Abstract
Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) is the causative agent of coronavirus disease 2019 (COVID-19). SARS-CoV-2 has been spreading worldwide since December 2019, resulting in the ongoing COVID-19 pandemic with 237 million infections and 4.8 million deaths by 11 October 2021. While there are great efforts of global vaccination, ending this pandemic has been challenged by issues of exceptionally high viral transmissibility, re-infection, vaccine-breakthrough infection, and immune escape variants of concern. Besides the record-breaking speed of vaccine research and development, antiviral drugs including SARS-CoV-2-specific human neutralizing antibodies (HuNAbs) have been actively explored for passive immunization. In support of HuNAb-based immunotherapy, passive immunization using convalescent patients’ plasma has generated promising evidence on clinical benefits for both mild and severe COVID-19 patients. Since the source of convalescent plasma is limited, the discovery of broadly reactive HuNAbs may have significant impacts on the fight against the COVID-19 pandemic. In this review, therefore, we discuss the current technologies of gene cloning, modes of action, in vitro and in vivo potency and breadth, and clinical development for potent SARS-CoV-2-specific HuNAbs.
Keywords: SARS-CoV-2, neutralizing antibody, vaccine, immunotherapy, passive immunization
Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) is the causative agent of coronavirus disease 2019 (COVID-19) [1]. SARS-CoV-2 infects target cells mainly through the interaction between its spike (S) glycoprotein and the host cellular angiotensin-converting enzyme 2 (ACE2) receptor. The S protein is comprised of the S1 subunit and the S2 subunit. Within the S1 subunit, the receptor-binding domain (RBD) of SARS-CoV-2 contains the key functional determinant for binding with ACE2, whereas the S2 subunit engages the subsequent virus-cell membrane fusion for viral entry [2]. After the COVID-19 outbreak, both soluble RBD and trimeric S proteins have been extensively used as the bait for fishing out RBD- and S-specific human neutralizing antibodies (HuNAbs) during the processes of single B cell-, deep-sequencing- and phage display-based antibody gene cloning. The majority of HuNAbs obtained, therefore, block primarily the interaction between RBD/S and ACE2 for viral neutralization. Besides vaccine development, antiviral drugs including SARS-CoV-2-specific HuNAbs have also been actively explored for passive immunization. To support HuNAb-based immunotherapy, passive immunization using convalescent patients’ plasma has generated promising evidence on clinical benefits for both mild and severe COVID-19 patients [3, 4]. Recently, significant progresses in the discovery of HuNAb have resulted in HuNAb-based clinical immunotherapy for COVID-19 patients. This review aims to highlight these progresses to facilitate the knowledge exchange and the fight against COVID-19.
Cloning of monoclonal HuNAb
B cell receptor (BCR) repertoires exhibit high sequence diversity due to the somatic recombination and hypermutation during the B cell development. BCR is defined as a transmembrane receptor located on the B cell surface and interacts with a specific antigen epitope through its variable region to initiate antibody response. This variable region, therefore, shares the identical gene sequence with the antibody that is produced by this B cell. The somatic recombination of three gene segments of the heavy (H) chain locus (V, D, J) and two gene segments of the light (L) chain locus (V, J) to diversify the variable region gene. The variable region of an antibody immunoglobulin (Ig) determines the specificity for interaction with a corresponding viral antigenic epitope. The somatic hypermutation involves the B cell proliferation in the germinal center with random mutations in the genes encoding the variable region of individual monoclonal antibody (mAb), essential for high-affinity binding to an antigenic epitope, so-called the antibody affinity maturation process. There are no identical BCRs between two different B cells. To ensure native pairing of antibody H and L chains, it is necessary to analyze one B cell at a time for cloning an mAb [5]. With the advancement of antibody gene cloning techniques, such as hybridoma technology, human B cell immortalization, antibody phage display, human immunoglobulin transgenic mice and single B cell antibody technology [6], cloning of a functional HuNAb is no longer a search of a needle in the ocean. During the ongoing COVID-19 pandemic, most of the SARS-CoV-2-specific antibodies have been obtained through the single B cell technology.
Single B cell-based antibody cloning technique
Single B cell-based antibody cloning is a technique to obtain the variable region of mAb H/L genes from individual memory B cell that has the capacity to produce the high-affinity antibody. It involves the amplification of auto-paired Ig H/L chain RNA sequences from the heterogeneous memory B cell population and in vitro construction into functional mAbs [7, 8]. This technique has been successfully used for isolating neutralizing mAbs from convalescent patients against various viral infections such as the Epstein-Barr virus (EBV), human immunodeficiency virus (HIV), dengue, and SARS-CoV-2 [9–12]. Since 2009, the combination of single-cell RT-PCR and single B-cell sorting has improved the success rate of antibody gene cloning greatly. The acquirement of single antigen-binding memory B cell by fluorescence-activated cell sorting (FACS) or optofluidics platform is the major technical improvement, allowing subsequent nested RT-PCR using primers to amplify naturally paired antibody H/L gene sequences from individual memory B cells [13–16]. After the COVID-19 outbreak, this method was quickly utilized to isolate HuNAb from SARS-CoV-2-infected patients [11, 17]. Moreover, there is the development of high-throughput single-cell RNA and VDJ deep sequencing of BCR repertoires accompanied by the bioinformatics analysis [18, 19]. This technique has outcompeted the single-cell RT-PCR in terms of high-throughput screening of a large pool of diverse memory B cells. Interestingly, immunization using RBD/S proteins or direct infection of mice with the genetically humanized immune system can also generate complete HuNAbs against SARS-CoV-2 [20]. This murine platform may simplify the source of antigen-specific human B cells although the antibody affinity maturation process remains to be improved in this model. Nevertheless, using this platform, some SARS-CoV-2-specific HuNAbs have been successfully cloned and screened [20]. The deep sequencing of variable regions and BCR repertoire has revealed H/L pairing of human antibody characteristics [20]. In addition, the combination of microfluidic-based technique and bioinformatics analysis can further improve the efficiency of identifying highly potent HuNAbs against specific viral antigens, which has implications for the fight against COVID-19 and other emerging infectious diseases.
