Abstract
Rapid identification of Escherichia coli O157:H7 is important for patient management and for prompt epidemiological investigations. We evaluated one in-house method and three commercially available kits for their ability to extract E. coli O157:H7 DNA directly from stool specimens for PCR. Of the 153 stool specimens tested, 107 were culture positive and 46 were culture negative. The sensitivities and specificities of the in-house enrichment method, IsoQuick kit, NucliSens kit, and QIAamp kit were comparable, as follows: 83 and 98%, 85 and 100%, 74 and 98%, and 86 and 100%, respectively. False-negative PCR results may be due to the presence of either inherent inhibitors or small numbers of organisms. The presence of large amounts of bacteria relative to the amount of the E. coli O157:H7 target may result in the lower sensitivities of the assays. All commercial kits were rapid and easy to use, although DNA extracted with the QIAamp kit did not require further dilution of the DNA template prior to PCR.
PCR has become a very rapid and reliable tool for the molecular biology-based diagnosis of a variety of infectious disease (9). PCR has been applied for the detection of microorganisms from microbial cultures and tissues and directly from clinical samples. Fecal specimens are among the most complex specimens for direct PCR testing due to the presence of inherent PCR inhibitors that are often coextracted along with bacterial DNA (2, 28, 33). Potential PCR inhibitors found in stool specimens include heme, bilirubins, bile salts, and complex carbohydrates (6, 19, 24, 28, 32). Few studies have evaluated or compared simple DNA extraction methods that would facilitate and improve the sensitivity of PCR detection of enteric pathogens from fecal specimens (3, 5, 18, 19, 21, 28, 32).
Conventional methods for the detection and identification of Escherichia coli O157:H7 include growth of non-sorbitol-fermenting E. coli colonies on sorbitol MacConkey agar culture (SMAC), followed by serological confirmation with O157- and H7-specific antisera (16). Culture alone can be insensitive, especially for the detection of small numbers of E. coli O157:H7, and it is unable to detect non-O157 verotoxin-producing E. coli (25, 30). Recent studies have shown that enzyme-linked immunosorbent assays may be more sensitive than culture and are able to detect verotoxins or specific serotype antigens (7, 25). Our previous work has documented the use of multiplex PCR for the detection of E. coli O157:H7 and verotoxin-producing, non-O157 E. coli strains that are not readily detected by conventional culture and has documented its ability to provide rapid same-day results (22, 23). In this study, three commercially available DNA extraction kits and one in-house method were evaluated for their abilities to yield amplifiable DNA for the PCR detection of E. coli O157:H7 directly from clinical stool specimens in comparison to the abilities of standard culture methods to detect E. coli O157:H7. Each method was also evaluated for its ease of use, cost per test, and overall sensitivity and specificity. A more rapid, simple, and sensitive technique for direct detection of E. coli O157:H7 from stool specimens would greatly expedite the laboratory detection of this important enteric pathogen.
(This paper was presented in part at the 39th Interscience Conference on Antimicrobial Agents and Chemotherapy, 26 to 29 September 1999, San Francisco, Calif. [J. L. Holland, L. Louie, A. E. Simor, and M. Louie, Abstr. 39th Intersci. Conf. Antimicrob. Agents Chemother., abstr. 1566, p. 224, 1999].)
MATERIALS AND METHODS
Bacterial strains and clinical stool specimens.
Strain CL8, a well-characterized E. coli O157:H7 strain, was used for the seeding experiments and as the PCR-positive control. It is known to harbor the VT1, VT2, and the eaeA genes (22). A non-O157:H7 E. coli strain, ATCC 25922, was used as the PCR-negative control.
Stool specimens (n = 107) which grew non-sorbitol-fermenting E. coli colonies on SMAC and which were suspected to be E. coli O157:H7 positive were collected from patient specimens submitted for workup for gastroenteritis. Stool specimens (n = 46) known to be culture negative for enteric pathogens including E. coli O157:H7 were also collected for testing. All samples, from two private community-based laboratories and a tertiary-care hospital-based laboratory, were received anonymously with no identifying patient demographic information and were inspected upon receipt for consistency and the presence of gross blood. Duplicate samples from the same patient on the same day were excluded. Each stool sample was gently mixed, aliquoted into 500-μl volumes, and stored at −70°C prior to testing. Of the stools tested, 36% had been frozen for up to 2 years, 49% had been frozen for up to 1 year, and 15% had been frozen for up to 3 months or were used immediately. A different aliquot of each frozen stool specimen was used for each DNA extraction method.
Sensitivity testing.
To estimate the sensitivity of each DNA extraction method, seeding experiments were performed in triplicate with three different liquid stool samples that were culture negative for E. coli O157:H7. Culture-negative, liquid stool specimens were seeded with dilutions of E. coli O157:H7 strain CL8 prepared in phosphate-buffered saline, ranging from 101 to 108 CFU/ml. A 500-μl volume of each dilution of strain CL8 was added to 100 μl of stool, and the components were mixed by vortexing. The seeded stool was centrifuged at 16,000 × g for 5 min, and the supernatant was removed. DNA was extracted from the pellet by the in-house enrichment method and the three commercial methods as described below. The dilutions of CL8 were inoculated onto blood agar plates, and the plates were incubated overnight to verify the bacterial inoculum used for seeding of the stools.
