Abstract
A pathogenic scotochromogenic Mycobacterium xenopi-like organism was isolated from the lung of an immunocompetent young woman. This pathogen caused severe bilateral cavitary lung disease, making two surgical interventions necessary after years of chronic disease. This case prompted us to characterize this mycobacterium by a polyphasic taxonomic approach. The isolate contained chemotaxonomic markers which were typical for the genus Mycobacterium, i.e., the meso isomer of 2,6-diaminopimelic acid, arabinose, and galactose as diagnostic whole-cell sugars, MK-9(H2) as the principal isoprenoid quinone, a mycolic acid pattern of α-mycolates, ketomycolates, and wax ester mycolates, unbranched saturated and unsaturated fatty acids plus a significant amount of tuberculostearic acid, and small amounts of a C20:0 secondary alcohol. On the basis of its unique 16S rRNA and 16S-23S spacer gene sequences, we propose that the isolate should be assigned to a new species, Mycobacterium heckeshornense. This novel species is phylogenetically closely related to M. xenopi. The type strain of M. heckeshornense is strain S369 (DSM 44428T). The GenBank accession number of the 16S rRNA gene of M. heckeshornense is AF174290.
The suborder Corynebacterineae of the order Actinomycetales, class Actinobacteria, constitutes a phylogenetically coherent group which includes the genera Corynebacterium, Dietzia, Rhodococcus, Nocardia, Skermania, Gordonia, Tsukamurella, Williamsia, and Mycobacterium (8, 27). These genera can easily be differentiated from other bacteria by a combination of chemical markers, such as meso-diaminopimelic acid in the cell wall and the heteropolysaccharide arabinogalactan, which connects the mycolic acids (alpha-branched, beta-hydroxylated long-chain fatty acids) with the cell wall. Individual genera of the Corynebacterineae can be differentiated via lipid cell wall analysis, e.g., the chain length and type of mycolic acids, the type of quinone, and the qualitative and quantitative differences in their fatty acid patterns.
The present study characterizes a novel scotochromogenic mycobacterium repeatedly isolated from the respiratory tract of an immunocompetent woman treated at the Heckeshorn Lung Clinic, Berlin, Germany. As shown by chemotaxonomic data, the isolates fell into a defined cluster of slowly growing scotochromogenic mycobacteria which includes Mycobacterium xenopi and Mycobacterium botniense. The phylogenetic analysis and biochemical tests showed that this isolate represents a new species of the genus Mycobacterium for which the name Mycobacterium heckeshornense is proposed.
MATERIALS AND METHODS
Case report.
In March 1993 a 30-year-old Caucasian woman with an inconspicuous medical history complained of cough and fatigue. Chest X ray revealed a cavitation in the right upper lobe as well as an infiltrate in the left upper lobe. The finding of Staphylococcus aureus and a negative tuberculin skin test led to nonspecific antibiotic treatment with doxycycline. The left-sided infiltrate then disappeared and the cavitary lesion shrank. At the end of 1993 the patient experienced a clinical relapse with additional weight loss. Another antibiotic treatment with a cephalosporin led again to temporary clinical and radiological improvement. In October 1994, when the patient was admitted to the Heckeshorn Lung Clinic for the first time, a chest X ray revealed an enlargement of the right-sided cavitary lesion and again an infiltrate in the left upper lobe. Sputum smears were negative for acid-fast bacilli, while bronchoscopy yielded acid-fast bacilli from the right upper lobe. Mycobacteria were cultured, and the isolate was identified as an M. xenopi-like organism by conventional biochemical methods. A biopsy from the right upper lobe histologically revealed epithelioid cell granulomatosis. Treatment with isoniazid, rifampin, protionamide, ethambutol, and ciprofloxacin was initiated. During this treatment, the patient improved clinically, but only a moderate regression of the bilateral lesions was observed radiologically. The organism was isolated repeatedly from microscopically positive sputum samples for 2 months; thereafter, sputum cultures remained negative. The treatment was continued for 1 year. The search for an underlying immunological disorder was negative. There was a normal distribution of cells in white blood cell counts as well as lymphocyte subsets. Immunoglobulin G subclass concentrations in serum were not decreased. Antibodies to human immunodeficiency virus were not found in serum. Esophagography and gastroscopy did not show any signs of reflux or hernia. No metabolic disorder was diagnosed.
