGUGGGG | 25 | Canonical | 85°C |
| MALAT1 rG4_06 | AGGGAUGGGAGGAGGGGGUGGGGC | 24 | Canonical | >85°C |
| MALAT1 rG4_09 | AGGGGAGGGAAAGGGGGAAAGCGGGC | 26 | Canonical | 81°C |
| MALAT1 rG4_10 | AGUGGCUGAGAGGGCUUUUGGGUGGG | 25 | Bulge | 73°C |
Note: All the MALAT1 rG4s are in parallel topology, as evidenced by the CD data in Figure 1.
Circular dichroism (CD) spectroscopy
CD assay was performed with a Jasco CD J-150 spectrometer and a 1 cm path length quartz cuvette. The reaction samples containing 5 μM oligos were prepared in 10 mM LiCac buffer (pH 7.0) and 150 mM KCl/LiCl to form total volume of 2 ml. The mixtures were then vortexed and heated at 95°C for 5 min and allowed to cool for 15 minutes at room temperature for renaturation. The samples were scanned at a 2 nm interval from 220 to 310 nm, and the data were blanked and normalised to mean residue ellipticity before smoothed over 10 nm. All the data were analyzed with Spectra Manager™ Suite and Microsoft Excel.
UV melting assay
UV melting assay was performed with a Cary 100 UV–vis spectrophotometer and a 1 cm path length quartz cuvette. Following previous studies (8,52), the reaction samples containing 5 μM oligos were prepared in 10 mM LiCac buffer (pH 7.0) and 150 mM KCl to form a total volume of 2 ml. The mixtures were vortexed and heated at 95°C for 5 min and allowed to cool for 15 min at room temperature for renaturation. In cuvettes sealed with Teflon tape, the samples were monitored at 295 nm from 5°C to 95°C with data collected over 0.5°C (53). The data were blanked and smoothed over 5 nm. All the data were analyzed with Microsoft Excel.
Fluorescence spectroscopy
Fluorescence spectra was performed with a HORIBA FluoroMax-4 fluorescence spectrophotometer (Japan) and a 1 cm path length quartz cuvette. The reaction samples containing 1 μM oligos were prepared in 10 mM LiCac buffer (pH 7.0) and 150 mM KCl/LiCl to form a total volume of 100 μl. The mixtures were vortexed and heated at 95°C for 5 min and allowed to cool for 15 min at room temperature for renaturation. ThT ligand (1 μM) was added to the mixture. The emission spectra were collected from 440 to 700 nm with an excitation wavelength of 425 nm. The entrance and exit slits were 5 and 2 nm respectively, with the data collected every 2 nm. All the data were analyzed with Microsoft Excel.
Comparative sequence analysis and gene conservation alignment
MALAT1 gene sequences from different organisms were obtained from Ensembl Genome Browser (http://www.ensembl.org) using human MALAT1 genes (Ensembl release 104) as reference (54). Multiple sequence alignments of the MALAT1 genes were carried out with Jalview (55).
G4 prediction analysis
G4 prediction analysis including cGcC (Consecutive G over consecutive C ratio) (56), G4H (G4Hunter) (57) and G4NN (G4 Neural Network) (58) was carried out by G4RNA screener v.0.2 (59) (http://scottgroup.med.usherbrooke.ca/G4RNA_screener/) with default setting. The G4 threshold is defined as cGcC >4.5, G4H >0.9, G4NN >0.5.
Electrophoretic mobility shift assay (EMSA)
Reaction mixtures containing 5 nM 5′ FAM-labeled RNA, 20 mM HEPES pH 7.5, 100 mM KCl, 0.12 mM EDTA, 25 mM Tris–HCl (pH 7.5), 10% glycerol and increasing concentrations of NONO (53–312) were prepared in 10 μl and incubated at 4°C for 1 h. For inhibition assay, NONO (53–312) was increased to and fixed at 200 nM, and increasing concentrations of RHAU53 mixed with 5 nM 5′ FAM-labeled RNA and fixed amount of NONO(53–312). The RNA was heated at 75°C for 5 minutes for denaturation before adding. The RNA–protein mixture were resolved by 4%, 37.5:1 (acrylamide: bis-acrylamide) non-denaturing polyacrylamide gel in 0.5X Tris/Borate/EDTA (TBE) at 4°C, 80 V for 30 min (60). The gel was scanned by FujiFilm FLA-9000 Gel Imager at 650 V and quantified by ImageJ. The curve fitting and Kd/IC50 determination were carried out by Graphpad Prism using the one site-specific binding model.
Filter-binding assay
Reaction mixtures containing 2 nM 5′ Biotin-labeled RNA, 20 mM HEPES pH 7.5, 100 mM KCl, 0.12 mM EDTA, 25 mM Tris–HCl (pH 7.5) and increasing concentrations of NONO (53–312) were prepared in 50 μl and incubated at 4°C for 1 h before loaded onto a Dot Blot apparatus (Bio-rad) containing a nitrocellulose membrane (top) and nylon membrane (bottom). For inhibition assay, increasing concentrations of PDS/L-Apt.4–1c was mixed with 2 nM 5′ biotin-labeled RNA and fixed amount of NONO(53–312). The nylon membrane was rinsed with 0.5X TBE and nitrocellulose membrane was treated with 0.5 M KOH for 15 minutes at 4°C and rinse with 0.5X TBE before use. The membranes were wash 3 times with binding buffer before and after the mixture applied to the apparatus. The cross-linking step was performed under UV irradiation at 254 nm, 120 000 microjoules/cm2 for 5 min. The results were detected using a chemiluminescent nucleic acid detection module kit (Thermo Scientific) and scanned by ChemiDoc™ Touch Imaging System and quantified by ImageJ. The curve fitting and Kd determination were carried out by Graphpad Prism using the one site-specific binding model.
Microscale thermophoresis (MST) binding assay
Reaction mixtures containing 30 nM 5′ FAM-labeled RNA, 150 mM KCl, 1 mM MgCl2, 25 mM Tris–HCl (pH 7.5) and increasing concentrations of protein, ligand or aptamer were prepared in 10 μl and incubated at 37°C for 1 h. For inhibition assay, Increasing concentrations of PDS/RHAU53/L-Apt.4–1c was mixed with 50 nM 5′ FAM-labeled RNA and fixed amount of NONO(53–312). The RNA and aptamer were heated at 75°C for 5 min for denaturation before adding. The reaction mixtures were then loaded to MST capillary tubes (Nano-Temper Monolith NT.115), the measurement were carried out at 25°C using blue light mode and the binding affinity mode. The data was analysed by MST nano temper analysis (nta) analysis software. The curve fitting and Kd/IC50 determination were carried out by Graphpad Prism using the one site-specific binding model.
RG4 pull-down assay
HEK293T (cell line authenticated and tested without mycoplasma contamination) nuclear lysate is extracted using NE-PER™ Nuclear extraction kit from Thermo Scientific™. Streptavidin C1 magnetic beads were blocked by 4 ng/ml yeast tRNA and 50 μg/ml BSA buffer for 30 minutes in room temperature and nuclear lysate were pre-cleared by incubating with 20 μl prewashed streptavidin C1 magnetic beads (Thermo Scientific™) for 1 h in 4°C. Three hundred pmol oligo and 60 μl prewashed streptavidin C1 magnetic beads were added for each reaction and the 5′ Biotin-labeled rG4 wildtype or mutant oligos were heated at 75°C for 5 min for denaturation before adding to the mixture containing buffer A (10 mM Tris–HCl pH 7.5, 100 mM KCl, 0.1 mM EDTA, with/without 20 μM PDS). The beads immobilization step is processed in room temperature for 30 minutes in buffer A. The beads were washed for 3 times with buffer A. And 500 μg nuclear lysate was added in each reaction to the incubation mixture containing buffer B (20 mM Tris–HCl pH 7.5, 50 mM KCl, 0.5 mM EDTA, 10% glycerol). The incubation step is processed in 4°C for 2 h and followed by 5 times stringent washes using buffer B. The enriched protein were eluted by heating at 95°C for 30 min in 1× Laemmli sample buffer (BioRad) and processed to western blot. The data was quantified by ImageJ and analyzed with Microsoft Excel.
Comprehensive identification of RNA-binding proteins followed by western blot (ChIRP-WB)
Three hundred million HEK293T cells were used per ChIRP-WB experiment. The cells were treated with 10 μM PDS (or DMSO as control) for 24 h before collecting. The cells were crosslinked for 30 min in 3% formaldehyde, followed by 0.125 M glycine quenching for 5 min. The cells were wash by cold PBS and collected by centrifuge at 2000 rcf for 5 min. Cell pellets were dissolved in 1 mL cell lysis buffer (50 mM Tris–HCl pH 6.8, 1% SDS, 10 mM EDTA, 1 mM PMSF, protease inhibitor cocktail) per 100 mg and sonicated in 4°C using Bioruptor (Diagenode) until lysate is clear, centrifuged the cells at 18 000 rcf at 4°C for 10 min and collected the supernatant. Ten μl of the control sample were saved as ‘input’. Each lysate sample was precleared with 30 μl streptavidin C1 magnetic beads (Thermo Scientific™). Two mL hybridization buffer and 100 pmol probes per 1 ml of lysate were added to the mixture with 5′ Biotin-labeled MALAT1 probes (Supplementary Table S3) and rotated in 37°C overnight. The hybridization buffer contains 750 mM NaCl, 50 mM Tris–HCl pH 6.8, 0.5% SDS, 1 mM EDTA, 15% formamide, 1 mM PMSF, protease inhibitor cocktail, RNase inhibitor. After the hybridization, 100 μl streptavidin C1 magnetic beads per 100 pmol probes were added and continue to mix samples at 37°C for 30 min. The enriched RNA and RNA binding protein are isolated and processed to western blot and qPCR separately following the protocol (61). The data was analyzed with Microsoft Excel.
