Summary
MV130 is an inactivated polybacterial mucosal vaccine that confers protection to patients against recurrent respiratory infections, including those of viral etiology. However, its mechanism of action remains poorly understood. Here, we find that intranasal prophylaxis with MV130 modulates the lung immune landscape and provides long-term heterologous protection against viral respiratory infections in mice. Intranasal administration of MV130 provides protection against systemic candidiasis in wild-type and Rag1-deficient mice lacking functional lymphocytes, indicative of innate immune-mediated protection. Moreover, pharmacological inhibition of trained immunity with metformin abrogates the protection conferred by MV130 against influenza A virus respiratory infection. MV130 induces reprogramming of both mouse bone marrow progenitor cells and in vitro human monocytes, promoting an enhanced cytokine production that relies on a metabolic shift. Our results unveil that the mucosal administration of a fully inactivated bacterial vaccine provides protection against viral infections by a mechanism associated with the induction of trained immunity.
Keywords: polybacterial immunomodulator, mucosal, vaccine, trained immunity, macrophages, Candida albicans, viral infection, influenza, respiratory infection, human monocytes
Graphical abstract
Highlights
-
•
MV130 reduces morbi-mortality in mouse models of viral respiratory infection
-
•
MV130 modulates mouse immunity complying with key features of trained immunity
-
•
MV130 promotes reprogramming of mouse bone marrow progenitors
-
•
MV130 induces trained immunity in human monocytes
Brandi et al. investigate the mechanism of action of an inactivated polybacterial mucosal immunomodulator, MV130, which confers protection in patients suffering from recurrent respiratory infections. They show that MV130 induces trained immunity both in mouse bone marrow progenitors, resulting in protection from viral infection, and in human monocytes in vitro.
Introduction
Respiratory tract infections (RTIs), an important economic and public health burden, are mainly caused by bacteria and viruses, including influenza virus, respiratory syncytial virus, rhinoviruses, and coronaviruses (Braido et al., 2007). Due to genetic characteristics, environmental factors, and immunological immaturity, children present an increased incidence of recurrent RTIs, which is a significant risk factor for developing asthma later in life (Esposito et al., 2018). Although recurrent RTIs are commonly of viral etiology, the traditional therapeutic strategies rely on antibiotic treatments to prevent secondary bacterial infections (van Houten et al., 2019). Therefore, the search for and development of alternative preventive treatments for viral infections have been priorities in recent years. In this direction, several studies have shown that bacterial preparations provide protection against recurrent RTI through the modulation of both cellular and humoral responses (Koatz et al., 2016; Laiño et al., 2018).
MV130 is a polybacterial mucosal vaccine manufactured by Inmunotek (Spain), composed of different proportions of whole heat-inactivated Gram-positive (90%) and Gram-negative (10%) bacteria including Streptococcus pneumoniae, Staphylococcus aureus, Staphylococcus epidermidis, Klebsiella pneumoniae, Moraxella catarrhalis, and Haemophilus influenzae (Alecsandru et al., 2011; Cirauqui et al., 2018). Clinical studies have shown that patients suffering from recurrent RTIs present a reduced number of viral and/or bacterial infectious episodes after MV130 treatment (Alecsandru et al., 2011; García González and Arrutia Díez, 2019). Moreover, in a phase 3 randomized, double-blind, placebo-controlled trial (NCT01734811), children treated with MV130 showed an important reduction in the incidence and severity of recurrent wheezing (Nieto et al., 2021), a condition associated with viral infections (Jartti et al., 2019; Le Souëf, 2018). The immunomodulatory properties of MV130 have been studied on dendritic cells (DCs) (Cirauqui et al., 2018), oral epithelial cells (Molero-Abraham et al., 2019), and mesenchymal stem cells (Vázquez et al., 2020). However, the mechanisms underlying the broad anti-infectious protection provided by MV130 remain largely unknown.
Trained immunity is the boost of innate immune responses induced by some stimuli, which provide a long-term protection against related or unrelated infections (Netea et al., 2011, 2016). Trained immunity inducers range from infectious agents, such as low doses of live Candida albicans (Quintin et al., 2012) and live vaccines such as Bacillus Calmette-Guerin (BCG) (Kleinnijenhuis et al., 2012), and also include specific microbial components, such as the polysaccharide β-glucan (Quintin et al., 2012) or muramyl dipeptide (Ifrim et al., 2014) from fungal and bacterial cell walls, respectively. Its independence from adaptive immunity has been demonstrated by studies performed on mice lacking B and T cells (Kleinnijenhuis et al., 2012; Quintin et al., 2012). Trained immunity is characterized by an enhanced cytokine production in response to a second challenge, relying on chromatin remodeling and metabolic rewiring (Cheng et al., 2014). In particular, during trained immunity, the promoter regions of genes encoding for some proinflammatory cytokines are enriched in specific histone modifications such as H3-histone-lysine-4 trimethylation (H3K4me3), which are responsible for increased chromatin accessibility with subsequent gene overexpression (Quintin et al., 2012; Saeed et al., 2014). These modifications, defined as epigenetic changes, are reversible and do not affect the DNA sequence but rather change the expression profile of the involved genes (Lacal and Ventura, 2018). Accordingly, the pan-methyltransferase inhibitor 5′-methylthioadenosine (MTA) abolishes these modifications and, thus, the induction of trained immunity (Bekkering et al., 2014; Ifrim et al., 2014; Kleinnijenhuis et al., 2012). Trained innate immune cells display a shift from oxidative phosphorylation to aerobic glycolysis, which requires the protein kinase B (Akt)/mammalian target of rapamycin (mTOR)/hypoxia inducible factor 1α (HIF-1α) pathway (Braza et al., 2018; Kumar et al., 2014). However, oxidative phosphorylation (OXPHOS) also contributes to the induction of trained immunity (Groh et al., 2021). To explain the long-lasting effect of trained immunity, the persistence of trained innate immune cells has been linked to the reprogramming of hematopoietic stem and progenitor cells in the bone marrow (Christ et al., 2018; Kaufmann et al., 2018; Mitroulis et al., 2018).
Here, we demonstrate that pretreatment with the polybacterial mucosal vaccine MV130 provides long-term protection in viral infection models, which is independent of inflammatory monocytes and dependent on the mTOR pathway. In addition, we found that MV130 protects against systemic C. albicans infection in a B and T cell-independent manner, promotes epigenetic rewiring of myeloid progenitors, and induces both epigenetic and metabolic changes in human monocytes in vitro. Thus, we conclude that MV130 can protect against a wide range of pathogens through induction of trained immunity.
Results
MV130 confers heterologous protection against unrelated viral respiratory infections in mice
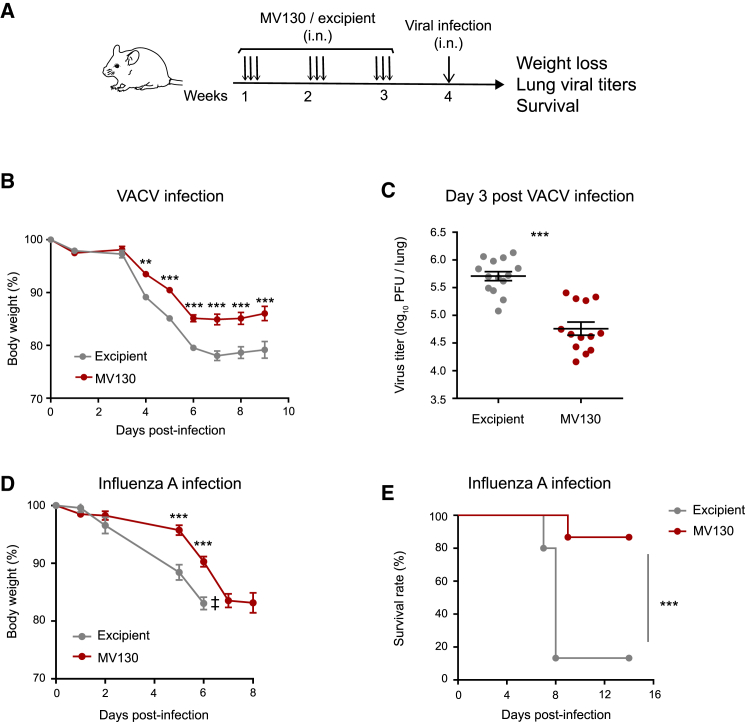
To assess the potential role of MV130 in cross-protecting against viral infections, we designed an in vivo experimental approach to mimic human respiratory infections in mice. The mucosal preparation MV130 or a control (excipient) was administered intranasally (i.n.) three times a week for 3 consecutive weeks (Figure 1A). Seven days after the last dose, mice were challenged with vaccinia virus (VACV) intranasally. Animals pretreated with MV130 showed less infection-related morbidity, measured as a weight loss, than the control group (Figure 1B). This was accompanied by a 10-fold decrease in viral load in the lungs of MV130-treated mice compared with control mice at day 3 post infection (Figure 1C). To decipher whether the control of the virus was exerted in the lung or in upper airways, we evaluated both trachea and lung viral load at day 2 post infection (Figure S1). At this time point no virus was detected in the trachea (Figure S1), consistent with previously published data showing that VACV preferentially replicates in the lung (Reading and Smith, 2003). Importantly, there were no differences in lung viral load between excipient- and MV130-treated mice at this time (Figure S1), suggesting that the same amount of virus reaches the lung and starts to be controlled locally at day 3 post infection. To test the breadth of the protective role of MV130 against antigenically unrelated viral infections, we pretreated mice with MV130 or excipient and infected them with influenza A/Puerto Rico/8/1934 (H1N1) (PR8) virus (Figures 1D and 1E). As shown, pretreatment with MV130 also reduced morbidity (Figure 1D) and mortality (Figure 1E) compared with controls against this human respiratory virus. Thus, these results indicate that MV130 induces heterologous protection against two unrelated respiratory viral infection models.
Figure 1.
MV130 protects against viral respiratory infections
(A) Graphical outline of the in vivo models of MV130 (109 bacteria/mL) treatment followed by i.n. viral infection.
(B) Weight loss after VACV i.n. infection (5 × 104 PFU/mouse). Mean ± SEM are presented.
(C) Lung viral load on day 3 post VACV infection. Individual data and the mean ± SEM are presented.
(D and E) Weight loss (D) and survival (E) of mice infected i.n. with influenza A virus (2 × 103 PFU/mouse = 2 × LD50). ‡ indicates that the excipient group had to be excluded from the graph from day 6 because mice started to die (D).
In (B) and (D), weights were recorded daily and compared using a two-way ANOVA test. In (C), lung viral titers were compared using an unpaired Student's t test. In (E), survival curves were compared with a log rank (Mantel-Cox) test. Results from a pool of two independent experiments with n = 16 (B) and n = 15 (D and E) mice per group. ∗∗p < 0.01; ∗∗∗p < 0.001.
