Abstract
The distribution of genes for an outer membrane protein (OmpW) and a regulatory protein (ToxR) in Vibrio cholerae and other organisms was studied using respective primers and probes. PCR amplification results showed that all (100%) of the 254 V. cholerae strains tested were positive for ompW and 229 (∼98%) of 233 were positive for toxR. None of the 40 strains belonging to other Vibrio species produced amplicons with either ompW- or toxR-specific primers, while 80 bacterial strains from other genera tested were also found to be negative by the assay. These studies were extended with representative number of strains using ompW- and toxR-specific probes in DNA dot blot assay. While the V. cholerae strains reacted with ompW probe, only one (V. mimicus) out of 60 other bacterial strains tested showed weak recognition. In contrast, several strains belonging to other Vibrio species (e.g., V. mimicus, V. splendidus, V. alginolyticus, V. fluvialis, V. proteolyticus, V. aestuarianus, V. salmonicida, V. furnissii, and V. parahaemolyticus) showed weak to strong reactivity to the toxR probe. Restriction fragment length polymorphism analysis and nucleotide sequence data revealed that the ompW sequence is highly conserved among V. cholerae strains belonging to different biotypes and/or serogroups. All of these results suggest that the ompW gene can be targeted for the species-specific identification of V. cholerae strains. The scope of this study was further extended through the development of a one-step multiplex PCR assay for the simultaneous amplification of ompW and ctxA genes which should be of considerable value in the screening of both toxigenic and nontoxigenic V. cholerae strains of clinical as well as environmental origin.
The diarrheal disease cholera in the epidemic form is caused by the organism Vibrio cholerae belonging to the O1 or O139 serogroup (10). V. cholerae organisms belonging to non-O1/non-O139 serogroups, which can be isolated in abundance from aquatic or estuarine sources, cause sporadic cases or limited outbreaks of diarrhea in humans (17). Identification of V. cholerae is usually achieved through a series of biochemical tests after their growth and isolation on a selective plating medium e.g., TCBS agar (25). The process, however, is laborious and time-consuming and may be quite expensive for a laboratory handling a large number of clinical and/or environmental samples. Further, close relatedness among V. cholerae and certain other members of the Vibrio spp. (e.g., V. mimicus) or Aeromonas spp. with respect to their biochemical properties has often made unambiguous identification of the organism quite difficult. Although the commercial (or otherwise) availability of O1 and/or O139 antisera has considerably helped in the identification of epidemic causing strains of V. cholerae, it may not be true for the non-O1/non-O139 strains which, until recently, were known to range from the O2 to O138 and from the O140 to O193 serogroups (32). The problem has been accentuated recently as the non-O1/non-O139 strains of diverse serogroups have been implicated as the causative agents of a large number of diarrheal cases in the Indian subcontinent (23, 28) and elsewhere (2, 4). As a matter of fact, the majority of these strains do not contain virulence markers such as cholera toxin, toxin-coregulated pilus (TCP), etc. (10, 17), that are known to be associated with the pathogenic strains of V. cholerae O1 or O139, thereby making their identification more difficult.
Attempts to identify V. cholerae strains on the basis of their 16S rRNA sequences have not been successful so far due to the lack of appreciable differences between these sequences occurring in V. cholerae and other members of Vibrionaceae family (13, 24). Other approaches were directed toward the identification of toxigenic strains of V. cholerae through the use of cholera toxin gene (ctxAB) probes or appropriate primers for the amplification of toxin genes by PCR assay (21). A multiplex PCR has also been developed to identify epidemic-causing strains of V. cholerae containing the ctxA and TCP protein subunit (tcpA) genes (11). V. cholerae strains belonging to the O1 or O139 serogroups could be detected by using probes or primers designed from their rfb regions responsible for “O” antigen biosynthesis (1, 7). However, none of these methods are applicable for the identification of all V. cholerae strains. Recently, a PCR based method targeted to the toxR gene was developed for the species-specific identification of V. parahaemolyticus (12). No information, however, is available in the literature regarding the identification of V. cholerae using toxR primers, although toxR genes (sharing sequence homology) are distributed among certain other Vibrio species, including V. cholerae (12, 15, 18, 22).
In the present study, we have evaluated the use of toxR primers and/or probes for the identification of V. cholerae strains. The study is extended using primers and/or probes targeted to a gene encoding an outer membrane protein OmpW of V. cholerae whose nucleotide sequence was first reported by Jalajakumari and Manning (8). We demonstrate that although the toxR primers are sensitive and specific for V. cholerae strains, the primers targeted to ompW are better suited for the purpose due to the unique presence of the gene with conserved sequence in V. cholerae. Finally, a multiplex-PCR assay has been developed for the detection of toxigenic strains of V. cholerae through simultaneous amplification of ompW and ctxA genes.
MATERIALS AND METHODS
Bacterial strains.
