Abstract
Population-level immune surveillance, which includes monitoring exposure and assessing vaccine-induced immunity, is a crucial component of public health decision-making during a pandemic. Serosurveys estimating the prevalence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) antibodies in the population played a key role in characterizing SARS-CoV-2 epidemiology during the early phases of the pandemic. Existing serosurveys provide infrastructure to continue immune surveillance but must be adapted to remain relevant in the SARS-CoV-2 vaccine era. Here, we delineate how SARS-CoV-2 serosurveys should be designed to distinguish infection- and vaccine-induced humoral immune responses to efficiently monitor the evolution of the pandemic. We discuss how serosurvey results can inform vaccine distribution to improve allocation efficiency in countries with scarce vaccine supplies and help assess the need for booster doses in countries with substantial vaccine coverage.
Keywords: COVID, 19, cross, sectional, SARS, CoV, 2, seroprevalence, vaccines
INTRODUCTION
As severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) vaccinations have decoupled coronavirus disease 2019 (COVID-19) cases from deaths, the path toward endemicity has become clearer. However, public health officials must respond to the pandemic’s evolving landscape to chart society’s course toward maximum population-level immunity.
Serosurveys are epidemiological studies that estimate population-level antibody prevalence. Before vaccines were broadly available, SARS-CoV-2 antibodies (ie, seropositivity) served as evidence of prior infection; accordingly, serosurvey data were used to characterize SARS-CoV-2 epidemiology [1] and identify vulnerable and disproportionately infected populations [2]. As vaccines also elicit antibodies, serosurveys must now be adapted to distinguish infection- and vaccine-induced immunity [3]. Additionally, vaccines have been distributed unevenly: some countries with scarce supply are forced to allocate vaccines judiciously [4], whereas countries with greater supply and broad uptake have begun considering the benefits of booster doses [5].
In this Perspective, we outline how serosurveys can be adapted for the SARS-CoV-2 vaccine era to (1) distinguish infection- and vaccine-induced immunity, (2) guide vaccine allocation in countries with scarce supply, and (3) evaluate the need for booster doses in regions with high vaccine coverage.
Box 1. Background on SARS-CoV-2 Serosurveys.
Serological assays measure antibody levels in blood samples. SARS-CoV-2 seroprevalence studies, also termed serosurveys, estimate the prevalence of SARS-CoV-2-seropositive individuals in a population by administering antibody tests to a sample of that population. Seropositivity is an indicator of past infection or vaccination and has implications for protection from future infection. Stratifying seroprevalence by geography, age, race, occupation, and other variables can illuminate how different factors shaped the pandemic’s trajectory and identify future public health priorities.
As of November 18, 2021, >2500 SARS-CoV-2 serosurveys have been conducted globally [6]. Serosurveys must be carefully conducted to produce credible seroprevalence estimates; however, many serosurveys used nonrandom sampling strategies, deployed tests with low sensitivity and/or specificity, or failed to correct estimates for sampling bias or imperfect test accuracy [7].
Abbreviation: SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
SEROSURVEYS SHOULD BE DESIGNED TO DISTINGUISH INFECTION- AND VACCINE-INDUCED HUMORAL IMMUNE RESPONSES
Before broad vaccination campaigns, serosurveys were relatively straightforward to interpret because SARS-CoV-2 antibodies were elicited primarily by symptomatic or asymptomatic infection, but also, in some instances, by seasonal human coronaviruses, especially if tests lacked specificity [8]. To maximize the value of serosurveys, it is now important to differentiate seropositivity due to past infection, past vaccination, or both. This stratification enables public health officials to monitor true infection rates, and in turn estimate infection fatality ratios (IFRs) and asymptomatic infections. In some countries, distinguishing infection- and vaccine-induced immunity may also guide the prioritization of vaccine distribution (Section 2).
SARS-CoV-2 humoral immune responses are commonly characterized by detection of antibodies against the spike glycoprotein (S), the receptor-binding domain (RBD) within the spike glycoprotein, and the nucleocapsid protein (N). Infection elicits antibodies against all 3 targets; however, many vaccines (eg, current mRNA and viral vector vaccines) only target S protein–derived antigens [9]. Thus, in countries using only S-targeting vaccines, seropositivity for anti-N antibodies indicates prior infection. In countries using vaccines that elicit both anti-S and anti-N seropositivity (eg, inactivated or attenuated virus vaccines), it is not possible to distinguish infection- and vaccine-induced immunity via serostatus alone.
