Abstract
With the outbreak of severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), coronaviruses have begun to attract great attention across the world. Of the known human coronaviruses, however, Middle East respiratory syndrome coronavirus (MERS-CoV) is the most lethal. Coronavirus proteins can be divided into three groups: nonstructural proteins, structural proteins, and accessory proteins. While the number of each of these proteins varies greatly among different coronaviruses, accessory proteins are most closely related to the pathogenicity of the virus. We found for the first time that the ORF3 accessory protein of MERS-CoV, which closely resembles the ORF3a proteins of severe acute respiratory syndrome coronavirus and SARS-CoV-2, has the ability to induce apoptosis in cells in a dose-dependent manner. Through bioinformatics analysis and validation, we revealed that ORF3 is an unstable protein and has a shorter half-life in cells compared to that of severe acute respiratory syndrome coronavirus and SARS-CoV-2 ORF3a proteins. After screening, we identified a host E3 ligase, HUWE1, that specifically induces MERS-CoV ORF3 protein ubiquitination and degradation through the ubiquitin–proteasome system. This results in the diminished ability of ORF3 to induce apoptosis, which might partially explain the lower spread of MERS-CoV compared to other coronaviruses. In summary, this study reveals a pathological function of MERS-CoV ORF3 protein and identifies a potential host antiviral protein, HUWE1, with an ability to antagonize MERS-CoV pathogenesis by inducing ORF3 degradation, thus enriching our knowledge of the pathogenesis of MERS-CoV and suggesting new targets and strategies for clinical development of drugs for MERS-CoV treatment.
Keywords: MERS-CoV, ORF3, apoptosis, ubiquitination, degradation, HUWE1
Abbreviations: BTM, bortezomib; CHX, cycloheximide; CQ, chloroquine; IFN, interferon; KEGG, Kyoto Encyclopedia of Genes and Genomes; M, membrane; MERS-CoV, Middle East respiratory syndrome coronavirus; N, nucleocapsid; NSPs, nonstructural proteins; PROTAC, protein hydrolysis targeting chimera; SARS-CoV, severe acute respiratory syndrome coronavirus; SARS-CoV-2, severe acute respiratory syndrome coronavirus-2
Coronaviruses are a group of related RNA viruses with envelope and a linear single positive strand genome (1, 2, 3, 4, 5) that infect a broad range of hosts, producing symptoms and diseases ranging from the common cold to severe and fatal illnesses. At present, seven coronaviruses can infect humans, including HCoV-229E, HCoV-OC43, HCoV-NL63, HCoV-HKU1, severe acute respiratory syndrome coronavirus (SARS-CoV), Middle East respiratory syndrome coronavirus (MERS-CoV), and severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) (3, 6, 7). With the emergence of SARS-CoV-2 at the end of 2019, public attention to coronaviruses has increased in an unprecedented manner (8, 9, 10). Its global spread has brought great threat to human health and social stability. As of October 8, 2021, the number of confirmed cases has reached nearly 214 million with a case fatality rate of 2% (https://covid19.who.int). While public health measures and immunization programs have been effective to control the pandemic, there remains a need for a more in-depth and systematic research efforts to understand the pathogenesis of coronaviruses.
Of the seven coronaviruses with ability to infect humans, SARS-CoV, MERS-CoV, and SARS-CoV-2 cause severe acute respiratory syndrome that could be fatal (11, 12, 13). Among these three coronaviruses, MERS-CoV has the highest pathogenicity and mortality of about 35%. Since the first MERS-CoV outbreak in the Middle East in 2012, there have been confirmed 2468 cases and 851 deaths (14, 15, 16, 17). Unlike SARS-CoV-2, the distribution of MERS-CoV had been limited with just few countries reporting laboratory-confirmed infections. But because of its high mortality, the pathogenicity of MERS-CoV proteins warrant further investigations since such efforts could provide new strategies and targets for the prevention and treatment of MERS-CoV and other emerging coronaviruses.
The typical genome of a coronavirus is around 30 kb. MERS-CoV genome is 30.12 kb, which can encode 25 viral proteins (18). Coronavirus proteins can be divided into three categories: nonstructural proteins (NSPs) that are translated by frameshift translation of polyproteins under the cleavage of proteases, structural proteins such as spike, envelope, membrane (M), and nucleocapsid (N), and accessory proteins of varying numbers in different coronavirus (19). NSPs are generally involved in virus replication, including multiple enzymes such as papain-like protease (20, 21), main protease (22, 23) and RNA polymerase (24). The structural proteins form the protein shell, protecting the genome of the virus and mediating the virus invasion in host cells (25). However, the number of accessory proteins in different coronaviruses varies. At present, the prevailing view is that accessory proteins are related to the pathogenicity of coronaviruses (26, 27, 28, 29).
When the coronavirus invades the host, the viral protein encoded by the viral genome can affect the normal physiological functions or antagonize the immune response of the host. This can be achieved through various mechanisms including inhibition of interferon (IFN) production, interference with host cell metabolism, or induction of cell apoptosis (30). Some studies have reported that SARS-CoV ORF3a protein activated nuclear factor kappa B and NACHT, LRR and PYD domains-containing protein 3 inflammasome by promoting TNF receptor-associated factor 3-dependent ubiquitination of p105 and apoptosis-associated speck-like protein containing a caspase recruitment domain (31). Another accessory protein of SARS-CoV, ORF9b, induced a strong intracellular autophagic effect (32). ORF6, ORF8, and N protein of SARS-CoV-2 could inhibit the production of IFN-β (33, 34, 35), while ORF3a protein induced apoptosis in cells (36). For MERS-CoV, accessory proteins ORF4a, ORF4b, and ORF5 and structural proteins M and N demonstrated anti-IFN ability (37), among which ORF4b antagonized type I IFN production in both cytoplasm and nucleoplasm (38, 39). In addition, MERS-CoV membrane protein triggered apoptosis by activating protein kinase R-like endoplasmic reticulum kinase signaling (40). In contrast to SARS-CoV and SARS-CoV-2, the pathogenesis of MERS-CoV is poorly studied despite its high mortality rate.
An evolutionary tug-of-war exists between a virus and its host. The host seeks to eliminate the pathogen, while the virus aims to achieve immune escape. During virus infection of host cells and disease development, ubiquitination plays a double-edged sword (41). On one hand, viral proteins can induce or promote the ubiquitination and degradation of certain key cytokines, thereby weakening the immune response of cells or tampering their normal functions. For example, N protein of SARS-CoV and SARS-Cov-2 could inhibit the ubiquitination of RIG-I, which in turn weaken its activation (42, 43). SASR-CoV ORF8b induced degradation of IRF3 by the ubiquitin–proteasome pathway (44), and SARS-CoV-2 M protein induced TBK1 ubiquitination and degradation (45). All of these viral proteins can lead to suppression of the innate immune pathway. Nonetheless, the host cell can also employ a multitude of mechanisms to achieve viral clearance. Host cells could use its own E3 ligases to specifically degrade certain viral proteins, thereby reducing virulence of the virus. For instance, NSP16 and ORF8b of SARS-CoV and ORF9c of SARS-CoV-2 could be degraded through ubiquitin–proteasome system in the cells (46, 47, 48).
In our current work, we discovered that MERS-CoV ORF3 protein can induce cell apoptosis. By mass spectrometry analysis and experimental verification, we showed that ORF3 was an unstable protein that could be degraded through the ubiquitin–proteasome pathway in the host cell. By examining ORF3 protein interactome, we found that the E3 ligase HUWE1 regulated the ubiquitin-mediated degradation of ORF3. Moreover, HUWE1 weakened the ability of ORF3 to induce apoptosis. Our work reveals a new mechanism for the host cells to antagonize MERS-CoV and provides a new target for drug treatment of MERS-CoV.
Results
The ORF3 protein of MERS-CoV induces apoptosis in cells
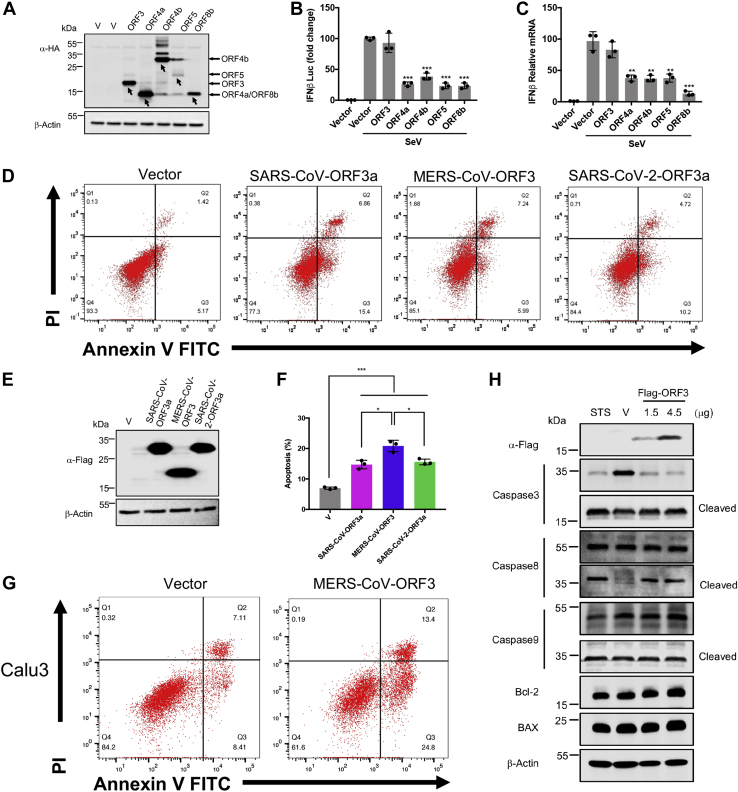
MERS-CoV encodes five accessory proteins: ORF3, ORF4a, ORF4b, ORF5, and ORF8b. To test whether these proteins are related to the pathogenicity of the virus, we tested their anti-IFN ability by luciferase reporter assay and qPCR. Under the condition of Sendai virus stimulation, the four accessory proteins—excluding ORF3—can inhibit the production of IFN-β (Fig. 1, A–C). This suggested that ORF3 may play another role that was distinct from the roles of the other accessory proteins. Because ORF3a protein of SARS-CoV and SARS-CoV-2 was reported to promote cell apoptosis (36, 49), we speculated whether the ORF3 protein of MERS-CoV had a similar function. We overexpressed ORF3a protein of SARS-CoV and SARS-CoV-2 as well as ORF3 protein of MERS-CoV in HEK293T cells. As shown in the results, ORF3 protein of MERS-CoV significantly increased the proportion of apoptotic cells, similar to ORF3a protein of SARS-CoV and SARS-CoV-2 (Fig. 1, D–F).