A total of 254 V. cholerae strains isolated from both clinical and environmental sources were included in this study. The majority of these were isolated locally over the years and kept in our collection. Others were standard or type (American Type Culture Collection [ATCC]) strains which had been initially isolated and used by various groups of workers around the world and made available to us. Organisms were grown on a selective medium (TCBS agar) and subsequently characterized by standard biochemical procedures (25). V. cholerae strains were subjected to serogroup analysis using O serogroup-specific antisera. Of the 254 V. cholerae strains included in this study, 36 and 22 strains belonged to serogroups O1 and O139, respectively. Two of these O1 strains had ATCC designations. A total of 176 strains belonged to non-O1/non-O139 serogroups on the basis of agglutination reactions using O2 to O141 specific antisera. Of these, 96 non-O1/non-O139 strains were, in fact, reference strains used to raise O-specific antisera (29), while two others were ATCC type strains. Of the remaining 20 V. cholerae strains which could not be typed by the existing O1 to O141 antisera, 16 belonged to the rough variety and were agglutinable by the rough antiserum. The remaining four strains were classified under O untypeable category.
A total of 40 strains belonging to other Vibrio species were also included in the study (Table 1). Of these, 17 were type strains with ATCC numbers. The sources of the others are indicated in Table 1. Other bacteria included in this study were Aeromonas spp., Escherichia coli, Shigella spp., Salmonella spp., Pseudomonas spp., Klebsiella spp., and Staphylococcus aureus.
TABLE 1.
Summarized results of PCR and DNA dot blot analyses of strains belonging to Vibrio spp. using primers or probes for ompW and toxR genes
| Analysis no(s). | Strain | Sourcea | Detectionb of:
|
|||
|---|---|---|---|---|---|---|
|
ompW by:
|
toxR by:
|
|||||
| PCR | Dot blot | PCR | Dot blot | |||
| 1 | V. cholerae O1 | 14035 (ATCC) | + | + | + | + |
| 2 | V. cholerae O1 | 39315 (ATCC) | + | + | + | + |
| 3–8 | V. cholerae O1 | Lab stock | + | + | + | + |
| 9–11 | V. cholerae O139 | Lab stock | + | + | + | + |
| 12–13 | V. cholerae (rough) | Lab stock | + | + | + | + |
| 14 | V. cholerae non-O1/non-O139 | 25872 (ATCC) | + | + | + | + |
| 15 | V. cholerae non-O1/non-O139 | 25874 (ATCC) | + | + | + | + |
| 16–18 | V. cholerae non-O1/non-O139 | Lab stock | + | + | + (2/3)c | + |
| 19 | V. mimicus | 33653 (ATCC) | − | +W | − | + |
| 20 | V. mimicus E13072 | NICED | − | − | − | − |
| 21 | V. mimicus 4208 | NICED | − | − | − | + |
| 22 | V. mimicus NT 2310 | NICED | − | − | − | − |
| 23 | V. mimicus 1 | NICED | − | − | − | − |
| 24 | V. mimicus 4053 | NICED | − | − | − | − |
| 25 | V. vulnificus | 33816 (ATCC) | − | − | − | − |
| 26 | V. vulnificus | 27562 (ATCC) | − | − | − | − |
| 27–28 | V. vulnificus | NICED | − | − | − | − |
| 29 | V. alginolyticus | 17749 (ATCC) | − | − | − | + |
| 30 | V. aestuarianus | 35048 (ATCC) | − | − | − | + |
| 31 | V. fluvialis | 33809 (ATCC) | − | − | − | + |
| 32 | V. furnissii | 35016 (ATCC) | − | − | − | +W |
| 33 | V. furnissii | NICED | − | − | − | − |
| 34 | V. proteolyticus | 15338 (ATCC) | − | − | − | +W |
| 35 | V. anguillarum | 19264 (ATCC) | − | − | − | − |
| 36 | V. anguillarum 943 | 43305 (ATCC) | − | − | − | − |
| 37 | V. anguillarum 944 | 43306 (ATCC) | − | − | − | − |
| 38 | V. anguillarum | NICED | − | − | − | − |
| 39 | V. carchariae | 35084 (ATCC) | − | − | − | − |
| 40 | V. tubiashii | 19109 (ATCC) | − | − | − | − |
| 41 | V. natriegens | 14048 (ATCC) | − | − | − | − |
| 42 | V. salmonicida | 43839 (ATCC) | − | − | − | +W |
| 43 | V. splendidus | 33125 (ATCC) | − | − | − | + |
| 44 | V. nereis | 25917 (ATCC) | − | − | − | − |
| 45 | V. parahaemolyticus | 2210001 (RIMD) | − | − | − | − |
| 46–51 | V. parahaemolyticus | NICED | − (6/6) | − (6/6) | − (6/6) | − (5/6)d |
| 52 | V. tyrogens | RS | − | − | − | − |
| 53 | V. tyrogens | RS | − | − | − | − |
| 54 | V. metschnikovii | RS | − | − | − | − |
| 55 | V. metschnikovii | NICED | − | − | − | − |
| 56 | V. metschnikovii | STM | − | − | − | − |
| 57 | V. hollisae | NICED | − | − | − | − |
| 58 | V. proteus | RS | − | − | − | − |
Lab stock, strains available as stock culture at the Bose Institute, Calcutta, India; NICED, National Institute of Cholera and Enteric Diseases, Calcutta, India; RIMD, Research Institute of Microbial Diseases, Osaka, Japan; RS, R. Sakazaki, Tokyo, Japan; STM, School of Tropical Medicine, Calcutta, India.