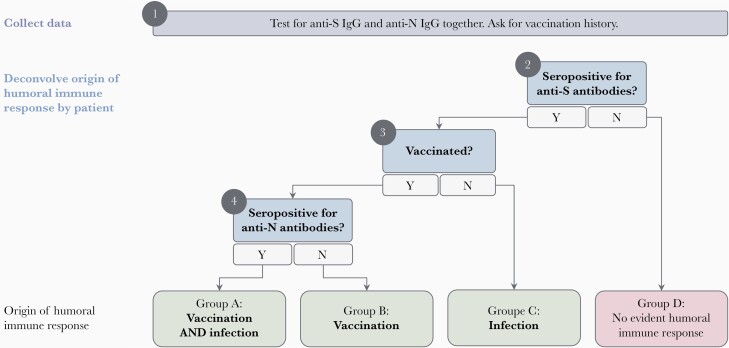
Figure 1 outlines a decision tree serosurvey investigators can use to identify the origin of humoral immune responses. Investigators should deploy tests for both anti-S (or fragments of the spike molecule, including anti-RBD) and anti-N antibodies and record individual history of vaccination and COVID-19 diagnosis when applicable. Assays measuring either total antibodies or immunoglobulin G can be deployed [10].
Figure 1.
Conceptual decision tree for distinguishing between an infection- and vaccine-induced humoral immune response. Serosurvey investigators should collect all data simultaneously (eg, deploy anti-S and anti-N assays together) before applying the conceptual decision tree. Assays measuring total antibodies or IgG can be used. Anti-RBD assays can be used in place of anti-S assays. A past diagnosis, if present, can support the conclusion that an individual should be assigned to group A or C. This decision tree is only applicable for S-targeting vaccines. Abbreviations: IgG, immunoglobulin; N, nucleocapsid protein; RBD, receptor-binding domain; S, spike glycoprotein.
If an individual is seronegative for anti-S antibodies, they have likely neither been infected nor been vaccinated (Group D); if the individual’s vaccination (or diagnosis) history suggests otherwise, they may have antibody levels lower than an assay’s limit of detection or have an immune defect causing insufficient antibody production [11]. Among unvaccinated individuals seropositive for anti-S antibodies (Group C), seropositivity is likely the result of prior infection. A positive anti-N test (or previous COVID-19 diagnosis) reinforces this result but is not required [12]. In vaccinated individuals who are seropositive for anti-S antibodies, investigators should determine whether individuals were previously infected based on anti-N seropositivity (Group A, previously infected and vaccinated; or Group B, vaccinated only). Diagnostic history can also confirm prior infection if available, but the absence of a past diagnosis does not preclude prior infection. Without access to vaccination or diagnosis history, investigators can still stratify participants into 4 groups: no available evidence of humoral immune response (no anti-S or anti-N antibodies), evidence of prior infection (only anti-N antibodies), evidence of previous vaccination (only anti-S antibodies), and evidence of prior infection with potential vaccination (both anti-S and anti-N antibodies).
This approach has important limitations. First, this algorithm does not apply where vaccines eliciting anti-N seropositivity are used, as discussed above. In these regions, investigators would need to rely on vaccination and diagnosis histories, which are not always available. Second, serological assays cannot identify individuals whose antibody levels have waned below the limit of detection: antibodies naturally degrade, and sensitivity in detecting prior infection after several months varies by immunoassay [10, 12]. The durability of infection-induced antibodies may also vary by disease severity [12]. Investigators should select assays with high analytical sensitivity and consider when quantitative assays may be more appropriate than qualitative assays (Section 3). Finally, participant-reported vaccination and diagnostic histories are subject to recall bias, and individuals with mild or asymptomatic infections may not have been diagnosed with COVID-19.
SEROSURVEYS COULD GUIDE EFFICIENT VACCINE ALLOCATION IN REGIONS WITH LIMITED VACCINE SUPPLY AND SUBSTANTIAL INFECTION UNDERREPORTING
Where vaccine supply is limited, it is crucial to distribute vaccine doses as efficiently as possible to protect the most vulnerable people and reduce opportunities for new variants of concern to emerge. Vaccine prioritization to date has largely been informed by mortality and exposure risk. To improve distribution efficiency, public health officials should also consider prioritizing regions with larger uninfected and unvaccinated populations, which may improve the marginal impact of each dose [13]. Additionally, previously infected individuals who receive 1 dose are likely to have similar immune responses to infection-naive individuals after 2 doses [14], suggesting that 1-dose regimens could be used in regions where many individuals have been infected, enabling second doses to be rerouted to virus hot spots.