Figure 1.
The ORF3 protein of MERS-CoV induces apoptosis in cells. A, HEK293T cells were transfected with the indicated plasmids expressing HA-tagged MERS-CoV accessory proteins. Forty-eight hours later, cells were collected for immunoblotting. B, HEK293T cells were cotransfected with empty vector or expression plasmids for MERS-CoV accessory proteins as indicated, together with IFNβ-luc and pRL-TK plasmids. Twenty-four hours after transfection, the cells were infected with Sendai virus (100 HAU/ml) for 12 h and lysed for dual luciferase assay. C, HEK293T cells transfected with indicated plasmids were infected with Sendai virus for 12 h. Total RNA was extracted, reverse transcribed, and analyzed by real-time PCR with primers specific for IFNβ. D and E, HEK293T cells were transfected with the indicated plasmids expressing Flag-tagged SARS-CoV ORF3a, MERS-CoV ORF3, and SARS-CoV-2 ORF3a. After 24 h, 90% cells were stained with Annexin V-FITC/PI for flow cytometric analysis (D), and the remaining cells were lysed for immunoblotting (E). F, the percentage of apoptotic cells were measured. Error bars indicate SD of technical triplicates. Statistical significance was calculated using unpaired, two-tailed Student’s t test. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001. G, Calu3 cells were transfected with the indicated plasmids expressing MERS-CoV ORF3. After 24 h, cells were stained with Annexin V-FITC/PI for flow cytometric analysis. H, HEK293T cells were transfected with empty vector or Flag-MERS-CoV ORF3 as indicated. After 24 h, cells were subjected for immunoblotting using the indicated antibodies. Cells treated with staurosporine (STS), an apoptosis inducer, for 5 h were used as a positive control. HAU, hemagglutinating unit; IFNβ-luc, interferon beta promoter-driven firefly luciferase reporter; MERS-CoV, Middle East respiratory syndrome coronavirus; pRL-TK, HSV-thymidine kinase promoter-driven Renilla luciferase reporter; SARS-CoV, severe acute respiratory syndrome coronavirus; SARS-CoV-2, severe acute respiratory syndrome coronavirus-2.
MERS-CoV is a virus that mainly infects the respiratory tract. To explore how ORF3 could affect the respiratory tract, we overexpressed ORF3 protein in normal lung cell BEAS-2B and lung cancer cells Calu3 and A549. Consistent with our results from HEK293T, ORF3 enhanced apoptosis in all of these cell lines (Figs. 1G and S1, A and B). To further elucidate this ORF3-induced apoptosis, we determined the levels and activation of proteins involved in apoptosis. We found that ORF3 induced the cleavage/activation of caspase-8 and caspase-3, whereas no apparent changes on BAX and Bcl-2 protein levels. This suggested that ORF3 induced cell death through the extrinsic pathway of apoptosis, not the mitochondrial pathway (Figs. 1H and S1C). Overall, the results implied that the ORF3 protein of MERS-CoV induced pathological changes in the host by inducing apoptosis in cells.
MERS-CoV ORF3 is an unstable protein
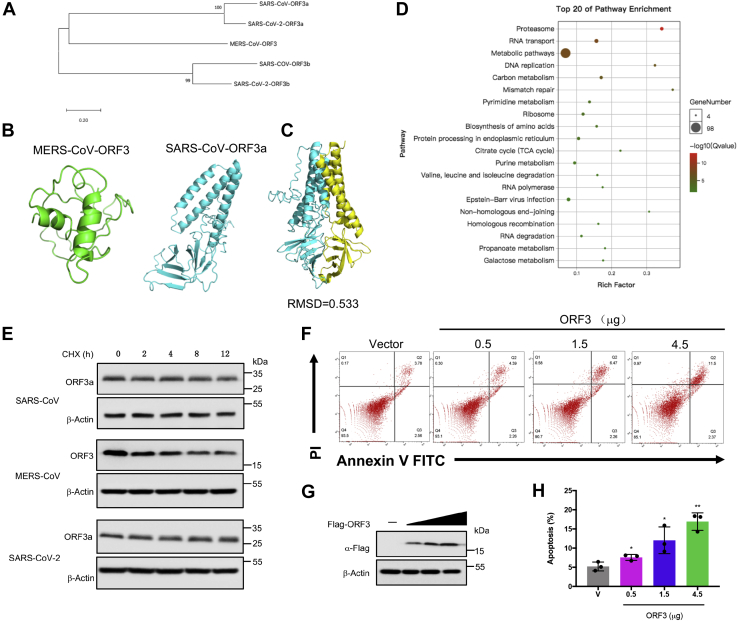
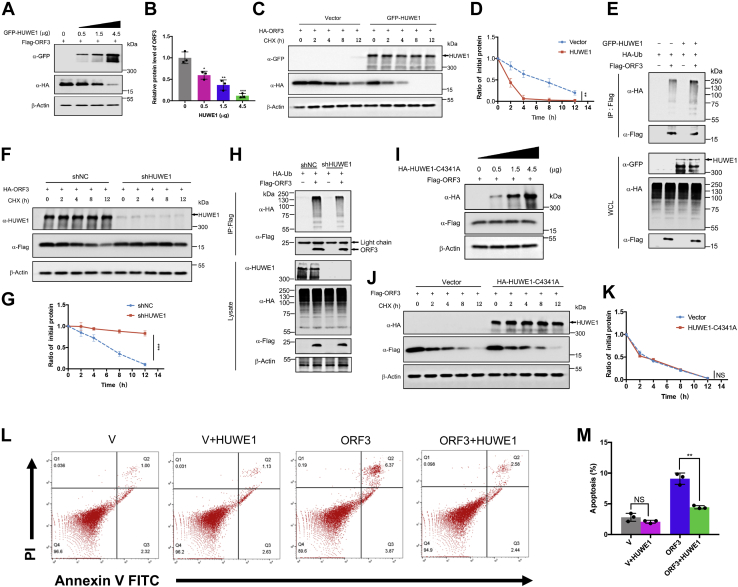
While ORF3 of MERS-CoV-2 and ORF3a of SARS-CoV and SARS-CoV-2 had similar function, they differed in terms of amino acid sequences. The amino acid sequence homology was higher between ORF3a/b of SARS-CoV and SARS-CoV-2, which is not surprising given the close evolutionary relatedness of the two viruses (5). But the ORF3 had low homology with ORF3a/b (Fig. 2A). To show structural homology of the three viral proteins, we utilized the resolved 3D structure of SARS-CoV-2 ORF3a deposited in public databases and used I-TASSER to predict the structures of the SARS-CoV ORF3a and MERS-CoV ORF3 (Fig. 2B). We found that the structure of both ORF3a were similar (Fig. 2C). But when the 3D structures of the MERS-CoV ORF3 protein and the two ORF3a proteins were aligned, the results showed they were quite different (Fig. S2, A and B). This implied that compared to the ORF3a protein of SARS-CoV and SARS-CoV-2, ORF3 protein of MERS-CoV had a distinct structure. We then pulled down the ORF3 protein in HEK293T cells (Fig. S2C). The proteins in the samples were identified by mass spectrometry (Table S1), and Gene Ontology enrichment (Fig. S2, D–G) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis (Fig. 2D) were performed. KEGG enrichment analysis showed that the interacting proteins of ORF3 were significantly enriched in the proteasome pathway which suggested that ORF3 might be an unstable protein. At this end, cells overexpressing ORF3 were treated with cycloheximide (CHX) to inhibit protein expression. When the synthesis of ORF3 protein was inhibited, the level of ORF3 protein gradually decreased over time, while ORF3a of SARS-CoV and SARS-CoV-2 was stable and had a longer half-life (Fig. 2E). Based on the earlier finding of proapoptotic effect of MERS-CoV ORF3, we overexpressed ORF3 in a dose-dependent manner and found that apoptosis was positively correlated with its expression (Fig. 2, F–H). These results suggested a new mechanism for the host to antagonize MERS-CoV by promoting ORF3 degradation to suppress its proapoptotic activity.
Figure 2.
MERS-CoV ORF3 is an unstable protein.A, the amino acid sequences of the specified genes were downloaded from NCBI and were compared by clustalW. The phylogenetic tree was constructed by MEGA. B, the protein structures of MERS-CoV ORF3 and SARS-CoV ORF3a were predicated by the iterative threading assembly refinement (I-TASSER) server. MERS-COV ORF3: green, SARS-CoV ORF3a: cyan. C, the structure alignment between ORF3a of SARS-CoV and SARS-CoV-2 was analyzed by PyMOL. SARS-CoV ORF3a: cyan, SARS-CoV-2 ORF3a: yellow. D, top 20 of KEGG terms enrichment. The interacting proteins of ORF3 were identified by mass spectrometry, and KEGG enrichment analysis was performed using the OmicShare tools, a free online platform. E, HEK293T cells transfected with the indicated plasmids were treated with cycloheximide (CHX, 100 μg/ml), and lysates were collected at the indicated times for immunoblotting. F and G, HEK293T cells were transfected with empty vector or increasing amounts of plasmids expressing MERS-CoV ORF3 as indicated. Twenty-four hours later, 90% cells were collected and stained with Annexin V-FITC/PI for flow cytometric analysis (F), and the remaining cells were lysed for immunoblotting (G). H, the percentage of apoptotic cells were measured. Error bars indicate SD of technical triplicates. Statistical significance was calculated using unpaired, two-tailed Student’s t test. ∗p < 0.05; ∗∗p < 0.01. KEGG, Kyoto Encyclopedia of Genes and Genomes; MERS-CoV, Middle East respiratory syndrome coronavirus; SARS-CoV, severe acute respiratory syndrome coronavirus; SARS-CoV-2, severe acute respiratory syndrome coronavirus-2.
