Abstract
The recently raised awareness of the threat of a new influenza pandemic has stimulated interest in the detection of influenza A viruses in human as well as animal secretions. Virus isolation alone is unsatisfactory for this purpose because of its inherent limited sensitivity and the lack of host cells that are universally permissive to all influenza A viruses. Previously described PCR methods are more sensitive but are targeted predominantly at virus strains currently circulating in humans, since the sequences of the primer sets display considerable numbers of mismatches to the sequences of animal influenza A viruses. Therefore, a new set of primers, based on highly conserved regions of the matrix gene, was designed for single-tube reverse transcription-PCR for the detection of influenza A viruses from multiple species. This PCR proved to be fully reactive with a panel of 25 genetically diverse virus isolates that were obtained from birds, humans, pigs, horses, and seals and that included all known subtypes of influenza A virus. It was not reactive with the 11 other RNA viruses tested. Comparative tests with throat swab samples from humans and fecal and cloacal swab samples from birds confirmed that the new PCR is faster and up to 100-fold more sensitive than classical virus isolation procedures.
Migratory birds and waterfowl are thought to serve as the reservoir for influenza A viruses in nature (24). To date, influenza A viruses representing 15 hemagglutinin (HA) and nine neuraminidase (NA) subtypes have been detected in wild birds and poultry throughout the world (19, 24). Since the general human population is serologically naive with respect to most avian HA and NA antigens, influenza A viruses of avian origin pose a threat that is at the basis of new pandemics in humans (4, 24). For some time it was thought that avian influenza viruses could be transmitted to humans only through coinfection and genetic reassortment of avian and swine or human influenza viruses in pigs (4, 13, 22, 24, 25). However, the recent zoonotic events in Hong Kong and mainland China caused by H5N1 and H9N2 influenza viruses suggest that avian influenza viruses can be transmitted directly to humans as well (5, 8–10, 15). The link between human influenza and the avian influenza virus reservoir has boosted the public health-related and scientific interest in the prevalence, variability, and zoonotic potential of avian influenza viruses.
Although the routine procedures for the detection of human influenza A viruses described to date, including in vitro virus isolation, immunofluorescence (IF), and PCR-based assays, are powerful tools, they may be less effective for the detection of influenza viruses of avian and porcine origin. The phenotypic and genetic heterogeneities of the latter viruses may result in a false-negative diagnosis of influenza A virus infection by in vitro cell culture or current protocols for PCR analysis. Importantly, sporadic zoonotic events of influenza A virus infection may remain undetected as a result of such false-negative diagnoses.
The aim of this study was to set up a rapid and sensitive PCR method for the screening of clinical specimens for the presence of phenotypically and genotypically diverse influenza A viruses. To this end, we have designed a primer set for PCR-based detection of influenza A viruses that was validated with clinical specimens and a panel of influenza A virus strains representing all known HA and NA subtypes obtained from a variety of host species and from different geographical locations. The efficacy of this PCR-based screening of samples from avian and human origin was compared with classical isolation of influenza A virus in embryonated chicken eggs or mammalian cell culture. We conclude that this PCR, based on the detection of gene segment 7 of influenza A virus, is fast, sensitive, and specific and is suitable for all genetic variants of influenza A virus known to date.
MATERIALS AND METHODS
Design of oligonucleotides.
PCR primers were designed on the basis of sequence information obtained from the Influenza Sequence Database at Los Alamos National Laboratories, Los Alamos, N.M. (http://www.flu.lanl.gov). To identify conserved sequences in the influenza virus gene segments, entropy plots were created with the Bioedit software package (available through http: //www.mbio.ncsu.edu/RNaseP/info/programs/BIOEDIT/bioedit.html). Because the HA and NA genes are genetically diverse and sequence information on the PA, PB1, and PB2 polymerase genes is limited (less than 100 sequence entries are available from the database, including partial sequences) only (partial) sequences representing gene segments 5, 7, and 8 encoding nucleoprotein, matrix, and nonstructural proteins, respectively, were analyzed. The degree of heterogeneity was expressed as entropy as defined by Shannon: H (1) = −Σf(b, 1) ln [f(b, 1)], where H (1) is the uncertainty at position 1, b represents a residue out of the allowed choices for the sequence in question (A, C, G, T, −), and f(b, 1) is the frequency at which residue b is found at position 1 (16, 21). Oligonucleotides M52C (5′-CTT CTA ACC GAG GTC GAA ACG-3′) and M253R (5′-AGG GCA TTT TGG ACA AAG/T CGT CTA-3′) were designed for PCR amplification of influenza A virus matrix gene sequences, and the biotinylated oligonucleotide Bio-M93C (5′-CCG TCA GGC CCC CTC AAA GCC GA-3′) was synthesized for hybridization purposes (Eurogentec, Seraing, Belgium).
Specimens.