Phage display-based antibody cloning technique
Phage display has been one of the widely used methods for cloning human antibodies. The two key steps of the phage display technique include the construction of an antibody gene library and the screening of antigen-specific antibodies. The human Fab library can be generated from peripheral blood mononuclear cells (PBMCs) derived from COVID-19 patients. A set of primers targeting the variable H/L chain regions are used to amplify the total antibody gene pool for the construction of the phage library [21]. In solution panning, SARS-CoV-2 RBD can be used as the bait to fish out the RBD-specific human Fabs, followed by construction of the Fabs into full-length IgG1 for subsequent biochemical and functional testing. Alternatively, the human Fab antibody library can also be synthesized using the selected human germline immunoglobulin variable segments. The diversity in the complementarity-determining regions (CDR) 3 of H/L chains (CDR-L3 and CDR-H3) can be introduced by well-designed mutagenic oligonucleotides. A competitive phage screening strategy has been successfully used to obtain RBD-specific HuNAbs against SARS-CoV-2 [22]. Interestingly, to obtain a cross-reactive HuNAb against both SARS-CoV and SARS-CoV-2. Mice immunized with SARS-CoV RBD were used for the establishment of the phage display library. Then, this library was screened using the SARS-CoV-2 RBD as the bait. Since these two RBDs share 75% similarity in their amino acid sequence, the cross-reactive murine mAbs were successfully obtained for subsequent humanization against both SARS-CoV and SARS-CoV-2 [23]. The drawbacks of the phage display-based antibody cloning technique include the unnatural pairing of VH/VL and a time-consuming panning procedure. Nevertheless, this technique remains very useful for obtaining antigen-specific antibodies including potent SARS-CoV-2-specific HuNAbs.
Mode of action and structural basis of SARS-CoV-2-specific HuNAbs
The S glycoprotein of SARS-CoV-2 exists in a prefusion trimer conformation that may rearrange during the fusion of the virus-cell membrane. This viral entry is a dynamic process beginning with the binding of the viral S1 portion to the host cell receptor ACE2. The interaction between S1 and ACE2 makes the prefusion trimer unstable, leading to the cleavage of S into S1 and S2 by cellular proteases (e.g., Furin) with the transition into a stable post-fusion conformation. The RBD in S1 transiently hides or exposes the determinants of receptor binding, due to its hinge-like conformational movement, displaying two structural states with ‘up’ and ‘down’ conformations. The ‘up’ conformation is an active state for ACE2 binding [24]. Once in the ‘down’ conformation, RBD is usually stable but is inaccessible to ACE2. Because of this unique property, the ‘up’ conformation serves as the major target for SARS-CoV-2 NAbs. Interestingly, two major conformational structural regions within the S1 have been identified as SARS-CoV-2 neutralizing domains. Besides RBD, the N-terminal domain (NTD) also possesses binding sites for HuNAbs.
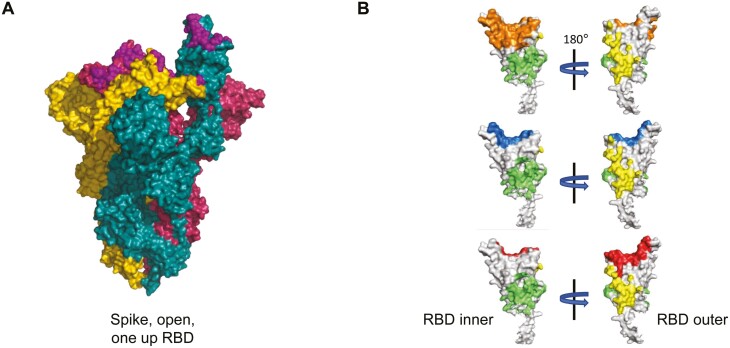
According to the CoV-AbDab, the coronavirus antibody database, there are amazingly a total number of 3152 SARS-CoV-2-specific antibodies by 23 September 2021, including 2693 mAbs and 459 nanobodies. One thousand hundred and forty-eight of the 2639 mAbs have neutralizing activities. In a separate report, among a total of 1584 RBD-specific antibodies, 902 have the neutralization ability [25]. By mapping the binding sites of a collection of mAbs against the S protein, NAbs bind to multiple S epitopes including ones in RBD, NTD, and quaternary regions [26, 27]. While RBD is the primary target for NAb, these antibodies can be classified into three main groups according to the distinct binding sites at the molecular level [28]. The first group of NAbs has epitopes within the RBD overlapping largely with the binding site of ACE2. These NAbs are described as receptor binding site (RBS) antibodies. Furthermore, due to different interacting angles with viral RBD, RBS NAbs are divided into three in-depth subclasses including RBS-A, RBS-B, and RBS-C [28]. As demonstrated in the crystal structures (Fig. 1), the binding sites and interaction direction of RBS-A antibodies with viral RBD shared the highest similarity with those of ACE2 binding. RBS-A antibodies bind to RBD like ACE2 mainly on the ‘left side’ of the ridge. Typically, both Fab-RBD and ACE2-RBD interactions show on one side of the ridge in the crystal complex. Compared to RBS-A antibodies, the epitope of RBS-B antibodies shows relatively less overlapping with the ACE2 binding site. RBS-B antibodies are positioned more upright and straddle the central of the ridge [28]. The RBS-C antibodies are the subclass of antibodies that have the least overlapping epitopes with ACE2 binding sites as they bind to viral RBD on the opposite side of the RBS-A antibodies. It should be emphasized that all three subclasses of RBS antibodies can compete with human ACE2 for SARS-CoV-2 neutralization. While several RBS-C antibodies can bind to the ‘up’ or ‘down’ RBD, the RBS-A antibodies can only interact with the ‘up’ RBD. The binding mode of RBS-B antibodies is in between as some can only bind to RBD in the ‘up’ state while others show in the ‘down’ state [28, 29]. The second group of NAb is represented by the cross-reactive CR3022 antibody