Abstract
Template-switching reverse transcription is widely used in RNA sequencing for low-input and low-quality samples, including RNA from single cells or formalin-fixed paraffin-embedded (FFPE) tissues. Previously, we identified the native eukaryotic mRNA 5′ cap as a key structural element for enhancing template switching efficiency. Here, we introduce CapTS-seq, a new strategy for sequencing small RNAs that combines chemical capping and template switching. We probed a variety of non-native synthetic cap structures and found that an unmethylated guanosine triphosphate cap led to the lowest bias and highest efficiency for template switching. Through cross-examination of different nucleotides at the cap position, our data provided unequivocal evidence that the 5′ cap acts as a template for the first nucleotide in reverse transcriptase-mediated post-templated addition to the emerging cDNA—a key feature to propel template switching. We deployed CapTS-seq for sequencing synthetic miRNAs, human total brain and liver FFPE RNA, and demonstrated that it consistently improves library quality for miRNAs in comparison with a gold standard template switching-based small RNA-seq kit.
INTRODUCTION
Small RNAs (<200 nucleotides) are for the most part non-coding regulatory elements and play a key role in gene expression. This diverse class of RNAs includes small interfering RNAs (siRNAs), microRNAs (miRNAs), Piwi-interacting RNAs (piRNAs), small nucleolar RNAs (snoRNAs), small nuclear RNAs (snRNAs), small Cajal body-specific RNAs (scaRNAs), and transfer RNAs (tRNAs) (1–3). An increasing number of small RNA species continue to be discovered as technologies become more sophisticated. Small RNAs regulate gene expression in plants, animals, and many fungi—including several roles in development, proliferation, differentiation, immune reaction, apoptosis, tumorigenesis and adaptation to stress (4,5). Given their importance in regulation, it is no surprise that miRNAs are candidates as biomarkers for several human diseases (6,7). miRNAs directly interact with target sites in the 3′ untranslated region of mRNA to repress expression. It has been estimated that >60% of mRNAs contain target sites for miRNAs, and many can target up to several hundred mRNAs, making miRNAs critical to a myriad of biological processes. In point of fact, a growing body of research has cast light on the association of aberrant miRNA expression with several human diseases (8). Therefore, developing accurate and reproducible ways to study these and other small RNAs is necessary to further decipher their biological consequences.
Microarray-based methods are often used for the analysis of miRNAs, but this technique suffers from low sensitivity, especially when targeting low abundant sequences (9). Newer methodologies based on next generation sequencing (NGS) have become available for small RNAs; however, there is still a lack of consistency and specificity compared with more mature mRNA sequencing workflows (10–16). Owing to the unique molecular characteristics of small RNAs, namely length, structure and chemical modifications, most sequencing strategies are riddled with bias. The main sources of bias in a typical library preparation workflow are the enzymatic ligations that introduce 5′ and 3′ sequencing adaptors to single-stranded templates (17). Poly-A or poly-C tailing the RNA 3′ end is a common approach to bypass the use of ligases. Nevertheless, biases can still emerge from end tailing depending on the RNA primary and secondary structures (18,19) and on the presence of modifications near the 3′ end (18). Adapting the RNA 5′ end is even more challenging because ligation at that position is often inefficient and highly dependent on the sequence and structure of both target and adapter (20–25). In addition, the formation of side products from intramolecular adapter ligation further complicates the analysis of the similarly sized adapted small RNAs (21). A great deal of effort has been made to overcome ligation issues. A very recent example is the use of randomized splint ligation, which was shown to allow detection of small RNAs with much lower bias and higher sensitivity (26).
Alternatives to traditional ligation-based workflows have gained popularity. One of them is the use of template switching, which permits ligation-free incorporation of the 5′ adapter during reverse transcription (27–29). Template switching-based methods depend upon the natural tendency of Moloney murine leukemia virus (MMLV)-type reverse transcriptases to add nontemplated nucleotides at the 3′ end of the emerging cDNA strand. These nontemplated additions serve as an anchoring unit for annealing complementary nucleotides in a provided template switching oligonucleotide (TSO); upon reaching the cDNA-TSO cross-junction, the reverse transcriptase effectively switches templates, continuing cDNA synthesis out of the TSO sequence. By incorporating the 5′ adapter sequence into the TSO, and using polyadenylation to prime reverse transcription, ligation steps can be avoided altogether. For applications where the total RNA input is limited, such as single-cell RNA sequencing, template switching offers a critical advantage as it reduces the number of steps and sample loss during library preparation. Several kits that utilize template switching are commercially available, including from Takara (SMARTer cDNA Synthesis kits), Diagenode (D-Plex RNA-seq kits) and NEB (NEBNext Single Cell/Low Input kits). These kits have been successfully deployed in mRNA workflows, providing an appreciable decrease of biases in terms of sequence and presence of 2′-OMe modifications. For small RNAs, however, detection sensitivity continues to be an issue. Even though improvements in the levels of bias are apparent with small RNA template switching-based kits, the high background caused by rRNA and the strong formation of side products significantly decrease the mapping rate relative to ligation-based methods (30–32).
Until very recently, it was widely accepted that MMLV-based reverse transcriptases added several nontemplated deoxycytidines to the 3′ end of the nascent cDNA (33–36). This has served as a guide for the design of TSOs featuring three terminal guanosines (rGrGrG-3′) in many commercially available kits. In our recent study (37), however, we found that (i) rather than multiple nontemplated deoxycytidines, only one deoxycytidine was required for efficient template switching, and (ii) important to the success of this process was the presence of a templating N7-methylguanosine (m7G) cap. Furthermore, we demonstrated that uncapped RNAs (e.g. RNAs whose 5′ end comprises an OH or monophosphate) suffer from high biases and lower template switching efficiencies. In fact, we had raised the concern that this may be one of the main reasons behind the reported underperformance of template switching-based small RNA kits.
Although capping strategies have been applied to enrich full-length mRNA transcripts for transcription start site (TSS) identification (11,34,38), they were never considered in the context of small or segmented RNAs. In this report, we introduce a novel sequencing library preparation workflow (CapTS-seq) that combines chemical capping—to enable the installation of a synthetic cap onto 5′-monophosphate RNAs—and template switching reverse transcription. The workflow targeted for small or fragmented RNAs was conceived based on the analysis of nontemplated additions and template switching efficiencies seen with RNAs featuring a variety of synthetic cap structures. Compared to a gold standard template switching-based workflow for small RNA sequencing, CapTS-seq consistently produced higher sequencing read quality and increased detection of miRNAs in a diverse range of RNA samples, including purified small RNA, total brain RNA and RNA extracts from formalin-fixed paraffin-embedded (FFPE) tissue. Finally, we showed that miRNAs and fragmented mRNAs can be detected simultaneously in FFPE RNA, highlighting the power of CapTS-seq not only to profile small RNA expression, but also to shed light on the network of interactions of miRNAs with their target gene transcripts.
MATERIALS AND METHODS
Materials
All reagents were from New England Biolabs (NEB), Ipswich, MA, USA, unless otherwise stated. Unless otherwise specified, all oligonucleotides were obtained from Integrated DNA Technologies (IDT), Coralville, IA, USA. RNA templates, primers, and TSO sequences used in this study are shown in Supplementary Table S1. Synthesis and characterization of the nucleoside phosphorimidazolides (NMP-, NDP-, and NTP-imidazolides) used for chemical capping are described in Supplementary Methods and Supplementary Tables S2 and S3. Synthesis of the nucleoside phosphorimidazolides was performed as described previously (39–41).
Several different pools of RNA were obtained for sequencing. The 16 RNA pool was created by individually mixing 16 discrete 5′-phosphate oligonucleotide sequences (25mers) that vary only in the first two 5′ nucleotides. Mix4v7 was created by individually mixing 13 unique 5′-phosphate sequences (23mers) that do not map to any known miRNAs (Supplementary Table S4). The RNA oligonucleotide pool with randomized N1–N4 positions (25mers) was synthesized using the ‘hand-mix’ option with an equimolar ratio of all four bases (IDT). The miRXplore Universal Reference library obtained from Miltenyi Biotec (Auburn, CA, USA) contained an equimolar mixture of 962 unique 5′-phosphate miRNA sequences. Human brain total RNA (#R1234035) and human liver FFPE total RNA (#R2234149) were obtained from Biochain Institute (Newark, CA, USA). Prior to use in library preparation, human brain total RNA was subjected to size selection using a Zymo RNA Clean & Concentrator kit (Zymo Research, Irvine, CA, USA; #R1013) following the protocol for separation of small from large RNAs.
Chemical capping
Chemical capping of synthetic 5′-monophosphate RNA oligonucleotides (5 nmol) was performed at a 250 μl reaction scale. On ice, a 5′-monophosphate oligonucleotide (100 μM, 50 μl) was combined with Bis–Tris buffer pH 6 (1 M, 50 μl), MnCl2 (1 M, 5 μl), and DMF (50 μl). For the generation of NppN-25mers, an NMP-imidazolide (100 mM, 95 μl) was added to this solution, and the reaction incubated at 50°C for 5 h. For the generation of NpppN-25mers, an NDP-imidazolide (100 mM, 95 μl) was added, and the reaction incubated at 37°C for 5 h. For the generation of NppppN-25mers, an NTP-imidazolide (100 mM, 95 μl) was added, and the reaction incubated at room temperature for 4 h. After this time, any unreacted imidazolide was removed from the reaction along with salts and organic solvent using a Sep-Pak C18 cartridge (Waters, #WAT051910). Briefly, the capping reaction was diluted to 5 ml in 0.1 M triethylammonium bicarbonate (TEAB). This diluted reaction was applied to the conditioned cartridge (0.1 M TEAB) and then washed with 15 ml of 0.1 M TEAB. The capped oligonucleotide was eluted from the column using 1:1 TEAB:Acetonitrile (2 ml). The presence of the oligonucleotide on eluates was confirmed by NanoDrop. The crude material was concentrated on the SpeedVac and purified by polyacrylamide gel electrophoresis (PAGE). The identity of the capped oligonucleotides was confirmed by intact mass spectrometry (MS) analysis on a Thermo Q-Exactive Plus operating under negative electrospray ionization mode (–ESI) (Appendix 1 and Supplementary Table S3). ESI-MS raw data was deconvoluted using ProMass HR (Novatia, LCC).