Cloacal swab specimens were collected from ducks (widgeon [Mareca penelope], gadwall [Mareca strepera], and mallard [Anas plathyrhynchos]) at a marshaling lake in Lekkerkerk, The Netherlands, and droppings as well as cloacal swab specimens were collected from geese (greylag goose [Anser anser], white-fronted goose [Anser albifrons albifrons], barnacle goose [Branta leucopsis], and brent goose [Branta bernicla]) in Groningen and Eemdijk, The Netherlands, between 1997 and 1999. Cloacal swab specimens and droppings were collected from shorebirds at Öland, Sweden, in the spring of 1999. Cotton swabs were used for sampling and were subsequently stored in transport medium (23). Throat swab specimens collected from humans were also stored in transport medium. The samples were stored at 4°C for a few days, at −20°C for less than a week, or at −70°C for extended periods of time. Transport medium consisted of Hanks balanced salt solution supplemented with 10% glycerol, 200 U of penicillin per ml, 200 μg of streptomycin per ml, 100 U of polymyxin B sulfate per ml, 250 μg of gentamicin per ml, and 50 U of nystatin per ml (all from ICN, Zoetermeer, The Netherlands).
RNA isolation.
RNA was isolated with a high pure RNA isolation kit (Roche Molecular Biochemicals) according to the instructions from the manufacturer, with minor modifications. A 0.2-ml sample was homogenized by vortexing and was subsequently lysed with 0.4 ml of lysis-binding buffer to which poly(A) (Roche Molecular Biochemicals) was added as a carrier to 1 μg/ml. After binding to the column, DNase I digestion, and washing, the RNA was eluted in 50 μl of nuclease-free double-distilled water preheated to 80°C.
PCR.
The reverse transcription (RT) and PCRs were optimized with respect to enzymes, primer sets, and concentrations of reagents as well as cycling parameters. Samples were amplified in a one-step RT-PCR in a final volume of 25 μl containing 50 mM Tris · HCl (pH 8.5), 50 mM NaCl, 7 mM MgCl2, 2 mM dithiothreitol, 1 mM each deoxynucleoside triphosphate at a concentration of 1 mM, each oligonucleotide at a concentration of 0.4 μM, 2.5 U of recombinant RNAsin, 10 U of avian myeloblastosis virus reverse transcriptase, 2.5 U of Ampli-Taq DNA polymerase (all enzymes were from Promega Benelux B.V., Leiden, The Netherlands), and 5 μl of RNA. Thermocycling was performed in an MJ PTC-200 apparatus with the following cycling conditions: 30 min at 42°C and 4 min at 95°C once and then 1 min at 95°C, 1 min at 45°C, 3 min at 72°C 40 times. Each reaction was analyzed by agarose gel electrophoresis and ethidium bromide staining (10 μl/sample), followed by Southern blot hybridization (2) or dot blot hybridization (5 μl/sample).
Dot blot hybridization.
Five microliters of each of the PCR products was incubated for 5 min at room temperature with 45 μl of 10 mM Tris · HCl (pH 8.0), 1 mM EDTA, and 50 μl of 1 M NaOH for denaturation. The samples were transferred to prewetted Hybond N+ membranes (Amersham Pharmacia Biotech Benelux, Roosendaal, The Netherlands) with a dot blot apparatus while applying vacuum. The samples were then treated for 3 min with 0.1 ml of 1 M Tris · HCl (pH 8.0), after which vacuum was again applied for 10 s and the membrane was removed from the apparatus. The blots were washed three times for 10 min each time with 0.3 M NaCl–30 mM sodium citrate (pH 7), dried, and stored at 4°C. The blots were prehybridized for 5 min at 55°C in 2× SSPE (0.3 M NaCl, 20 mM NaH2PO4, 2 mM EDTA [pH 7.4]) and 0.1% sodium dodecyl sulfate (SDS), after which biotinylated oligonucleotide probe Bio-M93C was added to 2 pmol/ml and hybridization was continued for 45 min at 55°C. The blots were washed twice for 10 min each time at 55°C with hybridization buffer and transferred to 2× SSPE with 0.5% SDS, after which streptavidin-peroxidase (Roche Molecular Biochemicals) was added to 0.125 U/ml and the mixture was incubated for 45 min at 42°C. The blots were washed for 10 min at 42°C in 2× SSPE–0.5% SDS, 10 min at 42°C in 2× SSPE–0.1% SDS, and 10 min at room temperature in 2× SSPE, after which the samples were visualized with enhanced chemiluminescence detection reagents and by exposure to hyperfilm (Amersham Pharmacia Biotech Benelux) for 5 to 60 s.
Virus isolation and propagation.
The influenza A viruses listed in Table 1 have been described earlier and were kindly provided by R. G. Webster (14, 19). All of these viruses had been isolated and propagated in the allantoic cavities of 11-day-old embryonated chicken eggs (12). Influenza virus A/Netherlands/18/94 has been described previously (18). Influenza A virus strains not listed in Table 1 were isolated and propagated in Madin-Darby canine kidney (MDCK) cells or tertiary monkey kidney (tMK) cells derived from cynomolgus macaques (Macaca fascicularis) (7, 17). Virus stocks were titrated by end point dilution in MDCK or tMK cells, and the 50% tissue culture infective doses (TCID50s) were calculated as described previously (17). The HA titers in the virus stocks were determined with turkey erythrocytes by standard procedures (17). Virus isolates were characterized by hemagglutination inhibition assays with subtype-specific hyperimmune rabbit antisera raised against HA and NA preparations of the virus isolates listed in Table 1 (20).
