Abstract
The pandemic due to the outbreak of 2019 coronavirus disease (COVID-19) caused by novel severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) has raised significant public health concerns. Rapid, affordable, and accurate diagnostic testing not only paves the way for the effective treatment of diseases, but also plays a crucial role in preventing the spreading of infectious diseases. Herein, a one-pot CRISPR/Cas13a-based visual biosensor was proposed and developed for the rapid and low-cost nucleic acid detection. By combining Cas13a cleavage and Recombinase Polymerase Amplification (RPA) in a one-pot reaction in a disposable tube-in-tube vessel, amplicon contamination could be completely avoided. The RPA reaction is carried out in the inner tube containing two hydrophobic holes at the bottom. After the completion of amplification reaction, the reaction solution enters the outer tube containing pre-stored Cas13a reagent under the action of centrifugation or shaking. Inner and outer tubes are combined to form an independent reaction pot to complete the nucleic acid detection without opening the lid. This newly developed nucleic acid detection method not only meets the need of rapid nucleic acid detection at home without the need for any specialized equipment, but also fulfils the requirement of rapid on-site nucleic acid detection with the aid of small automated instruments. In this study, CRISPR/Cas13a and CRISPR/Cas12a were used to verify the reliability of the developed one-pot nucleic acid detection method. The performance of the system was verified by detecting the DNA virus, i.e., African swine fever virus (ASFV) and the RNA virus, i.e., SARS-Cov-2. The results indicate that the proposed method possesses a limit of detection of 3 copy/μL. The negative and positive test results are consistent with the results of real-time fluorescence quantitative polymerase chain reaction (PCR), but the time required is shorter and the cost is lower. Thus, this study makes this method available in resource-limited areas for the purpose of large-scale screening and in case of epidemic outbreak.
Keywords: CRISPR/Cas13a, One-pot reaction, Nucleic acid diagnostics, Contamination-free detection, Low-cost detection
1. Introduction
Frequent and large-scale infectious diseases exhibit a significant performance impact on the social stability and economic development, leading to the international or regional health crises. In particular, coronavirus disease (COVID-2019), which broke out at the end of 2019, has spread in more than 200 countries around the world, with more than 250 million confirmed cases (Sieber et al., 2021). Almost all infectious diseases are caused by nucleic acid pathogens (except in rare cases such as prions); therefore, pathogen-specific nucleic acids (RNA or DNA) are often used as the “gold standard” biomarker for the diagnosis of infectious diseases. Sensitive, distinctive, and rapid diagnostic testing not only paves the way for effective treatment of diseases, but also plays a crucial role in preventing the spreading of infectious diseases (Wei et al., 2021). The current epidemic of COVID-2019 is fierce with high infection rates and high requirements for the turnover of nucleic acid detection. Moreover, the strong latent characteristics of COVID-2019 indicate that highly sensitive rapid diagnosis and on-site screening of suspected patients and close contacts need to be carried out in communities, ports, and other places.
To meet the requirements of detection sensitivity and throughput analysis, the currently available diagnostic methods, e.g., real-time fluorescence quantitative PCR, are time consuming and require expensive laboratory settings and well-trained personnel (Mukama et al., 2020a). The existing rapid nucleic acid detection equipment based on microfluidics and PCR technology, such as GeneXpert, Filmarray, and other systems, have high testing cost (the cost of a single test is 100–500$). Therefore, such tests are not available in resource-limited areas, for large-scale screenings, and in case of outbreaks of epidemics. Regularly clustered interval short palindrome repeated sequence (CRISPR) nucleic acid detection technology is based on CRISPR-related proteins, which non-specifically cleave non-target nucleic acids after recognition of the target sequence to finally detect the sequence. This technology has been used to detect the nucleic acids of a variety of pathogens (Gootenberg et al., 2017; Myhrvold et al., 2018). In 2017, Zhang et al. from the Broad Institute of Harvard University pioneered SHERLOCK nucleic acid detection technology by using the cleavage activity of the CRISPR-Cas13a enzyme and a unique fluorescent reporter to successfully detected the Zika virus (Gootenberg et al., 2017). In 2018, Chen et al. developed DETECTR nucleic acid detection platform based on the excellent properties of Cas12a enzyme and successfully identified the two genes HPV16 and HPV18 (Chen et al., 2021). In the same year, this group developed DETECTR-Cas14 platform based on the Cas14 enzyme (Harrington et al., 2018) and applied it successfully for SNP genotyping and molecular diagnosis. Since then, a variety of nucleic acid detection methods based on CRISPR enzymes, such as Cas12 (Broughton et al., 2020; Li et al., 2018, 2019), Cas13a (Chang et al., 2020; Gootenberg et al., 2018; Shen et al., 2020), and Cas14 (Harrington et al., 2018), have been reported.
Compared to other traditional nucleic acid detection methods, CRISPR technology offers overwhelming advantages in the aspects of detection cost, efficiency, portability, and distinctiveness (Li et al., 2021; Wu et al., 2021). However, to improve the detection sensitivity, nucleic acid amplification should be carried out before the CRISPR nucleic acid detection reaction. Therefore, most of the current CRISPR nucleic acid detection strategies involve two steps (Kanitchinda et al., 2020; Lu et al., 2020; Mukama et al., 2020b) and require opening the lid and adding the CRISPR reaction system after nucleic acid amplification, which may cause contamination. The corresponding target nucleic acid sequence can also be detected by CRISPR technology alone, but its minimum detection limit can only reach the femtomolar level (Gootenberg et al., 2017), which is far from meeting the needs of clinical detection. Nucleic acid amplification reagents are mixed with Cas enzyme to perform nucleic acid amplification and detection at the same time and avoid opening the lid (Wang et al., 2021a; Wu et al., 2020; Zhang et al., 2020). However, this reduces the nucleic acid detection sensitivity, mainly because some components of the CRISPR reagent buffer have a certain inhibitory effect on nucleic acid amplification (Arizti-Sanz et al., 2020). For example, magnesium ion (Wang et al., 2019) contained in the CRISPR Cas buffer can affect the RPA reaction, reduce the amplification efficiency of the RPA reaction, or inhibit the occurrence of the RPA reaction. Furthermore, during the RPA reaction combined with CRISPR/Cas12a detection, Cas12 enzyme can recognize and specifically cleave sample nucleic acid and thus reduce the efficiency of the nucleic acid amplification reaction when a certain sample nucleic acid concentration is reached after amplification, which reduces the sensitivity and detection efficiency of this method (Wang et al., 2019). The same principle applies to the detection reaction of RPA combined with CRISPR/Cas13a. Notably, T7 RNA polymerase contained in the CRISPR/Cas13a reagent can convert double-stranded DNA amplified by RPA into single-stranded RNA and reduce the efficiency of the nucleic acid amplification reaction (Arizti-Sanz et al., 2020). This also fundamentally reduces the sensitivity and detection efficiency of the method.