Template-switching reverse transcription assays
To estimate the template switching efficiency, a template-switching reverse transcription reaction (10 μl total volume) was carried out in the presence of 0.1 μM RNA template (1 pmol), 1× Template Switching RT Buffer (NEB #B0466), 1 mM dNTP solution mixture (NEB #N0447), 30 nM 5′-FAM V5 primer, 1 μM TSO and 100 units of Template Switching RT (NEB #M0466). The reaction was performed at 42°C for 90 min, followed by a 10-min heat-denaturation step at 72°C. The reverse transcription reaction was directly analyzed by capillary electrophoresis (CE) without purification. The template switching efficiency was calculated by quantifying all template switching products, including concatemers that formed from multiple template switching events that occured on the same cDNA. The template switching products were summed and compared with the corresponding primer elongation products.
To access the nature and extent of the nontemplated addition, the reverse transcription reaction (30 μl total volume) was carried out without a TSO and in the presence of 1 μM RNA template (30 pmol), 1× Template Switching RT Buffer, 2 mM dNTP solution mixture, 0.5 μM 5′-FAM V5 primer, and 400 units of Template Switching RT. The reaction was performed at 42°C for 90 min, followed by a 10-min heat-denaturation step at 72°C. The template RNA was hydrolyzed by adding sodium hydroxide (1 M, 10 μl) and EDTA pH 8.0 (0.5 M, 10 μl) to the reaction and heating to 65°C for 15 min. The cDNA was purified using a Oligo Clean & Concentrator (Zymo Research #D4061) and analyzed by mass spectrometry (MS) as previously described (37).
To investigate the effect of chemical capping in miRNA sequence coverage, the miRXplore reference was first poly(A)-tailed and then either G-capped or left uncapped. The RNA (capped or uncapped) was quantified using a Qubit microRNA assay. A 0.5 pg RNA input was subjected to template switching and PCR amplification as described below.
Library construction using CapTS-seq
Phosphorylation of FFPE total RNA
FFPE Total RNA (250 ng) was dissolved in nuclease-free water (up to final volume of 50 μl), followed by 10× T4 PNK Reaction Buffer (NEB #B0201), ATP (final concentration 1 mM; NEB #P0756), and T4 PNK (1 μl; NEB #M0201). The reaction was incubated for 30 min at 37°C after which time the material was subjected to purification using a Zymo RNA Clean & Concentrator Kit following the standard protocol.
Chemical capping and XRN-1 treatment
5′-Phosphorylated RNA (125 ng of human total brain or liver FFPE RNA spiked with 0.2 ng of Mix4v7; or 20 ng of miRXplore reference, or 16 RNA pool, or randomized pool) was dissolved in 5× Bis–Tris buffer, pH 6 (final concentration 200 mM), MnCl2 (final concentration 20 mM), DMF (20% final volume), and nuclease-free water (up to final volume of 60 μl). To this reaction was added GDP-imidazolide (for Gppp cap) or NPn-imidazolide (for caps of varying nucleotide or phosphate length) dissolved in nuclease-free water (final concentration 30 mM). The capping reaction was incubated at 37°C for 4 h. The reaction was then diluted with nuclease-free water (180 μl), and then XRN-1 was added (5.5 μl; NEB #M0388). The reaction was incubated for additional 1 h at 37°C. The crude reaction was purified using the Oligo Clean-up and Concentration Kit (Norgen Biotek, Ontario, Canada; #34100).
Poly(A) tailing
RNA (0.2–200 ng) was dissolved in nuclease-free water (up to final volume of 20 μl), followed by 10× Escherichia coli Poly(A) Polymerase Reaction Buffer (2 μl; NEB #B0278), 10 mM ATP (2 μl; NEB #P0756) and E. coli Poly(A) Polymerase (1 μl; NEB #M0276). The reaction was incubated for 5 min at 16°C after which time the material was subjected to purification using a Zymo RNA Clean & Concentrator Kit.
Template-switching reverse transcription
The purified poly(A)-tailed RNA was dissolved in the Template Switching RT Buffer (5 μl; NEB #B0466), followed by Deoxynucleotide Solution Mix (2.5 μl; NEB #N0447), 10 μM template switching oligonucleotide containing the universal PCR primer sequence for Illumina (2.5 μl; IDT), 10 μM Poly(dT) VN SR RT primer for Illumina (1.25 μl, IDT), and nuclease-free water (up to 21 μl). The mixture was heated to 50°C for 10 min and slowly cooled to 25°C. To this mixture was added Template Switching RT Enzyme Mix (4 μl; NEB M0466). The reverse transcription reaction was incubated for 10 min at 25°C, 90 min at 42°C, 10 min at 70°C, and cooled to 4°C. The reaction was diluted with nuclease-free water (35 μl), vortexed with isopropanol (46.2 μl) and NEBNext Sample Purification Beads (47.7 μl; NEB #E7767). The cDNA product was allowed to bind to the beads for 10 min after which time the tube was placed on a magnetic rack to separate the beads, and the supernatant was removed. The beads were washed twice with 80% ethanol and allowed to dry for 10 min. To elute the cDNA, the beads were resuspended in 13 μl of nuclease-free water.
PCR amplification and barcoding of cDNA
The cDNA (10 μl) was dissolved in LongAmp Taq 2X Master Mix (25 μl; NEB #E7309), followed by Universal Primer for Illumina (2.5 μl; NEB #E6861), one primer from NEBNext Multiplex Oligos for Illumina (2.5 μl; NEB #E7335), and nuclease-free water (10 μl). The cDNA was denatured at 94°C for 30 s, and then cycled 6–9 times through 94°C (15 s), 62°C (30 s), 70°C (15 s), before a final extension at 70°C for 5 min. The reaction was purified using NEBNext Sample Purification Beads. Briefly, the PCR reaction (50 μl) was resuspended with beads (65 μl) to remove larger fragments. PCR products were allowed to bind for 5 min after which time, the beads were separated on a magnetic rack, and the supernatant removed to a fresh tube. More beads (185 μl) were added to this supernatant to remove smaller fragments and allowed to bind for 5 min, after which time the supernatant was removed via magnetic rack. The beads were washed twice with 80% ethanol, dried for 10 min, and resuspended in 15 μl of nuclease-free water. The purified PCR library was analyzed on a High Sensitivity DNA Chip (1 μl; Agilent Technologies #5067-4626) using a 2100 Bioanalyzer Instrument (Agilent Technologies, Santa Clara, CA, USA).
Library construction using SMARTer, TruSeq and NEBNext Ultra II
Libraries for small RNA sequencing were prepared using a SMARTer smRNA-Seq Kit (Takara Bio USA, Mountain View, CA, USA; #635029) or a TruSeq Small RNA Library Preparation Kit (Illumina, San Diego, CA, USA; #RS-200-0012) according to the manufacturer's instructions. Libraries for transcriptome analysis were prepared using a NEBNext Ultra II Directional RNA Library Prep Kit for Illumina (NEB; #E7760S), including purification by NEBNext Sample Purification Beads to isolate PCR products from 250 to 350 nt, according to the manufacturer's instructions. A NEBNext rRNA Depletion Kit (Human/Mouse/Rat) (NEB; #E6310) was used to deplete human rRNA for directional RNA library preparation using NEBNext Ultra II.
Library sequencing
cDNA libraries were sequenced on an Illumina MiSeq or NextSeq instrument (1 × 50 for synthetic and total brain RNA or 1 × 75 for FFPE liver RNA). Libraries were sequenced with the addition of 50% PhiX Control spike-in (Illumina #FC-110-3001) to improve diversity around the TSO 3′ end.
Data analysis
Processing of Illumina sequencing reads and control transcripts analysis
Cutadapt (42) was used to remove (i) the preceding nucleotides of template switching libraries (four nucleotides for capped libraries and three nucleotides for uncapped libraries) from their 5′ end, (ii) the adapter sequences, (iii) the low-quality bases (q < 20) and (iv) the poly(A) tails from the 3′ end of all the raw Illumina sequencing reads. This step also removed reads that became too short (<15 nt) after trimming. The trimmed reads were first mapped to the reference sequences of the Mix4v7 control using the Bowtie2 program (43). To assess biases among different libraries, the mapped reads were quantified based on the theoretical counts of spike-in control transcripts. The remaining non-control reads were then mapped to human ribosomal RNA and their associated spacer sequences using Bowtie2; only unmapped reads were kept for downstream analysis.
miRXplore analysis
Reads were trimmed using cutadapt as described above and mapped to the miRXplore reference sequences using bbmap (https://jgi.doe.gov/data-and-tools/bbtools/). Mapped reads were randomly downsampled using BBTools (https://sourceforge.net/projects/bbmap/) so that every sample included 1 million mapped reads. Transcript counts were obtained using idxstats from samtools (44). Considering the 962 miRNAs in miRXplore were present in equimolar amounts, an expected read count was generated by dividing the total number of mapped reads for each library by 962. Reads were then normalized by dividing the raw read counts by the expected read counts. A miRNA represented with the exact expected read count would have a normalized value of 1; over- and underrepresented sequences would have values greater and lower than 1, respectively. Statistical analyses were performed on the log transformed, normalized values using a mixed effects model with fixed effects for library preparation method and random effects for miRNA identity using the Lme4 package in R (R Core Team, version 3.6.3; https://www.r-project.org/) (45). Post hoc comparisons were performed where appropriate using least square means (R package, version 1.5.2-1; https://CRAN.R-project.org/package=emmeans). P-values were corrected for multiple comparisons using the Tukey correction. Difference logos were generated for enriched nucleotide motifs while controlling for the known distribution of miRNA sequences using DiffLogo (46).
microRNA analysis
miRDeep2 (47) was used to identify and quantify novel and known miRNAs from the processed good quality non-control-non-rRNA reads based on the human reference genome (hg38) and the known human mature miRNAs and their precursor sequences downloaded from the miRBase database (miRBase release 22). Two filtering criteria were applied to call high-confident mature miRNAs from the mirDeep2 results: (i) randfold P-value <0.05 and (ii) miRDeep2 score ≥0. For each identified miRNA species, the mean count of the reads that map to multiple genomic locations were calculated and used as the expression level for that miRNA. Comparative analysis of miRNAs between different methods (e.g. overlapping analysis and expression correlation analysis) were carried out in R.