In 1996, the patient complained of hemoptysis. Sputum cultures again became positive for mycobacteria. With persisting cavitation in the right upper lobe, additional aspergillus infection was suspected, which was confirmed by culture in another hospital. Additionally, a cavitation in the left upper lobe had been documented since November 1996. Antimycobacterial treatment plus itraconazole was administered for 12 months, and then right upper lobe resection was performed in October 1997. Resection of the left upper lobe was planned for January 1998 but was delayed because of repeated positive sputum smears postoperatively. After continued antimycobacterial treatment, resection was performed in January 1999 at our hospital. This lung specimen again contained masses of acid-fast bacilli. In view of the clinically obvious virulence of this mycobacterium, antimycobacterial treatment was continued, and mycobacterial cultures have remained negative to the present.
Culture, biochemical tests, and drug susceptibility.
Among several isolates recovered from this patient, two isolates (isolates S369T and S532) were chosen for detailed analysis. Of these, S369T was isolated from the first sputum specimen in October 1994 and S532 was obtained in January 6 years later from a lung biopsy specimen. Strain S504 was found in May 1999 in one of two sputum specimens from a second patient with chronic obstructive lung disease. The latter finding was interpreted as a transient colonization according to the recommendations of the American Thoracic Society (30). Specimens were stained, processed, and cultured by standard procedures in mycobacteriology (22). For fatty and mycolic acid analyses, isolates S369T, S532, and S504 were cultivated on Middlebrook 7H10 agar, supplemented with Middlebrook oleic acid-albumin-dextrose-catalase (Difco Laboratories, Detroit, Mich.) at 37°C for 2 weeks. The isolates were cultured for 4 weeks on Löwenstein-Jensen (L-J) medium at 37°C and tested for growth rate, for pigment production, and by all of the biochemical tests listed in Table 1 by standard methods (9, 15). Acetamidase, allantoinase, benzamidase, nicotinamidase, pyrazinamidase, succinamidase, and urease activities were determined by the method of Bönicke (1). Susceptibility to isoniazid, streptomycin, rifampin, ethambutol, p-aminosalicylic acid, prothionamide, capreomycin, cycloserine, ciprofloxacin, and clarithromycin was determined on L-J medium and was interpreted by the modified proportion method (3, 4).
TABLE 1.
Growth and biochemical characteristics of isolates assigned to M. heckeshornense sp. nov. compared to those of M. xenopia
| Characteristic |
M. heckeshornense sp. nov. isolates
|
M. xenopi DSM 43995T | ||
|---|---|---|---|---|
| S369T | S532 | S504 | ||
| Growth at: | ||||
| 30°C | − | − | − | − |
| 37°C | (+) | (+) | + | + |
| 40°C | + | + | + | + |
| 45°C | + | + | + | + |
| 50°C | − | − | − | − |
| 3-day arylsulfatase | − | − | − | + |
| 10-day arylsulfatase | − | − | − | + |
| Catalase (>45 mm) | − | − | − | − |
| Catalase (heat stable) | + | + | + | + |
| Niacin | − | − | − | − |
| Nitrate reduction | − | − | − | − |
| Tween 80 hydrolysis | − | − | − | − |
| 5% NaCl tolerance | − | − | − | − |
| Acetamidase | − | − | − | − |
| Benzamidase | − | − | − | − |
| Urease | − | − | − | − |
| Nicotinamidase | − | − | − | + |
| Pyrazinamidase | − | − | − | + |
| Allantoinase | − | − | − | − |
−, negative; (+), weakly positive; +, positive.
Determination of chemotaxonomic properties.
