Abstract
Background
Although an immunosuppressive tumor microenvironment (TME) is key for tumor progression, the molecular characteristics associated with the immunosuppressive TME remain unknown in triple-negative breast cancer (TNBC). Our previous functional proteomic study of TNBC tumors identified that C-JUN N-terminal kinase (JNK) pathway–related molecules were enriched in a cluster associated with the inflammatory pathway. However, the role of the JNK pathway in the TNBC TME is still unclear.
Methods
Transcriptomic analysis was conducted using The Cancer Genome Atlas datasets. The effect of JNK-IN-8, a covalent pan-JNK inhibitor, on TNBC tumor growth, lung metastasis, and the TME was measured in TNBC syngeneic mouse models (n = 13 per group). Tumor (n = 43) or serum (n = 46) samples from TNBC patients were analyzed using multiplex immunohistochemistry or Luminex assay. All statistical tests were 2-sided.
Results
CIBERSORT analysis revealed that TNBC patients with high phosphorylated JNK level (n = 47) had more regulatory T cell (Treg) infiltration than those with a low phosphorylated JNK level (n = 47) (P = .02). Inhibition of JNK signaling statistically significantly reduced tumor growth (P < .001) and tumor-infiltrating Tregs (P = .02) while increasing the infiltration of CD8+ T cells in TNBC mouse models through the reduction of C-C motif ligand 2 (CCL2). Tumor-associated macrophages were the predominant cells secreting CCL2, and inhibition of JNK signaling reduced CCL2 secretion of human primary macrophages. Moreover, in patients with TNBC (n = 43), those with high levels of CCL2+ tumor-associated macrophages had more Treg and less CD8+ T cell infiltration (P = .04), and the serum CCL2 level was associated with poor overall survival (hazard ratio = 2.65, 95% confidence interval = 1.29 to 5.44, P = .008) in TNBC patients (n = 46).
Conclusions
The JNK/C-JUN/CCL2 axis contributes to TNBC aggressiveness via forming an immunosuppressive TME and can offer novel therapeutic strategies for TNBC.
An immunosuppressive tumor microenvironment (TME) is critical for tumor progression, and regulatory T cells (Tregs) play central roles in developing an immunosuppressive TME (1,2) and inhibiting cytotoxic function and proliferation of effector T cells, including CD8+ T cells (3). The frequency of Treg infiltration into tumors is increased in correlation with the aggressiveness of breast cancer type. It is particularly prominent in triple-negative breast cancer (TNBC) (4). The depletion of Tregs in the T11 TNBC mouse model delayed tumor growth (5), suggesting the essential role of Tregs in TNBC tumor progression. However, the molecular mechanisms that establish the Treg-inflamed TME of TNBC are largely unknown. Although chemokines mediate chemotaxis of immune cells, including Tregs (5-8), to shape the TME (9), the regulatory mechanisms of chemokines involved in recruiting Tregs into the TME remain unclear.
C-JUN N-terminal kinases (JNKs) are members of the mitogen-activated protein kinase family and mediate various types of cellular signaling processes, such as cell proliferation (10), migration (11), and apoptosis (12). Growing evidence suggests that JNK signaling is involved in tumor progression, including TNBC (13–16). We previously reported that JNK contributes to TNBC tumorigenesis by promoting the cancer stem cell phenotype (17). In addition, JNK signaling is also known to be involved in inflammation and the immune system (18,19). However, the role of JNK signaling in regulating the TNBC TME remains largely unknown. Our recent functional proteomics (reverse phase protein array [RPPA]) study of 129 TNBC tumors identified that JNK pathway–related molecules, such as phosphorylated JNK (pJNK) and phosphorylated C-JUN (pC-JUN), were enriched in an inflammation-related cluster (20). These findings give rise to the hypothesis that JNK signaling drives TNBC aggressiveness by regulating not only the stemness of tumor cells themselves but also the inflammatory and immune process in the TME. In this study, we sought to decipher the role of JNK signaling in the TNBC TME, focusing on inflammation and the immune system.
Methods
Methods for cell culture, flow cytometry, tissue histology, enzyme-linked immunosorbent assay (ELISA), cell viability assays, cytokine/chemokine array, JNK-IN-8 treatment for THP-1 human monocytes and human primary macrophages and C-C motif ligand 2 (CCL2) assays, small interfering RNA (siRNA) transfection in primary macrophages, quantitative polymerase chain reaction (qPCR), siRNA transfection in primary macrophages, immunoblot analysis, chromatin immunoprecipitation assay, tumor-infiltrating lymphocyte (TIL) analysis, and the bioinformatics analysis are described in detail in the Supplementary Methods (available online).
Mice
C57BL/6 wild-type and BALB/c mice were purchased from Envigo. Ccl2-RFPflox/flox mice (21) and LyzM-Cre mice (22) were purchased from the Jackson Laboratory and bred in the specific pathogen-free animal facility of MD Anderson. All animal procedures were conducted under guidelines approved by the Institutional Animal Care and Use Committee at MD Anderson. Further details of the experiments are described in the Supplementary Methods (available online ).
Patient Samples
A tumor microarray from TNBC patients was purchased from US Biomax. Serum samples from 46 TNBC patients and 29 healthy donors were analyzed as previously described (23), and the MD Anderson Institutional Review Board approved this study. Further details of the experiments are described in the Supplementary Methods (available online).
Statistical Analysis
The Kaplan-Meier plots and log-rank test were used to evaluate survival statistics. The Wilcoxon 2-sample test was used to compare the difference in final tumor volume in animal studies. Group differences were evaluated using the Mann-Whitney test or Student t test to compare 2 groups and 1-way analysis of variance followed by the Tukey multiple comparison test to compare multiple groups. The correlation between the mRNA level of CCL2 and immune cell lineage markers was evaluated using Spearman’s correlation coefficient. The Kruskal-Wallis H test was used to compare the CCL2 concentration of sera. Univariate and multivariable Cox proportional hazards models were used to test for the statistical significance of the comparison for patients’ survival according to the log-transformed CCL2 concentration of sera. The analysis of statistical values was performed using GraphPad Prism 8 and R. P values less than .05 were considered statistically significant, and all statistical tests were 2-sided.
Results
Correlation Between pJNK and an Immunosuppressive TME in TNBC
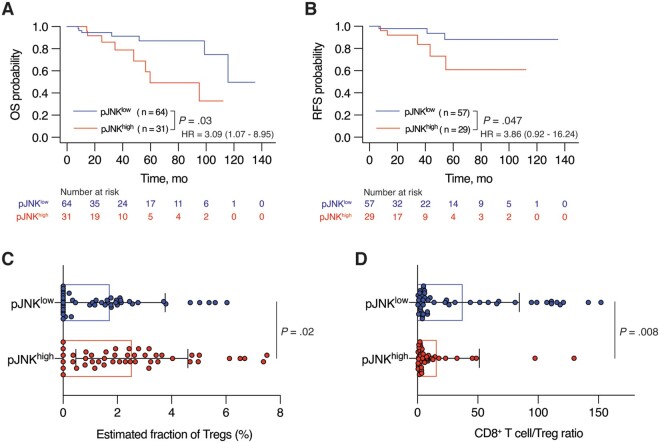
Our previous RPPA-based proteomics study revealed an inflammation-related TNBC cluster enriched with 17 proteins, including pJNK and pC-JUN (20). We found a correlation between high pJNK expression levels and short overall survival (hazard ratio [HR] = 3.09, 95% confidence interval [CI] = 1.07 to 8.95, P = .03; Figure 1, A) and relapse-free survival (HR = 3.86, 95% CI = 0 .92 to 16.24, P = .047; Figure 1, B) and a trend between high pC-JUN expression level and short overall survival (HR = 1.94, 95% CI = 0.62 to 6.13, P = .25; Supplementary Figure 1, A, available online) in 95 TNBC patients using The Cancer Genome Atlas (TCGA) RPPA data. We next analyzed gene expression in patients with high expression levels of both pJNK and pC-JUN (JNKhigh) and patients with low expression levels of both pJNK and pC-JUN (Supplementary Figure 1, B, available online). DESeq2 analysis revealed 50 differentially highly expressed genes in the JNKhigh subgroup (Supplementary Figure 1, C and Supplementary Table 1, available online), and Gene Ontology analysis showed that several pathways related to immune system signaling were highly enriched compared with pathways related to cell proliferation in the JNKhigh subgroup (Supplementary Figure 1, D, available online).