Some countries have faced the double burden of limited vaccine supply and high case loads [4]. However, case counts in many of these countries substantially underestimate infections [6, 7], making it challenging to use case surveillance to implement efficient vaccine distribution strategies. In these contexts, serosurveys that measure the prevalence of infection- and vaccine-induced immunity could be crucial tools to inform vaccine allocation on a population level and tailor infection control measures. Using convenience samples (eg, residual sera or banked blood) can be a convenient and economical way to estimate seroprevalence, keeping in mind potential sampling biases and the challenges of not having access to diagnostic and vaccination histories [7]. Communities interested in using individual-level serological testing to allocate vaccines should consider using multiple serological assays to assess seropositivity accurately, particularly when community seroprevalence is low [12].
ONGOING SEROSURVEILLANCE SHOULD GUIDE CONTINUED VACCINATION EFFORTS IN REGIONS WITH HIGH VACCINE COVERAGE
In regions with high vaccine coverage, SARS-CoV-2 is becoming an endemic pathogen [15]. Ongoing surveillance remains necessary—to identify and vaccinate susceptible populations, especially those requiring booster doses, and to monitor the rate of decay of protective antibodies.
A measurable correlate of protection (CoP) against SARS-CoV-2 infection would help identify the individuals most vulnerable to (re)infection; however, there is no widely recognized CoP threshold at present [16]. Binding and neutralizing antibody levels appear to correlate with vaccine efficacy [16, 17], suggesting that serosurveys measuring these antibodies could estimate population-level protection. However, the presence of neutralizing antibodies is transient, undermining the opportunity to use it as a basis for a CoP. Vaccinated individuals who initially mounted a robust neutralizing antibody response that eventually waned may still be protected by cellular immune mechanisms, though the relative contributions of humoral and cellular immunity in protection against infection, symptomatic infection, and severe disease remain to be established [17].
Two potential serosurvey designs might be suitable for populations with high vaccine coverage. First, investigators can deploy total antibody anti-S and anti-N assays, correct for assay sensitivity (including waning antibody levels) and specificity, and estimate cumulative infections and vaccinations. Second, investigators can use quantitative assays that permit comparison of antibody levels to a protective threshold, once one is defined, and estimation of population-level protection. Investigators can also measure the prevalence and relative levels of neutralizing antibodies in a subsample of participants, using scalable and cost-effective surrogate neutralization assays [18]. Finally, antibody levels should be calibrated to an international standard [19], enabling comparison with a protective threshold across studies. Importantly, cell-mediated immunity plays an important protective role [20], but such assays are not amenable to population-level studies because they are generally costly and labor-intensive.
CONCLUSIONS
We are in the midst of the largest vaccination campaign in history. Serosurveys can provide crucial insight into population-level immunity, if appropriately adapted (Box 2). By understanding the prevalence of infection- and vaccine-induced SARS-CoV-2 antibodies, decision-makers can closely monitor key epidemiological indicators and improve vaccine distribution strategies. Continued immune surveillance is warranted to ensure efficient and equitable progress toward sufficient immunity to return to normal.
Box 2. Recommendations.
Well-conducted SARS-CoV-2 serosurveys can distinguish which participants are seropositive as a result of past infection, past vaccination, or both. Doing so involves (1) testing for both anti-S and anti-N antibodies and (2) collecting vaccination history.
In countries with scarce vaccine supply, serosurveys could be used to inform efficient vaccine allocation, identifying susceptible groups that urgently need a first vaccine dose and previously infected groups in which a single vaccine dose may initially suffice.
In countries with high vaccine coverage, ongoing serosurveillance is crucial to inform continued vaccination efforts. In these studies, investigators can closely monitor potential correlates of protection using quantitative serological assays and testing a subsample with surrogate neutralization assays.
Abbreviation: SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Acknowledgments
We would like to thank Mel Krajden, Emily Boucher, and Jordan Van Wyk for their valuable insights and revisions to this manuscript.
Financial support. This work was not directly funded by any institution. SeroTracker is funded by the Public Health Agency of Canada through Canada’s COVID-19 Immunity Task Force, as well as by the Health Emergencies Program of the World Health Organization.