E. coli O157:H7 identification and confirmation.
Approximately 50 μl from both clinical and seeded stool specimens was inoculated and streaked onto SMAC plates for semiquantitative analysis. The growth of non-sorbitol- and sorbitol-fermenting colonies was quantified as ±, 1+, 2+, or 3+, depending on the degree of growth by quadrant (±, growth in primary inoculum; 1+, growth in first quadrant, etc.). Several isolated, non-sorbitol-fermenting colonies were picked and subcultured onto 5% sheep blood agar plates for overnight incubation at 37°C. Organisms which were identified as E. coli with the Vitek GNI card (bioMérieux Vitek Inc., Hazelwood, Mo.) were then tested with O157 and H7 antisera (Difco, Detroit, Mich.) according to the manufacturer's instructions. For PCR-positive but culture-negative specimens, several sorbitol-fermenting colonies were similarly selected and tested serologically for the presence of O157 and H7 antigens.
DNA extraction.
The commercial kits included the IsoQuick nucleic acid extraction kit (Orca Research, Bothell, Wash.), the NucliSens kit (Organon Teknika, Boxtel, The Netherlands), and the QIAamp DNA mini kit (Qiagen, Hilden, Germany). A 500-μl aliquot of the stored stool specimen was thawed at room temperature, and all DNA extractions were performed according to the manufacturers' instructions, with minor adjustments as described below. Subsequent PCR amplification with the DNA templates was performed blindly to minimize bias from culture results.
In-house enrichment method.
A stool specimen (100 μl) was inoculated into 2 ml of tryptic soy broth and was incubated by shaking at 250 rpm and 37°C for 4 h. A volume of 1.8 ml of the mixture was transferred to a clean tube, and the tube was centrifuged at 14,000 × g for 5 min. The resultant pellet was resuspended in a volume of 150 μl of Triton X-100 lysis buffer (100 mM NaCl, 10 mM Tris-HCl [pH 8.3], 1 mM EDTA [pH 9], 1% Triton X-100 [Sigma Chemical Co., St. Louis, Mo.]) (27). The sample was then boiled for 10 min, cooled, and centrifuged at 16,000 × g for 1 min. A 50-μl aliquot of the supernatant was removed and reserved as the PCR template.
IsoQuick nucleic acid extraction kit.
A stool specimen (100 μl) was centrifuged at 16,000 × g for 5 min, and the supernatant was removed. Reagent buffer was added to the stool pellet to a volume of 100 μl, and the pellet was resuspended by vortexing and pipetting. The specimen was lysed for 20 min, followed by DNA separation and alcohol precipitation. The final DNA pellet was dried and resuspended in 50 μl of sterile, distilled H2O for use as the PCR template.
NucliSens kit.
A stool specimen (100 μl) was added to 900 μl of lysis buffer and the components were mixed by vortexing. The sample was then allowed to lyse for 1.5 h, with inversion of the microcentrifuge tube every 0.5 h, and was centrifuged at 300 × g for 1 min to separate unlysed and coarse material from the rest of the specimen. Free silica was then added to the sample, and the bound DNA was washed. A 50-μl volume of elution buffer was added, and after centrifugation, a 5-μl aliquot was removed and reserved as the PCR template.
QIAamp DNA mini kit: tissue protocol.
A stool sample (100 μl) was lysed with buffer ATL (Qiagen) and proteinase K for 2 h at 55°C and then with buffer AL for 10 min at 70°C. The sample was then centrifuged and anhydrous ethanol was added to the supernatant. The sample mixture was then passed through the QIAamp kit column, followed by two washes with buffers AW1 and AW2 (Qiagen). The DNA was eluted in a volume of 200 μl of elution buffer, which was passed through the same column twice.
PCR amplification.
The primer sequences used for PCR are shown in Table 1. The VT1 primers amplify DNA from VT1-producing E. coli strains, while the VT2 primers amplify DNA from VT2- and VT2 variant-producing E. coli strains (11). The O157 serotype-specific eaeA primers are based on the unique 3′ end of the E. coli O157 eaeA gene sequence (22). Multiplex PCRs were performed in a 25-μl final reaction volume, which was as follows: primers VT1, VT2, and eaeA (Life Technologies, Gibco BRL, Burlington, Ontario, Canada) at concentrations of 0.2, 0.2, and 0.8 μM, respectively; each deoxynucleoside triphosphate at a concentration of 200 μM; 1× PCR buffer (10 mM Tris-HCl [pH 8.3], 50 mM KCl, 0.001% gelatin); 1.5 mM MgCl2; 2.5 U of Taq polymerase (Roche Molecular Systems, Mississauga, Ontario, Canada); and 1 μl of template DNA. Cycling conditions in a GeneAmp 9600 Thermocycler (Perkin-Elmer Cetus, Norwalk, Conn.) were as follows: 94°C for 2 min; 30 cycles of 94°C for 30 s, 56°C for 30 s, and 72°C for 30 s; and extension at 72°C for 10 min. PCR amplicons were run on a 1% agarose gel, stained with ethidium bromide, and visualized under UV illumination.
TABLE 1.