Figure 1.
The correlation between phosphorylated C-JUN N-terminal kinase (pJNK) and an immunosuppressive tumor microenvironment in triple-negative breast cancer (TNBC). A and B) Kaplan -Meier survival analyses of A) overall survival (OS) and B) relapse-free survival (RFS) of TNBC patients from The Cancer Genome Atlas cohort (n = 95 for OS and n = 86 for RFS) stratified based on high vs low expression of pJNK (pJNKhigh and pJNKlow). The hazard ratio (HR) with 95% confidence interval (CI) is shown on each graph. C and D) CIBERSORT analysis of C) estimated fraction of regulatory T cells (Tregs) and D) the ratio of estimated fraction of cluster of differentiation 8(CD8+) T cells to that of Tregs in pJNKhigh and pJNKlow subgroups of TNBC patients from The Cancer Cenome Atlas (TCGA ) cohort (n = 94). Data are summarized as mean and error bars represent the SD. The log-rank test (A and B) and Mann -Whitney test (C and D) were used to calculate P values. All statistical tests were 2-sided.
Using CIBERSORT (24) (Supplementary Figure 1, E, available online), we next found that the subgroup expressing high pJNK levels had more Tregs compared with the subgroup expressing low pJNK levels (n = 47 in each subgroup, P = .02; Figure 1, C). Furthermore, the ratio of CD8+ T cells to Tregs was statistically significantly lower in the pJNKhigh subgroup (mean [SD] = 15.98 [35.31] vs 37.56 [46.77], P = .008; Figure 1, D), reflecting impaired antitumor immunity in TNBC TME with high pJNK levels (25,26). Together, these results confirm that the JNK signaling pathway has a pivotal role in fostering an immunosuppressive TME through regulating the immune cell profile, especially Treg infiltration, in TNBC tumors.
Effect of JNK Inhibitor Treatment on TNBC Tumor Growth, Metastasis, and the Immune TME
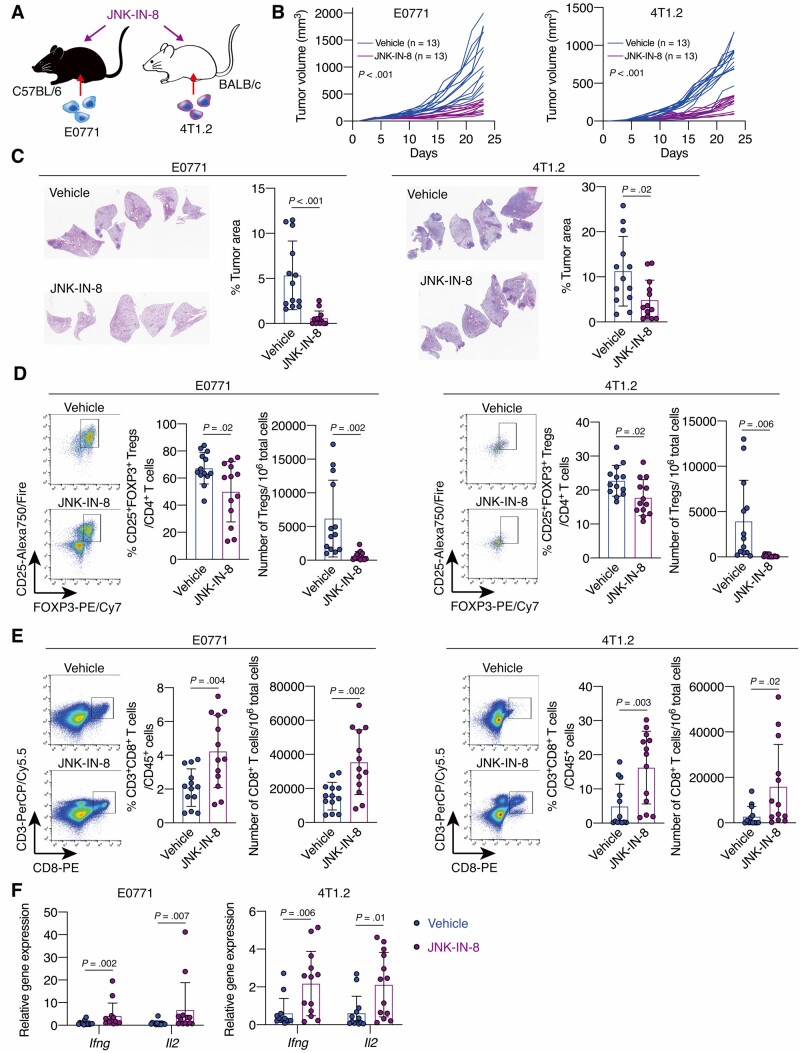
Next, we used E0771 and 4T1.2 syngeneic mouse models and treated the mice with JNK-IN-8, a covalent pan-JNK inhibitor (27), which can inhibit tumor cell proliferation in vitro (Figure 2, A;Supplementary Figure 2, A, available online). We observed that JNK-IN-8 decreased pJNK expression in E0771 tumors (Supplementary Figure 2, B, available online) and suppressed tumor growth (mean [SD] = 1239 [477] vs 412 [101] mm3, P < .001) (Figure 2, B) and lung metastasis (P < .001) (Figure 2, C). Flow cytometry analysis revealed that JNK-IN-8 treatment reduced the subset of CD25+Foxp3+ Tregs within the CD4+ T cell subset (P = .02) and the number of CD4+CD25+Foxp3+ Tregs in tumors (P = .002) (Figure 2, D;Supplementary Figure 2, C, available online), and JNK-IN-8 treatment increased the population of CD3+CD8+ T cells among CD45+ lymphocytes (P = .004) and the number of CD3+CD8+ T cells in tumors (P = .002) (Figure 2, E). Also, mRNA expression levels of Ifng and Il2, which indicate cytotoxic T-cell activation (28,29), were upregulated in tumors by JNK-IN-8 treatment (Figure 2, F). Similar results were observed in the 4T1.2 models (Figure 2, B-F). These results confirm that inhibition of JNK signaling can reverse the immunosuppressive TME and suppress TNBC tumor progression.
Figure 2.
The effect of C-JUN N-terminal kinase (JNK) inhibitor treatment on triple-negative breast cancer tumor growth, metastasis, and the immune tumor microenvironment. A) Schematic showing the experimental design: mouse breast cancer E0771 or 4T1.2 cells were injected into C57BL/6 or BALB/c mice, respectively. Mice were treated with JNK-IN-8 when the tumors became palpable. B) Tumor growth curves for vehicle- or JNK-IN-8–treated E0771 (n = 13 per group) and 4T1.2 (n = 13 per group) mouse models. C) Hematoxylin-eosin staining and quantification of the percentage tumor area for the lungs of vehicle- or JNK-IN-8–treated E0771 (n = 13 per group) and 4T1.2 (n = 13 per group) mice. D and E) Flow cytometry analysis for D) regulatory T cells and E) cluster of differentiation 8(CD8+)T cells in tumors from vehicle- or JNK-IN-8–treated E0771 (n = 13 per group) and 4T1.2 (n = 13 per group) mouse models. LIVE/DEAD Aqua– and CD45+ cells were gated. Shown are quantification of D) the percentage of CD25+ forkhead box p3 (FOXP3+) cells in CD3+CD4+ cells and the number of CD3+CD4+CD25+FOXP3+ cells in 106 tumor cells and E) the percentage of CD3+CD8+ cells in CD45+ cells and the number of CD3+CD8+ cells in 106 tumor cells. F) Ifng and Il2 mRNA expression in tumors from vehicle- or JNK-IN-8–treated E0771 (n = 13 per group) and 4T1.2 (n = 13 per group) models were evaluated by quantitative reverse transcription-polymerase chain reaction. Data are summarized as mean and error bars represent the SD. The Wilcoxon 2-sample test (B) and a 2-tailed Student t test were used to calculate P values. All statistical tests were 2-sided.