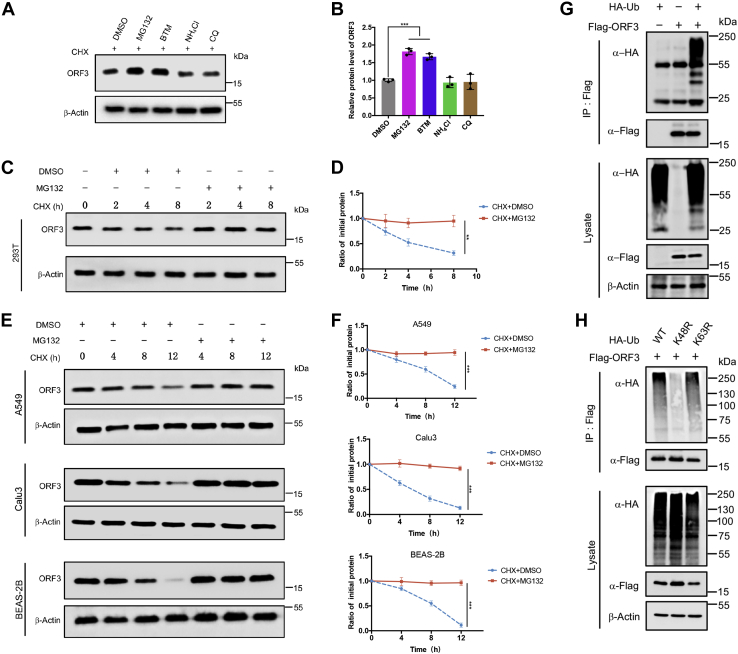
ORF3 is degraded by ubiquitin–proteasome system
There are two common ways to clear damaged and potentially toxic proteins in cells: the ubiquitin–proteasome pathway and the lysosomal pathway. Based on KEGG enrichment analysis, we found that ORF3 might be degraded through the proteasome pathway. To validate this, cells expressing ORF3 were treated with proteasome inhibitors MG132 and bortezomib (BTM) and lysosomal inhibitors NH4Cl and chloroquine for 12 h, and then the changes of ORF3 protein levels were analyzed. The treatment with proteasome inhibitors MG132 and BTM significantly increased the level of ORF3 protein as compared to the control group and the lysosomal inhibitor treatment groups (Fig. 3, A and B). Next, we overexpressed ORF3 in HEK293T and cotreated with CHX and the various inhibitors to analyze ORF3 half-life. After treatment with MG132 and BTM, the protein level of ORF3 remained stable without degradation (Fig. 3, C and D, Fig. S3A), while the treatment of NH4Cl and chloroquine failed to stabilize ORF3 (Fig. S3, B and C). Similarly, the stability of ORF3 under the influence of MG132 was also seen in A549, Calu3, and BEAS-2B cells (Fig. 3, E and F). If a protein is degraded by the proteasome, it often needs to be ubiquitinated before it can be recognized by the proteasome prior to hydrolysis by proteolytic enzymes. We sought to examine the possibility that ORF3 could be ubiquitinated. When ubiquitin and ORF3 were co-overexpressed, the in vivo ubiquitination assay showed that ORF3 could conjugate with ubiquitin chains (Fig. 3G). Also, ORF3 could conjugate with endogenous ubiquitin to form polyubiquitin chains (Fig. S3D). Ubiquitin can form ubiquitin chains through their own lysine and the first methionine. The ubiquitin chains formed by K48 and K63 were most widely studied, in which the K48-linked ubiquitin chain regulated protein degradation. Therefore, we co-expressed ORF3 and WT ubiquitin or mutated ubiquitin (K48R and K63R) for in vivo ubiquitination experiments. The results showed that ORF3 mainly conjugated with K48 ubiquitin chain in cells (Fig. 3H). These results proved that ORF3 can undergo ubiquitination, resulting to its degradation in host cells.
Figure 3.
ORF3 is degraded by ubiquitin–proteasome system.A, HEK293T cells overexpressing ORF3 protein were treated with DMSO, MG132 (20 μM), bortezomib (BTM, 10 μM), chloroquine (CQ, 20 μM), and NH4Cl (10 mM) for 8 h before collection. Then, ORF3 protein level was detected by immunoblotting. B, ORF3 protein levels normalized to β-actin were quantified by ImageJ. Results were shown as mean ± SD, n = 3 independent experiments. ∗∗∗p < 0.001, Student’s t test. C, HEK293T cells were transfected with the plasmids expressing ORF3 protein. Twelve hours later, cells were split equally into a 12-well plate. After 12 h, cells were then cotreated with MG132 (20 μM) and CHX (100 μg/ml) or DMSO and CHX (100 μg/ml) for 0, 2, 4, or 8 h. The protein level of ORF3 was analyzed by immunoblotting. D, ORF3 protein levels normalized to β-actin were quantified by ImageJ. Data were analyzed by GraphPad Prism 7 as mean ± SD, n = 3 independent experiments. ∗∗p < 0.01, two-way ANOVA test. E, A549, Calu3, and BEAS-2B cells were transfected with the plasmid containing HA-ORF3, split, and cotreated with MG132 (20 μM) and CHX (100 μg/ml) or DMSO and CHX (100 μg/ml) for 0, 4, 8, or 12 h. The protein level of ORF3 were analyzed by immunoblotting. F, ORF3 protein levels normalized to β-actin were quantified by ImageJ. Data were analyzed by GraphPad Prism 7 as mean ± SD, n = 3 independent experiments. ∗∗∗p < 0.001, two-way ANOVA test. G and H, HEK293T cells transfected with the indicated plasmids were treated with MG132 for 8 h before collection. The whole cell lysates were subjected to pulldown with anti-Flag beads and immunoblotting with anti-HA antibody to detect the polyubiquitin chains of ORF3. CHX, cycloheximide; DMSO, dimethyl sulfoxide.
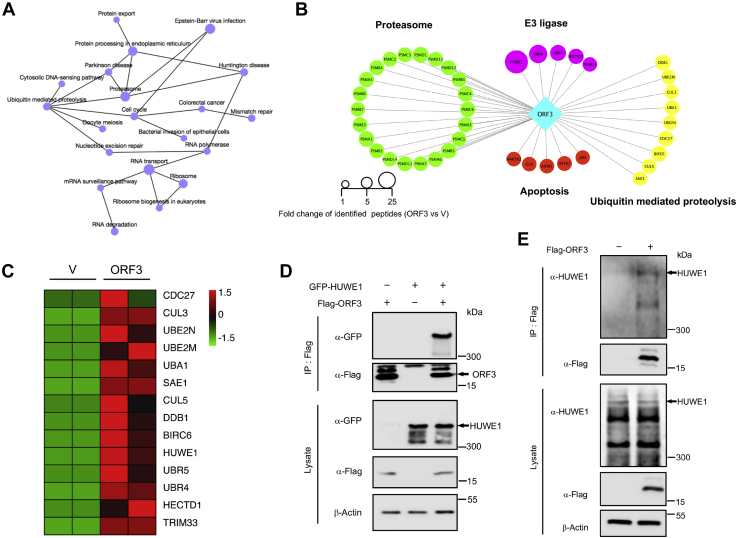
ORF3 interacts with E3 ligase HUWE1
In the KEGG pathway enrichment analysis of ORF3 interacting proteins, we created a network map of the first 20 pathways to acquire an intuitive understanding of their relationship (Fig. 4A). Because we sought to demonstrate the ubiquitination-mediated degradation of ORF3, we took out the proteins related to the ubiquitin–proteasome pathway, such as proteasome subunits, ubiquitin-mediated proteolysis, and E3 ligases to create a protein interaction network diagram (Fig. 4B). Meanwhile, based on the number of peptides of the interacting proteins identified by mass spectrometry, we made a heat map of the E3 ligases and ubiquitin-mediated proteolysis identified in the control group and the experimental group (Fig. 4C). As a key link in the ubiquitin–proteasome system, E3 ligase can specifically recognize the target protein and conjugate ubiquitin to the protein to form polyubiquitin chains and finally make it recognized by the proteasome and degraded (50). In our results, five E3 ligases were identified to interact with ORF3 protein. Among them, HUWE1 had the most identified peptides and was most likely to be the key E3 ligase for ORF3 ubiquitination. To verify whether ORF3 can interact with HUWE1 at the cellular level, we overexpressed ORF3 and HUWE1 in HEK293T cells. ORF3 could bind to both exogenous and endogenous HUWE1 (Fig. 4, D and E). These results confirmed the interaction of ORF3 and HUWE1 in cells, suggesting that HUWE1 was the key E3 ligase that regulated the ubiquitination and degradation of ORF3.
Figure 4.
ORF3 interacts with E3 ligase HUWE1. A, KEGG network was constructed based on KEGG enrichment analysis. B, the interacting proteins related to proteasome pathway, apoptosis, E3 ligase, and ubiquitin-mediated proteolysis were clustered as a protein–protein network. C, a heatmap of the E3 ligases and ubiquitin-mediated proteolysis were plotted based on the interacting proteins of MERS-CoV ORF3 protein. D, HEK293T cells were cotransfected with plasmids containing Flag-ORF3 and GFP-HUWE1. WCLs were precipitated with the anti-Flag beads. WCLs and precipitated proteins were detected by immunoblotting with indicated antibodies to analyze the interaction between ORF3 and HUWE1. E, HEK293T cells were transfected with plasmids containing Flag-ORF3. WCLs were precipitated with the anti-Flag beads to analyze the interaction between ORF3 and endogenous HUWE1. MERS-CoV, Middle East respiratory syndrome coronavirus; WCLs, whole cell lysates.
Proapoptotic activity of ORF3 is weakened by HUWE1-mediated degradation
Since our previous result confirmed the interaction of ORF3 with HUWE1, we next investigated whether HUWE1 was indeed the E3 ligase that regulated the ubiquitination and degradation of ORF3. As the expression of HUWE1 increased, the expression level of ORF3 protein gradually decreased (Fig. 5, A and B), while the other identified E3 ligases (UBR5, TRIM33, Cullin5, Cullin3, and UBR4) had no effect on the stability of ORF3 (Fig. S4, A–E). HUWE1 overexpression shortened the half-life of ORF3 protein, and ORF3 degraded faster than the control group (Fig. 5, C and D). Moreover, when HUWE1 was overexpressed, the ubiquitin chains of ORF3 were significantly enhanced (Fig. 5E). To further corroborate these results, HUWE1 was knocked down in HEK293T cells using shRNAs. The knockdown of HUWE1 increased ORF3 protein level (Fig. S4F). Next, we tested the stability of ORF3 in control cells and stable HUWE1-knockdown cell lines, respectively. We found that compared with the control group, the reduction of HUWE1 stabilized ORF3 with no apparent degradation (Fig. 5, F and G), and HUWE1 knockdown significantly reduced the ubiquitin chains (Fig. 5H). In addition, we overexpressed HUWE1-C4341A, the ubiquitin ligase activity–deficient mutant, to test its effect on the stability of ORF4b. With the increase of HUWE1-C4341A expression, the protein level of ORF4b was not reduced (Fig. 5I). Also, overexpression of C4341A did not shorten the half-life of ORF4b (Fig. 5, J and K). Furthermore, we performed an in vitro ubiquitination assay using GST-ORF3, HUWE1-HECT, and the enzyme-deficient HUWE1-HECT-C4341A purified from bacteria (Fig. S4G). These results confirmed that HUWE1 mediated ORF3 ubiquitination to direct its degradation by the proteasome. Unlike MERS-CoV ORF3, neither the ORF3a protein of SARS-CoV nor SARS-CoV-2 could interact with HUWE1 (Fig. S4H). Moreover, the overexpression of HUWE1 attenuated ORF3-induced apoptosis, while overexpression of HUWEI alone had little effect on apoptosis (Fig. 5, L and M). All in all, these results strongly indicated that HUWE1 was an E3 ligase that specifically regulated the degradation of ORF3 via ubiquitination.