+, Detectable; +W, weakly detectable; −, not detectable.
One strain gave a negative result.
One strain gave a weakly positive result.
PCR assay.
Amplification of the target gene was carried out by PCR assay using bacterial cell lysate as the source of template DNA. Briefly, bacterial cells were grown overnight at 37°C on Luria agar (LA) plates. For strains belonging to certain Vibrio species, the LA medium was supplemented with 3% NaCl. Next, isolated colonies were picked up and mixed with 100 μl of normal saline, and bacterial cells were pelleted by centrifugation. The cell pellet was resuspended in 100 μl of double-distilled water and boiled for 10 min. Cell debris was removed by centrifugation, and the supernatant containing the template DNA was taken into a fresh microfuge tube for PCR assay.
Four different primers were designed and synthesized on the basis of the available nucleotide sequence data of ompW (8). The primer sequences are indicated in Table 2. Three combinations of ompW primer pairs (1-2, 1-4, and 2-3) were used to generate amplicons of three different sizes in separate PCR tubes. Primers used for the amplification of the toxR gene are also indicated in Table 2. The sense primer corresponds to nucleotides 1 to 18 of the toxR gene, while the antisense primer was complementary to nucleotides 865 to 884 of the toxR gene (6, 15). The primers used for the amplification of ctxA are listed in Table 2.
TABLE 2.
Sequences of primers used in this study
| Primer no. | Target gene | Sequence (size) | Source or reference |
|---|---|---|---|
| 1 | ompW (terminal sense) | 5′-CACCAAGAAGGTGACTTTATTGTG-3′ (24-mer) | This study |
| 2 | ompW (terminal antisense) | 5′-GAACTTATAACCACCCGCG-3′ (19-mer) | This study |
| 3 | ompW (internal sense) | 5′-CCACCTACCTTTATGGTCC-3′ (19-mer) | This study |
| 4 | ompW (internal antisense) | 5′-GGTTTGTCGAATTAGCTTCACC-3′ (22-mer) | This study |
| 5 | toxR (sense) | 5′-ATGTTCGGATTAGGACAC-3′ (18-mer) | 6, 15 |
| 6 | toxR (antisense) | 5′-TACTCACACACTTTGATGGC-3′ (20-mer) | 6, 15 |
| 7 | ctxA (sense) | 5′-CTCAGACGGGATTTGTTAGGCACG-3′ (24-mer) | 11 |
| 8 | ctxA (antisense) | 5′-TCTATCTCTGTAGCCCCTATTACG-3′ (24-mer) | 11 |
PCR amplification of the target DNA was carried out in a thermal cycler (Perkin-Elmer) using 200-μl PCR tubes with a reaction mixture volume of 25 μl. Each of the reaction mixtures contained 3 μl of template DNA (lysate), 2.5 μl of each primer (10 pmol/μl), 2.5 μl of 2.5 mM deoxynucleoside triphosphates, 0.3 μl (5 U/μl) of Taq DNA polymerase (Takara Shuzo Co., Ltd.), 2.5 μl of 10× reaction buffer containing 20 mM MgCl2 (Extaq; Takara), and 11.8 μl of distilled water. The reaction mixture was subjected to an amplification of 30 cycles, each of which consisted of three steps in the following order: denaturation of template DNA at 94°C for 30 s, annealing of the template DNA at 64°C for 30 s, and extension of the primers at 72°C for 30 s. Before initiation of the first cycle, the reaction mixture was heated at 94°C for 5 min to allow complete denaturation of the template. PCR products, thus obtained, were electrophoresed through 1.5% (wt/vol) agarose gel to resolve the amplified products which were visualized under UV light after ethidium bromide staining.
A multiplex PCR assay was carried out by the simultaneous addition of primer pairs for ompW (primers 1 and 2, Table 2) and ctxA (primers 7 and 8, Table 2) in the same reaction mixture. In initial experiments, the ctxA primer concentration was varied between 1.0 and 0.15 pmol/μl, keeping the ompW primer concentration fixed either at 1.0 or at 1.2 pmol/μl in the final reaction mixture of 25 μl. Optimum results were obtained with primer concentrations of 1.2 and 0.25 pmol/μl for ompW and ctxA, respectively. Other conditions for PCR amplification remained as described earlier.
DNA dot blot assay.