Transcriptome analysis
For transcriptome profiling, processed good quality non-control-non-rRNA reads were mapped to the human reference genome sequence (GRCh38) and the gene annotation on the primary assembly downloaded from GENCODE (release 36) (48) plus the human tRNA annotation downloaded from tRNAscan-SE Genomic tRNA Database (49) using the STAR aligner (50). Gene count matrices were then generated from the STAR alignment results by the htseq-count function of the HTseq tool (51). Raw gene counts were converted to transcripts per kilobase million (TPM) by the effective transcript length and the sequencing depth (total read counts) to make them amenable for comparison among different libraries. Statistical analysis and correlation plotting were conducted in R.
microRNA target prediction and analysis
Predicted biological targets of miRNAs in the human genome were downloaded from the TargetScan database Release 7.2 (52). Predicted targets with a context++ score less than −0.5 (lower value indicates more significant) were then matched and assigned to high-confident mature miRNAs that are identified by the sequencing methods studied in this work. Network analysis of microRNA and mRNA interactions were conducted using the ggraph and tidygraph packages in R. Functional annotation and enrichment analysis of target genes were carried out using the DAVID functional annotation tools (53), of which the Functional Annotation Clustering program was used with the selection of GO Ontology, INTERPRO and SMART annotations.
RESULTS
A strategy for chemically capping small RNAs
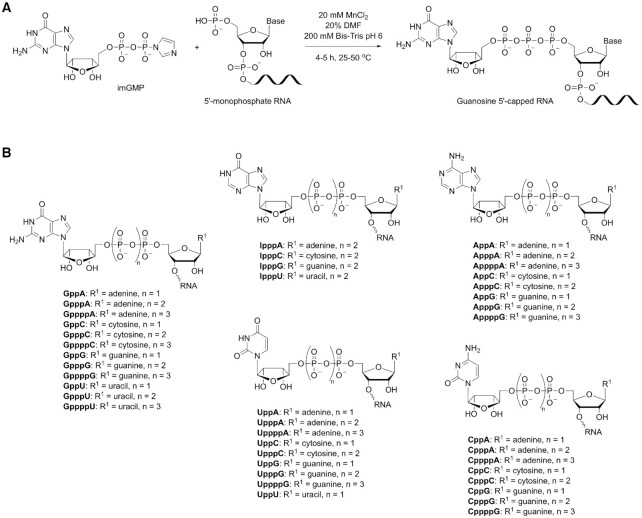
Our previous study showed that the m7G cap increases the efficiency by which a given RNA undergoes template switching during reverse transcription (37). The 5′ m7G cap is an evolutionarily conserved modification of eukaryotic mRNA that is installed co-transcriptionally in the nucleus (54). 5′-Triphosphate RNAs can be capped in vivo or in vitro using eukaryotic or viral RNA capping enzymes. While cytoplasmic capping of 5′-phosphate RNAs has been reported (54), this has yet to be translated into practice. To overcome the challenge of introducing a cap structure to small RNAs presenting a 5′-monophosphate, we took inspiration from literature and built upon an earlier report of nonenzymatic capping of 5′-monophosphorylated oligonucleotides in aqueous solution (55). To achieve efficient conversion of 5′-p RNAs into the desired capped forms, we performed extensive investigation of reaction buffers and pH, concentration and identity of divalent metal catalysts, concentration of imidazolides, presence of additives (such as co-solvents or polyethylene glycol), and finally reaction time and temperature (Supplementary Figure S1). Having a set of optimal chemical capping conditions (20 mM MnCl2, 20% DMF, 200 mM Bis–Tris buffer pH 6, 4–5 h, 24–50°C), we synthesized a collection of 41 distinct RNA templates (each 25 nucleotides long) that varied the nucleoside cap, polyphosphate linker length, and identity of 5′ starting nucleotide (Figure 1). Different reaction temperatures were required for the synthesis of tetraphosphate, triphosphate, and diphosphate caps to balance reaction yield with rate of hydrolytic decomposition of the imidazolides. The more reactive triphosphate imidazolides performed well at room temperature, while the less reactive diphosphate and monophosphate imidazolides required reactions at 37°C and 50°C, respectively. Capping yields were generally high, yielding >80% conversion for diphosphate and triphosphate caps. To ensure complete removal of any unreacted 5′-monophosphate RNAs, crude chemically capped products were purified either through PAGE (for terminal transferase and template switching profiling assays) or enzymatic digestion with the 5′-monophosphate-dependent exonuclease XRN-1 (for sequencing assays).
Figure 1.
Overview of chemical capping. (A) Representative example of a 5′-monophosphate RNA capped through reaction with a guanosine 5′-phosphoroimidazolide forming a 5′-5′ phosphodiester linkage. (B) Structure of 41 distinct capped 25mer RNAs varying the nucleotide cap (guanosine, adenosine, cytidine, thymidine or inosine) and the length of the polyphosphate bridge (di-, tri- or tetraphosphate).
Unmethylated guanosine cap leads to the highest template switching efficiency
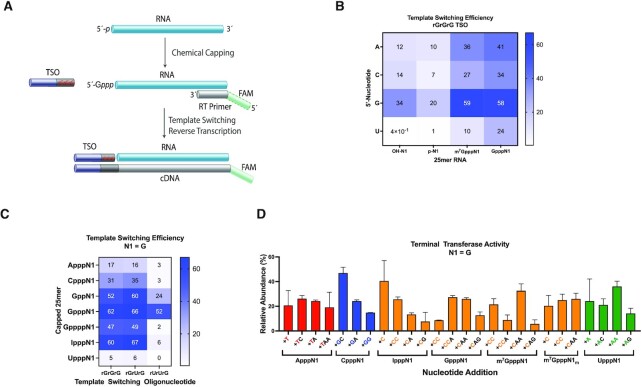
Chemical capping enables equipping RNAs with cap structures that are inaccessible by standard enzymatic capping. Taking advantage of this unique feature of our approach, we set out to explore how non-native caps affect the efficiency and bias in template switching reactions. For comparative purposes, we obtained the m7G-capped analogues, m7GpppN- and m7GpppNm-25mer, through enzymatic capping and 2′-O-methylation of 5′-triphosphate RNAs as described previously (37). We carried out template-switching reverse transcription, as shown in Figure 2A, using a 5′-FAM labeled DNA primer, an rGrGrG-3′ TSO, and RNA templates whose 5′ end was either capped (m7Gppp- or Gppp-) or uncapped (HO- or p-). Capillary electrophoresis (CE) was used to quantify primer elongation and template switching products, including concatemers that formed when multiple template-switching events occurred on the same cDNA. In line with what we have observed for m7G-capped RNAs (m7GpppN1) (37), the presence of an unmethylated guanosine cap (GpppN1) enhanced the template switching efficiency about two- to fourfold for most templates (Figure 2B). Remarkably, G-capping provided a greater than 20-fold increase in efficiency for an RNA template starting with uridine (as a note, in this particular template both N1 and N2 = U). m7G-Capping, in contrast, was not as effective as G-capping at enhancing 5′-U RNA detection. This is especially relevant because the majority of the known human miRNA sequences have a 5′-U (26).
Figure 2.
The effect of the cap structure on template switching and terminal transferase activities. (A) Schematic of cDNA synthesis through chemical capping of a monophosphate RNA template followed by template-switching reverse transcription. An unmethylated guanosine cap is shown. Red shades in the template switching oligonucleotide (TSO) represent variable 3′ end nucleotides. Green shades in the DNA RT primer represent either a fluorescent label (CE assays) or a P7 sequence index (sequencing assays). (B) Heat map showing how the template switching efficiency is affected by the first nucleotide of the RNA template and 5′ end capping. Color gradient indicates percentage of template switching products as determined by CE analysis. (C) Heat map of the template switching efficiency for various non-native RNA caps in the presence of three different TSOs. (D) Nontemplated deoxynucleotide addition profile in absence of a TSO as determined by mass spectrometry analysis. Only the most abundant nontemplated additions (represented in successive order of incorporation to the 3′ end of the cDNA) are shown. Data represent mean ± SD of n = 2 independent experiments. All assays were performed with discrete synthetic 25mer RNAs.
The more uniform yield distribution elicited by unmethylated G-caps led us to anticipate that G-capping could potentially lower sequencing biases in the template switching step of small RNA sequencing workflows. We thus expanded the analysis to a set of 16 RNAs varying the first two nucleotides at the 5′ end. These RNAs were either uncapped 5′-monophosphates (emulating naturally processed small RNAs) or subjected to chemical capping in the form of GpppNN. Not surprisingly, uncapped 5′-p RNAs led to a significantly lower template switching efficiency than did G-capped templates (Supplementary Figure S2A versus S2B). Only 5′-p RNAs starting with a guanosine nucleotide (5′-pGN) produced any substantial template switching, mirroring our previous findings (37). Similar results were obtained using a different TSO (rGrUrG-3′ TSO) (Supplementary Figure S2C versus S2D).
Next, we determined how the identity of the 5′ cap nucleoside and the length of the 5′–5′ phosphate linker affected the template switching efficiency. To our knowledge, only the native m7G cap has been tested in template switching reactions. For our initial screen, we selected a set of capped RNAs with the starting nucleotide N1 = G and varied the nature of the cap (guanosine, adenosine, cytidine, thymidine or inosine) and the polyphosphate bridge (di-, tri- or tetraphosphate for a guanosine cap). Three TSOs, rGrGrG-3′, rGrUrG-3′ and rUrUrG-3′, were used in these experiments. We found that template switching was significantly boosted by G- and I-capping RNAs (Figure 2C). We then extended this study to other nucleotides at position N1, covering the whole set of chemically capped RNAs of Figure 1B (Supplementary Figure S3A through S3E; the rUrUrG-3′ TSO was omitted from these experiments). G- and I-capped RNAs performed well across most templates, whereas A- and U-capped RNAs proved to be very poor substrates for template switching. C-capped RNAs showed a somewhat intermediate effect. Caps featuring triphosphate bridges consistently outperformed di- and tetraphosphate ones, suggesting that a balance between cap rigidity and distance from the first nucleotide at the 5′ end is required for optimal reaction.
5' Cap acts as a template for the first post-templated nucleotide addition
The template-independent terminal transferase activity of reverse transcriptases is inherently associated with template switching. We therefore sought to determine the nature and extent of nontemplated additions in the context of chemically capped RNA templates. To study the terminal transferase activity alone (disconnecting it from the template switching process), reverse transcription reactions were performed in the absence of a TSO. The resulting cDNA was analyzed by intact mass spectrometry as previously described (37). As with m7G-capped RNAs (this work and ref. 37), the first nontemplated nucleotide incorporated across all G-capped RNAs was almost always dC, confirming an earlier notion that the cap nucleoside contributes to ‘templating’ the protraction of cDNA (Figure 2D).
