Abstract
Ubc13-catalyzed K63 ubiquitination is a major control point for immune signaling. Recent evidence has shown that the control of multiple immune functions, including chronic inflammation, pathogen responses, lymphocyte activation, and regulatory signaling, is altered by K63 ubiquitination. In this review, we detail the novel cellular sensors that are dependent on K63 ubiquitination for their function in the immune signaling network. Many pathogens, including severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), can target K63 ubiquitination to inhibit pathogen immune responses; we describe novel details of the pathways involved and summarize recent clinically relevant SARS-CoV-2-specific responses. We also discuss recent evidence that regulatory T cell (Treg) versus T helper (TH) 1 and TH17 cell subset regulation might involve K63 ubiquitination. Knowledge gaps that merit future investigation and clinically relevant pathways are also addressed.
Keywords: ubiquitin, K63-linked ubiquitination, Ubc13, E3 ligase, pattern recognition receptor (PRR), SARS-CoV-2, immune response, immune tolerance
Biological significance of K63 ubiquitination
K63-linked ubiquitination is involved in the regulation of signal transduction by multiple receptors of innate and adaptive immunity in eukaryotes, including tumor necrosis factor (TNF) and members of the TNF-receptor (TNFR) family, T and B cell receptor (TCR/BCR), Toll-like receptor (TLR; see Glossary), Nod-like receptor (NLR), RIG-I-like receptor (RLR), and the Interleukin-1 receptor (IL-1R) pathways [1,2]. In this review, we focus on the emerging understanding of the role of K63-linked ubiquitination in an array of immune response mechanisms. We also highlight the important regulatory role of K63 ubiquitination in striking a balance between immune activating versus immune tolerance mechanisms. We review prominent examples of key signaling adaptors, including cellular inhibitor of apoptosis proteins (cIAPs), TNF receptor-associated factors (TRAFs), NLRs, RLRs, and stimulator of interferon genes [STING, also referred to as Mediator of IRF-3 activation (MITA) or endoplasmic reticulum IFN stimulator (ERIS)], that serve as docking platforms for K63-linked ubiquitin chain assembly during immune cell activation and tolerance mechanisms (Figure 1, Key figure). The complementary role of deubiquitinating enzymes is also briefly mentioned, as well as the emerging putative role of K63-linked ubiquitination in SARS-CoV-2 immune dysregulation. In addition, we summarize emerging breakthrough discoveries on SARS-CoV-2-induced specific immune responses that might be associated with K63 ubiquitination.
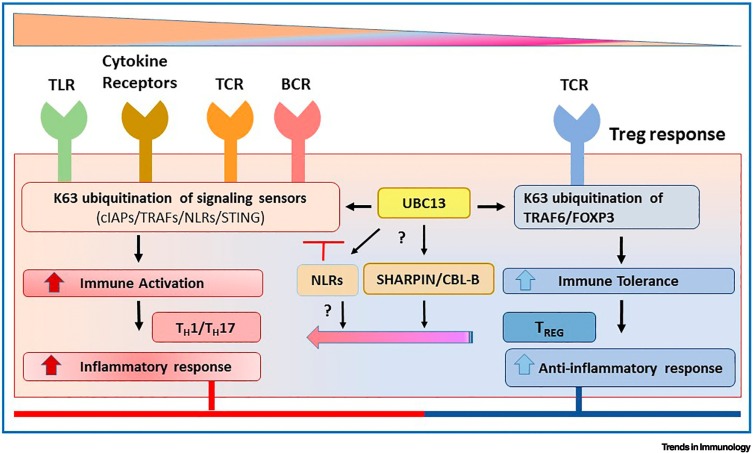
Figure 1.
Key figure. Ubc13-catalyzed K63-linked ubiquitination can regulate immune cell activation and immune tolerance mechanisms.
Signaling sensors of K63-linked ubiquitination [including cellular inhibitor of apoptosis proteins (cIAPs), TNF receptor-associated factors (TRAFs), Nod-like receptors (NLRs), and stimulator of interferon genes (STING)] activate innate and adaptive inflammatory responses (left). FOXP3-dependent K63 ubiquitination of target substrates regulates immune tolerance mechanisms that counteract the inflammatory phenotype (right). While some NLRs negatively regulate K63-linked ubiquitination to blunt inflammatory responses, the role of NLRs in regulatory T cell (Treg)- versus TH1/TH17-induced T cell responses is unknown. The mechanistic role of Ubc13 on the modulators of K63 ubiquitination [including SHARPIN or Casitas B lineage lymphoma b (CBL-B)], deficiency of which blunts immune tolerance mechanisms and causes a shift from Treg-induced immunosuppression to TH1/TH17-induced immune activation, needs further investigation. Abbreviations: BCR, B cell receptor; TCR, T cell receptor; TLR, Toll-like receptor.
Ubiquitination involves covalent binding of ubiquitin to target substrate proteins in an ATP-dependent manner, a mechanism that involves isopeptide linkage between the C terminus of ubiquitin and the epsilon amino group of an internal lysine residue of the target substrate [3,4]. Ubiquitin chain formation can involve any of the possible seven lysine (LYS, K) residues (K6, K11, K27, K29, K33, K48, and K63) or the N-terminal methionine (Met1) on ubiquitin, depending on various factors, including the type of E2, E3 ligases, and the target substrates, which ultimately dictate the functional specificity of any particular ubiquitination event [5] (Box 1, Box 2, Box 3 ). The K48-mediated ubiquitin linkages target proteins for proteasomal degradation and are typically referred to as canonical. The K63-linked ubiquitin chains do not target to the proteasome, but instead have important roles in eukaryotes, including DNA damage repair, cell signaling, and autophagy [1,5., 6., 7.]. Ubc13 is an unusual E2 that, in complex with its non-enzymatic cofactors (either Mms2 or Uev1a), catalyzes K63-specific linkages [8,9]. Ubc13-Mms2 is essential for cellular mechanisms involving DNA damage repair, whereas the Ubc13-Uev1a complex regulates cell signaling pathways that drive immune, inflammatory, and cell proliferation/survival responses [8,9,10,11]. K63-linked ubiquitin chains can be formed anchored to the target protein or can exist as free or unanchored chains and serve as docking sites for signaling molecules that drive immune pathways, such as TLRs, IL1R, and RLRs [12., 13., 14., 15.]. Heterogeneous K63 ubiquitin chains or K48/K63 branched ubiquitin chains can preferentially target the substrates for proteasome degradation [16]. K63 ubiquitin-conjugated E3 ligases and their target substrates undergo deubiquitination in the presence of deubiquitinases (DUBs) and have an important role in modulating K63-dependent cell signaling [17,18] (Box 4 ). Research on Ubc13-catalyzed K63 ubiquitination has rapidly advanced over the years and the functional implications of this non-proteolytic post-translational mechanism that are of relevance to innate and adaptive immunity are summarized herein. While the focus is largely on K63-linked ubiquitination and the role of Ubc13 in immune cell signaling, it is noteworthy that other important modulators of immune responses, including other unusual E2s, UbcH5, UbcH7, other E3 ligases, and LUBAC, are not addressed and are detailed elsewhere [2,19., 20., 21., 22., 23.].
Box 1. Ubiquitination pathways.
Depending on the signaling mechanisms and the target E3 ligase/substrate involved, ubiquitin chains of varying topologies conjugate to the RING domains of IAPs. A good example is polyubiquitination of RIPK1 by the TRAF2-cIAP1-Ubc13-UbcH5 complex, which modulates K63-linked ubiquitination of RIPK1 [100,101] (see Figure 2 in the main text). K63-linked ubiquitination of RIPK1 is a key mediator of the two co-regulatory but opposing downstream TNFR1 signaling pathways and delineates the dual regulatory role of RIPK1 [102,103]. One pathway includes formation of TNFR1 Complex I, which activates NF-κB and MAPK-driven transcriptional activation of prosurvival genes, and the other pathway either turns on RIPK1 kinase activity and involves TNFR1 Complex II, which drives classical apoptosis or signals RIPK1-RIPK3-mediated necroptotic cell death [104,105].
Alt-text: Box 1
Box 2. BCR activation and K63-linked ubiquitination.
In the context of BCR activation, TRAF3 essentially functions as a bridge to transfer K48-linked ubiquitin chains to NF-κB-inducing kinase (NIK) via the ubiquitin ligase complex (TRAF2-TRAF6-cIAP1/2). When TRAF3 itself undergoes degradation upon CD40 or BAFF stimulation, NIK is rescued from proteasomal degradation, which then activates the alternate NF-κB pathway [35]. TRAF2-mediates K63-linked ubiquitination of cIAP1/2, which is followed by K48 ubiquitin chain-mediated degradation of the adaptor TRAF3, which causes stabilization of NIK and activation of NF-κB in CD40- or BAFF-induced B cells.
TRAF3 has distinct phenotypic traits and is crucial in multiple facets of B cell proliferative responses [35,106., 107., 108.]. While TRAF3 negatively regulates CD40-, BAFF- MyD88-induced NF-κB and MAPK activation because of its propensity to undergo K48 ubiquitination, TRAF3 is indispensable for TRIF-dependent interferon (IFN) response upon viral sensing, which involves K63-linked ubiquitination of TRAF3 on its RING domain [109]. TLR3 receptor–TRIF adaptor–TRAF3-mediated events activate the TBK1-IRF3-IFN-β pathway.
Alt-text: Box 2
Box 3. NLRs and STING activation.
Some NLRs regulate STING activity (see Figure 3 in the main text). Stimulation of the immune response to viral infections involves many of the same immune pathways as immune responses to tumor cells, including Type 1 and 3 IFNs and cytokine secretion. While NLRC3 binds to STING and blocks Type 1 IFN responses [110], NLRC5 attenuates Type 1 IFN responses due to its ability to interfere with the binding of the RLRs, RIG-I and MDA-5, to MAVS [111]. NLRX1 also binds to mitochondrial adaptor MAVS and prevents its interaction with RIG-I, thus inhibiting not only NF-κB, but also IRF3 pathway-induced antiviral immune responses [112]. NLRP12 abrogates K63 ubiquitination of RIG-I, prevents RIG-I from activating MAVS, and inhibits RIG-I-induced antiviral immune responses [113,114]. NLRP12 reduces K63 ubiquitination of RIG-I, while enhancing K48-linked ubiquitination of RIG-I, as demonstrated both in an overexpressing system using HEK293 cells and endogenously in dendritic cells (derived from WT and Nlrp12 –/– mice).
Alt-text: Box 3
Box 4. Deubiquitinases that reverse K63-linked ubiquitination.