Potential conflicts of interest. The authors declare no relevant financial or nonfinancial competing interests. All authors: no reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Author contributions. N.D., M.Y.L., R.A., and N.B. contributed to the conceptualization; N.D., M.Y.L., R.A., N.B., J.P., B.D.M., and M.A.L. contributed to the investigation; J.P., B.D.M., and M.A.L. contributed to supervision; N.D., M.Y.L., R.A., N.B., J.P., B.D.M., M.P.C., M.B., C.H., T.Y., M.L., C.G., A.C.G., and M.A.L. contributed to the validation; N.D., M.B., and C.C. contributed to visualization; N.D., M.Y.L., R.A., T.Y., and N.B. contributed to writing the original draft; N.D., M.Y.L., R.A., N.B., J.P., B.D.M., M.P.C., M.B., C.C., C.H., T.Y., M.L., C.G., A.C.G., and M.A.L. contributed to writing, review, and editing.
Patient consent. Our study does not include factors necessitating patient consent.
References
- 1. Cheng MP, Yansouni CP, Basta NE, et al. Serodiagnostics for severe acute respiratory syndrome-related coronavirus 2: a narrative review. Ann Intern Med 2020; 173:450–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Anand S, Montez-Rath M, Han J, et al. Prevalence of SARS-CoV-2 antibodies in a large nationwide sample of patients on dialysis in the USA: a cross-sectional study. Lancet 2020; 396:1335–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Shrotri M, Navaratnam AMD, Nguyen V, et al. ; Virus Watch Collaborative. Spike-antibody waning after second dose of BNT162b2 or ChAdOx1. Lancet 2021; 398:385–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. So AD, Woo J.. Reserving coronavirus disease 2019 vaccines for global access: cross sectional analysis. BMJ 2020; 371:m4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Planas D, Veyer D, Baidaliuk A, et al. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature 2021; 596:276–80. [DOI] [PubMed] [Google Scholar]
- 6. Arora RK, Joseph A, Wyk JV, et al. SeroTracker: a global SARS-CoV-2 seroprevalence dashboard. Lancet Infect Dis 2021; 21:e75–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bobrovitz N, Arora RK, Cao C, et al. Global seroprevalence of SARS-CoV-2 antibodies: a systematic review and meta-analysis. PLoS One 2021; 16:e0252617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Galipeau Y, Greig M, Liu G, et al. Humoral responses and serological assays in SARS-CoV-2 infections. Front Immunol 2020; 11:610688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dai L, Gao GF.. Viral targets for vaccines against COVID-19. Nat Rev Immunol 2021; 21:73–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Peluso M, Takahashi S, Hakim J, et al. SARS-CoV-2 antibody magnitude and detectability are driven by disease severity, timing, and assay. Sci Adv 2021; 7:eabh3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Prévost J, Gasser R, Beaudoin-Bussières G, et al. Cross-sectional evaluation of humoral responses against SARS-CoV-2 spike. Cell Rep Med 2020; 1:100126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ripperger TJ, Uhrlaub JL, Watanabe M, et al. Orthogonal SARS-CoV-2 serological assays enable surveillance of low-prevalence communities and reveal durable humoral immunity. Immunity 2020; 53:925–33.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bubar KM, Reinholt K, Kissler SM, et al. Model-informed COVID-19 vaccine prioritization strategies by age and serostatus. Science 2021; 371:916–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Planas D, Veyer D, Baidaliuk A, et al. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature 2021; 596:276–80. [DOI] [PubMed] [Google Scholar]
- 15. Telenti A, Arvin A, Corey L, et al. After the pandemic: perspectives on the future trajectory of COVID-19. Nature 2021; 596:495–504. [DOI] [PubMed] [Google Scholar]
- 16. Krammer F. A correlate of protection for SARS-CoV-2 vaccines is urgently needed. Nat Med 2021; 27:1147–8. [DOI] [PubMed] [Google Scholar]
- 17. Khoury DS, Cromer D, Reynaldi A, et al. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat Med 2021; 27:1205–11. [DOI] [PubMed] [Google Scholar]
- 18. Abe KT, Li Z, Samson R, et al. A simple protein-based surrogate neutralization assay for SARS-CoV-2. JCI Insight 2020; 5:e142362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kristiansen PA, Page M, Bernasconi V, et al. WHO international standard for anti-SARS-CoV-2 immunoglobulin. Lancet 2021; 397:1347–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dan JM, Mateus J, Kato Y, et al. Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science 2021; 371:eabf4063. [DOI] [PMC free article] [PubMed] [Google Scholar]