Abstract
A multiplex PCR assay was developed by using primers to the fiber gene that could differentiate human adenovirus (Ad) species A through F in a single amplification reaction. The assay correctly identified the species of all 49 recognized Ad prototype strains as well as 180 geographically and temporally diverse Ad field isolates. Ad serotype 6 (Ad6) (species C), Ad16 (species B), Ad31 (species A), and Ad40 and Ad41 (species F) could also be distinguished by amplicon size within each respective species. In comparison, a previously described Ad species-specific multiplex PCR assay that used primers to the Ad hexon gene gave equivocal results with several serotypes of species B, whereas our multiplex assay amplified all species B serotypes equally well. Our multiplex PCR assay will permit rapid, accurate, and cost-effective classification of Ad isolates.
Human adenoviruses (Ads) have been recovered from virtually every human organ system and have been associated with a wide spectrum of clinical disease (20). There are presently 49 recognized Ad serotypes defined by neutralization with type-specific animal antisera, and candidate serotypes 50 and 51 have recently been reported (11). Ad serotypes can be classified into six species (formerly subgenera [5]), designated species A through F. This classification scheme is generally consistent with subgroupings of Ads on the basis of their physiocochemical, biological, and genetic properties and is diagnostically important in that infections with different Ad species and serotypes are often associated with distinct clinical outcomes and epidemiologic features (for a review, see reference 46).
For type-specific identification of Ads, a provisional grouping is usually first obtained by determining the agglutination patterns of the isolate with human and animal erythrocytes (19, 44). Differentiation of Ads into smaller, more manageable groups makes subsequent typing more convenient and conserves antisera; in many cases, species identification is sufficient for clinical investigations. If type-specific identification is desired, hemagglutination inhibition can be performed with animal antisera prepared to each Ad serotype. However, hemagglutination patterns can sometimes be difficult to interpret and do not readily differentiate all Ad species, and erythrocytes with acceptable agglutination properties may be difficult to obtain from some animals (20). Type-specific neutralization can provide a definitive identification, but results may not be available for weeks, and unless the hemagglutination properties of the isolate are known or clinical or epidemiologic data that can narrow the choice of antisera to be tested are available, neutralization assays can be exceedingly work intensive. Ad serotype-specific rabbit immune sera (15) and monoclonal antibodies (53) have successfully been used to classify Ads, but these reagents are not widely available; and Ad genomic DNA restriction fragment analysis (1), although of proven value for molecular epidemiologic studies, poses the same problems of work intensity and required expertise as classic methods. Consequently, few diagnostic laboratories identify Ads beyond the genus Mastadenovirus.
The PCR assay has become a popular alternative for Ad detection, offering the potential for rapid, sensitive, and precise molecular identification. PCR assays with Ad group-specific primers individually (4, 8, 10, 12, 18, 24, 30, 32, 36, 37, 43) or combined in assays for multiple human pathogens (16, 25, 34) have proved to be comparable or better than classic cell culture or immunodiagnostic methods for detection of Ads in clinical samples. PCR has also been used for classification of Ads to the species and serotype levels on the basis of tests with primers to the pIX (3), VA RNA (29), and hexon (9, 24, 37, 40, 42) genes.
The diagnostic utility of the fiber gene, whose product mediates cellular attachment and hemagglutination and, together with the hexon, confers Ad serotype specificity (33, 52), as a suitable site for molecular discrimination has not been evaluated. Therefore, our objective in this study was to develop a nonnested multiplex PCR for one-step amplification and identification of human Ad species on the basis of tests with primers designed to the fiber gene. The efficacy of our assay was compared with that of a recently described multiplex PCR that used primers to the Ad hexon gene (42).
MATERIALS AND METHODS
Ads.
The Ads used in this study included (i) prototype strains Ad1 to Ad47 obtained from the American Type Culture Collection (Rockville, Md.) or the National Institute of Allergy and Infectious Diseases (National Institute of Health, Bethesda, Md.); (ii) prototype strains Ad48 (T85-884) and Ad49 (T87-677), kindly provided by David Schnurr, Berkeley, Calif.; and (iii) 180 temporally and geographically diverse field isolates obtained from Centers for Disease Control and Prevention (CDC) archives.
Ad identification.
All archived Ad strains were originally isolated in HEp-2, primary human embryonic kidney (HEK), and/or Graham-293 cells and were typed at CDC by hemagglutination inhibition (23) and/or neutralization (21) assays with Ad-specific reference horse antisera (22). PCR testing was performed directly with the original Ad isolate. Ads giving discrepant PCR results were passaged once in A-549 or Graham-293 cells in Eagle's minimal essential medium supplemented with 2% fetal bovine serum and antibiotics and were retested by neutralization assay and PCR. All presumptive Ad type 40 (Ad40) and Ad41 isolates were also tested by a commercial enzyme immunoassay with Ad40/41 serotype-specific monoclonal antibodies (Premier Adenoclone-Type 40/41; Meridian Diagnostics, Inc., Cincinnati, Ohio).
PCR assays. (i) Ad species-specific primers.
Candidate species-specific oligonucleotide primers complementary to the fiber gene of the respective Ad species were designed from alignments prepared by the program PILEUP (Genetics Computer Group) of 34 previously submitted fiber gene sequences available from GenBank (GenBank sequence accession numbers are given in parentheses): species A, Ad12 (x73487) and Ad31 (x76548); species B, Ad3 (m12411), Ad7 (z48954), Ad7 (m23696) Ad11 (l08232), Ad16 (u06106), Ad21 (u06107), Ad34 (u10271), and Ad35 (u32664, u10272); species C, Ad2 (j01917 and x00049), and Ad5 (m18369); species D, Ad8 (x72934 and x74660), Ad9 (x74659), Ad15 (x74658, x74669, and s75136), Ad17 (af108105 and y14241), Ad19 (u69130, u69131, and x94485), Ad28 (y14242), and Ad37 (u69132, x94484); species E, Ad4 (x76547 and l19194); and species F, Ad40 (l19443 and m28822), Ad41 (x16583), and Ad41 (m60327). Sequence data from a limited region of the fiber genes of 12 strains of Ad40 and Ad41 published by Kidd et al. (28) were also included in the analysis. The six species-specific primer pairs, corresponding to species A, B, C, D, E, and F, respectively, that gave the best results individually and in multiplex PCR assays are listed in Table 1.
