Abstract
Background
Pheochromocytoma and paraganglioma (PPGL) are neuroendocrine tumors with frequent mutations in genes linked to the tricarboxylic acid cycle. However, no pathogenic variant has been found to date in succinyl-CoA ligase (SUCL), an enzyme that provides substrate for succinate dehydrogenase (SDH; mitochondrial complex II [CII]), a known tumor suppressor in PPGL.
Methods
A cohort of 352 patients with apparently sporadic PPGL underwent genetic testing using a panel of 54 genes developed at the National Institutes of Health, including the SUCLG2 subunit of SUCL. Gene deletion, succinate levels, and protein levels were assessed in tumors where possible. To confirm the possible mechanism, we used a progenitor cell line, hPheo1, derived from a human pheochromocytoma, and ablated and re-expressed SUCLG2.
Results
We describe 8 germline variants in the guanosine triphosphate–binding domain of SUCLG2 in 15 patients (15 of 352, 4.3%) with apparently sporadic PPGL. Analysis of SUCLG2-mutated tumors and SUCLG2-deficient hPheo1 cells revealed absence of SUCLG2 protein, decrease in the level of the SDHB subunit of SDH, and faulty assembly of the complex II, resulting in aberrant respiration and elevated succinate accumulation.
Conclusions
Our study suggests SUCLG2 as a novel candidate gene in the genetic landscape of PPGL. Large-scale sequencing may uncover additional cases harboring SUCLG2 variants and provide more detailed information about their prevalence and penetrance.
Pheochromocytoma and paraganglioma (PPGL) are rare neuroendocrine tumors derived from neural crest cells (1). A pioneering study of Neumann and colleagues and recent comprehensive molecular analysis of these tumors identified, to this date, germline variants in more than 20 PPGL-susceptibility genes present in up to 40% of cases (2-6). Moreover, an additional 25%-30% of PPGL patients carry a somatic mutation (3), with up to 39% in 13 genes in some cohorts (5). These and other genetic, epigenetic, and relevant mechanistic studies on PPGL have increased our understanding of their pathogenesis and clinical behavior and uncovered potential new therapeutic targets in these hard-to-treat tumors (3,7,8).
Succinyl-CoA ligase (SUCL), a tricarboxylic acid (TCA) cycle enzyme, catalyzes reversible conversion of succinyl-CoA and adenosine diphosphate (ADP) or guanosine diphosphate (GDP) to succinate and ATP or guanosine triphosphate (GTP), respectively (9). SUCL is a heterodimer comprising α subunit encoded by SUCLG1 and a β subunit encoded by either the ATP-forming SUCLA2 or the GTP-forming SUCLG2. SUCL is the only TCA cycle enzyme that can produce ATP- or GTP-linking the enzyme to mitochondrial DNA synthesis (10,11). Besides its important function in the TCA cycle, SUCL also plays a role in a number of anabolic processes, such as heme or lipid synthesis. Alterations in SUCLG1 and SUCLA2 activity because of their mutations cause encephalomyopathy presenting as hypotonia, dystonia and Leigh-like syndrome, seizure, psychomotor retardation, deafness, methylmalonic aciduria, and lactic acidosis (12-14). Kacso et al. (11) noted that the Suclg2 gene could be essential for embryogenesis in mice as Suclg2-/- mice were embryonically lethal. Yet, the role of SUCLG2 mutations in pathogenesis of human diseases has not been established.
Methods
Adrenomedullary tissue separation, isolation of mitochondria, Western blot analysis, in-gel succinate dehydrogenase (SDH) activity, succinate quinone reductase (SQR) activity, native blue gel electrophoresis, mitochondrial DNA (mtDNA) assay, respiration assay, prediction of functional impact, and a detailed description of listed methods are included in the Supplementary Methods (available online).
Patient Characteristics and Genetic Analysis
Patients were evaluated using a protocol approved by the Eunice Kennedy Shriver National Institute of Child Health and Human Development Institutional Review Board. All patients provided written informed consent approved by the National Institutes of Health ethics committee. In total, 352 patients with apparently sporadic PPGL underwent genetic testing of blood leukocyte DNA using a panel of 54 genes developed at the National Institutes of Health. Germline SUCLG2 variants were validated by Sanger sequencing. Additional data for SUCLG2 variant descriptions come from public databases including www.cbioportal.org, www.varsome.com, cancer.sanger.ac.uk/cosmic databases, and gnomad.broadinstitute.org from November 25, 2020. Seven family members of 4 patients were recruited and gave consent to analyze their saliva sample for presence of SUCLG2 variants. Personal and family history were obtained by clinicians, and the available data were evaluated by clinical staff to record each patient’s description, including evaluation of family incidence of PPGL.
Immunohistochemistry
Immunohistochemistry of SUCLG2 was performed as described in the Supplementary Methods (available online).
FISH Analysis
For interphase fluorescence in situ hybridization (FISH) analysis, bacterial artificial chromosome DNA probes RP11-146E16 and RP11-927D18 labeled with orange fluorescence were purchased from Empire Genomics (Buffalo, NY). For chromosome quantification, we used Vysis CEP 3 (alpha satellite) Spectrum Green-labeled probes (Abbott Molecular, Abbott Park, IL). FISH assays were performed on 5 µm formalin-fixed-paraffin-embedded tumor sections using standardized protocol with slight modifications (15), provided in the Supplementary Methods (available online).
Cell Lines and In Vitro Gene Manipulation
For in vitro experiments, a progenitor cell line derived from a human pheochromocytoma (hPheo1) (16) was maintained in the RPMI medium (Sigma-Aldrich) supplemented with 10% fetal bovine serum, antibiotics, and 1 mM pyruvate at 37°C under 5% CO2. Genomic deletion of SUCLG2 in hPheo1 cells was performed using the CRISPR/AsCas12a (also known as AsCpf1) system (17), as well as the chimeric Cas12a and the guide RNA-expression plasmid pX AsCpf1-Venus-NLS. crRNA sequences were selected using the Crispor software (http://crispor.tefor.net/). Oligonucleotide design, transfection, and primers design for genomic DNA polymerase chain reaction are detailed in the Supplementary Methods (available online).