TABLE 1.
Virus isolates used for the validation of PCR-based detection of influenza A virus
| Influenza A virus strain | HA subtype | NA subtype | HA titer | Lane no. (Fig. 2) |
|---|---|---|---|---|
| A/Puerto Rico/8/34 | 1 | 1 | 384 | 1 |
| A/Fort Monmouth/1/47 | 1 | 1 | 384 | 2 |
| A/Swine/Shope/56 | 1 | 1 | 512 | 3 |
| A/Duck/Alberta/35/76 | 1 | 1 | 768 | 4 |
| A/Singapore/1/57 | 2 | 2 | 256 | 5 |
| A/Hong Kong/1/68 | 3 | 2 | 512 | 6 |
| A/Equine/Miami/1/63 | 4 | 8 | 256 | 7 |
| A/Duck/Ukraine/1/63 | 5 | 8 | 512 | 8 |
| A/Duck/Czechoslovakia/1/56 | 6 | 6 | 256 | 9 |
| A/Tern/South Africa/61 | 5 | 3 | 256 | 10 |
| A/Duck/Hong Kong/205/77 | 5 | 3 | 128 | 11 |
| A/Turkey/Massachusetts/65 | 6 | —a | 512 | 12 |
| A/Shearwater/Australia/1/72 | 6 | 5 | 192 | 13 |
| A/Equine/Prague/1/56 | 7 | 7 | 1024 | 14 |
| A/Seal/Massachusetts/1/80 | 7 | 7 | 128 | 15 |
| A/Turkey/Ontario/6118/68 | 8 | 4 | 128 | 16 |
| A/Turkey/Wisconsin/1/66 | 9 | 2 | 384 | 17 |
| A/Chicken/Germany/49 | 10 | 7 | 384 | 18 |
| A/Duck/England/1/56 | 11 | 6 | 256 | 19 |
| A/Duck/Memphis/546/76 | 11 | 9 | 768 | 20 |
| A/Duck/Alberta/60/76 | 12 | 5 | 128 | 21 |
| A/Gull/Maryland/704/77 | 13 | 6 | 256 | 22 |
| A/Mallard/Gurjev/263/82 | 14 | — | 768 | 23 |
| A/Duck/Australia/341/83 | 15 | 8 | 256 | 24 |
| A/Shearwater/West Australia/2576/79 | 15 | 9 | 512 | 25 |
—, NA subtype unknown.
Human respiratory syncytial virus (HRSV) was grown in HEp-2 cells, mumps and measles viruses were grown in Vero cells, human parainfluenza virus (PIV) types 1 through 4 (PIV-1 through PIV-4) and influenza B virus were grown in tMK cells, and Sendai virus, simian parainfluenza virus type 5 (SV5), and Newcastle disease virus (NDV) were grown in embryonated chicken eggs. The virus titers of these stocks typically ranged from 104 to 106 TCID50s/ml.
RESULTS
Design of oligonucleotides for PCR detection of influenza A viruses.
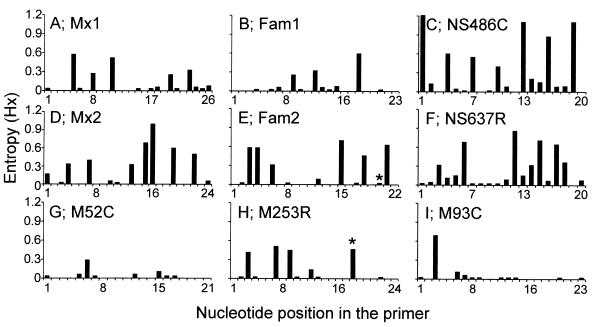
Avian and mammalian influenza A virus nucleotide sequences available from the influenza sequence database (http://www.flu.lanl.gov) were compared to the sequences of previously described primer sets Mx1 and Mx2 (3), Fam1 and Fam2 (1), and NS486C and NS637R (6, 7) to analyze their potential for the detection of genetically diverse influenza A viruses. The variability between the influenza A virus nucleotide sequences and each position in the potential PCR primers was calculated by using the entropy algorithm available from the Bioedit software package (16, 21). Although each of the primer sequences was based on a relatively conserved domain of gene segments 7 and 8 of influenza A virus, considerable heterogeneity was observed for each of the oligonucleotide sets (Fig. 1). The 3′ ends of oligonucleotides are of the greatest importance for the successful amplification by PCR. Of all three published primer sets (Fig. 1A to F), at least one of the oligonucleotides displayed considerable numbers of mismatches with the sequences in the database. Since such mismatches may lead to false-negative PCR results, we designed new primer sets based on segment 7 of influenza A virus, which is relatively conserved compared to the other segments. Within the M1 coding sequence of gene segment 7, several regions (positions 32 to 93, 149 to 204, and 218 to 276) were identified that are relatively conserved among influenza A virus strains obtained from a variety of host species and from different geographical regions. Oligonucleotides M52C (nucleotide positions 32 to 52), M93C (positions 71 to 93), and M253R (positions 253 to 276) (Fig. 1) were designed on the basis of these conserved regions of the influenza A virus genome. Although other conserved regions were identified in the NS2 coding sequence of gene segment 8 and the M1 coding sequence of segment 7, we found primers based on these sequences to be less suitable for PCR amplification of selected influenza A virus strains (data not shown).