Figure 5.
Proapoptotic activity of ORF3 is weakened by HUWE1-mediated degradation. A, HEK293T cells overexpressing the ORF3 protein were transfected with increasing amounts of plasmids containing GFP-HUWE1. Cells were collected 48 h after transfection, and the protein level of ORF3 was analyzed by immunoblotting. B, ORF3 protein levels normalized to β-actin were quantified by ImageJ. Data were analyzed by GraphPad Prism 7 as mean ± SD, n = 3 independent experiments. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, Student’s t test. C, HEK293T cells expressing ORF3 protein were transfected with vector or GFP-HUWE1 plasmids, treated with CHX, and collected at indicated time for immunoblotting to test the protein level of ORF3. D, quantification of ORF3 protein level was normalized to β-actin. Results were shown as mean ± SD, n = 3 independent experiments. ∗∗p < 0.01, two-way ANOVA test. E, HEK293T cells transfected with the indicated plasmids were treated with MG132 for 8 h before collection. The whole cell lysates were subjected to pulldown with anti-Flag beads and to detect the polyubiquitin chains of ORF3 by immunoblotting. F, HEK293T cells stably expressing shNC or shHUWE1 were transfected with ORF3-containing plasmid, treated with CHX, and collected at indicated time to test the protein level of ORF3. G, quantification of ORF3 protein level was normalized to β-actin. Results were shown as mean ± SD, n = 3 independent experiments. ∗∗∗p < 0.001, two-way ANOVA test. H, HEK293T cells stably expressing shNC and shHUWE1 were transfected with the ORF3 expressing plasmid and treated with MG132 for 8 h before collection. The whole cell lysates were subjected to pulldown and detect the polyubiquitin chains of ORF3. I, HEK293T cells overexpressing the ORF3 protein were transfected with increasing amounts of plasmids containing HA-HUWE1-C4341A. Cells were collected 48 h after transfection, and the protein level of ORF3 was analyzed by immunoblotting. J, HEK293T cells expressing ORF3 protein were transfected with vector or HA-HUWE1-C4341A plasmids, treated with CHX, and collected at indicated time for immunoblotting to test the protein level of ORF3. K, quantification of ORF3 protein level was normalized to β-actin. Results were shown as mean ± SD, n = 3 independent experiments. NS, not significant, two-way ANOVA test. L, HEK293T cells were transfected with indicated plasmids. Twenty-four hours later, cells were stained with Annexin V-FITC/PI for flow cytometric analysis. M, the percentage of the apoptotic cells was measured. Error bars indicate SD of technical triplicates. Statistical significance was calculated using unpaired, two-tailed Student’s t test. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, NS, not significant.
Ubiquitination-resistant mutant could increase the stability of ORF3
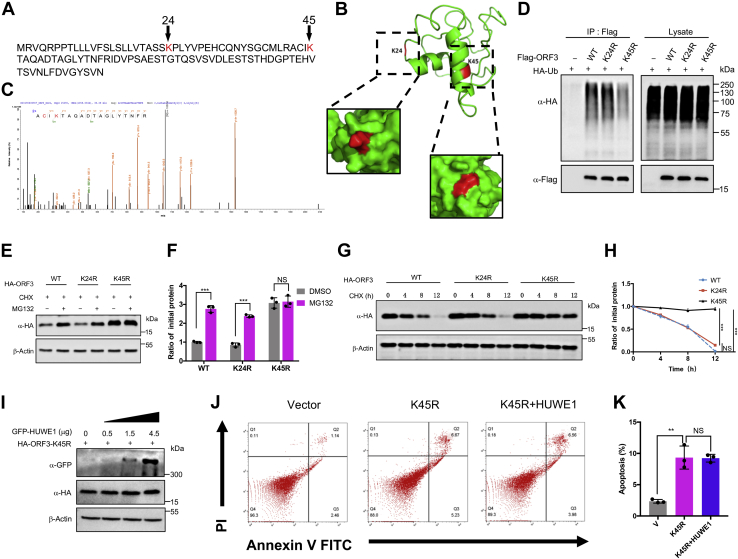
Ubiquitination generally occurs on the lysine residues of the substrate protein (51). ORF3 is a small accessory protein (only103 amino acids long) with only two lysine residues: K24 and K45 (Fig. 6A). Using PyMOL to analyze the predicted structure of the ORF3 protein, we found that K24 was located on the random coil while K45 was located on the β-fold; both of which were located on the surface of the protein (Fig. 6B). The tandem mass spectrometry results further showed that K45 could be a ubiquitination site (Fig. 6C). To validate the MS data, we constructed ubiquitination-resistant mutants of ORF3-K24R and ORF3-K45R. The polyubiquitin chains conjugated on the ORF3-K45R was significantly weaker than ORF3-WT and ORF3-K24R (Fig. 6D). Further, ORF3-WT and ORF3-K24R protein levels with MG132 treatment were significantly higher than that of the control group without MG132, while the protein level of ORF3-K45R was not affected by MG132 (Fig. 6, E and F). Moreover, we went on to analyze the half-lives of ORF3-WT, ORF3-K24R, and ORF3-K45R. With CHX treatment, the protein levels of ORF3-WT and ORF3-K24R gradually decreased with time, while K45R remained relatively stable with a longer half-life (Fig. 6, G and H). To further determine that lysine 45 was the site in which HUWE1 conjugated ubiquitin, we overexpressed HUWE1 in a gradient and tested its effect on the ubiquitination-resistant mutant ORF3-K45R. The protein level of ORF3-K45R was not affected by the overexpression of HUWE1 (Fig. 6I). Also, ORF3-K45R could still induced apoptosis and was not affected by HUWE1 (Fig. 6, J and K). These results demonstrated that K45 was the site regulated by HUWE1 to facilitate ubiquitination that affected ORF3 stability.
Figure 6.
Ubiquitination-resistant mutant could increase the stability of ORF3. A, the amino acid sequence of MERS-CoV ORF3 was downloaded from NCBI, and all the lysine residues were marked in red. B, the structure of ORF3 protein was visualized by PyMOL with two lysine residues highlighted in red. C, HEK293T cells were transfected with ORF3-expressing plasmid for 48 h. ORF3 protein was purified and analyzed by tandem mass spectrometry. One peptide containing glycine residue was identified, K45 (in red). D, HEK293T cells were transfected with the indicated plasmids and treated with MG132 (20 μM) for 8 h before collection. The whole cell lysates were subjected to pulldown with anti-Flag beads and later immunoblotting with indicated antibodies to detect the polyubiquitin chains of ORF3-WT and ORF3 mutants. E, single ubiquitination-resistant mutants, including K24R and K45R were constructed. HEK293T cells were transfected with the indicated plasmids and treated with MG132 (20 μM) for 8 h before collection. Cell lysates were used to detect the indicated protein level by immunoblotting. F, Quantification of ORF3 protein level was normalized to β-actin. Results were shown as mean ± SD, n = 3 independent experiments. ∗∗∗p < 0.001, NS, not significant, Student’s t test. G, HEK293T cells were transfected with plasmids expressing ORF3-WT, ORF3-K24R, and ORF3-K45R. Twelve hours later, cells were evenly divided into 12-well plates. After 24 h, cells were treated with CHX (100 μg/ml) and collected at indicated time to detect the protein level of ORF3. H, quantification of ORF3 protein level was normalized to β-actin. Results were shown as mean ± SD, n = 3 independent experiments. ∗∗∗, p < 0.001, NS, not significant, two-way ANOVA test. I, HEK293T cells overexpressing the ORF3-K45R protein were transfected with increasing amounts of plasmids containing GFP-HUWE1. Cells were collected 48 h after transfection of the GFP-HUWE1 plasmid, and the protein level of ORF3-K45R was analyzed by immunoblotting. J, HEK293T cells were transfected with indicated plasmids. Twenty-four hours later, cells were stained with Annexin V-FITC/PI for flow cytometric analysis. K, the percentage of the apoptotic cells was measured. Error bars indicate SD of technical triplicates. Statistical significance was calculated using unpaired, two-tailed Student’s t test. ∗∗p < 0.01, NS, not significant. CHX, cycloheximide; MERS-CoV, Middle East respiratory syndrome coronavirus.
Discussion
The SARS-CoV-2 outbreak is so far the most serious global health crisis in the recent history. Because of its cosmopolitan distribution as well as its detrimental impact to the health-care system and economies, SARS-CoV-2 has received an unparalleled attention. In a way, the COVID-19 pandemic somehow led to increasing concerns on coronaviruses. Since 2003, several coronaviruses that cause severe respiratory diseases have emerged in a cyclical outbreak trend. Among them, MERS-CoV was by far the most lethal of all the human coronaviruses. First identified in the Middle East in 2012, the mortality rate of MERS-CoV is as high as 35%, which is several folds higher than the mortality of SARS-CoV and SARS-CoV-2. There is neither specific treatment nor approved vaccine for MERS-CoV. Although MERS-CoV was predominantly endemic in the Middle East, there was a trend for MERS-CoV to cause a global epidemic as well, since cases were reported in Europe, North America, and Asia, especially in South Korea, where a major outbreak occurred in 2015 (52, 53). While the infection rate of MERS-CoV is lower than SARS-CoV-2, its reemergence in the future still constitutes a huge threat to public health. Thus, further research on the transmission route, pathogenesis, and development of antiviral drugs for MERS-CoV also warrant attention.