Contributions of CCL2 to JNK-Mediated Immunosuppressive TME Formation and Tumor Progression in TNBC
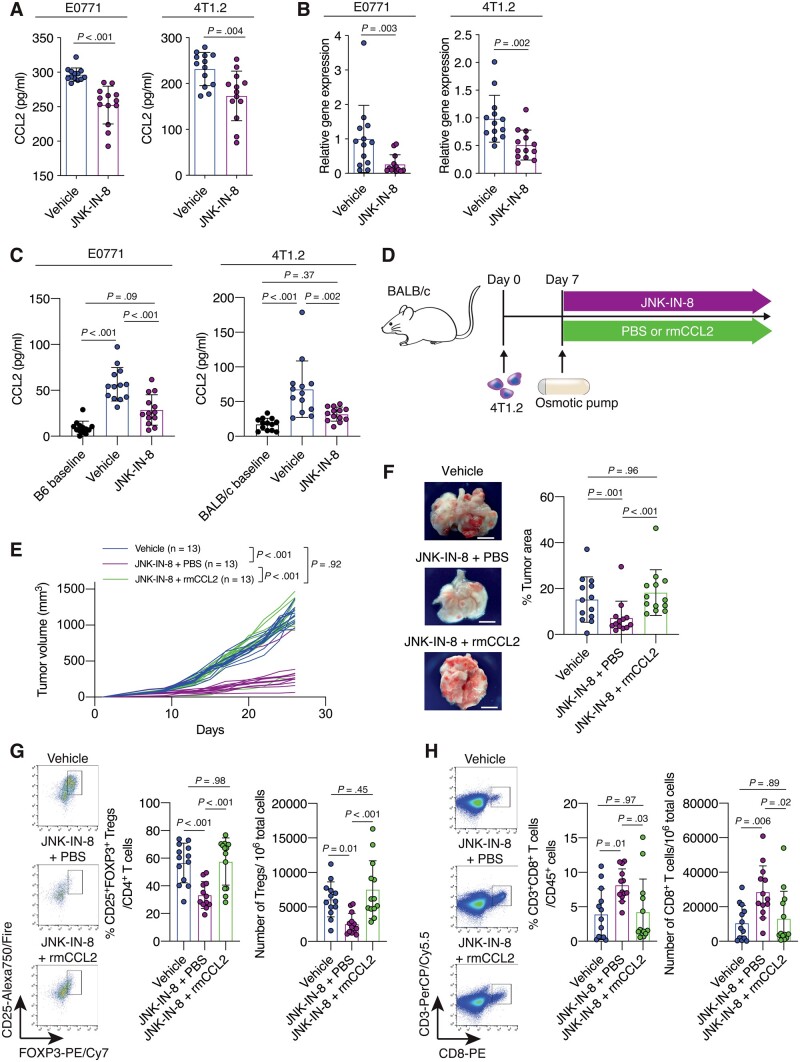
To determine which chemokines (30) in the tumors from mouse TNBC models were essential for Treg infiltration into the tumors, we conducted a chemokine antibody array screening of E0771 and 4T1.2 tumors from vehicle- and JNK-IN-8-treated mice (Supplementary Figure 2, D, available online). We found that expression levels of CCL2, CCL5, C-X-C motif ligand 1 (CXCL1), CXCL5, and C-X3-C motif ligand 1 (CX3CL1) were reduced in JNK-IN-8-treated E0771 and 4T1.2 tumors (Supplementary Figure 2, E, available online). Among these chemokines, CCL5, CXCL5, and CX3CL1 were low, and CCL2, but not CXCL1, was statistically significantly reduced by JNK-IN-8 (mean [SD] = 296.5 [9.6] vs 252.4 [27.4] pg/mL, P < .001) (Figure 3, A;Supplementary Figure 2, F, available online). We also confirmed that JNK-IN-8 reduced the mRNA expression level in tumors (P = .003) and the serum concentration of CCL2 (P < .001) (Figure 3, B and C). Similar results were observed in the 4T1.2 models (Figure 3, A-C). Based on these findings, we decided to further study JNK’s regulation of CCL2, which was consistently affected by JNK-IN-8 treatment and known to recruit immunosuppressive cells, such as myeloid-derived suppressor cells (31,32) and Tregs (6,33–36) .
Figure 3.
The contributions of C-C motif ligand 2 (CCL2) to C-JUN N-terminal kinase (JNK)-mediated immunosuppressive tumor microenvironment (TME) formation and tumor progression in triple-negative breast cancer (TNBC). A) Quantification of CCL2 concentration in tumors from the vehicle- or JNK-IN-8–treated E0771 (n = 13 per group) and 4T1.2 (n = 13 per group) models. B) Quantification of Ccl2 mRNA expression in tumors from vehicle- or JNK-IN-8-treated E0771 (n = 13 per group) and 4T1.2 (n = 13 per group) models as evaluated by quantitative reverse transcription-–polymerase chain reaction (qRT-PCR). C) Quantification of CCL2 concentration in sera from the vehicle- or JNK-IN-8–treated E0771 (n = 13 per group) and 4T1.2 (n = 13 per group). The CCL2 levels in sera collected before tumor injection are indicated as the B6 baseline for the E0771 model (n = 13) and the BALB/c baseline for the 4T1.2 model (n = 13). D) Schematic showing the experimental design: 4T1.2 cells were injected into BALB/c mice. Seven days after tumor injection, mice were treated with vehicle (not shown) or with JNK-IN-8. JNK-IN-8–treated mice were administered either phosphate-buffered saline (PBS) or recombinant murine CCL2 (rmCCL2) via an osmotic pump implanted in the backs of the mice. E) Tumor growth curves for the vehicle, JNK-IN-8 + PBS, and JNK-IN-8 + rmCCL2 groups (n = 13 per group). F) Gross appearance of the lungs and quantification of the percentage tumor area for the lungs of mice from each group. G and H) Flow cytometry analysis of tumors from each group for (G) regulatory T cells (Tregs) and (H) cluster of differentiation 8 (CD8+) T cells. LIVE/DEAD Aqua– and CD45+ cells were gated. Quantification of (G) the percentage of CD25+ forkhead box p3 (FOXP3+) cells in CD3+CD4+ cells and the number of CD3+CD4+CD25+FOXP3+ cells in 106 tumor cells and (H) the percentage of CD3+CD8+ cells in CD45+ cells and the number of CD3+CD8+ cells in 106 tumor cells is shown. Data are summarized as mean and the error bars represent the SD. A 2-tailed Student t test (A and B), 1-way analysis of variance (ANOVA) followed by Tukey multiple comparison test (C, and F–H), and the Wilcoxon 2-sample test (E) were used to calculate P values. All statistical tests were 2-sided. FOXP3 = forkhead box p3.
To test whether CCL2 is the key regulator of the JNK-mediated immunosuppressive TME and tumorigenesis, we treated 4T1.2 tumor-bearing mice with JNK-IN-8 and either PBS or murine recombinant CCL2 (rmCCL2), which was administered by an osmotic pump implanted subcutaneously in the backs of mice (37) (Figure 3, D). The administration of rmCCL2 into 4T1.2-tumor-bearing mice treated with JNK-IN-8 was validated to increase the CCL2 serum concentration to the same level of vehicle-treated mice (Supplementary Figure 2, G, available online). We observed that rmCCL2 administration rescued tumor growth (P < .001) (Figure 3, E) and the extent of lung metastasis (P < .001) (Figure 3, F) of JNK-IN-8–treated mice to the same level of vehicle-treated mice. rmCCL2 administration also rescued the reduced Treg infiltration into tumors of JNK-IN-8–treated mice (P < .001) (Figure 3, G), whereas the infiltration of CD3+CD8+ T cells into tumors of JNK-IN-8–treated mice was suppressed by rmCCL2 administration (P = .03) (Figure 3, H). These results indicate that CCL2 contributes to JNK-mediated immunosuppressive TME formation and tumor progression.