For SUCLG2 re-expression, SUCLG2 DNA constructs were subcloned into the pCDH-CMV-MCS-EF1-Puro vector (CD510B-1; System Biosciences, Palo Alto, CA). Recombinant lentiviral particles were obtained from calcium phosphate-transfected HEK 293T cells using the packaging plasmid psPAX2 (12260; Addgene, Watertown, MA) and pMD2.G (12259; Addgene) and the SUCLG2 pCDH construct. Target cells were transduced at the multiplicity of infection of 5 to 10 and selected by puromycin (2 μg/mL; InvivoGen, San Diego, CA). Detailed protocol and information can be found in the Supplementary Methods (available online).
Succinate-to-Fumarate Ratio Evaluation
Cell extracts were prepared and processed as detailed in the Supplementary Methods (available online). Silylated extracts were analyzed using 2-dimensional gas chromatography with mass spectrometric detection (GC × GC-MS; Pegasus 4D; LECO Corp, St Joseph, MO) with more specific details in Supplementary Methods (available online). We used ChromaTOF software (v.4.51; LECO Corporation) for instrument control, data acquisition, and data processing. Succinic and fumaric acids were detected as tert-butyl silyl derivatives, with their identities confirmed by co-elution with standards. Analytes were quantified using masses of m/z 289 (succinic acid) and m/z 287 (fumaric acid).
Metabolomics of Patient Tissues
Quantification of organic acids was performed by liquid chromatography tandem mass spectrometry on a Thermo Quantiva instrument with Dionex 3000 high-performance liquid chromatography system (Thermo Fisher Scientific, Waltham, MA) using isotope dilution. Briefly, 40 µl of the tissue homogenate (100 mg/mL) was transferred to a microcentrifuge tube to which 10 µl of internal standard mixture was added. Derivatization was performed by adding 50 µl of 0.4 M o-benzylhydroxylamine (methanol/200 nM ammonium formate; 1:1) and 10 µl of N-(3-dimethylaminopropyl)-N'-ethylcarbodiimide hydrochloride in water to each tube. The samples were incubated for 10 minutes, followed by the addition of 100 µl water to each tube and 600 µl of ethyl acetate. The tubes were mixed and centrifuged at 18 000 g for 5 minutes, after which 100 µl of the ethyl acetate layer was transferred and dried under nitrogen. Each sample was reconstituted in 1 mL of a 1:1 methanol: water mixture for analysis. The standard operating procedure is available from the metabolomics workbench (www.metabolomicsworkbench.org).
Statistical Analysis
Data are expressed as the mean (SD) of at least 3 independent experiments. Data were evaluated by 1-way analysis of variance with multiple comparison post hoc Tukey correction in GraphPad Prism (v.7.03; GraphPad Software, La Jolla, CA) and a Mann-Whitney nonparametric test for patient metabolomic data. All statistical tests were 2-sided. Data were considered statistically significant at a P value less than .05.
Results
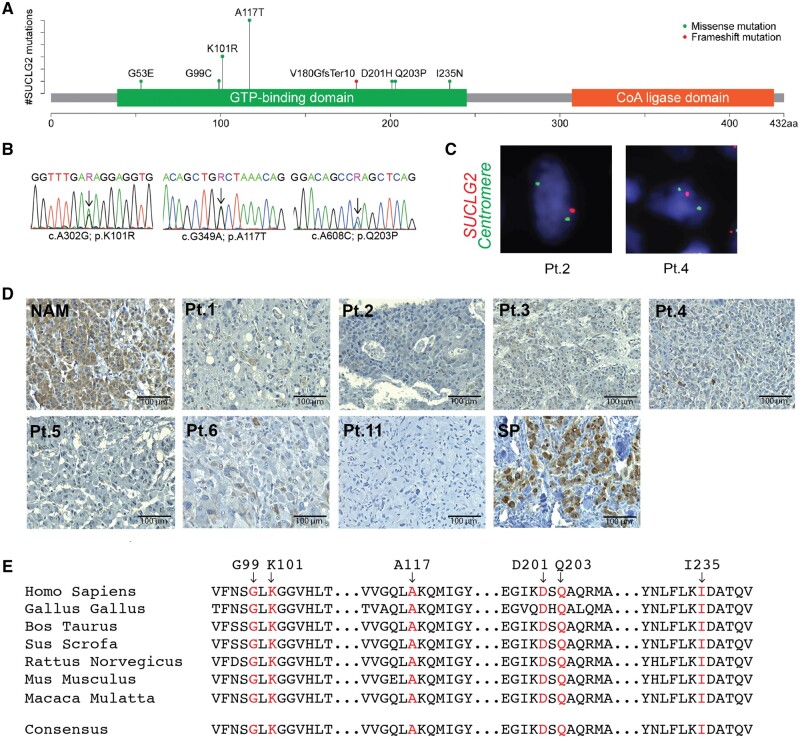
We identified 15 individuals with germline SUCLG2 variants in our cohort of 352 patients (15 of 352, 4.3%) with apparently sporadic PPGL (Figure 1, A and B; Table 1). Seven men and 8 women were found to carry 7 missense SUCLG2 variants (c.704T>A in 1 patient; c.302A>G in 3 patients; c.349G>A in 6 patients; c.296G>T in 1 patient; c.608A>C in 1 patient; c.158G>A in 1 patient; c.601G>C in 1 patient) and 1 deletion (c.539_540delTG) within the GTP-binding pocket of the protein (Figure 1, A;Supplementary Figure 1, available online). Nine patients presented with pheochromocytoma (PHEO), 5 with paraganglioma (PGL), and 1 with primary tumor of unknown origin. Seven patients developed metastatic disease. Most patients presented with the noradrenergic biochemical phenotype. However, 4 patients with PHEO presented with both elevated plasma metanephrine and normetanephrine levels (Table 1).