that recognizes a cryptic site in RBD. CR3022 is an antibody isolated from a phage library of a convalescent SARS patient in 2003. CR3022 binds to a cross-reactive epitope on SARS-CoV-2 RBD but it does not neutralize SARS-CoV-2. The position of the CR3022 cryptic site is highly conserved near the ‘tail’ part of an ‘up’RBD. This binding site is common for cross-reactive antibodies against both SARS-CoV and SARS-CoV-2. Different from CR3022, however, several CR3022-like antibodies present various neutralizing activities against both SARS-CoV and SARS-CoV-2 [28]. The third group of NAb is represented by the S309 antibody that neutralizes both SARS-CoV-2 and SARS-CoV pseudoviruses as well as authentic SARS-CoV-2 by engaging the RBD of the S glycoprotein through binding to an epitope containing the N343 glycan, without competing with ACE2 interaction. The S309 antibody was identified from a SARS convalescent patient in 2003 as well. It binds to either the ‘up’ or the ‘down’RBD. The binding site of S309 is not as conserved as the one for the CR3022 antibody [28]. Besides three groups of RBD-specific NAbs, the mode of action for NTD-directed NAbs remains incompletely revealed [26, 27]. It is possible that the binding of these NAbs to NTD may affect indirectly the conformational interaction between RBD and ACE2. Since the S2 subunit is more conserved among coronaviruses than S1, some cross-reactive neutralizing antibodies specific to S2 of the S protein have been reported [30, 31]. Interestingly, SARS survivors have developed potent cross-clade pan sarbecovirus neutralizing antibodies after immunization with the BioNTech mRNA vaccine [32]. The findings of multiple NAb targets of vulnerability on SARS-CoV-2 S protein are useful not only for designing vaccine and antiviral but also for building the basis for HuNAb-cocktail treatment.
Figure 1.
Categorized recognition sites of SARS-CoV-2 RBD-specific mAbs. (A) Conformational structure of SARS-CoV-2 trimer spike glycoprotein (PDB: 7BNM, 1 up RBD) with the ACE2 binding footprints (purple) on RBDs. (B) The crystal structure of RBD (white) is displayed in two orientations (PDB:7BNM, amino acid position: 320–540). The binding sites of different subclasses of mAbs are labelled in orange for mAb CB6 (RBS-A, PDB ID: 7C01), blue for mAb 2-4 (RBS-B, PDB ID: 6XEY), red for mAb P2B-2F6 (RBS-C, PDB ID: 7BWJ), green for mAb CR3022 (PDB ID: 6W41) cryptic site, and yellow for mAb S309 (PDB ID: 6WPS) proteoglycan site.
In vitro potency of SARS-CoV-2-specific HuNAbs
To study the neutralizing potency of HuNAbs targeting the S glycoprotein of SARS-CoV-2, both pseudovirus and authentic virus neutralization assays have been developed. For the pseudovirus assay, two systems have been established through co-transfection with the functional S glycoprotein of SARS-CoV-2. One system employs the luciferase reporter in the backbone of lentivirus (e.g., HIV-1) [33, 34]. The other system employs the luciferase reporter in the backbone of the vesicular stomatitis virus (VSV) [26, 35]. The authentic neutralization assay uses primary SARS-CoV-2 strains isolated from COVID-19 patients directly. It should be noted that different research groups may use various types of target cells for in vitro neutralization assays, which may generate incomparable data to compare the neutralization potency. For example, various Vero cell lines and 293T cells stably expressing human ACE2 have been extensively used for viral neutralization assays. Bearing this in mind, one may refer to some representative HuNAbs in (Table 1). All these HuNAbs are generated in the native form of IgG1. Usually, HuNAbs with the half-maximal inhibitory concentrations (IC50) less than 0.1 µg/ml are categorized as potent neutralizers whereas those with IC50 values of 0.1–1 µg/ml and 1–10 µg/mL are considered as moderate and weak neutralizers, respectively [36]. The most potent SARS-CoV-2 HuNAb may display IC50 values at the single-digit ng/ml. For example, one of the most potent HuNAbs, namely 2–15, displays the IC50 value of as low as 0.7 ng/ml [26].
Table 1.
Characteristics of human neutralizing antibodies (HuNAbs)
| Antibody | Pseudovirus neutralization IC50(ng/ml) | Live virus neutralization IC50 (ng/ml) | Cross with SARS-CoV | Subclass | Reference |
|---|---|---|---|---|---|
| REGN10933 (casirivimab, CAS) | 10.4 | 5.61 | No | RBS-A | Hansen et al. (2020) [20] |
| REGN10987 (imdevimab, IMD) | 6.09 | 6.32 | No | RBS-C | Hansen et al. (2020) [20] |
| BD-368-2 | 1.2 | 15 | na∗ | RBS-C | Cao et al. (2020) [19] |
| P2C-1F11 (BRII196) | 30 | 30 | No | RBS-A | Ju et al. (2020) [68] |
| 2-15 | 5 | 0.7 | na | RBS-A | Liu et al. (2020) [26] |
| S309 | na | 79 | Yes | S309 proteoglycan site | Pinto et al. (2020) [69] |
| CV07-250 | na | 3.5 | No | RBS-B | Kreye et al. (2020) [70] |
| 47D11 | 61 | 570 | Yes | na | Wang et al. (2020) [71] |
| C105 | 26.1 | na | No | RBS-A | Barnes et al. (2020) [72] |
| COVA1-18 | 8 | 7 | No | RBS-A | Brouwer et al. (2020) [27] |
| COVA2-39 | 36 | 54 | No | RBS-B | Brouwer et al. (2020) [27] |
| CC6.29 | 2.0 | 7.1 | No | RBS-A | Rogers et al. (2020) [73] |
| CB6 (LY-CoV016, etesevimab, ETE) | 41 | 36 | No | RBS-A | Shi et al. (2020) [52] |
| Ab169 (LY-CoV555, bamlanivimab, BAM) | 0.012 | 0.02 | na∗ | RBS-B | Jones et al. (2021) [53] |
| H014 | 450 | 5700 | Yes | CR3022 cryptic site | Lv et al. (2020) [74] |
| ZB8 | 9.5 | 13 | No | RBS-B | Zhou et al. (2021) [46] |
∗Not available.