Role of Tumor-Associated Macrophages (TAMs) in Producing CCL2 in TME and TNBC Tumor Progression
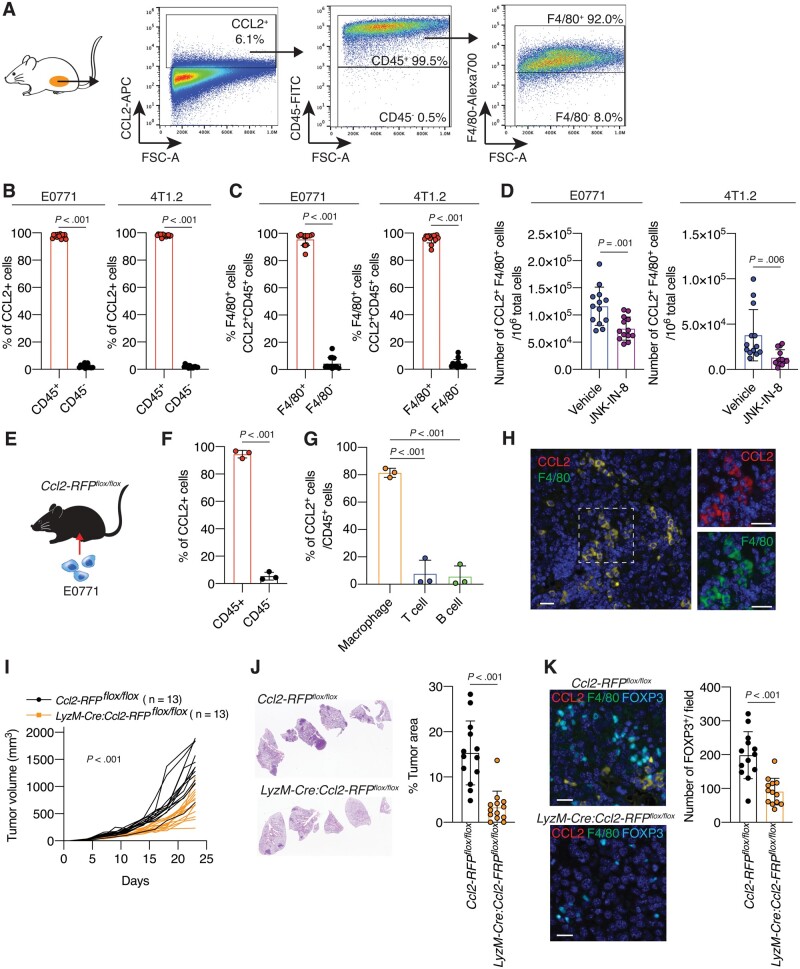
To determine the cells responsible for CCL2 production in TNBC tumors, we examined CCL2-expressing cells in tumors from E0771 and 4T1.2 models by flow cytometry (Figure 4, A). We observed that CCL2-expressing cells were composed of predominantly CD45+ cells (97.6% in E0771) (Figure 4, B). Furthermore, most of the CCL2-expressing CD45+ cells were F4/80+ TAMs (95.6% in E0771) (Figure 4, C), and JNK-IN-8 treatment statistically significantly reduced CCL2-expressing F4/80+ TAMs in tumors from the E0771 model (mean [SD] = 1.16 × 105 [3.50 × 104] vs 7.50 × 104 [2.13 × 104] cells, P = .001) (Figure 4, D). We observed similar results in the 4T1.2 models (Figure 4, B-D). To further confirm these findings, we used Ccl2-RFPflox/flox mice (21) harboring a Ccl2 gene modified with a red fluorescent protein (RFP) gene. E0771 cells were inoculated into mammary fat pads of Ccl2-RFPflox/flox mice (Figure 4, E), and tumors were subjected to further analyses. Again, CD45+ cells were the predominant CCL2-expressing (RFP+) cells in tumors (P < .001) (Figure 4, F). F4/80+ TAMs accounted for more than 81.5% of CCL2-expressing CD45+ cells, whereas CD3+ T cells or CD19+ B cells occupied only small percentages of CCL2-expressing CD45+ cells (P < .001) (Figure 4, G;Supplementary Figure 3, A, available online). The CCL2 expression in TAMs was also validated using multiplex immunohistochemistry (IHC) (Figure 4, H).
Figure 4.
The role of tumor-associated macrophages (TAMs) in producing C-C motif ligand 2 (CCL2) in the tumor microenvironment (TME) and triple-negative breast cancer (TNBC) tumor progression. A) Schematic of gating strategy of flow cytometry analysis. Tumors from E0771 or 4T1.2 mouse models were dissociated to obtain a single-cell suspension and stained with antibodies. Cells were first gated to exclude debris and dead cells (not shown), and CCL2+ cells were selected. Cells were further gated by cluster of differentiation 45 (CD45) and then F4/80 expression. Quantification of the percentage of (B) CD45+ or CD45– cells within the CCL2+ cells and (C) F4/80+ or F4/80– cells within the CCL2+CD45+ cells in tumors from 4T1.2 or E0771 models (n = 13). D) Quantification of the number of CCL2+CD45+F4/80+ cells in 106 tumor cells from vehicle- or JNK-IN-8–treated 4T1.2 and E0771 mouse models (n = 13 per group). E) Schematic of experimental design: E0771 cells were injected into Ccl2-RFPflox/flox mice. At 28 days after cell injection, mice were killed and tumors were subjected to further analysis. Quantification of the percentages of (F) CD45+ or CD45– cells within the CCL2-red fluorescent protein (RFP+) cells and (G) CCL2-RFP+F4/80+ cells (macrophages), CCL2-RFP+CD3+ cells (T cells), and CCL2-RFP+CD19+ cells (B cells) within the CD45+ cells in tumors from E0771-inoculated Ccl2-RFPflox/flox mice (n = 3). Also, see Supplementary Figure 4, A (available online). H) Multiplex immunohistochemistry (IHC) staining for CCL2-RFP (red) and F4/80 (green) of an E0771 tumor section from a Ccl2-RFPflox/flox mouse. Scale bar = 50 μm. I) Tumor growth curves for E0771 tumors in Ccl2-RFPflox/flox and LyzM-Cre: Ccl2-RFPflox/flox mice groups (n = 13 per group). J) H&E staining and quantification of the percentage tumor area within the lungs of mice from each group (n = 13). K) (left) Representative images of multiplex IHC of E0771 tumor sections from Ccl2-RFPflox/flox and LyzM-Cre: Ccl2-RFPflox/flox mice for CCL2-RFP (red), F4/80 (green), and forkhead box p3 (FOXP3) (cyan). Scale bar = 50 μm. (Right) Quantification of the number of FOXP3+ cells in tumors from each group (n = 13). Data are summarized as mean and error bars represent the SD. A 2-tailed Student t test (B-D, F, J, and K), 1-way analysis of variance (ANOVA) followed by Tukey multiple comparison test (G), and the Wilcoxon 2-sample test (I) were used to calculate P values. All statistical tests were 2-sided.
Furthermore, we crossed Ccl2-RFPflox/flox mice with LyzM-Cre mice (22) to generate mice with CCL2 deficiency, specifically in myeloid cells, including macrophages (Supplementary Figure 3, B, available online). We then inoculated E0771 cells into mammary fat pads of LyzM-Cre: Ccl2-RFPflox/flox mice and Ccl2-RFPflox/flox mice (Supplementary Figure 3, C, available online). The tumor growth (mean [SD] = 1362 [326] vs 690 [233] mm3, P < .001) and lung metastasis (P < .001) of E0771 cells were markedly suppressed in LyzM-Cre: Ccl2-RFPflox/flox mice compared with Ccl2-RFPflox/flox mice (Figure 4, I and J). Moreover, the number of tumor-infiltrating FOXP3+ Tregs was also reduced in LyzM-Cre: Ccl2-RFPflox/flox mice (P < .001) (Figure 4, K). Together, these results confirm that TAMs are the main source of CCL2 in the TNBC TME and contribute to attracting Tregs into the TME and driving the aggressiveness of TNBC.