Quantitative PCR (qPCR)
The RNA samples were resuspend in 100 μl proteinase K (pK) buffer containing 100 mM NaCl, 10 mM Tris–HCl pH 7.0, 1 mM EDTA, 0.5% SDS and 5% v/v pK and incubated in 50°C for 45 min and heated at 95°C for 10 min (61). The RNA was isolated using Trizol:chloroform and ethanol and purified using RNeasy plus mini Kit (Qiagen). Reverse transcription was performed using PrimeScript™ RT Master Mix (TAKARA) and qPCR was carried out using SYBR Green Supermix (BioRad) and the SYBR program in CFX Connect Real-Time PCR Detection System. The primers used are listed in Supplementary Table S2.
Western blot
Samples for western blotting were resolved on 4–10% SDS-PAGE gels (BioRad) and transferred to immobilon-P PVDF membrane (Millipore). After blocking with 5% skim milk for 1 h at room temperature, the membrane was immunoblotted with primary antibodies diluted 1:1000 in blocking buffer for 1 h at room temperature and washed 3 times with TBS-T for 15 min. Secondary antibodies were diluted 1:1000 in blocking buffer and applied for 1 h at room temperature. The results were visualized using ChemiDoc™ Touch Imaging System and quantified by ImageJ. Primary antibodies used are as follows: Anti-nmt55/p54nrb antibody (Abcam ab70335), Anti-GAPDH (Bio-station limited sc-32233).
RESULTS
Identification and characterization of G-quadruplex structures in MALAT1 lncRNA
To explore rG4 in ncRNAs, we first examined our recently published in vitro rG4-seq dataset (5). While the rG4-seq dataset was collected using purified polyadenylated (polyA) RNA isolated from human HeLa cells, we have managed to identify more than 350 rG4s in a total of 203 ncRNA on the list (Supplementary Table S1), such as lncRNAs NEAT1 and MALAT1. We also employed the G4 prediction tools such as cGcC (56), G4hunter (57), and G4NN (58) on this ncRNA rG4 list. We found that 328 rG4 sequences showed at least one score higher than the threshold set for those programs to score for G4, and 226 rG4 sequences showed all three scores above the threshold set (Supplementary Table S1), suggesting the G4 prediction and rG4-seq data result are largely consistent. Using rG4-seq data and rG4 motif analysis, we have identified 15 and 11 rG4 forming sequences in lncRNA NEAT1 and MALAT1, respectively (Supplementary Table S1). Noteworthy, an independent study has recently predicted NEAT1 to contain rG4 motifs and verified their formation using biophysical and biochemical assays (33), which further support our rG4-seq data. In this work, we focus on studying the formation of G-quadruplex structure in another important lncRNA MALAT1, and we selected six representative MALAT1 rG4s according to the rG4 length and rG4 structural subtype. Overall, we selected three canonical rG4s and three non-canonical rG4s, including two long-loop rG4s and one bulge rG4, and the length is from 24 nt to 37 nt (Table 1).
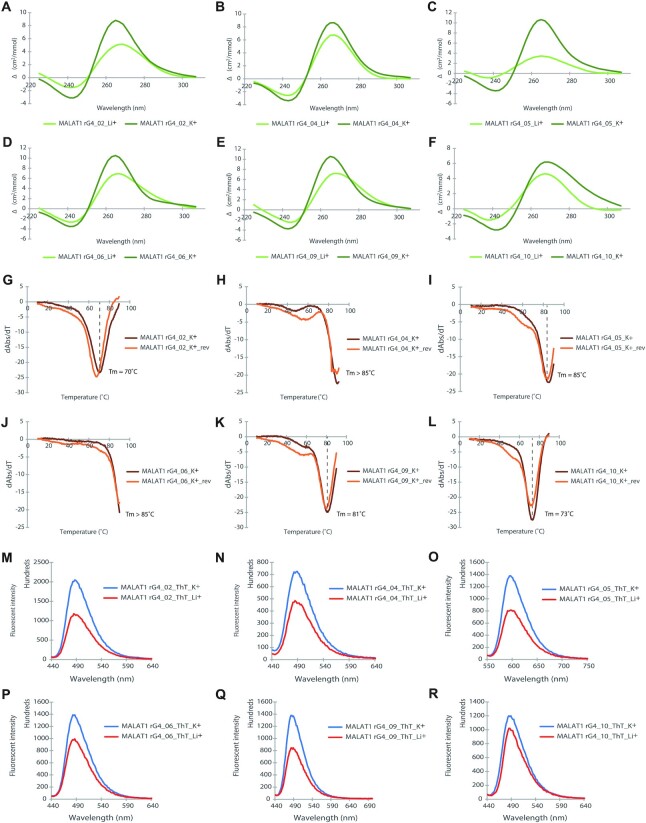
To verify rG4 formation in MALAT1 lncRNA, we have altogether carried out three spectroscopy assays. First, we conducted circular dichroism (CD) assay to determine the secondary structures and topologies of the six selected RNA oligonucleotides from MALAT1 (Figure 1A-F). The results showed that all six RNAs displayed a negative peak at 240 nm and a positive peak at 260 nm under physiologically relevant K+ concentration (150 mM) (Figure 1A–F), which suggests the formation of rG4 with parallel topology (62). Importantly, the CD signal is K+-dependent. The CD signature of rG4 disappeared when the reaction was performed under the 150 mM Li+ conditions (Figure 1A–F), supporting the distinctive CD profiles caused by the formation of rG4 structure. Next, we performed UV melting assay under 150 mM K+ to verify rG4 formation, and to examine the thermostability of these six rG4s. Our data displayed a hypochromic shift at 295 nm (Figure 1G–L), which is a hallmark of rG4 formation (53). In addition, the UV melting results revealed that the melting temperature (Tm) of all six rG4 oligonucleotides are larger than 70 °C (Figure 1G–L), which indicated that they are highly thermostable and folded into rG4 conformation under physiological temperature conditions. Last, we carried out the fluorescent turn-on spectroscopy assay using G-quadruplex-specific ligand thioflavin T (ThT) (63). Substantial fluorescence enhancement was observed in 150 mM K+ compared to 150 mM Li+ conditions for all six rG4 oligonucleotides (Figure 1M–R), which further substantiated the formation of rG4 structures. Collectively, these spectroscopic assays verified that MALAT1 lncRNA harbors several thermostable rG4 structures with parallel topology under physiological relevant potassium ion and temperature conditions.
Figure 1.
Biophysical characterization of MALAT1 rG4 structures. (A–F) CD spectrum under K+ and Li+ conditions. The CD pattern becomes stronger in the presence of K+ rather than Li+, suggesting rG4 formation. The negative peak at 240 nm and the positive peak at 262 nm under K+ condition suggest a parallel topology of rG4. (G–L) UV melting. Hypochromic shift is observed at 295 nm, a strong sign of rG4 formation. The melting temperatures (Tms) for the MALAT1 rG4s are indicated. (M–R) ThT enhanced fluorescence spectroscopy under K+ and Li+ conditions with an emission maximum wavelength of 490 nm. The fluorescence intensity of ThT is higher in rG4 under K+ compared to Li+, verifying the formation of rG4.