To solve the problems of complex operation, aerosol pollution, and low sensitivity in CRISPR nucleic acid detection, some researchers have proposed to add amplification reagents to the bottom of the tube and CRISPR reagents to the cap or wall of the tube in advance to realize nucleic acid detection without opening the lid (Ali et al., 2020; Chen et al., 2020; Wang et al., 2019, Wang et al., 2020a, Wang et al., 2020b, 2021b; Zhang et al., 2020). Through this method, Ali et al. constructed iSCAN system for the rapid detection of COVID-19 based on the combination of LAMP amplification, CRISPR, and lateral flow immunochromatography technology (Ali et al., 2020). Researchers from the University of Connecticut solved the above-mentioned problems based on the principle of density stratification. They used different concentrations of sucrose solutions to physically separate the RPA reaction system and the Cas12a reaction system in one tube to avoid laboratory aerosol contamination (Yin et al., 2020). The innovation of experimental consumables is also an important approach to solve the above-mentioned problems. For example, Wang et al. separated the two reaction systems by pre-adding the rapid PCR reagent and CRISPR reagent to the bottom and top of the capillary tube, respectively, to realize nucleic acid detection without opening the cap (Wang et al., 2021a). Wu et al. proposed a Cas12a-PB sensor composed of a chamber for CRISPR detection, opened on one side of the PMMA plate; and an amplification reaction tube, connected to the other side to realize the Cas12a-PB detection based on CRISPR reaction and RPA reaction (Wu et al., 2020). Moreover, some researchers also directly mixed RPA reagents and CRISPR reagents to create one-pot SHERLOCK (Kellner et al., 2020). Although the detection efficiency is improved and the problem of pollution caused in the two-step detection process is avoided, this method reduces the detection sensitivity, brings more challenges to the optimization of experimental conditions, increases false negatives, and reduces the stability of system reaction. All these methods provide new ideas for solving aerosol pollution and improving detection accuracy and sensitivity, and interestingly, they can be further optimized in terms of stability, ease of use, and cost.
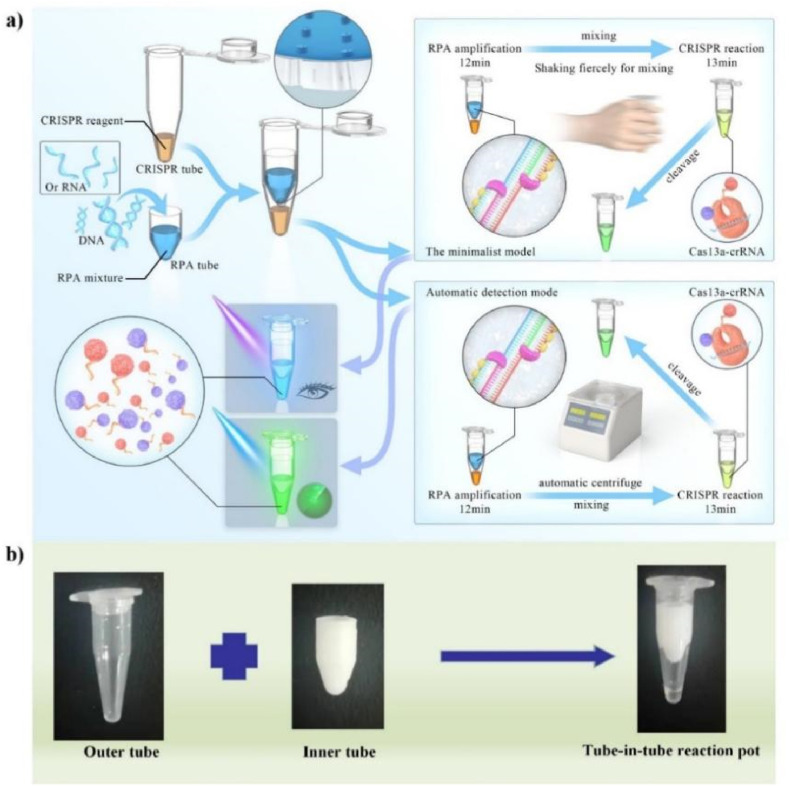
In this study, a one-pot CRISPR Cas13a-based contamination-free biosensor was proposed for the rapid and low-cost nucleic acid detection, and the method is named as ECS-CRISPR (Easy to operate, Contamination-free, and Stable CRISPR) nucleic acid detection. Fig. 1 a and b illustrate the detailed tube-in-tube vessel design and working principle of the nucleic acid detection. The main disposable material of the visual sensor is a tube-in-tube structure. The inner tube contains two hydrophobic holes and is used to complete the RPA, i.e., nucleic acid amplification reaction. The CRISPR Cas13a fluorescence detection reagent is pre-stored in the inner tube. The outer tube and the inner tube are combined into one structure, which is referred to as the tube-in-tube vessel. After completion of the amplification reaction, the product is transferred to the outer tube via the hydrophobic hole by centrifugation, and the fluorescent visual detection of target nucleic acid is realized under blue light emitting diode (LED) excitation. The sensor can detect pathogen nucleic acids within 25 min without any special equipment, meeting the needs of point-of-care nucleic acid detection in areas with limited resources. Moreover, it can also be used in conjunction with an automated miniaturized instrument. This instrument can automatically perform the contamination-free detection of eight nucleic acid samples within 25 min. The entire reaction process is simple to operate and suitable for nucleic acid analysis in community hospitals and ports. The external structure of the tube-in-tube vessel is consistent with that of an ordinary PCR tube, which is convenient for experimental operation. By expanding the size of the centrifugal device, the detection throughput can be further improved for large hospitals and central laboratories. In the study, CRISPR/Cas13a and CRISPR/Cas12a were used to investigate and verify the reliability of the developed one-pot nucleic acid detection method. The DNA virus, namely ASFV, was selected for verification experiments. The experimental results were found to be consistent with the results of traditional fluorescence quantitative PCR detection, and the limit of detection of the proposed method was 3 copy/μL. Rapid contamination-free on-site nucleic acid detection is a convenient and low-cost method, which may provide an opportunity for mass population screening and field deployable diagnostics.
Fig. 1.
CRISPR/Cas13a-based contamination-free biosensor for rapid nucleic acid diagnostics: a) Schematic showing the working principle of the nucleic acid biosensor and b) Tube-in-tube vessel structure composed of an inner tube and an outer tube.
2. Experimental
Details about all the required materials, reagents, instrumentation, and methods are provided in the Supplementary Information.
3. Results and discussion
3.1. Overview of ECS-CRISPR detection
In this study, the two methods, namely, isothermal RPA and CRISPR/Cas13a detection were combined. The target detection sequence was amplified by RPA and detected based on the non-specific shearing effect after specific activation of CRISPR/Cas13a. According to the literature, the RPA process can be disturbed by the CRISPR/Cas13a reagent system, if both the reagents are directly mixed (Gootenberg et al., 2017). Therefore, the detection was performed via two separated reactions. Opening the lid after amplification can cause aerosol cross contamination, which leads to a significant challenge for automation. The other challenge is the direct coupling of amplification with CRISPR-Cas13a detection in a one-pot approach. In this study, the innovative design of a disposable tube-in-tube reaction vessel was presented for the one-pot amplification reaction and CRISPR/Cas13a detection. The RPA reaction was carried out in the inner tube containing two hydrophobic holes at the bottom. After the completion of amplification reaction, the reaction solution was allowed to enter the outer tube containing pre-stored Cas13a reagent under the action of centrifugation or shaking. Inner and outer tubes were combined to form an independent reaction pot to complete the nucleic acid detection without opening the lid. To meet the needs of nucleic acid detection in a variety of application scenarios, a minimalist model requiring no equipment was developed herein. Moreover, small instruments were invented to assist the rapid automatic on-site detection mode.