Amino acids and sugars of whole-cell hydrolysates were analyzed by thin-layer chromatography (TLC) as described previously (28). Isoprenoid quinones were extracted by the small-scale integrated procedure described by Minnikin et al. (18). Menaquinones were studied by high-performance liquid chromatography (HPLC) as previously described in detail (11, 21, 26). Fatty acid methyl esters were obtained by saponification, methylation, and extraction, separated by gas-liquid chromatography (GLC), and determined by using the Microbial Identification System standard software package (21; M. Sasser, Identification of bacteria by gas chromatography of cellular fatty acids, MIDI technical note 101, MIDI, Newark, Del., 1990). Freeze-dried bacteria (50 mg) were degraded by treatment at 75°C with a mixture (3 ml) of methanol-toluene-sulfuric acid (30:15:1; vol/vol) for 16 h, and the hexane extracts were examined by TLC and two-dimensional chromatography (16, 19, 21, 26). For mycolic acid analysis by HPLC, 40 mg (wet weight) of cells was harvested from petri dishes. The cells were suspended in an alkaline solution (25% KOH) and saponified by heating to cleave the mycolic acids bound to the cell walls. The mycolic acids were then obtained by acidification and extraction into chloroform and then converted to their p-bromophenacyl esters as described previously (2; J. L. Miller, Sherlock mycobacteria identification by high-performance liquid chromatography, A training manual, MIDI, 1997). Low- and high-molecular-weight internal standards (Ribi ImmunoChem Research, Hamilton, Mont.) were added to the samples. The mycolic acid p-bromophenacyl ester mixtures were separated by HPLC fitted with a C18 Ultrasphere-XL cartridge column (Beckman Instruments Inc., Berkeley, Calif.) at 35°C. The chromatograph for HPLC was operated by Sherlock System software (MIDI). The same procedure used for preparation of fatty acids was used for preparation of the mycolic acid methyl esters. Ten microliters of 0.2 M methanolic trimethylsulfonium hydroxide was added to the extract to enhance the pyrolysis of the mycolic acid esters in the injector block of the chromatograph for GLC heated at 350°C. For the analysis of the mycolic acid cleavage products, GLC conditions were used as described previously (20).
Analysis of ribosomal gene sequences.
PCR-based amplification of strains S369T and S504 and sequencing of the nearly complete 16S rRNA gene (rDNA) (both strands) were performed as described before (21, 26). Isolate S532 was identified by partial sequencing of the variable regions A and B only (23). The sequence of the new strain was aligned with 48 mycobacterial 16S rDNA reference sequences by using the Genetic Data Environment software, version 2.2 (12). No gaps were removed, and probable sequencing errors within the reference sequences were not weighted. A phylogenetic tree was constructed by using the neighbor-joining method (25) and was applied to distances corrected for multiple hits and for unequal transition and transversion rates according to Kimura's two-parameter model (10), thus omitting parts of uncertain alignment at both ends of the gene. Tree positions were confirmed by parsimony analysis. Bootstrapping was not performed due to the high degree of resemblance of mycobacterial 16S rDNA.
Additionally, we sequenced the 16S-23S spacer of strain S369T and S504 by the protocol used previously (23). Sequences were compared with known spacer sequences by applying a BLAST search. The closest hits were selected for sequence similarity analysis by using the maximum matching option within the DNASIS software (version 2.5; Hitachi Software Engineering Co., Ltd., San Bruno, Calif.).
Nucleotide sequence accession number.
The nucleotide sequence of the 16S rDNA of strain S369T has been deposited in GenBank database under accession number AF174290.
RESULTS AND DISCUSSION
Microbiological and clinical findings.
Growth of isolates S369T, S532, and S504 was observed on L-J slants and Middlebrook media (7H10, 7H11 agar, and 12B broth), whereas the bacteria preferred Middlebrook to the egg-based medium (L-J was a very poor supporter of growth for isolate S532). Small scotochromogenic, yellow, round, smooth colonies (diameter, 0.5 mm) were visible after 4 weeks of growth at temperatures ranging from 37 to 45°C. Microscopically, the cells presented as gram-positive, non-spore-forming, acid-alcohol-fast, nonmotile, pleomorphic rods. These findings, together with the biochemical properties shown in Table 1, indicated that the isolates phenotypically resembled M. xenopi. The lack of arylsulfatase, nicotinamidase, and pyrazinamidase enzyme activities, however, was in discordance with the characteristics of M. xenopi. Reactions at variance to those for M. botniense, a saprophytic mycobacterium closely related to M. xenopi recently found in stream water (29), were tests for 10-day arylsulfatase, heat-stable catalase, and pyrazinamidase activities. Strains S369T and S504 showed resistance to isoniazid, while isolate S532 was also resistant to rifampin, probably due to acquired resistance.
The mycobacteria described in this report were isolated from two patients suffering from lung disease. Although we cannot with complete certainty exclude an underlying unrecognized immunological disorder in the first patient, this case clearly meets criteria that strongly suggest the high potential pathogenicity of M. heckeshornense for patients with or without preexisting lung disease. Similar to other nontuberculous mycobacteria that cause pulmonary infections, including M. xenopi, which is a well-recognized opportunistic pathogen of the lung (6), M. heckeshornense shows the ability to appear both as a transient colonizer and as a severe pathogen causing multiple lung cavities. Unfortunately, precise identification of nontuberculous mycobacteria, especially by molecular biology-based methods, is not widely established in routine microbiological laboratories, and M. xenopi is often considered nonpathogenic (7). It may be that this novel species has previously been misidentified as M. xenopi on the basis of poor phenotypic markers (or a tendency to assign uncertain species to one of the well-established taxa) and occurs in human specimens more frequently than expected. This assumption is supported by the fact that, during this study, we have found a 16S rDNA sequence entry in the GenBank database of an organism provisionally named M. xenopi (submitted January 2000, accession number AJ243481) which is identical to the sequence of M. heckeshornense. To our knowledge this isolate caused extrapulmonary infection. A detailed report on this case will follow shortly.