MV130 modulates the lung immune landscape and induces long-term heterologous protection
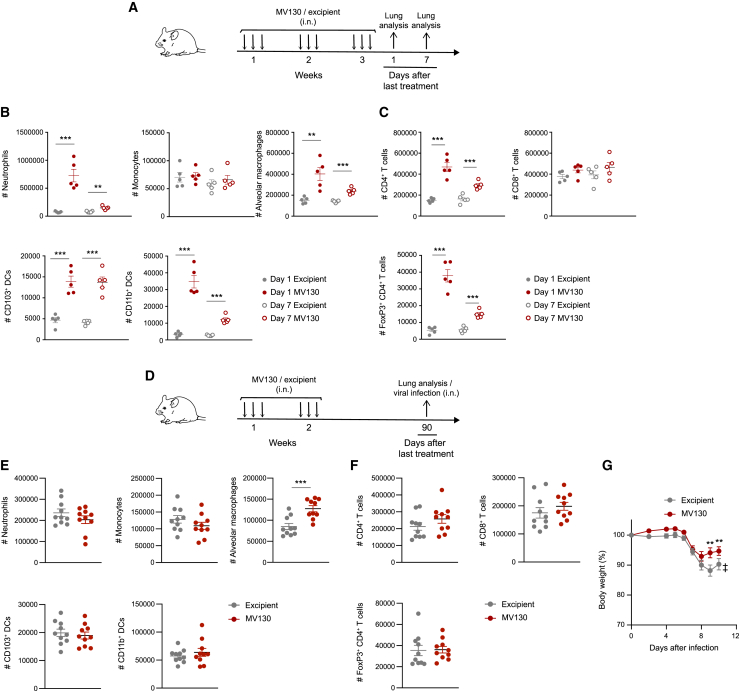
To further understand the mechanisms underlying the protective effects of MV130, we next characterized the lung immune cells after MV130 treatment following the scheme shown in Figure 2A. Lung cell subsets were analyzed following the flow cytometry gating strategy detailed in Figure S2. Myeloid and lymphoid cell populations were increased on day 1, and even on day 7, after the last dose of MV130 (Figures 2B and 2C). In particular, neutrophils, alveolar macrophages (AMs), and both CD11b+ and CD103+ DCs were expanded within the myeloid compartment (Figure 2B). Similarly, a significant increase was seen in the total numbers of CD4+ T cells, including the minor subset of FOXP3+ regulatory CD4+ T cells, while the number of CD8+ T cells remained unchanged (Figure 2C). The magnitude of these changes on day 7, i.e., at the time of virus challenge, was milder compared with day 1, suggesting a quick return to steady-state levels. These results indicated that MV130 is able to promote a transient influx of selected immune cells to the lung.
Figure 2.
MV130 modulates lung immune landscape and promotes long-term protection against viral infections
(A) Graphical outline for data in (B) and (C).
(B and C) Absolute numbers of the indicated lung immune cell populations, myeloid (B) and lymphoid (C), on day 1 and day 7 after the last MV130 (109 bacteria/mL) administration, were analyzed by flow cytometry. Individual data (n = 5) and the mean ± SEM are shown. ∗∗p < 0.01; ∗∗∗p < 0.001 (unpaired Student's t test comparing excipient and MV130 in the same time point).
(D) Graphical outline for data in (E), (F), and (G).
(E and F) Absolute numbers of the indicated lung immune cell populations, myeloid (E) and lymphoid (F), 3 months after the last MV130 (109 bacteria/mL) administration, were analyzed by flow cytometry. Individual data (n = 10) and the mean ± SEM are shown. ∗∗∗p < 0.001 (unpaired Student's t test comparing excipient and MV130).
(G) Weight loss of mice infected i.n. with influenza A virus (103 PFU/mouse). Results from a pool of two independent experiments (mean ± SEM) are shown (n = 16 in excipient group and 16 in MV130 group). ‡ indicates that the excipient group had to be excluded from the graph from day 10 because mice started to die. Weights were compared using a two-way ANOVA test comparing MV130 and excipient groups. ∗∗p < 0.01.
To test the long-term effects of MV130 treatment, we analyzed lung cell subsets 3 months after the last MV130 challenge following the scheme in Figure 2D. There were no differences among either myeloid (Figure 2E) or lymphoid (Figure 2F) cell subsets analyzed, except for a significant but mild increase in AMs of MV130-treated mice, compared with excipient-treated mice (Figure 2E). Of note, pretreatment of mice with MV130 3 months before infection with PR8 influenza A virus reduced infection-related morbidity compared with excipient-pretreated mice (Figure 2G). This result demonstrates that MV130 confers long-term protection against viral infections.
MV130 acts as an inducer of trained immunity in vivo
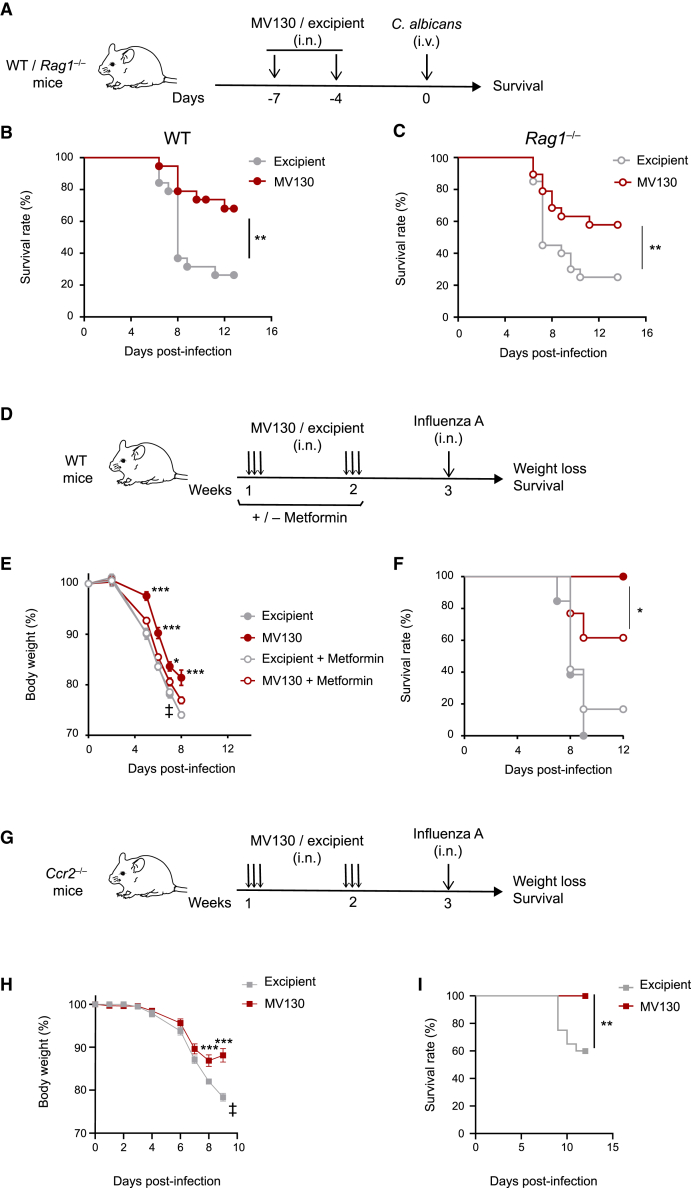
Given the above results, we were prompted to assess whether i.n. MV130 administration was able to induce systemic heterologous protection in a classical trained immunity model. The resistance to systemic infection by C. albicans is a standard model to test innate training in vivo (Quintin et al., 2012). With this aim, mice were pretreated i.n. with MV130 or excipient and, 4 days after the last dose, were intravenously (i.v.) infected with C. albicans (Figure 3A). As shown in Figure 3B, MV130-treated mice displayed reduced mortality compared with excipient-treated mice following systemic C. albicans infection, confirming the ability of mucosal pretreatment with only two doses of MV130 to induce heterologous and systemic protection (Figures 3A and 3B). MV130-mediated protection in this infection model was also seen in Rag1-deficient mice (Figure 3C), which lack mature B and T lymphocytes, thus supporting the notion that this protection mainly relies on innate immunity (Kleinnijenhuis et al., 2012, 2014; Quintin et al., 2012).
Figure 3.
MV130 shows key features of trained immunity
(A) Graphical outline: wild-type C57BL/6 and Rag1−/− mice were treated with MV130 (109 bacteria/mL) or excipient at day −7 and day −4, followed by intravenous infection with 3 × 105 (B) or 1.5 × 105 (C) C. albicans at day 0.
(B and C) Survival of wild-type C57BL/6 (B) and Rag1−/− (C) mice in two pooled independent experiments with n = 19 per group (B) and with n = 20 for excipient-treated and n = 19 for MV130-treated (C) mice. Survival curves were compared with log rank (Mantel-Cox) test. ∗∗p < 0.01.
(D) Graphical outline of the effect of in vivo inhibition with metformin (or not) in mice pretreated with MV130 (109 bacteria/mL) or excipient and subsequently challenged with influenza A virus (2 × 103 PFU/mouse = 2 × LD50).
(E and F) Weight loss (E) and survival (F) of mice following the treatments indicated in (D). Results from a pool of two independent experiments are shown, with n = 13 for excipient, n = 11 for MV130, n = 12 for excipient + metformin, and n = 13 for MV130 + metformin treated mice. Weights were compared using a two-way ANOVA test comparing MV130 and metformin + MV130 groups. Survival curves were compared with log rank (Mantel-Cox) test. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.
(G) Graphical outline for data in (H) and (I).
(H and I) Weight loss (H) and survival (I) of mice following 2 weeks of treatment with MV130/excipient as shown in (G). Results from a pool of two independent experiments are shown (n = 20 in excipient group and 18 in MV130 group). (E and H) data are shown as mean ± SEM. Weights were compared using a two-way ANOVA test comparing MV130 and excipient groups. Survival curves were compared with log rank (Mantel-Cox) test. ∗∗p < 0.01; ∗∗∗p < 0.001.
Activation of mTOR is a key signaling step in the induction of innate immune cell training, and mTOR inhibition completely abrogates the protection conferred by either β-glucan both in vitro and in vivo (Cheng et al., 2014) or BCG (Arts et al., 2016). Metformin, a pharmacological inhibitor of trained immunity, activates the AMPK pathway, leading to mTOR inhibition (Arts et al., 2016; Braza et al., 2018; Cheng et al., 2014). Thus, to further support that MV130-mediated protection against heterologous viral infections relies on training of innate immune cells, mice received metformin in the drinking water, following the scheme shown in Figure 3D. One week later, mice were infected i.n. with influenza A virus. Notably, coadministration of metformin reduced the protection against influenza A infection provided by MV130, in terms of both weight loss (Figure 3E) and survival (Figure 3F). These data indicate that the mTOR pathway is required for the optimal protective effects of MV130, further indicating that trained immunity is involved in its mechanism of action.
Since either AMs or Ccr2-dependent inflammatory monocytes infiltrating the lung can show features of innate immune memory (Quintin et al., 2012; Yao et al., 2018), we addressed the potential involvement of lung-infiltrating monocytes in the local response mediated by MV130. With this aim we used Ccr2-deficient mice, which are impaired in the release of monocytes from the bone marrow to the blood and the recruitment of inflammatory monocytes to the tissues (Tsou et al., 2007). Indeed, we checked that Ccr2 deficiency impaired the presence of monocytes in blood not only in the steady state but also after administration of MV130 (Figure S3). Of note, pretreatment of Ccr2-deficient mice with MV130 1 week before infection with PR8 influenza A virus (Figure 3G) prevented morbidity and mortality compared with excipient-treated mice (Figures 3H and 3I). This result indicates that the protection mediated by MV130 is independent of inflammatory monocytes.