K63 ubiquitin-conjugated E3 ligases and their target substrates are also subjected to deubiquitination in the presence of DUBs [17,18]. The zinc-finger (ZnF) protein A20 and cylindramotosis (CYLD) protein are DUBs that catalyze the hydrolysis of K63-linked ubiquitin chains conjugated to target E3 ligases or substrates [115]. CYLD–/– T cells are hypersensitive to TCR/CD3 and TCR/CD28 activation of T cells. A20 negatively regulates NF-κB signaling by inhibiting E3 ligase or substrate activity (TRAF2, TRAF6, cIAP1, RIPK1, and MALT1) [116,117] (see Figure 2 in the main text). A20 inhibits the interaction between E2s Ubc13 or UbH5c and, by doing so, A20 along with Tax1-binding protein 1 (TAX1BP1) targets Ubc13 or UbcH5 for K48-ubiquitin chain-conjugated proteasomal degradation [118]. A20 functions to deubiquitinate K63-ubiquitin linkages conjugated to MALT1 of CBM-Ubc13-TRAF6 complex and impairs TCR-induced NF-κB signaling; A20 also undergoes proteasomal degradation to restore TCR/CD28-induced IKK activation [119]. OTUD7B is a DUB that deubiquitinates K63-assembled ubiquitin chains from GβL, a substrate for TRAF2 E3 ligase [120]. Ubiquitin-specific protease 5 (USP5) specifically targets the free C-terminal diglycine motif of the unanchored polyubiquitin chain and deubiquitinates ubiquitin monomers from its proximal end [121]. It would be interesting to investigate whether USP5 targets the unanchored K63-linked ubiquitin chains catalyzed by Ubc13, for example in the context of RLR signaling and antiviral immunity [12., 13., 14., 15.,121].
Given the importance of K63-linked ubiquitination in mediating several innate and adaptive immune signaling pathways, it is not surprising that pathogens have evolved mechanisms to counteract this post-translational modification. For example, some bacterial virulence factors have DUB-like activity, such as SseL (from Salmonella enterica serovar typhimurium), YopJ (from Yersinia pestis), and ChlaDUB1 (from Chlamydia trachomatis). These bacterial proteins modulate host immune responses [122., 123., 124.]. Macrophages infected with SseL-deficient Salmonella show increases in IκBα degradation and ubiquitination followed by transcriptional activation of NF-κB-dependent gene expression [122]. Although not a member of the DUB family of cysteine proteases, SARS-CoV-1 encodes a Papain-like protease protein (PLpro; derived from the larger nsp3) that inhibits STING by deubiquitination of K63 linkages [81]. USP49, a ubiquitin-specific protease, interacts with and deubiquitinates STING after HSV-1 infection. USP49 decreases antiviral responses, and knockdown or knockouts of USP49 potentiate not only HSV-1, but also cytoplasmic DNA and cGAMP-induced Type 1 IFN production [125]. The SARS-CoV-2 PLPro protein inhibits TLR-7 signaling by deubiquitinating K63 linkages from TRAF3 and TRAF6 in human promonocyte cells that were stably transfected [126]. These findings further demonstrate that K63 ubiquitination is important for Type 1 IFN production and antiviral responses due to both STING and cGAMP, helping to explain why pathogens may target STING for deubiquitination. In summary, DUBs function as a counter-regulatory mechanism to either shut down or promote signaling.
Alt-text: Box 4
Ubc13 turns on the inflammatory response
Studies using gene ablation in mice demonstrated the key roles of Ubc13 in pathways for host defense, autoimmunity, and inflammatory diseases [24., 25., 26.]. Given that the homozygous Ubc13 knockout is embryonic lethal, haploinsufficient Ubc13 +/– mice were generated on a C57BL/6 and 129SV background by retroviral insertional mutagenesis that inactivated one Ubc13 allele; these heterozygotic Ubc13 +/– mice showed a normal phenotype [24].
Comparisons of murine splenocytes and macrophages of homozygous wild-type (WT) Ubc13 +/+ and heterozygous Ubc13 +/– cells stimulated with lipopolysaccharide (LPS) or TNF revealed reduced cytokine production by Ubc13 +/– immune cells relative to those from WT controls [24]. In culture, splenocytes of Ubc13 +/– mice, upon stimulation with LPS, exhibited reduced IκBα degradation, phospho-JNK and phospho-p38 MAPK expression, which were associated with diminished TRAF6 polyubiquitination relative to WT controls [24]. In addition, Ubc13 +/– mice exhibited reduced Ubc13 protein in most tissues, along with significantly impaired inflammatory signaling in response to TNF and LPS stimulation in vivo [24]. This decreased signaling was associated with reduced in vivo TRAF6 ubiquitination and reduced in vivo activation of NF-κB and stress kinases (p38 MAPK and JNK), demonstrating a key role for Ubc13 in inflammatory responses [24].
However, in other independent studies, tissue-specific gene-knockout studies reported in B cells, T cells, and macrophages derived from Ubc13-deficient mice (Lck_Cre Ube2n fl/fl mice generated on a mixed C57BL/6 and 129P2/OlaHsd background) showed different results [25,26]. While homozygous Ubc13 –/– B cells and macrophages demonstrated essential roles for Ubc13 in BCR-, TLR/IL-1R-, or CD40-mediated activation of MAPKs, Ubc13 had a minor role in BCR-, TLR-, IL-1R-, or CD40-mediated NF-κB activation and IL-1β-mediated TAK1 phosphorylation in these same cells [25]. Nevertheless, conditional deletion of Ubc13 in T cells (Lck_Cre Ube2n fl/fl thymocytes) showed reduced NF-κB essential modulator (NEMO, also known as IKK-γ) ubiquitination, IκBα degradation/phosphorylation, and NF-κB activation upon phorbol 12-myristate 13-acetate (PMA)/Ionophore and anti-CD3/CD28 antibody stimulation relative to WT [26]. The authors attributed this discrepancy to Ubc13 deletion efficiency in each conditional model and/or to a cell type-specific role of Ubc13 in the activation of NF-κB [26]. Comparison of the results from homozygous versus heterozygous deletion of Ubc13 also suggested gene dose-dependency effects and a cell type-specific role for Ubc13 [24., 25., 26.]. In summary, these studies confirmed that Ubc13 could regulate immune cell signaling responses in a cell type-specific manner [24., 25., 26.].
Recently, male and female haploinsufficient Ubc13 +/– mice (on a C57BL/6 background [24]) were shown to be protected against age-related insulin resistance under normal diet (ND) and high-fat diet (HFD) conditions compared with WT controls, suggesting an additional role for K63 ubiquitination in chronic inflammation-induced metabolic syndromes [27]. Histological analysis of visceral adipose tissue (VAT) showed the adipocyte cell size to be smaller in female HFD-fed Ubc13 +/−mice compared with WT mice [27]. Moreover, 18-week-old female Ubc13 +/– mice showed lower inflammatory expression of the cytokine genes TNFA, IL6, and IL1B in VAT, secondary to reduced weight gain, compared with male Ubc13 +/− and WT mice [27]. These results indicated that the effects of Ubc13 haploinsufficiency were more prominent in female HFD mice compared with male mice; thus, further investigations are needed to understand the mechanism behind these mouse gender differences.
K63 ubiquitination in immune signaling
K63-linked ubiquitination involving cIAPs and TRAFs in immune cells: implications for immunomodulation
cIAPs are phylogenetically conserved proteins that have up to three Baculovirus IAP repeat (BIR) motifs. Some cIAPs are implicated in the suppression of apoptosis by virtue of their ability to bind active caspase-family proteases [28,29]. The IAP family of proteins, including cIAP1, cIAP2, and XIAP, have E3 ubiquitin protein ligase activity [30., 31., 32.]. TRAF2 and TRAF6 form a complex with cIAP1/2 to conjugate K63-linked ubiquitin chains catalyzed by Ubc13, followed by NF-κB or MAPK activation in human immune cell activation via antigen and pattern recognition receptors (PRRs) [33,34] (Figure 2 , Box 2). While IAPs activate the classical NF-κB pathway, relying on K63 ubiquitination, these E3 ligases repress the non-canonical NF-κB cascade by promoting K48-mediated degradation of NF-κB-inducing kinase (NIK); both pathways proceed in a TRAF-dependent manner. The immunomodulatory role of TRAFs is reviewed elsewhere [35] (Figure 2).
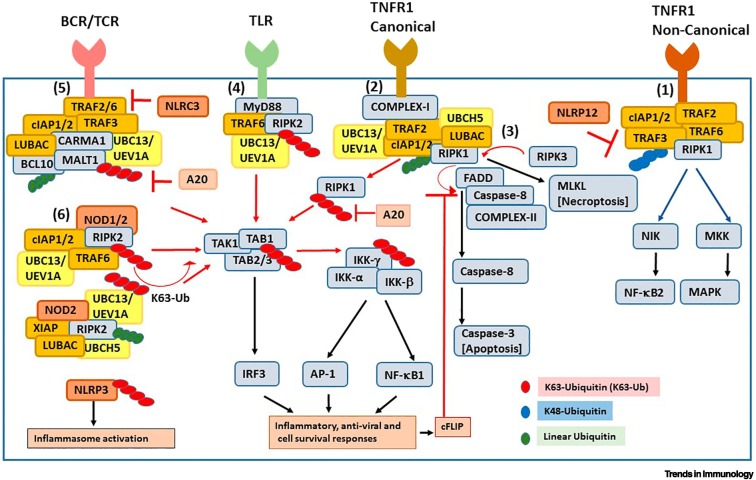
Figure 2.
K63 ubiquitination in immune cell signaling cascades.