TABLE 1.
Oligonucleotide primers for PCR amplification of adenovirus species
| Species | Primer | Polarity | Gene | Gene region | Position (nt no.)a | Sequence (5′→3′) | Amplicon size (bp)b |
|---|---|---|---|---|---|---|---|
| A to F | Ad1 | + | Hexon | 1834–1853 | TTCCCCATGGCICAYAACAC | 482 | |
| Ad2 | − | 2315–2296 | CCCTGGTAKCCRATRTTGTA | ||||
| A | AdA1 | + | Fiber | Tail | 29392–29414 | GCTGAAGAAMCWGAAGAAAATGA | 1444–1537 |
| AdA2 | − | Knob | 30928–30908 | CRTTTGGTCTAGGGTAAGCAC | |||
| B | AdB1 | + | Fiber | Tail | 4573–4595 | TSTACCCYTATGAAGATGAAAGC | 670–772 |
| AdB2 | − | Knob | 5242–5220 | GGATAAGCTGTAGTRCTKGGCAT | |||
| C | AdC1 | + | Fiber | Flank | 333–354 | TATTCAGCATCACCTCCTTTCC | 1988–2000 |
| AdC2 | − | Flank | 2320–2301 | AAGCTATGTGGTGGTGGGGC | |||
| D | AdD1 | + | Fiber | Flank | 5–25 | GATGTCAAATTCCTGGTCCAC | 1205–1221 |
| AdD2 | − | Flank | 1219–1198 | TACCCGTGCTGGTGTAAAAATC | |||
| E | AdE1 | + | Fiber | Tail | 48–67 | TCCCTACGATGCAGACAACG | 967 |
| AdE2 | − | Knob | 1014–994 | AGTGCCATCTATGCTATCTCC | |||
| F | AdF1 | + | Fiber | Shaft | 1734–1754 | ACTTAATGCTGACACGGGCAC | 541–586 |
| AdF2 | − | Shaft | 2274–2253 | TAATGTTTGTGTTACTCCGCTC |
Nucleotide (nt) numbering for Ad group-specific primers based on published hexon gene sequence of Ad3 (41); nucleotide numbering for Ad species-specific primers based on published fiber gene sequences of Ad12 (49), Ad7 (26), Ad5 (7), Ad8 (39), Ad4 (17), and Ad40 (27).
Predicted amplicon size range based on previously published nucleotide sequences of representative Ad serotypes for each species.
(ii) Ad group-specific primers.
Ad group-specific primers complementary to regions of the hexon gene conserved among all recognized human Ad serotypes were designed from alignments of 23 previously published hexon gene sequences (GenBank accession numbers are given in parentheses): species A, Ad12 (x73487) and Ad31 (x74661); species B, Ad3 (x76549), Ad7 (x76551, z48571, af065065, af053086, af053087, af053085, af065067, af065068, and af065066), and Ad16 (x74662); species C, Ad2 (j01917) and Ad5 (j01966 and x02997); species D, Ad48 (u20821); species E, Ad4 (x84646, af065062, af065063, and af065064); and species F, Ad40 (x51782) and Ad41 (x51783) (Table 1). All DNA extracts were tested with the Ad-specific group primers to confirm successful extraction of Ad DNA.
(iii) DNA extraction.
For Ad DNA extraction, 50 μl of culture lysate was added to a 1.5-ml microcentrifuge tube containing 150 μl of distilled H2O, 60 μl of 5× TNE buffer (50 mM Tris-HCl, 5 mM EDTA, 50 mM NaCl [pH 8.0]), 30 μl of 10% sodium dodecyl sulfate, and 10 μl of 10 mg of proteinase K (Boehringer Mannheim) per ml, and the tube was heated at 50°C for 60 min. The DNA was then extracted once with equal volumes of phenol, once with phenol-chloroform-isoamyl alcohol (25:24:1), and once with chloroform-isomyl alcohol (24:1).
(iv) DNA amplification and detection.
PCR was performed in 50-μl volumes containing 45 μl of reaction mixture (10 mM Tris-HCl [pH 8.3], 1.5 mM MgCl2, 50 mM KCl, 200 μM each deoxynucleoside triphosphate, 0.2 mM each primer, 1 U of Taq DNA polymerase [Boehringer Mannheim]) per target species and 5 μl of DNA extract. The amplification reaction was carried out in a GeneAmp PCR System 2400 thermal cycler (Applied Biosystems) with (i) preliminary denaturation for 5 min at 94°C, followed by (ii) 30 cycles of denaturation at 94°C for 1 min, annealing at 54°C for 45 s, and primer extension at 72°C for 2 min and (iii) a final product extension at 72°C for 5 min. Ten microliters of each reaction product was then visualized by ethidium bromide staining and UV transillumination following electrophoretic separation (1.5 h, 120V) on 1% agarose gels.
Sensitivity and specificity studies.
The sensitivity of the multiplex PCR assay was evaluated with representative serotypes for each of the six Ad species (Ad12, Ad7, Ad5, Ad8, Ad4, Ad40). Ad genomic DNA was purified by the method of Elsom and Herzog (14) and was quantified spectrophotometrically at an optical density at 260 nm. Serial 10-fold dilutions of purified Ad DNA were amplified in the multiplex PCR assay, and the detection limits were determined for each serotype. The minimum number of Ad genome copies amplified was then estimated from the formula of Uchio et al. (51). To exclude the possibility that our Ad primers might cross-react with other DNA viruses, DNA extracts of herpes simplex virus types 1 and 2, Epstein-Barr virus, cytomegalovirus, varicella-zoster virus, simian vacuolating virus 40, and human parvovirus B19 were also tested.