Figure 1.
Detection of SUCLG2 variants in PPGL patients. A) Schematic illustration of SUCLG2 germline variants in the GTP-binding site. B) Sanger sequencing showing SUCLG2 germline variants in patient leukocyte DNA. C) FISH assay showing apparent heterozygous deletion of SUCLG2 in tumor specimens. D) Immunohistochemistry analysis revealing a substantial decrease in SUCLG2 level in PPGL with SUCLG2 germline variants (patient number refers to Table 1) relative to that in normal adrenal medulla (NAM) and sporadic PPGL (SP). E) Sequence alignment of SUCLG2 residues 85 to 240 (human SUCLG2 nomenclature) showing conservation of the mutated residues. FISH = fluorescence in situ hybridization; GTP = guanosine triphosphate; PPGL = pheochromocytoma and paraganglioma;.
Table 1.
Clinical characteristics of patients
| ID | Sex | Age at initial diagnosis, y | Mutation in SUCLG2 | Protein change | mtDNA depletion | Biochemistry at initial diagnosisa | Initial tumor type | Primary tumor size, cm | Metastatic | Recurrent |
|---|---|---|---|---|---|---|---|---|---|---|
| 1b | Male | 22 | c.704T>A | I235N | Yes | NE, NMN, CgA | sPGL | 3.5 | No | No |
| 2 | Male | 43 | c.302A>G | K101R | Yes | NE, NMN, MN, CgA | PHEO | 3.8 | No | No |
| 3 | Female | 45 | c.302A>G | K101R | Yes | NE, NMN | PHEO | 4.3 | No | No |
| 4b | Male | 25 | c.349G>A | A117T | Yes | NEc, NMNc | PHEO | 7.5 | No | Yes |
| 5 | Male | 51 | c.295G>T | G99C | Yes | NEc, NMNc | PHEO | 6.0 | Yes | Yes |
| 6 | Female | 41 | c.349G>A | A117T | Yes | NE, NMN, EPI, MN | PHEO | 5.8 | No | No |
| 7 | Female | 10 | c.349G>A | A117T | NA | NMN | PHEO | 4.1 | No | No |
| 8b | Female | 29 | c.349G>A | A117T | NA | CgAc | pPGL | NA | Yes | No |
| 9 | Male | 32 | c.608A>C | Q203P | NA | EPI, NE | PHEO | NA | No | No |
| 10 | Female | 39 | c.302A>G | K101R | NA | NE, DA | PGL | 4.6 | Yes | No |
| 11 | Female | 38 | c.539_540delTG | Val180fs | Yes | NMN, MN | PHEO | 6.5 | Yes | Yes |
| 12 | Male | 32 | c.158G>A | G53E | NA | NE, NMN, CgA | PGL | 7.0 | Yes | No |
| 13 | Female | 70 | c.349G>A | A117T | NA | NE, NMN | PGL | 4.5 | No | No |
| 14b | Male | 56 | c.349G>A | A117T | NA | MTX, CgA | UNP | NA | Yes | No |
| 15b | Female | 25 | c.601G>C | D201H | NA | NEc, NMNc, EPIc, MNc, DAc | PHEO | 12.4 | Yes | No |
Increased analytes displayed. For primary tumor size, only the largest tumor diameter is shown. CgA = chromogranin A; DA: dopamine; EPI = epinephrine; MN = metanephrine measured either in plasma or urine; MTX = methoxytyramine; NA = not available; NE = norepinephrine; NMN = normetanephrine; PHEO = pheochromocytoma; PGL = paraganglioma; pPGL = parasympathetic PGL; sPGL = sympathetic paraganglioma; UNP = unknown primary.
Indicates death of the patient.
Biochemistry done at the National Institutes of Health, at the time of initial diagnosis not done or no data available.
In PPGL-related gene panel testing, we did not identify any known pathogenic variants in SUCLG2-mutated patients (Supplementary Table 1, available online). No patient reported any family history of PPGL. We collected saliva samples from 7 first-degree relatives of 4 patients (Table 2). For 3 patients—patients 2 (c.302A>G), 10 (c.302A>G), and 12 (c.158G>A)—1 of the parents was a carrier of a particular variant. In one case, a sample from a sibling was collected and presented with the same variant. For patient 11 (c.539_540delTG), only 1 parental DNA sample was available and was negative for the variant (Table 2).
Table 2.
Detection of SUCLG2 variants in patients’ relativesa
| ID sample | SUCLG2 variant | Sample | Variant status | Cancer history |
|---|---|---|---|---|
| 2 | c.302A>G | Paternal | Present | NR |
| Maternal | ND | NR | ||
| Sibling | Present | Pancreatic neuroendocrine tumor | ||
| 10 | c.302A>G | Paternal | ND | Lung |
| Maternal | Present | Breast | ||
| 11 | c.539_540delTG | Paternal | NA | Prostate, colon, lung |
| Maternal | Not detected | NR | ||
| 12 | c.158G>A | Paternal | NA | Suspected liver |
| Maternal | Present | NR |
Available saliva samples from first-degree relatives were tested for presence of particular SUCLG2 variants by Sanger Sequencing. NA = sample not available; ND = not detected; NR = not reported.