Efficacy of SARS-CoV-2-specific HuNAbs in animal models
Several animal models have been established to mimic the natural course of SARS-CoV-2 infection in humans. Since ACE2 contains natural variation in different animal species, their susceptibility to live SARS-CoV-2 infection may vary significantly. Based on the structural analysis, there are 29 amino acid residues at the interface of ACE2 and SARS-CoV-2 RBD, which determines the binding affinity of these two proteins. Sequence alignment of these 29 amino acid residues reveals high similarity between human ACE2 with homologues of Syrian golden hamster, rhesus macaque and common marmoset [37]. Since hamster ACE2 exhibits a high binding affinity with the S glycoprotein of SARS-CoV-2 by in silico prediction, this animal model exhibits acute clinical and histopathological manifestations that model the upper and lower respiratory tract infection in humans [37]. Non-human primates share 100% identity of these 29 amino acid residues with human ACE2 but only display a transient and mild clinical manifestation after the live SARS-CoV-2 challenge [38–40]. The amount of ACE2 and TMPRSS2 expression in upper and lower respiratory tracts of rhesus monkeys likely affects their susceptibility to live SARS-CoV-2 infection, which explains why the intratracheal viral inoculation has been used for a viral challenge in the experiments. Mouse is generally resistant to wild-type SARS-CoV-2 challenge because mouse ACE2 does not effectively bind the viral S glycoprotein [41]. However, after transduction with adenovirus- or adeno-associated virus-vectored human ACE2 (Ad5-hACE2 or AAV-hACE2), the mouse becomes susceptible to SARS-CoV-2 infection [42]. Meantime, transgenic mice with human ACE2 expression lead to mild or lethal SARS-CoV-2 infection [43]. Subsequently, SARS-CoV-2 with the N501Y mutation can interact with murine ACE2 and infect mouse species directly [44, 45]. These animal models have been widely used to evaluate the in vivo efficacy of various HuNAbs and vaccines against SARS-CoV-2 (Table 2). We and others reported that most of these HuNAbs can suppress viral loads in the lungs and alleviate lung injury in animal models. We, however, found that systemic HuNAb injection or DNA vaccination has limited efficacy in preventing SARS-CoV-2 nasal infection in Syrian hamsters likely due to insufficient biodistribution of antibody at the site of viral transmission [21]. Using the same model, we recently found that potent neutralizing dimeric IgA may enhance SARS-CoV-2 nasal infection probably by engaging alternative cellular entry and cell-to-cell transmission mechanisms [46]. In fact, few animal studies have shown significant reduction of nasal viral loads in various animal models post HuNAb injection and systemic vaccination [26, 47, 48], implicating that an exceptionally high dose is likely required for improved protection. Some published studies claim sterile protection without actual evaluation of SARS-CoV-2 nasal infection. These results explain the rising number of re-infections and over thousands of vaccine-breakthrough infections in humans [49–51]. The data generated in animal models, therefore, may have significant implications to human vaccine and passive immunization studies.
Table 2.
In vivo efficacy of HuNAbs determined in animal models
| Antibody | Dosage (mg/kg) | Animal model | Experiments | Efficacy | Reference |
|---|---|---|---|---|---|
| REGN-COV2 | 50, 5, 0.5 | Syrian hamster | Prophylactic (−2 day) | Alter the course of infection | Baum et al. (2020) [75] |
| Treatment (1 dpi∗) | Alter the course of infection | ||||
| 50, 0.3 | Rhesus macaque | Prophylactic (−3 day) | 50 mg/kg could minimize virus replication, but 0.3 mg/kg could not | ||
| 150, 25 | Treatment (1 dpi) | Accelerate viral clearance at both dosage | |||
| BD-368-2 | 20 | hACE2 transgenic mice | Prophylactic (−1 day) | Completely inhibit infection in lung | Cao et al. (2020) [19] |
| Therapeutic (2h after infection) | 3–4 logs decrease in virus copies in lung | ||||
| H014 | 50 | hACE2-humanized mice | Prophylactic (−12 hour) plus therapeutic (4 hours after infection) | 1–2 logs decrease in virus copies in lung | Lv et al. (2020) [74] |
| Therapeutic (4 hours after infection) | 0.5–1 log decrease in virus copies in lung | ||||
| 2-15 | 1.5 | Syrian hamster | Prophylactic (−1 day) | 4 logs or more decrease in virus copies in lung | Liu et al. (2020) [26] |
| 1B07 | 10 | AdV-hACE2- transducedBALB/c mice | Prophylactic (−1 day) | About 1 log decrease in virus copies in lung | Hassan et al. (2020) [20] |
| CB6 | 50 | Rhesus macaque | Prophylactic (−1 day) | 4–5 logs decrease in virus copies in lung | Shi et al., (2020) [52] |
| Treatment (1 dpi and 3 dpi) | 3–4 logs decrease in virus copies in lung | ||||
| COV2-2196/2130 | 0.2mg/mice | AdV-hACE2- transducedBALB/c mice | Prophylactic (−1day) | About 1 log decrease in virus copies in lung | Zost et al. (2020) [76] |
| 50 | Prophylactic (−3 day) | Completely inhibit infection in lung | |||
| Rhesus macaque | |||||
| CC12.1/C12.23 | 16.5, 4.2, 0.9, 0.2, 0.06 | Syrian hamster | Prophylactic (−1day) | 16.5, 4.2, 0.9 mg/kg could minimize virus replication, 0.2, but 0.06 mg/kg could not | Rogers et al. (2020) [73] |
∗dpi: day post infection.