Primers used in multiplex PCR for amplification of VT1, VT2, and eaeA gene sequences and in PCR inhibition studies
| Primer | Oligonucleotide sequence (5′→3′) | Size of amplicon product (bp) | Reference |
|---|---|---|---|
| VT1 (SLT1)-F | ACA CTG GAT GAT CTC AGT GG | 614 | 11 |
| VT1 (SLT1)-R | CTG AAT CCC CCT CCA TTA TG | ||
| VT2 (SLT2)-F | CCA TGA CAA CGG ACA GCA GTT | 779 | 11 |
| VT2 (SLT2)-R | CCT GTC AAC TGA GCA CTT TG | ||
| eaeAO157:H7-F | AAG CGA CTG AGG TCA CT | 450 | 22 |
| eaeAO157:H7-R | ACG CTG CTC ACT AGA TGT | ||
| 16S-F | AGA GTT TGA TCA TGG CTC AG | 798 | 8 |
| 16S-R | GGA CTA CCA GGG TAT CTA AT |
Inhibition studies.
E. coli O157:H7 culture-positive and PCR-negative specimens were analyzed for stool-derived PCR inhibitors by a PCR assay which amplified a conserved 16S bacterial RNA gene (rDNA) sequence. Primers 16S-F and 16S-R (Table 1) were designed on the basis of the 16S rDNA sequence of E. coli (Genbank accession no. J01859) (8). PCR was performed by using each primer at a concentration of 0.2 μM and the cycling conditions described above. DNA templates that yielded a strong positive 16S rDNA gene product but that were multiplex PCR negative for E. coli O157:H7 were assumed to be negative for reasons other than inhibition and were not tested further. DNA templates that yielded negative 16S rDNA PCRs were diluted 1:10 with sterile distilled H2O, and PCRs for E. coli O157:H7 and 16S rDNA were repeated.
PCR inhibition by normal enteric flora.
To see if the overgrowth of enteric flora interfered with the yield of amplifiable E. coli O157:H7 DNA, the sensitivity of each method was determined for specimens grouped by the relative growth of sorbitol-fermenting colonies compared to that of non-sorbitol-fermenting colonies as follows: growth of sorbitol-fermenting colonies > growth of non-sorbitol-fermenting colonies, growth of sorbitol-fermenting colonies < growth of non-sorbitol-fermenting colonies, growth of sorbitol-fermenting colonies = growth of non-sorbitol-fermenting colonies, and presence of non-sorbitol-fermenting colonies only. Since seeding studies showed that successful PCR detection required at least growth of 1 + non-sorbitol-fermenting colonies (∼103 to 104 /g of stool) on SMAC, those specimens that had only ± growth of non-sorbitol-fermenting colonies on SMAC were removed from this analysis.
RESULTS
Seeding experiments.
By multiplex PCR, the limit of detection of seeded E. coli O157:H7 strain CL8 was 104 CFU/g of stool for the in-house enrichment, IsoQuick kit, and QIAamp kit and 105 CFU/g of stool for the NucliSens kit (Fig. 1). None of the PCRs with seeded specimens were inhibited, as each sample amplified the 16S rDNA product when the 16S rDNA-specific primers were used. Stool samples seeded with CL8 at concentrations of 101 to 102, 103 to 104, 105 to 106, and 107 to 108 CFU/g of stool correlated with the ±, 1+, 2+, and 3+ growth of non-sorbitol-fermenting colonies on SMAC, respectively.
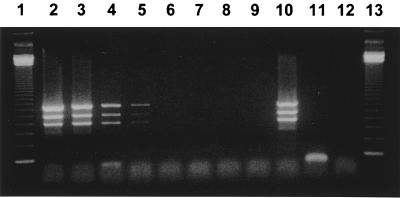
FIG. 1.
Representative gel showing multiplex PCR amplification of DNA extracted with the QIAamp kit from dilutions of E. coli O157:H7 strain CL8 seeded into culture-negative stool specimens. Lanes 1 and 13, 123-bp DNA size marker (Life Technologies, Gibco BRL); lanes 2 to 9, CL8-seeded stool at serial, 10-fold, decreasing concentrations of 107 to 100 CFU/g of stool; lane 10, positive control strain E. coli O157:H7 strain CL8; lane 11, negative control strain E. coli ATCC 25922; lane 11, PCR assay reagent control (no template DNA).
Clinical specimens.
All 153 stool specimens were received in liquid form, of which 10 specimens were visibly stained with blood. Of the 153 stool specimens tested, 104 were E. coli O157:H7 culture positive, 3 were E. coli O157:H− culture positive, and 46 were culture negative, as determined by O157 and H7 serology. Sorbitol-fermenting colonies picked from culture-negative but PCR-positive specimens were negative by O157 and H7 serology.
PCR detection of E. coli O157:H17.
Multiplex PCRs were considered positive for E. coli O157:H7 when either one or both of the verotoxin genes was amplified along with the eaeA gene. The detection of only verotoxin genes might indicate the presence of other verotoxin-producing E. coli serotypes, which would otherwise not be detected by conventional culture. The same combination of amplified genes was displayed when two or more DNA extraction methods yielded a positive multiplex PCR result. Of the 107 culture-positive specimens, 83% (89 of 107) were PCR positive for the VT1, VT2, and eaeA genes, 8% (9 of 107) were PCR positive for the VT2 and eaeA genes, and 3% (3 of 107) were PCR positive for the VT1 and eaeA genes only. Of the six remaining culture-positive specimens, all except one were PCR negative; the one exception was PCR positive only for the VT1 and VT2 genes. All except one of these PCR-negative specimens had only ± growth of non-sorbitol-fermenting colonies; the one exception had 2+ growth of non-sorbitol-fermenting colonies.