Genomic DNA of bacterial strains was isolated using a miniscale preparation method (31) with minor modifications. For this, organisms were grown overnight at 37°C in 5 ml of Luria broth (supplemented with 3% NaCl to support the growth of certain noncholera vibrios). Next, 1.5 ml of the bacterial culture was centrifuged, and the pellet thus obtained was resuspended in 567 μl of TE buffer (10 mM Tris-HCl containing 1 mM EDTA; pH 8.0) followed by the addition of 30 μl of 10% (wt/vol) sodium dodecyl sulfate and 3 μl of a proteinase K solution (20 μg/μl) (Sigma Chemical Co.). The mixture was incubated at 50°C for 90 min to obtain a clear lysate. Next, 100 μl of 5 M NaCl and 100 μl of 10% (wt/vol) cetyltrimethylammonium bromide (CTAB) in 0.7 M NaCl were added to the lysate and kept at 65°C for 10 min. Thereafter, the DNA material was extracted by adding an equal volume of a mixture of chloroform-isoamyl alcohol (24:1). After centrifugation, the aqueous phase containing nucleic acids was collected and reextracted with phenol-chloroform-isoamyl alcohol (25:24:1) to remove proteinaceous material. The aqueous phase was transferred to a fresh 1.5-ml microfuge tube, and DNA was precipitated by the addition of 0.6 volume of isopropanol at room temperature. The precipitate was collected by centrifugation, washed with 70% (vol/vol) ethanol, and finally reconstituted in 100 μl of TE buffer. The concentration of DNA was measured spectrophotometrically (26).
Purified bacterial DNA preparations were used for dot blot assay using ompW and/or toxR probes. The probes were prepared by PCR amplification of target genes ompW and/or toxR using primer pairs 1 and 2 (for ompW) and 5 and 6 (for toxR) (Table 2). Appropriate amplicons thus obtained were purified by using the QIAQuick purification kit (Qiagen) and labeled with horseradish peroxidase by the glutaraldehyde conjugation method using the Direct Nucleic Acid Labeling Kit ECL (Amersham Lifesciences). Briefly, 50 ng of DNA sample (amplicon) was taken in 10 μl of water and denatured by heating for 10 min in a boiling water bath, followed by immediate chilling. The cooled DNA was mixed with equal volumes of the labeling reagent and glutaraldehyde solution (supplied with the kit). Following incubation at 37°C for 20 min, the labeled probe was taken in hybridization buffer (5 ml) and used. For the dot blot assay, the target DNA was denatured by boiling in a water bath for 10 min and spotted onto a nylon membrane presoaked with 2 N NaOH and 2× SSC (30 mM trisodium citrate plus 0.3 M NaCl) buffer. The spotted DNA was linked to the membrane by UV irradiation, prehybridized at 42°C for 1 h, and hybridized with the labeled probe (10 ng/ml) for 14 h at 42°C. After hybridization, the membrane was washed thoroughly with primary (twice at 42°C) and secondary (twice at room temperature) wash buffers under highly stringent conditions. Next, the membrane was exposed to the detection solution and autoradiographed using X-ray film.
Restriction fragment length polymorphism (RFLP) analysis of ompW PCR amplicons.
The 588-bp PCR amplicons of ompW obtained from representative V. cholerae strains using primers 1 and 2 (Table 2) were purified by the Gel Extraction Purification kit (Qiagen) and subsequently digested with the three restriction enzymes HindIII, NdeI, and HpaI (Gennei) in separate reactions. Digested materials were run on a 2% (wt/vol) agarose gel, stained with ethidium bromide, and viewed under UV light.
Sequencing of ompW amplicons.
The nucleotide sequence of ompW amplicons obtained by PCR using the primer pair 1 and 2 (Table 2) from V. cholerae strains was determined using an automated DNA sequencer (Perkin-Elmer 310).
RESULTS
PCR assay using ompW and toxR primers.
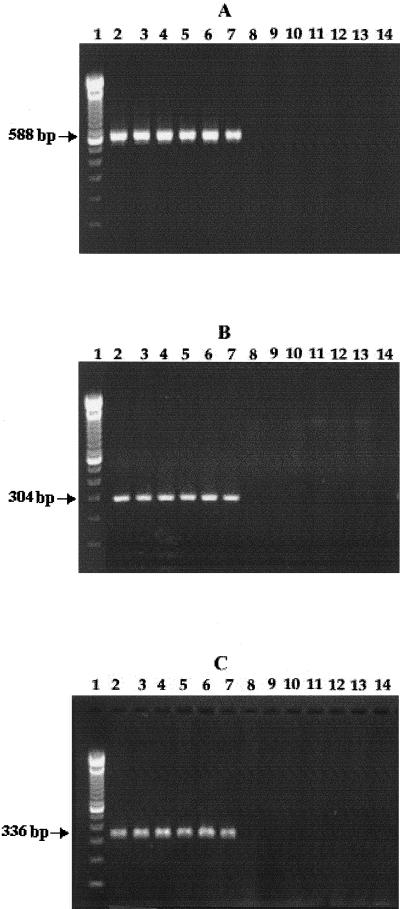
PCR amplification of ompW gene of V. cholerae using three combinations of primers (primers 1 and 2, primers 1 and 4, and primers 2 and 3; Table 2) yielded amplicons of 588, 304, and 336 bp, respectively. Initial experiments were carried out with about 50 V. cholerae and other strains. Representative data are shown in Fig. 1. It should be noted that the bacterial strains other than V. cholerae did not produce any amplified product under the experimental conditions used.
FIG. 1.