First, the non-equipment minimalist working mode (Supplementary Fig. S2) is suitable for detection at home and in places with extremely poor and limited resources. The visual detection of the target sequences was achieved using a battery-powered ultraviolet (UV) light and compatible disposable reagents. In this detection mode, there is no need to open the lid after amplification. The reaction tube was shaken manually and violently to drain the reagents in the inner reaction tube into the outer reaction tube and complete the mixing. After completion of the CRISPR detection reaction, negative and positive samples were recognized by the naked eye under UV light or blue LED. Complex instruments were not needed in this detection process, meeting the needs of testing at home and in regions with extreme lack of resources. Furthermore, the total cost of disposable reagents is expected to not exceed 1$ (Supplementary Table S2).
Second, the miniaturized instrument-assisted working mode (Supplementary Fig. S3) is suitable for application scenarios such as clinical diagnosis in community hospitals, emergency departments, or immediate diagnosis and screening for disease prevention and control. We developed a portable automated instrument that matches the tube-in-tube vessel, provides the required temperature of 39 °C, and performs functions such as centrifugation, mixing, automatic fluorescence detection, and automatic judgment of experimental results during the reaction process. In this detection mode, no tedious manual operation is required, and the process contains no lid-opening operation step. In this study, an equipment that allows testing of eight samples at a time was developed, and the test results are automatically output within 25 min. In this working mode, the limitations of cross-contamination and automation are overcome. Moreover, this miniaturized instrument (18 × 27 × 18.5 cm3) meets the portability requirements as well.
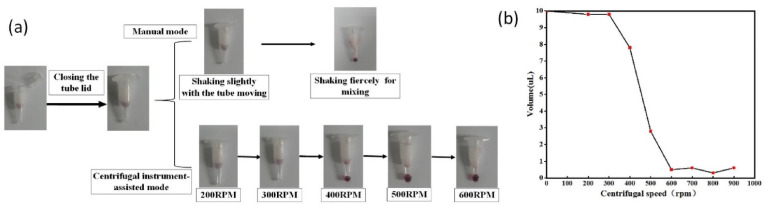
To complete the detection, the amplified nucleic acid in the inner tube needs to pass through the small hole of the inner tube to enter the outer tube. Therefore, it is necessary to verify whether the liquid is transferred into the outer tube under the action of centrifugation or vigorous shaking, while the liquid in the inner tube should not flow out under normal conditions. In this study, the liquid's surface shape and pressure at the small hole of the inner tube were calculated based on fluid mechanics, followed by simulation of the calculation results by using the Matlab software (calculation and simulation process and the corresponding results are shown in the Supporting Information). The calculation and simulation results revealed that the position and size of the small hole affect the state of the fluid in the inner tube. The smaller the radius of the small hole, the higher the surface tension of the liquid, the higher the resistance the solution needs to overcome to flow out from the small hole, and the higher the required centrifugal rotation speed or vibration acceleration. Moreover, the closer the small hole is to the bottom of the chamber, the higher the pressure under static conditions, and the easier is the transfer of liquid through the small hole. However, when the position of the small hole was very close to the position of the centrifugal axis during centrifugation, the liquid in the inner tube did not flow out independently of the rotation speed. To avoid this situation, two small holes were evenly distributed in a circle above the bottom of the inner tube to ensure the flowing out of the liquid in the inner tube under any assembly condition. At the same time, as the solution in the inner tube continuously flowed out through the small hole, the pressure at the small hole gradually decreased due to the decrease in the volume of the remaining solution in the inner tube. When this pressure and the surface tension of the small hole reached an equilibrium, the flow slowed down and stopped. The remaining volume in the small cavity was the residual volume. To avoid any residual volume, the centrifugal or shaking rate should be increased, and all the remaining solution should be discharged under the action of inertia after the liquid pressure and surface tension are balanced.
The above-mentioned conclusions were verified under experimental conditions. For an improved visualization of the experimental results, magenta solution (2%) was added to the RPA reaction solution in a ratio of 1:50 to adjust the red experimental liquid. Then, the red liquid (10 μL) was added into the inner tube, and the experiment was carried out both under centrifugation as well as under vigorous manual shaking. The radius of the small hole was 0.5 mm (determined by the lower limit of the laboratory three-dimensional printer), and the height from the bottom of the inner tube to the center of the hole was 1 mm. The image of the liquid state after centrifugation was recorded, and the volume of the liquid remaining in the inner tube was measured (Fig. 2 ). In the centrifugal mode, when the centrifugal rotation speed reached 400 rpm, the liquid in the inner tube flowed out, but a residual volume of liquid remained in the inner tube. When the rotation speed was increased to 600 rpm, the liquid in the inner tube almost completely entered the outer tube within 30 s, as confirmed by Fig. 2b, and the residual liquid volume in the inner tube was less than 0.5 μL. The above-mentioned experimental results are consistent with the simulation results (Supplementary S8). In the manual mode, the fluid in the inner tube did not flow out during low-speed shaking, capping, and moving the test tube. However, when the fluid was shaken at high speed, the fluid in the inner tube rapidly flowed out from the small hole and entered the outer tube without the appearance of any dead volume.
Fig. 2.
Validation of the fluid in the disposable tube-in-tube reaction vessel: a) The experimental results confirming that the liquid in the inner tube does not flow out under capping and slight shaking, as well as under low-speed centrifugation. However, when the rotation speed exceeds 400 rpm, the solution in the inner tube flows out, and no observable solution residue remains in the inner tube when the rotation speed reaches 600 rpm and b) Volume of the red liquid remaining in the inner tube after centrifuging for sufficient time at different centrifugation speeds. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.2. Optimization of ECS-CRISPR assay
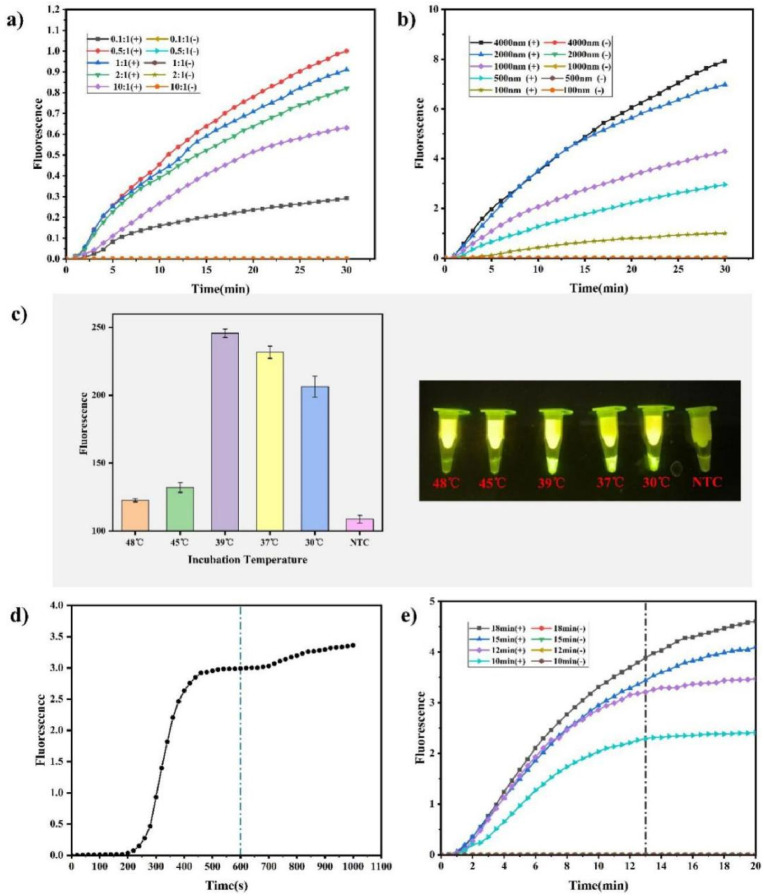
Notably, the CRISPR activity determines the sensitivity of this assay; therefore herein, systematic studies of the CRISPR reaction conditions were conducted to optimize the assay performance (Huang et al., 2020). The enzymes crRNA and Cas form a complex that can identify the corresponding target sequence and can non-specifically cut the reporter. The amount of complex formed by crRNA and Cas enzyme determines the speed of the shearing reaction. Therefore, the concentration ratio of Cas enzyme to crRNA that forms the complex is critical for the reaction. For the systematic exploration of the optimal concentration ratio of crRNA to Cas enzyme, a concentration ratio gradient of crRNA to Cas enzyme (0.1:1, 0.5:1, 1:1, 2:1, and 10:1) was studied, and the concentration of Cas enzyme was fixed at 100 nM. Experiments to verify CRISPR nucleic acid detection were performed separately, while all other conditions were maintained constant. The experimental results (Fig. 3 a) show that when the ratio of crRNA to Cas enzyme was 0.5:1, the fluorescence signal was generated faster, while the reaction rate decreased and the fluorescence intensity became lower when the ratio of crRNA to Cas enzyme was greater than 0.5:1. This is consistent with previous research results (Hsieh et al., 2020). It was thus speculated that this result is attributed to the fact that a large amount of remaining crRNA resulted in cleavage of the crRNA sequence by the activated Cas enzyme, which competes with the cleavage of the fluorescent reporter, causing a decrease in the rate of cutting the fluorescent reporter and affecting the reaction speed. When the ratio of crRNA to Cas enzyme was lower than 0.5:1, the reaction speed decreased, probably because a large number of Cas enzymes could not combine with crRNA to form a complex, resulting in only a small part of functioning Cas enzymes and thus a reduced reaction efficiency.