K63-linked polyubiquitin chains (red ovals) and linear polyubiquitin chains (green ovals) catalyzed by Ubc13-Uev1a and UbcH5/linear ubiquitin chain assembly complex (LUBAC) are necessary for efficient activation of immune and antiviral cell signaling pathways. (1) The tumor necrosis factor receptor 1 (TNFR1)-induced non-canonical mechanism involves K48-ubiquitin chain-mediated degradation of TNF receptor-associated factor 3 (TRAF3) by cellular inhibitor of apoptosis proteins 1/2 (cIAP1/2), release of TRAF2/TRAF6 for K63 ubiquitination, and activation of non-canonical nuclear factor KappaB (NF-κB)/MAPK. (2,3) TNFR1-induced K63-ubiquitination of RIPK1 drives two opposing TNFR1 canonical signaling pathways: canonical NF-κB activation modulated by TRAF2-cIAP1-UbcH5-LUBAC-Ubc13-RIPK1 (shown as Complex-I) and classical apoptosis mediated by RIPK1, Caspase-8 and FADD (shown as Complex-II), which is regulated by RIPK1 kinase activity or necroptosis mediated by RIPK1-RIPK3 activity. (4) Stimulation of Toll-like receptors (e.g., TLR4) triggers MyD88-dependent TRAF6- cIAP1/2-Ubc13-catalyzed K63 ubiquitination of downstream substrates followed by NF-κB/MAPK activation. (5) T/B cell receptor (TCR/BCR) activation recruits the caspase recruitment domain-containing membrane-associated guanylate kinase protein1–B cell lymphoma 10–mucosa-associated lymphoid tissue protein 1 (paracaspase) [CARMA1-BCL10-MALT1 (CBM)] complex and activates E2-E3 ligase-mediated K63 ubiquitination-dependent downstream pathways. (6) Stimulation of Nod-like receptors (NLRs) recruits the adaptor proteins to trigger K63 ubiquitination of E3 ligases and their target substrates (including RIPK2) for activation of downstream pathways. The deubiquitinase A20 and some NLR family proteins [e.g., Nod-like receptor Card domain 3 (NLRC3) and NOD-, LRR-, and pyrin domain-containing protein 12 (NLRP12)] negatively regulate K63 ubiquitination-mediated immune cell activation.
Of clinical relevance, several cIAPs are overexpressed in various types of cancer [36., 37., 38., 39.]. Human MALT lymphoma samples with the frequently observed t(11;18)(q21;q21)-positive translocation express cIAP2/MALT1 fusion protein, which shows an increase in polyubiquitinated NEMO compared with human tonsil samples, suggesting that the c-IAP2/MALT1 fusion protein drives constitutive NF-κB activation in MALT tumor-bearing cells [40]. The cIAP2/MALT1 fusion gene construct overexpressed in 293T cells as cIAP2/MALT1 fusion protein has been shown to interact with Ubc13, suggesting that this fusion protein is also involved in K63 ubiquitination [40].
In the context of TCR activation, TRAF2/TRAF6-dependent K63-ubiquitination of MALT1 in the Caspase recruitment domain-containing membrane-associated guanylate kinase protein1-B cell lymphoma 10-mucosa-associated lymphoid tissue protein 1 (paracaspase) [ CARMA1 - BCL 10 - MALT1 (CBM)] complex is required for NF-κB activation [32,41] (Figure 2). MALT1 K63 ubiquitination, in response to PMA/Ionomycin- and/or anti-CD3/CD28 antibody-induced human Jurkat and murine CD4+ T cell activation, was associated with IκBα degradation and NF-κB activation compared with uninduced cells [41]. TRAF2-binding (WT) cIAP2/MALT1 versus TRAF2-nonbinding (mutant) cIAP2/MALT1 in Jurkat T cells showed elevated NF-κB induction for WT, but not mutant complexes [32]. Ubc13-Uev1a- (and UbcH5c-)catalyzed TRAF6-mediated K63 ubiquitination of MALT1 was confirmed using a cell-free system [41,42]. MALT1-deficient T cells isolated from Malt1 –/– mice showed impaired IκBα degradation and NF-κB activation upon PMA/Ionomycin stimulation compared with WT controls [41]. These observations implied that ubiquitinated MALT1 of the CBM complex could serve as a docking platform for subsequent ubiquitination, followed by activation of the IKK complex of the classical NF-κB cascade upon PKC-theta-dependent TCR engagement (Figure 2).
Additionally, SHARPIN [a subunit of the linear ubiquitin chain assembly complex (LUBAC), which promotes M1-linear ubiquitin chains] has been shown to bind K63-ubiquitinated cIAP1/2 in activated B cell-like diffuse large B cell lymphoma (ABC DLBCL) cells [43,44]. cIAP1/2 interacts with the CBM complex and inactivation of cIAP1/2 using the CRISPR/Cas9 gene-targeting system in human HBL1 ABC DLBCL cells showed a decrease in IκBα degradation and NF-κB activation relative to controls, highlighting an immunomodulatory role of cIAP1/2 in classical NF-κB activation [43,44]. Mechanistically, these results suggest that K63 ubiquitination of BCL10 and MALT1 is needed for LUBAC-mediated recruitment of IKK into the CBM complex, as well as subsequent ubiquitination of NEMO for activation of NF-κB signaling in lymphocytes [43., 44., 45.] (Figure 2).
Immunomodulatory functions of K63-linked ubiquitination in T cells
K63-linked ubiquitination has a dual regulatory role in both immune cell activation and immunosuppression, as part of immune surveillance mechanisms involving T cells (Figure 1). K63 ubiquitination of TRAF6 regulates FOXP3-dependent TGF-β-induced peripheral Treg function [46]. Traf6 fl/fl Foxp3Cre + mice with FOXP3+ Treg-restricted deletion of TRAF6 (Traf6 –/–) either failed to support the growth of implanted B16 melanoma cells or delayed the implantation of MC38 colon cancer cells compared with WT mice [46]. Impaired Treg function and increased anti-tumor immunity were also associated with impaired K63 ubiquitination of FOXP3, observed in TRAF6- and FOXP3-overexpressing HEK293T cells along with a robust increase in the production of IFN-γ and IL-17 cytokines from tumor-infiltrating leukocytes or tumor-draining lymph nodes upon ex vivo stimulation with PMA/Ionomycin [46]. In summary, these studies show a possible role of K63 ubiquitination in immune tolerance, as evidenced by the lack of tolerance and a strong T cell activation associated with a shift toward enhanced antitumor immunity in Traf6 fl/fl Foxp3Cre + mice compared with WT controls [46].
Treg-specific Ube2n-deficient mice (Ube2n fl/fl Foxp3 GFP-hCre; known as Ube2n TregKO), generated upon conditional deletion of the Ubc13 coding gene (Ube2n), displayed autoimmune symptoms with a simultaneous large increase in TH1, TH2, and TH17 effector CD4+ T cells compared with age-matched Ube2n +/+ Foxp3 GFP-hCre WT controls [47]. An autoimmune disease phenotype was observed in Rag1 –/– mice upon adoptive transfer of Tregs from Ube2n TregKO; these transferred Ubc13-deficient Tregs acquired a TH1/TH17 phenotype and were IFN-γ-IL17 double-positive under lymphopenic conditions compared with WT controls [47]. Compared with WT cells, Ubc13-deficient Tregs retained FOXP3 expression and had lower expression of the genes encoding IL10 (Il10) and suppressor of cytokine signaling 1 (Socs1, which prevents conversion of Tregs into TH1 and TH17-like effector T cells) [47]. Thus, the regulatory role of K63 ubiquitination is thought to be due to the maintenance of T cell homeostasis, as evidenced from the essential role of Ubc13 in the in vivo immunosuppressive function of Tregs, maintenance of Treg stability, and prevention of T cell transformation into an inflammatory CD4+ TH1- or TH17-like phenotype [47] (Figure 1).
There are several other modulators of K63 ubiquitination (Figure 3). For instance, SHARPIN-deficient Cpdm –/– mice show a significant reduction in the generation of Tregs, as evidenced by the inefficient induction of Foxp3 in Cpdm –/– CD4+ T cells upon anti-CD3 antibody stimulation, compared with Cpdm +/– mice [48]. The autoimmune and inflammatory phenotype of Cpdm –/– mice was overcome by replenishing Cpdm –/– neonatal mice with SHARPIN-sufficient Tregs from Foxp3 YFPCre Cpdm +/+ via adoptive transfer. This was shown by a reduction in the lung inflammatory effector T cell subsets (TH2 and TH17-like) and their cytokines (including IL-3, IL-5, and IL-17), compared with Cpdm –/– mice that received no Treg cells [48]. Human Jurkat T cells that stably expressed SHARPIN and endogenous SHARPIN in mouse WT CD4+ T cells assembled K63-ubiquitin chains, attenuated TCR signaling, and showed an increase in FOXP3 expression relative to controls [48]. However, CD4+ T cells from Cpdm –/– mice that lacked K63-ubiquitinated SHARPIN showed greater association of TCRζ with the signaling kinase Zap70 upon TCR stimulation compared with WT CD4+ T cells. Thus, K63 ubiquitination-deficient SHARPIN activated the TCR, inhibited FOXP3 induction, negatively modulated Tregs, and skewed the effector CD4+ T cell population toward the inflammatory TH17 phenotype to induce an inflammatory response upon TCR stimulation [48]. This suggests that aberrant ubiquitination can prevent lymphocyte populations from becoming self-tolerant and that, in turn, might contribute to autoimmunity.
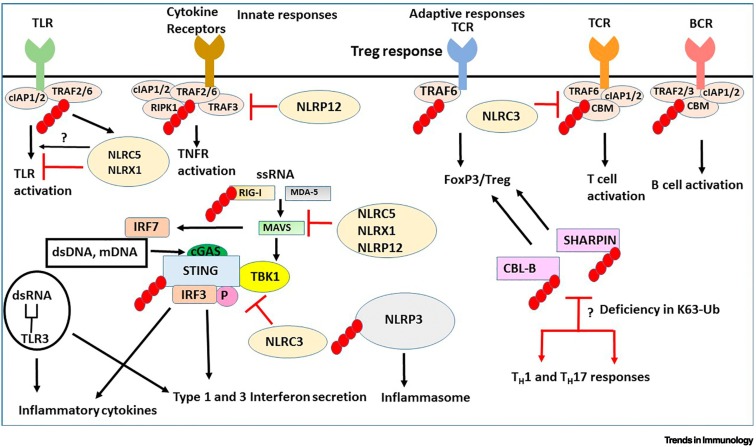
Figure 3.
Ubiquitination signaling pathways in innate and adaptive immunity.