Of the culture-positive specimens, 67 (63%) specimens were PCR positive and 6 (5%) were PCR negative by all four DNA extraction methods. Of the remaining 34 (32%) culture-positive specimens, not all the extraction methods yielded positive PCR results for the same specimens. For six specimens, PCR results were positive by only one method (four were positive by the in-house enrichment method and one each was positive with the IsoQuick and QIAamp kits). PCR was positive by three methods for 21 specimens, and PCR was positive by two methods for 7 specimens (Table 2).
TABLE 2.
Culture-positive stool specimens with variability in PCR results by DNA extraction method
| No. of different stool samples | Multiplex PCR result
|
|||
|---|---|---|---|---|
| In-house enrichment | IsoQuick kit | QIAamp kit | NucliSens kit | |
| 1 | − | − | + | + |
| 5 | − | + | + | − |
| 2 | + | + | − | − |
| 10 | + | + | + | − |
| 2 | + | + | − | + |
| 4 | + | − | + | + |
| 4 | − | + | + | + |
Of the 46 culture-negative specimens, one specimen with growth of 2+ sorbitol-fermenting and 3+ non-sorbitol-fermenting colonies was PCR positive for VT2 and eaeA by use of DNA extracted by the in-house enrichment method and with the NucliSens kit. Six other specimens with ± growth of sorbitol-fermenting colonies were positive for verotoxin but not eaeA genes (VT1 in three specimens and VT1 and VT2 in three specimens).
Table 3 shows the calculated sensitivities and specificities of PCR with DNA extracted by each of the study methods before and after dilution of the template DNA. All culture-positive, PCR-negative specimens yielded a 16S rDNA product by PCR either with undiluted DNA template (neat) or with template DNA diluted 1:10. The sensitivity for the QIAamp kit was not affected by further dilution of the DNA template. The sensitivity for the IsoOuick kit showed the most improvement when the template DNA of false-negative specimens was diluted 1:10 and the PCR was repeated. For the IsoOuick kit, 15% of PCR-positive specimens required dilution of the DNA template, while 8 and 4% of the DNA templates needed dilution before PCR for the NucliSens kit and the in-house enrichment methods, respectively. Overall, after dilution of the DNA templates, the sensitivities of PCR detection of E. coli O157:H7 following DNA extraction by the in-house enrichment method and with the IsoQuick and QIAamp kits were comparable (83 to 86%). The specificities were 98 to 100% for the PCR-based assays when culture was used as the “gold standard.”
TABLE 3.
Comparison of E. coli O157:H7 multiplex PCR results with standard culture and serology results
| Multiplex PCR, PCR result, and dilution (n = 153) | No. of specimens
|
||||
|---|---|---|---|---|---|
| Culture positive | Culture negative | Sensitivity (%)a
|
Specificity (%) | ||
| Neat | 1:10 | ||||
| In-house enrichment | |||||
| Positive | |||||
| Neat | 85 | 1 | 79 | 83 | 98 |
| 1:10 | 4 | ||||
| Negative | 18 | 45 | |||
| IsoQuick kit | |||||
| Positive | |||||
| Neat | 77 | 0 | 72 | 85 | 100 |
| 1:10 | 14 | ||||
| Negative | 16 | 46 | |||
| QIAamp kit | |||||
| Positive | |||||
| Neat | 91 | 0 | 85 | 86 | 100 |
| 1:10 | 1 | ||||
| Negative | 15 | 46 | |||
| NucliSens kit | |||||
| Positive | |||||
| Neat | 73 | 1 | 68 | 74 | 98 |
| 1:10 | 6 | ||||
| Negative | 28 | 45 | |||
Neat, undiluted template; 1:10, template diluted 1 in 10.
Of the 10 blood-stained, culture-positive stool samples, 7 were PCR positive by all four methods without the need to dilute the DNA template. All seven stool specimens had 3+ growth of both sorbitol-fermenting and non-sorbitol-fermenting colonies by culture. Three blood-stained, culture-positive specimens were PCR negative for E. coli O157:H7. For one blood-stained stool sample with 3+ growth of non-sorbitol-fermenting colonies and ± growth of sorbitol-fermenting colonies, the IsoQuick and QIAamp kits were able to detect all three genes, the VT1, VT2, and eaeA genes. The in-house enrichment method and the NucliSens kit detected only the verotoxin genes and not the eaeA genes for the same sample. We have found that, in general, the multiplex PCR assay described here yielded more intense bands for the VT2 gene, followed by respectively less intense bands for the VT1 gene and the eaeA gene (data not shown). For the third blood-stained stool specimen with 1+ growth of non-sorbitol-fermenting colonies and ± growth of sorbitol-fermenting colonies on SMAC, multiplex PCR for E. coli O157:H7 was positive only by the in-house enrichment extraction method.
PCR inhibition by normal enteric flora.
For each of the DNA extraction methods, the highest sensitivities were obtained when non-sorbitol-fermenting colonies were present in pure culture (Table 4). Dilution of the template DNA was not necessary for the QIAamp kit, regardless of the relative growth of sorbitol-fermenting and non-sorbitol-fermenting colonies. However, for all methods, dilution of the template was not required when non-sorbitol-fermenting colonies were present alone. The sensitivity was the lowest when the number of sorbitol-fermenting colonies was greater than the number of non-sorbitol-fermenting colonies by each of the methods. When the growth of the sorbitol-fermenting colonies was equal to the growth of the non-sorbitol-fermenting colonies, dilution of the template DNA was required only for the IsoQuick and the NucliSens kits.