Fig. 3.
ECS-CRISPR assay optimization: a) The ratio of crRNA to Cas enzyme, the concentration of Cas enzyme was 100 nM, b) Substrate RNA concentration. The concentration of Cas enzyme was 100 nM and the concentration of crRNA was 50 nM, c) Temperature dependency. The concentration of Cas enzyme was 100 nM, the concentration of crRNA was 50 nM, and the RNA substrate concentration was 2000 nM. The green value of the fluorescence image was analyzed by using the ImageJ software (NTC, non-template control reaction), d) Real-time fluorescence curve of RPA combined with fluorescent dyes, e) Influence of different RPA times on the real-time fluorescence curves of the detection. The concentration of Cas enzyme was 100 nM, the concentration of crRNA was 50 nM, the RNA substrate concentration was 2000 nM, and the incubation temperature was 39 °C. Bar graph data represented as the mean ± SD of three experimental replicates. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
The fluorescence signal of CRISPR nucleic acid detection was produced by the non-specific cleavage of the fluorescent reporter of the complex formed by crRNA and Cas enzyme. Therefore, the concentration of the fluorescent reporter also affects the rate of fluorescence generation and the final fluorescence intensity (Ding et al., 2020). In order to explore the optimal reporter concentration, CRISPR nucleic acid detection experiments were performed with a concentration gradient of fluorescent reporters set as 100, 500, 1000, 2000, and 4000 nM, respectively. Except for the reporter concentration, all other reaction conditions were maintained constant. The generation rate and intensity of the reporter's fluorescence signal were detected by real-time fluorescence PCR, and the results are shown in Fig. 3b. At reporter concentrations above 2000 nM, the concentration exhibited little effect on the generation rate and intensity of the final fluorescence signal. Based on these results and in order to save costs, herein, 2000 nM was selected as the reporter concentration of the final reaction system.
RPA reaction and CRISPR reaction are enzymatic reactions that require an appropriate temperature for the corresponding enzyme to get activated. Too high or too low temperatures adversely affect the entire reaction. According to the literature, most RPA and CRISPR reactions require temperatures between 37 and 39 °C. Considering that the selected temperature must meet the temperature requirements of both reactions at the same time, in this study, temperatures between 37 and 39 °C were identified as the optimal temperature range. To further explore the effect of temperatures other than this optimal temperature range on the entire reaction, the reactions at a low temperature of 30 °C and high temperatures of 45 and 48 °C were investigated herein. Therefore, the ECS-CRISPR reaction was performed at temperatures of 30, 37, 39, 45, and 48 °C, the concentration ratio of crRNA to Cas enzyme of 0.5:1, and the reporter concentration of 2000 nM. In order to ensure the completion of the reaction, both RPA time and CRISPR detection time were set to 20 min. The detection outcome was observed using the portable blue LED transilluminator, and the captured image of the transilluminator was treated by using the ImageJ software. Fig. 3c exhibits that temperatures of 30, 37, and 39 °C produced visible fluorescence signals, while 45 and 48 °C did not produce obvious fluorescence signals. The temperature of the human palm is approximately 36–37 °C, thus it was verified that the developed minimalist model could operate stably in the palm under any condition. The temperature environment provided by the developed automated instrument was set to 39 °C to ensure that the ECS-CRISPR reaction was carried out under appropriate conditions. To verify the temperature control performance of the automated instrument, temperature heating and constant temperature capability tests were conducted, and the results are shown in Fig. S4 (Supplementary Information).
The nucleic acid detection time of current standard PCR technology is about 1.5–2.5 h, which is not satisfactory. For the ECS-CRISPR detection proposed in this study, initially, the total detection time was optimized and verified. First, the target sequence was detected by performing RPA and using fluorescent dyes. Fig. 3d shows the recorded fluorescence curve, which begins to reach saturation after 10 min of RPA. It was assumed that after RPA for 10 min, the RPA reaction could cooperate with the CRISPR reaction for the corresponding target detection. To explore the optimal reaction time for the combination of both reactions and observe whether an extended reaction time of the RPA amplification would affect the final experimental results, it was decided to combine the RPA reaction system with CRISPR after 10 min, for nucleic acid detection. The experiment was performed at RPA reaction times of 18, 15, 12, and 10 min at a constant sample concentration of 3 × 104 copy/mL. After completion of the amplification reaction, the samples were stored in ice water at 0 °C. Then, all samples were subjected to the CRISPR reaction, and real-time fluorescence curves were simultaneously recorded using a real-time fluorescence PCR instrument to determine the CRISPR reaction time that allows distinguishing between negative and positive response (Fig. 3e). The fluorescence curves showed that the sample with the lowest concentration of 3 × 104 copy/mL could also clearly distinguish between negative and positive response after 12 min of CRISPR reaction. Moreover, the increase in the RPA response time exhibited a positive influence on CRISPR/Cas13a signals. The fastest fluorescence response and the shortest CRISPR detection time were obtained for a total amplification time of 18 min. However, considering the total time required for the two-step reaction, the RPA time of 12 min and the CRISPR nucleic acid detection time of 13 min were selected. For a total reaction time of 25 min, a distinct fluorescence signal that could distinguish between negative and positive response was obtained for high- and low-concentration samples.