The black arrows indicate stimulation of a pathway and the red arrows denote either inhibition or functional implications due to inhibition. The small red ovals attached to proteins are K63-linked ubiquitin chains. Membrane Toll-like receptors (TLRs) and cytokine receptors [including canonical tumor necrosis factor (TNF) receptors] activate ubiquitinated TNF receptor-associated factor 2/6 (TRAF2/6) pathways to stimulate cytokine and interferon (IFN) production. Single-strand (ss)RNA, double-strand (ds)RNA and mitochondrial (mt)DNA activate cytosolic receptors that are also controlled by ubiquitination. Stimulator of interferon genes (STING) is an intermediate for activation of downstream IFN-activating genes, IFN regulatory factor 3/7 (IRF3/7) by retinoic acid inducible gene-I (RIG-I), mitochondrial antiviral-signaling protein (MAVS), melanoma differentiation-associated protein 5 (MDA-5) through TANK-binding kinase 1 (TBK1), cyclic GMP-AMP (cGAMP) sensing dsRNA. K63 ubiquitination of SHANK-associated RH domain interacting protein in postsynaptic density (SHARPIN) and Casitas B-lineage Lymphoma-b (CBL-B) promotes regulatory T cell (Treg) lineage commitment and deficiency in K63-ubiquitinated SHARPIN or CBL-B stimulates helper T cell (TH) 1 and TH17 inflammatory responses. K63 ubiquitination of NOD-, LRR-, and pyrin domain-containing protein 3 (NLRP3) has a role in the activation of the inflammasome. Nod-like receptor card domain 3 (NLRC3) blocks both STING activation and TNF receptor-associated factor 6 (TRAF6) activation. NLRC5 and NLRX1 inhibit IKK activation, enhancement of nuclear factor KappaB (NF-κB) and inflammation signaling by TLRs. NLRP12 negatively regulates non-canonical NF-κB pathway. NLRC5, NLRX1, and NLRP12 negatively regulate RIG-induced antiviral immune responses. Inhibition of NF-κB, RIG-I, and MDA-5 signaling (by ssRNA) is blocked when NLRC5 is K63-ubiquitinated.
The E3 ligase Casitas B lineage lymphoma b (CBL-B) is another example of a modulator of K63 ubiquitination (Figure 3). Deficiency in CBL-B protein expression, recently observed in patients with systemic lupus erythematosus (SLE), correlated with a decrease in K63 ubiquitination specifically in CD4+CD25+ Tregs but not in CD4+CD25– effector T cells isolated from patients with SLE, suggesting that dysfunctional CBL-B associated with an aberrant K63-ubiquitination profile of Tregs from such patients contributes to impaired immunosuppression [49]. This is thought to be due to the ability of CBL-B to ubiquitinate the TCRζ chain to terminate anti-CD3/CD28 antibody-mediated TCR activation [49]. Additionally, phosphorylated STAT3 is increased in CD4+CD25+ Tregs from patients with SLE, but not from healthy controls [49]. IL-6-activated STAT3 has been found to be a transcriptional repressor of Ubc13, as demonstrated by chromatin immunoprecipitation of STAT3 association with the Ubc13 promoter [from Stat3-sufficient bone marrow-derived macrophages (BMDMs) in the presence and absence of IL-6] [50]. Deficiency in CBL-B that prevents STAT3 degradation might cause downregulation of Ubc13 expression, indicating that CBL-B might indirectly regulate Ubc13 function [50]. These reports suggest that Ubc13 maintains the immunosuppressive phenotype of Tregs and that CBL-B or SHARPIN deficiency contributes toward skewing the balance from a Treg to an inflammatory effector T cell phenotypic subset. This is relevant because Ubc13 also has a key role in immune cell activation pathways. However, further studies are required to assess these pathways and functional putative immunomodulatory outcomes.
Role of K63-linked ubiquitination in NLR-mediated signaling
NLR family members can form a variety of ‘inflammasomes’, some that stimulate activation of proinflammatory caspase and others that stimulate the MAPK, NF-κB, and endoplasmic reticulum (ER) stress pathways [51., 52., 53., 54., 55., 56., 57., 58., 59.] (Figure 2). The NLRs, NOD1/NOD2, undergo oligomerization upon recognition of pathogen-associated molecular patterns (PAMPs), recruit RIPK2 via CARD-CARD domain interactions to activate K63 ubiquitination of RIPK2, mediated by TRAF2, TRAF5, TRAF6, CARD9, cIAP1/2, and XIAP (which also interacts with RIPK2, recruits the E3 ligase LUBAC and promotes linear polyubiquitination on RIPK2) and the Ubc13-Uev1a complex (which catalyzes K63 ubiquitination) to drive NF-κB activation [60., 61., 62.] (Figure 2). Of note, TRAF3 mediates K63-linked ubiquitination of the inflammasome adaptor apoptosis-associated speck-like protein containing a CARD (ASC), which, in turn, also induces inflammasome formation [63].
Recently, Ubc13 was also shown to associate with NLRP3, catalyze K63-linked ubiquitination of NLRP3 and activate the inflammasome [64] (Figure 2, Figure 3 ). Upon stimulation with LPS and ATP, BMDMs from Ubc13 deltaMye mice (which specifically lack Ubc13 in myeloid cells; Ubc13 +/–) showed diminished secretion of IL-1β and impaired caspase-1 maturation compared with WT (Ubc13 +/+) controls [64]. Endogenous Ubc13 interacted with NLRP3 in BMDMs, correlating with inflammasome activation, NLRP3 was K63-ubiquitinated in the presence of WT Ubc13 but not mutant Ubc13C87A.
Some NLRs inhibit rather than enhance inflammatory responses induced by other PRRs by modulating K63 ubiquitination (Figure 3). BMDMs from Nlrc3-/- mice, upon stimulation with LPS and Nlrc3 –/– mouse CD4+ T cells stimulated with anti-CD3/CD28 antibodies, had more K63-ubiquitinated TRAF6, which also formed faster, compared with WT controls [65,66]. This was correlated with higher amounts of IFN-γ/TNF-α secreted upon stimulation of CD4+ T cells from Nlrc3 –/– mice and a rapid loss of IκBα, suggesting that NLRC3, a non-inflammasome-forming NLR, attenuated the E3 ligase function of TRAF6 [65,66]. Accordingly, adoptive transfer of CD4+ T cells isolated from either WT or Nlrc3 –/– mice into Rag1 –/– mice followed by induction of experimental autoimmune encephalitis (EAE, a model of multiple sclerosis) demonstrated that Rag1 –/– mice that received Nlrc3 –/– CD4+ T cells exhibited worse EAE symptoms compared with those that received WT control CD4+ T cells [66]. The exacerbation of EAE was associated with more IFN-γ and IL-17A, as part of TCR-induced NF-κB activation of its target genes in Nlrc3 –/– CD4+ T cells. This indicated a crucial checkpoint function of NLRC3 in preventing downstream NF-κB-induced inflammatory responses in this autoimmune model [66].
NLRC5 undergoes K63 ubiquitination in the presence of TRAF2 or TRAF6 and ubiquitinated NLRC5 regulates the catalytic function of the IKK complex [67]. Upon treatment with LPS, HEK293Ts overexpressing NLRC5, RAW264.7 macrophages, and mouse embryonic fibroblasts (MEFs) show increased K63 ubiquitination of NLRC5 [67]. Small interfering (si)RNA-mediated knockdown of endogenous Traf2 or Traf6 in primary macrophages followed by stimulation with LPS attenuated NLRC5 polyubiquitination, suggesting NLRC5 K63 ubiquitination as a component of LPS signaling responses [67].
NLRX1 is another non-inflammasome NLR that binds to TRAF6 and when stimulated with LPS, undergoes K63-linked ubiquitination; ubiquitinated NLRX1 inhibits IKK and NF-κB activation, as shown in overexpressed 293Ts [68,69] (Box 3). Consistent with its inhibitory role, Nlrx1 knockdown in mice (NLRX1-KD) show to increased plasma IL-6 and TNF-α and enhanced susceptibility to LPS-induced septic shock, as observed in peritoneal macrophages and MEFs from NLRX1-KD mice compared with WT controls [69]. NLRP12 can also suppress inflammation [70]. For example, colon organ cultures from Nlrp12 –/– mice subjected to acute experimental colitis (EC) showed increased amounts of NIK and p52, with little difference in pIκBα, pp65, IL-1β, and TNF-α compared with WT controls, indicating that Nlrp12 –/– mice showed increased susceptibility to inflammation in an acute EC model compared with WT control mice [70]. Myeloid dendritic cells (mDCs) from Nlrp12 –/– mice primed with Pam3Cys4 and stimulated with CD40L showed decreased TRAF3 amounts but no change in TRAF6 compared with WT controls, suggesting that, by binding to TRAF3, NLRP12 prevents the degradation of TRAF3 and negatively regulates the non-canonical NF-κB pathway [70].
Roles of K63-linked ubiquitination in signaling by STING in viral infections
STING was initially identified as a key downstream coordinating protein for the sensing of bacterial and viral DNA, promoting IFN-γ secretion [71]. STING, in collaboration with RIG-I, mitochondrial antiviral-signaling protein (MAVS), melanoma differentiation-associated protein 5 (MDA-5), cyclic GMP-AMP (cGAMP) and virus inhibitory protein, endoplasmic reticulum-associated, interferon (IFN)-inducible (Viperin) has a central coordinating role in cellular sensing of cytosolic DNA and pathogen-derived RNA [72]. While STING does not bind directly to foreign or self-DNA or RNA, STING operates as a central control point for antiviral responses from PRRs, such as TLR-7/8 [for single-strand (ss)RNA], TLR-3 [for double-strand (ds)RNA] and TLR-9 [72] (Figure 4 ). In addition to pathogen responses, host mitochondrial and dsDNA that enter the cytosol (e.g., during chromosomal nondisjunction events in aneuploid cancer cells) can also activate STING [73]. Thus, STING responds downstream to PRRs that recognize cytosolic viral ssRNA, dsRNA, and DNA, in addition to also responding to damage-associated molecular pattern s (DAMPs), such as mitochondrial and double-stranded host DNA [72,73].
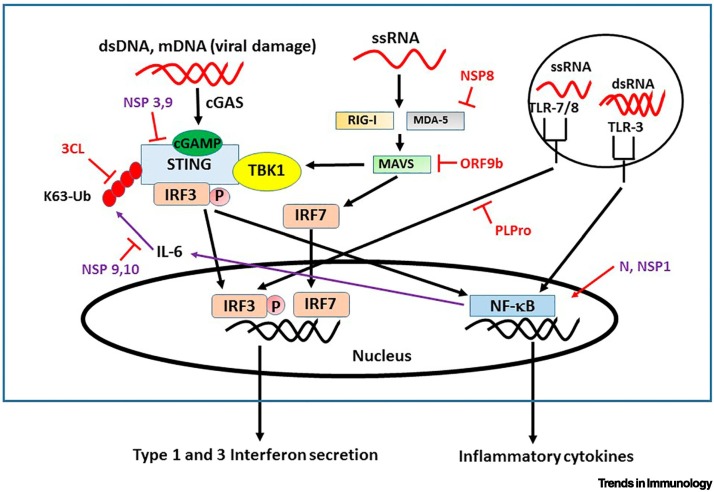
Figure 4.