TABLE 4.
Influence of competing enteric flora on sensitivity of PCR detection of E. coli O157:H7
| Relative growth of SF and NSF on SMACb | Sensitivitya (%) by method of DNA extraction
|
|||||||
|---|---|---|---|---|---|---|---|---|
| In-house method
|
IsoQuick kit
|
QIAamp kit
|
NucliSens kit
|
|||||
| Neat | 1:10 | Neat | 1:10 | Neat | 1:10 | Neat | 1:10 | |
| SF > NSF (nc = 4) | 25 | 50 | 50 | 75 | 25 | 25 | 25 | 25 |
| SF < NSF (n = 21) | 71 | 76 | 67 | 90 | 86 | 86 | 62 | 67 |
| SF = NSF (n = 62) | 90 | 94 | 76 | 89 | 90 | 92 | 71 | 79 |
| NSF (n = 14) | 93 | 93 | 93 | 93 | 93 | 93 | 93 | 100 |
Sensitivity was determined with undiluted (neat) and diluted (1:10) DNA template.
Non-sorbitol-fermenting colonies (NSF) were all identified as E. coli O157:H7 or E. coli O157:H−. SF, sorbitol-fermenting colonies.
n, number of specimens.
All DNA extraction methods were evaluated on the basis of their sample processing time and cost, as shown in Table 5. The tests with all three commercial kits were performed fairly quickly, and they were simple to perform, although further dilution of the DNA template was not required for the QIAamp kit. The cost per test differed for each extraction kit. The in-house enrichment method took longer to complete due to the enrichment growth step, but it had the shortest hands-on time and the lowest associated cost.
TABLE 5.
Overall comparison of the DNA extraction kits with respect to time and cost
| Test | Time to completion (h) | Hands-on time (min) | List price/specimena |
|---|---|---|---|
| IsoQuick kit | 1.5 | 40 | 2.00 |
| NucliSens kit | 1.5 | 40 | 17.45 |
| QIAamp kit | 1 | 30 | 2.30 |
| In-house enrichment | 4.5 | 20 | 0.50 |
Does not include the cost of the PCR; prices are in Canadian dollars.
DISCUSSION
In this study, several commercial methods and one in-house method were evaluated systematically for their yield of amplifiable DNA. The IsoQuick kit uses the chaotropic properties of guanidine thiocyanate for cell lysis and for inactivation of cell nucleases. DNA is then extracted with a proprietary, noncorrosive reagent and alcohol precipitation. The QIAamp kit uses proteinase K lysis followed by a silica column-based isolation of DNA. The NucliSens kit uses guanidine thiocyanate–Triton X-100 lysis followed by free silica-based isolation of DNA. The in-house enrichment method includes a 4-h tryptic soy broth enrichment of stool, followed by heat lysis with Triton X-100 lysis buffer.
The nonuniformity of stool samples in terms of physical matter, target organisms, and associated background fecal flora makes extraction of DNA from fecal specimens highly variable from specimen to specimen in terms of both the yield and the purity of the DNA (13, 19, 28, 33). False-negative PCR results may be due to the presence of a small number of target organisms in the volume of stool sampled and the decreased stability of cells with storage (5). In this study, efforts were taken to thoroughly mix each aliquot of stool for each DNA extraction method. Studies in our laboratory have shown no difference in PCR results when stool specimens were used fresh or frozen at −70°C (data not shown). The limit of detection for each of the methods studied was influenced to some degree by the relative growth of normal fecal flora. Sensitivities were much higher when non-sorbitol-fermenting colonies were present alone or at a higher density than sorbitol-fermenting colonies. Coextraction of other bacterial DNA from fecal samples may interfere with the efficiency of the PCR assay. Similarly, Thomas et al. (31) found that the availability of verotoxin genes for primer annealing was restricted by the relatively large amount of nonspecific DNA derived from coliforms present in large amounts in the the original specimen.
The multiplex PCR detection limits of 104 to 105 E. coli O157:H7 cells/g of stool, determined by using DNA extracted by these methods, were comparable to those reported by others for enteric pathogens (4, 17, 18, 20, 28). The in-house enrichment, the IsoQuick kit, and the QIAamp kit all shared comparable sensitivities, which were in the range of 83 to 86%. The NucliSens kit had the lowest sensitivity. Subsequent dilution of template DNA was required for 4, 15, 1, and 8% of specimens tested by the in-house enrichment method and with the IsoQuick kit, the QIAamp kit, and the NucliSens kit, respectively. For the IsoQuick kit, the overall sensitivity increased by 13% after dilutions were made. The QIAamp kit did not require dilution of DNA templates to control for inhibition of the PCR. This is in contrast to Monteiro et al. (24), who found that the selective binding characteristics of the QIAamp kit only partially removed PCR inhibitors. In their study, PCR inhibitors persisted in 63% (10 of 16) of samples and dilutions of at least 1:2 and 1:10 were necessary to remove their effects for the detection of Helicobacter pylori. In our study, the incorporation of an additional wash buffer as recommended by Qiagen might have aided in the removal of inhibitors. Other studies have also observed that the direct PCR detection of organisms from fecal specimens required greater dilution of the original DNA template (2, 14, 28).