Chemical capping provided us with a singular opportunity to assert this hypothesis by contrasting results from diverse cap structures. With that in mind, we examined RNA templates containing A-, C-, G-, I- and U-caps with varying 5′ end nucleotides and 5′-5′ phosphate linker lengths (Figure 2D and Supplementary Figure S4). As predicted, the 5′ cap dictated the first deoxynucleotide addition, acting indeed as a template. A-capped RNAs led to cDNA products with primarily thymidine as the first post-template (beyond the RNA template sequence) deoxynucleotide addition (which included + T, +TC, +TA and + TAA) (Figure 2D), regardless of the nucleotide at position N1 or the 5′-5′ phosphate linker length (Supplementary Figure S4A). C-capped RNAs led to cDNA products with primarily deoxyguanosine as the first post-template addition (+G, +GC and + GA) (Figure 2D and Supplementary Figure S4B), G- and I-capped RNAs led to deoxycytidine addition (+C, +CC and + CAA) (Figure 2D and Supplementary Figure S4C). U-capped RNAs led to deoxyadenosine addition (+A, +AA and + AAA) (Figure 2D and Supplementary Figure S4D). Once the cap-templated deoxynucleotide was incorporated to the cDNA strand, a subsequent addition of one or more dAs was often observed. The latter is commonly seen with many DNA polymerases displaying terminal transferase activity. The cap-templated addition was largely independent of the RNA nucleotides at positions N1 or N2, and completely absent in uncapped RNAs (Supplementary Figure S5).
In an attempt to enhance cap-specific template switching, we designed TSOs that matched the corresponding cap-templated addition profiles. TSOs were constructed with the final three 3′ end nucleotides comprising a 2′-fluoro modification to increase their templating power. To isolate the effect of cap-specific templating, only the outermost TSO nucleotide was varied (FrGFrGFrA-3′, FrGFrGFrC-3′, FrGFrGFrG-3′ and FrGFrGFrU-3′ TSOs). The expectation was that each capped RNA template would be more effectively paired with the 3′-TSO matching the addition pattern shown in Figure 2D (i.e. A-capped RNAs with the FrGFrGFrA-3′ TSO, C-capped RNAs with FrGFrGFrC-3′, and so on). Interestingly, however, only the FrGFrGFrG-3′ TSO promoted any substantial template switching for each of the five caps tested (Supplementary Figure S6A). With the clear exception of the FrGFrGFrG-3′/G- or I- cap pairs, only modest yields of template-switched cDNA products were obtained for the other matching TSO/cap pairs. This observation was even more pronounced when the TSOs were combined with unbalanced ratios of dNTPs intended to favor cap-templated addition (i.e. A-capped RNAs with 10× dTTP, C-capped RNAs with 10x dGTP, and so on) (Supplementary Figure S6B). Only the unbalanced 10x dCTP formulation showed any improvement to the template switching efficiency for the matching cap structure. We have shown previously that template switching and nontemplated addition are concurrent processes (37). The data on TSO/cap pairs add a new layer of complexity to that theory, suggesting the reverse transcriptase has a preference for guanine at the 3′ end of the incoming TSO, regardless of the cap structure and nucleotide addition profile.
Unmethylated guanosine cap reduces biases for template switching in synthetic miRNA pools
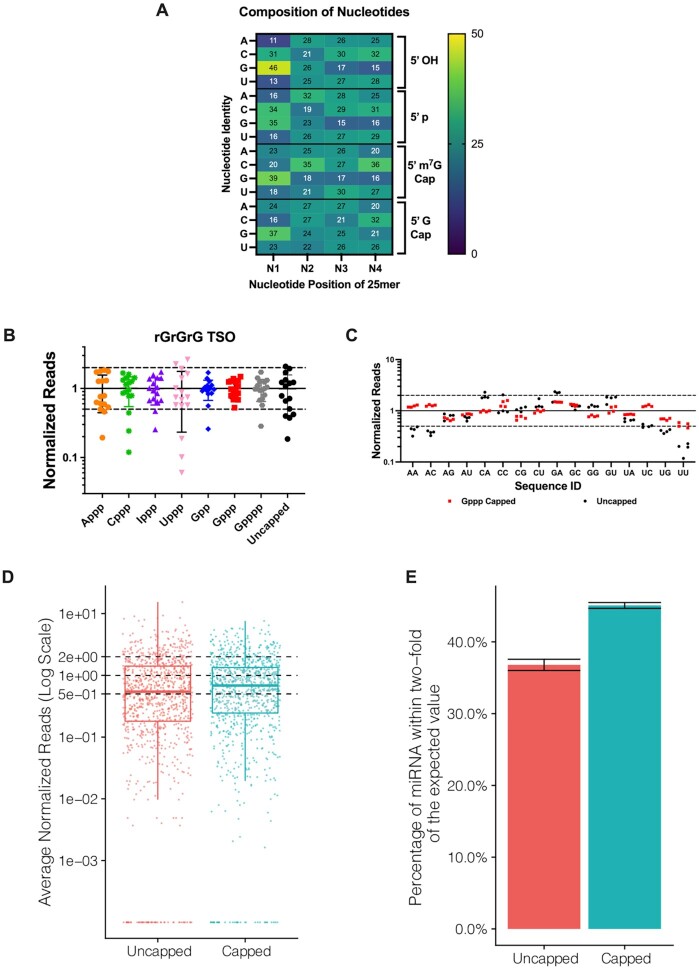
We next analyzed the effect of the various cap structures on sequence representation biases. First, template-switching reverse transcription was performed on a set of synthetic RNA oligonucleotides with the first four nucleotides randomized and either a 5′ OH, 5′ p, 5′ m7G cap, or 5′ G cap modification. Illumina libraries were constructed to determine the extent of biases. The sequencing data from template switching experiments were normalized to the relative composition of the first four ribonucleotides in the synthetic templates as previously described (37). The prevailing bias observed, regardless of the 5′ modification, was an overrepresentation of sequences with the first nucleotide N1 = G (Figure 3A). Uncapped templates (5′ OH or 5′ p) exhibited a marked underrepresentation of sequences with N1 = A or U. In general, the least sequence representation bias was found for RNA templates containing an unmethylated guanosine cap (5′ G cap) (Figure 3A). Although the overall bias decreased along positions N2–N4 for both capped and uncapped templates, it was almost negligible for unmethylated G-capped RNAs. Interestingly, a non-negligible underrepresentation of sequences with guanosine at positions N2-N4 and overrepresentation of cytidine at position N2 were observed for m7G-capped templates.
Figure 3.
Chemical capping reduces template switching biases. (A) Heat map of the contribution of the first four nucleotides (N1–N4) and the RNA 5′ end capping to template switching bias. Color gradient indicates normalized representation of nucleotide N in the sequencing reads: yellow (overrepresented), dark blue (underrepresented) and teal (neutral). The presence of an unmethylated guanosine cap minimizes overall nucleotide representation bias in the library. Heat map data represent means of n = 3–4 independent experiments, ±SD < 1. (B) Normalized read distribution of libraries prepared from a pool of 16 discrete RNA templates varying the first two nucleotides N1 and N2. The RNA pool was subjected to chemical capping as indicated. Template switching was performed with the rGrGrG-3′ TSO. Reads were normalized to 1 (solid line) and the interval of 2-fold above and below this boundary was denoted by dashed lines. (C) 5′-Gppp capped or uncapped RNA libraries shown in (B) arranged by the identity of the first two 5′ end nucleotides. Data shown in (B) and (C) represent mean ± SD of n = 4 experiments. Assays were performed with 25mer RNAs, either comprising synthetic oligonucleotides with randomized positions (A) or a pool of discrete sequences (B and C). (D) Normalized read counts for 0.5 pg input uncapped and capped libraries prepared from an equimolar pool of 962 synthetic miRNAs (miRXplore reference). Each miRNA is expected to have a normalized read value of 1 (central dashed line). The upper and lower dashed lines correspond to the interval of 2-fold above and below the expected value. (E) Percentage of miRNAs from (D) falling within 2-fold of their expected values shown as the average of three technical replicates per method. Error bars represent the standard deviation.
In the ensuing experiments, we examined a pool of 16 discrete RNA templates varying the first two nucleotides (N1 and N2). Differently from the randomized set above, each RNA in this pool was individually synthesized and purified. This pool was either left uncapped (in the form of 5′-monophosphate) or capped with one of guanosine, adenosine, cytidine, thymidine or inosine. A triphosphate bridge was chosen for all capped templates, except for guanosine caps, where di- and tetraphosphate bridges were also investigated. Template-switching reverse transcription, amplification and sequencing were performed as above. A template-specific DNA primer containing the Illumina P7 sequencing adapter was utilized in these experiments along with the TSOs rGrGrG-3′ (Figure 3B and C) or rGrUrG-3′ (Supplementary Figure S7). Reads were organized by the identity of the first two RNA 5′ end nucleotides. Normalized read counts above or below the interval of 2-fold of the expected value were considered over- or underrepresented, respectively. The results were in line with the trends seen above. The lowest sequencing bias was found for Gppp-capped templates, whereas the highest for Uppp-capped templates (Figure 3B). Guanosine caps performed better than other nucleotide caps, irrespective of the polyphosphate linker length. Critically, the presence of a Gppp cap enabled lowering the systematic underrepresentation of uncapped reads starting with uridine and adenosine, such as 5′-UU, 5′-UG, 5′-AA and 5′-AC templates (Figure 3C). In contrast to some promising results from template switching efficiency with individual RNA templates (Figure 2C, middle column), the rGrUrG-3′ TSO led to consistently higher sequencing biases (Supplementary Figure S7) and was not pursued further.
To further investigate whether a Gppp cap would also reduce the sequence representation bias in a more diverse miRNA pool, we performed template-switching reverse transcription on a synthetic reference, miRXplore, containing 962 unique human, mouse, rat, and viral miRNA sequences in equimolar ratio (24,26,31). To do so, the miRNA pool was first polyadenylated, then either G-capped or left uncapped, and subsequently subjected to library preparation and sequencing as described above. In line with our observations, the chemically capped libraries indeed provided a more uniform miRNA coverage (Figure 3D and E). From this dataset, about 37% of the detected miRNAs were within 2-fold of their expected value in uncapped libraries relative to 45% in capped libraries, confirming that the Gppp cap plays a critical role in reducing biases in template switching reactions.