Efficacy of SARS-CoV-2-specific HuNAbs in human trials
Several anti-SARS-CoV-2 HuNAbs have been developed for clinical studies. Paired HuNAbs have been formulated to improve the breadth of neutralization due to emerged SARS-CoV-2 variants and to minimize HuNAb escape variants. Most neutralizing antibodies under Emergency Use Authorization (EUA) showed live virus neutralization IC50 below 0.1 µg/ml. The paired antibodies often include one RBS-A HuNAb plus either one RBS-B or one RBS-C HuNAb. For example, the pair from the Eli Lily company included the RBS-B LY-CoV555 (bamlanivimab, BAM) and the RBS-A LY-CoV016 (etesevimab, ETE) with live virus-neutralizing IC50 around 0.02 µg/ml and 0.036 µg/ml, respectively [52, 53]. LY-CoV555 and LY-CoV016 were probably the first paired HuNAbs administered to COVID-19 patients in early June 2020 [54]. The randomized phase 2/3 trial evaluated the efficacy of combined LY-CoV555 (2800 mg) and LY-CoV016 (2800 mg) as compared with the LY-CoV555 monotherapy. A statistically significant reduction in viral load was found at day 11 among non-hospitalized patients with mild to moderate illness by the combination therapy but not by the monotherapy [54]. Subsequently, another clinical trial reported the REGN-CoV2 HuNAbs for prophylaxis and immunotherapy of COVID-19 patients. This phase 1/2/3 trial showed that both 2400 and 8000 mg of REGN-COV2 antibody cocktail showed an approximately 2-log reduction of viral loads in patients with baseline viral load higher than 107 copies/mL compared with the placebo group [55]. It should be noted that these clinical dosages are indeed much higher than 10–20 mg/kg tested in small animal models. Then, both companies have acquired permission from the U.S. Food and Drug Administration (FDA) for EUA of their HuNAbs. Now, these HuNAbs are used for emergency treatment of mild-to-moderate adult and pediatric patients (age 12 and older with body weight at least 40 kg) who have positive viral loads and are at high risk for progressing to severe COVID-19. Seven hundred milligram LY-CoV555 is recommended as monotherapy, or it can be combined with 1400 mg LY-CoV016. As for the REGN-CoV2, it consisted of paired RBS-A REGN10933 (casirivimab, CAS) and RBS-C REGN10987 (imdevimab, IMD) at a 1:1 ratio. REGN10933 and REGN10987 displayed live virus-neutralizing IC50 around 0.0056 µg/ml and 0.0063 µg/ml, respectively [20]. The recommended dosage of REGN-CoV2 is 2400 mg (1200 mg each) [56]. Recently, the combination of BRII-196 and BRII-198 was developed by the BRII Biosciences including one RBS-A and one RBS-C antibody, both with live virus neutralizing IC50 around 0.03 µg/ml. This antibody combination showed 78% efficacy in trials and has been approved for clinical use on 9 December 2021 by China FDA. Since antibody combination therapy may face reduced neutralizing abilities against SARS-CoV-2 variants of concerns (VOCs), more broadly reactive HuNAbs and next-generation antibodies, such as bispecific antibodies and engineered antibodies are on the list for clinical trials in different regions and countries.
Impact of SARS-CoV-2 VOCs on HuNAbs
SARS-CoV-2 is an RNA virus that is usually prone to mutate during the natural course of infection in humans. SARS-CoV-2, however, has a self-proof-correcting machinery system that maintains the relative genomic fidelity during viral replication [57]. Analysis of global phylogenies indicates a slow mutation rate of approximately two mutations per month in the viral genome [58]. Unlike SARS in 2003, since COVID-19 has not been eliminated till now, there is a growing concern on viral escapes due to immune pressure conferred by natural infection or vaccination. Indeed, the increasing and long-lasting COVID-19 pandemic has already resulted in several VOCs with various resistance profiles to HuNAbs.
A total number of 4,230,187 genome sequences of SARS-CoV-2 have been submitted to the hCoV-19 database of the Global initiative on sharing all influenza data (GISAID) since the outbreak of COVID-19. Several VOCs have had significant impacts on the trend of the pandemic. The top four noticeable VOCs include B.1.1.7 variant (Alpha, United Kingdom), B.1.351 (Beta, South Africa), P1 (Gamma, Japan/Brazil) and B.1.617.2 (Delta, India) [59]. Since late November 2021, a new VOC, B.1.1.529 (Omicron, South Africa) has been predicted to become another dominated strain. The Omicron variant was first discovered in South Africa with heavily mutated S glycoprotein containing 32 mutations including 15 in the RBD region, in addition to three deletions and one insertion [60]. The mutations or deletions of amino acids in the S glycoprotein of VOCs have led to not only HuNAb resistance but also probably increased the affinity binding to human ACE2 for higher viral transmissibility [61, 62]. Critically, all these five VOCs contain the D614G mutation, which enhances viral transmissibility and infectivity [62]. Moreover, the E484 position in RBD is an important binding site for many RBD-specific HuNAbs [61]. Unfortunately, VOCs of B.1.351 and B.1.1.529 have the E484K/A mutation. Moreover, some amino acid deletions in the S protein, such as the 242-244del in B.1.351 variants and the 144del in B.1.1.7 variants reduced more than the 1000-fold neutralizing activity of the NAbs that target the supersite of NTD domain [35]. To date, the impact of S mutations in Omicron variants on the ongoing pandemic remains unknown. Various combinations of these mutations and deletions in emerging VOCs, however, have substantially reduced the potency of HuNAbs elicited by vaccines and passive immunization, and therefore, becomes new challenges to public health and vaccine responses. Hopefully, the boost vaccine and immunotherapy using cocktailed HuNAbs can overcome these VOCs.