In conclusion, our study complements reports on the potential pathogenicity of nontuberculous mycobacteria, including many of those in the increasing list of new species described in recent years, and again points to the importance of precise identification of mycobacteria, irrespective of the common belief that they mainly occur as contaminants.
Chemotaxonomy.
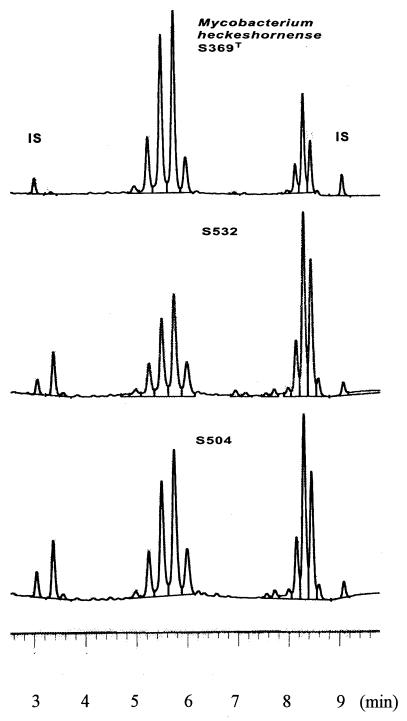
The amino acid and sugar analyses of whole-cell hydrolysates of the three isolates revealed meso-diaminopimelic acid, arabinose, and galactose. This combination of chemical markers grouped these isolates into the type IV cell wall actinomycetes (14). The occurrence of mycolic acids in whole-cell methanolysates classified S369T, S532, and S504 in the order Actinomycetales, suborder Corynebacterineae (27). The combination of long-chain mycolic acids and the isoprenoid quinone MK-9(H2) identified the strains as members of the genus Mycobacterium. A further differentiation within the genus Mycobacterium was obtained by separating mycolic acids by TLC in two directions. The analyses revealed four spots which could be identified as α-mycolates, ketomycolates, and ω-carboxymycolates plus alcohols (wax ester mycolates). This pattern is widely distributed among mycobacteria (5, 15). The gas chromatographic analyses of whole-cell methanolysates of the three isolates showed similar elution profiles (Table 2). The pattern was mainly composed of unbranched saturated and unsaturated fatty acid esters with chain lengths of 16 and 18 carbon atoms plus 10-methyl branched tuberculostearic acid methyl ester. Substantial amounts of the secondary alcohol 2-docosanol (not shown in Table 2) and smaller amounts of 2-eicosanol could also be found. This combination of fatty acids and alcohols classified the three isolates to the M. xenopi-M. botniense taxon (13, 29). The pyrolysis of the mycolic acid methyl esters released a saturated fatty acid methyl ester with a chain length of 26 carbon atoms. This is in accordance with the data reported for M. xenopi and M. botniense (17, 29). S369T, S532, and S504 could be differentiated from M. xenopi by the lack of decanoic acid (10:0) and dodecanoic acid (12:0) and from M. botniense by the missing multimethyl-eicosanoic acids (Table 2). Dodecanoic acid had previously been found to be a significant marker of M. xenopi, although 1 of 25 clinical strains in the report of Torkko et al. (29) contained this compound. Interestingly, this one isolate was found to have a 16S rDNA sequence that differed from the M. xenopi rDNA sequence. The HPLC mycolic acid elution profiles of the three isolates shown in Fig. 1 are undistinguishable from that of M. xenopi (2). The HPLC mycolic acid elution profile of M. botniense is very similar to that of M. xenopi (29).
TABLE 2.