MV130 reprograms multipotent progenitors
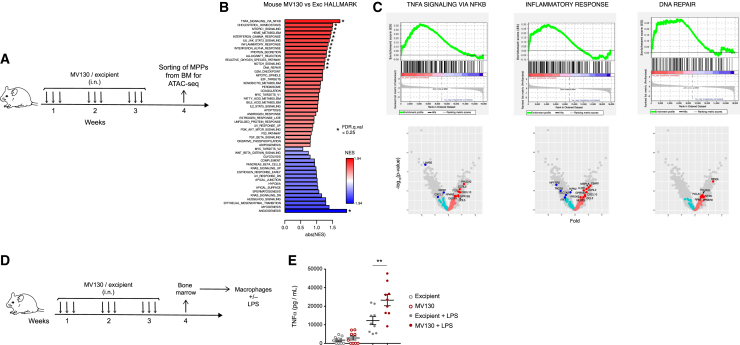
Trained immunity acts via modulation of hematopoietic stem and progenitor cells, and this is translated into functional changes of mature myeloid cells in the periphery (Kaufmann et al., 2018; Mitroulis et al., 2018). We thus hypothesized that MV130 could confer systemic long-term protection through reprogramming of hematopoietic precursors. To test this, we treated mice with MV130 or with its excipient following the scheme in Figure 4A and, after 1 week of resting, multipotent progenitors (MPPs; c-kit+ sca-1+ CD48+ CD150−) were sorted from the bone marrow (Figure S4A) and subsequently analyzed by assay for transposase-accessible chromatin with high-throughput sequencing (ATAC-seq). A consensus peak set representing 58,474 accessible chromatin regions was defined after processing ATAC-seq data from two treated and two control samples. Chromatin accessibility in treated samples, relative to control samples, was calculated for each peak and expressed as a logFC value. A number of 246 differentially accessible regions (DARs; false discovery rate [FDR] < 0.1) were identified in the MPPs from MV130-pretreated mice, compared with the excipient-pretreated mice, by the R package DiffBind (Figure S4B). Correlation analysis confirmed that peaks related to excipient-treated sample 1 and sample 2 clustered more closely to each other compared with peaks from MV130-treated mice and vice versa; thus, samples coming from mice that have received the same treatment are functionally equivalent (Figure S4C). Because the number of DARs was relatively low, we decided to base functional inferences on information from all detected accessible regions. To this end, the closest gene to each consensus peak was identified and the resulting collection of genes was ranked according to the logFC value of associated peaks; preranked gene set enrichment analysis (GSEA) was then used to identify Hallmark gene sets from the Molecular Signatures database (MSigDB) that were enriched in genes that had been associated with open chromatin regions with positive or negative logFC values. GSEA results indicated that gene sets related to inflammatory responses, DNA repair, and tumor necrosis factor α (TNFα) signaling, among others, are enriched in genes that are close to open chromatin regions from MPPs upon pretreatment with MV130 (Figures 4B and 4C), consistent with previous results for other trained immunity inducers (Mitroulis et al., 2018).
Figure 4.
MV130 trains bone marrow myeloid progenitors
(A) Graphical outline of the in vivo model of MV130 treatment followed by myeloid progenitor (MPP) sorting and subsequent ATAC-seq analysis.
(B and C) ATAC-seq analysis in MPPs sorted from bone marrow of mice 1 week after the last challenge with MV130 or its excipient. Bar plot showing enrichment results for all gene sets in the Hallmark collection. Asterisks denote significant enrichment (FDR q value <0.25). Positive enrichment score (in red) suggests association between gene sets and regions with increased chromatin accessibility in MV130-treated samples. Negative enrichment score is in blue (B). GSEA for TNFα signaling, inflammatory response, and DNA repair with their equivalent volcano plots showing biological pathways and genes differentially regulated in excipient-treated and MV130-treated mice (C).
(D) Graphical outline of the in vivo model of MV130 treatment followed by BMDM differentiation and challenge.
(E) TNFα production by ELISA in the supernatants of BMDM in response to LPS treatment. Individual data (n = 9) and the mean ± SEM from two independent experiments. Unpaired Student's t test between MV130 and excipient conditions. ∗p < 0.05; ∗∗p < 0.01.
To test for functional consequences of the reprogramming in bone marrow progenitor cells (Kaufmann et al., 2018; Mitroulis et al., 2018), we obtained bone marrow-derived macrophages (BMDMs), challenged them with lipopolysaccharide (LPS), and measured TNFα production 24 h later in the supernatant (Figure 4D). As shown in Figure 4E, BMDMs generated from MV130-treated mice produced higher levels of TNFα than those derived from excipient-treated control mice. On the contrary, no differences were found in the production of TNFα between MV130 and control, when whole bone marrow was challenged with LPS (Figure S4D). These data indicate that i.n. treatment with MV130 acts systemically to induce epigenetic changes in MPPs, which acquire the capability to generate trained mature macrophages.
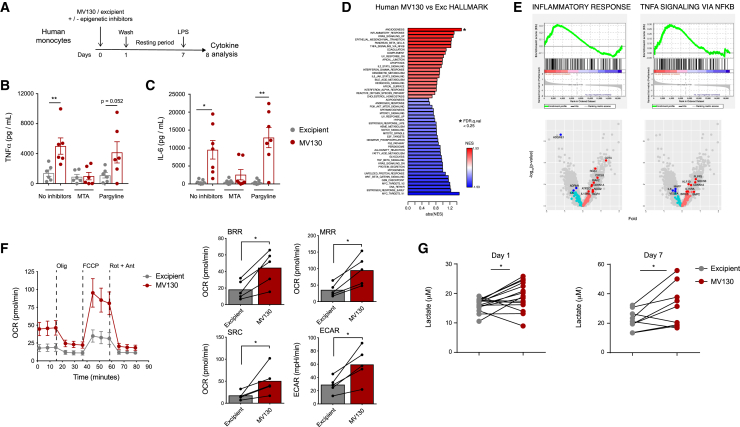
MV130 trains human monocytes in vitro
All the foregoing results were performed in mice; therefore, we were prompted to test whether MV130 may induce training in human monocytes. Different hallmarks that characterize trained immunity, such as an enhanced cytokine production, epigenetic reprogramming, and metabolic rewiring, were assessed in a well-established in vitro model of human monocytes (Netea et al., 2015a, 2015b). Human monocytes were challenged with MV130 or excipient, washed the following day, and stimulated with LPS at day 7. Twenty-four hours later, cytokines were measured in cell culture supernatants (Figure 5A). Monocyte pretreatment with MV130 enhanced the production of TNFα (Figure 5B) and interleukin-6 (IL-6) (Figure 5C) compared with excipient. To evaluate the contribution of epigenetic reprogramming to this increase, we treated monocytes during training with 5′-deoxy-5′-(methylthio)adenosine (MTA), a methyltransferase inhibitor that prevents training, or with pargyline, a demethylase inhibitor with no effects on the training process (Bekkering et al., 2014; Ifrim et al., 2014; Kleinnijenhuis et al., 2012). MTA abolished the cytokine increase, in strong contrast to pargyline (Figures 5B and 5C). These data indicate that pretreatment of human monocytes with MV130 results in increased inflammatory cytokine production that relies on epigenetic rewiring.
Figure 5.
MV130 trains human monocytes in vitro
(A) Schematic representation of the in vitro human monocyte training with MV130.
(B and C) Measurement of both TNFα (B) and IL-6 (C) levels in the supernatants of human monocytes treated as indicated in (A) by ELISA. Each dot represents an independent experiment using monocytes from different donors (n = 6) in independent experiments; mean ± SEM are presented. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001 using paired Student's t test.
(D and E) ATAC-seq analysis in human monocytes 5 days after challenge with MV130 or its excipient. Bar plot showing enrichment results for all gene sets in the Hallmark collection. Asterisks denote significant enrichment (FDR q value <0.25). Positive enrichment score (in red) suggests association between gene sets and regions with increased chromatin accessibility in MV130-treated samples. Negative enrichment score is in blue (D). GSEA for inflammatory response and TNFα signaling with their equivalent volcano plots showing biological pathways and genes differentially regulated in excipient- and MV130-treated cells (E).
(F) Oxygen consumption rate (OCR) of human monocytes incubated with excipient or MV130 was analyzed by Seahorse extracellular flux assay at day 5 post challenge. Basal respiration rate (BRR) was defined as OCR before inhibitor addition. Maximal respiration rate (MRR) was defined as the OCR after addition of carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone (FCCP). Spare respiration capacity (SRC) was defined as the difference between MRR and BRR. Extracellular acidification rate (ECAR) was measured in the absence of inhibitor. Error bars represent SEM. ∗p < 0.05.
(G) Analysis of lactate production in the supernatant of human monocytes 24 h (left, n = 15) and 7 days (right, n = 9) after training with MV130 or excipient. Each dot represents an independent experiment using monocytes from different donors; dots from the same experiment are paired. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001 using paired Student's t test.
To further test the role of epigenetic changes in MV130-mediated trained immunity induction in human monocytes, we performed global epigenetic profiling of open chromatin by ATAC-seq, as we had done in the mouse model. The number of DARs with FDR < 0.1 was 649 (Figure S5A), and the expected correlation was found among samples from the two different conditions (Figure S5B). Although the number of DARs was higher than that detected in mouse MPPs, we again used GSEA to make functional inferences using information from all detected accessible regions. GSEA results indicated that open chromatin regions are associated with inflammatory responses and TNFα signaling genes in human monocytes upon pretreatment with MV130 (Figures 5D and 5E). These results suggest that MV130 induces epigenetic modifications that promote open chromatin in regions that are proximal to proinflammatory pathway genes, which is a hallmark for trained immunity.
Trained immunity is also characterized by changes in cellular metabolism, with both glycolysis and oxidative phosphorylation being involved in this process (Cheng et al., 2014; Groh et al., 2021). Human monocytes pretreated 5 days before with MV130 showed higher basal respiration rate, maximal respiration rate, and spare respiration capacity than control ones, indicating more mitochondrial activity and higher oxidative metabolism (Figure 5F). Moreover, the extracellular acidification rate was found to be upregulated in MV130-pretreated monocytes (Figure 5F). Consistent with this, MV130-pretreated human monocytes increased the production of lactate, both at day 1 and at day 7 (Figure 5G), suggesting increased glycolytic metabolism. Thus, MV130 modulates the function of human monocytes, leading to a metabolic shift and increased cytokine production upon restimulation, which relies on epigenetic changes.
Discussion
The protective role of bacterial immunotherapy in recurrent RTIs has been demonstrated in different clinical studies (Esposito et al., 2018; Feleszko et al., 2019). However, their potential immune mechanisms remain unclear (Esposito et al., 2018) given the frequent viral etiology of these infections, especially in children (Huang et al., 2018; Li et al., 2019). Along the same lines, the polybacterial mucosal vaccine MV130 has been shown to be effective in preventing RTI recurrences in either adults (Alecsandru et al., 2011; García González and Arrutia Díez, 2019) or children (Nieto et al., 2021). Although the action of MV130 has been studied in different cell types (Cirauqui et al., 2018; Molero-Abraham et al., 2019; Vázquez et al., 2020), a clear mechanism of protection is still unidentified. Prevention of RTI recurrences in children points to an antiviral effect, as wheezing attacks in young children are predominantly linked to respiratory virus infections (Jartti et al., 2019; Meissner, 2016; Le Souëf, 2018). Therefore, we set out to investigate the potential antiviral mechanisms of MV130 in mouse experimental models of respiratory virus infections. Here, we found that pretreatment of mice with MV130 greatly reduces their morbidity and mortality when subsequently challenged with either vaccinia or influenza A viruses. These results concur with other experimental studies in which bacterial-derived products, also administered through mucosal routes, protected mice against influenza virus infection by putative non-specific effects on innate immune cells (Tuvim et al., 2009; Bessler et al., 2010; Norton et al., 2010).
Of note, the preventive effect of MV130 in children lasted at least 6 months after treatment interruption (Nieto et al., 2021), which points to a fairly long-lasting protection. We found that protection against viral infections persists for 3 months after the last MV130 challenge in mice. As the concept of trained immunity relies on the long-term adaptation of innate immune cells by their functional reprogramming (Netea et al., 2020), we were prompted to address whether MV130 protection was related to the induction of trained immunity. In this sense, trained immunity inducers, such as BCG or β-glucan, have been shown to promote protection against viral infections in animal models and humans (Arts et al., 2018; Geller and Yan, 2020). The current results clearly indicate that MV130 induces trained immunity in myeloid cells, as it fulfills the main hallmarks described for this phenomenon. MV130 treatment confers protection to mice challenged with C. albicans i.v., which is considered a gold-standard protection model of systemic trained immunity (Quintin et al., 2012). This is also seen in Rag1−/− mice, lacking mature B and T lymphocytes, which is another key feature of trained immunity (Kleinnijenhuis et al., 2014). Finally, the involvement of trained immunity in the antiviral effect of MV130 is supported by the lack of protection in influenza virus-challenged mice treated concomitantly with metformin, an mTOR inhibitor that prevents a proper induction of trained immunity (Cheng et al., 2014).