Future perspectives
With continuously emerging VOCs, vaccine-induced correlates of immune protection remain to be comprehensively investigated. Due to the urgency of the COVID-19 pandemic, huge global efforts have been placed for vaccine development. Till now, six vaccines have been approved by regulatory agencies for emergency use within just one year of the pandemic including (1) two mRNA-based vaccines, namely BNT162b2 (by Pfizer Inc. and BioNTech SE) and mRNA-1273 (by Moderna), expressing full S glycoprotein with an efficacy rate of 95% [63, 64] (2) the chimpanzee adenovirus-vectored vaccine, named ChAdOx1 nCoV-19 (by the Oxford University and AstraZeneca Inc.), encoding the full S glycoprotein with an efficacy rate of 70.4% [65] (3) the human adenovirus-vectored vaccine, namely Ad26.COV2.S (by Johnson & Johnson Inc.), encoding the full S glycoprotein with an efficacy rate of 73.1%, and (4) two inactivated vaccines CoronaVac and BIBP (by Sinovac Biotech and SinoPharm) with an efficacy rate of 83.5% and 78.1%, respectively. Since these efficacy rates were mainly determined for the prevention of severe diseases, their potency in preventing SARS-CoV-2 nasal infection and eliciting broadly reactive HuNAbs against VOCs remains to be carefully investigated. In recent studies, the B.1.351 variant was significantly resistant (10.3–12.4-fold) to neutralization by sera derived from vaccinated individuals (Moderna or BioNTech) compared to the D614G strain [35]. Although vaccinations reduced death rates, vaccine-induced attenuation of peak viral burden has decreased for the B.1.617.2 variant (absolute difference of 10–13% for BNT162b2 and 16% for ChAdOx1) compared to the B.1.1.7 variant in the UK [66]. These results are in line with the increasing number of breakthrough infections among the fully vaccinated population [49]. Nevertheless, extensive vaccination programs among general populations have contributed greatly to reduced numbers of hospitalization, mortality, and infections in countries like Israel, the UK, and the USA even when some VOCs have displayed reduced neutralization sensitivity [35]. Future boost vaccination and HuNAb-based immunotherapy should focus on broadly reactive immune protection [67]. Since systemic HuNAb did not completely prevent SARS-CoV-2 nasal infection, the role of mucosal immunity especially tissue-resident memory T-cells in the upper respiratory tract should be studied for long-term protection. Lastly, antibody-mediated enhancement of SARS-CoV-2 infection and immunopathogenesis should still be carefully monitored in the context of human infections and clinical care.
Acknowledgments
This review was submitted on behalf of the research group from the AIDS Institute and Department of Microbiology, Li Ka Shing Faculty of Medicine, the University of Hong Kong and Centre for Virology, Vaccinology and Therapeutics, Hong Kong Science and Technology Park. The Editor-in-Chief, Tim Elliott, and handling editor, Tao Dong, would like to thank the following reviewer, Ricardo Fernandes, and an anonymous reviewer, for their contribution to the publication of this article.
Glossary
Abbreviations
- AAV-hACE2
Adeno-associated virus-vectored human ACE2
- ACE2
Angiotensin-converting enzyme 2
- BAM
Bamlanivimab
- BCR
B cell receptor
- CAS
Casirivimab
- CDR
Complementarity-determining regions
- COVID-19
Coronavirus disease 2019
- EBV
Epstein-Barr virus
- ETE
Etesevimab
- EUA
Emergency Use Authorization
- FACS
Fluorescence-activated cell sorting
- FDA
The U.S. Food and Drug Administration
- GISAID
Global initiative on sharing all influenza data
- H
Heavy chain
- HIV
Human immunodeficiency virus
- HuNAbs
human neutralizing antibodies
- Ig
Immunoglobulin
- IMD
Imdevimab
- L
Light chain
- mAb
Monoclonal antibody
- NTD
N-terminal domain
- PBMC
Peripheral blood mononuclear cell
- RBD
Receptor-binding domain
- RBS
Receptor binding site
- S
Spike
- SARS-CoV-2
Severe acute respiratory syndrome coronavirus 2
- TMPRSS2
transmembrane serine protease 2
- VOC
Variants of concern
- VSV
Vesicular stomatitis virus
Author contributions
D.Z. and Z.C. wrote the manuscript. R.Z. contributed to the data and revision of the manuscript.
Funding
This work was funded by the Research Grants Council Collaborative Research Fund (C7156-20G to Z.C.), Hong Kong Special Administrative Region; the University Development Fund and Li Ka Shing Faculty of Medicine Matching Fund from the University of Hong Kong to the AIDS Institute; Health@InnoHK, Innovation and Technology Commission, the Government of the Hong Kong Special Administrative Region; and generous donations including the Friends of Hope Education Fund. Z.C.’s team was also partly supported by the Theme-Based Research Scheme (T11-706/18-N to Z.C.).
Conflict of interest
D.Z. and Z.C. are co-inventors of the ZDY HuNAbs.
Data availability
No data are available as this is a review article.