Regulatory Role of JNK/C-JUN Signaling Pathway on CCL2 Production in Human Macrophages
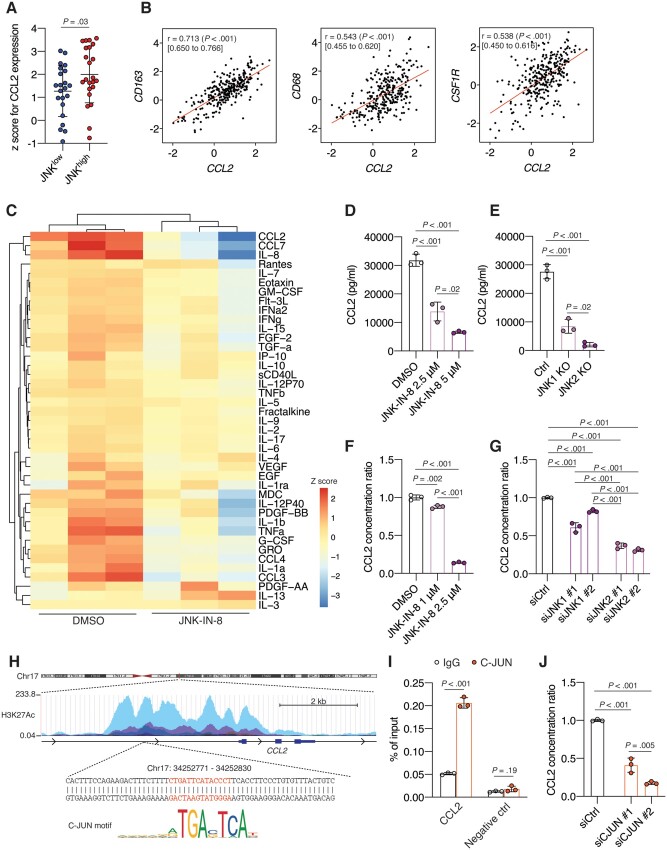
We observed that the CCL2 mRNA level was higher in the JNKhigh subgroup (n = 22) than in the subgroup of patients with low expression levels of both pJNK and pC-JUN (n = 23; P = .03) in the TCGA TNBC cohort (Figure 5, A). Next, we found a strong positive correlation between the mRNA level of CCL2 and that of CD163 (r = .71, 95% CI = 0.65 to .77, P < .001), CD68 (r = .54, 95% CI = 0.46 to .62, P < .001), and CSF1R (r = .54, 95% CI = 0.46 to .62, P < .001), which are lineage markers for macrophages and TAMs (38), in the TCGA and Molecular Taxonomy of Breast Cancer International Consortium (METABRIC ) datasets (39) (Figure 5, B;Supplementary Figure 4, A, available online), while no or only weak correlation was observed between CCL2 mRNA level and representative lineage marker genes for other TME components (40) (Supplementary Figure 4, A, available online), supporting a predominant role of TAMs in producing CCL2 in the TME of TNBC.
Figure 5.
The regulatory role of C-JUN N-terminal kinase (JNK)/C-JUN signaling pathway on C-C motif ligand 2 (CCL2) production in human macrophages. A) CCL2 mRNA expression in the JNKhigh (n = 22) and the JNKlow (n = 23) subgroups of triple-negative breast cancer (TNBC) patients from the The Cancer Genome Atlas (TCGA) cohort (n = 45). B) Scatterplots showing Spearman’s correlation coefficient r with 95% confidence interval (CI) of expression levels for CCL2 and cluster of differentiation 163 (CD163), CD68, or colony stimulation factor 1 recepto (CSF1R) mRNA expression in 299 TNBC patients from the Molecular Taxonomy of Breast Cancer International Consortium (METABRIC) cohort. C) Heatmap shows unsupervised hierarchical clustering of each cytokine or chemokine concentration in conditioned medium (CM) from dimethyl sulfoxide (DMSO) or 2.5 μM JNK-IN-8–treated M2 THP-1 cells as measured by Luminex assay. D) Quantification of the concentration of CCL2 in CM from M2 THP-1 cells treated with DMSO or 2.5 μM or 5 μM JNK-IN-8 (n = 3). E) Quantification of the fold change in CCL2 levels of CM from control (Ctrl), JNK1-knockout (KO), or JNK2-KO THP-1 cells (n = 3). F) Quantification of the fold change in CCL2 levels of CM from human primary macrophages treated with DMSO or 1 μM or 2.5 μM JNK-IN-8 (n = 3). G) Quantification of the fold change in CCL2 levels of CM from human primary macrophages in which JNK1 and JNK2 were silenced by siRNAs (n = 3). H) Schematic of the map of human chromosome 17, the genomic region upstream of CCL2, and the sequence with the C-JUN binding motif obtained from the University of California, Santa Cruz (UCSC) genome browser and Japan Automotive Software Platform and Architecture (JASPAR). I) Quantification of precipitated DNA by quantitative polymerase chain reaction (qPCR) in human primary macrophages following chromatin immunoprecipitation (ChIP) using an anti-C-JUN antibody or immunoglobulin G (IgG) antibody; the qPCR primer was specific to the CCL2 promoter region (CCL2) or intergenic region (negative ctrl). J) Quantification of the fold change in CCL2 level of CM from human primary macrophages in which siRNAs silenced C-JUN. Data are summarized as mean and error bars represent the SD. A 2-tailed Student t test (A and I) and 1-way analysis of variance (ANOVA) followed by Tukey multiple comparison tests (B, D-G, and J) were used to calculate P values. All statistical tests were 2-sided.
Next, we confirmed the regulatory role of the JNK/C-JUN pathway on CCL2 production in macrophages. Because TAMs have been linked with the M2-like phenotype (41), we first used THP-1 human monocytes, which were differentiated into macrophages with phorbol 12-myristate 13-acetate followed by polarization into M2 macrophages (M2 THP-1) with interleukin-4 and interleukin-13. We screened soluble factors in conditioned medium (CM ) from dimethyl sulfoxide (DMSO)- or JNK-IN-8–treated M2 THP-1 cells. We found that CCL2 was one of the most abundantly secreted chemokines among 41 cytokines or chemokines in the CM from M2 THP-1 cells. JNK inhibition by JNK-IN-8 statistically significantly reduced CCL2 in M2 THP-1 cells (mean [SD] CCL2: DMSO = 3.18 × 104 [2.13 × 103] pg/mL, 2.5 μM JNK-IN-8 = 1.39 × 104 [3.31 × 103] pg/mL, P < .001) (Figure 5, C and D; Supplementary Figure 4, B, available online). We also observed that even macrophages with phorbol 12-myristate 13-acetate cells secreted a considerable amount of CCL2, which was much higher than that produced by TNBC cell lines (Supplementary Figure 4, C, available online). Consistent with the results from pharmacological inhibition of JNK, we observed a statistically significant decrease of CCL2 production from phorbol 12-myristate 13-acetate–treated JNK1- and JNK2-knockout (KO) THP-1 cells compared with control THP-1 cells (mean [SD] CCL2: Ctrl = 2.76 × 104 [2.52 × 103] pg/mL; JNK1 KO = 8.49 × 103 [2.43 × 103] pg/mL; JNK2 KO = 2.04 × 103 [7.81 × 102] pg/mL; Ctrl vs JNK1 KO: P < .001; Ctrl vs JNK2 KO: P < .001) (Figure 5, E;Supplementary Figure 4, D, available online). Furthermore, CCL2 secretion from primary macrophages isolated from healthy donor’s buffy coats (Supplementary Figure 4, E, available online) was reduced by JNK-IN-8 treatment (DMSO vs 1 μM JNK-IN-8: P = .002) (Figure 5, F;Supplementary Figure 4, F, available online) or knockdown of the JNK1 or JNK2 gene (siCtrl vs siJNK1: P < .001; siCtrl vs siJNK2: P < .001) (Figure 5, G;Supplementary Figure 4, G, available online).
Moreover, we found that the CCL2 gene has a C-JUN binding motif on its promoter region (Figure 5, H), and chromatin immunoprecipitation analysis showed the binding of C-JUN to the promoter of CCL2 in human primary macrophages (P < .001) (Figure 5, I). Inhibition of C-JUN by siRNA knockdown (Supplementary Figure 4, H, available online) successfully reduced CCL2 secretion from human primary macrophages (P < .001) (Figure 5,J). Together, these data confirm that the JNK/C-JUN pathway plays a pivotal role in regulating CCL2 expression in human macrophages.