Epigenetic rewiring of bone marrow progenitors is another critical hallmark linked to trained immunity, as it may explain its long-term effect despite the relatively short life span of most myeloid cells (Kaufmann et al., 2018; Mitroulis et al., 2018). We find that, upon pretreatment of mice with MV130, bone marrow MPPs show open chromatin regions that are close to inflammatory responses, DNA repair, and TNFα, interferon, IL-1, and IL-6 signaling pathway genes, together with upregulation of cholesterol homeostasis genes, according to GSEA results. These findings are in line with previously published works showing the role of both cholesterol and IL-1 signaling in the reprogramming of hematopoietic stem cells and progenitors following oxidized low-density lipoprotein and β-glucan treatment (Christ et al., 2018; Mitroulis et al., 2018). Furthermore, BMDMs from MV130-treated mice displayed an enhanced production of proinflammatory cytokines upon restimulation with LPS, confirming that MV130 reprograms myeloid progenitors also at a functional level (Kaufmann et al., 2018). In addition, human monocytes pretreated with MV130 produced higher levels of proinflammatory cytokines in response to LPS as a second challenge. This effect is mediated by epigenetic rewiring, as demonstrated by the increased enrichment of open chromatin regions related to inflammatory responses and TNFα signaling genes in human monocytes pretreated with MV130, detected by ATAC-seq analysis. Consistently, increased TNFα production upon restimulation in MV130-pretreated monocytes is blocked with MTA, an epigenetic inhibitor known to prevent trained immunity (Kleinnijenhuis et al., 2012; Quintin et al., 2012). The described epigenetic changes are accompanied by the metabolic reprogramming of MV130-treated monocytes that show a higher energetic metabolic state than control treated cells. This is reflected by both increased OXPHOS and glycolytic metabolism (Cheng et al., 2014; Groh et al., 2021; Ifrim et al., 2013).
MV130 is a polybacterial mixture containing a high percentage of whole-cell Gram-positive bacteria with a minor proportion of Gram-negative bacteria. Systems biology combined with functional analyses indicated that both Toll-like receptor (TLR) and NOD-like receptor (NLR) pathways are activated by MV130 on human monocyte-derived DCs, using MyD88 and RIPK2-mediated signaling pathways, respectively (Cirauqui et al., 2018). While microbial ligands engaging NLRs (NOD1 and NOD2) are well-established training inducers, TLR ligands may have opposite effects depending on their type and concentration (Domínguez-Andrés et al., 2019; Ifrim et al., 2014). Although this adds a high level of complexity to the training stimuli derived from MV130, it is the final net result of their combination that determines their biological behavior, as has been noted for either single or multiple pattern recognition receptors (Brown et al., 2007). Another level of complexity when considering bacterial ligands as training inducers is the fact that some inactivated bacterial vaccines (e.g., diphtheria-tetanus-pertussis vaccine) seem detrimental for trained immunity induction (Blok et al., 2020). In fact, epidemiological data suggest that live vaccines (BCG, measles, oral polio) provide long-lasting beneficial effects against a variety of unrelated infections ascribed to a presumed trained immunity induction (Benn et al., 2020; Goodridge et al., 2016). While it is widely accepted that viable pathogens do stimulate the innate immune system better than their dead counterparts by promoting higher inflammation (Blander and Sander, 2012), no clear reasons have been found to explain the superior performance of vaccines containing replicating agents for inducing trained immunity (Arts et al., 2015). We show here that MV130, a fully inactivated mucosal bacterial vaccine, is able to induce trained immunity in mice both in vitro and in vivo. It is worth noting that MV130 is administered daily during several weeks or months, in contrast to single doses of conventional vaccines. Such repetitive doses could induce a sustained immunostimulatory effect for an optimal trained immunity in vivo induction, which might in some way resemble a replicating agent. The demonstration that mucosal mesenchymal stem cells may be acting as reservoirs for MV130 bacteria over time for their subsequent transfer to DCs (Vázquez et al., 2020) may point in the same direction. Nevertheless, examples of trained immunity induced in vivo by few or single doses of both products derived from microorganisms such as β-glucan (Saz-Leal et al., 2018) and adjuvant-free inactivated influenza vaccines (Debisarun et al., 2020) do exist.
Our analysis of the immune landscape of lungs from mice treated with MV130 revealed an expansion of both myeloid and lymphoid compartments. Interestingly, CD103+ DCs and CD4+ T cells, including the FOXP3+ regulatory T cell subset, were increased at day 7 after the last dose. A remarkably similar cell expansion has been previously described in the mesenteric lymph nodes of mice treated orally with a fairly similar bacterial preparation in the form of lysates (Navarro et al., 2011). These changes fit with the ability of MV130 to stimulate DCs to drive T helper 1 (Th1)/Th17 and IL-10 responses both in vitro and in vivo (Cirauqui et al., 2018). Our evidence indicates that AMs could be playing a main role in the trained immunity against viral infections, since they are the cell subset more affected in the long term by MV130 treatment and the protection is independent of inflammatory monocytes that could be recruited to the lung upon inflammation. This is consistent with previous reports showing that AMs can be trained (Yao et al., 2018). In this regard, it is known that metformin inhibits the transcription factor peroxisome proliferator-activated receptor γ (PPAR-γ) in AMs (Sozio et al., 2011), which, in turn, is critical in AMs to control the exaggerated antiviral and inflammatory responses by these cells, thus limiting the development of host disease (Huang et al., 2019). We cannot rule out that part of the observed effects after metformin treatment are due to the inhibition of PPAR-γ, resulting in increased lung inflammation and morbidity following influenza A infection.
Taken together, our results demonstrate that a fully inactivated mucosal bacterial vaccine, used to prevent recurrent respiratory viral (and bacterial) infections, induces trained immunity and may be protective through this mechanism. As these vaccines are generally formulated with the species of bacteria that cause frequent infections, they intrinsically contain bacterial antigens together with properly trained immunity inducers. Therefore, they may be ascribed to the recent notion of trained immunity-based vaccines (TIbV) (reviewed by Sánchez-Ramón et al., 2018). These vaccines may confer broad protection against different pathogens also enhancing antigen-specific responses (Cirauqui et al., 2018). While trained immunity may be modulated by particular host-pathogen interactions (Peignier and Parker, 2020), the potential of mucosal TIbV may be of especial interest in mucosal infections where concomitant pathogens, such as viruses and bacteria, are frequently involved (Mallia et al., 2012).
Limitations of the study
Although this study focused on assessing the ability of MV130 to induce trained immunity, MV130 is a complex mixture of bacteria in which the relevance of each bacterial species has not been explored. This is a limitation of the present study in terms of understanding the true contribution of individual bacteria and the mechanisms of induction that may be operating in this model. Moreover, we have not dissected the molecular moieties that could be mediating trained immunity in this polybacterial preparation or the immune cell receptor(s) on which they are acting. Further characterization of the system deserves future research.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| APC anti-mouse CD45.2 | eBioscience | Cat#558702; RRID: AB_1645215 |
| FITC anti-mouse CD11b | BD Biosciences | Cat#11-0112-85; RRID: AB_464936 |
| PE anti-mouse Ly6G | BD Pharmingen | Cat#551461; RRID: AB_394208 |
| PerCP-Cy 5.5 anti-mouse Ly6C | eBioscience | Cat#45-5932-82 RRID: AB_2723343 |
| V450 anti-mouse CD45.2 | BD Biosciences | Cat#560697; RRID: AB_1727495 |
| FITC Rat anti-mouse I-A/I-E | BD Pharmingen | Cat#553623; RRID: AB_394958 |
| PE/Cy7 anti-mouse CD11b | BD Pharmingen | Cat#552850; RRID: AB_394491 |
| PerCP-Cy™5.5 anti-mouse CD11c | BD Biosciences | Cat#560584; RRID: AB_1727422 |
| AlexaFluor 657 anti-mouse SiglecF | BD Pharmingen | Cat#562680; RRID: AB_2687570 |
| FITC anti-mouse CD3e | Tonbo | Cat#35-0031-U100; RRID: AB_2621659 |
| PE/Cy7 anti-mouse CD4 | Biolegend | Cat#100528; RRID: AB_312729 |
| V450 Rat anti-mouse CD8a | Beckton Dickinson | Cat#560469; RRID: AB_1645281 |
| APC anti-mouse CD25 | TonboBio | Cat#20-0251-U100; RRID: AB_2621567 |
| PE anti-mouse CD4 (GK1.5) | TonboBio | Cat#50-0041-U100; RRID: AB_2621736 |
| PE-Cyanine7 anti-mouse CD45R (B220) | Thermofisher | Cat# 14-0452-82; RRID: AB_467254 |
| Brilliant Violet 605™ anti-mouse/human CD11b | eBioscience (LabClinics) | Cat# 101237; RRID: AB_11126744 |
| FITC anti-mouse Ly6C | BD Biosciences | Cat# 561085; RRID: AB_10584332 |
| APC anti-mouse/rat Ly6g | BD Biosciences | Cat# 560599; RRID: AB_1727560 |
| PE Rat anti-mouse CD117 | BD Pharmigen | Cat# 553355; RRID: AB_394806 |
| PE-Cy7 Rat anti-mouse Ly-6A/E | BD Pharmigen | Cat# 558162; RRID: AB_647253 |
| Biotin Mouse Lineage Panel | BD Pharmigen | Cat# 559971; RRID: AB_10053179 |
| APC-Cy7 anti-mouse CD48 | Biolegend | Cat# 103431; RRID: AB_2561462 |
| Brilliant Violet 605™ anti-mouse CD150 | Biolegend | Cat# 115927; RRID: AB_11204248 |
| Brilliant Violet 650™ anti-mouse CD11c | Biolegend | Cat# 117339; RRID: AB_2562414 |
| BV421 Rat anti-mouse CD103 | BD Biosciences | Cat# 562771; RRID: AB_2737783 |
| Brilliant Violet 711™ anti-mouse Ly-6C | Biolegend | Cat# 128037; RRID: AB_2562630 |
| Anti-Mouse CD45 APC-eFluor® 780 | eBioscience | Cat# 47-0451-82; RRID: AB_1548781 |
| PERCP/Cy5.5 anti-mouse CD8a | Biolegend | Cat# 100734; RRID: AB_2075238 |
| PE/Cy7 anti-mouse CD4 | Biolegend | Cat# 100421; RRID: AB_312706 |
| BV786 Rat anti-mouse CD45R/B220 | BD Biosciences | Cat# 563894;RRID: AB_2738472 |
| BV786 Rat Anti-Mouse CD90.2 | BD Biosciences | Cat#740841; RRID: AB_2740495 |
| PE/Cy7 anti-mouse CD24 | Biolegend | Cat# 101822; RRID: AB_756048 |
| PerCP/Cy5.5 anti-mouse F4/80 | Biolegend | Cat# 123128;RRID: AB_893484 |
| Alexa Fluor® 700 anti-mouse I-A/I-E [M5/114.15.2] | BioLegend | Cat# 107622; RRID: AB_493727 |
| FITC anti-mouse/rat Foxp3 FJK-16s | eBioscience | Cat#11-5773-82; RRID: AB_465243 |