Composition of fatty acid methyl esters derived from whole-cell hydrolysates of M. heckeshornense sp. nov. isolates and M. xenopi DSM 43995T
| Fatty acida | Composition (%)
|
|||
|---|---|---|---|---|
| S369T | S532 | S504 | M. xenopi | |
| 10:0 | −b | − | − | 1.76 |
| 12:0 | − | 0.49 | − | 6.09 |
| 14:0 | 6.62 | 5.64 | 7.50 | 2.76 |
| 15:0 | 0.42 | 0.69 | 0.53 | 0.37 |
| 16:1 cis-7 | 1.33 | 1.45 | 1.09 | 0.39 |
| 16:1 cis-9 | − | − | − | 0.49 |
| 16:1 cis-10 | 2.16 | 2.20 | 2.00 | 1.08 |
| 16:0 | 45.09 | 44.66 | 46.23 | 46.57 |
| 16:0 10-methyl | 0.85 | 0.95 | 1.54 | 0.78 |
| 17:1 cis-10 | 0.90 | 0.59 | 0.90 | 0.61 |
| 17:0 | 0.66 | 0.68 | 0.69 | 0.54 |
| 18:2 cis-9, 10 | 0.27 | − | 0.43 | 1.90 |
| 18:1 cis-9 | 8.681 | 9.60 | 7.44 | 8.87 |
| 18:0 | 7.39 | 7.76 | 7.26 | 7.00 |
| 18:0 10-methyl | 20.56 | 21.20 | 19.25 | 19.10 |
| 20:0 | 0.72 | 1.12 | 0.64 | 0.35 |
| 20:0 2,4,6,x-methyl | − | − | − | − |
| 22:0 2,4,6,x,x-methyl | − | − | − | − |
| 20:0 2-OH | 4.35 | 2.76 | 4.32 | 3.85 |
Including secondary alcohols which were released from wax esters. Examples of abbreviations: 16:0, hexadecanoic acid (palmitic acid); 18:1 cis-9, cis-9-octadecenoic acid (oleic acid); 18:0 10-methyl, 10-methyl-octadecanoic acid (tuberculostearic acid); 20:0 2,4,6,x-methyl, 2,4,6,x-teramethyl-eicosanoic acid; 22:0 2,4,6,x,x-methyl, 2,4,6,x,x-pentamethyl-dodecosanoic acid; 20:0 2-OH, 2-eicosanol.
−, less than 0.1%.
FIG. 1.
HPLC mycolic acid patterns of the three M. heckeshornense sp. nov. isolates. IS, internal standards.
Phylogenetic analysis.
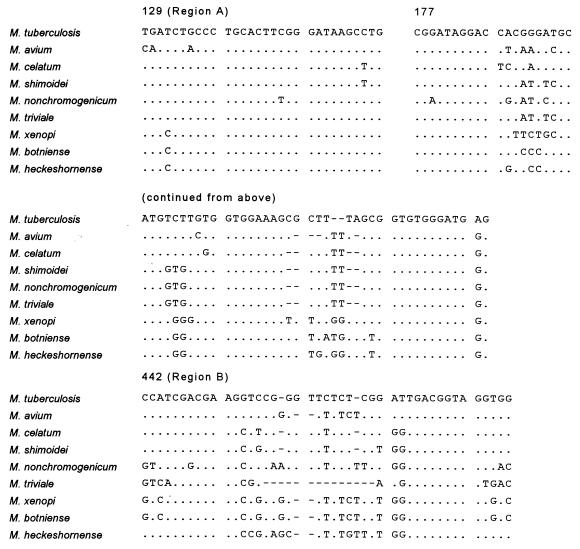
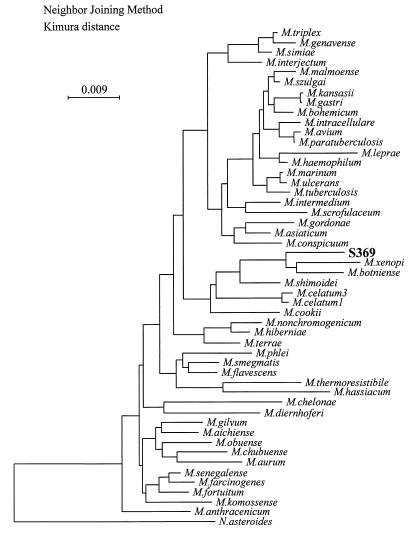
The complete 16S rDNA sequences of two isolates and the partial rDNA sequence of one strain of this novel species were identical and differed from all available 16S rDNA sequences. An alignment of relevant variable regions with those of a selection of other mycobacteria is shown in Fig. 2. Sequence similarity analysis revealed that the sequence was closest to M. xenopi (accession numbers X52929 and M61664) and M. botniense (accession number AJ012756), showing as many as 41 mismatches with the former and 40 mismatches plus 3 gaps with the latter (sequence homology, 97% over 1,484 bases). A distance matrix tree inferred from analysis of the 16S rDNA sequences of 48 mycobacteria is shown in Fig. 3. Both 16S-23S rDNA spacer sequences obtained from strains S369T and S504 were also identical. Although comparison with spacer sequences from M. xenopi and M. botniense revealed a very high degree of sequence divergence (Fig. 4), the spacer data further confirmed the relationship to M. xenopi, both by size (256 nucleotides compared to 235 and 273 nucleotides for M. xenopi and M. botniense, respectively) and by sequence similarity (84% sequence similarity to M. xenopi and less than 72% sequence similarity to M. botniense, M. shimoidei, or M. celatum). Thus, these data