FIG. 1.
Entropy plots of oligonucleotide-annealing sites in human and animal influenza A virus sequences available from the influenza virus sequence database. The sequences recognized by oligonucleotides Mx1, Fam1, NS486C, Mx2, Fam2, NS637R, M52C, M253R, and M93C were compared to all available influenza A virus sequences (n = 189, 189, 234, 203, 204, 249, 175, 215, and 189, respectively), and their heterogeneities are displayed in panels A through I, respectively. Oligonucleotide positions are given in the 5′ to 3′ direction, with position 1 being the extreme 5′ nucleotide. Asterisks indicate primer positions with degeneracy in the designed oligonucleotides. Oligonucleotides M52C, M253R, and M93C were designed in the present study.
Sensitivity and specificity of influenza A virus PCR.
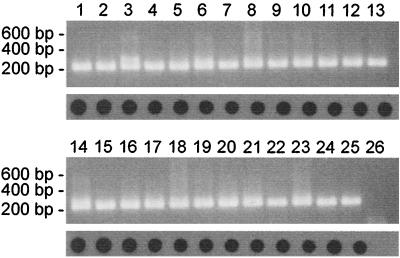
RNA was isolated from 0.2 ml of allantoic fluid containing the influenza A viruses shown in Table 1, and the equivalent of 4 μl of allantoic fluid was used for amplification by PCR with primer set M52C-M253R. For each of the virus strains tested, a band of 244 bp was amplified and was easily visualized on a 1% agarose gel stained with ethidium bromide (Fig. 2). Hybridization of dot blots with the internal biotinylated oligonucleotide probe M93C also resulted in clear signals for each of the influenza A virus strains tested.
FIG. 2.
PCR analysis of the influenza A viruses, listed in Table 1, which originated from different hosts and geographical locations. RNA was isolated from influenza A viruses grown in embryonated chicken eggs, followed by PCR analysis and agarose gel electrophoresis (top panels) or dot blot analysis (bottom panels). Lanes 1 to 25, see Table 1; lane 26, negative control.
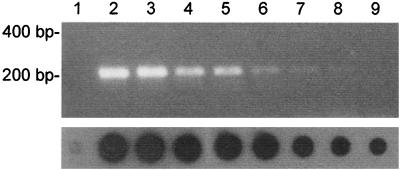
We next compared the sensitivity of this PCR with virus propagation in cell cultures. A stock of influenza virus A/Netherlands/18/94 (H3N2) was generated in tMK cells. This virus stock contained 107 TCID50s of influenza A virus per ml of culture supernatant, as determined with tMK and MDCK cells (17). Serial 10-fold dilutions of virus were made in transport medium, and RNA was isolated for use in PCR analysis, agarose gel electrophoresis, or dot blot hybridization. The expected DNA fragment of 244 bp was visible on an agarose gel stained with ethidium bromide when the RNA equivalent of 0.2 TCID50 of influenza A virus was used as input in the PCR (Fig. 3, lane 8). By using dot blots and hybridization, 0.02 TCID50 of influenza A virus was found to be the detection limit of the assay (Fig. 3, lane 9, and data not shown). Similar results were obtained with a second influenza A virus isolate, and such results were found to be reproducible (data not shown). These data indicate that our PCR procedure is up to 100-fold more sensitive than virus propagation in MDCK and tMK cells.
FIG. 3.
Sensitivity of detection of influenza A virus RNA by PCR. RNA was isolated from 0.2 ml of 10-fold serial dilutions of influenza virus A/Netherlands/18/94 (107 TCID50s/ml) and was used for PCR analysis followed by agarose gel electrophoresis and ethidium bromide staining (top panel) or dot blot analysis (bottom panel). Lane 1, negative control; lanes 2 to 9, dilution series representing the equivalent of 2 × 105 to 0.02 TCID50s per sample. Samples containing less than 0.02 TCID50 were negative by PCR and dot blot analysis (data not shown).