MALAT1 rG4s are conserved and interact with NONO protein in vitro and in nuclear lysate
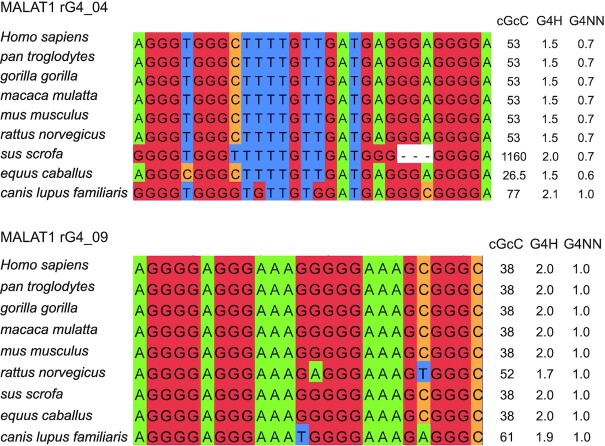
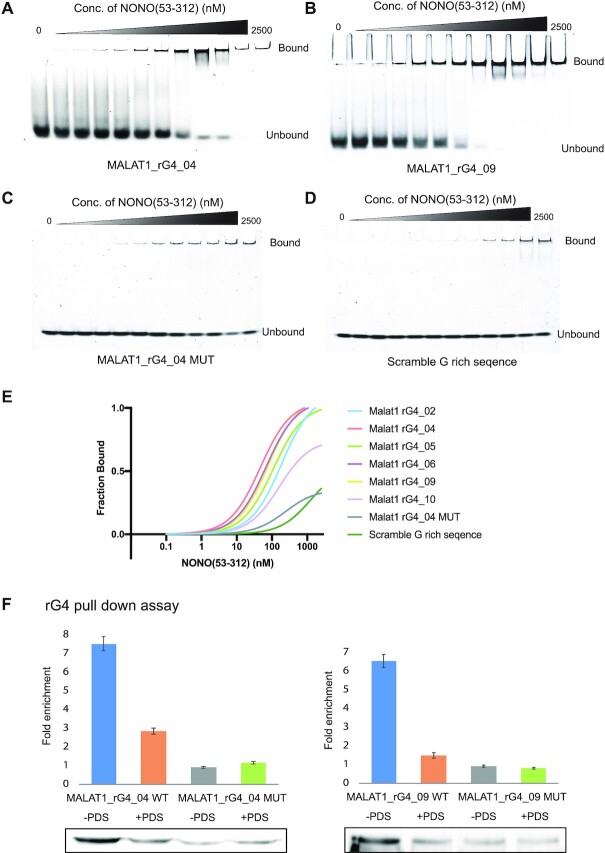
Using comparative sequence analysis, we found that some of these MALAT1 rG4s were highly conserved in eight other species, including primates and mammals (Figure 2 and Supplementary Figure S1). We also performed the G4 predictions on these rG4s and found they have similar scores as the human ones and passed the threshold set by the G4 prediction programs (Figure 2), suggesting these conserved MALAT1 rG4s are likely to be formed and functional. Next, we attempted to identify proteins that can interact with the MALAT1 rG4s. Studies have shown that RNA helicase DEAH box polypeptide 36 (DHX36) can bind rG4 with high affinity and selectivity (51,64), and it is found to localize both in the cytoplasm and nucleus (65). Therefore, we first performed the binding assay with MALAT1 rG4_04 and DHX36, and the result indicated that the MALAT1 rG4_04 binds to DHX36 strongly with a dissociation constant (Kd) value of 130 ± 16 nM (Supplementary Figure S2). Besides DHX36 protein, we hope to discover a new MALAT1 rG4 binding protein in this study. MALAT1 and NEAT1 are both well conserved and nuclear-enriched lncRNAs. MALAT1 lncRNA is localized in nuclear speckles, which are nuclear domains enriched in pre-mRNA splicing factors, while NEAT1 lncRNA is part of the paraspeckles, which are ribonucleoprotein bodies found in the interchromatin space of mammalian cell nuclei (66). Although MALAT1 and NEAT1 locate in different nuclear domains, they were found to localize adjacent to each other in the nucleus periodically and co-enrich many trans genomic binding sites and protein factors, such as NONO protein, indicating potential cooperation or complementary function between these two lncRNAs in regulating the formation of nuclear bodies (67). While NONO is one of the canonical paraspeckle proteins, it was also found in nuclear speckles, which suggest a diverse role of NONO (68,69). Recently, NEAT1 was found to harbour several conserved rG4s, which mediate the NONO-NEAT1 association (33). To study the association of NONO protein with MALAT1 rG4s, we first performed the electrophoretic mobility shift assay (EMSA) with purified recombinant protein NONO (53–312). NONO (53–312) contains two putative RNA-binding domain RRMs, the NONA/paraspeckle (NOPS) domain and half of the coiled-coil domain (33). The NEAT1_22619 works as a positive control to confirm that NONO (53–312) functions properly (Supplementary Figure S3 and Supplementary Table S2). We next tested the binding of MALAT1 rG4s with NONO (53–312), and the EMSA results showed that NONO (53–312) exhibited strong binding to all 6 MALAT1 rG4s, especially for MALAT1 rG4_04 and MALAT1 rG4_09 (Figure 3A-B and Supplementary Figure S4) The Kd values were determined to be 46.9 ± 12.1 nM and 80.9 ± 18.1 nM, respectively. To verify the binding is rG4-specific, we also designed two negative controls, an rG4 mutant MALAT1 rG4_04 MUT and a non-G-quadruplex-forming scramble G-rich sequence (Supplementary Table S2), and EMSA results showed both exhibited very weak binding to NONO (53–312) (Figure 3C-D). Besides, we also performed the filter-binding assay with biotin-labelled MALAT1 rG4_04 and MALAT1 rG4_09, both wildtypes and mutant (Supplementary Figure S5), and the results are consistent with EMSA (Figure 3E), illustrating MALAT1 rG4 wildtypes, but not mutants, interact with NONO protein. Interestingly, we found that NONO (53–312) can compete with DHX36 and disrupt the binding between MALAT1 rG4_04 and DHX36 (Supplementary Figure S6).
Figure 2.
rG4 conservation analysis and predicted G4 scores for MALAT1 rG4s. Comparative sequence analysis of MALAT1 rG4_04 and MALAT1 rG4_09 in different species and their predicted G4 scores. All G4 scores are higher than the threshold set in corresponding G4 prediction programs, i.e. cGcC > 4.5, G4H > 0.9, G4NN >0.5, suggested the high likelihood of G4 formation. The Us in the rG4 sequences are replaced by Ts in the comparative sequence analysis and G4 prediction.
Figure 3.
MALAT1 rG4-NONO protein interactions in vitro and in nuclear lysate. (A) EMSA with recombinant NONO (53–312) and MALAT1 rG4_04. (B) EMSA with recombinant NONO (53–312) and MALAT1 rG4_09. (C) EMSA with recombinant NONO (53–312) and MALAT1 rG4_04 MUT. (D) EMSA with recombinant NONO (53–312) and scramble G-rich sequence. (E) Binding curves of recombinant NONO (53–312) and MALAT1 rG4_02, MALAT1 rG4_04, MALAT1 rG4_05, MALAT1 rG4_06, MALAT1 rG4_09, MALAT1 rG4_10, MALAT1 rG4_04 MUT and scramble G-rich sequence. The bindings of NONO to rG4s are stronger than rG4 mutant and scramble G-rich sequence, suggesting the binding is rG4-specific. (F) rG4 pull-down assay with HEK293T nuclear lysate. Western blot result of rG4 pulldown shows that NONO is enriched by MALAT1_rG4_04 and MALAT1_rG4_09 wildtype rG4s but not rG4 mutants, and the interaction can be disrupted by adding 20 μM PDS. In each blot, the rG4 mutant with no PDS treatment is normalized to one. Three independent experiments are performed. Error bars represent the standard error of the mean (S.E.M.). The representative blot is shown here.
To strengthen the result and verify the interaction occur in more physiological-relevant settings, we next conducted pull-down experiments using biotin-labelled MALAT1 rG4 oligos and attempted to pull down endogenous NONO protein from cell lysate obtained from HEK293T cells. Given that NONO protein and MALAT1 lncRNA are both localized in the nucleus, we carefully extracted the nuclear lysate using NE-PER™ Nuclear extraction kit. We used two pairs of RNA oligonucleotides, MALAT1 rG4_04 WT and MALAT1_rG4_04 MUT, as well as MALAT1_rG4_09_WT and MALAT1_rG4_09_MUT. We showed that under normal conditions, the wildtype versions (both MALAT1 rG4_04 WT and MALAT1_rG4_09_WT) were able to pull down endogenous NONO protein more than the mutant version (MALAT1 rG4_04 MUT and MALAT1_rG4_09_MUT), with a ratio of 9:1 and 7:1, respectively (Figure 3F). G4 ligands, such as pyridostatin (PDS), have been previously reported to inhibit and display native G4-protein interaction (70,71). To support the binding relies on MALAT1 rG4 structure, PDS was added to the reaction to a final concentration of 20 μM, and a drop in the ratio was observed, indicating PDS binds to MALAT1 rG4 and displace the rG4-protein interaction. Altogether, the above results provided clear evidence that MALAT1 rG4s can interact with NONO protein in vitro and in nuclear cell lysate.