provide additional evidence that 16S-23S spacer sequences can be implemented as an adjunct in mycobacterial taxonomy (23). Besides this, the importance of the spacer lies in its potential to be used as a molecular tool for the identification of mycobacteria without the need for sequencing. We recently described such a method based on restriction fragment length polymorphism (RFLP) analysis of PCR-amplified 16S-23S spacer sequences (24). Strains S369T and S504 were included in that study and possessed unique RFLP patterns distinguishable from those of all other mycobacteria. Therefore, M. heckeshornense can easily be detected without the need for sophisticated molecular biology-based methods due to its unique spacer sequence.
FIG. 2.
Comparison of 16S rDNA signature sequences (hypervariable regions A and B) of selected species of the genus Mycobacterium including the novel species M. heckeshornense. Dots indicate identity, and hyphens represent alignment gaps. The corresponding positions of the Escherichia coli 16S rDNA are shown for reference. The sequence accession numbers are as follows: M. tuberculosis, X58890; M. avium, X52918; M. celatum, L08170; M. shimoidei, AJ005005; M. nonchromogenicum, X52928; M. triviale, X88924; M. xenopi, M61664; and M. botniense, AJ012756.
FIG. 3.
Phylogenetic tree indicating the relationship of M. heckeshornense sp. nov. strain S369T to other mycobacterial species. The tree was inferred by the neighbor-joining method applied to distances corrected for multiple hits and for unequal transition and transversion rates by Kimura's two-parameter model (10). The tree was routed by using Nocardia asteroides as an outgroup. The bar indicates the expected number of substitutions per site.
FIG. 4.
Alignment of 16S-23S rDNA spacer sequences of M. heckeshornense sp. nov., M. xenopi DSM 43995T (23), and M. botniense ATCC 700701T (29). Dots indicate identity, and hyphens represent alignment gaps. The lengths of the spacers (in nucleotides) are indicated at the ends of the sequences.
Differentiation of M. heckeshornense sp. nov. from other slowly growing mycobacteria.
The results of physiological and phylogenetic analyses and some of the chemotaxonomic results indicate that strain S369T represents a new species of the genus Mycobacterium. The phylogenetic position of this so far unclassified organism is within the cluster defined by M. xenopi and M. botniense. All three species synthesize α-mycolates, ketomycolates, and wax ester mycolates and release a hexacosanoic pyrolysis ester from their mycolic acids. Key features are negative tests for arylsulfatase and pyrazinamidase and susceptibility to antimycobacterial drugs. Since M. heckeshornense closely resembles M. xenopi in fatty acid and mycolic acid analyses, a definite separation from M. xenopi is obtained by its unique 16S or 16S-23S rDNA sequences.
Description of M. heckeshornense sp. nov.