Stimulator of interferon genes (STING) in interferon (IFN) responses.
STING has a central role in stimulating innate immunity, Type 1 and 3 IFN production, and also inflammation via nuclear factor KappaB (NF-κB). IFN production by STING is dependent on K63 ubiquitination (red ovals). STING acts as a linker for activation of downstream IFN activating genes (IFN regulatory factor 3; IRF3,) by retinoic acid inducible gene-1 (RIG-I), mitochondrial antiviral-signaling protein (MAVS), melanoma differentiation-associated protein 5 (MDA-5) through TANK-binding kinase 1 (TBK1) and cyclic GMP-AMP (cGAMP), which senses RNA viruses. Additionally, TLR7/8 and TLR3 signal IFN production by single-strand (ss)RNA and double-strand (ds)RNA and these signal through K63-ubiquitinated TNF receptor-associated factor 3/6 (TRAF3/6; not shown here). The four identified severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) open reading frame proteins that inhibit K63 ubiquitination and innate responses are 3CL, ORF9, NSP8, and Papain-like protease (PLPro) (in red). Unknown mechanisms are in purple. The purple arrows show a feedback loop that turns off STING and is inhibited by the NSP9,10 proteins, which can block the K63 ubiquitination of STING that is key for IFN 1 and 3 production but not for NF-κB activation.
RIG-I binds to viral RNA and activates MAVS via the CARD-like domain to aggregate and bind E3 ubiquitin ligases, which include TRAF2, TRAF5, TRAF6, and LUBAC [74]. Similarly, MDA-5 can also activate MAVS via the CARD domain [75]. The DNA and RNA sensors RIG-I and MDA-5, pair with MAVS while cGAS and nucleic acid-sensing TLRs pair with their signaling molecules STING and TRIF, respectively, activating the kinase TANK-binding kinase 1 (TBK1) to phosphorylate IRF3 and stimulate Type 1 IFN production [76]. Further research has shown that Ube2D3, together with the ubiquitin ligase Riplet, activates proteins via K63-ubiquitin anchored chains, while Ube2N preferentially activates via unanchored K63 polyubiquitin chains [13].
C-GAS recognizes dsDNA [from both viral and mitochondrial (mt)DNA] and increases the production of the novel intracellular second messenger cGAMP [77]. cGAMP directly binds and activates STING [78], which leads not only to both TBK-1 binding and IRF3 binding (and ultimately Type 1 IFN induction and secretion), but also NF-κB activation (and cytokine secretion), similar to RIG-I and TLR7/8 activation (Figure 4). Myb Like, SWIRM and MPN Domains 1 (MYSM1) is a metalloproteinase that deubiquitinates and cleaves the K63-linked ubiquitinated STING suppressing cGAS-STING signaling; Mysm1-deficient C57BL/6 mice exhibit a hyperinflammatory response, acute tissue damage, and high mortality when infected by viruses such as HSV-1 [79]. Additionally, MYSM1 has been reported to act as a suppressor for SLE [79].
K63 ubiquitination of STING at the K224 residue is essential for the delivery of STING to TBK1 and the binding to IFN regulatory factor 3 (IRF3) that initiates Type 1 IFN production and secretion [80]. TRAF E3 ligases catalyze the formation of K63-conjugated ubiquitin chains on STING; such K63-linked ubiquitination is involved in STING-TRAF6-TBK1/STING-TRAF3-TBK1 complex formation [81., 82., 83.]. The E3 ligases belonging to the TRIM family, TRIM56 and TRIM32, also mediate K63-linked ubiquitination of STING [84,85]. The non-proteolytic E2 Ubc13-catalyzes TRAF6-mediated K63-linked ubiquitin chains on STING and UbcH5c catalyzes TRIM56-mediated mono-ubiquitination of cGAS [82,86].
Viruses, such as Human T cell lymphotropic virus type 1 (HTLV-1), can impair host immunity by inhibiting host cell IFN-γ responses by producing the Tax protein [87]. HTLV-1 Tax protein decreases K63 ubiquitination of STING and inhibits STING interaction with TBK1 [88]. Other RNA viruses, such as coronaviruses and flaviviruses, also interfere with STING to inhibit the antiviral IFN response [89]. Whereas activation of STING turns on innate immunity, antiviral responses, and Type 1 IFNs, its overactivation can also lead to autoimmunity. There is strong evidence linking excessive Type 1 IFN to the promotion and exacerbation of certain autoimmune diseases, especially SLE (although autoimmune diseases clearly have multifactorial causes and inputs) [90]. Of note, some NLRs that are activated by bacterial PAMPs also regulate STING activity as do viral PAMPs (Figure 3, Box 3).
SARS-CoV-2 infection and K63-linked ubiquitination in immune responses
Recent evidence has highlighted the crucial role of STING and Type 1 and 3 IFN secretion in the innate immune response to SARS-CoV-2 infection. Decreased Type 1 IFN (specifically low IFN-α and no IFN-β) has been strongly associated with more serious coronavirus 2019 (COVID-19) symptoms and death, while the amount of NF-κB-associated cytokines (e.g., TNF-α, IL-1 and IL-6) are elevated in patients with severe COVID-19 [91]. In human lung epithelial cells and in an animal model of COVID (K18-ACE2-transgenic mice), a STING agonist (diABZI-4) strongly inhibited SARS-CoV-2 replication for both the alpha and delta variants (B.1.135.1) [92]. The study also showed that SARS-CoV-2 could induce a delayed immune response that could be overcome with exogenous IFN administration [92]. Two other research groups also reported that this same STING agonist (diABZI-4) can significantly inhibit SARS-CoV-2 replication in human lung epithelial cells [93,94].
Since K63 ubiquitination is pivotal in STING signaling (as described in the preceding text), several studies have examined the potential inhibitory role of SARS-CoV-2 open-reading frame proteins in K63 ubiquitination and in diminishing Type 1 IFN secretion. There are four SARS-CoV-2 proteins that have been shown to alter K63 ubiquitination, affect IFN signaling and may lead to decreased Type 1 and 3 interferon secretion (Figure 4): 3CL, ORF9b, NSP8 , PLpro. 3CL is an important protease in SARS-CoV-2, which directly inhibits STING K63 ubiquitination and causes a decrease in downstream signaling in human, mouse, and chicken lung epithelial cells [95]. The ORF9b SARS-CoV-2 protein antagonizes Type 1 IFN production after activation of RIG-I-MAVS by blocking K63 ubiquitination of NEMO in human primary epithelial cells [96]. The SARS-CoV-2 nucleocapsid (N) protein also inhibits Type 1 IFN secretion by inhibition of RIG-I, probably due to inhibition of RIG-I K63 ubiquitination by TRIM25 E3 ligase [as has been shown for Middle East respiratory syndrome (MERS)] in human embryonic kidney cells (HEK-293) [97,98]. NSP8 from SARS-CoV-2 suppresses MDA-5-stimulated IFN signaling by binding to MDA-5 CARD and blocking K63-linked ubiquitination in human embryonic kidney cells [99]. In summary, SARS-CoV-2 inhibition of Type 1 and 3 IFNs appears to be important for evading the innate immune response. Multiple mechanisms of inhibition of interferon signaling by SARS-CoV-2 proteins have been elucidated and alterations in K63 ubiquitination are prominent in this evasion of the host response (Figure 4).
Concluding remarks
K63 ubiquitination is important as a proximal event in signal transduction pathways involved in innate and adaptive immunity. Ubc13-catalyzed K63 ubiquitination has a key role in striking a balance between immune cell activation and immune tolerance mechanisms, as evidenced from murine studies. Recent findings demonstrated that the control of multiple immune functions, such as chronic inflammation, pathogen responses, lymphocyte activation, regulatory signaling pathways, and lymphocyte subset development, can be altered by K63 ubiquitination. While we have discussed some details of K63 ubiquitination-dependent regulation of immune signaling and described some of the novel cellular sensors that are dependent on K63 ubiquitination during immune signaling, numerous questions remain (see Outstanding questions). For instance, mechanistic details of SARS-CoV-2 infection and K63 ubiquitination in immune responses remain far from being understood and require robust experimentation. Therefore, although advances have been made regarding K63 ubiquitination in the immune system, the field eagerly awaits much-needed further research in this important area, particularly when considering the relevance to coronavirus infections, such as from SARS-CoV-2.
Outstanding questions.
What is the mechanistic role of cIAP/TRAF-mediated K63 ubiquitination in regulating a balance between TH1, TH17 versus Treg immune cell functions? K63-linked ubiquitination can balance on/off immune responses to preserve homeostasis. The redundant and non-redundant functions of cIAP and TRAF members mediating K63 ubiquitination in immunostimulatory versus immunosuppressive phenotypes needs further research.
What are the relevant E3 ligases involved in the regulation of chronic inflammation-induced metabolic syndrome? Ubc13 +/– mice are protected from age-related insulin resistance and obesity. The involvement of relevant E3 ligases may provide further evidence of the explicit role of Ubc13-catalyzed K63 ubiquitination in chronic inflammation-induced metabolic syndrome.
What is the role of Ubc13 in K63-ubiquitinated SHARPIN- and CBL-B-mediated regulation of immunological homeostasis? Direct mechanistic information as to how K63-ubiquitinated SHARPIN and CBL-B control immunological homeostasis may be obtained from investigations on Ube2n TregKO mice, which acquire TH1/TH17 phenotypes under lymphopenic and inflammatory conditions.
Is there putative coordinated regulation between IAPs and TRAFs in K63-linked ubiquitination of the inflammasome? Some NLRs regulate TRAF-mediated K63 ubiquitination. The coordinated regulation between NLRs and TLRs, including the likely E2-E3 pairs, in K63 ubiquitination-mediated inflammasome activation needs further investigation.
What is the role of K63-linked ubiquitination mediated by NLRs in the SARS-CoV-2-induced IFN response? Deciphering K63 ubiquitination in SARS-CoV-2 infection may help characterize pathogen-specific induced signaling pathways. For example, NLRP12 reduces binding between RIG-I and an E3 ligase and functions as an attenuating factor in SARS-CoV-2-induced RIG-I-mediated IFN-γ expression.
Does viral/bacterial DNA directly interact with K63-linked ubiquitin chains to trigger an immune response? Recent studies reported that K63-linked ubiquitin chains specifically interacted with DNA. Investigations to assess whether K63-linked ubiquitin chains interact directly with viral/bacterial DNA in triggering immune response may be informative.
Alt-text: Outstanding questions
Acknowledgments
Acknowledgment
The authors acknowledge Catarina Sacristan for editorial help with the manuscript.