SUCLG2 variant sites were located within highly evolutionarily conserved positions (Figure 1, E). Consistently, GERP++ (Genomic Evolutionary Rate Profiling) returned scores above 5 (Table 3). Five missense variants (c.704T>A, c.295G>T, c.608A>C, c.158G>A, c.601G>C) were not listed in ClinVar and occurred at a very low frequency (<10-3) with no homozygotes in the general population (Genome Aggregation Database). The combination of protein function prediction tools led to inconclusive results for functional outcomes of variant c.704T>A and c.601G>C and damaging functional outcomes for variants c.295G>T, c.608A>C, and c.158G>A (Table 3). The deletion variant c.539_540delTG could not be classified by the used prediction tools. Variants c.302A>G and c.349G>A were listed in ClinVar database for germline variants, both with benign and likely benign prediction, with frequency in the general population of 0.0047 and 0.0076, respectively. Evaluation from protein function prediction tools led to inconclusive outcomes (Table 3). The variant c.349G>A was recorded in the Catalogue Of Somatic Mutations In Cancer database in 3 cases of lung cancer and 2 cases of upper aerodigestive tract cancer. Overall, prediction of functional impact was rather inconclusive (Table 3). Thus, we performed functional studies in the available material (Table 3; Figures 1, C and D, and 2, A and B; Supplementary Table 2, available online).
Table 3.
SUCLG2 variants (chr.3, NM_003848.4), in silico prediction and summary of functional outcomesa
| SUCLG2 variants | ID sample |
|||||||
|---|---|---|---|---|---|---|---|---|
| 1 | 2, 3, 10 | 4, 6-8, 13, 14 | 5 | 9 | 11 | 12 | 15 | |
| Nucleotide change | c.704T>A | c.302A>G | c.349G>A | c.295G>T | c.608A>C | c.539_540delTG | c.158G>A | c.601G>C |
| Amino acid change | I235N | K101R | A117T | G99C | Q203P | Val180GlyfsTer10 | G53E | D201H |
| Exon | 7 | 3 | 4 | 3 | 6 | 5 | 2 | 6 |
| Position in Hg19 | 67 559 284 | 67 579 535 | 67 578 624 | 67 579 542 | 67 568 723 | 67 570 936—7 | 67 659 947 | 67 568 730 |
| gnomAD allele frequency (No. of homozygotes) | 0.000004029 = 1/248 218 (0) | 0.004736 = 1326/279 992 (7) | 0.007629 = 2141/280 634 (57) | 0.0001643 = 46/280 026 (0) | 0.0001465 = 41/279 854 (0) | 0.00001781 = 5/280 762 (0) | not described | 0.000004035 = 1/247 828 (0) |
| ClinVar ID | — | 257 601 benign | 774 168 benign/likely benign | — | — | — | — | — |
| dbSNP | rs1433609554 | rs74675534 | rs13318110 | rs200916187 | rs201652728 | — | — | rs902797961 |
| Protein function prediction | ||||||||
| Mutation assessor (score) | Neutral (−0.66) | Medium (2.36) | Medium (2.04) | High (3.75) | High (3.91) | — | Medium (2.97) | Medium (2.13) |
| SIFT | Tolerated | Damaging | Tolerated | Damaging | Tolerated/damaging | — | Damaging | Tolerated/damaging |
| Polyphen-2 (score) | Probably damaging (0.51) | Benign (0.19) | Benign (0.03) | Probably damaging (1) | Probably damaging (0.98) | — | Probably damaging (0.94) | Benign (0.06) |
| Functional impact (damaging/benign)b | 5/7 | 6/5c | 6/5c | 11/1 | 9/3 | — | 11/1 | 8/4 |
| Evolutionary conservation | ||||||||
| GERP++ (score) | Conserved (5.8) | Conserved (5.01) | Conserved (5.95) | Conserved (5.01) | Conserved (5.87) | Conserved (5.7; 5.09) | Conserved (5.78) | Conserved (5.87) |
| Functional outcomes | ||||||||
| IHC SUCLG2 | Negative | #2; 3 negative | #4; 6 negative | Negative | — | Negative | — | — |
| Western blotd | Decrease | #2 decrease | #4 decrease | — | — | — | — | — |
| FISH SUCLG2 | — | #2 deletion (78% cells) | #4 deletion (44% cells) | — | — | Deletion (48% cells) | — | — |
| SDH activity | Decrease | #2 decrease | #4 decrease | — | — | — | — | — |
| Succ/Fum ratio | Increase | #2; 3 increase | #4; 6 increase | Increase | — | — | — | — |
Data come from www.cbioportal.org, www.varsome.com, and gnomad.broadinstitute.org. Functional studies data are summarized from experiments and results reported in Figures 1 and 2 and Supplementary Table 2 (available online). FISH = fluorescence in situ hybridization; Fum = fumarate; gnomad = Genome Aggregation Database; IHC = immunohistochemistry; SDH = succinate dehydrogenase; Succ = succinate.
Includes in silico predictive programs available on www.varsome.com used in the American College of Medical Genetics and Genomics (ACMG) classification—BayesDel_addAF, DEOGEN2, EIGEN, MVP, MutationAssesor, PrimateAI, SIFT, DANN, FATHMM-MKL, LIST-S2, M-Cap, MutationTaster.
M-Cap not available.
Western blot of SUCLG2 in mitochondria extract from tumor tissue.
FISH detected gene deletion in available tumor tissues (Figure 1, C), that is, archival material from 3 patients: patients 2 (c.302A>G), 4 (c.349G>A), and 11 (c.539_540delTG). All 3 patients demonstrated loss of 1 copy of the SUCLG2 gene. The SUCLG2 gene deletion was detected in 77.2%, 47.8%, and 44.2% of cells in c.302A>G (patient 2), c.349G>A (patient 4), and c.539_540delTG (patient 11) tumors, respectively. Loss of the entire chromosome 3 copy was the most common pattern in the tumor cell population, although a hemizygous deletion of the SUCLG2 gene with the retention of 2 chromosome 3 centromeres was observed in a subpopulation of the tumor cells as well. Furthermore, immunohistochemistry revealed a substantial decrease in the SUCLG2 protein level in tumor tissue from patients 1-6 and 11 listed in Table 1 (Figure 1, D).