Mycobacterium heckeshornense (he.ckes.hor.nen′se; L.n. adj., a peninsula in Berlin, Germany, referring to the place where the hospital in which the strain was found is situated). The cells are gram-positive, non-spore-forming, nonmotile short rods that are partially coccoid without branching. Smooth, scotochromogenic colonies with a yellow color appear after 4 weeks of culture. The pigment formation is weaker than the intense pigmentation usually seen in M. xenopi. The cells are able to grow in a range of 37 to 45°C, but growth was best supported at 42°C. The cell wall of the strain contains arabinose and galactose as major cell wall sugars. meso-Diaminopimelic acid is the only cell wall diamino acid. The only isoprenoid quinone is MK-9(H2). The fatty acid pattern from whole-cell methanolysates is composed of tetradecanoic acid (5.6 to 7.5%), palmitoleic acid (1%), cis-10-hexadecenoic acid (2%), palmitic acid (45%), oleic acid (7.4 to 9.6%), stearic acid (7.5%), and tuberculostearic acid (19 to 21%). The secondary alcohols 2-eicosanol and 2-docosanol are also present. TLC of mycolic acid methanolysates revealed α-mycolates, ketomycolates, wax carboxymycolates, and 2-eicosanol (wax ester mycolates). The mycolic acid HPLC elution profile of M. heckeshornense does not differ from that of the closely related species M. xenopi. M. heckeshornense can safely be identified by its unique ribosomal sequences and by negative 3-day arylsulfatase and pyrazinamidase tests in conjunction with its thermophilic growth behavior. Isolates S369T, S532, and S504 have been deposited in the Deutsche Sammlung von Mikroorganismen und Zellkulturen, Braunschweig, Germany, under accession numbers DSM 44428T, DSM 44483, and DSM 44482, respectively.
ACKNOWLEDGMENTS
We thank Gabriele Pötter and Michaela Schmidt for expert technical assistance during the study.
REFERENCES
- 1.Bönicke R. Identification of mycobacteria by biochemical methods. Bull Int Union Tuberc. 1962;32:13–86. [Google Scholar]
- 2.Butler W R, Thilbert L, Kilburn J O. Identification of Mycobacterium avium complex strains and some similar species by high-performance liquid chromatography. J Clin Microbiol. 1992;30:2698–2704. doi: 10.1128/jcm.30.10.2698-2704.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deutsches Institut für Normung. DIN 58943, parts 3, 8, and 9. Berlin, Germany: Beuth Verlag; 1991. [Google Scholar]
- 4.Deutsches Zentralkomitee zur Bekämpfung der Tuberkulose. Die Bakteriologie der Tuberkulose. Pneumologie. 1991;45:753–774. [Google Scholar]
- 5.Hinrikson H P, Pfyffer G E. Mycobacterial mycolic acids. Med Microbial Lett. 1994;3:49–57. , 97–106. [Google Scholar]
- 6.Hoffner S E. Pulmonary infections caused by less frequently encountered slow-growing environmental mycobacteria. Eur J Clin Microbiol Infect Dis. 1994;3:937–941. doi: 10.1007/BF02111495. [DOI] [PubMed] [Google Scholar]
- 7.Jiva T M, Jacoby H M, Weymouth L A, Kaminski D A, Portmore A C. Mycobacterium xenopi: innocent bystander or emerging pathogen? Clin Infect Dis. 1997;24:226–232. [PubMed] [Google Scholar]
- 8.Kämpfer P, Andersson M A, Rainey F A, Kroppenstedt R M, Salkinoja-Salonen M. Williamsia muralis gen. nov., sp. nov., isolated from indoor environment of children's day care centre. Int J Syst Bacteriol. 1999;49:681–687. doi: 10.1099/00207713-49-2-681. [DOI] [PubMed] [Google Scholar]
- 9.Kent P T, Kubica G P. Public health mycobacteriology—a guide for the level III laboratory. U.S. Department of Health and Human Services Publication (CDC) 86-8230. Atlanta, Ga: Centers for Disease Control; 1985. [Google Scholar]
- 10.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 11.Kroppenstedt R M. Fatty acid and menaquinone analysis of actinomycetes and related organisms. Soc Appl Bacteriol Tech Ser. 1985;20:173–199. [Google Scholar]
- 12.Larsen N, Olsen G J, Maidak B L, McCaughey M J, Overbeek R, Macke T, Marsh T L, Woese C. The ribosomal database project. Nucleic Acids Res. 1993;21:3021–3023. doi: 10.1093/nar/21.13.3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Larsson L, Jimenez J, Valero-Guillen P, Martin-Luengo F, Kubin M. Establishment of 2-docosanol as a cellular marker compound in the identification of Mycobacterium xenopi. J Clin Microbiol. 1989;27:2388–2390. doi: 10.1128/jcm.27.10.2388-2390.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lechevalier H A, Lechevalier M P. A critical evaluation of the genera of aerobic actinomycetes. In: Prauser H, editor. The Actinomycetales. Jena, Germany: Gustav Fischer Verlag; 1970. pp. 393–405. [Google Scholar]