A universal workflow for sequencing of RNAs with uncapped ends
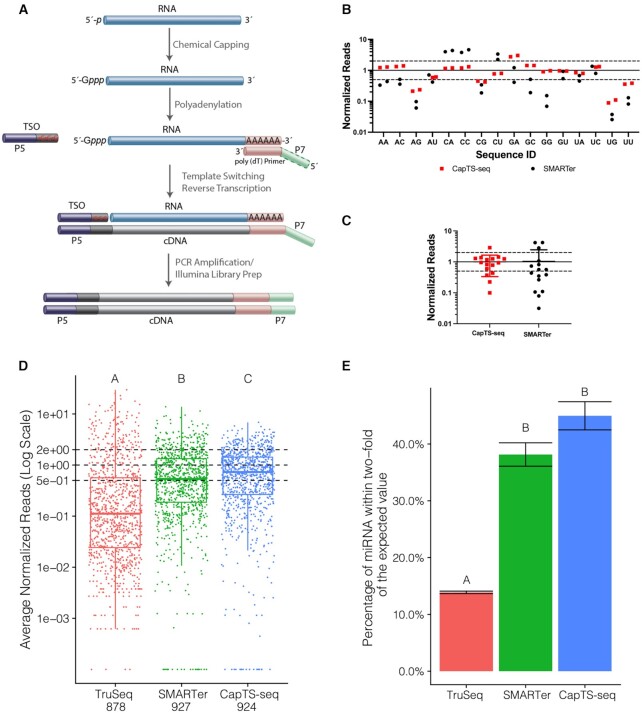
Having demonstrated that the installation of a Gppp cap reduced bias and enhanced efficiency in template switching, we set out to broaden our strategy, hereinafter referred to as CapTS-seq, to small RNAs with uncapped 5′ ends. To allow template-independent reverse transcription priming, RNAs were 3′ polyadenylated. A poly(dT) primer permits a reliable and largely sequence-independent incorporation of a 3′ end sequencing adapter to the cDNA strand in a ligation-free fashion. A synthetic 25mer RNA was utilized as a model for establishing conditions for controlled polyadenylation. Under these conditions, a short stretch of ∼13 A’s was appended to the control RNA 3′ end, as determined by mass spectrometry (Supplementary Figure S8), although the extent of polyadenylation may vary for different cellular RNA inputs. The whole workflow comprising chemical capping, poly(A) tailing, and template-switching reverse transcription is shown in Figure 4A. The Takara SMARTer smRNA-Seq kit (here referred to as SMARTer)—which is the gold standard kit for small non-coding RNA sequencing whose adaptation strategy combines 5′ template switching and 3′ poly(A) tailing—was chosen for side-by-side method comparison. SMARTer was used according to the manufacturer instructions. It is important to note that individual steps in CapTS-seq and SMARTer may differ in terms of buffer composition, enzymes, and TSO(s). Furthermore, SMARTer does not employ a capping step, and thus template switching is carried out with uncapped RNAs. Hence, only the overall performance between these two methods was accessed.
Figure 4.
CapTS-seq improves small RNA sequence coverage representation. (A) Schematic of CapTS-seq library preparation workflow using a poly(dT) primer for reverse transcription. (B) Normalized sequencing reads of CapTS-seq and SMARTer libraries generated from a pool of 16 discrete RNA templates (25mers) varying the first two nucleotides N1 and N2. (C) Box plot depicting read average of two technical replicates for each sequence represented in (B). Reads were normalized to 1 (solid line) and the interval of 2-fold above and below this boundary was denoted by dashed lines. (D) Normalized read counts of TruSeq, SMARTer and CapTS-seq libraries prepared from an equimolar mix of 962 synthetic miRNAs (miRXplore reference). Each miRNA is expected to have a normalized read value of 1 (central dashed line). The upper and lower dashed lines correspond to the interval of 2-fold above and below the expected value. (E) Percentage of miRNAs falling within 2-fold of their expected values shown as the average of two technical replicates per method. Error bars represent the standard deviation. Letter codes indicate groups that are significantly different from each other with Tukey corrected P < 0.01.
CapTS-seq was first deployed for sequencing the pool of 16 RNAs with variable N1 and N2 positions. CapTS-seq greatly reduced sequencing representation biases relative to SMARTer. The vast majority of CapTS-seq reads were within the 2-fold interval of expected values. Compared with data obtained using a sequence-specific primer (Figure 3C, red squares), there was a slight increase in underrepresentation of sequences starting with 5′-AG, 5′-UG and 5′-UU, and overrepresentation of 5′-GA (Figure 4B, red squares). The added bias introduced by 3′ polyadenylation, while not ideal, was sufficiently low that it did not require further optimization beyond the scope of this study. Many more sequences were underrepresented (5′-AA, 5′-AG, 5′-CG, 5′-GC, 5′-GG, 5′-UC and 5′-UU) and overrepresented (5′-CA, 5′-CC and 5′-CU) when libraries were prepared with SMARTer (Figure 4B, black circles). The overall departure from the expected read representation for both methods is summarized in Figure 4C. Altogether, these results echoed what we had found when the capping step was omitted from the library preparation protocol (Figure 3C), providing further evidence that chemically equipping RNA templates with a guanosine cap does improve coverage accuracy during template switching.
Next, we tested the feasibility of CapTS-seq to detect miRNAs in the miRXplore reference. As a further point of comparison, miRXplore libraries were prepared either using SMARTer or an Illumina TruSeq Small RNA Library Prep Kit (here referred to as TruSeq). TruSeq is a benchmark method for small RNA sequencing that relies on standard enzymatic ligation for 5′ adaptation (instead of template switching). After trimming, sorting, and normalizing reads to the theoretical counts in the miRXplore pool, we found that CapTS-seq provided the most accurate miRNA coverage, i.e. more miRNA sequences were detected closer to their expected abundance (Figure 4D and E). From this dataset, about 15% of the detected miRNAs were within 2-fold of their expected value in TruSeq, compared with 40% in SMARTer and 45% in CapTS-seq libraries (Figure 4E). Significant statistical differences among the three methods were observed in regard to their mean transcript levels (Figure 4D, ANOVA c2 = 1021.4, P < 2 × 10−16). This was further confirmed by post hoc analysis of all pairwise comparisons. CapTS-seq and SMARTer libraries, however, were statistically similar to each other, but different from TruSeq in regard to the percentage of miRNA reads within the expected range (Figure 4E). All three methods had excellent technical reproducibility with Pearson's correlations between replicates greater than 0.9. Differences in bias levels between CapTS-seq and SMARTer became apparent by inspection of sequence logos generated for sets of the topmost overrepresented miRNAs in each library (Supplementary Figure S9). SMARTer libraries showed a strong overrepresentation of reads starting with 5′-C along with depletion of reads starting with 5′-A and 5′-U, which is similar to what we have seen for the control 16 RNA library (Figure 4B, black circles). CapTS-seq did exhibit some variable degree of bias at the first six positions, however, the average nucleotide divergence was not as pronounced (Supplementary Figure S9, lower panels). Interestingly, although much fewer TruSeq reads fell within 2-fold of their expected values (Figure 4D), no bias correlation with a particular nucleotide was observed among the most under- or overrepresented sequences (Supplementary Figure S9). Overall, CapTS-seq appears to improve read coverage representation relative not only to traditional ligation-based methods, such as TruSeq, but also to existing template switching-based methods, such as SMARTer.
Applying CapTS-seq to total RNA
CapTS-seq produces better quality libraries
Finally, we tested CapTS-seq in two distinct human total RNA samples: frozen brain tissue and liver FFPE tissue. Total RNA from adult normal brain tissue was first subjected to size selection to capture small RNAs and remove most of rRNA content. Brain tissue libraries were prepared as described above using either CapTS-seq, a variation of CapTS-seq in the absence of the chemical capping (here referred to as ‘TS-seq (uncapped)’), or SMARTer. For FFPE total liver RNA, a slight modification in the library preparation protocol was required. Because the RNA extracted from FFPE samples may be highly degraded, the phosphorylation state at its 3′ and 5′ ends is variable (56). To repair the 3′ end before polyadenylation and make a uniform ‘cappable’ 5′-monophosphate end, FFPE total RNA was treated with T4 polynucleotide kinase (T4 PNK). T4 PNK is a multifunctional enzyme that displays both 5′-kinase and 2′,3′-cyclic phosphodiesterase activities, and has been widely used for DNA and RNA end healing (57). Due to the fragmented nature of FFPE RNA, the small RNA enrichment step was omitted from library preparation. Only CapTS-seq and SMARTer were used for preparing libraries from FFPE total liver RNA. We found that CapTS-seq improved the quality of the libraries made from both total RNA samples (Supplementary Tables S5 and S6). As a general trend, CapTS-seq consistently reduced the rRNA content and yielded more useful sequencing reads than TS-seq (uncapped) or SMARTer did (Supplementary Figure S10A and S10B). This is particularly meaningful because rRNA makes up the largest fraction of the reads in a typical RNA library (16). Moreover, comparing the read length distribution before and after 3′ and 5′ end trimming, we found that CapTS-seq significantly boosts the detection of reads at ∼20 nt (miRNA reads) relative to TS-seq (uncapped) or SMARTer (Supplementary Figure S10C and S10D).
CapTS-seq enables detection of more unique miRNA species
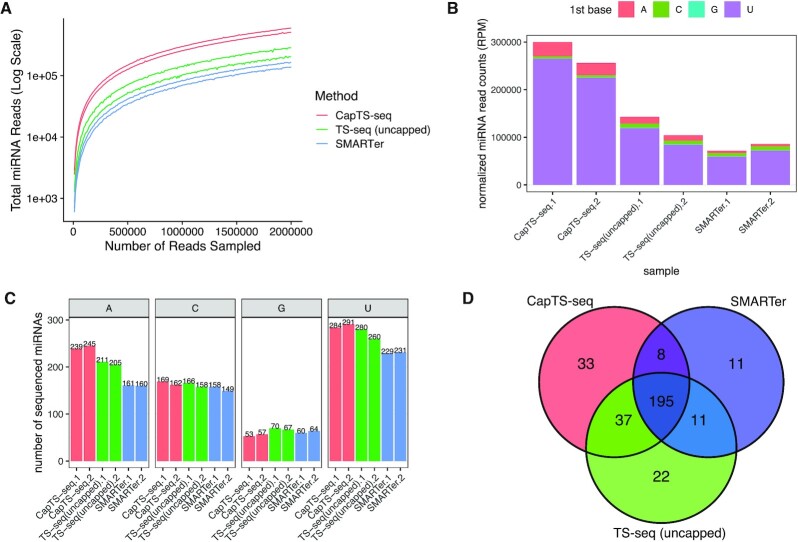
Brain tissue and liver FFPE libraries were analyzed for miRNA content by mapping reads to a human reference genome (hg38). By plotting incremental subsets of randomly selected reads from each of total brain RNA (Figure 5A) and FFPE liver RNA libraries (Figure 6A), we found that CapTS-seq consistently detected more miRNAs than either TS-seq (uncapped) (in total brain RNA) or SMARTer (in both total brain and liver FFPE RNA), independently of the sequencing depth. The total number of mapped reads (4 million for total brain and 19 million for FFPE libraries) was normalized and sorted according to the miRNA starting nucleotide (Figures 5B and 6B). All three human brain libraries revealed that the vast majority of miRNA sequences start with a 5′-U, which is consistent with results presented in other studies (26). This is further confirmed when considering the number of sequenced miRNAs in each library (Figure 5C and Supplementary Figure S11C). In line with data obtained from synthetic RNA libraries (Figures 3C and 4B), CapTS-seq detected more 5′-U and 5′-A miRNAs than either TS-seq (uncapped) or SMARTer did in total brain RNA. Conversely, TS-seq (uncapped) and SMARTer libraries were comparatively enriched in 5′-G and 5′-C miRNAs (a slight overrepresentation of 5′-G and 5′-C sequences was also seen for these methods in libraries made from synthetic RNAs as shown in Figures 3C and 4B). Collectively, the overall correlation of the common miRNAs among the three methods was highest for miRNAs starting with 5′-U and lowest for miRNAs starting with 5′-G, likely reflecting their disproportionate abundance (Supplementary Figure S11D).