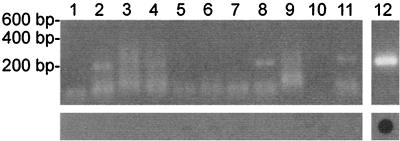
To test the specificities of our PCR primers, RNA was isolated from stocks of a number of RNA viruses, followed by PCR amplification and gel electrophoresis or dot blot hybridization. RNA was isolated from 0.2 ml of virus stocks containing either influenza B virus, HRSV, PIV-1 through PIV-4, simian parainfluenza virus type 5 (SV5), NDV, mumps virus, measles virus, or Sendai virus. One-tenth of the RNA, representing the equivalent of 20 μl of virus stock ranging in titer from 104 to 106 TCID50s/ml, was used for PCR. Upon agarose gel electrophoresis, weak bands and smears of bands ranging from 150 to 400 bp in length were observed after PCR amplification of some of the virus samples (PIV-1, -2, and -3, NDV, mumps virus, and influenza B virus), presumably as a result of nonspecific amplification of the high levels of viral RNA present in these samples. However, upon hybridization of dot blots with the biotinylated oligonucleotide M93C, all RNA virus samples except for that with influenza A virus were negative (Fig. 4).
FIG. 4.
Specificity of detection of influenza A virus RNA by PCR. RNA was isolated from virus stocks and was used for PCR analysis and subsequent agarose gel electrophoresis (top panel) or dot blot hybridization (bottom panel). Lanes: 1, HRSV; 2, PIV-1; 3, PIV-2; 4, PIV-3; 5, PIV-4; 6, Sendai virus; 7, SV5; 8, NDV; 9, mumps virus; 10, measles virus; 11, influenza B virus; 12, influenza A virus.
Detection of influenza A virus in human throat swab samples.
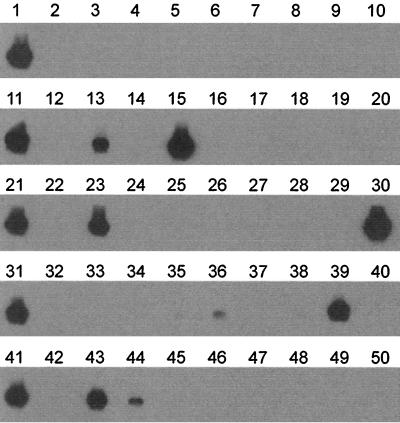
Throat swab samples sent to the virus diagnostic laboratory at Erasmus University Medical Center are routinely tested for the presence of influenza A virus by direct IF (DIF) and inoculation in MDCK or tMK cell cultures in combination with IF (7). For a selection of influenza A virus-positive throat swab samples obtained in the 1994-1995 influenza season, influenza A virus titers were determined by end point dilution and inoculation of tMK cells. A selection of influenza A virus-positive (n = 13) and influenza A virus-negative (n = 26) samples was coded and tested blindly by PCR and dot blot hybridization. All influenza A virus-positive samples, with titers ranging from 0 to 105.75 TCID50s per ml of throat swab sample, were positive upon agarose gel electrophoresis and dot blot hybridization (Fig. 5). One of the influenza A virus PCR-positive samples (lane 6) tested negative upon inoculation of mammalian cell cultures (hence, 0 TCID50). This sample had been found to be influenza A virus positive by DIF with the cells present in the throat swab sample (7), but no virus could be isolated. Of 26 negative control samples (13 were influenza B virus positive and 13 were influenza A and B virus negative in mammalian cell cultures), 24 were negative upon PCR and dot blot analyses. Two of the swabs were negative for influenza A virus in mammalian cell culture and by IF but yielded very weak signals after PCR and dot blot hybridization (lanes 9 and 30). These weak dot blot signals may be due to background hybridization or the presence of very small amounts of influenza A virus RNA in the throat swabs.
FIG. 5.
PCR-based detection of influenza A virus in 39 human throat swab samples. Throat swab samples that were tested previously for the presence of influenza A virus by classical screening methods (7) were randomized and tested blindly by PCR. RNA was isolated from 0.2 ml of a throat swab sample and was used for PCR and dot blot analysis. Lanes 1, 4, 7, 8, 13, 16, 18, 23, 24, 30, 34, 35, and 38, influenza virus-negative samples; lanes 2, 5, 9, 10, 12, 14, 15, 20, 21, 22, 25, 29, and 31, influenza B virus-positive samples; lane 40, 10 TCID50s of influenza virus A/Netherlands/18/94 as a positive control; lanes 3, 6, 11, 17, 19, 26, 27, 28, 32, 33, 36, 37, and 39, influenza A virus-positive samples in which virus titers determined in MDCK cells were 105.75, 0, 103.5, 102.25, 100.75, 104.25, 100.75, 103.75, 104.25, 105.25, 104.5, 105.75, and 103.5 TCID50s/ml respectively.
Detection of influenza A virus in bird samples.