| Hoechst 33258 | ThermoFisher | Cat# H3569; RRID: AB_2651133 |
| anti-mouse CD16/CD32, clone 2.4G2 | Tonbo | Cat# 70-0161; RRID: AB_2621487 |
| Bacterial and virus strains | ||
| Western Reserve (WR) strain Vaccinia virus (VACV) | (Iborra et al., 2016) | N/A |
| Influenza A/Puerto Rico/8/34 | (Iborra et al., 2016) | N/A |
| Candida albicans (strain SC5314, clinical isolate) | (Saz-Leal et al., 2018) | N/A |
| Biological samples | ||
| Buffy coats | Andalusian Biobank | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Metformin hydrochloride | Sigma-aldrich | PHR1084 |
| Pargyline hydrochloride | Sigma-aldrich | P8013 |
| 5′-Deoxy-5′-(methylthio)adenosine | Sigma-aldrich | D5011 |
| LPS-EK | Invivogen | tlrl-eklps |
| Oligomycin | Sigma | Cat# O4876 |
| FCCP | Sigma | Cat# C2920 |
| Rotenone | Sigma | Cat# R8875 |
| Antimycin A | Sigma | Cat# A8674 |
| DMEM | Gibco, Life Technologies | Cat#D5030 |
| RPMI 1640 Media | Gibco, Life Technologies | Cat#61870150 |
| PBS 1X pH 7.2 | Gibco, Life Technologies | Cat#20012-019 |
| Liberase TL | Roche | Cat#5401020001 |
| TMB Stop Solution | SeraCare | Cat#5150-0020 |
| IFNγ | BD Pharmingen™ | Cat#554587 |
| Critical commercial assays | ||
| Mouse TNFα Elisa Kit | BD OptEIA | Cat#555268 |
| Human IL-6 DuoSet Elisa | R&D Systems | Cat#DY206-05 |
| Human TNFα DuoSet Elisa | R&D Systems | Cat#DY210 |
| Lactate assay kit | Sigma | Cat#MAK064 |
| Illumina Tagment DNA Enzyme and Buffer | Illumina | Cat# 20034197 |
| MinElute PCR Purification Kit (250) | Quiagen | Cat# 28006 |
| AMPure XP | Beckman | Cat#A63880 |
| Streptavidin Microbeads | Miltenyi | Cat# 130-048-101 |
| LS Columns | Miltenyi | Cat# 130-042-401 |
| NEBNext® High-Fidelity 2X PCR Master Mix | New Englands BioLabs | M0541S |
| Deposited data | ||
| Raw data files for ATAC sequencing | NCBI Gene Expression Omnibus | GEO: GSE183721 |
| Experimental models: Cell lines | ||
| CV1 | CSIC/Autónoma University | N/A |
| L929 | (Saz-Leal et al., 2018) | N/A |
| Experimental models: Organisms/strains | ||
| C57BL/6 | The Jackson Laboratory | RRID: IMSR_JAX:000664 |
| B6.129S7-Rag1tm1Mom/J | The Jackson Laboratory | RRID: IMSR_JAX:002216 |
| Ccr2-/- mice (B6 background) | Kindly supplied by Dr. F. Tacke (RW-TH-University Hospital Aachen, Germany) | N/A |
| Oligonucleotides | ||
| Adaptors for ATAC-Seq, see Table S1 | (Buenrostro et al., 2015) | N/A |
| Software and algorithms | ||
| FlowJo | Flowjo | https://www.flowjo.com/ |
| Graph Prism | GraphPad | https://www.graphpad.com/ |
| Microsoft Excel | Microsoft | https://products.office.com/ |
| FastQC | Babraham Bioinformatics | https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ |
| Cutadapt | (Martin, 2011) | https://cutadapt.readthedocs.io/en/stable/ |
| Bowtie2 | (Langmead and Salzberg, 2012) | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
| MACS3 | (Zhang et al., 2008) | https://github.com/macs3-project/MACS |
| HOMER | (Heinz et al., 2010) | http://homer.ucsd.edu/homer/ |
| DiffBind | (Stark and Brown, 2011) | https://bioconductor.org/packages/release/bioc/html/DiffBind.html |
| GSEA | (Subramanian et al., 2005) | https://www.gsea-msigdb.org |
| MSigDB (Hallmark gene set collection) | (Liberzon et al., 2015) | https://www.gsea-msigdb.org/gsea/msigdb/ |
| Other | ||
| Excipient | Inmunotek | N/A |
| MV130 | Inmunotek | N/A |
| Canto HTS | BD Biosciences | N/A |
| Fortessa | BD Biosciences | N/A |
| Aria Cell Sorter | BD Biosciences | N/A |
| Qubit® fluorometer | Life Technologies | N/A |
| Bioanalyzer | Agilent | N/A |
| NextSeq 2000 System | Illumina | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, David Sancho (dsancho@cnic.es).
Materials availability
This study did not generate new unique reagents. All requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact author. All reagents will be made available on request after completion of a Materials Transfer Agreement.
Experimental model and subject details
Mouse strains
Mice were bred at the CNIC and UCSF specific pathogen-free conditions. Mouse strains include C57BL/6 mice, Rag1−/− mice (B6.129S7-Rag1tm1Mom/J) both from The Jackson Laboratory and Ccr2 −/− mice (B6 background) that were kindly supplied by Dr. F. Tacke (RW-TH-University Hospital Aachen, Germany). We used 7- to 10-week-old female mice for viral infection models as sex-associated phenotypical differences have been described in response to viral infections (Geurs et al., 2012). For the other experimental procedures both male and female age-matched mice were equally used. All animal procedures were approved by the local ethics committee and complied with the EU Directive 2010/63EU and Recommendation 2007/526/EC regarding the protection of animals used for experimental research, enforced in Spanish law under Real Decreto 1201/2005.
Cell lines
The CV-1 cell line, used for the determination of the lung viral titer, was a gift from M. del Val (CSIC/Autónoma University, Madrid, Spain). It was cultured in Dulbecco's Modified Eagle's Medium (DMEM, Sigma) supplemented with 10% heat-inactivated fetal calf serum (FCS, Sigma), 100 U/mL penicillin and 100 μg/mL streptomycin (both from Lonza) at 37°C and 5% CO2. CV-1 cells were seeded in monolayers on 24-well plates (Corning) and left 24 h before viral infection.
The L929 cell line (ATCC CCL-1TM), used for the production of the M-CSF supernatant, was grown on 175 cm2 cell culture flasks (Stemcell) and cultured in RPMI 1640 (Sigma) supplemented with 10% heat-inactivated fetal calf serum (FCS, Sigma), 1 mM pyruvate (Lonza), 100 μM non-essential amino acids (Thermo Fisher Scientific), 2 mM L-glutamine, 100 U/ml penicillin, 100 μg/mL streptomycin (all three from Lonza) and 50 μM 2-mercaptoethanol (Merck, Darmstad, Germany), herein called R10. Supernatants were obtained by filtering 15-days long cultures through 0.22 μm Stericup Filter units (Merck Millipore) and were used to subsequently supplement the medium for the generation of bone marrow derived macrophages.
Microorganisms
Western Reserve (WR) strain Vaccinia virus (VACV) was kindly provided by J.W. Yewdell and J.R. Bennink (NIH, Bethesda, Maryland, USA). Stocks were generated in CV-1 cell line monolayers and subsequently used as clarified sonicated cell extracts.
Influenza A/Puerto Rico/8/34 (flu PR8) virus was a gift from E. Nistal-Villán (San Pablo CEU University, Madrid, Spain).
Candida albicans (strain SC5314, clinical isolate) was kindly provided by Prof. C. Gil (Complutense University, Madrid, Spain). The fungus was grown on yeast extract-peptone-dextrose (YPD)-agar plates (Sigma) at 30°C for 48 h to maintain the degree of virulence.
Human samples
Buffy coats from healthy adult volunteers, of unspecified age and gender, were obtained from the Andalusian Biobank after approval by the local Instituto de Salud Carlos III (ISCIII) Research Ethics Committee (PI 47_2018). Human peripheral blood mononuclear cells (hPBMCs) were isolated by differential centrifugation using Biocoll Separating Solution (Cultek). Cells were washed twice with phosphate-buffered saline (PBS, Gibco) and total hPBMCs were resuspended in Roswell Park Memorial Institute (RPMI) 1640 (Sigma). Cells were counted in a CASY cell counter (Innovatis AG) and 5 × 10 5 cells were plated in 96-well plates (Corning, 100 μL final volume) whereas 15 × 106 cells were plated in 10cm Petri dishes (Greiner), incubated at 37°C and 5% CO2 for 1 h and then washed with warm PBS. Remaining adherent cells, mainly monocytes (10% approximately, 5 × 104 cells/well and 1.5 × 106 cells/plate) were kept in RPMI 1640 (Sigma) supplemented with 10% FCS, 2 mM L-glutamine (Lonza), 100 U/mL penicillin and 100 μg/mL streptomycin and used for the experiments described below.
Methods details
Murine infection models
For viral infection models, WT mice were intranasally (i.n.) challenged with 50 μL of MV130 (300 Formazin Turbidity Units [FTU]/mL ∼ 109 bacteria/mL) or excipient 3 times a week for 3 weeks. One week after the last challenge, mice were i.n. infected with 50 μL of VACV WR (5 × 104 plaque-forming units -PFU- per mouse) or influenza virus A/Puerto Rico/8/1934 (PR8) (2 × 103 PFU per mouse), both diluted in sterile PBS, and mice monitored daily for weight, general health, and survival, following the institutional guidance. For metformin inhibition experiments, WT mice were intranasally (i.n.) challenged as described above 3 times a week for 2 weeks, and the pharmacological inhibitor metformin hydrochloride (Sigma) was administered ad libitum in drinking water (0.3 mg/mL of water, corresponding to an approximate dose of 72 mg/Kg per day) from 1 day before the first MV130/excipient challenge to four days after the last dose.
In the case of Candida albicans infection model, mice were i.n. trained with 50 μL of MV130 (300 FTU/mL ∼ 109 bacteria/mL) or excipient on days −7 and −4. At day 0, mice were intravenously infected with 3 × 105 (WT mice) or 105 (Rag1-deficient mice) C. albicans and monitored daily for general health, and survival, following the institutional guidance.
Lung and trachea viral titer
In order to determine lung and trachea viral titers, mice were sacrificed with a lethal dose of Dolethal (100 mg/Kg) and whole lungs were perfused with cold PBS. Both Lungs and trachea were harvested, kept in DMEM (1mL for lung and 0.5mL for trachea) and cut in small pieces before freezing. Both organs were frozen and thawed twice, sonicated for 3 min and serially diluted in DMEM. Culture medium was removed from CV-1-containing wells and 200 μL of dilution was added. After the first hour, 500 μL of fresh CV-1 medium was additionally provided. 36 h later, supernatant was removed, and wells were stained for 5 min with crystal violet solution (0.5% crystal violet [Sigma], 10% methanol [Sigma] and 1% paraformaldehyde [Merck] in deionized water) and washed. Viral plaques were counted, and data are shown as log10.
Lung characterization
Mice were sacrificed with a lethal dose of Dolethal (100 mg/Kg) and perfused with cold PBS. Harvested lungs were collected in RPMI, cut into small pieces, and enzyme digested with Liberase TM (Sigma-Aldrich) for 30 min at 37°C. Cells were passed through a 70 μm cell strainer (Falcon) and washed with flow cytometry buffer. After red blood cell lysis, cells were centrifuged, resuspended in cold flow cytometry buffer and stained for flow cytometry analysis.
Blood analysis
Murine blood was collected via submandibular bleeding, in EDTA-coated tubes both before and 2 days after the last challenge with MV130/excipient. Cells were lysed with red blood cell lysis twice for 5 min. After centrifugation cells were resuspended in cold flow cytometry buffer and stained for flow cytometry analysis of CD11b+ Ly6Chi monocytes.