Figure 5.
CapTS-seq improves miRNA detection in total brain RNA. (A) Number of miRNAs detected with a mirDeep2 score >0 at various subsampled read depths. Datasets were randomly sampled with the number of reads increasing by 5000 in each sample. Values represent the mean number detected in two technical replicates individually sampled at each read depth. (B) Normalized miRNA read counts (number of miRNA reads per 1 million human alignments) for each method. Colored bars represent the miRNA 5′-nucleotide. (C) Total miRNA species counts sorted by the identity of the 5′ end nucleotide. Both miRNA strands (miRNA-5p and miRNA-3p) are counted for this analysis. (D) Overlap of unique miRNAs identified in two combined replicates between CapTS-seq, TS-seq (uncapped) and SMARTer libraries. CapTS-seq detected more unique miRNAs. RPM, reads per million mapped reads.
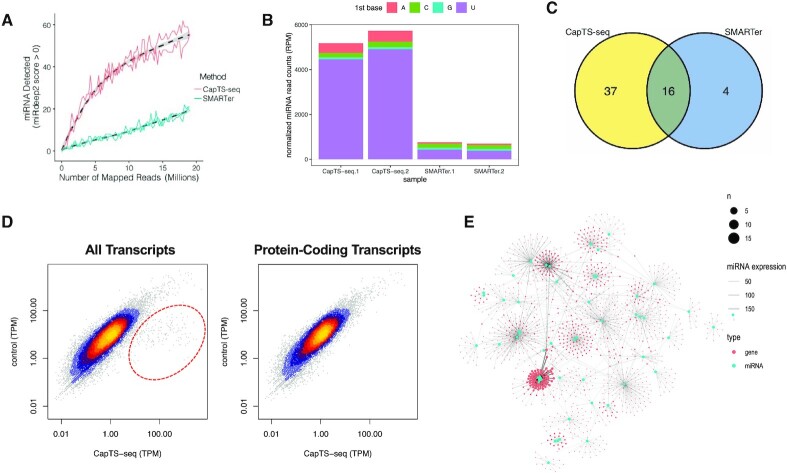
Figure 6.
CapTS-seq improves miRNAs detection in FFPE RNA and enables simultaneous transcriptome analysis. (A) Number of miRNAs detected with a mirdeep2 score >0 at various subsampled read depths. Datasets were randomly sampled with the number of reads increasing by 5000 in each sample. Values represent the mean number detected in two technical replicates individually sampled at each read depth. (B) Normalized miRNA read counts (number of miRNA reads per 1 million human alignments) for each method. Colored bars represent the miRNA 5′-nucleotide. (C) Overlap of unique miRNAs identified between CapTS-seq and SMARTer libraries. CapTS-seq detected more unique miRNAs. (D) Correlation of normalized read counts between CapTS-seq and a control RNA-seq library (NEBNext Ultra II). Left panel, all transcripts; right panel, protein-coding transcripts only. TPM, transcripts per kilobase million. TPM values were averaged between two technical replicates. Spearman's rank correlation coefficients for protein-coding reads (0.92) and for all transcripts (0.76). Reads within the red circle highlight small RNAs that are uniquely detected in CapTS-seq but not in a standard transcriptome RNA-seq library. (E) Interaction network of miRNAs (cyan dots) and their target genes (red dots) as detected in CapTS-seq libraries. The size of the dots represents the number of miRNAs that target this particular gene. The width of the connecting lines reflects the expression level of individual miRNAs.
In terms of unique miRNAs detected, CapTS-seq libraries rendered 273 unique mature miRNA species from total brain RNA—21% more than SMARTer libraries did (225) from the same 4 million sequencing reads—of which 203 were shared between the two methods and 70 were unique to CapTS-seq (threefold more unique miRNAs relative to SMARTer) (Figure 5D and Supplementary Figure S11A). The difference in uniquely detected miRNAs is more subtle in comparison with TS-seq (uncapped) (265) from the same 4 million sequencing reads. It is noteworthy that CapTS-seq had a higher overall miRNA read count in total brain RNA than either TS-seq (uncapped) or SMARTer did (Figure 5B). Although fewer miRNAs were detected in FFPE, similar trends were observed. CapTS-seq FFPE RNA libraries rendered 53 unique mature miRNA species from 19 million sequencing reads, which is 65% more than SMARTer did (20). More unique miRNAs were found with CapTS-seq (37) than with SMARTer (4) (Figure 6C and Supplementary Figure S12A). Again, a similar pattern of miRNA sequence representation was observed in the FFPE liver libraries, with CapTS-seq showing a marked improvement in the detection of miRNAs starting with 5′-A and 5′-U (Supplementary Figure S12C and S12D). It is important to note that replicates within each of CapTS-seq, TS-seq (uncapped), and SMARTer libraries consistently showed a high read count correlation (Supplementary Figures S11B and S12B), indicating that all methods were reproducible and robust.
CapTS-seq provides a door to decoding dynamic miRNA regulation of target genes
As a final measure of the power of CapTS-seq, we performed integrated transcriptome analysis of human liver FFPE libraries in the interest of detecting both miRNA and mRNA in a single experiment. The reads were trimmed and aligned to a human reference using STAR aligner and compared with those of a standard RNA-seq library (NEBNext Ultra II kit) (Supplementary Table S7). Aside from a small subset of transcripts that were enriched in CapTS-seq libraries, read counts at the transcript level correlated well between the two methods (Figure 6D and Supplementary Figure S13A, left panels). Consistent with our expectations, most of the unique transcripts detected by CapTS-seq corresponded to non-coding small RNA transcripts (Supplementary Figure 14). In fact, by considering protein-coding transcripts only, CapTS-seq normalized read counts (TPM) highly correlated with those of the control RNA-seq library with a Spearman's rank correlation coefficient of 0.92 (Figure 6D and Supplementary Figure S13A, right panels). Interestingly, we found a significant number of reads of 30–40 nucleotides long, of which nearly half map to mature tRNAs (Supplementary Figures S13C and S15). The formation of small tRNA fragments has been associated with specific cleavage patterns suggesting the occurrence of independent pathways for tRNA processing rather than random degradation (58).
The ability to detect small RNAs and mRNAs simultaneously has been recently demonstrated using thermostable group II intron reverse transcriptase sequencing (59). That prompted us to ask whether CapTS-seq could be used to investigate interactions between miRNAs and their target gene transcripts. We found, indeed, that nearly one third of the predicted target genes were associated with more than one miRNA (with similar or different seed sequences) in the human liver FFPE sample (Figure 6E and Appendix 2). In striking comparison, only a small portion of the predicted target genes (15%) was found to be associated with multiple miRNAs by SMARTer (Supplementary Figure 13B). Moreover, CapTS-seq detected many more genes targeted by multiple miRNAs of different seed sequences and possibly at different loci (90) than SMARTer did (32) (Appendix 2). It has been shown that the interaction of miRNAs with their target genes is complex and that the expression of some genes is determined by a combination of multiple miRNA activities (60,61). CapTS-seq provides a potentially useful platform to decode these interactions. The functional annotation of genes that are predicted to be targets of miRNAs in CapTS-seq libraries revealed, for instance, that the KRAB-containing proteins—many of which function as transcriptional repressors—and chemotactic cytokines—which are often induced during immune response—were overrepresented in the human liver FFPE sample (Supplementary Table S8). While we see these correlational results as promising, significant work still remains to fully validate the application of CapTS-seq for interaction network analysis.
DISCUSSION
CapTS-seq is a new tool for small RNA sequencing that utilizes chemical capping as a key step in library preparation. Drawing upon our previous study into template switching and nontemplated additions by MMLV-based reverse transcriptases (37), here we leverage a chemical capping strategy by which to equip RNAs with a non-native nucleotide cap and shed light on important aspects of the template switching mechanism. To explain the incorporation of ‘nontemplated’ deoxycytidines at the cDNA 3′ end, several studies have suggested that the native m7G-cap acts as a template for the reverse transcriptase (34,37). Our study provides definitive evidence for a cap-mediated post-template deoxynucleotide addition. By varying the nucleotide cap, we were able to anticipate and then unambiguously confirm the identity of this first post-template deoxynucleotide. Furthermore, we show that the cap-templated addition is largely independent of the RNA sequence and completely absent in uncapped RNAs. To our surprise, however, attempts to tune the efficiency of post-templated additions were essentially fruitless for non-guanosine caps. The widely-used rGrGrG-3′ TSO was still the most effective with all of the chemical caps tested here, despite a lack of post-template + dC with most non-G caps. We speculate that the reverse transcriptase, while still bound to the RNA template and nascent cDNA, can better accommodate a guanine at the 3′ end of the incoming TSO, and this drives the template switching reaction forward. The interplay between TSO, RNA template and reverse transcriptase is fairly complex and will certainly continue demanding innovative experimental design to clarify the subtleties of its mechanism.