Declaration of interests
J. Reed is an employee of Sanofi. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
Glossary
- Caspase recruitment domain-containing membrane-associated guanylate kinase protein1-B cell lymphoma 10-mucosa-associated lymphoid tissue protein 1 (paracaspase) [CARMA1-BCL10-MALT1 (CBM)] complex
forms a signalosome in the classical NF-κB activation pathway.
- Damage-associated molecular patterns (DAMPs)
include aged, dead, and damaged self-structures released from host cells in response to necrosis or apoptosis and are endogenous alarmins that trigger an immune response.
- Inflammasome
multiprotein oligomeric complex assembled by some NLRs upon sensing intracellular PAMPs/DAMPs; serves as a signaling platform for the recruitment of caspase-1, which activates IL-1β from its precursor form (pro-IL-1β) to induce innate immunity.
- Linear ubiquitin chain assembly complex (LUBAC)
comprises the catalytically active HOIP and accessory proteins HOIP-L and SHANK-associated RH domain interacting protein SHARPIN; functions as an E3 ligase that forms linear ubiquitin chains from Met1 of ubiquitin.
- Pattern recognition receptors (PRRs)
present on the host cell surface, in the endosome, or in the cytoplasm; recognize a variety of conserved structural motifs present on PAMPs and DAMPs, activating immune signaling pathways that target pathogens/alarmins for their eventual clearance by the host immune system.
- Pathogen-associated molecular patterns (PAMPs)
conserved molecular structures present on microbes or pathogens; specific to the pathogen of interest, are carbohydrates, lipoproteins, or nucleic acids (bacterial or viral DNA and RNA) sensed by host cells to trigger an immune response.
- NLRP3
also known as cryopyrin; a cytosolic PRR that forms the inflammasome upon sensing PAMPs/DAMPs to trigger an induced innate immune response.
- Nod-like receptors (NLRs)
a family of cytoplasmic PRR sensors that detect cytosolic PAMPs or DAMPs via LRR domains and assemble to form inflammasomes, which trigger innate immunity in response to infection or endogenous alarmin signals.
- Regulatory T cell (Treg)
subpopulation of CD4+ helper T cells with immunosuppressive properties that help maintain self-tolerance and immune cell homeostasis.
- Retinoic acid-inducible gene I (RIG-I)-like receptors (RLRs)
family of RIG-I-like receptors, RNA helicases, cytoplasmic PRRs, sense cytosolic viral RNA, induce transcriptional activation of Type 1 IFN (IFN-α/β) responses, and trigger antiviral immunity.
- Toll-like receptors (TLRs)
family of PRRs homologous to the Drosophila Toll receptor; form dimers with extracellular leucine-rich repeat (LRR) domains, sense the presence of pathogenic microbes, bind to PAMP ligands and, upon recognition of PAMPs, activate downstream signaling cascades contributing to innate/inflammatory responses.
References
- 1.Chen Z.J. Ubiquitination in signaling to and activation of IKK. Immunol. Rev. 2012;246:95–106. doi: 10.1111/j.1600-065X.2012.01108.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ebner P., et al. Ubiquitin enzymes in the regulation of immune responses. Crit. Rev. Biochem. Mol. Biol. 2017;52:425–460. doi: 10.1080/10409238.2017.1325829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ciechanover A. The unravelling of the ubiquitin system. Nat. Rev. Mol. Cell Biol. 2015;16:322–324. doi: 10.1038/nrm3982. [DOI] [PubMed] [Google Scholar]
- 4.Hershko A., et al. The ubiquitin system. Nat. Med. 2000;6:1073–1081. doi: 10.1038/80384. [DOI] [PubMed] [Google Scholar]
- 5.Yau R., Rape M. The increasing complexity of the ubiquitin code. Nat. Cell Biol. 2016;18:579–586. doi: 10.1038/ncb3358. [DOI] [PubMed] [Google Scholar]
- 6.Liu P., et al. K63-linked polyubiquitin chains bind to DNA to facilitate DNA damage repair. Sci. Signal. 2018;11 doi: 10.1126/scisignal.aar8133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kwon Y.T., Ciechanover A. The ubiquitin code in the ubiquitin-proteasome system and autophagy. Trends Biochem. Sci. 2017;42:873–886. doi: 10.1016/j.tibs.2017.09.002. [DOI] [PubMed] [Google Scholar]
- 8.Deng L., et al. Activation of the IkappaB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell. 2000;103:351–361. doi: 10.1016/s0092-8674(00)00126-4. [DOI] [PubMed] [Google Scholar]
- 9.Hofmann R.M., Pickart C.M. Noncanonical MMS2-encoded ubiquitin-conjugating enzyme functions in assembly of novel polyubiquitin chains for DNA repair. Cell. 1999;96:645–653. doi: 10.1016/s0092-8674(00)80575-9. [DOI] [PubMed] [Google Scholar]
- 10.Andersen P.L., et al. Distinct regulation of Ubc13 functions by the two ubiquitin-conjugating enzyme variants Mms2 and Uev1A. J. Cell Biol. 2005;170:745–755. doi: 10.1083/jcb.200502113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu X., Karin M. Emerging roles of Lys63-linked polyubiquitylation in immune responses. Immunol. Rev. 2015;266:161–174. doi: 10.1111/imr.12310. [DOI] [PubMed] [Google Scholar]
- 12.Parvatiyar K., Harhaj E.W. Cell signaling. Anchors away for ubiquitin chains. Science. 2010;328:1244–1245. doi: 10.1126/science.1192296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shi Y., et al. Ube2D3 and Ube2N are essential for RIG-I-mediated MAVS aggregation in antiviral innate immunity. Nat. Commun. 2017;8:15138. doi: 10.1038/ncomms15138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xia Z.P., et al. Direct activation of protein kinases by unanchored polyubiquitin chains. Nature. 2009;461:114–119. doi: 10.1038/nature08247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zeng W., et al. Reconstitution of the RIG-I pathway reveals a signaling role of unanchored polyubiquitin chains in innate immunity. Cell. 2010;141:315–330. doi: 10.1016/j.cell.2010.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ohtake F., et al. K63 ubiquitylation triggers proteasomal degradation by seeding branched ubiquitin chains. Proc. Natl. Acad. Sci. U. S. A. 2018;115:E1401–E1408. doi: 10.1073/pnas.1716673115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harhaj E.W., Dixit V.M. Deubiquitinases in the regulation of NF-κB signaling. Cell Res. 2011;21:22–39. doi: 10.1038/cr.2010.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harhaj E.W., Dixit V.M. Regulation of NF-κB by deubiquitinases. Immunol. Rev. 2012;246:107–124. doi: 10.1111/j.1600-065X.2012.01100.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oikawa D., et al. Linear ubiquitin code: its writer, erasers, decoders, inhibitors, and implications in disorders. Int. J. Mol. Sci. 2020;21:3381. doi: 10.3390/ijms21093381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peltzer N., Walczak H. Cell death and inflammation - a vital but dangerous liaison. Trends Immunol. 2019;40:387–402. doi: 10.1016/j.it.2019.03.006. [DOI] [PubMed] [Google Scholar]
- 21.Spit M., et al. Linear ubiquitination at a glance. J. Cell Sci. 2019;132 doi: 10.1242/jcs.208512. [DOI] [PubMed] [Google Scholar]
- 22.Zinngrebe J., et al. Ubiquitin in the immune system. EMBO Rep. 2014;15:28–45. doi: 10.1002/embr.201338025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iwai K. Discovery of linear ubiquitination, a crucial regulator for immune signaling and cell death. FEBS J. 2021;288:1060–1069. doi: 10.1111/febs.15471. [DOI] [PubMed] [Google Scholar]
- 24.Fukushima T., et al. Ubiquitin-conjugating enzyme Ubc13 is a critical component of TNF receptor-associated factor (TRAF)-mediated inflammatory responses. Proc. Natl. Acad. Sci. U. S. A. 2007;104:6371–6376. doi: 10.1073/pnas.0700548104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamamoto M., et al. Key function for the Ubc13 E2 ubiquitin-conjugating enzyme in immune receptor signaling. Nat. Immunol. 2006;7:962–970. doi: 10.1038/ni1367. [DOI] [PubMed] [Google Scholar]
- 26.Yamamoto M., et al. Cutting edge: pivotal function of Ubc13 in thymocyte TCR signaling. J. Immunol. 2006;177:7520–7524. doi: 10.4049/jimmunol.177.11.7520. [DOI] [PubMed] [Google Scholar]
- 27.Joo E., et al. Ubc13 haploinsufficiency protects against age-related insulin resistance and high-fat diet-induced obesity. Sci. Rep. 2016;6:35983. doi: 10.1038/srep35983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deveraux Q.L., Reed J.C. IAP family proteins--suppressors of apoptosis. Genes Dev. 1999;13:239–252. doi: 10.1101/gad.13.3.239. [DOI] [PubMed] [Google Scholar]
- 29.Vandenabeele P., Bertrand M.J. The role of the IAP E3 ubiquitin ligases in regulating pattern-recognition receptor signalling. Nat. Rev. Immunol. 2012;12:833–844. doi: 10.1038/nri3325. [DOI] [PubMed] [Google Scholar]
- 30.Yang Y., et al. Ubiquitin protein ligase activity of IAPs and their degradation in proteasomes in response to apoptotic stimuli. Science. 2000;288:874–877. doi: 10.1126/science.288.5467.874. [DOI] [PubMed] [Google Scholar]
- 31.Samuel T., et al. Distinct BIR domains of cIAP1 mediate binding to and ubiquitination of tumor necrosis factor receptor-associated factor 2 and second mitochondrial activator of caspases. J. Biol. Chem. 2006;281:1080–1090. doi: 10.1074/jbc.M509381200. [DOI] [PubMed] [Google Scholar]
- 32.Garrison J.B., et al. TRAF2-binding BIR1 domain of c-IAP2/MALT1 fusion protein is essential for activation of NF-kappaB. Oncogene. 2009;28:1584–1593. doi: 10.1038/onc.2009.17. [DOI] [PubMed] [Google Scholar]
- 33.Dhillon B., et al. The evolving role of TRAFs in mediating inflammatory responses. Front. Immunol. 2019;10:104. doi: 10.3389/fimmu.2019.00104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karin M., Gallagher E. TNFR signaling: ubiquitin-conjugated TRAFfic signals control stop-and-go for MAPK signaling complexes. Immunol. Rev. 2009;228:225–240. doi: 10.1111/j.1600-065X.2008.00755.x. [DOI] [PubMed] [Google Scholar]
- 35.Arkee T., Bishop G.A. TRAF family molecules in T cells: Multiple receptors and functions. J. Leukoc. Biol. 2020;107:907–915. doi: 10.1002/JLB.2MR1119-397R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krajewska M., et al. Analysis of apoptosis protein expression in early-stage colorectal cancer suggests opportunities for new prognostic biomarkers. Clin. Cancer Res. 2005;11:5451–5461. doi: 10.1158/1078-0432.CCR-05-0094. [DOI] [PubMed] [Google Scholar]