Sorting of mouse MPPs
Mice were sacrified in a CO2 chamber to collect femurs and tibia that were subsequently flushed with cold PBS to harvest bone marrow. RBC lysis was performed once for 3 min followed by enrichment of Lineage negative (Lin−) cells. For that, cells were incubated in cold flow cytometry buffer with a panel of biotinylated antibodies against lineage committed cells (CD3, CD45R, CD11b, Ter-119, Gr-1 all biotinylated and from BD Pharmigen. After washing, cells were incubated with streptavidin magnetic beads (Miltenyi) and passed through LS columns (Miltenyi). Thus, collected cells were stained with Sca-1, c-kit (CD117), CD48, CD150 antibodies and with the viability marker Hoechst 33258 prior to be sorted by FACS Aria II for further ATAC-Sequencing analysis. MPPs were identified as Lin-c-kit+ sca-1+ CD48+ CD150- cells.
Trained immunity in BMDM
MV130 and excipient pretreated mice were sacrificed in a CO2 chamber to collect femurs and tibia that were subsequently flushed with cold PBS to harvest bone marrow. RBC lysis was performed once for 3 min at room temperature (RT). To obtain BMDM, following an already published paper (Saz-Leal et al., 2018), cell suspensions were plated in non-treated cell culture plates (Corning) in R10, plus M-CSF (30% mycoplasma-free L929 cell supernatant) at 37°C for 5 days. At day 5, BMDM were detached in phosphate buffered saline (PBS, Gibco) supplemented with 5 mM EDTA (PBS/EDTA, Life Technologies), counted and subsequently plated in R10, at equal number (105 cells per well) in 96-well plates (200-μL final volume, Corning) and rested overnight. Then, cells were primed with 25 ng/mL IFNγ (BD Biosciences, San Jose, CA) for 24h. After that, cells were washed and stimulated with R10 or 1 μg/mL check Escherichia coli LPS (EK, Invivogen, San Diego, CA) overnight. Supernatants were subsequently collected for TNFα measurement by ELISA (Mouse TNFα Opteia ELISA kit, BD Biosciences).
Ex vivo stimulation of BM cells
Bone marrow single cell suspension was obtained from femurs of MV130 or excipient pretreated mice, as described above. Afterward, cells were cultured in R10, counted and plated at equal number (105 cells/well) in 96-well flat bottom wells. Following an already published protocol (Christ et al., 2018) cells were incubate for 6 h with LPS (10ng/ml; TLR4 ligand) or with R10 as control and TNF-α levels were subsequently measured in the supernatants by ELISA (Mouse TNFα Opteia ELISA kit, BD Biosciences).
Trained immunity in human monocytes
Plated human monocytes were stimulated at day 0 with MV130 (2 × 104 bacteria/well) or excipient in a final volume of 200 μL for 24 h, washed with fresh medium and rested. On day 7, cells were washed again and further stimulated with 1 μg/mL LPS for 24 h and supernatants were collected for TNF-α and IL-6 measurement by ELISA, following manufacturer's instructions (Human TNF-α DuoSet and Human IL-6 DuoSet, both from R&D Systems). When required, cells were pre-treated with the epigenetic inhibitors 5′-Deoxy-5′-(methylthio) adenosine (MTA) (1 mM) or pargyline (3 μM) 60 min prior to MV130/excipient stimulation. To explore the metabolic status of the cells, after the challenge with MV130 or excipient, and always prior to LPS stimulation, supernatants were collected at days 1 and 7 and lactate concentration was determined by Lactate Assay Kit (Sigma-Aldrich), following manufacturer's instructions.
Cell preparation for ATAC-Seq
For ATAC-sequencing, 30.000 flow sorted mouse MPPs, obtained pooling two biological replicates for every condition and 50.000 human monocytes, were collected in ice-cold Flow cytometry buffer, and immediately processed following previously published protocols (Buenrostro et al., 2015). In particular, cells were resuspended and spun down in 25 μL cold lysis buffer at 500 × g for 20 min at 4°C. Afterward, the transposition reaction was started by adding Nextera's Tn5 Transposase in reaction buffer. The transposition reaction mix was incubated for 30 min at 37°C, and DNA was purified using a Quiagen MinElute PCR purification kit. ATAC-seq libraries were generated using different adaptors (Table S1) from a previously published paper (Buenrostro et al., 2015) and were purified using a PCR purification MinElute kit (Qiagen). Afterward, AMPure XP beads (Beckman) were used for size selection of the libraries, which have been subsequently quantified using a Qubit fluorometer (ThermoFisher Scientific). Libraries size and quality were assessed using a 2100 Bioanalyzer instrument (Agilent). Finally, Libraries were sequenced 2 × 50 in a paired end flowcell (PE) run on Illumina NextSeq 2000 System. (Illumina). NGS experiments were performed in the Genomics Unit of the CNIC.
ATAC-seq data analysis
Sequencing reads were pre-processed by means of a pipeline that used FastQC, to assess read quality, and Cutadapt to trim sequencing reads, eliminating Illumina and Nextera transposase adapter contaminations, and to discard reads that were shorter than 30 bp. Resulting reads were then mapped against reference genomes GRCm38/MM10 or GRCh38/HG38, for mouse and human samples, respectively, with a pipeline that used bowtie2 as aligner, Piccard to mark duplicate alignments, and samtools to eliminate duplicate, chimeric and sub-optimally multi-mapped alignments, keeping only properly paired and mapped reads. Alignments against the mitochondrial genome or the chromosome Y were also removed. The final number of correctly aligned, filtered read pairs was between 8 and 17 million for mouse samples, and between 17 and 24 million for human samples. TSS enrichment values, calculated with HOMER's annotatePeaks function had values between 21 and 25 for mouse samples, and between 15 and 22 for human samples. Once filtered alignments had been obtained, peaks (accessible DNA regions) were called with MACS3, using parameters "--nomodel --shift −100 --extsize 200″, and "-q 0.05″ as the false discovery rate cut-off. The numbers of peaks detected were between 47 and 63 thousand for mouse samples, and between 24 and 37 thousand for human samples. Next, filtered alignments and peaks, in bam and bed formats, respectively, were processed with the R package DiffBind to define consensus sets of 58,474 peaks for mouse samples, and 34,049 peaks for human samples. DiffBind was also used to recalculate read coverage for consensus peaks and to normalize peak coverage signal intensity across samples, to calculate logFC values between condition-averaged intensities, and to identify differentially accessible regions (DARs) in the activated condition versus the non-activated one, using EdgeR as analysis method, and taking into account that human samples were paired. The fraction of reads in peaks (FRiP score), as calculated by DiffBind, was around 0.25 for mouse samples, and around 0.12 for human samples. Correlation analyses based on the signal intensity of consensus peaks indicated that samples clustered correctly by condition (Figures S4C and S5B). The number of DARs (with FDR <0.1) was 246 for mouse samples, and 649 for human samples (Figures S4B and S5A). Because the number of DARs detected in both mouse and human assays was relatively low, we decided to rely on GSEA to obtain functional information from ATAC-Seq results. To this end, we used HOMER to identify the closest gene to each consensus peak; then, the resulting gene lists were ranked according to the logFC value of associated peaks; finally, GSEA preranked was used to identify Hallmark gene sets from the Molecular Sigatures database (MSigDB) that were significantly enriched in genes that had been associated to open chromatin regions with extreme logFC values (positive or negative). Other data manipulations and graphical representations (bar and volcano plots) were produced with R.
Seahorse assay of human monocytes
Real-time Oxygen Consumption Rate (OCR) and Extracellular Acidification Rate (ECAR) in human monocytes were determined with an XF-96 Extracellular Flux Analyzer (Seahorse Bioscience). Seahorse cell culture plates (Agilent) were coated with 22.4μg/ml Cell-Tak for 20 min at room temperature. Human monocytes, obtained as previously described, were plated in 10cm Petri dishes (Greiner) and stimulated at day 0 with MV130 (3 × 106 bacteria/plate) or excipient in a final volume of 10 mL. After 24 h they were washed and left resting. At day 5 cells were detached, resuspended in Seahorse assay medium (DMEM, 100μg/ml penicillin, 100 μg/mL streptomycin, 2mM glutamine, 25mM glucose, 1mM pyruvate) pH 7.4 and subsequently plated onto Seahorse cell culture plates in triplicates. Seahorse cartridges were hydrated overnight in distilled water and calibrated for 1 h in Seahorse XF Calibrant solution in a non-CO2-corrected incubator at 37°C. Seahorse assay was run using the Mito Stress protocol. Briefly, the following inhibitors were added to the seahorse cartridge consecutively: Port A - 1μM oligomycin; Port B - 1μM FCCP; Port C - 1μM rotenone and 1μM antimycin A. OCR and ECAR were analyzed using the Mito Stress protocol with 3 measurements before inhibitor injection, and 3 measurements upon each subsequent injection. Basal respiration rate (BRR) was defined as OCR before inhibitor addition. Maximal respiration rate (MRR) was defined as the OCR after addition of FCCP. Spare respiration capacity (SRC) was defined as the difference between MRR and BRR. ECAR was measured in the absence of inhibitor.
Quantification and statistical analysis
Statistical analysis
Mice were included in the studies in a blind manner and randomly assigned to receive MV130 or excipient (1:1 simple randomization). The same procedure was used to allocate mice to different groups in the presence of metformin.
Sample size was calculated according to a previously performed pilot study to determine the infective dose of different pathogens and the protective ability of MV130 vaccine. For the Influenza infection experiments, we considered two independent study groups with a dichotomous primary endpoint represented by mortality. As statistical parameters, we chose an anticipated incidence of 50% for one group and 5% for another one, an alpha error of 0.05 and a power of 0.8. For the experiments involving Vaccinia infection, we considered two independent study groups with a continuous primary endpoint represented by the percentage of weight loss. As statistical parameters we chose an anticipated means of 20 and 15% of weight decrease with a standard deviation of 2, an alpha error of 0.05 and a power of 0.8.
For C. albicans experiments, we considered two independent study groups with a dichotomous primary endpoint represented by mortality. As statistical parameters we chose an anticipated incidence of 60% for one group and 20% for another one, an alpha error of 0.05 and a power of 0.8. In the experimental models, the distribution normality of data was evaluated with the Kolmogorov-Smirnov (KS) test. Differences in weight loss between MV130 and excipient groups were compared using a two-way ANOVA test, followed by multiple comparisons corrected using Bonferroni statistical hypothesis testing. Survival curves were compared with log rank (Mantel-Cox) test. Two-tailed unpaired Student's t test was generally used to evaluate statistical significance between two conditions, except for the Seahorse assay of human monocytes in which paired Student's t test has been applied. In vitro experiments with human monocytes were evaluated using RM one-way ANOVA with the Geisser-Greenhouse correction with Fisher's LSD test. Differences were considered significant at P < 0.05 (∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001). (Prism 9, Graph-Pad Software). The statistical test used and the definition of n, are indicated in figure legends.
Acknowledgments
We are grateful to members of the D.S. laboratory for discussions and critical reading of the manuscript. We thank the CNIC facilities and personnel for assistance. P.B. is funded by grant BES-2014-069933 (“Ayudas para Contratos Predoctorales para la Formación de Doctores 2014”) from the Spanish Ministry of Economy, Industry and Competitiveness (MINECO). L.C. was a recipient of a European Respiratory Society Fellowship (RESPIRE2-2013-3708). G.D. is supported by a European Molecular Biology Organization Long-term Fellowship (ALTF 379-2019). This project has received funding from the European Union’s Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie grant agreement No. 892965. Work in the D.S. laboratory is funded by the CNIC; by the European Research Council (ERC-2016-consolidator grant 725091); by the European Commission (635122-PROCROP H2020); by Ministerio de Ciencia e Innovación (MICINN), Agencia Estatal de Investigación (AEI), and Fondo Europeo de Desarrollo Regional (FEDER) (SAF2016-79040-R); by AEI (PID2019-108157RB); by Comunidad de Madrid (B2017/BMD-3733 Immunothercan-CM); by FIS-Instituto de Salud Carlos III, MICINN and FEDER (RD16/0015/0018-REEM); by a collaboration agreement with Inmunotek; by Atresmedia (Constantes y Vitales prize); by Fundació La Marató de TV3 (201723); and by Fondo Solidario Juntos (Banco Santander). The CNIC is supported by the Instituto de Salud Carlos III, the MICINN, and the Pro CNIC Foundation.