Comparison of multiplex PCR assays.
As a means of validating our assay and comparing our primers to primers that target other regions of the Ad genome, selected Ads were simultaneously tested by a previously described multiplex PCR assay that used species-specific primers to the Ad hexon gene (42).
RESULTS
Development and optimization of multiplex PCR assay.
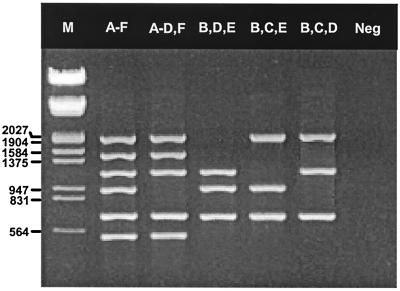
The primers designed in this study were evaluated in multiple combinations under various amplification conditions to identify primer pairs that give optimal results with representative Ad serotypes of each species (species A, Ad12; species B, Ad7; species C, Ad5; species D, Ad8; species E, Ad4; and species F, Ad40). When the final optimized assay was tested with purified Ad DNA, approximately 165, 10, 100, 185, 5, and 20 genome copies of the six respective serotypes were detected. The multiplex PCR assay could amplify all six representative serotypes of each species in a single reaction, or selected primer subsets could be combined for isolates associated with particular clinical presentations, e.g., combinations of primers specific for species A, B, C, D, and F, species B, D, and E, species B, C, and E, or species B, C, and D for isolates associated with gastroenteritis, conjunctivitis, respiratory infections, and genital-urinary tract infections, respectively (Fig. 1). No cross-reactions were observed with the aforementioned non-Ad DNA viruses, and cultures for two Ad isolates that contained contaminating adeno-associated virus were correctly identified to the species level without apparent interference (data not shown).
FIG. 1.
Ethidium bromide-stained agarose gel showing PCR products from five different combinations of Ad species-specific primers. Lanes, from left to right:, respectively: M, molecular weight marker III (Boehringer Mannheim); species A-F, species A to F; A-D, F, species A, B, C, D, and F; B, D, E, species B, D, and E; B, C, E, species B, C, and E; B, C, D, species B, C, and D; Neg, template-free negative control. Numbers on the left are in base pairs.
Evaluation of multiplex PCR with Ad prototype strains.
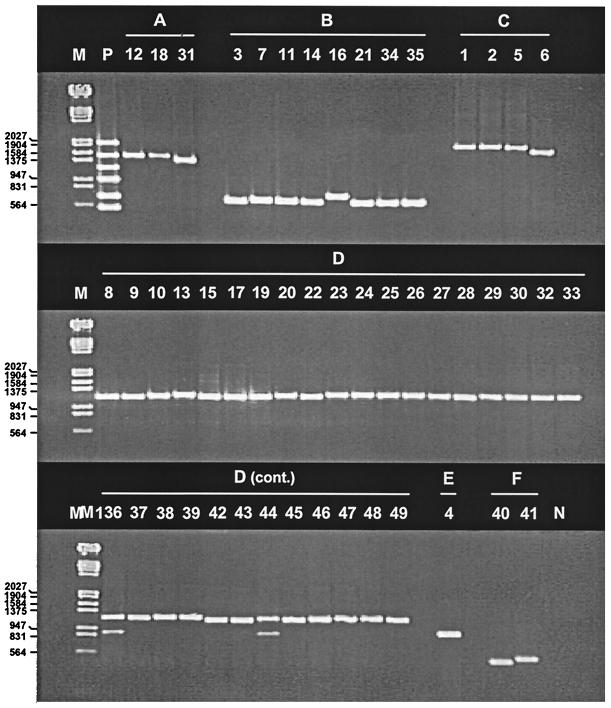
When tested against each of the 49 Ad prototype strains, the multiplex PCR assay generally gave intense, discrete bands of the expected size (Fig. 2). Variations in amplicon size between serotypes were noted for some species, as predicted from published sequences: for species A, Ad31 amplicons (1,444 bp [40]) were smaller than Ad12 amplicons (1,537 bp [49]); for species B, Ad16 amplicons (772 bp [47]) were larger than Ad3 and Ad7 amplicons (670 bp [26, 48]); and for species F, Ad40 amplicons (541 bp [27]) were smaller than Ad41 amplicons (586 bp [35]). Ad6 amplicons were smaller (∼50 bp, as estimated by gel electrophoresis) than the amplicons of isolates of other species C serotypes. In addition to size differences, secondary amplification products of smaller molecular size (∼900 bp, as estimated by gel electrophoresis) were present for Ad36 and Ad44 of species D. However, the secondary bands had lower intensities and their sizes did not interfere with interpretation of the assay results. Ad18 amplicons were less intense than the amplicons of other species A isolates in the multiplex reaction but were still detectable; when the species A-specific primers were tested individually with Ad18, the amplicon yield was comparable to those obtained with the other species-specific primers.
FIG. 2.
Ethidium bromide-stained agarose gel showing PCR products of 49 Ad prototype strains. Lanes, from left to right, respectively: M, molecular weight marker III (Boehringer Mannheim); P, pooled control DNAs of representative Ad serotypes of species A to F; 1 to 49, individual Ad serotypes; N, pooled control DNA without indicated Ad species. Numbers on the left are in base pairs.
Evaluation of multiplex PCR with Ad field isolates.