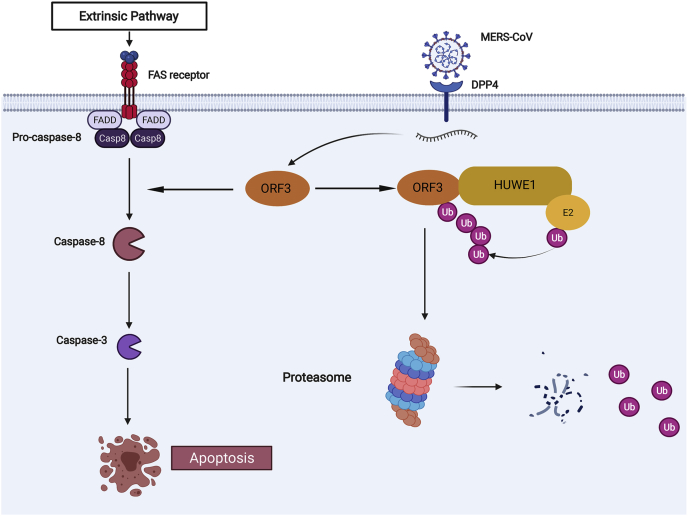
Previous studies on the pathogenicity of MERS-CoV have mostly focused on IFN-resistant accessory proteins, such as ORF4a, ORF4b, and ORF5, or viral proteases, such as papain-like protease and main protease (37, 54, 55). In particular, our study has several highlights. First, we have identified for the first time that the viral protein ORF3 of MERS-CoV could induce apoptosis in host cells, revealing a new pathogenic mechanism that can be capitalized as new viral target protein for clinical treatment of MERS-CoV. Second, we found that ORF3 protein was unstable in host cells and could be degraded through the ubiquitin–proteasome pathway. Third, among the interacting proteins of ORF3, HUWE1 was an E3 ligase with the most peptides identified by mass spectrometry, implying that HUWE1 was likely to be the specific E3 ligase. The interaction of HUWE1 and ORF3 was further validated experimentally, and HUWE1 ubiquitinated ORF3 at lysine 45 to tag it for proteasomal degradation. Fourth, overexpression of HUWE1 can effectively attenuate the induction of apoptosis by ORF3-WT, but not the ubiquitination-resistant mutant ORF3-K45R. This suggested that HUWE1 was an anti-MERS-CoV factor in host cells that could effectively antagonize the pathogenicity of ORF3 (Fig. 7). Whereas HUWE1 had been reported to play roles in various pathologies (56, 57, 58, 59, 60, 61) and was rarely studied as antiviral factor in host immunity (62, 63, 64), this was the first time that HUWE1 was found to play an antagonistic role in the pathogenesis of coronavirus infection.
Figure 7.
A pattern diagram of HUWE1 mediating the ubiquitination and degradation of ORF3 to weaken its proapoptotic activity. DPP4, dipeptidyl peptidase 4; FADD, Fas-associated death domain; FAS, tumor necrosis actor receptor superfamily member 6.
Apoptosis is a highly organized type of programmed cell death that enables cells to maintain the stability of the internal environment in normal physiological processes or under pathological conditions (65). However, excessive apoptosis can disrupt the structure and integrity of the bronchoalveolar network and lead to associated acute respiratory distress syndrome (66). Coronavirus-induced apoptosis was described in multiple tissues (67, 68, 69, 70, 71), and diverse components were shown to trigger apoptosis (36, 40, 49, 72, 73). Nonetheless, the role and significance of apoptosis in the pathogenesis of MERS-CoV remains largely unexplored. During long-term evolution, viruses have also developed a self-defense mechanism that induces or inhibits apoptosis by viral proteins. For example, in the early stages of infection, viruses inhibited apoptosis until active viral particles were produced. In the late stage of infection, the virus encoded another protein that induced apoptosis and wrapped the virus particles in apoptotic vesicles, which were later absorbed by surrounding cells (74). Taking this perspective, virus-induced apoptosis facilitates the release and spread of the virus (75, 76, 77, 78). Here, we showed that similar to ORF3a of SARS-CoV and SARS-CoV-2, MERS-CoV ORF3 protein also induced apoptosis in the host cells, but it had a shorter half-life compared to that of ORF3a and could be degraded by the proteasome. This could at least partially explain the less infectivity of MERS-CoV compared to SARS-CoV and SARS-CoV-2.
Our discovery of ORF3 degradation by HUWE1 might offer therapeutic implications. One is, regulating ORF3 by ubiquitination and degradation may confer better resistance to viral release during its life cycle. This can be achieved by designing small compounds to enhance the binding between HUWE1 and ORF3, thereby increasing the degradation of ORF3. In recent years, an emerging protein hydrolysis targeting chimera (PROTAC) strategy has provided a new approach to drug development (79, 80, 81). PROTAC has been widely used in drug development in the field of cancer and is expanding into areas such as immune abnormalities and neurodegenerative disease (82). However, PROTAC strategies for viral infections are rarely addressed (83). Since viral proteins are exogenous, degradation of viral proteins by PROTACs has higher targeting efficiency and specificity. Moreover, the antigenic fragments generated by the degradation of proteins induced by PROTACs also effectively promotes the immune response of the organism (84). Therefore, targeting the MERS-CoV ORF3 protein by PROTAC small molecule drugs will have more advantages and better prospects for clinical applications.
There were several limitations in this study that requires follow-up research. First, ORF3 induced apoptosis through the extrinsic pathway, and the KEGG enrichment analysis identified several host proteins in the apoptosis pathway to interact with ORF3.Whether ORF3 protein affects the functions of these proteins to induce apoptosis remains to be investigated. In addition, it has not been addressed whether the replicative capacity of MERS-CoV could be enhanced when expression of HUWE1 in host cell is repressed or when mutation is introduced at lysine K45 of ORF3. This would require experiments in BSL3. Live virus assays can also further clarify the antagonistic effect of HUWE1 as an antiviral host factor against MERS-CoV.
Our study reveals a novel mechanism for host antagonism to the pathogenicity of MERS-CoV. We found that MERS-CoV ORF3 protein can induce apoptosis in host cells. Further, ORF3 protein was unstable and can be degraded by the ubiquitin–proteasome system, resulting in the inhibition of its ability to induce apoptosis. We uncovered the E3 ligase HUWE1 mediated the ubiquitination of ORF3 protein to facilitate its degradation. This was the first time that HUWE1 had been shown to play an antiviral role in host immunity. Current therapeutic antibodies or small molecule drugs against MERS-CoV mostly target the spike protein and are based on the inhibition of virus entry into cells and virus replication. There are fewer therapies related to the inhibition of cytopathic effects. This study described the pathogenic mechanism of MERS-CoV and identified a new host protein against MERS-CoV. Our findings provided a new target protein for treatment of MERS-CoV infection with great prospects for clinical application.
Experimental procedures
Cell culture
HEK293T, A549, Calu3, and BEAS-2B cells were purchased from ATCC. HEK293T, A549, and BEAS-2B were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Hyclone). Calu3 cells were cultured in minimal essential medium (Hyclone). All these cell lines were supplemented with 10% fetal bovine serum (ZATA), penicillin (100 U/ml), and streptomycin (100 μg/ml) and maintained at 37 °C in a humidified atmosphere of 5% CO2.
Plasmids and antibodies
The MERS-CoV pCAG-Flag-ORF3 was a gift from Wenjie Tan lab. The SARS-CoV and SARS-CoV-2 pCAG-Flag-ORF3a was purchased from Tsingke. All the mutants and target gene with different tags were cloned into pCAGGS vector by us. HUWE1-WT and HUWE1-C4341A expressing plasmid was a gift from Genze Shao lab. Antibodies used in immunoblotting are as follows: anti-FLAG (Sigma), anti-HA (CST), anti-Ubiquitin (LifeSensors), anti-HUWE1 (Abcam), anti-β-actin (Sigma), anti-GAPDH (Sigma).
Cell transfections, immunoprecipitation, and immunoblotting
Cells were transfected with various plasmids using Lipofectamine 3000 (Invitrogen) reagent or PEI according to the manufacturer’s protocol. Forty-eight hours later, cells were harvested and lysed with NP40 buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1% NP-40, 5 mM EDTA, 5% glycerol) supplemented with a protease inhibitor cocktail (Bimake). Whole cell lysates were sonicated and centrifuged at 12,000 rpm for 15 min. Supernatant was harvested and then incubated with anti-Flag agarose beads at 4 °C for 4 h. The agarose beads were washed extensively, and samples were eluted by boiling at 95 °C for 10 min with protein loading buffer. Both lysates and immunoprecipitate were examined using the indicated primary antibodies followed by detection with the related secondary antibodies and the Immobilon Western Chemiluminescent HRP Substrate (Millipore).
Dual luciferase reporter gene assay
The IFN beta promoter–driven firefly luciferase reporter plasmid was transfected into HEK293T cells seeded in 12-well plates with Renilla luciferase expression vectors at a ratio of 10:1 (Firefly: Renilla). Twenty-four hours after transfection, the cells were infected with Sendai virus (100 HAU/ml) for 12 h, and the medium was removed. After washing once with PBS, the cells were lysed and used to measure luciferase activity (Dual Luciferase Reporter Gene Assay Kit, 11402ES60, Yeasen). The relative firefly luciferase activity levels were normalized to the luciferase activity of Renilla control plasmid. Data represent the mean ± SD of three independent experiments.
Mass spectrometry
To identified the interacting proteins and ubiquitination sites of MERS-CoV ORF3 protein, HEK293T cells were transfected with plasmids containing Flag-ORF3. Transfected cells were harvested at 48 h after transfection, and the ORF3 proteins were purified with Flag-conjugated agarose beads (Sigma) from whole cell lysates. The proteins on the beads were eluted by 0.2 M glycine–HCl buffer (pH 3.0) and neutralized with 1.0 M Tris-HCl buffer (pH 9.0). The proteins were digested by trypsin, and each fraction was injected for nanoLC-MS/MS analysis. The LC-MS/MS analysis and data analysis were performed by Shanghai Applied Protein Technology. Gene Ontology enrichment and KEGG enrichment analyses were performed using the OmicShare tools, a free online platform for data analysis (http://www.omicshare.com/tools).
Protein half-life assay
For the ORF3 half-life assay, plasmids encoding ORF3 WT were transfected into HEK293T, A549, Calu3, and BEAS-2B cells when these cells in 6 cm dishes reached about 90% confluence. Twelve hours after transfection, the cells were evenly divided into 12-well plates. Twenty-four hours later, the cells were treated with the protein synthesis inhibitor CHX (Sigma, 100 μg/ml) for the indicated durations before collection. ORF3 mutants expressing plasmids were used in transfection as indicated in individual experiments. The cells stably expressing the indicated shRNA were also transfected with ORF3 expressing plasmid and then treated with CHX. For the proteasome inhibitor, MG132 (Sigma, 20 μM) was added into the cells at the same time with CHX.
In vivo ORF3 ubiquitination assay
For in vivo ORF3 ubiquitination assay, Flag-ORF3 and HA-ubiquitin were transfected into HEK293T cells. Twenty-four hours, the cells were treated with 20 μM of the proteasome inhibitor MG132 (Sigma) for 8 h. The cells were washed with PBS, pelleted, and lysed with NP40 lysis buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1% NP-40, 5 mM EDTA, 5% glycerol) plus 0.1% SDS, 10 mM DTT, and 20 μM MG132. The lysates were centrifuged to obtain cytosolic proteins and incubated with anti-Flag agarose beads for 4 h at 4 °C. Then, the beads were washed three times with PBS. The proteins were released from the beads by boiling in protein loading buffer and analyzed by immunoblotting.