Improvement of the limit of detection by each of these methods might still be enhanced by a number of other variables. PCR sensitivities may have been improved for the commercial kits if broth enrichment for E. coli O157:H7 in stools was performed prior to DNA extraction. Incorporation of selective and inhibitory agents for E. coli O157:H7 and other fecal flora, respectively, might further increase the yield of the desired target DNA upon extraction. Studies have also shown that selective enrichment by culture of the target microorganism, with or without immunomagnetic separation, increases the overall yield of desirable DNA and consequently improves the recovery by PCR (18, 29, 32, 34). Another consideration is the addition of carrier nucleic acid, such as yeast tRNA, during the extraction process to increase the recovery of small amounts of DNA (10). Other studies have shown that the limit of detection of amplified PCR products was improved 10- to 100-fold when Southern blot hybridization with labeled probes was used compared to the limit of detection by visualization of amplified products by ethidium bromide staining of agarose gels (17, 20). In another study, Shetab et al. (26) used a phenol-chloroform extraction method followed by PCR and detected 102 to 103 Bacteroides fragilis cells/g of stool, using a DNA template volume of 10 μl instead of 1 μl. Similarly, Caeiro et al. (3) detected 102 to 103 enterotoxigenic E. coli cells/g of stool when 10 μl of DNA template was used in a 100-μl PCR mixture. One limitation of our study was that the sensitivity of each method was not evaluated with a larger aliquot of the diluted template and an increased volume of the PCR mixture.
The presence of blood did not appear to inhibit the PCR according to the results of our study. Blood has been implicated by others as a major inhibitor of PCR amplification; however, certain Taq polymerases can amplify DNA in the presence of blood should this be a problem (1). Of the 10 blood-stained stool samples that we tested, PCR was able to amplify either E. coli O157:H7 targets or the 16S rDNA product, suggesting that the DNA extraction methods were sufficient for removal of the inhibitory heme elements. Three blood-stained, culture-positive, but PCR-negative specimens yielded 16S rDNA when the neat DNA templates were used. For one specimen, negative results by all four methods of DNA extraction may be attributed to small numbers of E. coli O157:H7 since culture on SMAC showed a ± growth of non-sorbitol-fermenting colonies. For another blood-stained, PCR-positive sample with only 1+ growth of non-sorbitol-fermenting colonies, prior enrichment by the in-house enrichment method may have increased the absolute numbers of E. coli O157:H7, giving a higher yield of target DNA.
As in other studies, DNA extracted from specimens culture positive and PCR negative for E. coli O157:H7 were amplified with bacterial 16S rDNA primers to ensure the presence of amplifiable DNA (12, 13). Our E. coli O157:H7 multiplex PCR assay for the VT1, VT2, and eaeA genes was not multiplexed with the 16S rDNA primers due to concern for decreased amplification signals for the verotoxin and eaeA gene products with multiple primers. Ibrahim et al. (15) found that coamplification of a 16S rDNA fragment was less efficient in the presence of the yst target for the detection of Yersinia, which was thought to be attributed to the differences in the melting temperatures of the primer pairs used. Although the occurrence of false-negative results for specimens due to inherent fecal inhibitors was expected to be uncommon, an improvement to our study would have been to include a standardized internal control in the multiplex PCR. The efficiency of the 16S rDNA PCR was greater than that of the E. coli O157:H7 PCR due to the expansive rDNA target provided by the coextracted, background microflora. As a result, inhibition might have interfered with amplification of the VT1, VT2, and eaeA genes but still allowed a successful 16S rDNA amplification.
Attempts were not made to determine what specific inhibitory components in the stool specimens contributed to false-negative PCRs overall. For a number of specimens dilution of the final DNA template was still required to reduce the effects of the inherent inhibitors. Continued work on the identification of these inhibitory substances and the development of methods for their removal are important. More work is also needed to optimize the extraction process to increase the yield of desired bacterial DNA and to reduce the presence of competing DNA from coexisting microflora.
Other considerations in choosing a fecal processing method for PCR include ease of use, cost, and time to completion. As shown in Table 5, the in-house enrichment method could be performed easily and economically with minimal hands-on time, although the 4-h broth enrichment step increased the total time to completion. Without this step, seeding studies yielded a sensitivity limit of only 107 CFU/g of stool (data not shown). The test with the QIAamp kit was very rapid, and its silica column was simple to use for washing and elution of bound DNA. The test with the IsoQuick kit was slightly more involved, requiring a DNA precipitation step and about 40 min of hands-on time. Given that 15% of PCR-positive samples required dilution when the IsoQuick kit was used, a resuspension volume greater than the recommended 100 μl would benefit the sensitivity of this method. Optimization of the elution or resuspension volume to give the greatest PCR sensitivity would eliminate the need for subsequent dilution of the template and repeat PCR assays. The poor sensitivity of the NucliSens kit might be the result of incomplete lysis of stool components, which interfered with the subsequent binding and elution of bacterial DNA. Lack of sufficient sample lysis was possibly due to the absence of an enzymatic digestion step in the protocol. Also, in comparison to the QIAamp kit, the washing steps required with the NucliSens kit were lengthy due to the frequent resuspension of free silica.