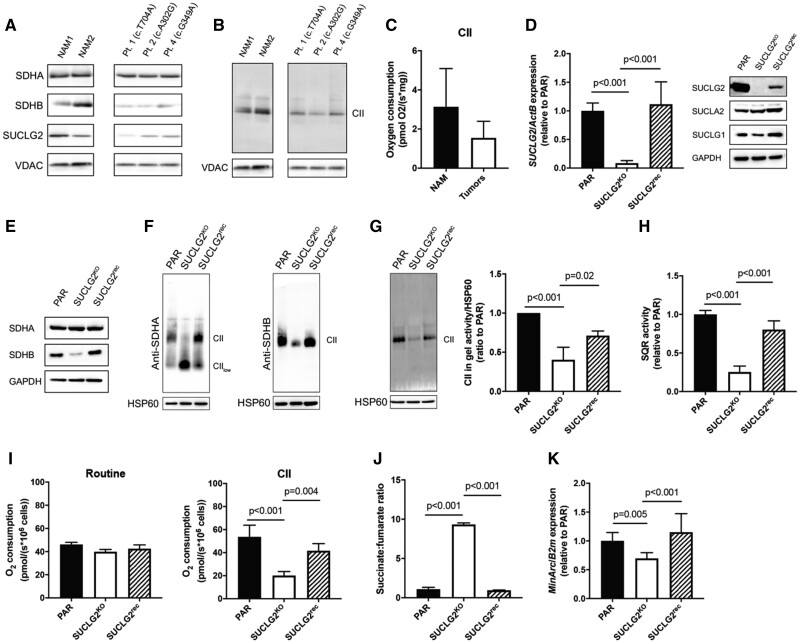
Metabolomic analysis of tumor biopsies from 6 patients with SUCLG2 variants (patients 1-6 in Table 1) showed increased succinate-to-fumarate ratio relative to normal adrenal medulla (mean [SD] = 12.25 [5.46] vs 2.07 [1.89]; P = .02), a shift to glycolytic metabolism, and dysregulation of the TCA cycle (Supplementary Table 2, available online), pointing to dysfunction of succinate dehydrogenase (SDH). Western blot analysis of tumor but not normal adrenal medulla mitochondria showed a substantially decreased level of the SDH subunit B (SDHB) protein and no effect on the SDH subunit A (SDHA) protein (Figure 2, A). Consistently , decreased SDH in-gel activity and mitochondrial complex II (CII)–dependent respiration were detected in tumor mitochondria (Figure 2, B and C), suggesting that SUCLG2 defects may compromise SDH.
Figure 2.
Mitochondrial complex II functions in SUCLG2-mutated tumors and SUCLG2-silented hPheo1 cell line. A) Western blot analysis of SDHB, SDHA, and SUCLG2; B) SDH in-gel activity; and C) CII-dependent respiration in mitochondrial lysate of normal adrenal medulla (NAM) and PPGL patients with mutant SUCLG2. Pt 1, 2, and 4 refers to patients 1, 2, and 4 in Table 1. Data are presented as mean [SD] and evaluated by a 2-sided Student t test. D) SUCLG2 mRNA (left) and SUCLG2, SUCLA2, and SUCLG1 protein levels (right) in parental (PAR), SUCLG2KO, and SUCLG2rec cells. E) SDHA and SDHB protein levels in PAR, SUCLG2KO, and SUCLG2rec cells. F) Native blue gels showing the assembly of mitochondrial CII in PAR, SUCLG2KO, and SUCLG2rec cells using antibodies against SDHA and SDHB. G) In-gel SDH activity in PAR, SUCLG2KO, and SUCLG2rec cells. H) SQR activity in PAR, SUCLG2KO, and SUCLG2rec cells. I) Routine (left) and CII-dependent (right) respiration in PAR, SUCLG2KO, and SUCLG2rec cells. J) Succinate-to-fumarate ratio in PAR, SUCLG2KO, and SUCLG2rec cells. K) Levels of mtDNA in PAR, SUCLG2KO, and SUCLG2rec cells. All in vitro experiments were performed at least 3 times independently. Data are presented as mean (SD) and evaluated by 1-way analysis of variance with post hoc Tukey test. All statistical tests were 2-sided. PAR = parental hPheo1 cells; NAM = normal adrenal medulla; CII = mitochondrial complex II.
To investigate the impact of SUCLG2 variants in more detail, we established SUCLG2-knockout (SUCLG2KO) hPheo1 cells in which we subsequently re-expressed the protein to prepare SUCLG2rec cells. Western blot revealed that absence of the SUCLG2 protein had little effect on SUCLG1 and no effect on SUCLA2 levels (Figure 2, D). Assessment of CII subunits indicated unchanged SDHA but reduced SDHB protein levels in SUCLG2KO cells relative to parental cells, which was recovered by SUCLG2 re-expression (Figure 2, E). Although there was some variability in the extent of SDHB downregulation in individual SUCLG2KO clones (data not shown), reversal of the phenotype by cDNA confirmed the specificity. Native blue gel electrophoresis revealed impaired CII assembly in SUCLG2KO cells, which was present as the CIIlow subassembly, consistent with decreased SDHB (18), and fully assembled CII was observed in parental and SUCLG2rec cells (Figure 2, F).
These data indicate a disruption of CII in SUCLG2-deficient cells, possibly affecting mitochondrial function. Therefore, we assessed CII activity in SUCLG2KO and SUCLG2rec cells, comprising SDH-dependent conversion of succinate to fumarate in the TCA cycle and SQR-mediated transfer of electrons from succinate to ubiquinone as part of OXPHOS. We found that both SDH and SQR activities were suppressed in SUCLG2KO cells to 40.3% (SD = 16.1, P < .001) and 25.3% (SD = 7.8, P < .001), respectively, and recovered in SUCLG2rec cells to parental cell values (Figure 2, G and H). Consistently, CII-dependent respiration was depressed in SUCLG2KO cells compared with parental cells (mean [SD] = 20.1 [3.5] pmol/(s*106 cells) vs 53.7 [10.2] pmol/(s*106 cells); P < .001) and restored in SUCLG2rec cells (Figure 2, I). Moreover, the succinate-to-fumarate ratio was increased by up to 10-fold in SUCLG2KO cells relative to that in control (mean [SD] = 9.3 [0.2] vs 1.1 [0.2]; P < .001) and SUCLG2rec cells (mean [SD] = 0.95 [0.04]; P < .001) (Figure 2, J), recapitulating the increase succinate-to-fumarate ratio in patient tumors (Supplementary Table 2, available online).