- 37.Carter B.Z., et al. Simultaneous activation of p53 and inhibition of XIAP enhance the activation of apoptosis signaling pathways in AML. Blood. 2010;115:306–314. doi: 10.1182/blood-2009-03-212563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Werner T.A., et al. IAPs cause resistance to TRAIL-dependent apoptosis in follicular thyroid cancer. Endocr. Relat. Cancer. 2018;25:295–308. doi: 10.1530/ERC-17-0479. [DOI] [PubMed] [Google Scholar]
- 39.Dizdar L., et al. Clinicopathological and functional implications of the inhibitor of apoptosis proteins survivin and XIAP in esophageal cancer. Oncol. Lett. 2018;15:3779–3789. doi: 10.3892/ol.2018.7755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou H., et al. Constitutive NF-kappaB activation by the t(11;18)(q21;q21) product in MALT lymphoma is linked to deregulated ubiquitin ligase activity. Cancer Cell. 2005;7:425–431. doi: 10.1016/j.ccr.2005.04.012. [DOI] [PubMed] [Google Scholar]
- 41.Oeckinghaus A., et al. Malt1 ubiquitination triggers NF-kappaB signaling upon T-cell activation. EMBO J. 2007;26:4634–4645. doi: 10.1038/sj.emboj.7601897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun L., et al. The TRAF6 ubiquitin ligase and TAK1 kinase mediate IKK activation by BCL10 and MALT1 in T lymphocytes. Mol. Cell. 2004;14:289–301. doi: 10.1016/s1097-2765(04)00236-9. [DOI] [PubMed] [Google Scholar]
- 43.Yang Y., et al. Targeting non-proteolytic protein ubiquitination for the treatment of diffuse large B cell lymphoma. Cancer Cell. 2016;29:494–507. doi: 10.1016/j.ccell.2016.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang Y.K., et al. Molecular determinants of scaffold-induced linear ubiquitinylation of B cell lymphoma/leukemia 10 (Bcl10) during T cell receptor and oncogenic caspase recruitment domain-containing protein 11 (CARD11) signaling. J. Biol. Chem. 2016;291:25921–25936. doi: 10.1074/jbc.M116.754028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Giardino Torchia M.L., et al. c-IAP1 and c-IAP2 redundancy differs between T and B cells. PLoS ONE. 2013;8 doi: 10.1371/journal.pone.0066161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ni X., et al. TRAF6 directs FOXP3 localization and facilitates regulatory T-cell function through K63-linked ubiquitination. EMBO J. 2019;38 doi: 10.15252/embj.201899766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chang J.H., et al. Ubc13 maintains the suppressive function of regulatory T cells and prevents their conversion into effector-like T cells. Nat. Immunol. 2012;13:481–490. doi: 10.1038/ni.2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Park Y., et al. SHARPIN controls regulatory T cells by negatively modulating the T cell antigen receptor complex. Nat. Immunol. 2016;17:286–296. doi: 10.1038/ni.3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Romo-Tena J., et al. Lys63-polyubiquitination by the E3 ligase casitas B-lineage lymphoma-b (Cbl-b) modulates peripheral regulatory T cell tolerance in patients with systemic lupus erythematosus. Clin. Exp. Immunol. 2018;191:42–49. doi: 10.1111/cei.13054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang H., et al. STAT3 restrains RANK- and TLR4-mediated signalling by suppressing expression of the E2 ubiquitin-conjugating enzyme Ubc13. Nat. Commun. 2014;5:5798. doi: 10.1038/ncomms6798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stehlik C., et al. The PAAD/PYRIN-family protein ASC is a dual regulator of a conserved step in nuclear factor kappaB activation pathways. J. Exp. Med. 2002;196:1605–1615. doi: 10.1084/jem.20021552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Martinon F., et al. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol. Cell. 2002;10:417–426. doi: 10.1016/s1097-2765(02)00599-3. [DOI] [PubMed] [Google Scholar]
- 53.Bruey J.M., et al. Bcl-2 and Bcl-XL regulate proinflammatory caspase-1 activation by interaction with NALP1. Cell. 2007;129:45–56. doi: 10.1016/j.cell.2007.01.045. [DOI] [PubMed] [Google Scholar]
- 54.Faustin B., et al. Reconstituted NALP1 inflammasome reveals two-step mechanism of caspase-1 activation. Mol. Cell. 2007;25:713–724. doi: 10.1016/j.molcel.2007.01.032. [DOI] [PubMed] [Google Scholar]
- 55.Hasegawa M., et al. A critical role of RICK/RIP2 polyubiquitination in Nod-induced NF-kappaB activation. EMBO J. 2008;27:373–383. doi: 10.1038/sj.emboj.7601962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Krieg A., et al. XIAP mediates NOD signaling via interaction with RIP2. Proc. Natl. Acad. Sci. U. S. A. 2009;106:14524–14529. doi: 10.1073/pnas.0907131106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gregory S.M., et al. Discovery of a viral NLR homolog that inhibits the inflammasome. Science. 2011;331:330–334. doi: 10.1126/science.1199478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gerlic M., et al. Vaccinia virus F1L protein promotes virulence by inhibiting inflammasome activation. Proc. Natl. Acad. Sci. U. S. A. 2013;110:7808–7813. doi: 10.1073/pnas.1215995110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.D'Osualdo A., et al. Transcription factor ATF4 induces NLRP1 inflammasome expression during endoplasmic reticulum stress. PLoS ONE. 2015;10 doi: 10.1371/journal.pone.0130635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gong Q., et al. Structural basis of RIP2 activation and signaling. Nat. Commun. 2018;9:4993. doi: 10.1038/s41467-018-07447-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pellegrini E., et al. RIP2 filament formation is required for NOD2 dependent NF-κB signalling. Nat. Commun. 2018;9:4043. doi: 10.1038/s41467-018-06451-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bednash J.S., Mallampalli R.K. Regulation of inflammasomes by ubiquitination. Cell. Mol. Immunol. 2016;13:722–728. doi: 10.1038/cmi.2016.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Guan K., et al. MAVS promotes inflammasome activation by targeting ASC for K63-linked ubiquitination via the E3 ligase TRAF3. J. Immunol. 2015;194:4880–4890. doi: 10.4049/jimmunol.1402851. [DOI] [PubMed] [Google Scholar]
- 64.Ni J., et al. Ubc13 promotes k63-linked polyubiquitination of NLRP3 to activate inflammasome. J. Immunol. 2021;206:2376–2385. doi: 10.4049/jimmunol.2001178. [DOI] [PubMed] [Google Scholar]
- 65.Schneider M., et al. The innate immune sensor NLRC3 attenuates Toll-like receptor signaling via modification of the signaling adaptor TRAF6 and transcription factor NF-kappaB. Nat. Immunol. 2012;13:823–831. doi: 10.1038/ni.2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Uchimura T., et al. The innate immune sensor NLRC3 acts as a rheostat that fine-tunes T cell responses in infection and autoimmunity. Immunity. 2018;49:1049–1061. doi: 10.1016/j.immuni.2018.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Meng Q., et al. Reversible ubiquitination shapes NLRC5 function and modulates NF-kappaB activation switch. J. Cell Biol. 2015;211:1025–1040. doi: 10.1083/jcb.201505091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Allen I.C., et al. NLRX1 protein attenuates inflammatory responses to infection by interfering with the RIG-I-MAVS and TRAF6-NF-kappaB signaling pathways. Immunity. 2011;34:854–865. doi: 10.1016/j.immuni.2011.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xia X., et al. NLRX1 negatively regulates TLR-induced NF-kappaB signaling by targeting TRAF6 and IKK. Immunity. 2011;34:8438–8453. doi: 10.1016/j.immuni.2011.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Allen I.C., et al. NLRP12 suppresses colon inflammation and tumorigenesis through the negative regulation of noncanonical NF-kappaB signaling. Immunity. 2012;36:742–754. doi: 10.1016/j.immuni.2012.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ran Y., et al. MITA/STING: a central and multifaceted mediator in innate immune response. Cytokine Growth Factor Rev. 2014;25:631–639. doi: 10.1016/j.cytogfr.2014.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Anwar S., et al. cGAS-STING-mediated sensing pathways in DNA and RNA virus infections: crosstalk with other sensing pathways. Arch. Virol. 2021;166:3255–3268. doi: 10.1007/s00705-021-05211-x. [DOI] [PubMed] [Google Scholar]
- 73.Hong Z. et al., cGAS-STING pathway: post-translational modifications and functions in sterile inflammatory diseases. FEBS J. Published online July 26, 2021. 10.1111/febs.16137. [DOI] [PubMed]
- 74.Liu S., et al. MAVS recruits multiple ubiquitin E3 ligases to activate antiviral signaling cascades. Elife. 2013;2 doi: 10.7554/eLife.00785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kawai T., et al. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat. Immunol. 2005;6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- 76.Liu S., et al. Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science. 2015;347 doi: 10.1126/science.aaa2630. [DOI] [PubMed] [Google Scholar]
- 77.Zhang X., et al. The cytosolic DNA sensor cGAS forms an oligomeric complex with DNA and undergoes switch-like conformational changes in the activation loop. Cell Rep. 2014;6:421–430. doi: 10.1016/j.celrep.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ablasser A., et al. cGAS produces a 2'-5'-linked cyclic dinucleotide second messenger that activates STING. Nature. 2013;498:380–384. doi: 10.1038/nature12306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tian M., et al. MYSM1 represses innate immunity and autoimmunity through suppressing the cGAS-STING pathway. Cell Rep. 2020;33 doi: 10.1016/j.celrep.2020.108297. [DOI] [PubMed] [Google Scholar]
- 80.Ni G., et al. Ubiquitination of STING at lysine 224 controls IRF3 activation. Sci. Immunol. 2017;2 doi: 10.1126/sciimmunol.aah7119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chen X., et al. SARS coronavirus papain-like protease inhibits the type I interferon signaling pathway through interaction with the STING-TRAF3-TBK1 complex. Protein Cell. 2014;5:369–381. doi: 10.1007/s13238-014-0026-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dunphy G., et al. Non-canonical activation of the DNA sensing adaptor STING by ATM and IFI16 mediates NF-κB signaling after nuclear DNA damage. Mol. Cell. 2018;71:745–760. doi: 10.1016/j.molcel.2018.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chen X., Chen Y. Ubiquitination of cGAS by TRAF6 regulates anti-DNA viral innate immune responses. Biochem. Biophys. Res. Commun. 2019;514:659–664. doi: 10.1016/j.bbrc.2019.05.022. [DOI] [PubMed] [Google Scholar]