Of 180 diverse Ad isolates tested by multiplex PCR, 173 correctly matched the species previously determined by hemaglutination inhibition and/or neutralization assay (Table 2). Seven “misidentified” Ads were passaged once in A549 cells and were retested by neutralization assay with horse antisera specific for the original Ad serotype and all serotypes within the newly designated species. In every case, the Ad serotypes identified differed from the original serotype designation and were consistent with the species identified by the multiplex PCR assay (Table 3). Amplicons of 19 field isolates previously serotyped as Ad31 (6), Ad6 (4), Ad40 (2), and Ad41 (3) were identical in size to the amplicons of the respective prototype strains. Although the amplicons of only 5 of 11 field isolates previously serotyped as Ad16 were identical to the amplicons of the prototype strains, the 6 isolates that gave discrepant results were identified as Ad3 on retesting by neutralization assay. Two isolates that were neutralized with both Ad12 and Ad31 antisera were consistent with Ad12 according to their amplicon sizes. Three isolates, previously designated Ad39 and Ad40 and subsequently reclassified as Ad31 by neutralization assay, were confirmed to be Ad31 according to their amplicon sizes; these isolates were obtained from children with acute gastroenteritis, consistent with the clinical profile for this serotype.
TABLE 2.
Comparison of types determined by species-specific multiplex PCR assays with types of 180 previously typed Ad field isolates
| Original identificationa
|
No. of isolates tested | No. of isolates with the indicated result:
|
||||
|---|---|---|---|---|---|---|
| Fiber multiplex PCR assay
|
Hexon multiplex PCR assayb
|
|||||
| Species | Serotype | Correct | Incorrect | Correct | Incorrect | |
| A | 12 | 2 | 2 | 0 | 2 | 0 |
| 18 | 10 | 9 | 1 | 9 | 1 | |
| 31 | 8 | 7 | 1 | 7 | 1 | |
| 31 + 12 | 2 | 2 | 0 | 2 | 0 | |
| Subtotal | 22 | 20 | 2 | 20 | 2 | |
| B | 3 | 4 | 4 | 0 | 4 | 0 |
| 7, 7a | 42 | 42 | 0 | 41 | 1 | |
| 11 | 3 | 3 | 0 | 1 | 2 | |
| 11/H14 | 1 | 1 | 0 | 1 | 0 | |
| 14 | 1 | 1 | 0 | 0 | 1 | |
| 16c | 11 | 11 | 0 | 11 | 0 | |
| 21/H21 + 35 | 2 | 2 | 0 | 0 | 2 | |
| 34/H11 | 1 | 1 | 0 | 1 | 0 | |
| 34 | 2 | 2 | 0 | 2 | 0 | |
| 35 | 2 | 2 | 0 | 1 | 1 | |
| Subtotal | 69 | 69 | 0 | 62 | 7 | |
| C | 1 | 8 | 7 | 1 | 7 | 1 |
| 2 | 10 | 10 | 0 | 10 | 0 | |
| 5 | 4 | 4 | 0 | 4 | 0 | |
| 6 | 5 | 5 | 0 | 5 | 0 | |
| Subtotal | 27 | 26 | 1 | 26 | 1 | |
| D | 8 | 2 | 2 | 0 | 2 | 0 |
| 9 | 1 | 1 | 0 | 1 | 0 | |
| 13 | 1 | 1 | 0 | 1 | 0 | |
| 15/H29 | 1 | 1 | 0 | 1 | 0 | |
| 17 | 1 | 1 | 0 | 1 | 0 | |
| 19 | 9 | 9 | 0 | 9 | 0 | |
| 22 | 1 | 1 | 0 | 1 | 0 | |
| 23 | 1 | 1 | 0 | 1 | 0 | |
| 28 | 1 | 1 | 0 | 1 | 0 | |
| 29 | 1 | 1 | 0 | 1 | 0 | |
| 30 | 1 | 1 | 0 | 1 | 0 | |
| 32/H27 | 1 | 1 | 0 | 1 | 0 | |
| 36 | 1 | 1 | 0 | 1 | 0 | |
| 37 | 2 | 2 | 0 | 2 | 0 | |
| 38 | 1 | 1 | 0 | 1 | 0 | |
| 39 | 1 | 0 | 1 | 0 | 1 | |
| 42 | 1 | 1 | 0 | 1 | 0 | |
| 43 | 1 | 1 | 0 | 1 | 0 | |
| 44/H13 | 1 | 1 | 0 | 1 | 0 | |
| 45 | 1 | 1 | 0 | 1 | 0 | |
| 46/H13 | 1 | 1 | 0 | 1 | 0 | |
| 47 | 1 | 1 | 0 | 1 | 0 | |
| Subtotal | 32 | 31 | 1 | 31 | 1 | |
| E | 4 | 12 | 12 | 0 | 12 | 0 |
| F | 40 | 6 | 3 | 3 | 3 | 3 |
| 41 | 4 | 4 | 0 | 4 | 0 | |
| 40 or 41 | 8 | 8 | 0 | 8 | 0 | |
| Subtotal | 18 | 15 | 3 | 15 | 3 | |
| Total | 180 | 173 | 7 | 166 | 14 | |
Original identification of Ad isolates by hemagglutination inhibition (23) and/or neutralization (21) assay. Eight Ad40/41 isolates originally identified by commercial Ad40/41 enzyme immunoassay only.
Ad species-specific hexon multiplex PCR procedure performed as described by Pring-Åkerblom et al. (42).
Six of 11 Ad16 field isolates were later identified as Ad3 on retesting by neutralization assay.
TABLE 3.