MALAT1 interacts with NONO via rG4 structure in cells
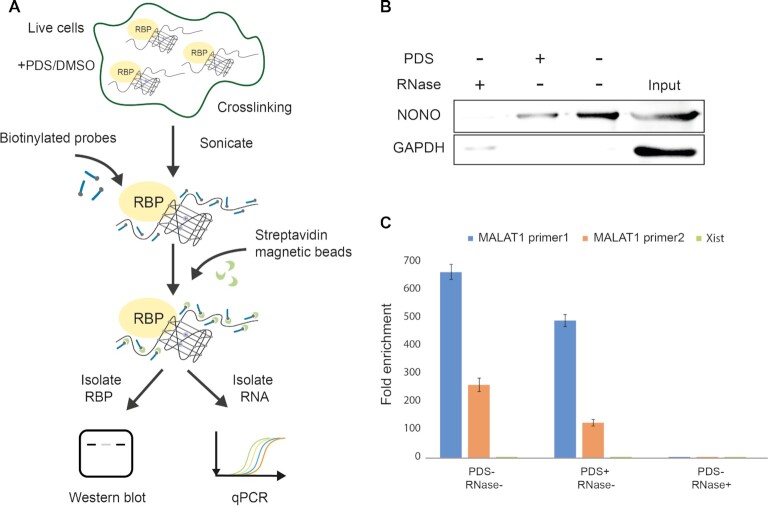
Having found that NONO recognizes MALAT1 rG4s both in vitro and in cell lysate, we aspired to ascertain whether the preference for MALAT1 rG4s detected is reflected in living cells as well. To examine the binding between MALAT1 and NONO in cells, we first designed and applied a total of 40 biotinylated MALAT1 probes to carry out an RNA binding protein detection method referred to as ChIRP-western blot (ChIRP-WB) (72) (Supplementary Table S3). Our rationale is that if NONO protein interacts with MALAT1 lncRNA in cells, then after crosslinking, sonication, biotinylated MALAT1 lncRNA probe capture and enrichment, the NONO protein should be enriched in the western blot assay (Figure 4A). Besides, in order to determine whether the rG4 structures in MALAT1 participate in the MALAT1-NONO interaction in cells, we also treated another set of HEK293T cells with 10 μM PDS (or DMSO as control) for 24 hours before crosslinking (Figure 4A). We also conducted negative control with RNase A treatment. Compared with the negative control with RNase treatment, NONO was significantly enriched in both groups (with/without PDS), demonstrating that MALAT1 interacts with NONO in cells. Importantly, we found that the MALAT1-NONO interaction was weakened by adding G4-specific ligand PDS, indicating NONO protein recognizes MALAT1 lncRNA through rG4 structures. This is also in agreement with our observation in the pull-down assay, where the addition of PDS dissociated the MALAT1 rG4-NONO protein binding in nuclear lysate (Figure 3F). GAPDH protein, which is not known to bind to MALAT1, was used as a negative control for ChIRP-WB experiments, and no or very weak band can be detected under all three conditions tested (Figure 4B). Furthermore, to validate the specificity and the efficiency of biotinylated MALAT1 probes, we carried out qPCR using two sets of MALAT1 primers and one set of Xist primers (Supplementary Table S2). The results showed that MALAT1 was greatly enriched while Xist was not, highlighting that the biotinylated MALAT1 probes can capture and enrich MALAT1 lncRNA successfully and efficiently (Figure 4C). Combining both in vitro, in nuclear lysate data earlier (Figure 3), and in cell data here (Figure 4), we have provided substantial evidence that MALAT1 rG4s can strongly interact with NONO both in vitro, in cell lysate, and in cells.
Figure 4.
MALAT1 rG4-NONO protein interaction in cells. (A) Scheme for the ChIRP-WB and qPCR. RNA binding protein (RBP) is crosslinked to lncRNA of interest in cells, followed by sonication. Biotinylated tiling probes are then hybridized to target lncRNA, and the complexes are purified using magnetic streptavidin beads, followed by stringent washes. The enriched lncRNA and RBP are isolated and processed by western blot and qPCR separately. (B) NONO protein is enriched by MALAT1 ChIRP, and their interaction can be disrupted by 10 μM G4 ligand PDS, indicating the binding is mediated by rG4. GAPDH is used as negative control in this experiment. Three independent experiments are performed. (C) MALAT1 RNA is pulled down by biotinylated probes, and PDS treatment do not have much effect on the pull-down efficiency. Both sets of MALAT1 qPCR primers shows great MALAT1 lncRNA enrichment. Xist lncRNA is used as a negative control in this experiment and no enrichment is observed. Three independent experiments are performed. Error bars represent the S.E.M.
MALAT1 rG4-NONO interaction can be suppressed by multiple G4 targeting tools
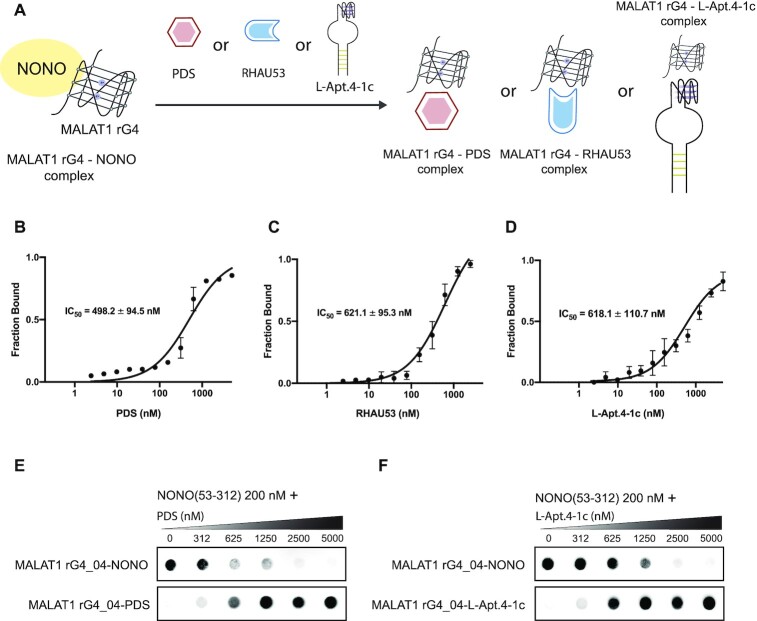
Motivated by the finding that MALAT1 lncRNA interacts with NONO via rG4 structure, we wonder if the interaction between MALAT1 rG4 and NONO can be disrupted by G4 targeting tools such as G4-specific small molecule, peptide, and aptamer (Figure 5A). Currently, targeting G4 with the small molecule is the most commonly employed strategy to recognize G4 motifs specifically and interfere with G4–protein interactions (73). We first tested whether MALAT1 rG4_04 can interact with G4-specific ligand PDS using MST and found the rG4-PDS complex to form at around 20 nM PDS, with a Kd value of 66 ± 8 nM (Supplementary Figure S7). To determine the ability of PDS to inhibit MALAT1 rG4–NONO interaction, we assembled the MALAT1 rG4_04–NONO (53–312) complex and titrated it with an increasing concentration of PDS (0–1000 nM). The MST result showed that PDS could effectively suppress MALAT1 rG4_04–NONO (53–312) interaction with a half-maximal inhibitory concentration (IC50) value of 498.2 ± 94.5 nM (Figure 5B), highlighting that PDS can successfully dissociate MALAT1 rG4_04–NONO (53–312) interaction even when the concentration of NONO (53–312) is as high as 200 nM. Noteworthy, this result further supports our above-mentioned data that PDS can interfere with the interaction between MALAT1 lncRNA and NONO in nuclear lysate and in cell (Figures 3 and 4). Next, we assessed the interference of MALAT1 rG4_04–NONO (53–312) interaction using the G4-specific peptide approach. As mentioned above, DHX36, or RHAU, is a family member of the ATP-dependent RNA helicase that specifically binds to and resolves parallel-stranded G-quadruplexes (74). Recently, the key protein region involved in G4 interactions was determined to be near the N-terminus of RHAU protein. One widely used and studied is RHAU53, which contains 53-amino acids long RHAU protein fragments (75). As such, we used the RHAU53 peptide as a G4 targeting peptide in this inhibition assay (Supplementary Table S2). Like PDS, we constructed the MALAT1 rG4_04–NONO (53–312) complex followed by an escalating concentration of RHAU53. From the MST data, the IC50 was determined to be 621.1 ± 95.3 nM (Figure 5C), which is comparable with the PDS result. Moreover, we demonstrated the MALAT1 rG4_04–NONO (53–312) interaction can be suppressed by rG4-targeting L-RNA aptamer, a new class of G4-targeting tool. L-Apt.4–1c is a novel L-RNA aptamer developed by our group recently (76). It was reported to generally bind to rG4s and interfere with rG4–protein interaction (76,77). In this study, we first tested the binding affinity between MALAT1 rG4_04 and L-Apt.4–1c and found the Kd to be 165 ± 45 nM (Supplementary Figure S8), indicating they strongly and directly interact with each other. To assess its capability of suppressing MALAT1 rG4_04–NONO (53–312) interaction, we performed MST inhibition assay and monitored the IC50 to be 618.1 ± 110.7 nM (Figure 5D), demonstrating that the inhibition ability of L-Apt.4–1c is analogous with PDS and RHAU53.
Figure 5.
Suppression of MALAT1 rG4-NONO interaction by multiple G4 targeting tools. (A) Schematic representation of MALAT1 rG4-NONO interaction that is interfered by various G4 targeting tools including small molecule, peptide, and L-aptamer. (B) Saturation plot of PDS for its inhibition of MALAT1 rG4-NONO interaction. Reaction mixture contains 50 nM FAM MALAT1 rG4_04, 200 nM NONO (53–312) and increasing concentrations of PDS (0.15–5000 nM). The IC50 is found to 498.2 ± 94.5 nM. (C) Similar set up as (B) except RHAU53 is used. The IC50 value is found to be 621.1 ± 95.3 nM. (D) Similar set up as (B) except L-Apt.4–1c was used. The IC50 value is found to be 618.1 ± 110.7 nM. (E) Filter-binding result of PDS for its inhibition of MALAT1 rG4-NONO interaction. Reaction mixture contains 2 nM Biotin MALAT1 rG4_04, 150 nM NONO (53–312) and increasing concentrations of PDS (0.15–5000 nM). (F) Similar set up as (E) except L-Apt.4–1c is used.