Because SUCLG1 and SUCLA2 germline variants are linked to the mtDNA-depletion syndrome (19), we evaluated mtDNA levels in patients with SUCLG2 germline variants and in SUCLG2KO cells. mtDNA was significantly reduced in tumor tissues (Table 1) and cells where it recovered on re-expression of SUCLG2 (Figure 2, K), suggesting that the integrity of SUCL protein plays a role in mtDNA maintenance. However, no patient from our cohort had distinctive symptoms of mitochondrial disorders seen in SUCLG1 and SUCLA2 variants.
Discussion
With respect to metabolic diseases, SUCL has a unique position in the TCA cycle for 2 major reasons. First, its products ATP or GTP connect the enzyme to mtDNA synthesis and maintenance via mitochondrial nucleoside diphosphate kinase (10,11), which links SUCL to inborn mitochondrial pathologies. Second, the other product of the enzyme, succinate, is an oncometabolite (20) and a substrate for CII that is linked to PPGL. Despite such close proximity, mutations in SUCL subunits have not been reported in cancer until now.
We evaluated 352 apparently sporadic PPGL patients and found 15 (4.3%) patients with 8 germline variants located within the GTP-binding domain of SUCLG2 (Table 1). Because GTP activity was proposed to be important for phosphorylation of active-site histidine residues to trigger the movement of the carboxylate of succinate into position to be phosphorylated in porcine SUCLG2 (21), we further analyzed these variants. Albeit, functional impact prediction for variant c.704T>A was inconclusive, and the tumor was negative for SUCLG2, showed decreased protein level in the mitochondrial extract, functional changes in SDH activity, and succinate accumulation, suggesting a damaging effect of the variant. Two most represented variants c.302A>G and c.349G>A in our cohort were evaluated in ClinVar as benign or likely benign; however, detailed information on the cohort and its specificity was not available to us, and the functional impact on protein function was inconclusive. Yet, tumors with these variants presented with negative immunohistochemistry staining for SUCLG2, protein level decrease in mitochondrial extract, gene deletion, altered SDH function, and TCA cycle metabolite disbalance, suggesting a damaging effect of the variants. Damaging functional impact from predictions was suggested for the variant c.295G>T that was reflected by negative SUCLG2 staining and succinate accumulation in available tumor sample. The deletion variant c.539_540delTG could not be classified by the used prediction tools, but we reported the gene deletion by the FISH assay and negative immunohistochemistry staining suggesting damaging impact of the variant. Variants c.608A>C, c.158G>A, and c.601G>C were predicted to have damaging impact on protein function, but no tissues for analysis were available to confirm this notion.
Our study revealed that SUCLG2 germline variants impacted TCA cycle function, resulting in increased succinate levels associated with decreased SDH activity in PPGL, suggesting a possible mechanism for tumor development. SDH links the TCA cycle directly to OXPHOS (22), and genes encoding SDH subunits are frequently mutated in PPGL (23). SDHx mutations result in accumulation of succinate, which acts as an oncometabolite that contributes to prolyl hydroxylases suppression associated with stabilization of hypoxia-inducible factors, DNA hypermethylation, and histone modifications, all presumably favoring tumorigenicity (7,20,24,25). Interestingly, in our cohort, the malignancy rate for SUCLG2-mutated PPGL (7 of 15 patients, 46.7%, in time of analysis) was similar to SDHx-mutated PPGLs (26-28), and the patients presented mostly with the noradrenergic biochemical phenotype that is generally connected to pseudohypoxia in PPGLs (TCA cycle enzymes and pseudohypoxia signaling mutations) (29,30). Importantly, SUCLG2 manipulation in hPheo1 cells confirmed a link between the SUCLG2 mutations and SDH function. Ablation of SUCLG2 affected the level of the SDHB protein and CII assembly, and consequently SDH functions, resulting in succinate accumulation. These changes mostly recovered by re-expression of SUCLG2 in the knock-out cells, pointing to specific effects of SUCLG2 deletion. We did not observe mtDNA-related disease symptoms often reported in patients with SUCLG1 and SUCLA2 variants in our cohort of SUCLG2 mutant patients even though mtDNA levels were decreased in the experimental cell model and patient tumor tissue, consistent with previous research on the link between SUCL and mtDNA maintenance (11).
Patients described here presented without any family history of PPGL, which is supportive of the low disease penetrance that was previously reported for another TCA cycle gene SDHA (31,32). We were able to recruit family members of 4 patients; however, for 2 of these patients, we had only 1 parental sample. We found parental carriers of variants c.302A>G and c.158G>A, and although various cancers were reported in these families, none of them were of PPGL etiology. Interestingly, we found that the sibling of patient 2, who is also a carrier of c.302A>G variant presented with pancreatic neuroendocrine tumor. Even though only a limited number of relatives were available, SUCLG2 variants seem to have incomplete penetrance that was repeatedly recorded for novel PPGL-related TCA genes like fumarate hydratase (33) or malate dehydrogenase (34).
From a pathophysiological viewpoint, it is possible that patients with SUCLG2 germline variants consequently develop CII dysfunction leading to PPGL formation. Previously, Rapizzi et al. (35) described impairment of SDH activity in non-SDHx–mutated PPGLs, possibly because of germline variants in other known (ie, MAX) or unknown susceptibility genes. It remains to be shown whether SUCLG2 mutations consistently lead to CII dysfunction and what determines the severity of the effect. Therefore, further studies to reveal the molecular mechanism associated with the link between SUCLG2 germline variants and CII function are needed and are subjects of our ongoing research.