Author contributions
P.B., L.C., and D.S. conceived and designed the laboratory experiments; P.B., L.C., F.J.C., S.M.-C., G.D., C.R., P.S.-L., M.E., C.d.F., and J.A.-I. performed the laboratory experiments; M.J.G., A.Q., and A.D. performed essential analyses; E.N.-V. and C.A. provided essential reagents; P.B., L.C., and D.S. analyzed and interpreted the laboratory experiments; P.B., L.C., F.J.C., A.N., M.C., J.L.S., and D.S. wrote the manuscript. All authors reviewed and revised the manuscript prior to submission.
Declaration of interests
J.L.S., M.C., L.C., S. M.-C., and P.S.-L. were employees of Inmunotek S.L. at the time of the work. The D.S. lab receives funds from a collaboration agreement between CNIC and Inmunotek. The other authors declare no competing interests.
Published: January 4, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2021.110184.
Supplemental information
Data and code availability
•ATAC-Seq data have been deposited at NCBI's Gene Expression Omnibus and are publicly available as of the date of publication. Accession number is listed in the key resources table.
•This paper does not report original code.
•Any additional information required to reanalyze the data reported in this work paper is available from the Lead Contact upon request.
References
- Alecsandru D., Valor L., Sánchez-Ramón S., Gil J., Carbone J., Navarro J., Rodríguez J., Rodríguez-Sainz C., Fernández-Cruz E. Sublingual therapeutic immunization with a polyvalent bacterial preparation in patients with recurrent respiratory infections: immunomodulatory effect on antigen-specific memory CD4+ T cells and impact on clinical outcome. Clin. Exp. Immunol. 2011;164:100–107. doi: 10.1111/j.1365-2249.2011.04320.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arts R.J.W., Blok B.A., Aaby P., Joosten L.A.B., de Jong D., van der Meer J.W.M., Benn C.S., van Crevel R., Netea M.G. Long-term in vitro and in vivo effects of γ-irradiated BCG on innate and adaptive immunity. J. Leukoc. Biol. 2015;98:995–1001. doi: 10.1189/jlb.4MA0215-059R. [DOI] [PubMed] [Google Scholar]
- Arts R.J.W., Carvalho A., La Rocca C., Palma C., Rodrigues F., Silvestre R., Kleinnijenhuis J., Lachmandas E., Gonçalves L.G., Belinha A., et al. Immunometabolic pathways in BCG-induced trained immunity. Cell Rep. 2016;17:2562–2571. doi: 10.1016/j.celrep.2016.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arts R.J.W., Moorlag S.J.C.F.M., Novakovic B., Li Y., Wang S.Y., Oosting M., Kumar V., Xavier R.J., Wijmenga C., Joosten L.A.B., et al. BCG vaccination protects against experimental viral infection in humans through the induction of cytokines associated with trained immunity. Cell Host Microbe. 2018;23:89–100.e5. doi: 10.1016/j.chom.2017.12.010. [DOI] [PubMed] [Google Scholar]
- Bekkering S., Quintin J., Joosten L.A.B., van der Meer J.W.M., Netea M.G., Riksen N.P. Oxidized low-density lipoprotein induces long-term proinflammatory cytokine production and foam cell formation via epigenetic reprogramming of monocytes. Arterioscler. Thromb. Vasc. Biol. 2014;34:1731–1738. doi: 10.1161/ATVBAHA.114.303887. [DOI] [PubMed] [Google Scholar]
- Benn C.S., Fisker A.B., Rieckmann A., Sørup S., Aaby P. Vaccinology: time to change the paradigm? Lancet Infect. Dis. 2020;20:e274–e283. doi: 10.1016/S1473-3099(19)30742-X. [DOI] [PubMed] [Google Scholar]
- Bessler W.G., Vor dem Esche U., Masihi N. The bacterial extract OM-85 BV protects mice against influenza and salmonella infection. Int. Immunopharmacol. 2010;10:1086–1090. doi: 10.1016/j.intimp.2010.06.009. [DOI] [PubMed] [Google Scholar]
- Blander J.M., Sander L.E. Beyond pattern recognition: five immune checkpoints for scaling the microbial threat. Nat. Rev. Immunol. 2012;12:215–225. doi: 10.1038/nri3167. [DOI] [PubMed] [Google Scholar]
- Blok B.A., de Bree L.C.J., Diavatopoulos D.A., Langereis J.D., Joosten L.A.B., Aaby P., van Crevel R., Benn C.S., Netea M.G. Interacting, nonspecific, immunological effects of bacille Calmette-Guérin and tetanus-diphtheria-pertussis inactivated polio vaccinations: an explorative, randomized trial. Clin. Infect. Dis. 2020;70:455–463. doi: 10.1093/cid/ciz246. [DOI] [PubMed] [Google Scholar]
- Braido F., Tarantini F., Ghiglione V., Melioli G., Canonica G.W. Bacterial lysate in the prevention of acute exacerbation of COPD and in respiratory recurrent infections. Int. J. COPD. 2007;2:335–345. [PMC free article] [PubMed] [Google Scholar]
- Braza M.S., van Leent M.M.T., Lameijer M., Sanchez-Gaytan B.L., Arts R.J.W., Pérez-Medina C., Conde P., Garcia M.R., Gonzalez-Perez M., Brahmachary M., et al. Inhibiting inflammation with myeloid cell-specific nanobiologics promotes organ transplant acceptance. Immunity. 2018;49:819–828.e6. doi: 10.1016/j.immuni.2018.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown K.L., Cosseau C., Gardy J.L., Hancock R.E.W. Complexities of targeting innate immunity to treat infection. Trends Immunol. 2007;28:260–266. doi: 10.1016/j.it.2007.04.005. [DOI] [PubMed] [Google Scholar]
- Buenrostro J.D., Wu B., Litzenburger U.M., Ruff D., Gonzales M.L., Snyder M.P., Chang H.Y., Greenleaf W.J. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature. 2015;523:486–490. doi: 10.1038/nature14590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S.-C., Quintin J., Cramer R.A., Shepardson K.M., Saeed S., Kumar V., Giamarellos-Bourboulis E.J., Martens J.H.A., Rao N.A., Aghajanirefah A., et al. mTOR- and HIF-1α-mediated aerobic glycolysis as metabolic basis for trained immunity. Science. 2014;345:1250684. doi: 10.1126/science.1250684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christ A., Günther P., Lauterbach M.A.R., Duewell P., Biswas D., Pelka K., Scholz C.J., Oosting M., Haendler K., Baßler K., et al. diet triggers NLRP3-dependent innate immune reprogramming. Cell. 2018;172:162–175.e14. doi: 10.1016/j.cell.2017.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirauqui C., Benito-Villalvilla C., Sánchez-Ramón S., Sirvent S., Diez-Rivero C.M., Conejero L., Brandi P., Hernández-Cillero L., Ochoa J.L., Pérez-Villamil B., et al. Human dendritic cells activated with MV130 induce Th1, Th17 and IL-10 responses via RIPK2 and MyD88 signalling pathways. Eur. J. Immunol. 2018;48:180–193. doi: 10.1002/eji.201747024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debisarun P.A., Struycken P., Domínguez-Andrés J., Moorlag S.J.C.F.M., Taks E., Gössling K.L., Ostermann P.N., Müller L., Schaal H., ten Oever J., et al. The effect of influenza vaccination on trained immunity: Impact on COVID-19. MedRxiv. 2020;2020 doi: 10.1101/2020.10.14.20212498. [DOI] [Google Scholar]
- Domínguez-Andrés J., Novakovic B., Li Y., Scicluna B.P., Gresnigt M.S., Arts R.J.W., Oosting M., Moorlag S.J.C.F.M., Groh L.A., Zwaag J., et al. The itaconate pathway is a central regulatory node linking innate immune tolerance and trained immunity. Cell Metab. 2019;29:211–220.e5. doi: 10.1016/j.cmet.2018.09.003. [DOI] [PubMed] [Google Scholar]
- Esposito S., Soto-Martinez M.E., Feleszko W., Jones M.H., Shen K.L., Schaad U.B. Nonspecific immunomodulators for recurrent respiratory tract infections, wheezing and asthma in children: a systematic review of mechanistic and clinical evidence. Curr. Opin. Allergy Clin. Immunol. 2018;18:198–209. doi: 10.1097/ACI.0000000000000433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feleszko W., Marengo R., Vieira A.S., Ratajczak K., Mayorga Butrón J.L. Immunity-targeted approaches to the management of chronic and recurrent upper respiratory tract disorders in children. Clin. Otolaryngol. 2019;44:502–510. doi: 10.1111/coa.13335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García González L.-A., Arrutia Díez F. Mucosal bacterial immunotherapy with MV130 highly reduces the need of tonsillectomy in adults with recurrent tonsillitis. Hum. Vaccin. Immunother. 2019;15:2150–2153. doi: 10.1080/21645515.2019.1581537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geller A., Yan J. Could the induction of trained immunity by β-glucan serve as a defense against COVID-19? Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.01782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geurs T.L., Hill E.B., Lippold D.M., French A.R. Sex differences in murine susceptibility to systemic viral infections. J. Autoimmun. 2012;38:J245–J253. doi: 10.1016/j.jaut.2011.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodridge H.S., Ahmed S.S., Curtis N., Kollmann T.R., Levy O., Netea M.G., Pollard A.J., van Crevel R., Wilson C.B. Harnessing the beneficial heterologous effects of vaccination. Nat. Rev. Immunol. 2016;16:392–400. doi: 10.1038/nri.2016.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groh L.A., Ferreira A.V., Helder L., van der Heijden C.D.C.C., Novakovic B., van de Westerlo E., Matzaraki V., Moorlag S.J.C.F.M., de Bree L.C., Koeken V.A.C.M., et al. oxLDL-induced trained immunity is dependent on mitochondrial metabolic reprogramming. Immunometabolism. 2021;3 doi: 10.20900/immunometab20210025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Houten C.B., Cohen A., Engelhard D., Hays J.P., Karlsson R., Moore E., Fernández D., Kreisberg R., Collins L.V., de Waal W., et al. Antibiotic misuse in respiratory tract infections in children and adults a prospective, multicentre study (TAILORED Treatment) Eur. J. Clin. Microbiol. Infect. Dis. 2019;38:505–514. doi: 10.1007/s10096-018-03454-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H.-S., Tsai C.-L., Chang J., Hsu T.-C., Lin S., Lee C.-C. Multiplex PCR system for the rapid diagnosis of respiratory virus infection: systematic review and meta-analysis. Clin. Microbiol. Infect. 2018;24:1055–1063. doi: 10.1016/j.cmi.2017.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S., Zhu B., Cheon I.S., Goplen N.P., Jiang L., Zhang R., Peebles R.S., Mack M., Kaplan M.H., Limper A.H., et al. PPAR-γ in macrophages limits pulmonary inflammation and promotes host recovery following respiratory viral infection. J. Virol. 2019;93 doi: 10.1128/JVI.00030-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iborra S., Martínez-López M., Khouili S.C., Enamorado M., Cueto F.J., Conde-Garrosa R., del Fresno C., Sancho D. Optimal generation of tissue-resident but not circulating memory T cells during viral infection requires crosspriming by DNGR-1+ dendritic cells. Immunity. 2016;45:847–860. doi: 10.1016/j.immuni.2016.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ifrim D.C., Joosten L.A., Kullberg B.J., Jacobs L., Jansen T., Williams D.L., Gow N.A., van der Meer J.W., Netea M.G., Quintin J. Candida albicans primes TLR cytokine responses through a Dectin-1/Raf-1-mediated pathway. J. Immunol. 2013;190:4129–4135. doi: 10.4049/jimmunol.1202611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ifrim D.C., Quintin J., Joosten L.A.B., Jacobs C., Jansen T., Jacobs L., Gow N.A.R., Williams D.L., van der Meer J.W.M., Netea M.G. Trained immunity or tolerance: opposing functional programs induced in human monocytes after engagement of various pattern recognition receptors. Clin. Vaccin. Immunol. 2014;21:534–545. doi: 10.1128/CVI.00688-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jartti T., Smits H.H., Bønnelykke K., Bircan O., Elenius V., Konradsen J.R., Maggina P., Makrinioti H., Stokholm J., Hedlin G., et al. Bronchiolitis needs a revisit: distinguishing between virus entities and their treatments. Allergy. 2019;74:40–52. doi: 10.1111/all.13624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann E., Sanz J., Dunn J.L., Khan N., Mendonça L.E., Pacis A., Tzelepis F., Pernet E., Dumaine A., Grenier J.C., et al. BCG educates hematopoietic stem cells to generate protective innate immunity against. Tuberculosis Cell. 2018;172:176–190.e19. doi: 10.1016/j.cell.2017.12.031. [DOI] [PubMed] [Google Scholar]