Confirmation of results for Ad isolates with discrepant results by species-specific multiplex PCR assays
| Isolate | Date collected (mo/day/yr) | Location | Original identificationa
|
Multiplex PCR assaysb
|
Repeat identificationc
|
|||
|---|---|---|---|---|---|---|---|---|
| Species | Serotype | Fiber | Hexon | Species | Serotype | |||
| V-1707 | 5/3/84 | Alabama | A | Ad18 | C | C | C | Ad2 |
| V-2362 | 8/16/91 | New Mexico | A | Ad31 | D | D | D | Ad36 |
| 99018072 | NDd | Brazil | B | Ad11 | B | Neg | B | Ad11 |
| V-2181 | 12/20/88 | New York | B | Ad11 | B | Neg | B | Ad11 |
| 98034069 | X/X/95 | Egypt | B | Ad14 | B | Neg | B | Ad14 |
| V-2167A | 9/8/85 | Tennessee | B | Ad21/H21 + 35 | B | Neg | B | Ad21 + 35 |
| RU-8176 | 2/24/77 | Tennessee | B | Ad21/H21 + 35 | B | Neg | B | Ad21 + 35 |
| V-2079A | 4/30/87 | Canada | B | Ad35 | B | Neg | B | Ad35 |
| V-2064A | 1/30/87 | Colorado | B | Ad7 | B | Neg | B | Ad7 |
| V-2215 | 7/26/89 | Pennsylvania | C | Ad1 | D | D | D | Ad19 |
| V-375-4 | 8/28/89 | El Salvador | D | Ad39 | A | A | A | Ad31 |
| V-2158B | 6/22/88 | Maryland | F | Ad40 | A | A | A | Ad31 |
| V-1533-18 | 11/24/81 | Arizona | F | Ad40 | A | A | A | Ad31 |
| V-2159A | 6/29/88 | Maryland | F | Ad40 | D | D | D | Ad9 |
Original identification of Ad isolates made by hemagglutination inhibition (23) and/or neutralization (21) assay.
Ad species-specific hexon multiplex PCR procedure performed as described by Pring-Åkerblom et al. (42).
Repeat identification of Ad isolates made by microneutralization assay with type-specific horse antisera.
ND, not determined.
Comparison of multiplex PCR assays.
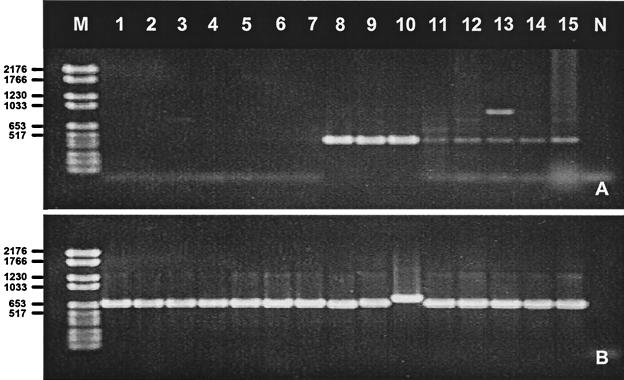
To further assess the specificity of our multiplex PCR assay, the 180 Ad field isolates were simultaneously tested by a previously described multiplex PCR assay that used subgenus-specific primers to the Ad hexon gene (42). In most cases, we obtained identical results by both assays (Tables 2 and 3). However, we were unable to amplify seven species B field isolates, including serotypes Ad7, Ad11, Ad14, Ad21, and Ad35, by the hexon multiplex PCR. When the eight prototype strains of species B were amplified by the hexon multiplex PCR, only prototype strains Ad3, Ad7, and Ad16 gave definitive results, whereas Ad11, Ad14, Ad21, Ad34, and Ad35 amplified poorly in both multiplex reactions and in reactions with the primers to the species B hexon gene alone. In contrast, our multiplex PCR successfully amplified all species B strains (Fig. 3).
FIG. 3.
Ethidium bromide-stained agarose gel showing PCR products of the hexon (A) and fiber (B) multiplex assays. Lanes: M, molecular weight marker VI (Boehringer Mannheim); 1, isolate 98034069; 2, V-2064A; 3, 99018072; 4, V-2181; 5, V-2079A; 6, V-2167A; 7, RU-8176; 8, prototype Ad3; 9, Ad7; 10, Ad16; 11, Ad21; 12, Ad11; 13 Ad14; 14, Ad34; 15, Ad35; N, negative control. Both assays were performed with the same Ad DNA extracts, and identical results were obtained in two separate amplification reactions. Numbers on the left are in base pairs.
DISCUSSION
Our objective in this study was to develop a multiplex PCR assay that could supplant hemagglutination for Ad species identification. Whereas PCR assays described for identification of Ad species and serotypes have targeted the hexon (42), pIX (3), and VA RNA (29) genes, we chose the Ad fiber gene which, along with the hexon gene products, confers Ad type specificity and forms the basis of the hemagglutination inhibition test (13, 52). Using previously published fiber gene sequences, we were able to identify regions that were suitable for primer design and that were conserved within but variable between Ad species. The six PCR amplicons (from species A to F) were readily distinguished by size, and therefore, species-specific identification could be obtained without reliance on a second nested PCR or restriction enzyme analysis. Our six subgenus-specific primer pairs could be combined in a single multiplex reaction, and reactions could be run with the primers individually or in various combinations, as dictated by the clinical presentation or virus isolation site. Mixed Ad infections involving different species could theoretically be detected, and fastidious Ads that may be difficult to grow to sufficient titer for identification by classic methods could readily be identified by PCR.
Our multiplex PCR assay correctly identified all 49 recognized Ad serotypes and 180 geographically and temporally diverse Ad field isolates to the species level, including several naturally occurring intermediate strains of species B and D. Several field isolates identified incorrectly by classic typing methods were correctly identified by multiplex PCR, as confirmed by repeat type-specific neutralization assay. Although a review of the original laboratory records did not identify any obvious explanation for these errors, misreading of the hemagglutination profile may have led to the selection of inappropriate subsets of antisera for testing by the hemagglutination inhibition and/or neutralization assay.