3.3. Sensitivity of the ECS-CRISPR platform
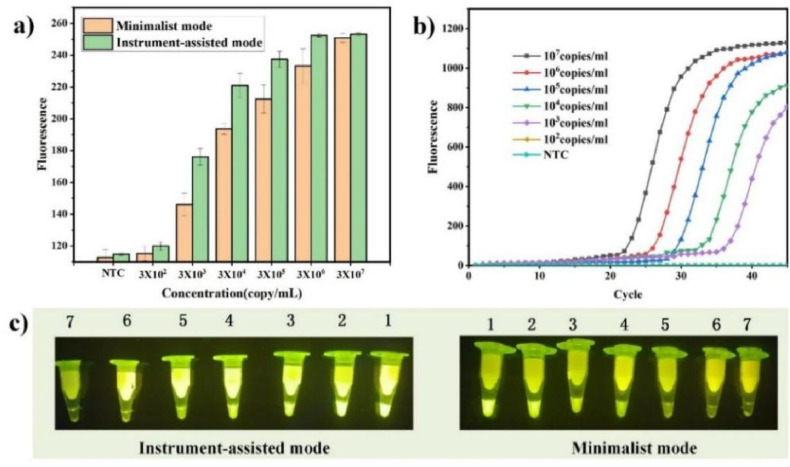
Initially, nucleic acid detection based on only CRISPR technology could reach a minimum detection limit of 50 fM (Gootenberg et al., 2017), which is far from meeting the needs of clinical testing. The CRISPR detection method was combined with pre-amplification technology for nucleic acid detection, and the minimum detection limit could reach the single molecule level (Ai et al., 2019; Lee et al., 2021; Qian et al., 2019). Furthermore, the sensitivity of this method even exceeded that of standard qPCR technology (Wang, X.Y. et al., 2020). To evaluate the feasibility and stability of the actual application of the ECS-CRISPR platform in the above-mentioned two working modes and to explore the detection sensitivity, ASFV nucleic acid standard product samples were tested herein at the concentrations of 3 × 107, 3 × 106, 3 × 105, 3 × 104, 3 × 103, and 3 × 102 copy/mL, both in the minimalist and automated modes. In the minimalist mode, the test results were observed by the naked eye and recorded using a mobile phone camera. Moreover, the samples were detected with automated instruments, and the test results were compared with those of standard fluorescent PCR. Fig. 4 a shows that the ASFV template concentration of 3 × 103 copy/mL or higher could be detected by an obvious fluorescence signal. The fluorescence signal intensity in the minimalist working mode was slightly lower than that in the automated mode; however, the fluorescence intensity could still be recognized by the naked eye. The low fluorescence signal intensity mainly resulted from the experimental conditions in the minimalist working mode, which were not stable enough. However, when the ASFV template concentration was as low as 3 × 102 copy/mL, no fluorescence signal was observed. This indicates that the minimum detection limit of ECS-CRISPR detection was at least 3 × 103 copy/mL, which is the same as or higher than the minimum detection limit measured by other CRISPR detection methods combined with preamplification technology (Jiao et al., 2021; Sullivan et al., 2019; Zhou et al., 2020). Under the same conditions, the same series of samples with different concentrations was subjected to standard PCR experiments, and the limit of detection of our proposed method was found to be 3 copy/μL (Fig. 4b). Although the actual limit of detection of the ultra-sensitive ASFV fluorescent PCR method could reach 150 copy/mL, this method achieved a high sensitivity and did not involve opening operations in the entire detection process, which effectively avoids laboratory aerosol pollution. Moreover, the required detection time is only 1/3 of the standard PCR time. Furthermore, detection limit experiments were also conducted for the new coronavirus nucleic acid and the obtained detection limit results were in the same order of magnitude (Supplementary Information S9).
Fig. 4.
Sensitivity assessment of CRISPR/Cas13a-assisted RPA for the ASFV. Ten gradual dilutions of the DNA template from 3 × 107 to 3 × 102 copy/mL and non-template control reaction (NTC): a) Visualization of CRISPR/Cas13a-assisted RPA by the miniaturized instrument-assisted working mode and the minimalist working mode. Analysis of the green value of the fluorescence image by using the ImageJ software (NTC, non-template control reaction), b) Real-time fluorescence curve of standard PCR amplification. Negative control without template addition (NTC), c) Under LED light irradiation, visualization of CRISPR/Cas13a-assisted RPA by the instrument-assisted working mode and minimalist working mode. Sample 1–6: DNA templates with ten gradual dilutions from 3 × 107 to 3 × 102 copy/mL; sample 7: negative control without template addition (NTC). Bar graph data represented as the mean ± SD of three experimental replicates (p < 0.05). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.4. Feasibility and reliability analysis of ECS-CRISPR
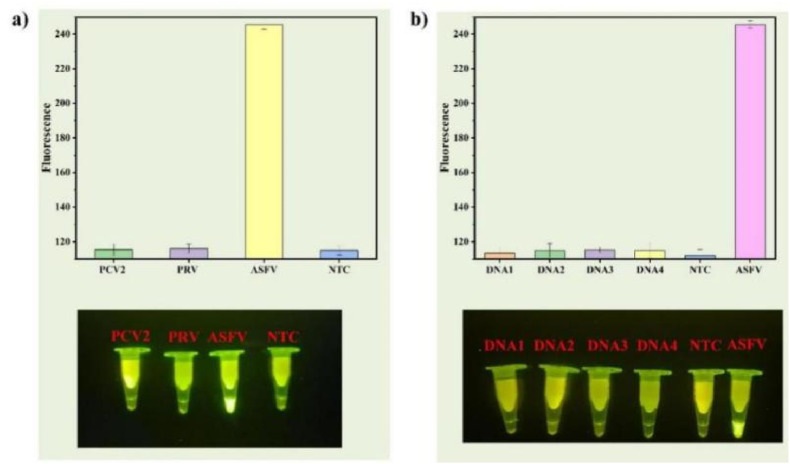
Specificity is a very significant problem in the process of nucleic acid detection. Good specificity helps to avoid the production of false positive results, reduce error judgement, and improve the detection accuracy. To assess whether the ECS-CRISPR nucleic acid detection method has a good specificity for virus detection, the ASFV virus target sequence among selected similar sequences of the double-stranded DNA virus PRV and the single-stranded DNA virus PCV2 as references were analyzed under optimized experimental conditions. The three virus sequences and double distilled water were analyzed under the two working modes of ECS-CRISPR nucleic acid detection. The detection reagents included specific ASFV sequence amplification primers and crRNA. Fig. 5 a demonstrates that the ASFV sample exhibits an obvious fluorescence signal, while all other samples show the absence of fluorescence signals. This experiment shows that the ECS-CRISPR method detects the target virus sequence among similar virus sequences with good specificity.
Fig. 5.
Specificity of the ECS-CRISPR method: a) ECS-CRISPR method for ASFV-associated viruses. Analysis of the green value of the fluorescence image by using the ImageJ software (NTC, non-template control reaction), b) ECS-CRISPR method for ASFV mismatched viruses. Analysis of the green value of the fluorescence image by using the ImageJ software (NTC, non-template control reaction). Bar graph data represented as the mean ± SD of three experimental replicates. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Furthermore, we wanted to confirm whether single-base mismatches could be detected by this method. Therefore, four more mismatch sequences of the ASFV virus were artificially synthesized. These sequences differed from the ASFV virus sequence by one to two bases, as shown in Table S3 (Supplementary Information). Samples of nucleic acids with these four mismatch sequences, as well as double distilled water and the ASFV target sequence, were detected by the same method, and the results are shown in Fig. 5b. Furthermore, in this study, the ASFV target sequence in these six samples was also successfully detected, as the other samples did not show any fluorescence signal. This experiment proved that single-base mismatches could be identified by our method. Therefore, ECS-CRISPR detection is a method with high specificity and the ability to distinguish single-base mismatches.