PCR amplification results obtained with bacterial strains using ompW-specific primer pairs 1 and 2 (A), 1 and 4 (B), and 2 and 3 (C). The bacterial strains used were V. cholerae O1 classical (lane 2), O1 El Tor (lane 3), O139 (lane 4), rough (lane 5), and non-O1/non-O139 (lanes 6 and 7). Other bacteria used were V. parahaemolyticus (lane 8), V. mimicus (lane 9), V. anguillarum (lane 10), V. alginolyticus (lane 11), V. furnissii (lane 12), Aeromonas spp. (lane 13), and enteroaggregative E. coli (lane 14). Lane 1 represents marker DNA of known molecular weights. The amplicon sizes are indicated by arrows.
Subsequent experiments were carried out with the ompW primer pair 1 and 2 because this produced an amplicon (588 bp) which differed considerably in size from the ctxA amplicon (301 bp) generated with the primers 7 and 8 (Table 2). PCR amplification data obtained with 254 strains of V. cholerae, 40 strains of other Vibrio spp., and 80 bacterial strains from other genera are presented in a summarized form (Table 3). While all V. cholerae strains were found to be positive by the ompW-based PCR assay, other organisms tested, including those belonging to other Vibrio species, were found to be negative. When these strains were subjected to PCR using toxR primers, only 4 of 233 V. cholerae strains tested were found to be negative. All four strains belonged to typeable non-O1/non-O139 serogroups. Noncholera vibrios and other bacterial species failed to yield any toxR amplicon when tested under comparable conditions.
TABLE 3.
Summarized PCR results obtained with V. cholerae and other bacteria using ompW and toxR-specific primers
| Strain | No. of strains positive/total no. of strains tested
|
|
|---|---|---|
| ompWa | toxRb | |
| V. cholerae O1c | 36/36 | 36/36 |
| V. cholerae O139 | 22/22 | 22/22 |
| V. cholerae non-O1/non-O139c | 176/176 | 155/159 |
| V. cholerae (rough/OUTd) | 20/20 | 16/16 |
| Other Vibrio sppe | 0/40 | 0/40 |
| Aeromonas spp. | 0/7 | 0/7 |
| E. colif,g | 0/28 | 0/28 |
| Shigella spp. | 0/19 | 0/19 |
| Salmonella spp. | 0/16 | 0/16 |
| Pseudomonas spp.g | 0/7 | 0/7 |
| Klebsiella spp. | 0/2 | 0/2 |
| S. aureusg | 0/1 | 0/1 |
DNA dot blot analysis using ompW and toxR probe.
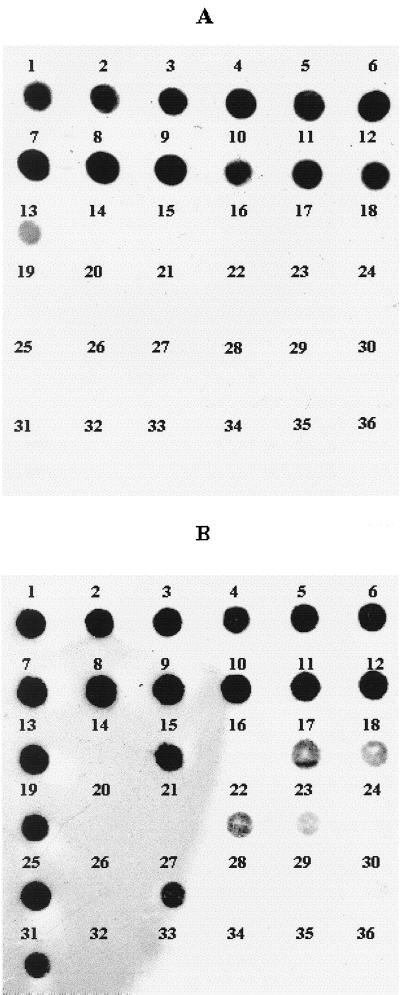
Strains belonging to different bacterial species were subjected to DNA dot blot analysis using ompW and toxR probes. The results presented in Table 1 show that all of the 18 V. cholerae strains tested produced positive signals with both of the probes. In contrast, of 40 strains belonging to other Vibrio species tested, only 1 V. mimicus strain gave a weak signal against the ompW probe, while 10 strains, e.g., V. mimicus (two strains), V. splendidus, V. alginolyticus, V. fluvialis, V. proteolyticus, V. aestuarianus, V. salmonicida, V. furnissii, and V. parahaemolyticus gave weak to strong signals with toxR probe (Table 1, Fig. 2). Twenty strains belonging to other bacterial species (e.g., Aeromonas spp., E. coli, Shigella spp., Salmonella spp., Klebsiella spp. Pseudomonas spp., and S. aureus) were found to be negative for both ompW and toxR genes in the dot blot assay (data not shown).
FIG. 2.