In vitro ORF3 ubiquitination assay
The MERS-CoV ORF3 and the HUWE1 HECT domain of WT and C4341A mutant were cloned and expressed as GST-fusion proteins. All the proteins were expressed in BL21 bacterial cells and purified with Glutathione Sepharose beads. The beads with ORF3 protein were incubated with HUWE1 HECT WT or C4341A mutant, together with 200 ng of E1 (UBE1, R&D system), 400 ng of E2 (UbcH5b, R&D system), and 2 μg of recombinant ubiquitin (R&D system) in reaction buffer (50 mM Tris [PH 7.4], 2 mM MgCl2, 4 mM ATP [Sigma]) at 37 °C for 1 h. The supernatant was removed, and the reaction was terminated by adding 2x protein loading buffer. The ubiquitin chain was detected by immunoblotting.
Lentivirus-mediated stable cell line construction
When the HEK293T cells reached 60% confluency, the lentiviral packaging plasmid and shRNA were cotransfected into the cells in a certain ratio (psPAX2: pMD2.G: shRNA = 5:7:10) through Lipofectamine3000 and replaced with new DMEM medium after 6 h of transfection. After 48 h of transfection, the culture medium was collected, centrifuged, and filtered with a 0.45 μm filter membrane to obtain the virus liquid. HEK293T cells were infected with the lentivirus in the presence of polybrene (5 μg/ml) with centrifugation at 1800 rpm for 40 min at 30 °C. Cells were selected at 48 h after infection and maintained in 10% fetal bovine serum DMEM supplemented with puromycin (1∼2 μg/ml). The knockdown efficiency of HUWE1 was detected by immunoblotting with anti-HUWE1 antibody. The shRNA sequences are below.
shHUWE1 #1: 5′-GCTAACTCGGCTACAACATTT-3’;
#2: 5′-CCTAGGCTGCAGGACTAATAT-3’;
#3: 5′-TTTGATGTCAAGCGCAAATAT-3’;
#4: 5′-TGCACAGCTAATGATTCAATG-3’.
RNA isolation and quantitative real-time PCR
Total cell RNA was extracted using TRIzol reagent (Invitrogen) following the manufacturer’s instructions. One microgram of total RNA was subjected to reverse transcription to synthesize cDNA using RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific). Quantitative real-time PCR was performed using SYBR Green Master Mix (Monad), and a 10 μl volume reaction consisted of 1 μl reverse transcription product and 300 nM of each primer. Relative gene mRNA levels for each target gene were calculated by the 2−△△Ct method using β-actin as an internal control. The primers used for IFNβ are as follows:
IFNβ-F: CTTTCGAAGCCTTTGCTCTG;
IFNβ-R: CAGGAGAGCAATTTGGAGGA.
Protein 3D structure prediction and alignment
The 3D structures of SARS-CoV-2 ORF3a protein (7KJR) was downloaded from Protein Data Bank (PDB, https://www.rcsb.org/). The protein structures of SARS-CoV ORF3a protein and MERS-CoV ORF3 protein were predicated by the iterative threading assembly refinement (I-TASSER) server which generated 3D atomic models from multiple threading alignments and iterative structure assembly simulations. The alignment between different protein structures was performed by PyMOL.
Phylogenetic tree construction
The target viral proteins’ amino acid sequences were downloaded from the NCBI, and multiple alignment of the sequences were performed using clustalW. The bootstrap method was chosen to calculate the test of hylogeny. The number of bootstrap replications was set to 1000, and the model selection was Poisson model. The root represented the origin of evolution, branches represented the relationship between these sequences, and the branch length represented evolutionary change. The bootstrap value marked on the node was used to evaluate the trustworthiness of the branch.
Apoptosis detection
The flow cytometry assay was used to detect cell apoptosis. The cells seeded on the 6-well plates were transfected with indicated plasmids. After 36 h, cells were washed with PBS for three times and collected by centrifugation at 1000g for 5 min. The apoptosis assay was performed using FITC Annexin V Apoptosis Detection Kit I (BD Pharmingen), and the steps were described as follows: Washed cells twice with cold PBS and then resuspend cells in 1x Annexin V binding buffer at 1 × 106 cells/ml. Transferred 150 μl of the solution to a 5 ml culture tube. Then added 5 μl of FITC Annexin V and 5 μl propidium iodide staining solution. Gently vortexed the cells and incubated for 15 min at room temperature (20–25 °C) in the dark. Added 300 μl of binding buffer to each tube and performed flow cytometry analysis within 1 h.
Caspase-3/8/9 activity detection assays
The cells seeded on the plates were transfected with indicated plasmids. Five hours before the collection, cells were treated with staurosporine (1 μM, Sigma) or dimethyl sulfoxide and then were lysed for Western blot. The indicated primary antibodies were used to detect the full-length and cleaved caspase3/8/9. Antibodies used in immunoblotting are as follows: anti-caspase3 (abcam, ab32351), anti-caspase8 (proteintech, 13423-1-AP), anti-caspase9 (proteintech, 10380-1-AP), anti-Bcl-2 (abcam, ab32124), and anti-BAX (abcam, ab32503).
Statistical analysis
Each experiment was repeated at least three times. All results were shown as the mean ± SD. Student’s t test (unpaired, two-tailed) was used to compare two independent groups, and two-way ANOVA test was performed for comparisons of multiple groups. All statistical analyses were performed with GraphPad Prism 7. p < 0.05 was considered statistically significant.
Data availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE repository with the dataset identifier PXD030024. All other data are included in this article and its supporting information.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We like to thank Professor Wenjie Tan for the plasmids containing coronavirus proteins. We also thank Professor Genze Shao for providing the HUWE1-expressing plasmid. Shanghai Applied Protein Technology is acknowledged for Mass spectrometry, and the Nursing Development, Central South University, is acknowledged for performing the flow cytometry assay.
Author contributions
Y. Z. investigation; Y. Z. writing-original draft; R. Z., Sixu Liu, Z. D., Y. Z., and Sijia Li data curation; R. Z., Sixu Liu, A. D., Shiqin Li, Z. C., P. L., A. R., and X. C. validation; C. D., Y. L., S. T., Y. L., and L. X. writing-review & editing; A. D., Shiqin Li, and Z. C. software; Z. D., Y. Z., Sijia Li, Y. L., S. T., Y. L., and L. X. resources; Q. Z., J. P., and X. D. visualization; Q. Z., J. P., and X. D. supervision; Z. X., Shanni Li, and T. J. project administration; Z. X. conceptualization; Z. X. methodology.
Funding and additional information
This work was supported by the National Key Research and Development Program of China (2021YFC2300103 and 2016YFD0500300), the National Natural Science Foundation of China (U21A20384 and 82072293), the Key Research and Development Program of Hunan Province (2020SK2054), Zhejiang University special scientific research fund for COVID-19 prevention and control (2020XGZX033), Changsha special scientific research fund for COVID-19 prevention and control (kq2001030), and the Central South University Graduate Research Innovation Project (2018zzts387).
Edited by George DeMartino
Supporting information
References
- 1.Lai M.M., Cavanagh D. The molecular biology of coronaviruses. Adv. Virus Res. 1997;48:1–100. doi: 10.1016/S0065-3527(08)60286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weiss S.R., Navas-Martin S. Coronavirus pathogenesis and the emerging pathogen severe acute respiratory syndrome coronavirus. Microbiol. Mol. Biol. Rev. 2005;69:635–664. doi: 10.1128/MMBR.69.4.635-664.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Su S., Wong G., Shi W., Liu J., Lai A.C.K., Zhou J., Liu W., Bi Y., Gao G.F. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016;24:490–502. doi: 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., Xiang J., Wang Y., Song B., Gu X., Guan L., Wei Y., Li H., Wu X., Xu J., et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: A retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu A., Peng Y., Huang B., Ding X., Wang X., Niu P., Meng J., Zhu Z., Zhang Z., Wang J., Sheng J., Quan L., Xia Z., Tan W., Cheng G., et al. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe. 2020;27:325–328. doi: 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan J.F., Kok K.H., Zhu Z., Chu H., To K.K., Yuan S., Yuen K.Y. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microbes Infect. 2020;9:221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cui J., Li F., Shi Z.L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guan W.J., Ni Z.Y., Hu Y., Liang W.H., Ou C.Q., He J.X., Liu L., Shan H., Lei C.L., Hui D.S.C., Du B., Li L.J., Zeng G., Yuen K.Y., Chen R.C., et al. Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y., Yuan M.L., Zhang Y.L., Dai F.H., Liu Y., Wang Q.M., et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Jiang R.D., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Drosten C., Günther S., Preiser W., van der Werf S., Brodt H.R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A., Berger A., Burguière A.M., Cinatl J., Eickmann M., Escriou N., et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 12.Guery B., Poissy J., el Mansouf L., Séjourné C., Ettahar N., Lemaire X., Vuotto F., Goffard A., Behillil S., Enouf V., Caro V., Mailles A., Che D., Manuguerra J.C., Mathieu D., et al. Clinical features and viral diagnosis of two cases of infection with Middle East respiratory syndrome coronavirus: A report of nosocomial transmission. Lancet. 2013;381:2265–2272. doi: 10.1016/S0140-6736(13)60982-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang X., Yu Y., Xu J., Shu H., Xia J., Liu H., Wu Y., Zhang L., Yu Z., Fang M., Yu T., Wang Y., Pan S., Zou X., Yuan S., et al. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: A single-centered, retrospective, observational study. Lancet Respir. Med. 2020;8:475–481. doi: 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D., Fouchier R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 15.Cotten M., Watson S.J., Kellam P., Al-Rabeeah A.A., Makhdoom H.Q., Assiri A., Al-Tawfiq J.A., Alhakeem R.F., Madani H., AlRabiah F.A., Al Hajjar S., Al-nassir W.N., Albarrak A., Flemban H., Balkhy H.H., et al. Transmission and evolution of the Middle East respiratory syndrome coronavirus in Saudi arabia: A descriptive genomic study. Lancet. 2013;382:1993–2002. doi: 10.1016/S0140-6736(13)61887-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cotten M., Watson S.J., Zumla A.I., Makhdoom H.Q., Palser A.L., Ong S.H., Al Rabeeah A.A., Alhakeem R.F., Assiri A., Al-Tawfiq J.A., Albarrak A., Barry M., Shibl A., Alrabiah F.A., Hajjar S., et al. Spread, circulation, and evolution of the Middle East respiratory syndrome coronavirus. mBio. 2014;5 doi: 10.1128/mBio.01062-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Azhar E.I., El-Kafrawy S.A., Farraj S.A., Hassan A.M., Al-Saeed M.S., Hashem A.M., Madani T.A. Evidence for camel-to-human transmission of MERS coronavirus. N. Engl. J. Med. 2014;370:2499–2505. doi: 10.1056/NEJMoa1401505. [DOI] [PubMed] [Google Scholar]