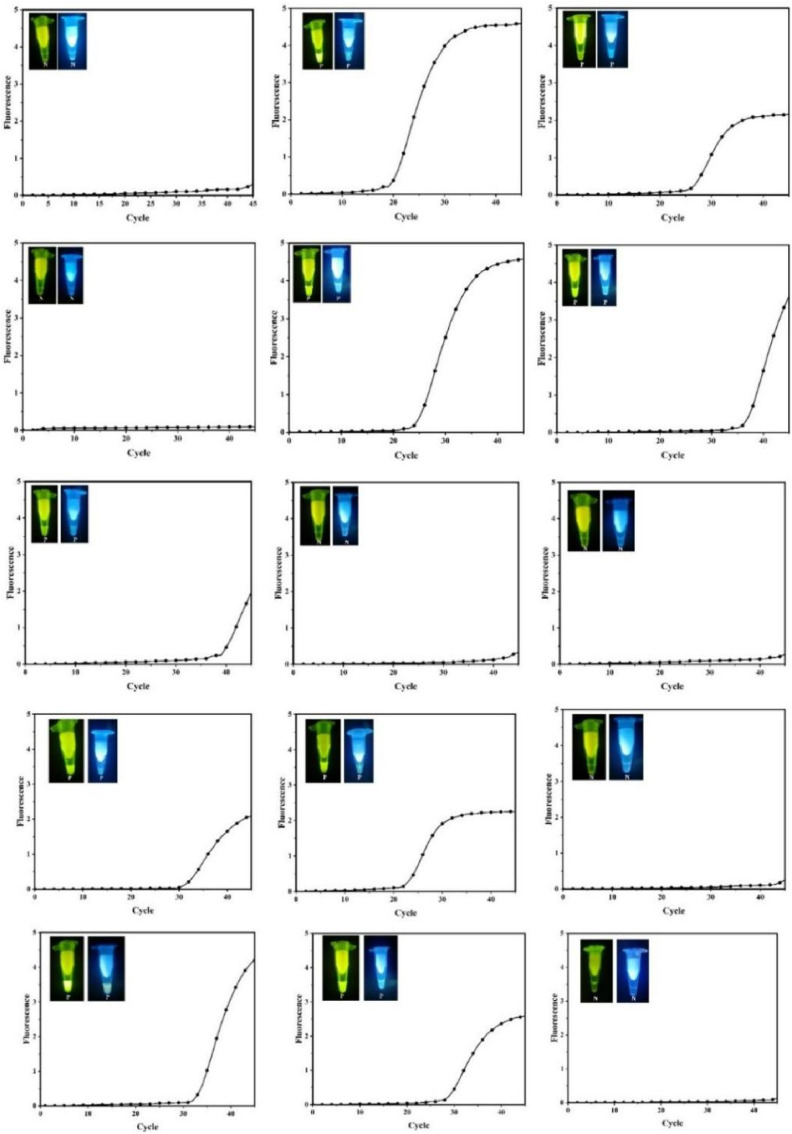
3.5. ECS-CRISPR detection of ASFV in real animal samples
Given the outstanding performance, ECS-CRISPR was further applied in the detection of ASFV in real animal samples. Pig specimens (nasal swabs, plasma) were obtained from Shanxi Xían Tianbo Medical Laboratory. Herein, traditional purification methods were used to purify the obtained samples, and the obtained ASFV virus was verified in the laboratory. Then, nine ASFV positive and six negative samples were detected by ECS-CRISPR (Fig. 6 ), showing good overall agreement with quantitative real-time PCR results obtained by state and hospital laboratories.
Fig. 6.
Fluorescence spectroscopy of CRISPR under blue LED irradiation during the detection of 15 ASFV samples photographed with a smart phone camera and using standard PCR as the control. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.6. ECS-CRISPR method involving CRISPR Cas12a nucleic acid detection
The above-mentioned tube-in-tube nucleic acid detection method established in this study was verified based on CRISPR-Cas13a. In this section, the versatility of the established method is demonstrated by applying the developed one-pot nucleic acid detection method to the RPA combined with CRISPR/Cas12a nucleic acid detection model system. In order to evaluate the feasibility and stability of the application of the ECS-CRISPR platform in the RPA combined with CRISPR/Cas12a detection model, herein, 3 × 106, 3 × 105, 3 × 104, 3 × 103, and 3 × 102 copy/mL ASFV targets were used for detection, and nuclease-free water was selected as a negative control experiment for ASFV targets. The detection method is described in S1.8. The test results were observed by the naked eye and recorded using a mobile phone camera. Then, the green value of the fluorescence image was analyzed by using the ImageJ software. Fig. S16 shows that the ASFV nucleic acid standard at concentrations of 3 × 103 copy/mL or higher could be detected by an obvious fluorescence signal. However, when the concentration of the ASFV nucleic acid standard was as low as 3 × 102 copy/mL and the target was nuclease-free water, no fluorescence signal was observed. The experimental results prove that both the CRISPR/Cas12a and CRISPR/Cas13a nucleic acid detection systems utilized in the ECS-CRISPR method have the same detection limit. This also confirms that the application of the ECS-CRISPR platform to the RPA combined CRISPR/Cas12a detection model has good feasibility and stability.
4. Conclusions
This study provides a fast, low-cost, and visual CRISPR/Cas13a nucleic acid detection method. The innovatively developed disposable tube-in-tube reaction vessel that combines RPA and CRISPR/Cas13a or Cas12a detection in one pot is easy to operate and avoids contamination. This research mainly solves the problems of aerosol pollution, complex operation, and high cost of CRISPR nucleic acid detection. In follow-up research, the automatic integration of nucleic acid extraction into the currently developed method needs to be addressed. It is speculated that possibly this method can be used in conjunction with nucleic acid extraction-free reagents to realize the “sample in-result out” detection of viral nucleic acids. The system was tested using the DNA virus ASFV and the RNA virus SARS-Cov-2. The limit of detection of the proposed method was 3 copy/μL, and the negative and positive test results were consistent with the fluorescence quantitative PCR results. In the minimalist mode, only disposables, reagents, and blue LED light excitation were needed for the visual detection of viral nucleic acid within 25 min, meeting the needs of nucleic acid detection in homes and areas with limited conditions, and the cost of a single sample detection was found to be less than $1. This system can also be used in conjunction with automated instruments for the simultaneous detection of multiple samples, meeting the needs of community hospitals and port screening for multi-sample testing. The centrifugal device can be further expanded to achieve high-throughput detection, meeting the screening needs of a large number of people in central laboratories. This new nucleic acid detection strategy may bridge the gaps that exist in the global health care system, providing an opportunity for mass population screening and field deployable diagnostics.
CRediT authorship contribution statement
Fei Hu: Conceptualization, Methodology, Visualization, Writing – original draft, Writing – review & editing. Yanfei Liu: Data curation, Software, Validation, Writing – original draft. Shuhao Zhao: Validation. Zengming Zhang: Writing – review & editing. Xichen Li: Software. Niancai Peng: Conceptualization, Supervision, Funding acquisition, Project administration. Zhuangde Jiang: Conceptualization, Resources.