There are several limitations of our study. First, we did not have a chance to collect and sequence DNA of all first-degree family members to fully confirm the prevalence and penetrance of the reported SUCLG2 germline variants. Second, we did not have access to or availability of all patient tumor samples to validate all parameters in each sample. We believe that information from future PPGL validation cohorts will help complete the picture and establish the clinical significance of SUCLG2 variants in PPGLs.
In summary, our analysis of SUCLG2-mutated tumors and in vitro experiments in SUCLG2-deficient cells identified association and mechanism between SUCLG2 variants and PPGL, and we propose SUCLG2 as a PPGL-associated candidate gene, which should be further evaluated to establish clinical significance in the genetic landscape of apparently sporadic PPGL cases. Future studies involving large-scale sequencing may discover additional cases harboring SUCLG2 variants and provide more detailed information about their prevalence and penetrance. These findings will further clarify the relationship between SUCLG2 and SDHx proteins, particularly SDHB, as well as their role in the disease etiology.
Funding
This work was supported by the Intramural Research Program of the National Institutes of Health: Eunice Kennedy Shriver National Institute of Child Health and Human Development and National Cancer Institute; and in part by the Czech Science Foundation (18-10832S, 19-20553S, 20-11724Y, and 20-05942S), the Czech Health Foundation (17-30138A), and the Australian Research Council. The project was also supported in part by the Program of Ministry of Education Youth and Sports of the Czech Republic (MEYS) grant LM2018130.
Notes
Role of the funders: The funders were not involved in the design, conduct, or reporting of the study as well as the writing of the manuscript.
Disclosures: The authors declare no competing interests.
Author contributions: Conceptualization and design: KHV, YP, JR, JN, CY, and KP. Methodology, investigation, data curation, and formal analysis: KHV, YP, LK, MK, ZN, SB, SDP, RZ, JZ, T-TH, IJ, OU, SH, SD, TJG, HKG, JC, and XW. Visualization: KHV, YP, and CY. Resources: KP, PEK, ZF, IH, BS, HKG, DT, and NN. Supervision and project administration: KHV, JN, CY, and KP. Funding acquisition: JN, CY, SB, JR, BS, KP. Writing—original draft: KHV, YP, JR, JN, CY, and KP. All authors reviewed and edited the original draft
Acknowledgments: We would like to acknowledge Dr Lukas Lacina for preparation of normal adrenal medulla (equipment of Center for Tumor Ecology, reg. no. CZ.02.1.01/0.0/0.0/16_019/0000785), Dr Yanqin Yang for help to analyze the gene sequencing data, Nidhi Kapoor for help with metabolomics, and Leah Meuter for tireless collection and evaluation of the patient data.
Data Availability
The authors confirm that the data supporting the findings of this study are available within the article and its Supplementary Material (available online). Data from consented patients are available in dbGaP with accession number phs002405.v1.p1.
Supplementary Material
References
- 1. Lenders JW, Eisenhofer G, Mannelli M, et al. Phaeochromocytoma. Lancet. 2005;366(9486):665–675. [DOI] [PubMed] [Google Scholar]
- 2. Neumann HP, Bausch B, McWhinney SR, et al. ; for the Freiburg-Warsaw-Columbus Pheochromocytoma Study Group. Germ-line mutations in nonsyndromic pheochromocytoma. N Engl J Med. 2002;346(19):1459–1466. [DOI] [PubMed] [Google Scholar]
- 3. Dahia PL. Pheochromocytoma and paraganglioma pathogenesis: learning from genetic heterogeneity. Nat Rev Cancer. 2014;14(2):108–119. [DOI] [PubMed] [Google Scholar]
- 4. Favier J, Amar L, Gimenez-Roqueplo AP.. Paraganglioma and phaeochromocytoma: from genetics to personalized medicine. Nat Rev Endocrinol. 2015;11(2):101–111. [DOI] [PubMed] [Google Scholar]
- 5. Fishbein L, Leshchiner I, Walter V, et al. ; for the Cancer Genome Atlas Research Network. Comprehensive molecular characterization of pheochromocytoma and paraganglioma. Cancer Cell. 2017;31(2):181–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Crona J, Lamarca A, Ghosal S, et al. Genotype-phenotype correlations in pheochromocytoma and paraganglioma: a systematic review and individual patient meta-analysis. Endocr Relat Cancer. 2019;26(5):539–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Letouze E, Martinelli C, Loriot C, et al. SDH mutations establish a hypermethylator phenotype in paraganglioma. Cancer Cell. 2013;23(6):739–752. [DOI] [PubMed] [Google Scholar]
- 8. de Cubas AA, Korpershoek E, Inglada-Perez L, et al. DNA methylation profiling in pheochromocytoma and paraganglioma reveals diagnostic and prognostic markers. Clin Cancer Res. 2015;21(13):3020–3030. [DOI] [PubMed] [Google Scholar]
- 9. Johnson JD, Mehus JG, Tews K, et al. Genetic evidence for the expression of ATP- and GTP-specific succinyl-CoA synthetases in multicellular eucaryotes. J Biol Chem. 1998;273(42):27580–27586. [DOI] [PubMed] [Google Scholar]
- 10. Miller C, Wang L, Ostergaard E, et al. The interplay between SUCLA2, SUCLG2, and mitochondrial DNA depletion. Biochim Biophys Acta. 2011;1812(5):625–629. [DOI] [PubMed] [Google Scholar]