- 18.Scobey T., Yount B.L., Sims A.C., Donaldson E.F., Agnihothram S.S., Menachery V.D., Graham R.L., Swanstrom J., Bove P.F., Kim J.D., Grego S., Randell S.H., Baric R.S. Reverse genetics with a full-length infectious cDNA of the Middle East respiratory syndrome coronavirus. Proc. Natl. Acad. Sci. U. S. A. 2013;110:16157–16162. doi: 10.1073/pnas.1311542110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Forni D., Cagliani R., Clerici M., Sironi M. Molecular evolution of human coronavirus genomes. Trends Microbiol. 2017;25:35–48. doi: 10.1016/j.tim.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shin D., Mukherjee R., Grewe D., Bojkova D., Baek K., Bhattacharya A., Schulz L., Widera M., Mehdipour A.R., Tascher G., Geurink P.P., Wilhelm A., van der Heden van Noort G.J., Ovaa H., Müller S., et al. Papain-like protease regulates SARS-CoV-2 viral spread and innate immunity. Nature. 2020;587:657–662. doi: 10.1038/s41586-020-2601-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bailey-Elkin B.A., Knaap R.C., Johnson G.G., Dalebout T.J., Ninaber D.K., van Kasteren P.B., Bredenbeek P.J., Snijder E.J., Kikkert M., Mark B.L. Crystal structure of the Middle East respiratory syndrome coronavirus (MERS-CoV) papain-like protease bound to ubiquitin facilitates targeted disruption of deubiquitinating activity to demonstrate its role in innate immune suppression. J. Biol. Chem. 2014;289:34667–34682. doi: 10.1074/jbc.M114.609644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anand K., Ziebuhr J., Wadhwani P., Mesters J.R., Hilgenfeld R. Coronavirus main proteinase (3CLpro) structure: Basis for design of anti-SARS drugs. Science. 2003;300:1763–1767. doi: 10.1126/science.1085658. [DOI] [PubMed] [Google Scholar]
- 23.Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., Zhang B., Li X., Zhang L., Peng C., Duan Y., Yu J., Wang L., Yang K., Liu F., et al. Structure of M(pro) from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582:289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 24.Gao Y., Yan L., Huang Y., Liu F., Zhao Y., Cao L., Wang T., Sun Q., Ming Z., Zhang L., Ge J., Zheng L., Zhang Y., Wang H., Zhu Y., et al. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science. 2020;368:779–782. doi: 10.1126/science.abb7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marra M.A., Jones S.J., Astell C.R., Holt R.A., Brooks-Wilson A., Butterfield Y.S., Khattra J., Asano J.K., Barber S.A., Chan S.Y., Cloutier A., Coughlin S.M., Freeman D., Girn N., Griffith O.L., et al. The genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- 26.Menachery V.D., Mitchell H.D., Cockrell A.S., Gralinski L.E., Yount B.L., Jr., Graham R.L., McAnarney E.T., Douglas M.G., Scobey T., Beall A., Dinnon K., 3rd, Kocher J.F., Hale A.E., Stratton K.G., Waters K.M., et al. MERS-CoV accessory ORFs play key role for infection and pathogenesis. mBio. 2017;8 doi: 10.1128/mBio.00665-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Redondo N., Zaldívar-López S., Garrido J.J., Montoya M. SARS-CoV-2 accessory proteins in viral pathogenesis: Knowns and unknowns. Front. Immunol. 2021;12:708264. doi: 10.3389/fimmu.2021.708264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu D.X., Fung T.S., Chong K.K., Shukla A., Hilgenfeld R. Accessory proteins of SARS-CoV and other coronaviruses. Antivir. Res. 2014;109:97–109. doi: 10.1016/j.antiviral.2014.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Michel C.J., Mayer C., Poch O., Thompson J.D. Characterization of accessory genes in coronavirus genomes. Virol. J. 2020;17:131. doi: 10.1186/s12985-020-01402-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weiss S.R., Leibowitz J.L. Coronavirus pathogenesis. Adv. Virus Res. 2011;81:85–164. doi: 10.1016/B978-0-12-385885-6.00009-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Siu K.L., Yuen K.S., Castaño-Rodriguez C., Ye Z.W., Yeung M.L., Fung S.Y., Yuan S., Chan C.P., Yuen K.Y., Enjuanes L., Jin D.Y. Severe acute respiratory syndrome coronavirus ORF3a protein activates the NLRP3 inflammasome by promoting TRAF3-dependent ubiquitination of ASC. FASEB J. 2019;33:8865–8877. doi: 10.1096/fj.201802418R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shi C.S., Qi H.Y., Boularan C., Huang N.N., Abu-Asab M., Shelhamer J.H., Kehrl J.H. SARS-coronavirus open reading frame-9b suppresses innate immunity by targeting mitochondria and the MAVS/TRAF3/TRAF6 signalosome. J. Immunol. 2014;193:3080–3089. doi: 10.4049/jimmunol.1303196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miorin L., Kehrer T., Sanchez-Aparicio M.T., Zhang K., Cohen P., Patel R.S., Cupic A., Makio T., Mei M., Moreno E., Danziger O., White K.M., Rathnasinghe R., Uccellini M., Gao S., et al. SARS-CoV-2 Orf6 hijacks Nup98 to block STAT nuclear import and antagonize interferon signaling. Proc. Natl. Acad. Sci. U. S. A. 2020;117:28344–28354. doi: 10.1073/pnas.2016650117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yuen C.K., Lam J.Y., Wong W.M., Mak L.F., Wang X., Chu H., Cai J.P., Jin D.Y., To K.K., Chan J.F., Yuen K.Y., Kok K.H. SARS-CoV-2 nsp13, nsp14, nsp15 and orf6 function as potent interferon antagonists. Emerg. Microbes Infect. 2020;9:1418–1428. doi: 10.1080/22221751.2020.1780953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li J.Y., Liao C.H., Wang Q., Tan Y.J., Luo R., Qiu Y., Ge X.Y. The ORF6, ORF8 and nucleocapsid proteins of SARS-CoV-2 inhibit type I interferon signaling pathway. Virus Res. 2020;286:198074. doi: 10.1016/j.virusres.2020.198074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ren Y., Shu T., Wu D., Mu J., Wang C., Huang M., Han Y., Zhang X.Y., Zhou W., Qiu Y., Zhou X. The ORF3a protein of SARS-CoV-2 induces apoptosis in cells. Cell. Mol. Immunol. 2020;17:881–883. doi: 10.1038/s41423-020-0485-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang Y., Zhang L., Geng H., Deng Y., Huang B., Guo Y., Zhao Z., Tan W. The structural and accessory proteins M, ORF 4a, ORF 4b, and ORF 5 of Middle East respiratory syndrome coronavirus (MERS-CoV) are potent interferon antagonists. Protein Cell. 2013;4:951–961. doi: 10.1007/s13238-013-3096-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang Y., Ye F., Zhu N., Wang W., Deng Y., Zhao Z., Tan W. Middle East respiratory syndrome coronavirus ORF4b protein inhibits type I interferon production through both cytoplasmic and nuclear targets. Sci. Rep. 2015;5:17554. doi: 10.1038/srep17554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matthews K.L., Coleman C.M., van der Meer Y., Snijder E.J., Frieman M.B. The ORF4b-encoded accessory proteins of Middle East respiratory syndrome coronavirus and two related bat coronaviruses localize to the nucleus and inhibit innate immune signalling. J. Gen. Virol. 2014;95:874–882. doi: 10.1099/vir.0.062059-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chu H., Shuai H., Hou Y., Zhang X., Wen L., Huang X., Hu B., Yang D., Wang Y., Yoon C., Wong B.H., Li C., Zhao X., Poon V.K., Cai J.P., et al. Targeting highly pathogenic coronavirus-induced apoptosis reduces viral pathogenesis and disease severity. Sci. Adv. 2021;7 doi: 10.1126/sciadv.abf8577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Isaacson M.K., Ploegh H.L. Ubiquitination, ubiquitin-like modifiers, and deubiquitination in viral infection. Cell Host Microbe. 2009;5:559–570. doi: 10.1016/j.chom.2009.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hu Y., Li W., Gao T., Cui Y., Jin Y., Li P., Ma Q., Liu X., Cao C. The severe acute respiratory syndrome coronavirus nucleocapsid inhibits type I interferon production by interfering with TRIM25-mediated RIG-I ubiquitination. J. Virol. 2017;91 doi: 10.1128/JVI.02143-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oh S.J., Shin O.S. SARS-CoV-2 nucleocapsid protein targets RIG-I-like receptor pathways to inhibit the induction of interferon response. Cells. 2021;10:530. doi: 10.3390/cells10030530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wong H.H., Fung T.S., Fang S., Huang M., Le M.T., Liu D.X. Accessory proteins 8b and 8ab of severe acute respiratory syndrome coronavirus suppress the interferon signaling pathway by mediating ubiquitin-dependent rapid degradation of interferon regulatory factor 3. Virology. 2018;515:165–175. doi: 10.1016/j.virol.2017.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sui L., Zhao Y., Wang W., Wu P., Wang Z., Yu Y., Hou Z., Tan G., Liu Q. SARS-CoV-2 membrane protein inhibits type I interferon production through ubiquitin-mediated degradation of TBK1. Front. Immunol. 2021;12:662989. doi: 10.3389/fimmu.2021.662989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu X., Chen S., Hou P., Wang M., Chen Y., Guo D. VHL negatively regulates SARS coronavirus replication by modulating nsp16 ubiquitination and stability. Biochem. Biophys. Res. Commun. 2015;459:270–276. doi: 10.1016/j.bbrc.2015.02.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Le T.M., Wong H.H., Tay F.P., Fang S., Keng C.T., Tan Y.J., Liu D.X. Expression, post-translational modification and biochemical characterization of proteins encoded by subgenomic mRNA8 of the severe acute respiratory syndrome coronavirus. FEBS J. 2007;274:4211–4222. doi: 10.1111/j.1742-4658.2007.05947.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dominguez Andres A., Feng Y., Campos A.R., Yin J., Yang C.C., James B., Murad R., Kim H., Deshpande A.J., Gordon D.E., Krogan N., Pippa R., Ronai Z.A. SARS-CoV-2 ORF9c is a membrane-associated protein that suppresses antiviral responses in cells. bioRxiv. 2020 doi: 10.1101/2020.08.18.256776. [preprint] [DOI] [Google Scholar]