Association of CCL2-Expressing TAMs With an Immunosuppressive TME, and the Correlation Between CCL2 Serum Concentration and Prognosis of TNBC Patients
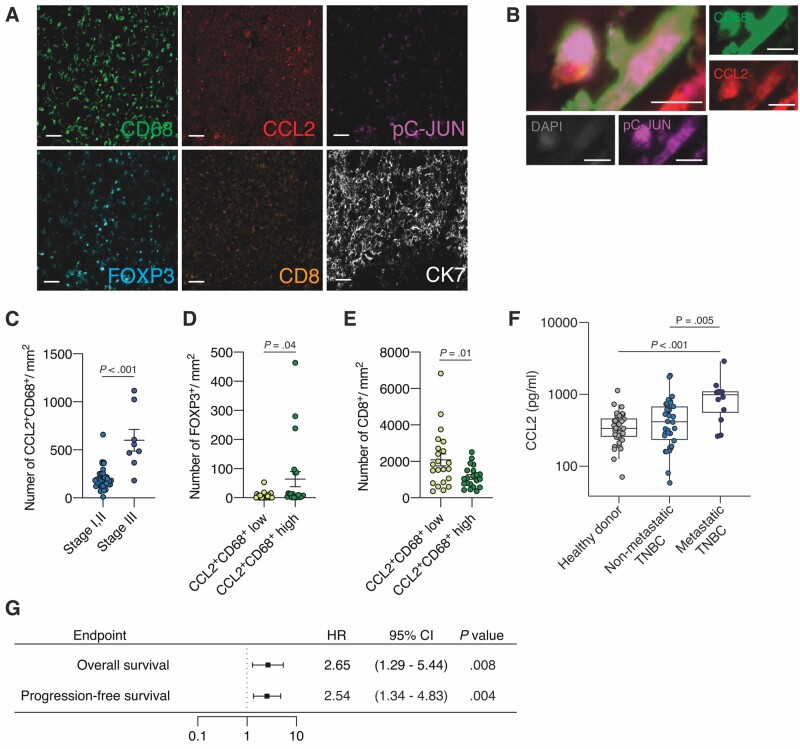
We conducted a fluorescence multiplex IHC study on a tumor tissue microarray of 43 TNBC patients from US Biomax (Figure 6, A) and found colocalization of CCL2 and pC-JUN on CD68+ macrophages (Figure 6, B), suggesting CCL2 secretion from JNK pathway–activated TAMs. More CCL2-expressing CD68+ TAMs were seen in tumors from stage III patients (n = 8) than in tumors from stage I and II patients (n = 35) (P < .001) (Figure 6, C). Furthermore, FOXP3+ Tregs were observed more frequently in the CCL2+CD68+ TAM-inflamed tumors (P = .04) (Figure 6, D), whereas CD8+ T cells were less frequently observed in such tumors (P = .01) (Figure 6, E).
Figure 6.
Association of C-C motif ligand 2 (CCL2)-expressing tumor-associated macrophages (TAMs) with an immunosuppressive tumor microenvironment (TME), and the correlation between CCL2 serum concentration and prognosis of TNBC patients. A) Representative images of a triple-negative breast cancer (TNBC) section subjected to multiplex immunohistochemistry (IHC) for the following markers: cytokeratin 7 (CK7) (white), cluster of differentiation 68 (CD68) (green), CD8 (orange), pC-JUN (magenta), CCL2 (red), and forkhead box p3 (FOXP3) (cyan). Scale bar = 50 μm. B) Representative images of multiplex IHC of CCL2+pC-JUN+CD68+ macrophages: CD68 (green), pC-JUN (magenta), CCL2 (red), and 4′,6-diamidino-2-phenylindole (DAPI) (gray). Scale bar = 10 μm. C) Quantitation of the number of CCL2+CD68+ macrophages per area (mm2) in patients with stage I and II (n = 35) and stage III (n = 8) TNBC. D and E) Quantitation of the number of D) FOXP3+ regulatory T cells (Tregs) and E) CD8+ T cells per area (mm2) in tumors with high (n = 22) and low (n = 21) levels of CCL2+CD68+ macrophages. F) Quantification of the CCL2 concentration of sera previously collected at MD Anderson from healthy donors (n = 29), patients with nonmetastatic TNBC (n = 34), and patients with metastatic TNBC (n = 12). G) Hazard ratios (HRs; represented by squares) and 95% confidence intervals (CIs; represented by horizontal lines) for overall survival and progression-free survival of TNBC patients (n = 46) according to log-transformed CCL2 level in serum. A univariate Cox proportional-hazards model was used to estimate hazard ratios and the statistical significance of the comparison for survival. Data are summarized as mean and the error bars represent the SD (C-F). The Mann-Whitney test C-E) and Kruskal-Wallis H test F) were used to calculate P values. All statistical tests were 2-sided.
By analyzing TILs in digital pathology images for 93 patients with TNBC in the TCGA database, we observed that pJNK expression was higher in patients with poor outcomes than in patients with good outcomes among TNBC patients in the TCGA cohort with similar amounts of TILs. The increased TILs have previously been associated with a good prognosis (42) (Supplementary Figure 5, A-E, available online). Furthermore, we observed fewer TILs in CCL2+CD68+ TAM-inflamed tumors than in tumors with few CCL2+CD68+ TAMs in 43 TNBC patient samples (P = .01) (Supplementary Figure 5, F, available online).
Moreover, we examined the serum level of CCL2 from 46 TNBC patients and 29 healthy donors at MD Anderson (23) and found that the median CCL2 level was statistically significantly higher in patients with metastatic TNBC (n = 12) than in patients with nonmetastatic TNBC (n = 34) (mean [SD] = 989.24 [692.05] vs 511.12 [403.72] pg/mL, P = .005) (Figure 6, F), and a higher log-transformed serum CCL2 level was associated with poor overall (HR = 2.65, 95% CI = 1.29 to 5.44, P = .008) and progression-free survival (HR = 2.54, 95% CI = 1.34 to 4.38, P = .004) of TNBC patients (Figure 6, G;Supplementary Figure 5, G-I, available online).
Discussion
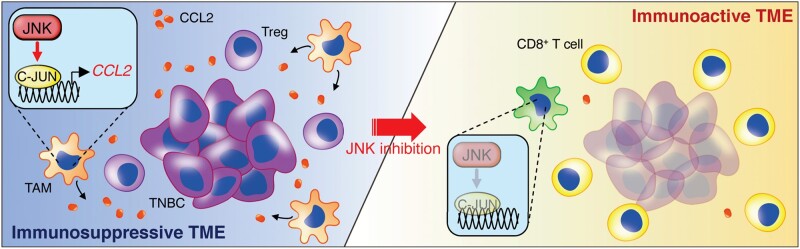
In this study, we showed for the first time, to our knowledge, that the JNK/C-JUN/CCL2 axis promotes the formation of an immunosuppressive TME in TNBC (Figure 7). Tregs play a pivotal role in developing an immunosuppressive TME, which accelerates tumor progression (1,2), and our transcriptome and proteome analysis of TCGA TNBC patient datasets revealed that JNK phosphorylation status is associated with high Treg infiltration and low CD8+ T cell/Treg ratio in the tumor as well as unfavorable prognosis of TNBC. We showed that several chemokines in tumors from TNBC mouse models were decreased by JNK-IN-8 treatment, including CCL2, CCL5, CXCL1, CXCL5, and CX3CL1. Notably, these chemokines have been reported to be involved in Treg recruitment and activation (6,33–36,43–46). Furthermore, we demonstrated that CCL2 administration mitigated the inhibitory effect of JNK-IN-8 on tumor-infiltrating Tregs as well as tumor growth and lung metastasis in a TNBC mouse model. Thus, CCL2 is a key chemokine involved in the JNK-mediated immunosuppressive TME of TNBC.
Figure 7.
Proposed mechanism. C-JUN N-terminal kinase (JNK)/C-JUN pathway-activated tumor-associated macrophages (TAMs) secrete C-C motif ligand 2 (CCL2) to foster an immunosuppressive tumor microenvironment (TME) in which Tregs are recruited into the triple-negative breast cancer (TNBC) tumor. JNK inhibition suppresses CCL2 production from TAMs and regulatory T cell (Treg) infiltration, leading to cluster of differentiation 8 (CD8+) T cell recruitment into tumors and cultivating an immunoreactive TME in TNBC.
Because we focused mainly on CCL2, we recognize that the assessment of other chemokines’ cause and effect contributions is limited. Therefore, further studies should address the complex JNK-regulated chemokine network in establishing an immunosuppressive TME in TNBC.