Declaration of competing interest
The authors declare that they have no competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant Nos. 62005209 and 61827827), the China Postdoctoral Science Foundation (Grant No. 2020M673417), the Key Research and Development Program of Shaanxi (Grant No. 2021GXLH-Z-045), and the Fundamental Research Funds for the Central Universities (Grant No. xzy012020095).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bios.2022.113994.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Ai J.W., Zhou X., Xu T., Yang M.L., Chen Y.Y., He G.Q., Pan N., Cai Y.W., Li Y.J., Wang X.R., Su H., Wang T., Zeng W.Q., Zhang W.H. CRISPR-based rapid and ultra-sensitive diagnostic test for Mycobacterium tuberculosis. Emerg. Microb. Infect. 2019;8(1):1361–1369. doi: 10.1080/22221751.2019.1664939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali Z., Aman R., Mahas A., Rao G.S., Tehseen M., Marsic T., Salunke R., Subudhi A.K., Hala S.M., Hamdan S.M., Pain A., Alofi F.S., Alsomali A., Hashem A.M., Khogeer A., Almontashiri N.A.M., Abedalthagafi M., Hassan N., Mahfouz M.M. iSCAN: an RT-LAMP-coupled CRISPR-Cas12 module for rapid, sensitive detection of SARS-CoV-2. Virus Res. 2020;288 doi: 10.1016/j.virusres.2020.198129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arizti-Sanz J., Freije C.A., Stanton A.C., Petros B.A., Boehm C.K., Siddiqui S., Shaw B.M., Adams G., Kosoko-Thoroddsen T.S.F., Kemball M.E., Uwanibe J.N., Ajogbasile F.V., Eromon P.E., Gross R., Wronka L., Caviness K., Hensley L.E., Bergman N.H., MacInnis B.L., Happi C.T., Lemieux J.E., Sabeti P.C., Myhrvold C. Streamlined inactivation, amplification, and Cas13-based detection of SARS-CoV-2. Nat. Commun. 2020;11(1) doi: 10.1038/s41467-020-19097-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broughton J.P., Deng X.D., Yu G.X., Fasching C.L., Servellita V., Singh J., Miao X., Streithorst J.A., Granados A., Sotomayor-Gonzalez A., Zorn K., Gopez A., Hsu E., Gu W., Miller S., Pan C.Y., Guevara H., Wadford D.A., Chen J.S., Chiu C.Y. CRISPR-Cas12-based detection of SARS-CoV-2. Nat. Biotechnol. 2020;38(7):870. doi: 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y.F., Deng Y., Li T.Y., Wang J., Wang T.Y., Tan F.F., Li X.D., Tian K.G. Visual detection of porcine reproductive and respiratory syndrome virus using CRISPR-Cas13a. Transboundary Emerg. Dis. 2020;67(2):564–571. doi: 10.1111/tbed.13368. [DOI] [PubMed] [Google Scholar]
- Chen J.S., Ma E., Harrington L.B., Da Costa M., Tian X., Palefsky J.M., Doudna J.A. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity (vol 71, pg 313, 2021) Science. 2021;(6531):371. doi: 10.1126/science.aar6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.J., Shi Y., Chen Y., Yang Z., Wu H., Zhou Z.H., Li J., Ping J.F., He L.P., Shen H., Chen Z.X., Wu J., Yu Y.S., Zhang Y.J., Chen H. Contamination-free visual detection of SARS-CoV-2 with CRISPR/Cas12a: a promising method in the point-of-care detection. Biosens. Bioelectron. 2020;169 doi: 10.1016/j.bios.2020.112642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X., Yin K., Li Z.Y., Lalla R.V., Ballesteros E., Sfeir M.M., Liu C.C. Ultrasensitive and visual detection of SARS-CoV-2 using all-in-one dual CRISPR-Cas12a assay. Nat. Commun. 2020;11(1) doi: 10.1038/s41467-020-18575-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gootenberg J.S., Abudayyeh O.O., Kellner M.J., Joung J., Collins J.J., Zhang F. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science. 2018;360(6387):439–+. doi: 10.1126/science.aaq0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gootenberg J.S., Abudayyeh O.O., Lee J.W., Essletzbichler P., Dy A.J., Joung J., Verdine V., Donghia N., Daringer N.M., Freije C.A., Myhrvold C., Bhattacharyya R.P., Livny J., Regev A., Koonin E.V., Hung D.T., Sabeti P.C., Collins J.J., Zhang F. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science. 2017;356(6336):438–+. doi: 10.1126/science.aam9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington L.B., Burstein D., Chen J.S., Paez-Espino D., Ma E., Witte I.P., Cofsky J.C., Kyrpides N.C., Banfield J.F., Doudna J.A. Programmed DNA destruction by miniature CRISPR-Cas14 enzymes. Science. 2018;362(6416):839–+. doi: 10.1126/science.aav4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh K.W., Zhao G.J., Wang T.H. Applying biosensor development concepts to improve preamplification-free CRISPR/Cas12a-Dx. Analyst. 2020;145(14):4880–4888. doi: 10.1039/d0an00664e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z., Tian D., Liu Y., Lin Z., Lyon C.J., Lai W.H., Fusco D., Drouin A., Yin X.M., Hu T., Ning B. Ultra-sensitive and high-throughput CRISPR-p owered COVID-19 diagnosis. Biosens. Bioelectron. 2020;164 doi: 10.1016/j.bios.2020.112316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao J., Kong K.K., Han J.M., Song S.W., Bai T.H., Song C.H., Wang M.M., Yan Z.L., Zhang H.T., Zhang R.P., Feng J.C., Zheng X.B. Field detection of multiple RNA viruses/viroids in apple using a CRISPR/Cas12a-based visual assay. Plant Biotechnol. J. 2021;19(2):394–405. doi: 10.1111/pbi.13474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanitchinda S., Srisala J., Suebsing R., Prachumwat A., Chaijarasphong T. CRISPR-Cas fluorescent cleavage assay coupled with recombinase polymerase amplification for sensitive and specific detection of Enterocytozoon hepatopenaei. Biotechnol. Rep. 2020;27 doi: 10.1016/j.btre.2020.e00485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellner M.J., Koob J.G., Gootenberg J.S., Abudayyeh O.O., Zhang F. SHERLOCK: nucleic acid detection with CRISPR nucleases (vol 41, pg 325, 2019) Nat. Protoc. 2020;15(3) doi: 10.1038/s41596-020-0302-z. 1311-1311. [DOI] [PubMed] [Google Scholar]
- Lee C.Y., Degani I., Cheong J., Lee J.H., Choi H.J., Cheon J., Lee H. Fluorescence polarization system for rapid COVID-19 diagnosis. Biosens. Bioelectron. 2021;178 doi: 10.1016/j.bios.2021.113049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Ye Q.H., Chen M.T., Zhou B.Q., Zhang J.M., Pang R., Xue L., Wang J., Zeng H.Y., Wu S., Zhang Y.X., Ding Y., Wu Q.P. An ultrasensitive CRISPR/Cas12a based electrochemical biosensor for Listeria monocytogenes detection. Biosens. Bioelectron. 2021;179 doi: 10.1016/j.bios.2021.113073. [DOI] [PubMed] [Google Scholar]