Serotype-specific identification of Ad6, Ad16, Ad31, Ad40, and Ad41 on the basis of differences in amplicon size was an added benefit of our assay. This could prove particularly useful for the diagnosis of infant gastroenteritis, in which the three most commonly associated Ad serotypes, Ad31, Ad40, and Ad41 (2, 6), could readily be distinguished in a single amplification reaction. However, rare isolates of Ad41 possess a deletion whose size is identical to that of the sequence gap that distinguishes Ad40 from Ad41, which would result in the misclassification of these strains (28), and the occurrence of amplicon length variants with DNA insertions or deletions among Ad field isolates should be anticipated. Therefore, confirmation of these serotypes by restriction fragment analysis, probe hybridization, or direct sequencing would be warranted.
Most PCR assays for the species or serotype identification of Ads have been limited in scope (3, 24, 37, 40) or dependent upon restriction fragment length polymorphism analysis (29, 45), which can complicate interpretation of assay results. Although serotyping of Ads on the basis of sequencing of PCR products (31, 50) is potentially more informative, this approach is beyond the scope of most diagnostic laboratories. A more promising assay recently described by Pring-Åkerblom et al. (42) was based on multiplex PCR amplification of Ad species with species-specific primers to the Ad hexon gene. However, in our hands this assay performed poorly with Ad11, Ad21, Ad34, and Ad35, probably because of species B primer mismatches; the lack of availability of hexon gene sequence data for these viruses may have compromised primer design. Nevertheless, we found that the hexon multiplex PCR assay generally performed well with other Ad species and provided a useful complement to our assay in the identification and genetic characterization of Ad isolates.
We were unable to evaluate our multiplex PCR assay for direct detection of Ads in clinical specimens because of the limited availability of suitable specimens and can therefore recommend our assay only for identification of virus isolates. To further evaluate the accuracy of our assay, extensive prospective testing of geographically diverse Ad isolates is in progress. Because of time and cost constraints, few laboratories in the United States offer Ad identification services, and as the hyperimmune animal antisera pools used for hemagglutination inhibition and neutralization tests become depleted, the need for molecular biology-based identification methods has increased. Our Ad species-specific multiplex PCR assay should help address this need.
ADDENDUM IN PROOF
After submission of this manuscript, we received prototype strains Ad50 and Ad51 from Jan de Jong, Erasmus University Rotterdam, Rotterdam, The Netherlands, for evaluation. Ad50 and Ad51 were correctly identified as species B and D, respectively, by our multiplex PCR assay.
REFERENCES
- 1.Adrian T, Wadell G, Hierholzer J C, Wigand R. DNA restriction analysis of adenovirus prototype 1 to 41. Arch Virol. 1986;91:277–290. doi: 10.1007/BF01314287. [DOI] [PubMed] [Google Scholar]
- 2.Adrian T, Wigand R. Genome type analysis of adenovirus 31, a potential causitive agent of infants' enteritis. Arch Virol. 1989;105:81–87. doi: 10.1007/BF01311118. [DOI] [PubMed] [Google Scholar]
- 3.Akalu A, Seidel W, Liebermann H, Bauer U, Döhner L. Rapid identification of subgenera of human adenovirus by serological and PCR assays. J Virol Methods. 1998;71:187–196. doi: 10.1016/s0166-0934(97)00213-9. [DOI] [PubMed] [Google Scholar]
- 4.Allard A, Girones R, Juto P, Wadell G. Polymerase chain reaction for detection of adenoviruses in stool samples. J Clin Microbiol. 1990;28:2659–2667. doi: 10.1128/jcm.28.12.2659-2667.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benkö M, Harrach B, Russell W C. Family Adenoviridae. In: Van Regenmortel M H V, Fauquet C M, Bishop D K L, Carstens E B, Estes M K, Lemon S M, Maniloff J, Mayo M A, McGeoch D J, Pringle C R, Wickner R B, editors. Virus taxonomy. Seventh report of the International Committee on Taxonomy of Viruses. New York, N.Y: Academic Press, Inc.; 1999. pp. 227–238. [Google Scholar]
- 6.Brandt C D, Kim H W, Rodriguez W J, Arrobo J Q, Jeffries B C, Stallings E P, Lewis C, Miles A J, Gardner M K, Parrott R H. Adenoviruses and pediatric gastroenteritis. J Infect Dis. 1985;151:437–443. doi: 10.1093/infdis/151.3.437. [DOI] [PubMed] [Google Scholar]
- 7.Chroboczek J, Jacrot B. The sequence of adenovirus fiber: similarities and differences between serotypes 2 and 5. Virology. 1987;161:549–554. doi: 10.1016/0042-6822(87)90150-4. [DOI] [PubMed] [Google Scholar]
- 8.Cooper R J, Yeo A C, Bailey A S, Tullo A B. Adenovirus polymerase chain reaction assay for rapid diagnosis of conjunctivitis. Investig Ophalmol Vis Sci. 1999;40:90–95. [PubMed] [Google Scholar]
- 9.Crawford-Miksza L K, Nang R, Schnurr D P. Strain variation in adenovirus serotypes 4 and 7a causing acute respiratory disease. J Clin Microbiol. 1999;37:1107–1112. doi: 10.1128/jcm.37.4.1107-1112.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dalapathy S, Lily T K, Roy S, Madhavan H N. Development and use of nested polymerase chain reaction (PCR) for the detection of adenovirus from conjunctivitis specimens. J Clin Virol. 1998;11:77–84. doi: 10.1016/s0928-0197(98)00021-x. [DOI] [PubMed] [Google Scholar]