In summary, although all the commercial kits were fairly easy to use, the QIAamp kit had the highest sensitivity and required the least manipulation of template DNA prior to PCR. The in-house enrichment protocol also performed well and at a much lower cost per test. Development of a rapid procedure for extraction of DNA from stool specimens that reduces hands-on time is needed to make PCR detection of enteropathogens more practical for routine laboratory use. The ideal DNA extraction protocol would provide the highest yield of DNA with minimal coextraction of potential inhibitors, coupled with a simple and rapid procedure.
ACKNOWLEDGMENTS
This work was supported by the Physicians' Services Incorporated Foundation (grant 98-25).
We thank MDS Laboratories (Toronto, Ontario, Canada) and Gamma-Dynacare Medical Laboratories (Toronto, Ontario, Canada) for kindly providing clinical specimens for the study.
REFERENCES
- 1.Al-soud W A, Rådström P. Capacity of nine thermostable DNA polymerases to mediate DNA amplification in the presence of PCR-inhibiting samples. Appl Environ Microbiol. 1998;64:3748–3753. doi: 10.1128/aem.64.10.3748-3753.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brian M J, Frosolono M, Murray B E, Miranda A, Lopez E L, Gomez H F, Cleary T G. Polymerase chain reaction for diagnosis of enterohemorrhagic Escherichia coli infection and hemolytic-uremic syndrome. J Clin Microbiol. 1992;30:1801–1806. doi: 10.1128/jcm.30.7.1801-1806.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caeiro J P, Estrada-Garcia M T, Jiang Z, Mathewson J J, Adachi J A, Steffen R, DuPont H L. Improved detection of enterotoxigenic Escherichia coli among patients with travelers' diarrhea, by the use of the polymerase chain reaction technique. J Infect Dis. 1999;180:2053–2055. doi: 10.1086/315121. [DOI] [PubMed] [Google Scholar]
- 4.Cavé H, Mariani P, Grandchamp B, Elion J, Denamur E. Reliability of PCR directly from stool samples: usefulness of an internal standard. BioTechniques. 1994;16:809–810. [PubMed] [Google Scholar]
- 5.Da Silva A J, Bornay-Llinares F J, Moura I N S, Slemenda S B, Tuttle J L, Pieniazek N J. Fast and reliable extraction of protozoan parasite DNA from fecal specimens. Mol Diagn. 1999;4:57–64. doi: 10.1016/s1084-8592(99)80050-2. [DOI] [PubMed] [Google Scholar]
- 6.Demeke T, Adams R P. The effects of plant polysaccharides and buffer additives on PCR. BioTechniques. 1992;12:332–334. [PubMed] [Google Scholar]
- 7.Dylla B L, Vetter E A, Hughes J G, Cockerill F R., III Evaluation of an immunoassay for direct detection of Escherichia coli O157 in stool specimens. J Clin Microbiol. 1995;33:222–224. doi: 10.1128/jcm.33.1.222-224.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ehresmann C, Stiegler P, Fellner P, Ebel J P. The determination of the primary structure of the 16S ribosomal RNA of Escherichia coli. 2. Nucleotide sequences of products from partial enzymatic hydrolysis. Biochimie. 1972;54:901–967. doi: 10.1016/s0300-9084(72)80007-5. [DOI] [PubMed] [Google Scholar]
- 9.Fredricks D N, Relman D A. Application of polymerase chain reaction of the diagnosis of infectious diseases. Clin Infect Dis. 1999;29:475–488. doi: 10.1086/598618. [DOI] [PubMed] [Google Scholar]
- 10.Gallagher M L, Burke W F, Jr, Orzech K. Carrier RNA enhancement of recovery of DNA from dilute solutions. Biochem Biophys Res Commun. 1987;144:271–276. doi: 10.1016/s0006-291x(87)80506-5. [DOI] [PubMed] [Google Scholar]
- 11.Gannon V P J, King R K, Kim J Y, Goldsteyn Thomas E J. Rapid and sensitive method for detection of Shiga-like toxin-producing Escherichia coli in ground beef using the polymerase chain reaction. Appl Environ Microbiol. 1992;58:3809–3815. doi: 10.1128/aem.58.12.3809-3815.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gramley W A, Asghar A, Frierson H F, Jr, Powell S M. Detection of Helicobacter pylori DNA in fecal samples from infected individuals. J Clin Microbiol. 1999;37:2236–2240. doi: 10.1128/jcm.37.7.2236-2240.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gumerlock P H, Tang Y J, Weiss J B, Silva J., Jr Specific detection of toxigenic strains of Clostridium difficile in stool specimens. J Clin Microbiol. 1993;31:507–511. doi: 10.1128/jcm.31.3.507-511.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hornes E, Wasteson Y, Olsvik Ø. Detection of Escherichia coli heat-stable enterotoxin genes in pig stool specimens by an immobilized, colorimetric, nested polymerase chain reaction. J Clin Microbiol. 1991;29:2375–2379. doi: 10.1128/jcm.29.11.2375-2379.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ibrahim A, Liesack W, Stackebrandt E. Polymerase chain reaction-gene probe detection system specific for pathogenic strains of Yersinia enterocolitica. J Clin Microbiol. 1992;30:1942–1947. doi: 10.1128/jcm.30.8.1942-1947.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karmali M A. Infection by verocytotoxin-producing Escherichia coli. Clin Microbiol Rev. 1989;2:15–38. doi: 10.1128/cmr.2.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kato N, Ou C, Kato H, Bartley S L, Luo C, Killgore G E, Ueno K. Detection of toxigenic Clostridium difficile in stool specimens by the polymerase chain reaction. J Infect Dis. 1993;167:455–458. doi: 10.1093/infdis/167.2.455. [DOI] [PubMed] [Google Scholar]