- 84.Tsuchida T., et al. The ubiquitin ligase TRIM56 regulates innate immune responses to intracellular double-stranded DNA. Immunity. 2010;33:765–776. doi: 10.1016/j.immuni.2010.10.013. [DOI] [PubMed] [Google Scholar]
- 85.Zhang J., et al. TRIM32 protein modulates Type I interferon induction and cellular antiviral response by targeting MITA/STING protein for K63-linked ubiquitination. J. Biol. Chem. 2012;287:28646–28655. doi: 10.1074/jbc.M112.362608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Seo G.J., et al. TRIM56-mediated monoubiquitination of cGAS for cytosolic DNA sensing. Nat. Commun. 2018;9:613. doi: 10.1038/s41467-018-02936-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yuen C.K., et al. Suppression of Type I interferon production by human T-cell leukemia virus type 1 oncoprotein Tax through inhibition of IRF3 phosphorylation. J. Virol. 2016;90:3902–3912. doi: 10.1128/JVI.00129-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang J., et al. HTLV-1 Tax impairs K63-linked ubiquitination of STING to evade host innate immunity. Virus Res. 2017;232:13–21. doi: 10.1016/j.virusres.2017.01.016. [DOI] [PubMed] [Google Scholar]
- 89.Maringer K., Fernandez-Sesma A. Message in a bottle: lessons learned from antagonism of STING signalling during RNA virus infection. Cytokine Growth Factor Rev. 2014;25:669–679. doi: 10.1016/j.cytogfr.2014.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sozzani S., et al. Type I interferons in systemic autoimmunity. Autoimmunity. 2010;43:196–203. doi: 10.3109/08916930903510872. [DOI] [PubMed] [Google Scholar]
- 91.Hadjadj J., et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science. 2020;369:718–724. doi: 10.1126/science.abc6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Li M., et al. Pharmacological activation of STING blocks SARS-CoV-2 infection. Sci. Immunol. 2021;6 doi: 10.1126/sciimmunol.abi9007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Humphries F., et al. A diamidobenzimidazole STING agonist protects against SARS-CoV-2 infection. Sci. Immunol. 2021;6 doi: 10.1126/sciimmunol.abi9002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhu Q., et al. Inhibition of coronavirus infection by a synthetic STING agonist in primary human airway system. Antivir. Res. 2021;187 doi: 10.1016/j.antiviral.2021.105015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rui Y., et al. Unique and complementary suppression of cGAS-STING and RNA sensing- triggered innate immune responses by SARS-CoV-2 proteins. Signal Transduct. Target Ther. 2021;6:123. doi: 10.1038/s41392-021-00515-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wu J., et al. SARS-CoV-2 ORF9b inhibits RIG-I-MAVS antiviral signaling by interrupting K63-linked ubiquitination of NEMO. Cell Rep. 2021;34 doi: 10.1016/j.celrep.2021.108761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chang C.Y., et al. Middle East Respiratory Syndrome coronavirus nucleocapsid protein suppresses Type I and Type III interferon induction by targeting RIG-I signaling. J. Virol. 2020;94 doi: 10.1128/JVI.00099-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Oh S.J., Shin O.S. SARS-CoV-2 Nucleocapsid protein targets RIG-I-like receptor pathways to inhibit the induction of interferon response. Cells. 2021;10:530. doi: 10.3390/cells10030530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yang Z., et al. Suppression of MDA5-mediated antiviral immune responses by NSP8 of SARS-CoV-2. bioRxiv. 2020 doi: 10.1101/2020.08.12.247767. Published online August 12, 2020. [DOI] [Google Scholar]
- 100.Xu M., et al. A ubiquitin replacement strategy in human cells reveals distinct mechanisms of IKK activation by TNFalpha and IL-1beta. Mol. Cell. 2009;36:302–303. doi: 10.1016/j.molcel.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.de Almagro M.C., et al. Cellular IAP proteins and LUBAC differentially regulate necrosome-associated RIP1 ubiquitination. Cell Death Dis. 2015;6 doi: 10.1038/cddis.2015.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Dillon C.P., et al. RIPK1 blocks early postnatal lethality mediated by caspase-8 and RIPK3. Cell. 2014;157:1189–1202. doi: 10.1016/j.cell.2014.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Weinlich R., Green D.R. The two faces of receptor interacting protein kinase-1. Mol. Cell. 2014;56:469–480. doi: 10.1016/j.molcel.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Tang Y., et al. K63-linked ubiquitination regulates RIPK1 kinase activity to prevent cell death during embryogenesis and inflammation. Nat. Commun. 2019;10:4157. doi: 10.1038/s41467-019-12033-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zhang X., et al. Ubiquitination of RIPK1 suppresses programmed cell death by regulating RIPK1 kinase activation during embryogenesis. Nat. Commun. 2019;10:4158. doi: 10.1038/s41467-019-11839-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Bishop G.A., et al. TRAF3 as a multifaceted regulator of B lymphocyte survival and activation. Front. Immunol. 2018;9:2161. doi: 10.3389/fimmu.2018.02161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wallis A.M., Bishop G.A. TRAF3 regulation of inhibitory signaling pathways in B and T lymphocytes by kinase and phosphatase localization. J. Leukoc. Biol. 2018;103:1089–1098. doi: 10.1002/JLB.2MIR0817-339RR. [DOI] [PubMed] [Google Scholar]
- 108.Walsh M.C., et al. Tumor necrosis factor receptor-associated factor 6 (TRAF6) regulation of development, function, and homeostasis of the immune system. Immunol. Rev. 2015;266:72–92. doi: 10.1111/imr.12302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Tseng P.H., et al. Different modes of ubiquitination of the adaptor TRAF3 selectively activate the expression of type I interferons and proinflammatory cytokines. Nat. Immunol. 2010;11:70–75. doi: 10.1038/ni.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zhang L., et al. NLRC3, a member of the NLR family of proteins, is a negative regulator of innate immune signaling induced by the DNA sensor STING. Immunity. 2014;40:329–341. doi: 10.1016/j.immuni.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Cui J., et al. NLRC5 negatively regulates the NF-kappaB and type I interferon signaling pathways. Cell. 2010;141:483–496. doi: 10.1016/j.cell.2010.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Moore C.B., et al. NLRX1 is a regulator of mitochondrial antiviral immunity. Nature. 2008;451:573–577. doi: 10.1038/nature06501. [DOI] [PubMed] [Google Scholar]
- 113.Chen S.T., et al. NLRP12 regulates anti-viral RIG-I activation via interaction with TRIM25. Cell Host Microbe. 2019;25:602–616. doi: 10.1016/j.chom.2019.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kouwaki T., et al. Ubiquitin ligase RIPLET mediates polyubiquitination of RIG-I and LGP2 and regulates the innate immune responses to SARS-CoV-2 infection. bioRxiv. 2021 doi: 10.1101/2021.01.25.428042. Published online January 28, 2021. [DOI] [Google Scholar]
- 115.Lork M., et al. CYLD, A20 and OTULIN deubiquitinases in NF-kappaB signaling and cell death: so similar, yet so different. Cell Death Differ. 2017;24:1172–1183. doi: 10.1038/cdd.2017.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wertz I.E., et al. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-kappaB signalling. Nature. 2004;430:694–699. doi: 10.1038/nature02794. [DOI] [PubMed] [Google Scholar]
- 117.Shembade N., et al. Inhibition of NF-kappaB signaling by A20 through disruption of ubiquitin enzyme complexes. Science. 2010;327:1135–1139. doi: 10.1126/science.1182364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Shembade N., et al. The human T-cell leukemia virus type 1 Tax oncoprotein requires the ubiquitin-conjugating enzyme Ubc13 for NF-kappaB activation. J. Virol. 2007;81:13735–13742. doi: 10.1128/JVI.01790-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Duwel M., et al. A20 negatively regulates T cell receptor signaling to NF-kappaB by cleaving Malt1 ubiquitin chains. J. Immunol. 2009;182:7718–7728. doi: 10.4049/jimmunol.0803313. [DOI] [PubMed] [Google Scholar]
- 120.Wang B., et al. TRAF2 and OTUD7B govern a ubiquitin-dependent switch that regulates mTORC2 signalling. Nature. 2017;545:365–369. doi: 10.1038/nature22344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Xie X., et al. Deubiquitylases USP5 and USP13 are recruited to and regulate heat-induced stress granules through their deubiquitylating activities. J. Cell Sci. 2018;131 doi: 10.1242/jcs.210856. [DOI] [PubMed] [Google Scholar]
- 122.Le Negrate G., et al. Salmonella secreted factor L deubiquitinase of Salmonella typhimurium inhibits NF-kappaB, suppresses IkappaBalpha ubiquitination and modulates innate immune responses. J. Immunol. 2008;180:5045–5056. doi: 10.4049/jimmunol.180.7.5045. [DOI] [PubMed] [Google Scholar]
- 123.Le Negrate G., et al. ChlaDub1 of Chlamydia trachomatis suppresses NF-kappaB activation and inhibits IkappaBalpha ubiquitination and degradation. Cell. Microbiol. 2008;10:1879–1892. doi: 10.1111/j.1462-5822.2008.01178.x. [DOI] [PubMed] [Google Scholar]
- 124.Zhou H., et al. Yersinia virulence factor YopJ acts as a deubiquitinase to inhibit NF-kappa B activation. J. Exp. Med. 2005;202:1327–1332. doi: 10.1084/jem.20051194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ye L., et al. USP49 negatively regulates cellular antiviral responses via deconjugating K63-linked ubiquitination of MITA. PLoS Pathog. 2019;15 doi: 10.1371/journal.ppat.1007680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Li S.W., et al. SARS coronavirus papain-like protease inhibits the TLR7 signaling pathway through removing Lys63-linked polyubiquitination of TRAF3 and TRAF6. Int. J. Mol. Sci. 2016;17:678. doi: 10.3390/ijms17050678. [DOI] [PMC free article] [PubMed] [Google Scholar]