- 11.De Jong J C, Wermenbol A G, Verweij-Uijterwall M W, Slaterus K W, Wertheim-Van Dillen P, Van Dornum G J J, Khoo S H, Hierholzer J C. Adenoviruses from human immunodeficiency virus-infected individuals, including two strains that represent new candidate serotypes Ad50 and Ad51 of species B1 and D, respectively. J Clin Microbiol. 1999;37:3940–3945. doi: 10.1128/jcm.37.12.3940-3945.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Echavarria M, Forman M, Tichehurst J, Dumler J S, Charache P. PCR method for detection of adenvirus in urine of healthy and human immunodeficiency virus-infected individuals. J Clin Microbiol. 1998;36:3323–3326. doi: 10.1128/jcm.36.11.3323-3326.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eiz B, Adrian T, Pring-Åkerblom P. Recombinant fibre proteins of human adenoviruses Ad9, Ad15 and Ad19: localization of the haemagglutination properties and the type-specific determinant. Res Virol. 1997;148:5–10. doi: 10.1016/s0923-2516(97)81905-x. [DOI] [PubMed] [Google Scholar]
- 14.Elsom B L, Herzog N K. Rapid method for preparing adenovirus DNA. BioTechniques. 1997;22:868–870. doi: 10.2144/97225bm18. [DOI] [PubMed] [Google Scholar]
- 15.Gerna G, Cattaneo E, Grazia Revello M, Battaglia M. Grouping of human adenoviruses by early antigen reactivity. J Infect Dis. 1982;145:678–682. doi: 10.1093/infdis/145.2.678. [DOI] [PubMed] [Google Scholar]
- 16.Grondahl B, Puppe W, Hoppe A, Kuhne I, Weigl J A, Schmitt H J. Rapid identification of nine microorganisms causing acute respiratory tract infections by single-tube multiplex reverse transcription-PCR: fesibility study. J Clin Microbiol. 1999;37:1–7. doi: 10.1128/jcm.37.1.1-7.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gruber W C, Russell D J, Tibbetts C. Fiber gene and genomic origin of human adenovirus type 4. Virology. 1993;196:603–611. doi: 10.1006/viro.1993.1516. [DOI] [PubMed] [Google Scholar]
- 18.Henderson Y C, Liu T-J, Clayman G L. A simple and sensitive method for detecting adenovirus in serum and urine. J Virol Methods. 1998;71:51–56. doi: 10.1016/s0166-0934(97)00189-4. [DOI] [PubMed] [Google Scholar]
- 19.Hierholzer J C. Further subgrouping of the human adenoviruses by differential hemagglutination. J Infect Dis. 1973;128:541–550. doi: 10.1093/infdis/128.4.541. [DOI] [PubMed] [Google Scholar]
- 20.Hierholzer J C. Adenoviruses. In: Lennette E H, Lennette A D, Lennette T E, editors. Diagnostic procedures for viral, rickettsial and chlamydial infections. 7th ed. Washington, D.C.: American Public Health Association, Inc.; 1995. pp. 169–188. [Google Scholar]
- 21.Hierholzer J C, Bingham P G. Vero microcultures for adenovirus neutralization tests. J Clin Microbiol. 1978;7:499–506. doi: 10.1128/jcm.7.6.499-506.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hierholzer J C, Stone Y O, Broderson J R. Antigenic relationships among the 47 human adenoviruses determined in reference horse antisera. Arch Virol. 1991;121:179–197. doi: 10.1007/BF01316753. [DOI] [PubMed] [Google Scholar]
- 23.Hierholzer J C, Suggs M T, Hall E C. Standardized viral hemagglutination and hemagglutination-inhibition tests. II. Description and statistical evaluation. Appl Microbiol. 1969;18:824–833. doi: 10.1128/am.18.5.824-833.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hussain, M. A. S., P. Costello, D. J. Morris, A. S. Bailey, G. Corbitt, R. J. Cooper, and A. B. Tullo. Comparison of primer sets for detection of fecal and ocular adenovirus infection using the polymerase chain reaction. J. Med. Virol. 49:187–194. [DOI] [PubMed]
- 25.Jackson R, Morris J, Cooper R J, Bailey A S, Klapper P E, Cleator G M, Tullo A B. Multiplex polymerase chain reaction for adenovirus and herpes simplex virus in eye swabs. J Virol Methods. 1996;56:41–48. doi: 10.1016/0166-0934(95)01903-0. [DOI] [PubMed] [Google Scholar]
- 26.Kajon A E, Wadell G. Sequence analysis of the E3 region and fiber gene of human adenovirus genome type 7h. Virology. 1996;215:190–196. doi: 10.1006/viro.1996.0022. [DOI] [PubMed] [Google Scholar]
- 27.Kidd A H, Erasmus M J. Sequence characterization of the adenovirus 40 fiber gene. Virology. 1989;172:134–144. doi: 10.1016/0042-6822(89)90115-3. [DOI] [PubMed] [Google Scholar]
- 28.Kidd A H, Erasmus M J, Tiemessen C T. Fiber sequence heterogeneity in subgroup F adenoviruses. Virology. 1990;179:139–150. doi: 10.1016/0042-6822(90)90283-w. [DOI] [PubMed] [Google Scholar]
- 29.Kidd A H, Jönsson M, Garwicz D, Kajon A E, Wermenbol A G, Verveij M W, De Jong J C. Rapid subgenus identification of human adenovirus isolates by a general PCR. J Clin Microbiol. 1996;34:622–627. doi: 10.1128/jcm.34.3.622-627.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kinchington P R, Turse S E, Kowalski R P, Gordon Y J. Use of polymerase chain amplification reaction for the detection of adenoviruses in ocular swab specimens. Investig Ophalmol Vis Sci. 1994;35:4126–4134. [PubMed] [Google Scholar]