Additionally, we demonstrate that the presence of an unmethylated G cap reduces bias in template switching even further than a native m7G cap does (37) relative to uncapped RNA templates. This result is central to the development of CapTS-seq. Deploying chemical capping to sequencing small RNAs that have a ‘cappable’ 5′-phosphate end proved to be an effective strategy. Even though an exhaustive optimization of CapTS-seq is yet a task ahead, our universal workflow—which combines RNA 5′ chemical capping, 3′ polyadenylation, and template switching—consistently outperformed the gold standard Takara's SMARTer smRNA-seq in terms of library quality and correlation to miRNA targets. Both methods had clear advantage over Illumina TruSeq, which is a traditional ligation-based method and whose biases have been well documented (30). Although recent improvements in ligation-based workflows have been reported (26), ligation-free workflows based on template switching are still the tool of choice for challenging sample types, such as single-cell and fragmented input RNAs. Given the low mapping rates associated with template switching-based methods (31), several aspects of library preparation need to be improved to allow detection of more miRNAs with a fewer number of reads. rRNA contamination is a well-known problem, particularly for FFPE samples (62), with one study finding that only 0.6–2.3% of sequencing reads represent miRNAs (63). Additionally, template switching-based methods are not ideal for identifying isomiRs due to the imprecise ends that arise from polyadenylation and template switching processes. RNA modifications such as a 2′-O-methylation are known to inhibit poly(A) polymerases (64), hence RNA species such as plant miRNAs or piRNAs may not be captured as efficiently by CapTS-seq in its current format. Fortunately, the chemical capping strategy can, in principle, be applied in conjunction with any 3′ adaptation strategy. As such, future iterations of CapTS-seq could aim to address some of these limitations. In an age where personalized medicine is gaining popularity, a wealth of information awaits to be captured from FFPE clinical samples. The ability to sequence these samples with ease offers invaluable possibilities for understanding the causes and implications of RNA aberrant expression in diseased states. By improving sequencing read quality and usability, CapTS-seq revealed more mature miRNA in both human brain and human liver FFPE samples, thus providing a more complete picture of the miRNAs expression profile in these tissues.
Many distinct approaches have been used to identify differentially expressed miRNA and mRNA species. Integrative analyses of miRNA and mRNA profiles have been reported by essentially three strategies: (i) two independent small RNA-seq and standard RNA-seq sequencing runs (65,66), (ii) two separate library preparations that are sequenced together (67,68), or (iii) one single run using group II intron reverse transcriptase sequencing (59). Furthermore, it is often possible to capture some level of miRNA reads in mRNA libraries, or vice versa, depending on the RNA-seq workflow and size-selection scheme utilized. The ability to detect miRNAs and their gene targets in a single experiment potentially allows for a better understanding of their relationship and functional annotation. We show in this study that both MMLV-based template switching methods—CapTS-seq and SMARTer—can detect fragmented mRNAs in FFPE with a high correlation to that of a widely used ligation-based RNA-seq workflow. By enabling the capture of a larger number of unique miRNA species, CapTS-seq provided a more comprehensive overview of miRNA-mRNA associations than SMARTer did. It is important to note that CapTS-seq is not a replacement for standard RNA-seq in the detection of mRNA alone. In practical terms, CapTS-seq primarily detects small RNAs, which will result in only a fraction of the reads containing fragmented mRNAs, thereby requiring a higher read depth for reliable quantification of low expression mRNAs. miRNAs have long been considered as potential cancer biomarkers. However, further research is still needed to uncover downstream effects of differentially expressed miRNAs in diseased versus normal tissues. We believe CapTS-seq could play a key role in helping elucidate the complex network of interactions of miRNAs, including those of circulating miRNAs, and thus advance their applications in cancer diagnosis or prognosis. Efforts toward this end are underway in our laboratory.
DATA AVAILABILITY
Sequencing data have been uploaded to the NCBI sequencing reads archive (SRA) and are available under project accession: GSE171049.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank William Jack and Larry McReynolds for their helpful discussions and critical feedback on the manuscript. We thank Ryan Fuchs for the Mix4v7 oligonucleotide pool. We also thank Laurie Mazzola, Danielle Fuchs, and Kristen Augulewicz for performing capillary electrophoresis and sequencing. We are grateful to Donald Comb, Jim Ellard and Rich Roberts for their support of research at NEB.
Contributor Information
Madalee G Wulf, New England Biolabs, Inc., 240 County Road, Ipswich, MA 01938, USA.
Sean Maguire, New England Biolabs, Inc., 240 County Road, Ipswich, MA 01938, USA.
Nan Dai, New England Biolabs, Inc., 240 County Road, Ipswich, MA 01938, USA.
Alice Blondel, New England Biolabs, Inc., 240 County Road, Ipswich, MA 01938, USA.
Dora Posfai, New England Biolabs, Inc., 240 County Road, Ipswich, MA 01938, USA.
Keerthana Krishnan, New England Biolabs, Inc., 240 County Road, Ipswich, MA 01938, USA.
Zhiyi Sun, New England Biolabs, Inc., 240 County Road, Ipswich, MA 01938, USA.
Shengxi Guan, New England Biolabs, Inc., 240 County Road, Ipswich, MA 01938, USA.
Ivan R Corrêa, Jr, New England Biolabs, Inc., 240 County Road, Ipswich, MA 01938, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
New England Biolabs, Inc. Funding for open access charge: New England Biolabs, Inc.
Conflict of interest statement. M.G.W., S.M., N.D., D.P., K.K., Z.S., S.G. and I.R.C.J. are employees of New England Biolabs, Inc. New England Biolabs is a manufacturer and vendor of molecular biology reagents, including several enzymes and buffers used in this study. This affiliation does not affect the authors’ impartiality, adherence to journal standards and policies, or availability of data. New England Biolabs has filed a patent application based on the inventions in this study. CapTS-seq is not a commercial product from New England Biolabs.
REFERENCES
- 1. Finnegan E.J. The small RNA world. J. Cell Sci. 2003; 116:4689–4693. [DOI] [PubMed] [Google Scholar]
- 2. Hombach S., Kretz M.. Non-coding RNAs: classification, biology and functioning. Adv. Exp. Med. Biol. 2016; 937:3–17. [DOI] [PubMed] [Google Scholar]
- 3. Nakanishi K. Anatomy of RISC: how do small RNAs and chaperones activate Argonaute proteins. Wiley Interdiscip. Rev. RNA. 2016; 7:637–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bushati N., Cohen S.M.. microRNA functions. Annu. Rev. Cell Dev. Biol. 2007; 23:175–205. [DOI] [PubMed] [Google Scholar]
- 5. An S., Song J.J.. The coded functions of noncoding RNAs for gene regulation. Mol. Cells. 2011; 31:491–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhao C., Zhang J., Zhang S., Yu D., Chen Y., Liu Q., Shi M., Ni C., Zhu M.. Diagnostic and biological significance of microRNA-192 in pancreatic ductal adenocarcinoma. Oncol. Rep. 2013; 30:276–284. [DOI] [PubMed] [Google Scholar]
- 7. Wang Z., Han J., Cui Y., Fan K., Zhou X.. Circulating microRNA-21 as noninvasive predictive biomarker for response in cancer immunotherapy. Med. Hypotheses. 2013; 81:41–43. [DOI] [PubMed] [Google Scholar]
- 8. Wen Z., Zhang J., Tang P., Tu N., Wang K., Wu G.. Overexpression of miR-185 inhibits autophagy and apoptosis of dopaminergic neurons by regulating the AMPK/mTOR signaling pathway in Parkinson's disease. Mol. Med. Rep. 2018; 17:131–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhao S., Fung-Leung W.P., Bittner A., Ngo K., Liu X.. Comparison of RNA-Seq and microarray in transcriptome profiling of activated T cells. PLoS One. 2014; 9:e78644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fox-Walsh K., Davis-Turak J., Zhou Y., Li H., Fu X.D.. A multiplex RNA-seq strategy to profile poly(A +) RNA: Application to analysis of transcription response and 3′ end formation. Genomics. 2011; 98:266–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ettwiller L., Buswell J., Yigit E., Schildkraut I.. A novel enrichment strategy reveals unprecedented number of novel transcription start sites at single base resolution in a model prokaryote and the gut microbiome. BMC Genomics. 2016; 17:199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Takahashi H., Kato S., Murata M., Carninci P.. CAGE (cap analysis of gene expression): a protocol for the detection of promoter and transcriptional networks. Methods Mol. Biol. 2012; 786:181–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Babski J., Haas K.A., Näther-Schindler D., Pfeiffer F., Förstner K.U., Hammelmann M., Hilker R., Becker A., Sharma C.M., Marchfelder A., Soppa J.. Genome-wide identification of transcriptional start sites in the haloarchaeon Haloferax volcanii based on differential RNA-Seq (dRNA-Seq). BMC Genomics. 2016; 17:629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schwartz S., Motorin Y.. Next-generation sequencing technologies for detection of modified nucleotides in RNAs. RNA Biol. 2017; 14:1124–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hu Y., Lin J., Hu J., Hu G., Wang K., Zhang H., Reilly M.P., Li M.. PennDiff: detecting differential alternative splicing and transcription by RNA sequencing. Bioinformatics. 2018; 34:2384–2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Liscovitch-Brauer N., Alon S., Porath H.T., Elstein B., Unger R., Ziv T., Admon A., Levanon E.Y., Rosenthal J.J.C., Eisenberg E.. Trade-off between Transcriptome Plasticity and Genome Evolution in Cephalopods. Cell. 2017; 169:191–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhuang F., Fuchs R.T., Sun Z., Zheng Y., Robb G.B.. Structural bias in T4 RNA ligase-mediated 3′-adapter ligation. Nucleic Acids Res. 2012; 40:e54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Munafo D.B., Robb G.B.. Optimization of enzymatic reaction conditions for generating representative pools of cDNA from small RNA. RNA. 2010; 16:2537–2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yehudai-Resheff S., Schuster G.. Characterization of the E. coli poly(A) polymerase: nucleotide specificity, RNA-binding affinities and RNA structure dependence. Nucleic Acids Res. 2000; 28:1139–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Raabe C.A., Hoe C.H., Randau G., Brosius J., Tang T.H., Rozhdestvensky T.S.. The rocks and shallows of deep RNA sequencing: Examples in the Vibrio cholerae RNome. RNA. 2011; 17:1357–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hafner M., Renwick N., Brown M., Mihailovic A., Holoch D., Lin C., Pena J.T.G., Nusbaum J.D., Morozov P., Ludwig J.et al.. RNA-ligase-dependent biases in miRNA representation in deep-sequenced small RNA cDNA libraries. RNA. 2011; 17:1697–1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jayaprakash A.D., Jabado O., Brown B.D., Sachidanandam R.. Identification and remediation of biases in the activity of RNA ligases in small-RNA deep sequencing. Nucleic Acids Res. 2011; 39:e141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Raabe C.A., Tang T.-H., Brosius J., Rozhdestvensky T.S.. Biases in small RNA deep sequencing data. Nucleic Acids Res. 2014; 42:1414–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fuchs R.T., Sun Z., Zhuang F., Robb G.B.. Bias in ligation-based small RNA sequencing library construction is determined by adaptor and RNA structure. PLoS One. 2015; 10:e0126049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Alon S., Vigneault F., Eminaga S., Christodoulou D.C., Seidman J.G., Church G.M., Eisenberg E.. Barcoding bias in high-throughput multiplex sequencing of miRNA. Genome Res. 2011; 21:1506–1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Maguire S., Lohman G.J.S., Guan S.. A low-bias and sensitive small RNA library preparation method using randomized splint ligation. Nucleic Acids Res. 2020; 48:e80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhu Y.Y., Machleder E.M., Chenchik A., Li R., Siebert P.D.. Reverse transcriptase template switching: A SMARTTM approach for full-length cDNA library construction. BioTechniques. 2001; 30:892–897. [DOI] [PubMed] [Google Scholar]