- 18.Kongmuang U, Luk J M C, Lindberg A A. Comparison of three stool-processing methods for detection of Salmonella serogroups B, C2, and D by PCR. J Clin Microbiol. 1994;32:3072–3074. doi: 10.1128/jcm.32.12.3072-3074.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lantz P G, Matsson M, Wadström T, Rådström P. Removal of PCR inhibitors from human faecal samples through the use of an aqueous two-phase system for sample preparation prior to PCR. J Microbiol Methods. 1997;28:159–167. [Google Scholar]
- 20.Lawson A J, Shafi M S, Pathak K, Stanley J. Detection of campylobacter in gastroenteritis: comparison of direct PCR assay of faecal samples with selective culture. Epidemiol Infect. 1998;121:547–553. doi: 10.1017/s0950268898001630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lou Q, Chong S K F, Fitzgerald J F, Siders J A, Allen S D, Lee C. Rapid and effective method for preparation of fecal specimens for PCR assays. J Clin Microbiol. 1997;35:281–283. doi: 10.1128/jcm.35.1.281-283.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Louie M, De Azavedo J, Clarke R, Borczyk A, Loir H, Richter M, Brunton J. Sequence heterogeneity of the eae gene and detection of verotoxin-producing Escherichia coli using serotype-specific primers. Epidemiol Infect. 1994;112:449–461. doi: 10.1017/s0950268800051153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Louie M, Read S, Simor A E, Holland J, Louie L, Ziebell K, Brunton J, Hii J. Application of multiplex PCR for detection of non-O157 verocytotoxin-producing Escherichia coli in bloody stools: identification of serogroups O26 and O111. J Clin Microbiol. 1998;36:3375–3377. doi: 10.1128/jcm.36.11.3375-3377.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Monteiro L, Bonnemaison D, Vekris A, Petry K G, Bonnet J, Vidal R, Cabrita J, Mégraud F. Complex polysaccharides as PCR inhibitors in feces: Helicobacter pylori model. J Clin Microbiol. 1997;35:995–998. doi: 10.1128/jcm.35.4.995-998.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Novicki T J, Daly J A, Mottice S L, Carroll K C. Comparison of sorbitol MacConkey agar and a two-step method which utilizes enzyme-linked immunosorbent assay toxin testing and a chromogenic agar to detect and isolate enterohemorrhagic Escherichia coli. J Clin Microbiol. 2000;38:547–551. doi: 10.1128/jcm.38.2.547-551.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shetab R, Cohen S H, Prindiville T, Tang Y J, Cantrell M, Rahmani D, Silva J., Jr Detection of Bacteroides fragilis enterotoxin gene by PCR. J Clin Microbiol. 1998;36:1729–1732. doi: 10.1128/jcm.36.6.1729-1732.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sritharan V, Barker R H. A simple method for diagnosing M. tuberculosis infection in clinical samples using PCR. Mol Cell Probes. 1991;5:385–395. doi: 10.1016/s0890-8508(06)80011-3. [DOI] [PubMed] [Google Scholar]
- 28.Stacy-Phipps S, Mecca J J, Weiss J B. Multiplex PCR assay and simple preparation method for stool specimens detect enterotoxigenic Escherichia coli DNA during course of infection. J Clin Microbiol. 1995;33:1054–1059. doi: 10.1128/jcm.33.5.1054-1059.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stewart D S, Tortorello M L, Gendel S M. Evaluation of DNA preparation techniques for detection of the SLT-1 gene of Escherichia coli O157:H7 in bovine faeces using the polymerase chain reaction. Lett Appl Microbiol. 1998;26:93–97. doi: 10.1046/j.1472-765x.1998.00279.x. [DOI] [PubMed] [Google Scholar]
- 30.Tarr P I. Escherichia coli O157:H7: clinical, diagnostic, and epidemiological aspects of human infection. Clin Infect Dis. 1995;20:1–10. doi: 10.1093/clinids/20.1.1. [DOI] [PubMed] [Google Scholar]
- 31.Thomas A, Jiggle B, Smith H R, Rowe B. The detection of vero cytotoxin-producing Escherichia coli and Shigella dysenteriae type 1 in faecal specimens using polymerase chain reaction gene amplification. Lett Appl Microbiol. 1994;19:406–409. doi: 10.1111/j.1472-765x.1994.tb00968.x. [DOI] [PubMed] [Google Scholar]
- 32.Widjojoatmodjo M N, Fluit A C, Torensma R, Verdonk G P H T, Verhoef J. The magnetic immuno polymerase chain reaction assay for direct detection of salmonellae in fecal specimens. J Clin Microbiol. 1992;30:3195–3199. doi: 10.1128/jcm.30.12.3195-3199.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wilde J, Eiden J, Yolken R. Removal of inhibitory substances from human fecal specimens for detection of group A rotaviruses by reverse transcriptase and polymerase chain reactions. J Clin Microbiol. 1990;28:1300–1307. doi: 10.1128/jcm.28.6.1300-1307.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang G, Weintraub A. Rapid and sensitive assay for the detection of enterotoxigenic Bacteroides fragilis. J Clin Microbiol. 1998;36:3545–3548. doi: 10.1128/jcm.36.12.3545-3548.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]