- 49.Chan C.M., Tsoi H., Chan W.M., Zhai S., Wong C.O., Yao X., Chan W.Y., Tsui S.K., Chan H.Y. The ion channel activity of the SARS-coronavirus 3a protein is linked to its pro-apoptotic function. Int. J. Biochem. Cell Biol. 2009;41:2232–2239. doi: 10.1016/j.biocel.2009.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zheng N., Shabek N. Ubiquitin ligases: Structure, function, and regulation. Annu. Rev. Biochem. 2017;86:129–157. doi: 10.1146/annurev-biochem-060815-014922. [DOI] [PubMed] [Google Scholar]
- 51.Swatek K.N., Komander D. Ubiquitin modifications. Cell Res. 2016;26:399–422. doi: 10.1038/cr.2016.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zumla A., Hui D.S., Perlman S. Middle East respiratory syndrome. Lancet. 2015;386:995–1007. doi: 10.1016/S0140-6736(15)60454-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chafekar A., Fielding B.C. MERS-CoV: Understanding the latest human coronavirus threat. Viruses. 2018;10:93. doi: 10.3390/v10020093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mielech A.M., Kilianski A., Baez-Santos Y.M., Mesecar A.D., Baker S.C. MERS-CoV papain-like protease has deISGylating and deubiquitinating activities. Virology. 2014;450-451:64–70. doi: 10.1016/j.virol.2013.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hilgenfeld R. From SARS to MERS: Crystallographic studies on coronaviral proteases enable antiviral drug design. FEBS J. 2014;281:4085–4096. doi: 10.1111/febs.12936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bernassola F., Karin M., Ciechanover A., Melino G. The HECT family of E3 ubiquitin ligases: Multiple players in cancer development. Cancer Cell. 2008;14:10–21. doi: 10.1016/j.ccr.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 57.Bose R., Sheng K., Moawad A.R., Manku G., O'Flaherty C., Taketo T., Culty M., Fok K.L., Wing S.S. Ubiquitin ligase Huwe1 modulates spermatogenesis by regulating spermatogonial differentiation and entry into meiosis. Sci. Rep. 2017;7:17759. doi: 10.1038/s41598-017-17902-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhao X., Heng J.I., Guardavaccaro D., Jiang R., Pagano M., Guillemot F., Iavarone A., Lasorella A. The HECT-domain ubiquitin ligase Huwe1 controls neural differentiation and proliferation by destabilizing the N-Myc oncoprotein. Nat. Cell Biol. 2008;10:643–653. doi: 10.1038/ncb1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Strappazzon F., Di Rita A., Peschiaroli A., Leoncini P.P., Locatelli F., Melino G., Cecconi F. HUWE1 controls MCL1 stability to unleash AMBRA1-induced mitophagy. Cell Death Differ. 2020;27:1155–1168. doi: 10.1038/s41418-019-0404-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Guo Y., Li L., Xu T., Guo X., Wang C., Li Y., Yang Y., Yang D., Sun B., Zhao X., Shao G., Qi X. HUWE1 mediates inflammasome activation and promotes host defense against bacterial infection. J. Clin. Invest. 2020;130:6301–6316. doi: 10.1172/JCI138234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Aleidi S.M., Howe V., Sharpe L.J., Yang A., Rao G., Brown A.J., Gelissen I.C. The E3 ubiquitin ligases, HUWE1 and NEDD4-1, are involved in the post-translational regulation of the ABCG1 and ABCG4 lipid transporters. J. Biol. Chem. 2015;290:24604–24613. doi: 10.1074/jbc.M115.675579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yamamoto S.P., Okawa K., Nakano T., Sano K., Ogawa K., Masuda T., Morikawa Y., Koyanagi Y., Suzuki Y. Huwe1, a novel cellular interactor of Gag-Pol through integrase binding, negatively influences HIV-1 infectivity. Microbes Infect. 2011;13:339–349. doi: 10.1016/j.micinf.2010.12.002. [DOI] [PubMed] [Google Scholar]
- 63.Singh S., Ng J., Sivaraman J. Exploring the “other” subfamily of HECT E3-ligases for therapeutic intervention. Pharmacol. Ther. 2021;224:107809. doi: 10.1016/j.pharmthera.2021.107809. [DOI] [PubMed] [Google Scholar]
- 64.Leung A., Trac C., Kato H., Costello K.R., Chen Z., Natarajan R., Schones D.E. LTRs activated by Epstein-Barr virus-induced transformation of B cells alter the transcriptome. Genome Res. 2018;28:1791–1798. doi: 10.1101/gr.233585.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kerr J.F., Wyllie A.H., Currie A.R. Apoptosis: A basic biological phenomenon with wide-ranging implications in tissue kinetics. Br. J. Cancer. 1972;26:239–257. doi: 10.1038/bjc.1972.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Matthay M.A., Zemans R.L. The acute respiratory distress syndrome: Pathogenesis and treatment. Annu. Rev. Pathol. 2011;6:147–163. doi: 10.1146/annurev-pathol-011110-130158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chau T.N., Lee K.C., Yao H., Tsang T.Y., Chow T.C., Yeung Y.C., Choi K.W., Tso Y.K., Lau T., Lai S.T., Lai C.L. SARS-associated viral hepatitis caused by a novel coronavirus: Report of three cases. Hepatology. 2004;39:302–310. doi: 10.1002/hep.20111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tao X., Hill T.E., Morimoto C., Peters C.J., Ksiazek T.G., Tseng C.T. Bilateral entry and release of Middle East respiratory syndrome coronavirus induces profound apoptosis of human bronchial epithelial cells. J. Virol. 2013;87:9953–9958. doi: 10.1128/JVI.01562-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yeung M.L., Yao Y., Jia L., Chan J.F., Chan K.H., Cheung K.F., Chen H., Poon V.K., Tsang A.K., To K.K., Yiu M.K., Teng J.L., Chu H., Zhou J., Zhang Q., et al. MERS coronavirus induces apoptosis in kidney and lung by upregulating Smad7 and FGF2. Nat. Microbiol. 2016;1:16004. doi: 10.1038/nmicrobiol.2016.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhu N., Wang W., Liu Z., Liang C., Wang W., Ye F., Huang B., Zhao L., Wang H., Zhou W., Deng Y., Mao L., Su C., Qiang G., Jiang T., et al. Morphogenesis and cytopathic effect of SARS-CoV-2 infection in human airway epithelial cells. Nat. Commun. 2020;11:3910. doi: 10.1038/s41467-020-17796-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chan J.F., Zhang A.J., Yuan S., Poon V.K., Chan C.C., Lee A.C., Chan W.M., Fan Z., Tsoi H.W., Wen L., Liang R., Cao J., Chen Y., Tang K., Luo C., et al. Simulation of the clinical and pathological manifestations of coronavirus disease 2019 (COVID-19) in a golden Syrian hamster model: Implications for disease pathogenesis and transmissibility. Clin. Infect. Dis. 2020;71:2428–2446. doi: 10.1093/cid/ciaa325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tan Y.X., Tan T.H., Lee M.J., Tham P.Y., Gunalan V., Druce J., Birch C., Catton M., Fu N.Y., Yu V.C., Tan Y.J. Induction of apoptosis by the severe acute respiratory syndrome coronavirus 7a protein is dependent on its interaction with the Bcl-XL protein. J. Virol. 2007;81:6346–6355. doi: 10.1128/JVI.00090-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tsoi H., Li L., Chen Z.S., Lau K.F., Tsui S.K., Chan H.Y. The SARS-coronavirus membrane protein induces apoptosis via interfering with PDK1-PKB/Akt signalling. Biochem. J. 2014;464:439–447. doi: 10.1042/BJ20131461. [DOI] [PubMed] [Google Scholar]
- 74.Collins M. Potential roles of apoptosis in viral pathogenesis. Am. J. Respir. Crit. Care Med. 1995;152:S20–S24. doi: 10.1164/ajrccm/152.4_Pt_2.S20. [DOI] [PubMed] [Google Scholar]
- 75.Pan D., Pan L.Z., Hill R., Marcato P., Shmulevitz M., Vassilev L.T., Lee P.W. Stabilisation of p53 enhances reovirus-induced apoptosis and virus spread through p53-dependent NF-κB activation. Br. J. Cancer. 2011;105:1012–1022. doi: 10.1038/bjc.2011.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Karuppannan A.K., Kwang J. ORF3 of porcine circovirus 2 enhances the in vitro and in vivo spread of the of the virus. Virology. 2011;410:248–256. doi: 10.1016/j.virol.2010.11.009. [DOI] [PubMed] [Google Scholar]
- 77.Garant K.A., Shmulevitz M., Pan L., Daigle R.M., Ahn D.G., Gujar S.A., Lee P.W. Oncolytic reovirus induces intracellular redistribution of Ras to promote apoptosis and progeny virus release. Oncogene. 2016;35:771–782. doi: 10.1038/onc.2015.136. [DOI] [PubMed] [Google Scholar]
- 78.Fernandes M.H.V., Maggioli M.F., Otta J., Joshi L.R., Lawson S., Diel D.G. Senecavirus A 3C protease mediates host cell apoptosis late in infection. Front. Immunol. 2019;10:363. doi: 10.3389/fimmu.2019.00363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sakamoto K.M., Kim K.B., Kumagai A., Mercurio F., Crews C.M., Deshaies R.J. Protacs: Chimeric molecules that target proteins to the skp1-cullin-F box complex for ubiquitination and degradation. Proc. Natl. Acad. Sci. U. S. A. 2001;98:8554–8559. doi: 10.1073/pnas.141230798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Verma R., Mohl D., Deshaies R.J. Harnessing the power of proteolysis for targeted protein inactivation. Mol. Cell. 2020;77:446–460. doi: 10.1016/j.molcel.2020.01.010. [DOI] [PubMed] [Google Scholar]
- 81.Burslem G.M., Crews C.M. Proteolysis-targeting chimeras as therapeutics and tools for biological discovery. Cell. 2020;181:102–114. doi: 10.1016/j.cell.2019.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sun X., Gao H., Yang Y., He M., Wu Y., Song Y., Tong Y., Rao Y. PROTACs: Great opportunities for academia and industry. Signal Transduct. Target. Ther. 2019;4:64. doi: 10.1038/s41392-019-0101-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.de Wispelaere M., Du G., Donovan K.A., Zhang T., Eleuteri N.A., Yuan J.C., Kalabathula J., Nowak R.P., Fischer E.S., Gray N.S., Yang P.L. Small molecule degraders of the hepatitis C virus protease reduce susceptibility to resistance mutations. Nat. Commun. 2019;10:3468. doi: 10.1038/s41467-019-11429-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jensen S.M., Potts G.K., Ready D.B., Patterson M.J. Specific MHC-I peptides are induced using PROTACs. Front. Immunol. 2018;9:2697. doi: 10.3389/fimmu.2018.02697. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE repository with the dataset identifier PXD030024. All other data are included in this article and its supporting information.