References
- 1. Wiersinga WJ, Rhodes A, Cheng AC, et al. Pathophysiology, transmission, diagnosis, and treatment of coronavirus disease 2019 (COVID-19): A review. JAMA 2020;324:782–93. [DOI] [PubMed] [Google Scholar]
- 2. Lan J, Ge J, Yu J, et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 2020;581:215–20. [DOI] [PubMed] [Google Scholar]
- 3. Shen C, Wang Z, Zhao F, et al. Treatment of 5 critically Ill patients with COVID-19 with convalescent plasma. JAMA 2020;323:1582–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chen L, Xiong J, Bao L, et al. Convalescent plasma as a potential therapy for COVID-19. Lancet Infect Dis 2020;20:398–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bassing CH, Swat W, Alt FW.. The mechanism and regulation of chromosomal V(D)J recombination. Cell 2002;109(Suppl):S45–55. [DOI] [PubMed] [Google Scholar]
- 6. Tiller T. Single B cell antibody technologies. N Biotechnol 2011;28:453–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. El Debs B, Utharala R, Balyasnikova IV, et al. Functional single-cell hybridoma screening using droplet-based microfluidics. Proc Natl Acad Sci USA 2012;109:11570–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Niu X, Zhao L, Qu L, et al. Convalescent patient-derived monoclonal antibodies targeting different epitopes of E protein confer protection against Zika virus in a neonatal mouse model. Emerg Microbes Infect 2019;8:749–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Walker LM, Phogat SK, Chan-Hui PY, et al. Protocol G Principal Investigators. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science 2009;326:285–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Corti D, Zhao J, Pedotti M, et al. Prophylactic and postexposure efficacy of a potent human monoclonal antibody against MERS coronavirus. Proc Natl Acad Sci USA 2015;112:10473–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Scheid JF, Mouquet H, Feldhahn N, et al. Broad diversity of neutralizing antibodies isolated from memory B cells in HIV-infected individuals. Nature 2009;458:636–40. [DOI] [PubMed] [Google Scholar]
- 12. Setthapramote C, Sasaki T, Puiprom O, et al. Human monoclonal antibodies to neutralize all dengue virus serotypes using lymphocytes from patients at acute phase of the secondary infection. Biochem Biophys Res Commun 2012;423:867–72. [DOI] [PubMed] [Google Scholar]
- 13. Tiller T, Meffre E, Yurasov S, et al. Efficient generation of monoclonal antibodies from single human B cells by single cell RT-PCR and expression vector cloning. J Immunol Methods 2008;329:112–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wardemann H, Yurasov S, Schaefer A, et al. Predominant autoantibody production by early human B cell precursors. Science 2003;301:1374–7. [DOI] [PubMed] [Google Scholar]
- 15. Wrammert J, Smith K, Miller J, et al. Rapid cloning of high-affinity human monoclonal antibodies against influenza virus. Nature 2008;453:667–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Liao HX, Levesque MC, Nagel A, et al. High-throughput isolation of immunoglobulin genes from single human B cells and expression as monoclonal antibodies. J Virol Methods 2009;158:171–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang L, Shi W, Chappell JD, et al. Importance of neutralizing monoclonal antibodies targeting multiple antigenic sites on the middle east respiratory syndrome coronavirus spike glycoprotein to avoid neutralization escape. J Virol. 2018;92:e02002–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zost SJ, Gilchuk P, Chen RE, et al. Rapid isolation and profiling of a diverse panel of human monoclonal antibodies targeting the SARS-CoV-2 spike protein. Nat Med 2020;26:1422–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cao Y, Su B, Guo X, et al. Potent neutralizing antibodies against SARS-CoV-2 identified by high-throughput single-cell sequencing of convalescent patients’ B cells. Cell 2020;182:73–84.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hansen J, Baum A, Pascal KE, et al. Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science 2020;369:1010–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhou D, Chan JF, Zhou B, et al. Robust SARS-CoV-2 infection in nasal turbinates after treatment with systemic neutralizing antibodies. Cell Host Microbe 2021;29:551–563.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zeng X, Li L, Lin J, et al. Isolation of a human monoclonal antibody specific for the receptor binding domain of SARS-CoV-2 using a competitive phage biopanning strategy. Antib Ther 2020;3:95–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lv Z, Deng YQ, Ye Q, et al. Structural basis for neutralization of SARS-CoV-2 and SARS-CoV by a potent therapeutic antibody. Science 2020;369:1505–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wrapp D, Wang N, Corbett KS, et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 2020;367:1260–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Raybould MIJ, Kovaltsuk A, Marks C, et al. CoV-AbDab: the coronavirus antibody database. Bioinformatics 2021;37:734–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liu L, Wang P, Nair MS, et al. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature 2020;584:450–6. [DOI] [PubMed] [Google Scholar]
- 27. Brouwer PJM, Caniels TG, van der Straten K, et al. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science 2020;369:643–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yuan M, Liu H, Wu NC, et al. Recognition of the SARS-CoV-2 receptor binding domain by neutralizing antibodies. Biochem Biophys Res Commun 2021;538:192–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Niu L, Wittrock KN, Clabaugh GC, et al. A structural landscape of neutralizing antibodies against SARS-CoV-2 receptor binding domain. Front Immunol 2021;12:647934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Song G, He WT, Callaghan S, et al. Cross-reactive serum and memory B-cell responses to spike protein in SARS-CoV-2 and endemic coronavirus infection. Nat Commun 2021;12:2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nguyen-Contant P, Embong AK, Kanagaiah P, et al. S Protein-reactive IgG and memory B cell production after human SARS-CoV-2 infection includes broad reactivity to the S2 subunit. mBio. 2020;11:e01991–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tan CW, Chia WN, Young BE, et al. Pan-Sarbecovirus neutralizing antibodies in BNT162b2-immunized SARS-CoV-1 survivors. N Engl J Med 2021;385:1401–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Liu L, To KK, Chan KH, et al. High neutralizing antibody titer in intensive care unit patients with COVID-19. Emerg Microbes Infect 2020;9:1664–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhou R, To KK, Wong YC, et al. Acute SARS-CoV-2 infection impairs dendritic cell and T cell responses. Immunity 2020;53:864–877.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang P, Nair MS, Liu L, et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature 2021;593:130–5. [DOI] [PubMed] [Google Scholar]