DNA dot blot hybridization test carried out with V. cholerae strains using ompW (A) and toxR (B) gene probes. The test strains used were V. cholerae O1 classical (O395 [blot 1], 569B [blot 2], and ATCC 14035 [blot 3]), O1 El Tor (PG27 [blot 4] and ATCC 39315 [blot 5]), O139 (Arg3 [blot 6] and SG25 [blot 7]), rough ALO46 (blot 8), non-O1/non-O139 (ATCC 25872 [blot 9], ATCC 25874 [blot 10], V5 [blot 11], and S7 [blot 12]), V. mimicus ATCC 33653 (blot 13), V. tyrogens (blot 14), V. alginolyticus ATCC 17749 (blot 15), V. anguillarum ATCC 19264 (blot 16), V. furnissii ATCC 35016 (blot 17), V. proteolyticus ATCC 15338 (blot 18), V. mimicus (blot 19), V. vulnificus (ATCC 33816 [blot 20] and ATCC 27562 [blot 21]), V. salmonicida ATCC 43839 (blot 22), V. parahaemolyticus (121 [blot 23] and RIMD 2210001 [blot 24]), V. splendidus ATCC 33125 (blot 25), V. carchariae ATCC 35084 (blot 26), V. aestuarianus ATCC 35048 (blot 27), V. nereis ATCC 25917 (blot 28), V. natriegens ATCC 14048 (blot 29), V. tubiashii ATCC 19109 (blot 30), V. fluvialis ATCC 33809 (blot 31), Aeromonas sp. (blot 32), E. coli ATCC 25922 (blot 33), Shigella sp. (blot 34), Salmonella sp. (blot 35), and P. aeruginosa ATCC 27853 (blot 36).
RFLP analysis of ompW amplicons.
The 588-bp ompW amplicons obtained from different V. cholerae strains using the primer pair 1 and 2 (Table 2) were digested with the restriction enzymes (HindIII, NdeI, HpaI). The RFLP patterns presented in Fig. 3 demonstrate identity among the V. cholerae strains with respect to these restriction sites in the ompW gene.
FIG. 3.
RFLP analysis of ompW amplicons of different V. cholerae strains using the restriction enzymes HindIII (A), NdeI (B), and HpaI (C). The bacterial strains used were V. cholerae O1 classical (lane 1), O1 El Tor (lane 2), O139 (lane 3), and non-O1/non-O139 (lanes 4 to 9). Lane 10 represents the uncut ompW amplicon shown for a comparison. The fragment sizes are indicated by arrows.
Nucleotide sequence analysis of ompW amplicons.
The ompW amplicons generated from five V. cholerae strains belonging to different serogroups and/or biotypes (O1/El Tor, O34, O37, O53, and O139) were subjected to nucleotide sequence analysis, and the data were compared with each other as well as with the published sequence data of an O1 classical strain 569B (8). The results (Table 4) showed only minimum variation (ranging between 0.2 and 2.2%) in the ompW sequence among these strains.
TABLE 4.
Variations in ompW sequences among V. cholerae strains
| V. cholerae strain (serogroups/biotypes) | Sequence variationsa
|
|||||
|---|---|---|---|---|---|---|
| O1/classical | O1/El Tor | O139 | O34 | O37 | O53 | |
| O1/classical | 12/588 | 13/588 | 13/588 | 7/588 | 11/588 | |
| O1/El Tor | 2.0 | 1/588 | 3/588 | 7/588 | 11/588 | |
| O139 | 2.2 | 0.2 | 2/588 | 6/588 | 10/588 | |
| O34 | 2.2 | 0.5 | 0.3 | 6/588 | 10/588 | |
| O37 | 1.2 | 1.2 | 1.0 | 1.0 | 4/588 | |
| O53 | 1.9 | 1.9 | 1.7 | 1.7 | 0.7 | |
Values in the upper-right triangle indicate the numbers of nucleotide differences/total number compared. The values in the lower-left triangle indicate the percentage of sequence variations.
Multiplex-PCR assay using ompW and ctxA primers.
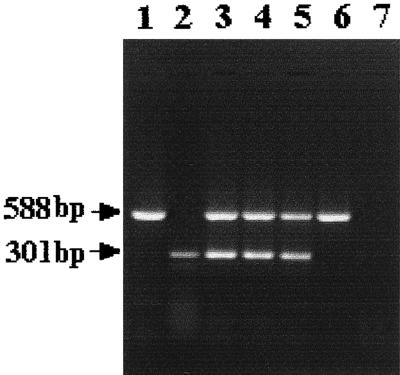
Simultaneous amplification of ompW and ctxA genes in a given reaction mixture generated amplicons of both ompW (588 bp) and ctxA (301 bp) in toxigenic V. cholerae strains belonging to the O1, O139, and non-O1/non-O139 serogroups (Fig. 4). As expected, a nontoxigenic strain yielded only the ompW amplicon (lane 6), while a V. mimicus strain failed to yield any amplicon under comparable conditions (lane 7).
FIG. 4.
Multiplex-PCR analysis of V. cholerae strains using primers 1 and 2 for ompW and primers 7 and 8 for ctxA. Bacterial strains used were V. cholerae O1 (lane 3), O139 (lane 4), non-O1/non-O139 (lane 5), a nontoxigenic non-O1/non-O139 (lane 6), and a V. mimicus strain (lane 7). The positions of the ompW and ctxA amplicons generated by the use of individual primer pairs are shown in lanes 1 and 2, respectively.