To support the MST results above, we carried out two independent assays, filter-binding assay and EMSA, and the results were consistent with our MST data. In filter-binding assay, by employing the reaction mixture to a Dot Blot apparatus containing a nitrocellulose membrane (top) and nylon membrane (bottom), rG4-protein bound would retain in the top membrane while the bottom membrane detained the rest. From the data, the MALAT1 rG4_04–NONO (53–312) complex was dissociated when increasing the concentration of PDS or L-Apt.4–1c (Figure 5E, F). Our EMSA result showed that as the concentration of RHAU53 escalates from 0 to 2500 nM, the MALAT1 rG4_04–NONO (53–312) complex band declined while the MALAT1 rG4_04–RHAU53 complex band increased (Supplementary Figure S9), suggesting the interference of RHAU53 to MALAT1 rG4_04–NONO (53–312) interaction. Taken the results together, we have demonstrated that the interaction between MALAT1 rG4 and NONO can be suppressed by multiple G4 targeting tools, including G4-specific small molecule, peptide, and L-RNA aptamer, which provide diverse approaches to targeting MALAT1 and its RNA–protein complex via its rG4 motifs.
DISCUSSION
Recent studies have shown that lncRNAs play vital roles in different biological processes (78–80). However, few works have elucidated RNA structural elements in lncRNAs (81–84). RG4s, being non-classical RNA structural motifs, have been studied more extensively in mRNAs but less so in ncRNAs such as lncRNAs (13,14,85). Besides the well-characterized rG4s in human telomerase RNA (hTR) (30,51,86) and telomeric repeat-containing RNA (TERRA) (31,32,87), so far only a handful of rG4 reported in lncRNAs, including FLJ39051(GSEC) (29), LUCAT1 (49) and NEAT1 (33). LncRNA MALAT1 is a highly conserved cancer-associated lncRNA that interacts with RNA-binding proteins to participate in transcription and post-transcriptional regulation in cells (88,89). It promotes cell proliferation, metastasis, and invasion in vitro and in vivo, thus playing a critical role in regulating cancer progression and metastasis (88–90). In this work, we have provided the first evidence that lncRNA MALAT1 contains multiple rG4 structures. By applying different biophysical and biochemical assays, we demonstrated that these rG4s are thermostable and formed in physiologically relevant conditions (Figure 1). Moreover, we found that the MALAT1 rG4s we studied in this work are conserved using comparative sequence analysis. Some of them are highly conserved across eight other species (Figure 2 and Supplementary Figure S1), including primates and mammals. Importantly, the G4 prediction scores were all above the G4 threshold (Figure 2), indicating that these sequences are likely to form rG4 structures. The conservation of the rG4 structure hints at the potential significance of the G4 structural element in the function of MALAT1 lncRNA.
MALAT1 and NEAT1 are both abundant and nuclear-enriched lncRNAs that localize adjacent to each other in the nucleus periodically and co-enrich many trans-genomic binding sites and protein factors, including NONO protein (67–69). The affinity of NONO for rG4 structures has been observed very recently for NEAT1 rG4s (33), and we have verified it as well (Supplementary Figure S3). In this study, we have demonstrated that NONO interacts with MALAT1 via rG4 structures both in vitro and in cells (Figures 3 and 4). MALAT1 and NEAT1 are predominately localized within nuclear speckles and paraspeckles (67), respectively, however, data has shown substantial overlaps between nuclear speckle and paraspeckle components associated with MALAT1 and NEAT1 (67). Moreover, knockout of MALAT1 affects the NEAT1 expression and paraspeckle formation in particular cell types, suggesting a potential co-regulatory function between this two lncRNAs (91,92). Taken together, we hypothesize that the rG4 structure motif may serve the role of potential cooperation or complementary function between these two lncRNAs in recruiting NONO and other nuclear proteins and regulating the formation of nuclear bodies. Our speculation is supported by data that suggested the canonical paraspeckle protein NONO was also found in nuclear speckles (68). In addition, SFPQ (Splicing factor proline- and glutamine-rich), a binding partner of NONO, was also reported to interact with NEAT1 rG4 (33) and MALAT1 (88,89), which further supports our hypothesis. Besides, our data showed that rG4 helicase DHX36 binds to MALAT1 rG4 strongly, with a comparable affinity as NONO protein (Supplementary Figures S2 and S6). Based on the proposed functions of DHX36 reported in other studies (29,51,93,94), we think that rG4 helicase DHX36 may also participate in the regulation of rG4-NONO complex formation in cells, potentially by competing for rG4 with NONO, followed by G4 unwinding. Our current data support the formation of MALAT1 rG4 and its interaction with NONO protein and DHX36 helicase. However, more needs to be done in order to decipher the underlying mechanism of MALAT1 rG4-protein regulation and function, as well as their participation and roles in regulating the formation of nuclear bodies in cells. While we have shown MALAT1 rG4s can form and are important in protein recognition, their folding dynamics in cells, as well as their biological consequences in cellular processes and cancer development remains elusive. The interaction between MALAT1 rG4 and NONO as well as DHX36 may be a promising target for MALAT1-related diseases and cancer therapy, which can be further investigated in future studies.
To explore the potential application, we have investigated the disruption ability of three classes of G4 targeting tools, including G4-specific small molecule, peptide and L-RNA aptamer. PDS is one of the popular G4 ligands used in recognizing G4 motifs and interfering with the G4–protein interactions (70,71). In this work, we have utilized PDS in vitro, in nuclear cell lysate, and in cell assays, underscoring its versatility and application in studying rG4s in lncRNA. RHAU53 peptide is an emerging G4 targeting tool that can specifically identify parallel G4, and several improved versions have been recently developed for specific G4 targeting (75,95–97). rG4-targeting L-RNA aptamer is a new class of G4-targeting tool introduced by us (60). L-Apt.4–1c is a novel L-RNA aptamer developed by our group recently (76). It was reported to generally bind to rG4s and interfere with the rG4–protein interaction in vitro and in cell lysate (76,77). Notably, L-Apt.4–1c is the shortest L-aptamer developed so far, with only 25 nt in length, making it easier to apply to experiments in cells in the near future. Based on our data, L-Apt.4–1c exhibited a similar ability to disrupt MALAT1 rG4-NONO interaction compared to PDS and RHAU53, highlighting the potential of L-aptamer in the G4 targeting field and the possibility of targeting lncRNA rG4-protein interactions. In addition, modification of the L-Apt.4–1c, such as cyclization (77), and optimization of the L-aptamer sequence should improve its performance further. It will be interesting and one of our future directions to apply these G4 targeting tools to explore the regulatory and mechanistic role of rG4s in MALAT1 lncRNA-associated cancers.
As described early in the result section, we have re-analyzed and identified more than 350 rG4 in about 200 ncRNAs using our published rG4-seq dataset. In this list (Supplementary Table S1), we also found rG4s in other biologically relevant lncRNAs such as TUG1 (taurine upregulated gene 1), HCG11 (HLA Complex Group 11), DGCR5 (DiGeorge Syndrome Critical Region Gene 5), HOTAIRM1 (HOXA Transcript Antisense RNA, Myeloid-Specific 1), which have been functionally characterized in other studies recently (98–105). Our view is that this list is likely an underestimation of the rG4s in ncRNAs as the rG4-seq was polyA-enriched (5), and that many ncRNAs do not have polyA tail or polyA region in their sequences. However, we have clearly illustrated in this study that the rG4 candidates obtained in this list are largely consistent with the G4 prediction tools results, suggesting that it is likely possible to yield valuable insights for experimentalists and look for ncRNA rG4s using computational approaches. Notably, based on our findings in this study on the rG4s in MALAT1 lncRNA, it is promising to apply the experimental strategies and methods reported here to investigate the rG4 structures and protein interactions in other important lncRNAs in vitro and in cells as well in the near future.
CONCLUSION
In sum, we have revealed novel rG4s in MALAT1 lncRNAs for the first time and showed that these rG4s are thermostable and conserved. We also uncovered these rG4s to have specific interactions with NONO protein both in vitro, in nuclear lysate and in cells. Importantly, we demonstrated that rG4 targeting tools including small molecule, peptide, and L-RNA aptamer could be used to suppress the MALAT1 rG4-NONO protein interaction, providing diverse strategies to targeting MALAT1 and its RNA–protein complex via its rG4 motifs. The findings reported in this study will motivate further exploration of the role of rG4s in MALAT1 lncRNA biology, and the approaches presented here should be applicable to the study of rG4 in other functional lncRNAs.
DATA AVAILABILITY
The data is available from the author upon request. The source data is also available at Mendeley Data (doi:10.17632/g5kh4gr398.1).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Omer Ziv, Eugene Yui-Ching Chow, Jia-Hao Yuan and Haizhou Zhao for the discussion and inputs in this manuscript.