- Li L.X., Li S.Y., Wu N., Wu J.C., Wang G., Zhao G.P., Wang J. HOLMESv2: a CRISPR-Cas12b-assisted platform for nucleic acid detection and DNA methylation quantitation. ACS Synth. Biol. 2019;8(10):2228–2237. doi: 10.1021/acssynbio.9b00209. [DOI] [PubMed] [Google Scholar]
- Li S.Y., Cheng Q.X., Wang J.M., Li X.Y., Zhang Z.L., Gao S., Cao R.B., Zhao G.P., Wang J. CRISPR-Cas12a-assisted nucleic acid detection. Cell Discov. 2018;4 doi: 10.1038/s41421-018-0028-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S.H., Li F., Chen Q.B., Wu J., Duan J.Y., Lei X.L., Zhang Y., Zhao D.M., Bu Z.G., Yin H. Rapid detection of African swine fever virus using Cas12a-based portable paper diagnostics. Cell Discov. 2020;6(1) doi: 10.1038/s41421-020-0151-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukama O., Nie C.R., Habimana J.D., Meng X.G., Ting Y., Songwe F., Al Farga A., Mugisha S., Rwibasira P., Zhang Y.H., Zeng L.W. Synergetic performance of isothermal amplification techniques and lateral flow approach for nucleic acid diagnostics. Anal. Biochem. 2020;600 doi: 10.1016/j.ab.2020.113762. [DOI] [PubMed] [Google Scholar]
- Mukama O., Wu J.H., Li Z.Y., Liang Q.X., Yi Z.J., Lu X.W., Liu Y.J., Liu Y.M., Hussain M., Makafe G.G., Liu J.X., Xu N., Zeng L.W. An ultrasensitive and specific point-of-care CRISPR/Cas12 based lateral flow biosensor for the rapid detection of nucleic acids. Biosens. Bioelectron. 2020;159 doi: 10.1016/j.bios.2020.112143. [DOI] [PubMed] [Google Scholar]
- Myhrvold C., Freije C.A., Gootenberg J.S., Abudayyeh O.O., Metsky H.C., Durbin A.F., Kellner M.J., Tan A.L., Paul L.M., Parham L.A., Garcia K.F., Barnes K.G., Chak B., Mondini A., Nogueira M.L., Isern S., Michael S.F., Lorenzana I., Yozwiak N.L., MacInnis B.L., Bosch I., Gehrke L., Zhang F., Sabeti P.C. Field-deployable viral diagnostics using CRISPR-Cas13. Science. 2018;360(6387):444–448. doi: 10.1126/science.aas8836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian C., Wang R., Wu H., Zhang F., Wu J., Wang L. Uracil-Mediated new photospacer-adjacent motif of Cas12a to realize visualized DNA detection at the single-copy level free from contamination. Anal. Chem. 2019;91(17):11362–11366. doi: 10.1021/acs.analchem.9b02554. [DOI] [PubMed] [Google Scholar]
- Shen J.J., Zhou X.M., Shan Y.Y., Yue H.H., Huang R., Hu J.M., Xing D. Sensitive detection of a bacterial pathogen using allosteric probe-initiated catalysis and CRISPR-Cas13a amplification reaction. Nat. Commun. 2020;11(1) doi: 10.1038/s41467-019-14135-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieber P., Flury D., Gusewell S., Albrich W.C., Boggian K., Gardiol C., Schlegel M., Sieber R., Vernazza P., Kohler P. Characteristics of patients with Coronavirus Disease 2019 (COVID-19) and seasonal influenza at time of hospital admission: a single center comparative study. BMC Infect. Dis. 2021;21(1) doi: 10.1186/s12879-021-05957-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan T.J., Dhar A.K., Cruz-Flores R., Bodnar A.G. Rapid, CRISPR-based, field-deployable detection of white spot syndrome virus in shrimp. Sci. Rep. 2019;9 doi: 10.1038/s41598-019-56170-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B., Wang R., Wang D.Q., Wu J., Li J.X., Wang J., Liu H.H., Wang Y.M. Cas12aVDet: a CRISPR/Cas12a-based platform for rapid and visual nucleic acid detection. Anal. Chem. 2019;91(19):12156–12161. doi: 10.1021/acs.analchem.9b01526. [DOI] [PubMed] [Google Scholar]
- Wang R., Chen R., Qian C., Pang Y.N., Wu J., Li F.Y. Ultrafast visual nucleic acid detection with CRISPR/Cas12a and rapid PCR in single capillary. Sensor. Actuator. B Chem. 2021;326 [Google Scholar]
- Wang R., Qian C.Y., Pang Y.A., Li M.M., Yang Y., Ma H.J., Zhao M.Y., Qian F., Yu H., Liu Z.P., Ni T., Zheng Y., Wang Y.M. opvCRISPR: one-pot visual RT-LAMP-CRISPR platform for SARS-cov-2 detection. Biosens. Bioelectron. 2021;172 doi: 10.1016/j.bios.2020.112766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.Y., He S., Zhao N., Liu X.H., Cao Y.C., Zhang G.H., Wang G., Guo C.H. Development and clinical application of a novel CRISPR-Cas12a based assay for the detection of African swine fever virus. Bmc Microbiology. 2020;20(1) doi: 10.1186/s12866-020-01966-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.Q., Ke Y.Q., Liu W.J., Sun Y.Q., Ding X.T. A One-Pot Toolbox Based on Cas12a/crRNA Enables Rapid Foodborne Pathogen Detection at Attomolar Level. Acs Sensors. 2020;5(5):1427–1435. doi: 10.1021/acssensors.0c00320. [DOI] [PubMed] [Google Scholar]
- Wei S., Suryawanshi H., Djandji A., Kohl E., Morgan S., Hod E.A., Whittier S., Roth K., Yeh R., Alejaldre J.C., Fleck E., Ferrara S., Hercz D., Andrews D., Lee L., Hendershot K.A., Goldstein J., Suh Y., Mansukhani M., Williams Z. Field-deployable, rapid diagnostic testing of saliva for SARS-CoV-2. Sci. Rep. 2021;11(1) doi: 10.1038/s41598-021-84792-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H., Chen Y.J., Shi Y., Wang L., Zhang M.Y., Wu J., Chen H. Carrying out pseudo dual nucleic acid detection from sample to visual result in a polypropylene bag with CRISPR/Cas12a. Biosens. Bioelectron. 2021;178 doi: 10.1016/j.bios.2021.113001. [DOI] [PubMed] [Google Scholar]
- Wu H., Qian C., Wu C., Wang Z., Wang D.C., Ye Z.Z., Ping J.F., Wu J., Ji F. End-point dual specific detection of nucleic acids using CRISPR/Cas12a based portable biosensor. Biosens. Bioelectron. 2020;157 doi: 10.1016/j.bios.2020.112153. [DOI] [PubMed] [Google Scholar]
- Yin K., Ding X., Li Z.Y., Zhao H., Cooper K., Liu C.C. Dynamic aqueous multiphase reaction system for one-pot CRISPR-Cas12a-based ultrasensitive and quantitative molecular diagnosis. Anal. Chem. 2020;92(12):8561–8568. doi: 10.1021/acs.analchem.0c01459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M.Y., Liu C.Z., Shi Y., Wu J., Wu J., Chen H. Selective endpoint visualized detection of Vibrio parahaemolyticus with CRISPR/Cas12a assisted PCR using thermal cycler for on-site application. Talanta. 2020;214 doi: 10.1016/j.talanta.2020.120818. [DOI] [PubMed] [Google Scholar]
- Zhou R.X., Li Y.Y., Dong T.Y., Tang Y.A., Li F. A sequence-specific plasmonic loop-mediated isothermal amplification assay with orthogonal color readouts enabled by CRISPR Cas12a. Chem. Commun. 2020;56(24):3536–3538. doi: 10.1039/d0cc00397b. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.