DISCUSSION
The toxR gene was shown to be involved in the regulation and expression of several genes of V. cholerae (19). Subsequent studies demonstrated the presence of toxR-related gene sequences in other organisms belonging to Vibrio spp., although their sequences showed considerable variations (14, 18, 22). As a matter of fact, the toxR gene was recently used as a probe for the species-specific identification of V. parahaemolyticus (12). Interestingly, this gene probe developed for V. parahaemolyticus failed to detect V. cholerae despite 52% identity in their toxR gene sequences. These results appear to be somewhat consistent with our data since the toxR probe for V. cholerae recognized only one, though weakly, of the seven V. parahaemolyticus strains tested (Table 1). However, as observed with the V. parahemolyticus toxR probe (12), the toxR probe for V. cholerae recognized organisms belonging to certain other Vibrio species with variable level of reactivities (Fig. 2). Incidentally, the organism V. alginolyticus was the common species recognized by both of the toxR probes.
The toxR primers used in this study were found to be quite specific (∼98%) for V. cholerae and were able to differentiate these from other Vibrio spp. (Table 3). These results suggest that one or both the primer sequences of V. cholerae toxR are likely to differ from the corresponding toxR sequences of other vibrios. That this may indeed be the case is supported by the information, though limited, available on toxR sequences (14, 18).
The ompW primers showed 100% specificity for all V. cholerae strains tested (Table 3). More importantly, the ompW gene probe did not hybridize with target DNAs of other bacteria except for showing a weak reaction with only one of six V. mimicus strains examined (Table 1, Fig. 2). This observation and the fact that ompW primers can differentiate between V. cholerae and V. mimicus strains assume considerable significance in view of the report that these two groups of organisms share common biochemical properties and serological markers (5). Therefore, the presence of ompW in V. cholerae strains, coupled with the fact that its nucleotide sequence remained practically unchanged among different V. cholerae strains, makes it a highly suitable genetic marker for the organism.
A literature survey showed that genes partially homologous to ompW of V. cholerae are present in certain other bacteria, e.g., E. coli, Aeromonas spp., etc. (9, 16). Several functions were proposed for the OmpW-related proteins in these bacteria, including their pore- or channel-forming (9) and colicin receptor properties (20). Although the precise function of the OmpW protein in V. cholerae is not yet known, it may play a role in the adherence process, which is likely to facilitate the survival of the organism within the host or in the environment or both (27). Preliminary genome data available (30) demonstrate the presence of two chromosomes in V. cholerae. It is important to note that while the ompW gene is present in the smaller chromosome, the toxR gene is located in the larger chromosome of the organism.
Epidemic-causing strains of V. cholerae belong to the O1 or O139 serogroups and produce cholera toxin, which is the major contributing factor for profuse diarrhea (cholera gravis) (10). However, genes related to ctxAB have also been demonstrated in a number of strains of non-O1/non-O139 V. cholerae (6, 17, 21) that are responsible for diarrheal episodes in humans, causing a considerable public health problem. The multiplex PCR described here is likely to facilitate the rapid detection of toxigenic V. cholerae strains and therefore play a key role in the cholera surveillance program.
The rRNA nucleotide sequences have provided valuable information for the identification and taxonomy of different bacterial species. Unfortunately, the 16S rRNA sequences of different Vibrio spp. show minimal differences, making species-specific identification difficult (13, 24). In a recent study, Chun et al. (3) were able to design primers based on subtle differences in the nucleotide sequences of 16S-23S rRNA intergenic spacer regions of V. cholerae and V. mimicus. PCR amplification using the primer pair was able to generate amplicons, though of variable sizes (295 to 310 bp), from several V. cholerae strains tested. The data presented in this study, however, demonstrate that PCR primers 1 and 2 designed on the basis of ompW sequence (uniquely present in V. cholerae) generate amplicons of identical sizes (588 bp) from all V. cholerae strains, which should provide a very rapid and reliable method for the species-specific identification of V. cholerae and for their differentiation from other bacteria. Further, identification of V. cholerae strains harboring genes for cholera toxin by one-step multiplex PCR assay (Fig. 4) is likely to enhance the scope of this method significantly toward the screening of both toxigenic and nontoxigenic V. cholerae strains of clinical as well as environmental origin.
ACKNOWLEDGMENTS
This study was supported by grants from the Indian Council of Scientific and Industrial Research (CSIR). The work was also supported, in part, by the Japan International Cooperation Agency (JICA/NICED, project 054-1061-EO).
The helpful technical assistance of Prabal Gupta is also acknowledged.