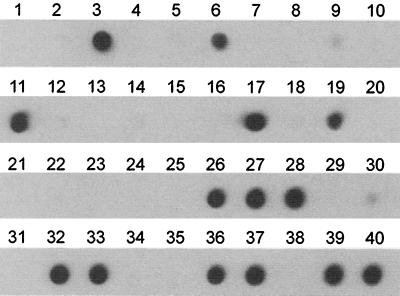
We next tested the suitability of the PCR for avian influenza A virus screening of cloacal swab and dropping samples from ducks, geese, and shorebirds collected in The Netherlands and Sweden. Because PCR screening appeared to be up to 100-fold more sensitive than virus isolation (see above) and to reduce cost and workload, the numbers of RNA isolations and PCR analyses were reduced by making pools of five samples each (40 μl per sample). Between each five pooled samples, a negative control consisting of transport medium was inserted to check for contamination during processing of the samples. Among the 235 pools of samples representing 1,175 individual specimens, RNA isolation, PCR, and Southern or dot blot hybridization revealed the presence of influenza A virus in 19 of them (the results of the analysis of 38 of these pools is shown in Fig. 6). RNA was then isolated from each of the individual samples present in these 19 pools, revealing that all except 1 pool contained a single positive bird sample; the one exception contained two positive samples.
FIG. 6.
PCR-based detection of influenza A virus in a representative set of avian cloacal swab and dropping samples. RNA was isolated from 0.2 ml of 38 pooled samples, each consisting of five individual bird samples, and was used for PCR and Southern blot analysis. Lanes 1, 11, 21, 31, and 41, positive controls representing 10 TCID50s of influenza virus A/Netherlands/18/94; lanes 7, 14, 20, 27, 34, 40, and 47, negative controls; lanes 2 to 5, duck cloacal swab samples; lanes 6, 8 to 10, 12, 13, 15 to 19, 22 to 26, and 28 to 30, goose dropping samples; lanes 32, 33, 35 to 39, 42 to 46, and 48 to 50, goose cloacal swab samples. Each of the pools represented in lanes 13, 15, 23, 30, 36, 39, 43, and 44 was found to contain a single positive individual bird sample. Virus was isolated in embryonated chicken eggs from samples represented in lanes 13, 15, 23, 30, 39, and 43 but not from those represented in lanes 35, 36, and 44.
Each of the 20 positive individual samples was used to inoculate two to four embryonated chicken eggs from which the allantoic fluids were collected, pooled, and inoculated a second time in duplicate in embryonated chicken eggs (blind passage). For 15 of 20 PCR-positive samples we were able to isolate influenza A virus in eggs. For the other five samples, which appeared to contain less virus, as judged by the intensity of the signals on dot blots (e.g., lanes 35, 36, and 44 in Fig. 6), no influenza A virus could be isolated even upon blind passage in embryonated chicken eggs.
To test the possibility that the PCR analysis would give false-negative results compared to virus isolation in eggs, 243 individual PCR-negative cloacal swab and dropping samples were inoculated into two to four embryonated chicken eggs each, followed by a blind passage of the pooled allantoic fluids in duplicate. We were unable to isolate influenza A virus from these PCR-negative samples, indicating that no false-negative results were obtained by PCR analysis. Inoculation of tMK and MDCK cell cultures with 212 random PCR-negative individual bird samples also did not reveal additional influenza A virus-positive samples. In fact, these cell lines were found to be less susceptible to avian influenza A virus than embryonated chicken eggs were (data not shown).
DISCUSSION
PCR-based methods for virus detection have been described for many clinically relevant viruses. The sensitivities and specificities of PCR-based methods are most critically determined by the choice of primer sequences. The sequences of the primer sets described earlier for PCR-based detection of influenza A virus may be appropriate for the detection of virus strains currently circulating in humans (1, 3, 6, 7) but display considerable numbers of mismatches when they are compared with the sequences of animal influenza A viruses. We have used an extensive amount of the sequence information available for influenza A virus to design a new PCR primer set for diagnostic purposes. Primers M52C and M253R and probe M93C span conserved sequences in gene segment 7 of influenza A virus and have no homology to nucleotide sequences from other species available from GenBank (http://www.ncbi.nlm.nih.gov). Our experimental data confirmed that PCR amplification and dot blot analyses with this set of primers does not pick up cross-reacting host-derived sequences or other RNA viruses and is suitable for detection of a wide variety of influenza A virus strains. The limited variability in influenza A virus sequences spanning the primer sequences is mostly confined to the 5′ ends of the oligonucleotides and therefore is unlikely to obscure PCR amplification. Indeed, we successfully amplified the genomes of virus isolates with mismatches in these primer sequences that were included in the viruses shown in Table 1 and Fig. 2.
On the basis of the results of titration experiments as well as on analyses of clinical specimens, we conclude that the PCR-based method is more sensitive (up to 100-fold) than virus isolation in eggs or mammalian cell cultures. This is not surprising in view of the sensitivity of PCR-based assays in general and the low ratio of infectious units to physical particles for RNA viruses such as influenza A virus. Perhaps as a result of the high sensitivity, we detected influenza A virus in a human throat swab sample from which no virus could be isolated. Individual cells isolated from this throat swab sample were positive upon DIF analysis, confirming influenza A virus infection.