- 15.Lévy-Frébault V V, Portales F. Proposed minimal standards for the genus Mycobacterium and for description of new slowly growing Mycobacterium species. Int J Syst Bacteriol. 1992;42:315–323. doi: 10.1099/00207713-42-2-315. [DOI] [PubMed] [Google Scholar]
- 16.Luquin M, Ausina V, Calahorra F L, Belda F, Barcelona M G, Celma C, Prats G. Evaluation of practical chromatographic procedures for identification of clinical isolates of mycobacteria. J Clin Microbiol. 1991;29:120–130. doi: 10.1128/jcm.29.1.120-130.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luquin M, Lopez F, Ausina V. Capillary gas chromatographic analysis of mycolic acid cleavage products, cellular fatty acids, and alcohols of Mycobacterium xenopi. J Clin Microbiol. 1989;27:1403–1406. doi: 10.1128/jcm.27.6.1403-1406.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Minnikin D E, O'Donnell A G, Goodfellow M, Alderson G, Athalye M, Schaal A, Parlett J H. An integrated procedure for the extraction of isoprenoid quinones and polar lipids. J Microbiol Methods. 1984;2:233–241. [Google Scholar]
- 19.Minnikin D E, Hutchinson I G, Caldicott A B, Goodfellow M. Thin-layer chromatography of methanolysates of mycolic acid-containing bacteria. J Chromatogr. 1980;188:221–233. [Google Scholar]
- 20.Müller K-D, Schmid E N, Kroppenstedt R M. Improved identification of mycobacteria by using the Microbial Identification System in combination with additional trimethylsulfonium hydroxide pyrolysis. J Clin Microbiol. 1998;36:2477–2480. doi: 10.1128/jcm.36.9.2477-2480.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reischl U, Emler S, Horak Z, Kaustova J, Kroppenstedt R M, Lehn N, Naumann L. Mycobacterium bohemicum sp. nov., a new slow-growing scotochromogenic mycobacterium. Int J Syst Bacteriol. 1998;48:1349–1355. doi: 10.1099/00207713-48-4-1349. [DOI] [PubMed] [Google Scholar]
- 22.Roberts E D, Koneman E W, Kim Y K. Mycobacterium. In: Balows A, Hausler W J Jr, Herrmann K L, Isenberg H D, Shadomy H J, editors. Manual of clinical microbiology. 5th ed. Washington, D.C.: American Society for Microbiology; 1991. pp. 304–339. [Google Scholar]
- 23.Roth A, Fischer M, Hamid H E, Ludwig W, Michalke S, Mauch H. Differentiation of phylogenetically related slowly growing mycobacteria based on 16S–23S rRNA gene internal transcribed spacer sequences. J Clin Microbiol. 1998;36:139–147. doi: 10.1128/jcm.36.1.139-147.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roth A, Reischl U, Streubel A, Naumann L, Kroppenstedt R M, Habicht M, Fischer M, Mauch H. Novel diagnostic algorithm for identification of mycobacteria using genus-specific amplification of the 16S–23S rRNA gene spacer and restriction endonucleases. J Clin Microbiol. 2000;38:1094–1104. doi: 10.1128/jcm.38.3.1094-1104.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 26.Schröder K-H, Naumann L, Kroppenstedt R M, Reischl U. Mycobacterium hassiacum sp. nov., a new rapidly growing thermophilic mycobacterium. Int J Syst Bacteriol. 1997;47:86–91. doi: 10.1099/00207713-47-1-86. [DOI] [PubMed] [Google Scholar]
- 27.Stackebrandt E, Rainey F A, Ward-Rainey N L. Proposal for a new hierarchic classification system, Actinobacteria classis nov. Int J Syst Bacteriol. 1997;47:479–491. [Google Scholar]
- 28.Stanek J L, Roberts G D. Simplified approach to identification of aerobic actinomycetes by thin-layer chromatography. Appl Microbiol. 1974;28:226–231. doi: 10.1128/am.28.2.226-231.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Torkko P, Suomalainen S, Iivavanen E, Suutari M, Tortoli E, Paulin L, Katila M-L. Mycobacterium xenopi and related organisms isolated from stream waters in Finland and description of Mycobacterium botniense sp. nov. Int J Syst Evol Microbiol. 2000;50:283–289. doi: 10.1099/00207713-50-1-283. [DOI] [PubMed] [Google Scholar]
- 30.Wallace R J, Jr, O'Brien R, Glassroth J, Raleigh J, Dutt A. Diagnosis and treatment of disease caused by nontuberculous mycobacteria. Am Rev Respir Dis. 1990;142:940–953. doi: 10.1164/ajrccm/142.4.940. [DOI] [PubMed] [Google Scholar]