- Kleinnijenhuis J., Quintin J., Preijers F., Joosten L.A.B., Ifrim D.C., Saeed S., Jacobs C., Van Loenhout J., De Jong D., Hendrik S., et al. Bacille Calmette-Guérin induces NOD2-dependent nonspecific protection from reinfection via epigenetic reprogramming of monocytes. Proc. Natl. Acad. Sci. U. S. A. 2012;109:17537–17542. doi: 10.1073/pnas.1202870109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinnijenhuis J., Quintin J., Preijers F., Benn C.S., Joosten L.A.B., Jacobs C., van Loenhout J., Xavier R.J., Aaby P., van der Meer J.W.M., et al. Long-lasting effects of BCG vaccination on both heterologous Th1/Th17 responses and innate trained immunity. J. Innate Immun. 2014;6:152–158. doi: 10.1159/000355628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koatz A.M., Coe N.A., Cicerán A., Alter A.J. Clinical and immunological benefits of OM-85 bacterial lysate in patients with allergic rhinitis, asthma, and COPD and recurrent respiratory infections. Lung. 2016;194:687–697. doi: 10.1007/s00408-016-9880-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar V., Giamarellos-Bourboulis E.J., Martens J.H.A., Rao N.A., Aghajanirefah A., Manjeri G.R., Li Y., Daniela C., Arts R.J.W., van der Meer B.M.J.W., et al. mTOR- and HIF1α-mediated aerobic glycolysis as metabolic basis for trained immunity. Science. 2014;345 doi: 10.1126/science.1250684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacal I., Ventura R. Epigenetic inheritance: concepts, mechanisms and perspectives. Front. Mol. Neurosci. 2018;11 doi: 10.3389/fnmol.2018.00292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laiño J., Villena J., Suvorov A., Zelaya H., Moyano R.O., Salva S., Alvarez S. Nasal immunization with recombinant chimeric pneumococcal protein and cell wall from immunobiotic bacteria improve resistance of infant mice to streptococcus pneumoniae infection. PLoS One. 2018;13 doi: 10.1371/journal.pone.0206661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Fu X., Ma J., Zhang J., Hu Y., Dong W., Wan Z., Li Q., Kuang Y.Q., Lan K., et al. Altered respiratory virome and serum cytokine profile associated with recurrent respiratory tract infections in children. Nat. Commun. 2019;10 doi: 10.1038/s41467-019-10294-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberzon A., Birger C., Thorvaldsdóttir H., Ghandi M., Mesirov J.P., Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1:417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallia P., Footitt J., Sotero R., Jepson A., Contoli M., Trujillo-Torralbo M.B., Kebadze T., Aniscenko J., Oleszkiewicz G., Gray K., et al. Rhinovirus infection induces degradation of antimicrobial peptides and secondary bacterial infection in chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 2012;186:1117–1124. doi: 10.1164/rccm.201205-0806OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal. 2011;17 doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- Meissner H.C. Viral bronchiolitis in children. N. Engl. J. Med. 2016;374:62–72. doi: 10.1056/NEJMra1413456. [DOI] [PubMed] [Google Scholar]
- Mitroulis I., Ruppova K., Wang B., Chen L.S., Grzybek M., Grinenko T., Eugster A., Troullinaki M., Palladini A., Kourtzelis I., et al. Modulation of myelopoiesis progenitors is an integral component of trained immunity. Cell. 2018;172:147–161.e12. doi: 10.1016/j.cell.2017.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molero-Abraham M., Sanchez-Trincado J.L., Gomez-Perosanz M., Torres-Gomez A., Subiza J.L., Lafuente E.M., Reche P.A. Human oral epithelial cells impair bacteria-mediated maturation of dendritic cells and render T cells unresponsive to stimulation. Front. Immunol. 2019;10 doi: 10.3389/fimmu.2019.01434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro S., Cossalter G., Chiavaroli C., Kanda A., Fleury S., Lazzari A., Cazareth J., Sparwasser T., Dombrowicz D., Glaichenhaus N., et al. The oral administration of bacterial extracts prevents asthma via the recruitment of regulatory T cells to the airways. Mucosal Immunol. 2011;4:53–65. doi: 10.1038/mi.2010.51. [DOI] [PubMed] [Google Scholar]
- Netea M.G., Quintin J., van der Meer J.W.M. Trained immunity: a memory for innate host defense. Cell Host Microbe. 2011;9:355–361. doi: 10.1016/j.chom.2011.04.006. [DOI] [PubMed] [Google Scholar]
- Netea M.G., Joosten L.A.B., van der Meer J.W.M., Kullberg B.-J., van de Veerdonk F.L. Immune defence against Candida fungal infections. Nat. Rev. Immunol. 2015;15:630–642. doi: 10.1038/nri3897. [DOI] [PubMed] [Google Scholar]
- Netea M.G., Latz E., Mills K.H.G., O’Neill L.A.J. Innate immune memory: a paradigm shift in understanding host defense. Nat. Immunol. 2015;16:675–679. doi: 10.1038/ni.3178. [DOI] [PubMed] [Google Scholar]
- Netea M.G., Joosten L.A.B., Latz E., Mills K.H.G., Natoli G., Stunnenberg H.G., O’Neill L.A.J., Xavier R.J. Trained immunity: a program of innate immune memory in health and disease. Science. 2016;352:427. doi: 10.1126/science.aaf1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netea M.G., Domínguez-Andrés J., Barreiro L.B., Chavakis T., Divangahi M., Fuchs E., Joosten L.A.B., van der Meer J.W.M., Mhlanga M.M., Mulder W.J.M., et al. Defining trained immunity and its role in health and disease. Nat. Rev. Immunol. 2020;20:375–388. doi: 10.1038/s41577-020-0285-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto A., Mazón A., Nieto M., Calderón R., Calaforra S., Selva B., Uixera S., Palao M.J., Brandi P., Conejero L., et al. Bacterial mucosal immunotherapy with MV130 prevents recurrent wheezing in children: a randomized, double-blind, placebo-controlled clinical trial. Am. J. Respir. Crit. Care Med. 2021;204:462–472. doi: 10.1164/rccm.202003-0520OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norton E.B., Clements J.D., Voss T.G., Cárdenas-Freytag L. Prophylactic administration of bacterially derived immunomodulators improves the outcome of influenza virus infection in a murine model. J. Virol. 2010;84:2983–2995. doi: 10.1128/JVI.01805-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peignier A., Parker D. Trained immunity and host-pathogen interactions. Cell. Microbiol. 2020;22 doi: 10.1111/cmi.13261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintin J., Saeed S., Martens J.H.A., Giamarellos-Bourboulis E.J., Ifrim D.C., Logie C., Jacobs L., Jansen T., Kullberg B.J., Wijmenga C., et al. Candida albicans infection affords protection against reinfection via functional reprogramming of monocytes. Cell Host Microbe. 2012;12:223–232. doi: 10.1016/j.chom.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reading P.C., Smith G.L. A kinetic analysis of immune mediators in the lungs of mice infected with vaccinia virus and comparison with intradermal infection. J. Gen. Virol. 2003;84:1973–1983. doi: 10.1099/vir.0.19285-0. [DOI] [PubMed] [Google Scholar]
- Saeed S., Quintin J., Kerstens H.H.D., Rao N.A., Aghajanirefah A., Matarese F., Cheng S.-C., Ratter J., Berentsen K., van der Ent M.A., et al. Epigenetic programming during monocyte to macrophage differentiation and trained innate immunity. Science. 2014;345:1251086. doi: 10.1126/science.1251086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Ramón S., Conejero L., Netea M.G., Sancho D., Palomares Ó., Subiza J.L. Trained immunity-based vaccines: a new paradigm for the development of broad-spectrum anti-infectious formulations. Front. Immunol. 2018;9:2936. doi: 10.3389/fimmu.2018.02936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saz-Leal P., Del Fresno C., Brandi P., Martínez-Cano S., Dungan O.M., Chisholm J.D., Kerr W.G., Sancho D. Targeting SHIP-1 in myeloid cells enhances trained immunity and boosts response to infection. Cell Rep. 2018;25:1118–1126. doi: 10.1016/j.celrep.2018.09.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Souëf P. Viral infections in wheezing disorders. Eur. Respir. Rev. 2018;27 doi: 10.1183/16000617.0133-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sozio M.S., Lu C., Zeng Y., Liangpunsakul S., Crabb D.W. Activated AMPK inhibits PPAR-α and PPAR-γ transcriptional activity in hepatoma cells. Am. J. Physiol. Gastrointest. Liver Physiol. 2011;301:739–747. doi: 10.1152/ajpgi.00432.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark R., Brown G. Bioconductor; 2011. DiffBind: Differential Binding Analysis of ChIP-Seq Peak Data. [DOI] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsou C.-L., Peters W., Si Y., Slaymaker S., Aslanian A.M., Weisberg S.P., Mack M., Charo I.F. Critical roles for CCR2 and MCP-3 in monocyte mobilization from bone marrow and recruitment to inflammatory sites. J. Clin. Invest. 2007;117:902–909. doi: 10.1172/JCI29919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuvim M.J., Evans S.E., Clement C.G., Dickey B.F., Gilbert B.E. Augmented lung inflammation protects against influenza A pneumonia. PLoS One. 2009;4 doi: 10.1371/journal.pone.0004176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vázquez A., Fernández-Sevilla L.M., Jiménez E., Pérez-Cabrera D., Yañez R., Subiza J.L., Varas A., Valencia J., Vicente A. Involvement of mesenchymal stem cells in oral mucosal bacterial immunotherapy. Front. Immunol. 2020;11:2954. doi: 10.3389/fimmu.2020.567391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y., Jeyanathan M., Haddadi S., Barra N.G., Vaseghi-Shanjani M., Damjanovic D., Lai R., Afkhami S., Chen Y., Dvorkin-Gheva A., et al. Induction of autonomous memory alveolar macrophages requires T cell help and is critical to trained immunity. Cell. 2018;175:1634–1650.e17. doi: 10.1016/j.cell.2018.09.042. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nussbaum C., Myers R.M., Brown M., Li W., et al. Model-based analysis of ChIP-seq (MACS) Genome Biol. 2008;9 doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
•ATAC-Seq data have been deposited at NCBI's Gene Expression Omnibus and are publicly available as of the date of publication. Accession number is listed in the key resources table.
•This paper does not report original code.
•Any additional information required to reanalyze the data reported in this work paper is available from the Lead Contact upon request.