- 28. Petalidis L., Bhattacharyya S., Morris G.A., Collins V.P., Freeman T.C., Lyons P.A.. Global amplification of mRNA by template-switching PCR: linearity and application to microarray analysis. Nucleic Acids Res. 2003; 31:e142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shi X., Kaminskyj G.W.. 5′ RACE by tailing a general template-switching oligonucleotide. BioTechniques. 2000; 29:2–4. [DOI] [PubMed] [Google Scholar]
- 30. Dard-Dascot C., Naquin D., d’Aubenton-Carafa Y., Alix K., Thermes C., van Dijk E.. Systematic comparison of small RNA library preparation protocols for next-generation sequencing. BMC Genomics. 2018; 19:118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wright C., Rajpurohit A., Burke E.E., Williams C., Collado-Torres L., Kimos M., Brandon N.J., Cross A.J., Jaffe A.E., Weinberger D.R.et al.. Comprehensive assessment of multiple biases in small RNA sequencing reveals significant differences in the performance of widely used methods. BMC Genomics. 2019; 20:513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Heinicke F., Zhong X., Zucknick M., Breidenbach J., Sundaram A.Y.M., T. Flåm S., Leithaug M., Dalland M., Farmer A., Henderson J.M.et al.. Systematic assessment of commercially available low-input miRNA library preparation kits. RNA Biol. 2020; 17:75–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zarlenga D. Greene J., Rao V.. cDNA Cloning and the Construction of Recombinant DNA. Recombinant DNA Principles and Methodologies. 1998; NY: Marcel Dekker, Inc. [Google Scholar]
- 34. Cumbie J.S., Ivanchenko M.G., Megraw M.. NanoCAGE-XL and CapFilter: an approach to genome wide identification of high confidence transcription start sites. BMC Genomics. 2015; 16:597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Arguel M.-J., LeBrigand K., Paquet A., Ruiz García S., Zaragosi L.-E., Barbry P., Waldmann R.. A cost effective 5′ selective single cell transcriptome profiling approach with improved UMI design. Nucleic Acids Res. 2017; 45:e48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tuschl T., Sharp P.A., Bartel D.P.. Selection in vitro of novel ribozymes from a partially randomized U2 and U6 snRNA library. EMBO J. 1998; 17:2637–2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wulf M.G., Maguire S., Humbert P., Dai N., Bei Y., Nichols N.M., Corrêa I.R., Guan S.. Non-templated addition and template switching by Moloney murine leukemia virus (MMLV)-based reverse transcriptases co-occur and compete with each other. J. Biol. Chem. 2019; 294:18220–18231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yan B., Boitano M., Clark T.A., Ettwiller L.. SMRT-Cappable-seq reveals complex operon variants in bacteria. Nat. Commun. 2018; 9:3676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jemielity J., Fowler T., Zuberek J., Stepinski J., Lewdorowicz M., Niedzwiecka A., Stolarski R., Darzynkiewicz E., Rhoads R.E.. Novel “anti-reverse” cap analogs with superior translational properties. RNA. 2003; 9:1108–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kowalska J., Lewdorowicz M., Zuberek J., Grudzien-Nogalska E., Bojarska E., Stepinski J., Rhoads R.E., Darzynkiewicz E., Davis R.E., Jemielity J.. Synthesis and characterization of mRNA cap analogs containing phosphorothioate substitutions that bind tightly to eIF4E and are resistant to the decapping pyrophosphatase DcpS. RNA. 2008; 14:1119–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Warminski M., Kowalska J., Buck J., Zuberek J., Lukaszewicz M., Nicola C., Kuhn A.N., Sahin U., Darzynkiewicz E., Jemielity J.. The synthesis of isopropylidene mRNA cap analogs modified with phosphorothioate moiety and their evaluation as promoters of mRNA translation. Bioorg. Med. Chem. Lett. 2013; 23:3753–3758. [DOI] [PubMed] [Google Scholar]
- 42. Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal. 2011; 17:10. [Google Scholar]
- 43. Langmead B., Salzberg S.L.. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012; 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R.. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009; 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bates D., Mächler M., Bolker B.M., Walker S.C.. Fitting linear mixed-effects models using lme4. J. Stat. Softw. 2015; 67: 10.18637/jss.v067.i01. [DOI] [Google Scholar]
- 46. Nettling M., Treutler H., Grau J., Keilwagen J., Posch S., Grosse I.. DiffLogo: a comparative visualization of sequence motifs. BMC Bioinformatics. 2015; 16:387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Friedländer M.R., MacKowiak S.D., Li N., Chen W., Rajewsky N.. MiRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012; 40:37–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Frankish A., Diekhans M., Ferreira A.M., Johnson R., Jungreis I., Loveland J., Mudge J.M., Sisu C., Wright J., Armstrong J.et al.. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. 2019; 47:D766–D773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Chan P.P., Lowe T.M.. GtRNAdb: A database of transfer RNA genes detected in genomic sequence. Nucleic Acids Res. 2009; 37:D93–D97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R.. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013; 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Anders S., Pyl P.T., Huber W.. HTSeq-A Python framework to work with high-throughput sequencing data. Bioinformatics. 2015; 31:166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Agarwal V., Bell G.W., Nam J.W., Bartel D.P.. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015; 4:e05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Huang D.W., Sherman B.T., Lempicki R.A.. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009; 37:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ramanathan A., Robb G.B., Chan S.-H.. mRNA capping: biological functions and applications. Nucleic Acids Res. 2016; 44:7511–7526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sawai H., Wakai H., Nakamura-Ozaki A.. Synthesis and reactions of nucleoside 5′-diphosphate imidazolide. A nonenzymatic capping agent for 5′-monophosphorylated oligoribonucleotides in aqueous solution. J. Org. Chem. 1999; 64:5836–5840. [Google Scholar]
- 56. Wimmer I., Tröscher A.R., Brunner F., Rubino S.J., Bien C.G., Weiner H.L., Lassmann H., Bauer J.. Systematic evaluation of RNA quality, microarray data reliability and pathway analysis in fresh, fresh frozen and formalin-fixed paraffin-embedded tissue samples. Sci. Rep. 2018; 8:6351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zhelkovsky A.M., McReynolds L.A.. Polynucleotide 3′-terminal phosphate modifications by RNA and DNA ligases. J. Biol. Chem. 2014; 289:33608–33616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kawaji H., Nakamura M., Takahashi Y., Sandelin A., Katayama S., Fukuda S., Daub C.O., Kai C., Kawai J., Yasuda J.et al.. Hidden layers of human small RNAs. BMC Genomics. 2008; 9:157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yao J., Wu D.C., Nottingham R.M., Lambowitz A.M.. Identification of protein-protected mrna fragments and structured excised intron rnas in human plasma by tgirt-seq peak calling. Elife. 2020; 9:e60743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wu S., Huang S., Ding J., Zhao Y., Liang L., Liu T., Zhan R., He X.. Multiple microRNAs modulate p21Cip1/Waf1 expression by directly targeting its 3′ untranslated region. Oncogene. 2010; 29:2302–2308. [DOI] [PubMed] [Google Scholar]
- 61. Xu P., Wu Q., Yu J., Rao Y., Kou Z., Fang G., Shi X., Liu W., Han H.. A systematic way to Infer the regulation relations of miRNAs on target genes and critical miRNAs in cancers. Front. Genet. 2020; 11:278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Lin X., Qiu L., Song X., Hou J., Chen W., Zhao J.. A comparative analysis of RNA sequencing methods with ribosome RNA depletion for degraded and low-input total RNA from formalin-fixed and paraffin-embedded samples. BMC Genomics. 2019; 20:831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Buitrago D.H., Patnaik S.K., Kadota K., Kannisto E., Jone D.R., Adusumilli P.S.. Small RNA sequencing for profiling MicroRNAs in long-term preserved formalin-fixed and paraffin-embedded non-small cell lung cancer tumor specimens. PLoS One. 2015; 10:e0121521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Yang Z., Ebright Y.W., Yu B., Chen X.. HEN1 recognizes 21-24 nt small RNA duplexes and deposits a methyl group onto the 2′ OH of the 3′ terminal nucleotide. Nucleic Acids Res. 2006; 34:667–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Yao Y., Jiang C., Wang F., Yan H., Long D., Zhao J., Wang J., Zhang C., Li Y., Tian X.et al.. Integrative analysis of miRNA and mRNA expression profiles associated with human atrial aging. Front. Physiol. 2019; 10:1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Liao J., Wang J., Liu Y., Li J., Duan L.. Transcriptome sequencing of lncRNA, miRNA, mRNA and interaction network constructing in coronary heart disease. BMC Med. Genomics. 2019; 12:124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Szeto C.Y.Y., Lin C.H., Choi S.C., Yip T.T.C., Ngan R.K.C., Tsao G.S.W., Li Lung M.. Integrated mRNA and microRNA transcriptome sequencing characterizes sequence variants and mRNA-microRNA regulatory network in nasopharyngeal carcinoma model systems. FEBS Open Bio. 2014; 4:128–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Röhr C., Kerick M., Fischer A., Kühn A., Kashofer K., Timmermann B., Daskalaki A., Meinel T., Drichel D., Börno S.T.et al.. High-throughput miRNA and mRNA sequencing of paired colorectal normal, tumor and metastasis tissues and bioinformatic modeling of miRNA-1 therapeutic applications. PLoS One. 2013; 8:e67461. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data have been uploaded to the NCBI sequencing reads archive (SRA) and are available under project accession: GSE171049.