- 11. Kacso G, Ravasz D, Doczi J, et al. Two transgenic mouse models for β-subunit components of succinate-CoA ligase yielding pleiotropic metabolic alterations. Biochem J. 2016;473(20):3463–3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Elpeleg O, Miller C, Hershkovitz E, et al. Deficiency of the ADP-forming succinyl-CoA synthase activity is associated with encephalomyopathy and mitochondrial DNA depletion. Am J Hum Genet. 2005;76(6):1081–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ostergaard E. Disorders caused by deficiency of succinate-CoA ligase. J Inherit Metab Dis. 2008;31(2):226–229. [DOI] [PubMed] [Google Scholar]
- 14. Maas RR, Marina AD, de Brouwer AP, et al. SUCLA2 deficiency: a deafness-dystonia syndrome with distinctive metabolic findings (report of a new patient and review of the literature). JIMD Rep. 2016;27:27–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pack SD, Zhuang Z.. Fluorescence in situ hybridization: application in cancer research and clinical diagnostics. Methods Mol Med. 2001;50:35–50. [DOI] [PubMed] [Google Scholar]
- 16. Ghayee HK, Bhagwandin VJ, Stastny V, et al. Progenitor cell line (hPheo1) derived from a human pheochromocytoma tumor. PLoS One. 2013;8(6):e65624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zetsche B, Heidenreich M, Mohanraju P, et al. Multiplex gene editing by CRISPR-Cpf1 using a single crRNA array. Nat Biotechnol. 2017;35(1):31–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bezawork-Geleta A, Wen H, Dong L, et al. Alternative assembly of respiratory complex II connects energy stress to metabolic checkpoints. Nat Commun. 2018;9(1):2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Carrozzo R, Verrigni D, Rasmussen M, et al. Succinate-CoA ligase deficiency due to mutations in SUCLA2 and SUCLG1: phenotype and genotype correlations in 71 patients. J Inherit Metab Dis. 2016;39(2):243–252. [DOI] [PubMed] [Google Scholar]
- 20. Selak MA, Armour SM, MacKenzie ED, et al. Succinate links TCA cycle dysfunction to oncogenesis by inhibiting HIF-alpha prolyl hydroxylase. Cancer Cell. 2005;7(1):77–85. [DOI] [PubMed] [Google Scholar]
- 21. Huang J, Fraser ME.. Structural basis for the binding of succinate to succinyl-CoA synthetase. Acta Crystallogr D Struct Biol. 2016;72(pt 8):912–921. [DOI] [PubMed] [Google Scholar]
- 22. Bezawork-Geleta A, Rohlena J, Dong L, et al. Mitochondrial complex II: at the crossroads. Trends Biochem Sci. 2017;42(4):312–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Amar L, Baudin E, Burnichon N, et al. Succinate dehydrogenase B gene mutations predict survival in patients with malignant pheochromocytomas or paragangliomas. J Clin Endocrinol Metab. 2007;92(10):3822–3828. [DOI] [PubMed] [Google Scholar]
- 24. Favier J, Brière J-J, Burnichon N, et al. The Warburg effect is genetically determined in inherited pheochromocytomas. PLoS One. 2009;4(9):e7094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hescot S, Curras-Freixes M, Deutschbein T, et al. ; for the European Network for the Study of Adrenal Tumors (ENS@T). Prognosis of Malignant Pheochromocytoma and Paraganglioma (MAPP-Prono Study): A European network for the study of adrenal tumors retrospective study. J Clin Endocrinol Metab. 2019;104(6):2367–2374. [DOI] [PubMed] [Google Scholar]
- 26. Amar L, Bertherat J, Baudin E, et al. Genetic testing in pheochromocytoma or functional paraganglioma. J Clin Oncol. 2005;23(34):8812–8818. [DOI] [PubMed] [Google Scholar]
- 27. Benn DE, Gimenez-Roqueplo A-P, Reilly JR, et al. Clinical presentation and penetrance of pheochromocytoma/paraganglioma syndromes. J Clin Endocrinol Metab. 2006;91(3):827–836. [DOI] [PubMed] [Google Scholar]
- 28. Brouwers FM, Eisenhofer G, Tao JJ, et al. High frequency of SDHB germline mutations in patients with malignant catecholamine-producing paragangliomas: implications for genetic testing. J Clin Endocrinol Metab. 2006;91(11):4505–4509. [DOI] [PubMed] [Google Scholar]
- 29. Kantorovich V, Pacak K.. Pheochromocytoma and paraganglioma. Prog Brain Res. 2010;182:343–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fliedner SMJ, Brabant G, Lehnert H.. Pheochromocytoma and paraganglioma: genotype versus anatomic location as determinants of tumor phenotype. Cell Tissue Res. 2018;372(2):347–365. [DOI] [PubMed] [Google Scholar]
- 31. Maniam P, Zhou K, Lonergan M, et al. Pathogenicity and penetrance of germline SDHA variants in pheochromocytoma and paraganglioma (PPGL). J Endocr Soc. 2018;2(7):806–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. van der Tuin K, Mensenkamp AR, Tops CMJ, et al. Clinical aspects of SDHA-related pheochromocytoma and paraganglioma: a nationwide study. J Clin Endocrinol Metab. 2018;103(2):438–445. [DOI] [PubMed] [Google Scholar]
- 33. Clark GR, Sciacovelli M, Gaude E, et al. Germline FH mutations presenting with pheochromocytoma. J Clin Endocrinol Metab. 2014;99(10):E2046–E2050. [DOI] [PubMed] [Google Scholar]
- 34. Cascón A, Comino-Méndez I, Currás-Freixes M, et al. Whole-exome sequencing identifies MDH2 as a new familial paraganglioma gene. J Natl Cancer Inst. 2015;107(5):djv053. [DOI] [PubMed] [Google Scholar]
- 35. Rapizzi E, Ercolino T, Canu L, et al. Mitochondrial function and content in pheochromocytoma/paraganglioma of succinate dehydrogenase mutation carriers. Endocr Relat Cancer. 2012;19(3):261–269. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article and its Supplementary Material (available online). Data from consented patients are available in dbGaP with accession number phs002405.v1.p1.