Contributor Information
Xi Mou, Department of Chemistry and State Key Laboratory of Marine Pollution, City University of Hong Kong, Kowloon Tong, Hong Kong SAR, China.
Shiau Wei Liew, Department of Chemistry and State Key Laboratory of Marine Pollution, City University of Hong Kong, Kowloon Tong, Hong Kong SAR, China.
Chun Kit Kwok, Department of Chemistry and State Key Laboratory of Marine Pollution, City University of Hong Kong, Kowloon Tong, Hong Kong SAR, China; Shenzhen Research Institute of City University of Hong Kong, Shenzhen, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Shenzhen Basic Research Project [JCYJ20180507181642811]; Research Grants Council of the Hong Kong SAR, China Projects [CityU 11100421, CityU 11101519, CityU 11100218, N_CityU110/17, CityU 21302317]; Croucher Foundation Project [9500030, 9509003]; State Key Laboratory of Marine Pollution Director Discretionary Fund; City University of Hong Kong projects [6000711, 7005503, 9667222, 9680261 to C.K.K.]. Funding for open access charge: Research Grants Council of the Hong Kong SAR [CityU 11101519].
Conflict of interest statement. None declared.
REFERENCES
- 1. Balasubramanian S., Neidle S. Quadruplex nucleic acids. Rsc Biomol Sci. 2006; 7:1–30. [Google Scholar]
- 2. Kwok C.K., Merrick C.J. G-Quadruplexes: prediction, characterization, and biological application. Trends Biotechnol. 2017; 35:997–1013. [DOI] [PubMed] [Google Scholar]
- 3. Huppert J.L., Balasubramanian S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005; 33:2908–2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chambers V.S., Marsico G., Boutell J.M., Di Antonio M., Smith G.P., Balasubramanian S. High-throughput sequencing of DNA G-quadruplex structures in the human genome. Nat. Biotechnol. 2015; 33:877–881. [DOI] [PubMed] [Google Scholar]
- 5. Kwok C.K., Marsico G., Sahakyan A.B., Chambers V.S., Balasubramanian S. rG4-seq reveals widespread formation of G-quadruplex structures in the human transcriptome. Nat. Methods. 2016; 13:841–844. [DOI] [PubMed] [Google Scholar]
- 6. Lightfoot H.L., Hagen T., Tatum N.J., Hall J. The diverse structural landscape of quadruplexes. FEBS Lett. 2019; 593:2083–2102. [DOI] [PubMed] [Google Scholar]
- 7. Banco M.T., Ferre-D’Amare A.R. The emerging structural complexity of G-quadruplex RNAs. RNA. 2021; 27:390–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chan C.Y., Umar M.I., Kwok C.K. Spectroscopic analysis reveals the effect of a single nucleotide bulge on G-quadruplex structures. Chem. Commun. (Camb). 2019; 55:2616–2619. [DOI] [PubMed] [Google Scholar]
- 9. Kwok C.K., Sherlock M.E., Bevilacqua P.C. Effect of loop sequence and loop length on the intrinsic fluorescence of G-quadruplexes. Biochemistry. 2013; 52:3019–3021. [DOI] [PubMed] [Google Scholar]
- 10. Spiegel J., Adhikari S., Balasubramanian S. The Structure and Function of DNA G-Quadruplexes. Trends Chem. 2020; 2:123–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Varshney D., Spiegel J., Zyner K., Tannahill D., Balasubramanian S. The regulation and functions of DNA and RNA G-quadruplexes. Nat. Rev. Mol. Cell Biol. 2020; 21:459–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Robinson J., Raguseo F., Nuccio S.P., Liano D., Di Antonio M. DNA G-quadruplex structures: more than simple roadblocks to transcription. Nucleic Acids Res. 2021; 49:8419–8431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tassinari M., Richter S.N., Gandellini P. Biological relevance and therapeutic potential of G-quadruplex structures in the human noncoding transcriptome. Nucleic Acids Res. 2021; 49:3617–3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lyu K., Chow E.Y., Mou X., Chan T.F., Kwok C.K. RNA G-quadruplexes (rG4s): genomics and biological functions. Nucleic Acids Res. 2021; 49:5426–5450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rouleau S., Glouzon J.S., Brumwell A., Bisaillon M., Perreault J.P. 3′ UTR G-quadruplexes regulate miRNA binding. RNA. 2017; 23:1172–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Stefanovic S., Bassell G.J., Mihailescu M.R. G quadruplex RNA structures in PSD-95 mRNA: potential regulators of miR-125a seed binding site accessibility. RNA. 2015; 21:48–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Subramanian M., Rage F., Tabet R., Flatter E., Mandel J.L., Moine H. G-quadruplex RNA structure as a signal for neurite mRNA targeting. EMBO Rep. 2011; 12:697–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Goering R., Hudish L.I., Guzman B.B., Raj N., Bassell G.J., Russ H.A., Dominguez D., Taliaferro J.M. FMRP promotes RNA localization to neuronal projections through interactions between its RGG domain and G-quadruplex RNA sequences. Elife. 2020; 9:e52621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Crenshaw E., Leung B.P., Kwok C.K., Sharoni M., Olson K., Sebastian N.P., Ansaloni S., Schweitzer-Stenner R., Akins M.R., Bevilacqua P.C. et al. Amyloid precursor protein translation is regulated by a 3′UTR guanine quadruplex. PLoS One. 2015; 10:e0143160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Murat P., Marsico G., Herdy B., Ghanbarian A.T., Portella G., Balasubramanian S. RNA G-quadruplexes at upstream open reading frames cause DHX36- and DHX9-dependent translation of human mRNAs. Genome Biol. 2018; 19:229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Beaudoin J.D., Perreault J.P. Exploring mRNA 3′-UTR G-quadruplexes: evidence of roles in both alternative polyadenylation and mRNA shortening. Nucleic Acids Res. 2013; 41:5898–5911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Marcel V., Tran P.L., Sagne C., Martel-Planche G., Vaslin L., Teulade-Fichou M.P., Hall J., Mergny J.L., Hainaut P., Van Dyck E. G-quadruplex structures in TP53 intron 3: role in alternative splicing and in production of p53 mRNA isoforms. Carcinogenesis. 2011; 32:271–278. [DOI] [PubMed] [Google Scholar]
- 23. Westmark C.J., Malter J.S. FMRP mediates mGluR(5)-dependent translation of amyloid precursor protein. PLoS Biol. 2007; 5:629–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Thandapani P., Song J., Gandin V., Cai Y., Rouleau S.G., Garant J.M., Boisvert F.M., Yu Z., Perreault J.P., Topisirovic I. et al. Aven recognition of RNA G-quadruplexes regulates translation of the mixed lineage leukemia protooncogenes. Elife. 2015; 4:e06234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Huang H., Zhang J., Harvey S.E., Hu X., Cheng C. RNA G-quadruplex secondary structure promotes alternative splicing via the RNA-binding protein hnRNPF. Genes Dev. 2017; 31:2296–2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shahid R., Bugaut A., Balasubramanian S. The BCL-2 5′ untranslated region contains an RNA G-quadruplex-forming motif that modulates protein expression. Biochemistry. 2010; 49:8300–8306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Beaudoin J.D., Perreault J.P. 5′-UTR G-quadruplex structures acting as translational repressors. Nucleic Acids Res. 2010; 38:7022–7036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mattick J.S., Makunin I.V. Non-coding RNA. Hum. Mol. Genet. 2006; 15:R17–R29. [DOI] [PubMed] [Google Scholar]
- 29. Matsumura K., Kawasaki Y., Miyamoto M., Kamoshida Y., Nakamura J., Negishi L., Suda S., Akiyama T. The novel G-quadruplex-containing long non-coding RNA GSEC antagonizes DHX36 and modulates colon cancer cell migration. Oncogene. 2017; 36:1191–1199. [DOI] [PubMed] [Google Scholar]
- 30. Lattmann S., Stadler M.B., Vaughn J.P., Akman S.A., Nagamine Y. The DEAH-box RNA helicase RHAU binds an intramolecular RNA G-quadruplex in TERC and associates with telomerase holoenzyme. Nucleic Acids Res. 2011; 39:9390–9404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Biffi G., Tannahill D., Balasubramanian S. An intramolecular G-quadruplex structure is required for binding of telomeric repeat-containing RNA to the telomeric protein TRF2. J. Am. Chem. Soc. 2012; 134:11974–11976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Deng Z., Norseen J., Wiedmer A., Riethman H., Lieberman P.M. TERRA RNA binding to TRF2 facilitates heterochromatin formation and ORC recruitment at telomeres. Mol. Cell. 2009; 35:403–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Simko E.A.J., Liu H., Zhang T., Velasquez A., Teli S., Haeusler A.R., Wang J. G-quadruplexes offer a conserved structural motif for NONO recruitment to NEAT1 architectural lncRNA. Nucleic Acids Res. 2020; 48:7421–7438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chan K.L., Peng B., Umar M.I., Chan C.Y., Sahakyan A.B., Le M.T.N., Kwok C.K. Structural analysis reveals the formation and role of RNA G-quadruplex structures in human mature microRNAs. Chem. Commun. (Camb.). 2018; 54:10878–10881. [DOI] [PubMed] [Google Scholar]