- 31.Li Q-G, Henningsson A, Juto P, Elgh F, Wadell G. Use of restriction fragment analysis and sequencing of a serotype-specific region to type adenovirus isolates. J Clin Microbiol. 1999;37:844–847. doi: 10.1128/jcm.37.3.844-847.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McDonough M, Kew O, Hierholzer J C. PCR detection of human adenviruses. In: Persing D H, Smith T F, Tenover F C, White T J, editors. Diagnostic molecular microbiology: principles and applications. Washington, D.C.: American Society for Microbiology; 1993. pp. 389–393. [Google Scholar]
- 33.Norrby E. The structural and functional diversity of adenovirus capsid components. J Gen Virol. 1969;5:221–236. doi: 10.1099/0022-1317-5-2-221. [DOI] [PubMed] [Google Scholar]
- 34.Osiowy C. Direct detection of respiratory syncytial virus, parainfluenza virus, and adenovirus in clinical respiratory specimens by a multiplex reverse transcription-PCR assay. J Clin Microbiol. 1998;36:3149–3154. doi: 10.1128/jcm.36.11.3149-3154.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pieniazek N J, Slemenda S B, Pieniazek D, Velarde J J, Luftig R B. Sequence of human enteric adenovirus type 41 Tak fiber protein gene. Nucleic Acids Res. 1989;17:9474. doi: 10.1093/nar/17.22.9474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Poddar S K. Detection of adenovirus using PCR and molecular beacon. J Virol Methods. 1999;82:19–26. doi: 10.1016/s0166-0934(99)00074-9. [DOI] [PubMed] [Google Scholar]
- 37.Pring-Åkerblom P, Adrian T. Type- and group-specific polymerase chain reaction for adenovirus detection. Res Virol. 1994;145:25–35. doi: 10.1016/s0923-2516(07)80004-5. [DOI] [PubMed] [Google Scholar]
- 38.Pring-Åkerblom P, Adrian T. Sequence characterization of the adenovirus 31 fibre and comparison with serotypes of subgenera A to F. Res Virol. 1995;146:343–354. doi: 10.1016/0923-2516(96)80597-8. [DOI] [PubMed] [Google Scholar]
- 39.Pring-Åkerblom P, Adrian T. Characterization of adenovirus subgenus D fiber genes. Virology. 1995;206:564–571. doi: 10.1016/s0042-6822(95)80073-5. [DOI] [PubMed] [Google Scholar]
- 40.Pring-Åkerblom P, Adrian T. PCR-based detection and typing of human adenoviruses in clinical samples. Res Virol. 1997;148:225–231. doi: 10.1016/s0923-2516(97)83992-1. [DOI] [PubMed] [Google Scholar]
- 41.Pring-Åkerblom P, Trijssenaar F E, Adrian T. Sequence characterization and comparison of human adenovirus subgenus B and E hexons. Virology. 1995;212:232–236. doi: 10.1006/viro.1995.1474. [DOI] [PubMed] [Google Scholar]
- 42.Pring-Åkerblom P, Trijssenaar F E, Adrian T, Hoyer H. Multiplex polymerase chain reaction for subgenus-specific detection of human adenoviruses in clinical samples. J Med Virol. 1999;58:87–92. [PubMed] [Google Scholar]
- 43.Raty R, Kleemola M, Melen K, Stenvik M, Julkunen I. Efficacy of PCR and other diagnostic methods for the detection of respiratory adenoviral infections. J Med Virol. 1999;59:66–72. [PubMed] [Google Scholar]
- 44.Rosen L. Hemagglutination by adenoviruses. Virology. 1958;5:574. doi: 10.1016/0042-6822(58)90050-3. [DOI] [PubMed] [Google Scholar]
- 45.Saitoh-Inagawa W, Oshima A, Aoki K, Itoh N, Isobe K, Uchio E, Ohno S, Nakajima H, Hata K, Ishiko H. Rapid diagnosis of adenoviral conjunctivitis by PCR and restriction fragment length polymorphism analysis. J Clin Microbiol. 1996;34:2113–2116. doi: 10.1128/jcm.34.9.2113-2116.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schmitz H, Wigand R, Heinrich W. Worldwide epidemiology of human adenovirus infections. Am J Epidemiol. 1983;117:455–466. doi: 10.1093/oxfordjournals.aje.a113563. [DOI] [PubMed] [Google Scholar]
- 47.Shieh W, Tibbetts C. Genetic variations and phylogenetic analysis of adenovirus fiber gene. Ph.D. thesis. Nashville, Tenn: Vanderbilt University; 1992. [Google Scholar]
- 48.Signäs C, Akusjärvi G, Pettersson U. Adenovirus 3 fiber polypeptide gene: implications for the structure of the fiber protein. J Virol. 1985;53:672–678. doi: 10.1128/jvi.53.2.672-678.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sprengel J, Schmitz B, Heuss-Neitzel D, Zock C, Doerfler W. Nucleotide sequence of human adenovirus type 12 DNA: comparative functional analysis. J Virol. 1994;68:379–389. doi: 10.1128/jvi.68.1.379-389.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Takeuchi S, Norihiko I, Uchio E, Aoki K, Ohno S. Serotyping of adenoviruses on conjunctival scrapings by PCR and sequence analysis. J Clin Microbiol. 1999;37:1839–1845. doi: 10.1128/jcm.37.6.1839-1845.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Uchio E, Aoki W, Saitoh W, Itoh N, Ohno S. Rapid diagnosis of adenoviral conjunctivitis on conjunctival swabs by 10 minute immunochromatography. Ophthalmology. 1997;104:1294–1299. doi: 10.1016/s0161-6420(97)30145-6. [DOI] [PubMed] [Google Scholar]
- 52.Watson G, Burdon M G, Russell W C. An antigenic analysis of the adenvirus type 2 fibre polypeptide. J Gen Virol. 1988;69:525–535. doi: 10.1099/0022-1317-69-3-525. [DOI] [PubMed] [Google Scholar]
- 53.Wood S R, Sharp I R, Caul E O, Paul I, Bailey A S, Hawkins M, Pugh S, Treharne J, Stevenson S. Rapid detection and serotyping of adenovirus by direct immunofluorescence. J Med Virol. 1997;51:198–201. doi: 10.1002/(sici)1096-9071(199703)51:3<198::aid-jmv9>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]