An additional advantage of the PCR-based method is its value in the identification of influenza A viruses from different species. Because of differences in cellular tropism between avian, human, and swine influenza A viruses, a single cell type for virus isolation for diagnostic purposes is not available. Continuous and primary cell lines obtained from a variety of animal species and embryonated chicken eggs are routinely used for isolation of influenza A viruses. Using the PCR-based method, we have detected many influenza A viruses in bird samples that could not be isolated in mammalian cell cultures and some that could not be isolated in embryonated chicken eggs. Presumably, this failure was due to a combination of low virus titers in the original specimens and the limited susceptibilities of the target cells to certain influenza A virus strains. As a national influenza center, we occasionally receive specimens from humans from which no virus can be isolated in mammalian cell cultures but that are readily found to be influenza A virus positive by this PCR approach (data not shown).
One disadvantage of PCR-based assays is that it is difficult to assess if weak positive PCR results (e.g., Fig. 5, lanes 9 and 30, and Fig. 6, lanes 35, 36, and 44) are the result of background hybridization or low virus titers in the original samples because of the lack of confirmation assays that are as sensitive as PCR-based methods. Therefore, it is of great importance that sufficient negative controls be included to determine a cutoff value for background hybridization. In addition, we routinely use 10-fold serial dilutions of a titrated influenza A virus stock as input material in our PCR-based assays to provide a semiquantitative estimate of variability between independent assays. Both sets of controls will aid in the determination of a cutoff value for background hybridization and weak positive samples.
By PCR-based assays, diagnosis of influenza A virus infection can be achieved within a single working day, which is significantly faster than the time to diagnosis of infection by classical methods. By virus culture approaches, positive results may be obtained in 24 h or more after inoculation, but a definite negative diagnosis may require culture for up to 2 weeks. The availability of NA inhibitors for the treatment of influenza virus infection may demand more rapid diagnosis of virus infection in the future. The benefit of these new drugs appears to depend heavily on the early start of treatment, i.e., within 2 days after the onset of disease (11).
Taken together, our data indicate that the newly designed PCR offers a more sensitive and faster tool for the diagnosis of human influenza A virus infection than virus isolation. Because of the better matching primers, it can be expected that for the detection of animal influenza A viruses this PCR is also more suitable than previous PCR protocols (1, 3, 7).
ACKNOWLEDGMENTS
We thank John de Boer, Hans Zantinge, Dick Jonkers, Björn Olsen, and their colleagues for collection of bird samples, Rob Webster for providing influenza A virus isolates, Jan Groen and Bernadette van den Hoogen for samples from RNA viruses, and Jan de Jong for critically reading the manuscript. R.A.M.F. is a fellow of the Royal Dutch Academy of Arts and Sciences.
This work was made possible in part through a grant from the Dutch Ministry of Agriculture and from the Foundation for Respiratory Virus Infections (SRVI).
REFERENCES
- 1.Atmar R L, Baxter B D, Dominguez E A, Taber L H. Comparison of reverse transcription-PCR with tissue culture and other rapid diagnostic assays for detection of type A influenza virus. J Clin Microbiol. 1996;34:2604–2606. doi: 10.1128/jcm.34.10.2604-2606.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown T. Analysis of DNA sequences by blotting and hybridization. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology, suppl. 45. New York, N.Y: John Wiley & Sons, Inc.; 2000. pp. 2.9.1–2.9.15. [Google Scholar]
- 3.Cherian T, Bobo L, Steinhoff M C, Karron R A, Yolken R H. Use of PCR-enzyme immunoassay for identification of influenza A virus matrix RNA in clinical samples negative for cultivable virus. J Clin Microbiol. 1994;32:623–628. doi: 10.1128/jcm.32.3.623-628.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Claas E C, Osterhaus A D. New clues to the emergence of flu pandemics. Nat Med. 1998;4:1122–1123. doi: 10.1038/2617. [DOI] [PubMed] [Google Scholar]
- 5.Claas E C, Osterhaus A D, van Beek R, De Jong J C, Rimmelzwaan G F, Senne D A, Krauss S, Shortridge K F, Webster R G. Human influenza A H5N1 virus related to a highly pathogenic avian influenza virus. Lancet. 1998;351:472–477. doi: 10.1016/S0140-6736(97)11212-0. [DOI] [PubMed] [Google Scholar]