- 36. Rogers TF, Zhao F, Huang D, et al. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science 2020;369:956–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chan JF, Zhang AJ, Yuan S, et al. Simulation of the clinical and pathological manifestations of coronavirus disease 2019 (COVID-19) in a Golden Syrian Hamster Model: Implications for disease pathogenesis and transmissibility. Clin Infect Dis 2020;71:2428–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rockx B, Kuiken T, Herfst S, et al. Comparative pathogenesis of COVID-19, MERS, and SARS in a nonhuman primate model. Science 2020;368:1012–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Munster VJ, Feldmann F, Williamson BN, et al. Respiratory disease in rhesus macaques inoculated with SARS-CoV-2. Nature 2020;585:268–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Speranza E, Williamson BN, Feldmann F, et al. Single-cell RNA sequencing reveals SARS-CoV-2 infection dynamics in lungs of African green monkeys. Sci Transl Med. 2021;13:eabe8146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhou P, Yang XL, Wang XG, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020;579:270–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sun J, Zhuang Z, Zheng J, et al. Generation of a broadly useful model for COVID-19 pathogenesis, vaccination, and treatment. Cell 2020;182:734–743.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Muñoz-Fontela C, Dowling WE, Funnell SGP, et al. Animal models for COVID-19. Nature 2020;586:509–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Niu Z, Zhang Z, Gao X, et al. N501Y mutation imparts cross-species transmission of SARS-CoV-2 to mice by enhancing receptor binding. Signal Transduct Target Ther 2021;6:284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sun S, Gu H, Cao L, et al. Characterization and structural basis of a lethal mouse-adapted SARS-CoV-2. Nat Commun 2021;12:5654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhou B, Zhou R, Chan JF-Wet al. SARS-CoV-2 hijacks neutralizing dimeric IgA for enhanced nasal infection and injury. bioRxiv. 2021. doi: 10.1101/2021.10.05.463282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. van Doremalen N, Lambe T, Spencer A, et al. ChAdOx1 nCoV-19 vaccine prevents SARS-CoV-2 pneumonia in rhesus macaques. Nature 2020;586:578–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Corbett KS, Edwards DK, Leist SR, et al. SARS-CoV-2 mRNA vaccine design enabled by prototype pathogen preparedness. Nature 2020;586:567–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hacisuleyman E, Hale C, Saito Y, et al. Vaccine breakthrough infections with SARS-CoV-2 variants. N Engl J Med 2021;384:2212–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Farinholt T, Doddapaneni H, Qin X, et al. Transmission event of SARS-CoV-2 delta variant reveals multiple vaccine breakthrough infections. BMC Med 2021;19:255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Blanquart F, Abad C, Ambroise J, et al. Characterisation of vaccine breakthrough infections of SARS-CoV-2 delta and alpha variants and within-host viral load dynamics in the community, France, June to July 2021. Eurosurveillance. 2021;26:2100824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Shi R, Shan C, Duan X, et al. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature 2020;584:120–4. [DOI] [PubMed] [Google Scholar]
- 53. Jones BE, Brown-Augsburger PL, Corbett KS, et al. The neutralizing antibody, LY-CoV555, protects against SARS-CoV-2 infection in nonhuman primates. Sci. Transl. Med. 2021;13:eabf1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Gottlieb RL, Nirula A, Chen P, et al. Effect of bamlanivimab as monotherapy or in combination with etesevimab on viral load in patients with mild to moderate COVID-19: a randomized clinical trial. JAMA 2021;325:632–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Weinreich DM, Sivapalasingam S, Norton T, et al. Trial investigators. REGN-COV2, a neutralizing antibody cocktail, in outpatients with Covid-19. N Engl J Med 2021;384:238–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Razonable RR, Pawlowski C, O’Horo JC, et al. Casirivimab-Imdevimab treatment is associated with reduced rates of hospitalization among high-risk patients with mild to moderate coronavirus disease-19. Eclinicalmedicine 2021;40:101102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Romano M, Ruggiero A, Squeglia F, et al. A structural view of SARS-CoV-2 RNA replication machinery: RNA synthesis, proofreading and final capping. Cells. 2020;9:1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Gupta RK. Will SARS-CoV-2 variants of concern affect the promise of vaccines? Nat Rev Immunol 2021;21:340–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Chakraborty C, Bhattacharya M, Sharma AR.. Present variants of concern and variants of interest of severe acute respiratory syndrome coronavirus 2: their significant mutations in S-glycoprotein, infectivity, re-infectivity, immune escape and vaccines activity. Rev Med Virol 2021:e2270. [Google Scholar]
- 60. Pulliam JRC, van Schalkwyk C, Govender N, et al. Increased risk of SARS-CoV-2 reinfection associated with emergence of the Omicron variant in South Africa. medRxiv. 2021. doi: 10.1101/2021.11.11.21266068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Jangra S, Ye C, Rathnasinghe R, et al. Personalized virology initiative study group. SARS-CoV-2 spike E484K mutation reduces antibody neutralisation. Lancet Microbe 2021;2:e283–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Plante JA, Liu Y, Liu J, et al. Spike mutation D614G alters SARS-CoV-2 fitness. Nature 2021;592:116–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Polack FP, Thomas SJ, Kitchin N, et al. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N Engl J Med 2020;383:2603–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Baden LR, El Sahly HM, Essink B, et al. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N Engl J Med 2021;384:403–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Voysey M, Clemens SAC, Madhi SA, et al. Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: an interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. Lancet 2021;397:99–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Pouwels KB, Pritchard E, Matthews PC, et al. Effect of delta variant on viral burden and vaccine effectiveness against new SARS-CoV-2 infections in the UK. Nat Med 2021;27:2127–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Baum A, Fulton BO, Wloga E, et al. Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies. Science 2020;369:1014–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Ju B, Zhang Q, Ge J, et al. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature 2020;584:115–9. [DOI] [PubMed] [Google Scholar]
- 69. Pinto D, Park YJ, Beltramello M, et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature 2020;583:290–5. [DOI] [PubMed] [Google Scholar]
- 70. Kreye J, Reincke SM, Kornau HC, et al. A therapeutic non-self-reactive SARS-CoV-2 antibody protects from lung pathology in a COVID-19 hamster model. Cell 2020;183:1058–1069.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Wang C, Li W, Drabek D, et al. A human monoclonal antibody blocking SARS-CoV-2 infection. Nat Commun 2020;11:2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Barnes CO, Jette CA, Abernathy ME, et al. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature 2020;588:682–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Rogers TF, Zhao F, Huang D, et al. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science. 2020:eabc7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Lv Z, Deng Y-Q, Ye Q, et al. Structural basis for neutralization of SARS-CoV-2 and SARS-CoV by a potent therapeutic antibody. Science. 2020:eabc5881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Baum A, Ajithdoss D, Copin R, et al. REGN-COV2 antibodies prevent and treat SARS-CoV-2 infection in rhesus macaques and hamsters. Science 2020;370:1110–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Zost SJ, Gilchuk P, Case JB, et al. Potently neutralizing and protective human antibodies against SARS-CoV-2. Nature 2020;584:443–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No data are available as this is a review article.