JNK signaling has been implicated in macrophage function, and JNK deficiency has been shown to cause reduced expression of cytokines or chemokines, including CCL2, in macrophages (47,48). Although CCL2 has been thought to be secreted mainly from tumor cells (49), growing evidence suggests CCL2 is also produced by nontumor cells in the TME (50–52). Indeed, our series of in vivo experiments showed that TAMs were the predominant cells producing CCL2 in the TME and that CCL2 depletion, specifically in myeloid cell lineages, including macrophages, inhibited Treg infiltration into tumors, tumor growth, and lung metastasis. Our study does not exclude the role of TNBC tumor cells themselves in secreting CCL2. Consistent with the previous report (51), we also observed CCL2 expression in tumor cells in TNBC patient samples. One possible explanation is that tumor cells express CCL2 in response to stimuli from the TME (53). Further studies are needed to clarify the molecular mechanisms involved in increasing CCL2 expression in TNBC tumor cells in vivo.
Because immunosuppressive TME can undermine the antitumor efficacy of immune checkpoint blockade therapy (54,55) and our work determines that JNK inhibition can reverse the immunosuppressive TME of TNBC, the strategy of combining JNK inhibition and immune checkpoint blockade has the possibility of showing a synergistic effect on TNBC tumor progression. Although anti-PD-L1 (atezolizumab) or anti-PD-1 (pembrolizumab) antibodies showed encouraging results in TNBC in recent clinical trials (56,57), there is still a need to enrich the population responses to immune checkpoint blockade. Our data showed that CCL2 expression is higher in the JNKhigh subgroup of TNBC with immunosuppressive TME. Moreover, the serum CCL2 level was higher in patients with advanced stage and associated with poor clinical outcomes of TNBC. Therefore, CCL2 is a potential biomarker for predicting response to JNK inhibition and immune checkpoint blockade therapies. Further studies are warranted to determine the clinical relevance of the JNK/CCL2 axis not only as a therapeutic target but also as a marker for selecting patients for JNK-targeted immunotherapies.
In summary, our study revealed that an active JNK signaling pathway cluster in TNBC represents immunosuppressive TME status. Furthermore, we confirmed that JNK-regulated CCL2 production from TAMs fosters an immunosuppressive TME by recruiting Tregs to promote TNBC aggressiveness. Thus, identification of immunosuppressive status in TNBC may justify targeting JNK, which has the potential to lead to novel strategies for TNBC treatment.
Funding
This work was funded by the Breast Cancer Research Foundation (award number BCRF-17–161); Morgan Welch Inflammatory Breast Cancer Research Program and Clinic; State of Texas Rare and Aggressive Breast Cancer Research Program; National Institutes of Health/National Cancer Institute through award numbers 1R01CA205043-01A1 (to N.T.U.), P30CA016672, and R50CA243707 (used the Flow Cytometry and Cellular Imaging Facility and the Research Animal Support Facility); Cancer Prevention and Research Institute of Texas (award number RP160657 to K.N.D.), and Welch Foundation (award number F-1390 to K.N.D.).
Notes
Role of the funder: The funder had no role in the research design, data acquisition, analyses, interpretation, authoring of the manuscript, or the determination to submit the manuscript for publication.
Disclosures: The authors declare no conflict of interest related to this work.
Author contributions: T.S., X.X., and N.T.U. designed the experiments. T.S., X.X., L.T.H.P., E.N.C., and X.W. performed the experiments, and T.S., X.X., J.P.L., E.N.C., and X.W. interpreted the results. T.S. and N.T.U. wrote the paper, and X.X., E.N.C., J.M.R., K.N.D., J.P.L., L.T.H.P., X.W., and D.T. reviewed and edited the paper.
Acknowledgements: LaKesla R. Iles and Geoffrey A. Bartholomeusz shared JNK1or JNK2 knockout THP-1 cells. Shan H. Shao developed and optimized the protocol for multiplex IHC. The authors thank Sunita Patterson of the Research Medical Library at MD Anderson for editorial assistance and Nalini B. Patel, and Jared K. Burks of the Flow Cytometry and Cellular Imaging Facility at MD Anderson for assistance with flow cytometry and multiplex IHC analysis.
Data Availability
The TCGA dataset is available from https://portal.gdc.cancer.gov/ and https://tcpaportal.org/tcpa/. The METABRIC dataset is available from https://www.cbioportal.org/. All other data associated with this study are present in the paper or the Supplementary Materials .
Supplementary Material
References
- 1. Zou W. Regulatory T cells, tumour immunity and immunotherapy. Nat Rev Immunol. 2006;6(4):295–307. [DOI] [PubMed] [Google Scholar]
- 2. Pitt JM, Marabelle A, Eggermont A, et al. Targeting the tumor microenvironment: removing obstruction to anticancer immune responses and immunotherapy. Ann Oncol. 2016;27(8):1482–1492. [DOI] [PubMed] [Google Scholar]
- 3. Ohue Y, Nishikawa H.. Regulatory T (Treg) cells in cancer: can Treg cells be a new therapeutic target? Cancer Sci. 2019;110(7):2080–2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Plitas G, Konopacki C, Wu K, et al. Regulatory T cells exhibit distinct features in human breast cancer. Immunity. 2016;45(5):1122–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Taylor NA, Vick SC, Iglesia MD, et al. Treg depletion potentiates checkpoint inhibition in claudin-low breast cancer. J Clin Invest. 2017;127(9):3472–3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chang AL, Miska J, Wainwright DA, et al. CCL2 produced by the glioma microenvironment is essential for the recruitment of regulatory T cells and myeloid-derived suppressor cells. Cancer Res. 2016;76(19):5671–5682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Halvorsen EC, Hamilton MJ, Young A, et al. Maraviroc decreases CCL8-mediated migration of CCR5(+) regulatory T cells and reduces metastatic tumor growth in the lungs. Oncoimmunology. 2016;5(6):e1150398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Olkhanud PB, Baatar D, Bodogai M, et al. Breast cancer lung metastasis requires expression of chemokine receptor CCR4 and regulatory T cells. Cancer Res. 2009;69(14):5996–6004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Roussos ET, Condeelis JS, Patsialou A.. Chemotaxis in cancer. Nat Rev Cancer. 2011;11(8):573–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gururajan M, Chui R, Karuppannan AK, et al. c-Jun N-terminal kinase (JNK) is required for survival and proliferation of B-lymphoma cells. Blood. 2005;106(4):1382–1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Huang Z, Yan DP, Ge BX.. JNK regulates cell migration through promotion of tyrosine phosphorylation of paxillin. Cell Signal. 2008;20(11):2002–2012. [DOI] [PubMed] [Google Scholar]
- 12. Dhanasekaran DN, Reddy EP.. JNK-signaling: a multiplexing hub in programmed cell death. Genes Cancer. 2017;8(9-10):682–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nasrazadani A, Van Den Berg CL.. c-Jun N-terminal kinase 2 regulates multiple receptor tyrosine kinase pathways in mouse mammary tumor growth and metastasis. Genes Cancer. 2011;2(1):31–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yeh YT, Hou MF, Chung YF, et al. Decreased expression of phosphorylated JNK in breast infiltrating ductal carcinoma is associated with a better overall survival. Int J Cancer. 2006;118(11):2678–2684. [DOI] [PubMed] [Google Scholar]
- 15. Semba T, Sammons R, Wang X, et al. JNK signaling in stem cell self-renewal and differentiation. Int J Mol Sci. 2020;21(7):2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang X, Chao L, Li X, et al. Elevated expression of phosphorylated c-Jun NH2-terminal kinase in basal-like and “triple-negative” breast cancers. Hum Pathol. 2010;41(3):401–406. [DOI] [PubMed] [Google Scholar]
- 17. Xie X, Kaoud TS, Edupuganti R, et al. c-Jun N-terminal kinase promotes stem cell phenotype in triple-negative breast cancer through upregulation of Notch1 via activation of c-Jun. Oncogene. 2017;36(18):2599–2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Arthur JS, Ley SC.. Mitogen-activated protein kinases in innate immunity. Nat Rev Immunol. 2013;13(9):679–692. [DOI] [PubMed] [Google Scholar]