- 6.Claas E C, Sprenger M J, Kleter G E, van Beek R, Quint W G, Masurel N. Type-specific identification of influenza viruses A, B and C by the polymerase chain reaction. J Virol Methods. 1992;39:1–13. doi: 10.1016/0166-0934(92)90120-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Claas E C, van Milaan A J, Sprenger M J, Ruiten-Stuiver M, Arron G I, Rothbarth P H, Masurel N. Prospective application of reverse transcriptase polymerase chain reaction for diagnosing influenza infections in respiratory samples from a children's hospital. J Clin Microbiol. 1993;31:2218–2221. doi: 10.1128/jcm.31.8.2218-2221.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Jong J C, Claas E C, Osterhaus A D, Webster R G, Lim W L. A pandemic warning? Nature. 1997;389:554. doi: 10.1038/39218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guan Y, Shortridge K F, Krauss S, Webster R G. Molecular characterization of H9N2 influenza viruses: were they the donors of the “internal” genes of H5N1 viruses in Hong Kong? Proc Natl Acad Sci USA. 1999;96:9363–9367. doi: 10.1073/pnas.96.16.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guo Y J, Krauss S, Senne D A, Mo I P, Lo K S, Xiong X P, Norwood M, Shortridge K F, Webster R G, Guan Y. Characterisation of the pathogenicity of members of the newly established H9N2 influenza virus lineage in Asia. Virology. 2000;267:279–288. doi: 10.1006/viro.1999.0115. [DOI] [PubMed] [Google Scholar]
- 11.Hayden F G, Osterhaus A D, Treanor J J, Fleming D M, Aoki F Y, Nicholson K G, Bohnen A M, Hirst H M, Keene O, Wightman K. Efficacy and safety of the neuraminidase inhibitor zanamivir in the treatment of influenzavirus infections. GG167 Influenza Study Group. N Engl J Med. 1997;337:874–880. doi: 10.1056/NEJM199709253371302. [DOI] [PubMed] [Google Scholar]
- 12.Hinshaw V S, Webster R G, Turner B. Novel influenza A viruses isolated from Canadian feral ducks: including strains antigenically related to swine influenza (Hsw1N1) viruses. J Gen Virol. 1978;41:115–127. doi: 10.1099/0022-1317-41-1-115. [DOI] [PubMed] [Google Scholar]
- 13.Ito T, Couceiro J N, Kelm S, Baum L G, Krauss S, Castrucci M R, Donatelli I, Kida H, Paulson J C, Webster R G, Kawaoka Y. Molecular basis for the generation in pigs of influenza A viruses with pandemic potential. J Virol. 1998;72:7367–7373. doi: 10.1128/jvi.72.9.7367-7373.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matrosovich M, Zhou N, Kawaoka Y, Webster R. The surface glycoproteins of H5 influenza viruses isolated from humans, chickens, and wild aquatic birds have distinguishable properties. J Virol. 1999;73:1146–1155. doi: 10.1128/jvi.73.2.1146-1155.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peiris M, Yuen K Y, Leung C W, Chan K H, Ip P L, Lai R W, Orr W K, Shortridge K F. Human infection with influenza H9N2. Lancet. 1999;354:916–917. doi: 10.1016/s0140-6736(99)03311-5. [DOI] [PubMed] [Google Scholar]
- 16.Pierce J R. An introduction to information theory: symbols, signals and noise. 2nd ed. New York, N.Y: Dover Publications, Inc.; 1980. [Google Scholar]
- 17.Rimmelzwaan G F, Baars M, Claas E C, Osterhaus A D. Comparison of RNA hybridization, hemagglutination assay, titration of infectious virus and immunofluorescence as methods for monitoring influenza virus replication in vitro. J Virol Methods. 1998;74:57–66. doi: 10.1016/s0166-0934(98)00071-8. [DOI] [PubMed] [Google Scholar]
- 18.Rimmelzwaan G F, Baars M, van Beek R, van Amerongen G, Lovgren-Bengtsson K, Claas E C, Osterhaus A D. Induction of protective immunity against influenza virus in a macaque model: comparison of conventional and iscom vaccines. J Gen Virol. 1997;78:757–765. doi: 10.1099/0022-1317-78-4-757. [DOI] [PubMed] [Google Scholar]
- 19.Rohm C, Zhou N, Suss J, Mackenzie J, Webster R G. Characterization of a novel influenza hemagglutinin, H15: criteria for determination of influenza A subtypes. Virology. 1996;217:508–516. doi: 10.1006/viro.1996.0145. [DOI] [PubMed] [Google Scholar]
- 20.Schild G C, Newman R W, Webster R G, Major D, Hinshaw V S. Antigenic analysis of influenza A virus surface antigens: considerations for the nomenclature of influenza virus. Brief review. Arch Virol. 1980;63:171–184. doi: 10.1007/BF01315024. [DOI] [PubMed] [Google Scholar]
- 21.Schneider T D, Stephens R M. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scholtissek C, Burger H, Kistner O, Shortridge K F. The nucleoprotein as a possible major factor in determining host specificity of influenza H3N2 viruses. Virology. 1985;147:287–294. doi: 10.1016/0042-6822(85)90131-x. [DOI] [PubMed] [Google Scholar]
- 23.Sharp G B, Kawaoka Y, Wright S M, Turner B, Hinshaw V, Webster R G. Wild ducks are the reservoir for only a limited number of influenza A subtypes. Epidemiol Infect. 1993;110:161–176. doi: 10.1017/s0950268800050780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Webster R G, Bean W J, Gorman O T, Chambers T M, Kawaoka Y. Evolution and ecology of influenza A viruses. Microbiol Rev. 1992;56:152–179. doi: 10.1128/mr.56.1.152-179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou N N, Senne D A, Landgraf J S, Swenson S L, Erickson G, Rossow K, Liu L, Yoon K J, Krauss S, Webster R G. Genetic reassortment of avian, swine, and human influenza A viruses in American pigs. J Virol. 1999;73:8851–8856. doi: 10.1128/jvi.73.10.8851-8856.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]