- 35. Kwok C.K., Sahakyan A.B., Balasubramanian S. Structural analysis using SHALiPE to reveal RNA G-quadruplex formation in human precursor microRNA. Angew. Chem. Int. Ed. Engl. 2016; 55:8958–8961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pandey S., Agarwala P., Jayaraj G.G., Gargallo R., Maiti S. The RNA stem-loop to G-quadruplex equilibrium controls mature microRNA production inside the cell. Biochemistry. 2015; 54:7067–7078. [DOI] [PubMed] [Google Scholar]
- 37. Vourekas A., Zheng K., Fu Q., Maragkakis M., Alexiou P., Ma J., Pillai R.S., Mourelatos Z., Wang P.J. The RNA helicase MOV10L1 binds piRNA precursors to initiate piRNA processing. Genes Dev. 2015; 29:617–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Balaratnam S., Hettiarachchilage M., West N., Piontkivska H., Basu S. A secondary structure within a human piRNA modulates its functionality. Biochimie. 2019; 157:72–80. [DOI] [PubMed] [Google Scholar]
- 39. Mestre-Fos S., Ito C., Moore C.M., Reddi A.R., Williams L.D. Human ribosomal G-quadruplexes regulate heme bioavailability. J. Biol. Chem. 2020; 295:14855–14865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mestre-Fos S., Penev P.I., Suttapitugsakul S., Hu M., Ito C., Petrov A.S., Wartell R.M., Wu R.H., Williams L.D. G-Quadruplexes in human ribosomal RNA. J. Mol. Biol. 2019; 431:1940–1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ivanov P., O’Day E., Emara M.M., Wagner G., Lieberman J., Anderson P. G-quadruplex structures contribute to the neuroprotective effects of angiogenin-induced tRNA fragments. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:18201–18206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lyons S.M., Gudanis D., Coyne S.M., Gdaniec Z., Ivanov P. Identification of functional tetramolecular RNA G-quadruplexes derived from transfer RNAs. Nat. Commun. 2017; 8:1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kharel P., Balaratnam S., Beals N., Basu S. The role of RNA G-quadruplexes in human diseases and therapeutic strategies. Wiley Interdiscip. Rev. RNA. 2020; 11:e1568. [DOI] [PubMed] [Google Scholar]
- 44. Sanchez de Groot N., Armaos A., Grana-Montes R., Alriquet M., Calloni G., Vabulas R.M., Tartaglia G.G. RNA structure drives interaction with proteins. Nat. Commun. 2019; 10:3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Corley M., Burns M.C., Yeo G.W. How RNA-binding proteins interact with RNA: molecules and mechanisms. Mol. Cell. 2020; 78:9–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Darnell J.C., Jensen K.B., Jin P., Brown V., Warren S.T., Darnell R.B. Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell. 2001; 107:489–499. [DOI] [PubMed] [Google Scholar]
- 47. Schaeffer C., Bardoni B., Mandel J.L., Ehresmann B., Ehresmann C., Moine H. The fragile X mental retardation protein binds specifically to its mRNA via a purine quartet motif. EMBO J. 2001; 20:4803–4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kenny P.J., Kim M., Skariah G., Nielsen J., Lannom M.C., Ceman S. The FMRP-MOV10 complex: a translational regulatory switch modulated by G-Quadruplexes. Nucleic Acids Res. 2020; 48:862–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wu R., Li L., Bai Y., Yu B., Xie C., Wu H., Zhang Y., Huang L., Yan Y., Li X. et al. The long noncoding RNA LUCAT1 promotes colorectal cancer cell proliferation by antagonizing Nucleolin to regulate MYC expression. Cell Death. Dis. 2020; 11:908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sexton A.N., Collins K. The 5′ guanosine tracts of human telomerase RNA are recognized by the G-quadruplex binding domain of the RNA helicase DHX36 and function to increase RNA accumulation. Mol. Cell. Biol. 2011; 31:736–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Booy E.P., Meier M., Okun N., Novakowski S.K., Xiong S., Stetefeld J., McKenna S.A. The RNA helicase RHAU (DHX36) unwinds a G4-quadruplex in human telomerase RNA and promotes the formation of the P1 helix template boundary. Nucleic Acids Res. 2012; 40:4110–4124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Mergny J.L., Lacroix L. UV melting of G-quadruplexes. Curr Protoc Nucleic Acid Chem. 2009; Chapter 17:Unit 17.1. [DOI] [PubMed] [Google Scholar]
- 53. Mergny J.L., Phan A.T., Lacroix L. Following G-quartet formation by UV-spectroscopy. FEBS Lett. 1998; 435:74–78. [DOI] [PubMed] [Google Scholar]
- 54. Howe K.L., Achuthan P., Allen J., Allen J., Alvarez-Jarreta J., Amode M.R., Armean I.M., Azov A.G., Bennett R., Bhai J. et al. Ensembl 2021. Nucleic Acids Res. 2021; 49:D884–D891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Procter J.B., Carstairs G.M., Soares B., Mourao K., Ofoegbu T.C., Barton D., Lui L., Menard A., Sherstnev N., Roldan-Martinez D. et al. Alignment of Biological Sequences with Jalview. Methods Mol. Biol. 2021; 2231:203–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Beaudoin J.D., Jodoin R., Perreault J.P. New scoring system to identify RNA G-quadruplex folding. Nucleic Acids Res. 2014; 42:1209–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Bedrat A., Lacroix L., Mergny J.L. Re-evaluation of G-quadruplex propensity with G4Hunter. Nucleic Acids Res. 2016; 44:1746–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Garant J.M., Perreault J.P., Scott M.S. Motif independent identification of potential RNA G-quadruplexes by G4RNA screener. Bioinformatics. 2017; 33:3532–3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wang H., Yang C., Tang H., Li Y. Stochastic collision electrochemistry from single G-quadruplex/hemin: electrochemical amplification and microRNA sensing. Anal. Chem. 2021; 93:4593–4600. [DOI] [PubMed] [Google Scholar]
- 60. Chan C.Y., Kwok C.K. Specific binding of a d-RNA G-quadruplex structure with an l-RNA aptamer. Angew. Chem. Int. Ed. 2020; 59:5293–5297. [DOI] [PubMed] [Google Scholar]
- 61. Chu C., Chang H.Y. ChIRP-MS: RNA-directed proteomic discovery. Methods Mol. Biol. 2018; 1861:37–45. [DOI] [PubMed] [Google Scholar]
- 62. Hardin C.C., Perry A.G., White K. Thermodynamic and kinetic characterization of the dissociation and assembly of quadruplex nucleic acids. Biopolymers. 2000; 56:147–194. [DOI] [PubMed] [Google Scholar]
- 63. Renaud de la Faverie A., Guedin A., Bedrat A., Yatsunyk L.A., Mergny J.L. Thioflavin T as a fluorescence light-up probe for G4 formation. Nucleic Acids Res. 2014; 42:e65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Creacy S.D., Routh E.D., Iwamoto F., Nagamine Y., Akman S.A., Vaughn J.P. G4 resolvase 1 binds both DNA and RNA tetramolecular quadruplex with high affinity and is the major source of tetramolecular quadruplex G4-DNA and G4-RNA resolving activity in HeLa cell lysates. J. Biol. Chem. 2008; 283:34626–34634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Zhang Z., Kim T., Bao M., Facchinetti V., Jung S.Y., Ghaffari A.A., Qin J., Cheng G., Liu Y.J. DDX1, DDX21, and DHX36 helicases form a complex with the adaptor molecule TRIF to sense dsRNA in dendritic cells. Immunity. 2011; 34:866–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Hutchinson J.N., Ensminger A.W., Clemson C.M., Lynch C.R., Lawrence J.B., Chess A. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics. 2007; 8:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. West J.A., Davis C.P., Sunwoo H., Simon M.D., Sadreyev R.I., Wang P.I., Tolstorukov M.Y., Kingston R.E. The long noncoding RNAs NEAT1 and MALAT1 bind active chromatin sites. Mol. Cell. 2014; 55:791–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Saitoh N., Spahr C.S., Patterson S.D., Bubulya P., Neuwald A.F., Spector D.L. Proteomic analysis of interchromatin granule clusters. Mol. Biol. Cell. 2004; 15:3876–3890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Fox A.H., Lam Y.W., Leung A.K., Lyon C.E., Andersen J., Mann M., Lamond A.I. Paraspeckles: a novel nuclear domain. Curr. Biol. 2002; 12:13–25. [DOI] [PubMed] [Google Scholar]