- 19. Zeke A, Misheva M, Remenyi A, et al. JNK signaling: regulation and functions based on complex protein-protein partnerships. Microbiol Mol Biol Rev. 2016;80(3):793–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Masuda H, Qi Y, Liu S, et al. Reverse phase protein array identification of triple-negative breast cancer subtypes and comparison with mRNA molecular subtypes. Oncotarget. 2017;8(41):70481–70495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shi C, Jia T, Mendez-Ferrer S, et al. Bone marrow mesenchymal stem and progenitor cells induce monocyte emigration in response to circulating toll-like receptor ligands. Immunity. 2011;34(4):590–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Clausen BE, Burkhardt C, Reith W, et al. Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res. 1999;8(4):265–277. [DOI] [PubMed] [Google Scholar]
- 23. Cohen EN, Fouad TM, Lee BN, et al. Elevated serum levels of sialyl Lewis X (sLe(X)) and inflammatory mediators in patients with breast cancer. Breast Cancer Res Treat. 2019;176(3):545–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12(5):453–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Preston CC, Maurer MJ, Oberg AL, et al. The ratios of CD8+ T cells to CD4+CD25+ FOXP3+ and FOXP3- T cells correlate with poor clinical outcome in human serous ovarian cancer. PLoS One. 2013;8(11):e80063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Baras AS, Drake C, Liu JJ, et al. The ratio of CD8 to Treg tumor-infiltrating lymphocytes is associated with response to cisplatin-based neoadjuvant chemotherapy in patients with muscle invasive urothelial carcinoma of the bladder. Oncoimmunology. 2016;5(5):e1134412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhang T, Inesta-Vaquera F, Niepel M, et al. Discovery of potent and selective covalent inhibitors of JNK. Chem Biol. 2012;19(1):140–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Slifka MK, Whitton JL.. Activated and memory CD8+ T cells can be distinguished by their cytokine profiles and phenotypic markers. J Immunol. 2000;164(1):208–216. [DOI] [PubMed] [Google Scholar]
- 29. Boyman O, Sprent J.. The role of interleukin-2 during homeostasis and activation of the immune system. Nat Rev Immunol. 2012;12(3):180–190. [DOI] [PubMed] [Google Scholar]
- 30. Viola A, Sarukhan A, Bronte V, et al. The pros and cons of chemokines in tumor immunology. Trends Immunol. 2012;33(10):496–504. [DOI] [PubMed] [Google Scholar]
- 31. Qian BZ, Li J, Zhang H, et al. CCL2 recruits inflammatory monocytes to facilitate breast-tumour metastasis. Nature. 2011;475(7355):222–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Huang B, Lei Z, Zhao J, et al. CCL2/CCR2 pathway mediates recruitment of myeloid suppressor cells to cancers. Cancer Lett. 2007;252(1):86–92. [DOI] [PubMed] [Google Scholar]
- 33. Loyher PL, Rochefort J, Baudesson de Chanville C, et al. CCR2 influences T regulatory cell migration to tumors and serves as a biomarker of cyclophosphamide sensitivity. Cancer Res. 2016;76(22):6483–6494. [DOI] [PubMed] [Google Scholar]
- 34. Ge Y, Bohm HH, Rathinasamy A, et al. Tumor-specific regulatory T cells from the bone marrow orchestrate antitumor immunity in breast cancer. Cancer Immunol Res. 2019;7(12):1998–2012. [DOI] [PubMed] [Google Scholar]
- 35. Yue Y, Lian J, Wang T, et al. Interleukin-33-nuclear factor-kappaB-CCL2 signaling pathway promotes progression of esophageal squamous cell carcinoma by directing regulatory T cells. Cancer Sci. 2020;111(3):795–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Vasanthakumar A, Chisanga D, Blume J, et al. Sex-specific adipose tissue imprinting of regulatory T cells. Nature. 2020;579(7800):581–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tateya S, Tamori Y, Kawaguchi T, et al. An increase in the circulating concentration of monocyte chemoattractant protein-1 elicits systemic insulin resistance irrespective of adipose tissue inflammation in mice. Endocrinology. 2010;151(3):971–979. [DOI] [PubMed] [Google Scholar]
- 38. Qiu SQ, Waaijer SJH, Zwager MC, et al. Tumor-associated macrophages in breast cancer: innocent bystander or important player? Cancer Treat Rev. 2018;70:178–189. [DOI] [PubMed] [Google Scholar]
- 39. Curtis C, Shah SP, Chin SF, et al. ; METABRIC Group. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486(7403):346–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Becht E, Giraldo NA, Lacroix L, et al. Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biol. 2016;17(1):218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sica A, Schioppa T, Mantovani A, et al. Tumour-associated macrophages are a distinct M2 polarised population promoting tumour progression: potential targets of anti-cancer therapy. Eur J Cancer. 2006;42(6):717–727. [DOI] [PubMed] [Google Scholar]
- 42. Salgado R, Denkert C, Demaria S, et al. ; International TILs Working Group 2014. The evaluation of tumor-infiltrating lymphocytes (TILs) in breast cancer: recommendations by an International TILs Working Group 2014. Ann Oncol. 2015;26(2):259–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tan MC, Goedegebuure PS, Belt BA, et al. Disruption of CCR5-dependent homing of regulatory T cells inhibits tumor growth in a murine model of pancreatic cancer. J Immunol. 2009;182(3):1746–1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lv M, Xu Y, Tang R, et al. miR141-CXCL1-CXCR2 signaling-induced Treg recruitment regulates metastases and survival of non-small cell lung cancer. Mol Cancer Ther. 2014;13(12):3152–3162. [DOI] [PubMed] [Google Scholar]
- 45. Shi G, Han J, Liu G, et al. Expansion of activated regulatory T cells by myeloid-specific chemokines via an alternative pathway in CSF of bacterial meningitis patients. Eur J Immunol. 2014;44(2):420–430. [DOI] [PubMed] [Google Scholar]
- 46. Hadis U, Wahl B, Schulz O, et al. Intestinal tolerance requires gut homing and expansion of FoxP3+ regulatory T cells in the lamina propria. Immunity. 2011;34(2):237–246. [DOI] [PubMed] [Google Scholar]
- 47. Han MS, Jung DY, Morel C, et al. JNK expression by macrophages promotes obesity-induced insulin resistance and inflammation. Science. 2013;339(6116):218–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Han MS, Barrett T, Brehm MA, et al. Inflammation mediated by JNK in myeloid cells promotes the development of hepatitis and hepatocellular carcinoma. Cell Rep. 2016;15(1):19–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yoshimura T. The production of monocyte chemoattractant protein-1 (MCP-1)/CCL2 in tumor microenvironments. Cytokine. 2017;98:71–78. [DOI] [PubMed] [Google Scholar]
- 50. Bussard KM, Venzon DJ, Mastro AM.. Osteoblasts are a major source of inflammatory cytokines in the tumor microenvironment of bone metastatic breast cancer. J Cell Biochem. 2010;111(5):1138–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Fujimoto H, Sangai T, Ishii G, et al. Stromal MCP-1 in mammary tumors induces tumor-associated macrophage infiltration and contributes to tumor progression. Int J Cancer. 2009;125(6):1276–1284. [DOI] [PubMed] [Google Scholar]
- 52. Li X, Loberg R, Liao J, et al. A destructive cascade mediated by CCL2 facilitates prostate cancer growth in bone. Cancer Res. 2009;69(4):1685–1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yoshimura T, Liu M, Chen X, et al. Crosstalk between tumor cells and macrophages in stroma renders tumor cells as the primary source of MCP-1/CCL2 in Lewis lung carcinoma. Front Immunol. 2015;6:332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Gajewski TF, Schreiber H, Fu YX.. Innate and adaptive immune cells in the tumor microenvironment. Nat Immunol. 2013;14(10):1014–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Su W, Han HH, Wang Y, et al. The polycomb repressor complex 1 drives double-negative prostate cancer metastasis by coordinating stemness and immune suppression. Cancer Cell. 2019;36(2):139–155 e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Schmid P, Adams S, Rugo HS, et al. Atezolizumab and Nab-paclitaxel in advanced triple-negative breast cancer. N Engl J Med. 2018;379(22):2108–2121. [DOI] [PubMed] [Google Scholar]
- 57. Schmid P, Cortes J, Pusztai L, et al. Pembrolizumab for early triple-negative breast cancer. N Engl J Med. 2020;382(9):810–821. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The TCGA dataset is available from https://portal.gdc.cancer.gov/ and https://tcpaportal.org/tcpa/. The METABRIC dataset is available from https://www.cbioportal.org/. All other data associated with this study are present in the paper or the Supplementary Materials .