- 70. Li L., Williams P., Ren W., Wang M.Y., Gao Z., Miao W., Huang M., Song J., Wang Y. YY1 interacts with guanine quadruplexes to regulate DNA looping and gene expression. Nat. Chem. Biol. 2021; 17:161–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Herviou P., Le Bras M., Dumas L., Hieblot C., Gilhodes J., Cioci G., Hugnot J.P., Ameadan A., Guillonneau F., Dassi E. et al. hnRNP H/F drive RNA G-quadruplex-mediated translation linked to genomic instability and therapy resistance in glioblastoma. Nat. Commun. 2020; 11:2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Chu C., Zhang Q.C., da Rocha S.T., Flynn R.A., Bharadwaj M., Calabrese J.M., Magnuson T., Heard E., Chang H.Y. Systematic discovery of Xist RNA binding proteins. Cell. 2015; 161:404–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Sun Z.Y., Wang X.N., Cheng S.Q., Su X.X., Ou T.M. Developing novel G-quadruplex ligands: from interaction with nucleic acids to interfering with nucleic acid(-)protein interaction. Molecules. 2019; 24:396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Abdelhaleem M., Maltais L., Wain H. The human DDX and DHX gene families of putative RNA helicases. Genomics. 2003; 81:618–622. [DOI] [PubMed] [Google Scholar]
- 75. Heddi B., Cheong V.V., Martadinata H., Phan A.T. Insights into G-quadruplex specific recognition by the DEAH-box helicase RHAU: Solution structure of a peptide-quadruplex complex. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:9608–9613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Umar M.I., Kwok C.K. Specific suppression of D-RNA G-quadruplex-protein interaction with an L-RNA aptamer. Nucleic Acids Res. 2020; 48:10125–10141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Ji D., Lyu K., Zhao H., Kwok C.K. Circular L-RNA aptamer promotes target recognition and controls gene activity. Nucleic Acids Res. 2021; 49:7280–7291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Kung J.T., Colognori D., Lee J.T. Long noncoding RNAs: past, present, and future. Genetics. 2013; 193:651–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Fang Y., Fullwood M.J. Roles, functions, and mechanisms of long non-coding RNAs in cancer. Genomics Proteomics Bioinformatics. 2016; 14:42–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Statello L., Guo C.J., Chen L.L., Huarte M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2021; 22:96–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Somarowthu S., Legiewicz M., Chillon I., Marcia M., Liu F., Pyle A.M. HOTAIR forms an intricate and modular secondary structure. Mol. Cell. 2015; 58:353–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Liu F., Somarowthu S., Pyle A.M. Visualizing the secondary and tertiary architectural domains of lncRNA RepA. Nat. Chem. Biol. 2017; 13:282–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Uroda T., Anastasakou E., Rossi A., Teulon J.M., Pellequer J.L., Annibale P., Pessey O., Inga A., Chillon I., Marcia M. Conserved pseudoknots in lncRNA MEG3 are essential for stimulation of the p53 pathway. Mol. Cell. 2019; 75:982–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Qian X., Zhao J., Yeung P.Y., Zhang Q.C., Kwok C.K. Revealing lncRNA structures and interactions by sequencing-based approaches. Trends Biochem. Sci. 2019; 44:33–52. [DOI] [PubMed] [Google Scholar]
- 85. Kamura T., Katsuda Y., Kitamura Y., Ihara T. G-quadruplexes in mRNA: A key structure for biological function. Biochem. Biophys. Res. Commun. 2020; 526:261–266. [DOI] [PubMed] [Google Scholar]
- 86. Martadinata H., Phan A.T. Formation of a stacked dimeric G-quadruplex containing bulges by the 5′-terminal region of human telomerase RNA (hTERC). Biochemistry. 2014; 53:1595–1600. [DOI] [PubMed] [Google Scholar]
- 87. Cusanelli E., Chartrand P. Telomeric repeat-containing RNA TERRA: a noncoding RNA connecting telomere biology to genome integrity. Front Genet. 2015; 6:143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Ji Q., Zhang L., Liu X., Zhou L., Wang W., Han Z., Sui H., Tang Y., Wang Y., Liu N. et al. Long non-coding RNA MALAT1 promotes tumour growth and metastasis in colorectal cancer through binding to SFPQ and releasing oncogene PTBP2 from SFPQ/PTBP2 complex. Br. J. Cancer. 2014; 111:736–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Ji Q., Cai G., Liu X., Zhang Y., Wang Y., Zhou L., Sui H., Li Q. MALAT1 regulates the transcriptional and translational levels of proto-oncogene RUNX2 in colorectal cancer metastasis. Cell Death. Dis. 2019; 10:378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Amodio N., Raimondi L., Juli G., Stamato M.A., Caracciolo D., Tagliaferri P., Tassone P. MALAT1: a druggable long non-coding RNA for targeted anti-cancer approaches. J. Hematol. Oncol. 2018; 11:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Nakagawa S., Ip J.Y., Shioi G., Tripathi V., Zong X., Hirose T., Prasanth K.V. Malat1 is not an essential component of nuclear speckles in mice. RNA. 2012; 18:1487–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Zhang B., Arun G., Mao Y.S., Lazar Z., Hung G., Bhattacharjee G., Xiao X., Booth C.J., Wu J., Zhang C. et al. The lncRNA Malat1 is dispensable for mouse development but its transcription plays a cis-regulatory role in the adult. Cell Rep. 2012; 2:111–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Chen X., Yuan J., Xue G., Campanario S., Wang D., Wang W., Mou X., Liew S.W., Umar M.I., Isern J. et al. Translational control by DHX36 binding to 5′UTR G-quadruplex is essential for muscle stem-cell regenerative functions. Nat. Commun. 2021; 12:5043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Sauer M., Juranek S.A., Marks J., De Magis A., Kazemier H.G., Hilbig D., Benhalevy D., Wang X., Hafner M., Paeschke K. DHX36 prevents the accumulation of translationally inactive mRNAs with G4-structures in untranslated regions. Nat. Commun. 2019; 10:2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Minard A., Morgan D., Raguseo F., Di Porzio A., Liano D., Jamieson A.G., Di Antonio M. A short peptide that preferentially binds c-MYC G-quadruplex DNA. Chem. Commun. (Camb.). 2020; 56:8940–8943. [DOI] [PubMed] [Google Scholar]
- 96. Wen C.J., Gong J.Y., Zheng K.W., He Y.D., Zhang J.Y., Hao Y.H., Tan Z. Targeting nucleic acids with a G-triplex-to-G-quadruplex transformation and stabilization using a peptide-PNA G-tract conjugate. Chem. Commun. (Camb.). 2020; 56:6567–6570. [DOI] [PubMed] [Google Scholar]
- 97. He Y.D., Zheng K.W., Wen C.J., Li X.M., Gong J.Y., Hao Y.H., Zhao Y., Tan Z. Selective targeting of guanine-vacancy-bearing G-quadruplexes by G-quartet complementation and stabilization with a guanine-peptide conjugate. J. Am. Chem. Soc. 2020; 142:11394–11403. [DOI] [PubMed] [Google Scholar]
- 98. Dumbović G., Braunschweig U., Langner H.K., Smallegan M., Biayna J., Hass E.P., Jastrzebska K., Blencowe B., Cech T.R., Caruthers M.H. et al. Nuclear compartmentalization of TERT mRNA and TUG1 lncRNA is driven by intron retention. Nat. Commun. 2021; 12:3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Lewandowski J.P., Dumbovic G., Watson A.R., Hwang T., Jacobs-Palmer E., Chang N., Much C., Turner K.M., Kirby C., Rubinstein N.D. et al. The Tug1 lncRNA locus is essential for male fertility. Genome Biol. 2020; 21:237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Zhang Q., Yang K., Li J., Chen F., Li Y., Lin Q. Long noncoding RNA HCG11 acts as a tumor suppressor in gastric cancer by regulating miR-942-5p/BRMS1 Axis. J. Oncol. 2021; 2021:9961189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Xu Y., Zheng Y., Liu H., Li T. Modulation of IGF2BP1 by long non-coding RNA HCG11 suppresses apoptosis of hepatocellular carcinoma cells via MAPK signaling transduction. Int. J. Oncol. 2017; 51:791–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Wang Y.G., Liu J., Shi M., Chen F.X. LncRNA DGCR5 represses the development of hepatocellular carcinoma by targeting the miR-346/KLF14 axis. J. Cell. Physiol. 2018; 234:572–580. [DOI] [PubMed] [Google Scholar]
- 103. Xue C., Chen C., Gu X., Li L. Progress and assessment of lncRNA DGCR5 in malignant phenotype and immune infiltration of human cancers. Am J Cancer Res. 2021; 11:1–13. [PMC free article] [PubMed] [Google Scholar]
- 104. Zhang L., Zhang J., Li S., Zhang Y., Liu Y., Dong J., Zhao W., Yu B., Wang H., Liu J. Genomic amplification of long noncoding RNA HOTAIRM1 drives anaplastic thyroid cancer progression via repressing miR-144 biogenesis. RNA Biol. 2021; 18:547–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Wang H., Li H., Jiang Q., Dong X., Li S., Cheng S., Shi J., Liu L., Qian Z., Dong J. HOTAIRM1 promotes malignant progression of transformed fibroblasts in glioma stem-like cells remodeled microenvironment via regulating miR-133b-3p/TGFbeta axis. Front. Oncol. 2021; 11:603128. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data is available from the author upon request. The source data is also available at Mendeley Data (doi:10.17632/g5kh4gr398.1).