REFERENCES
- 1.Albert M J, Islam D, Nahar S, Qadri F, Falklind S, Weintraub A. Rapid detection of Vibrio cholerae O139 Bengal from stool specimens by PCR. J Clin Microbiol. 1997;35:1663–1635. doi: 10.1128/jcm.35.6.1633-1635.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bagchi K, Echeverria P, Arthur J D, Sethabutr O, Serichantalergs O, Hoge C W. Epidemic diarrhoea caused by Vibrio cholerae non-O1 that produced heat-stable toxin among Khmers in a camp in thailand. J Clin Microbiol. 1993;31:1315–1317. doi: 10.1128/jcm.31.5.1315-1317.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chun J, Huq A, Colwell R R. Analysis of 16S–23S rRNA intergenic spacer regions of Vibrio cholerae and Vibrio mimicus. Appl Environ Microbiol. 1999;65:2202–2208. doi: 10.1128/aem.65.5.2202-2208.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dalsgaard A, Albert M J, Taylor D N, Shimada T, Meza R, Serichantalergs O, Echeverria P. Characterization of Vibrio cholerae non-O1 serogroup obtained from an outbreak of diarrhea in Lima, Peru. J Clin Microbiol. 1995;33:2715–2722. doi: 10.1128/jcm.33.10.2715-2722.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davis B R, Fanning G R, Madden J M, Steigerwalt A G, Bradford H B, Jr, Smith H L, Jr, Brenner D J. Characterization of biochemically atypical Vibrio cholerae strains and designation of a new pathogenic species Vibrio mimicus. J Clin Microbiol. 1981;14:631–639. doi: 10.1128/jcm.14.6.631-639.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ghosh C, Nandy R K, Dasgupta S K, Nair G B, Hall R H, Ghose A C. A search for cholera toxin (CT), toxin coregulated pilus (TCP), the regulatory element ToxR and other virulence factors in non-O1/non-O139 Vibrio cholerae. Microb Pathog. 1997;22:199–208. doi: 10.1006/mpat.1996.0105. [DOI] [PubMed] [Google Scholar]
- 7.Hoshino K, Yamasaki S, Mukhopadhyay A K, Chakraborty S, Basu A, Bhattacharya S K, Nair G B, Shimada T, Takeda Y. Development and evaluation of a multiplex PCR assay for rapid detection of toxigenic Vibrio cholerae O1 and O139. FEMS Immunol Med Microbiol. 1998;20:201–207. doi: 10.1111/j.1574-695X.1998.tb01128.x. [DOI] [PubMed] [Google Scholar]
- 8.Jalajakumari M B, Manning P A. Nucleotide sequence of the gene, ompW, encoding a 22 kDa immunogenic outer membrane protein of Vibrio cholerae. Nucleic Acids Res. 1990;18:2180. doi: 10.1093/nar/18.8.2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jeanteur D, Gletsu N, Pattus F, Buckley J T. Purification of Aeromonas hydrophila major outer membrane proteins: N-terminal sequence analysis and channel-forming properties. Mol Microbiol. 1992;22:3355–3363. doi: 10.1111/j.1365-2958.1992.tb02203.x. [DOI] [PubMed] [Google Scholar]
- 10.Kaper J B, Morris J G, Jr, Levine M M. Cholera. Clin Microbiol Rev. 1995;8:48–86. doi: 10.1128/cmr.8.1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keasler S P, Hall R H. Detecting and biotyping Vibrio cholerae O1 with multiplex polymerase chain reaction. Lancet. 1993;341:1661. doi: 10.1016/0140-6736(93)90792-f. [DOI] [PubMed] [Google Scholar]
- 12.Kim Y B, Okuda J, Matsumoto C, Takahashi N, Hashimoto S, Nishibuchi M. Identification of Vibrio parahaemolyticus strains at the species level by PCR targeted to the toxR gene. J Clin Microbiol. 1999;37:1173–1177. doi: 10.1128/jcm.37.4.1173-1177.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kita-Tsukamoto K, Oyaizu, Nanba K, Shimidu U. Phylogenetic relationships of marine bacteria, mainly members of the family Vibrio cholerae, determined on the basis of 16S rRNA sequences. Int J Syst Bacteriol. 1993;43:8–19. doi: 10.1099/00207713-43-1-8. [DOI] [PubMed] [Google Scholar]
- 14.Lin Z, Kumagai K, Baba K, Mekalanos J J, Nishibuchi M. Vibrio parahaemolyticus has a homolog of the Vibrio cholerae ToxRS operon that mediates environmentally induced regulation of the thermostable direct hemolysin gene. J Bacteriol. 1993;175:3844–3855. doi: 10.1128/jb.175.12.3844-3855.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller V L, Taylor R K, Mekalanos J J. Cholera toxin transcriptional activator ToxR is a transmembrane DNA binding protein. Cell. 1987;48:271–279. doi: 10.1016/0092-8674(87)90430-2. [DOI] [PubMed] [Google Scholar]
- 16.Molloy M P, Herbert B R, Walsh B J, Tyler M I, Traini M, Sanchez J C, Hochstrasser D F, Williams K L, Gooley A A. Extraction of membrane proteins by differential solubilization for separation using two-dimensional gel electrophoresis. Electrophoresis. 1998;19:837–844. doi: 10.1002/elps.1150190539. [DOI] [PubMed] [Google Scholar]
- 17.Morris J G., Jr Non-O group 1 Vibrio cholerae: a look at the epidemiology of an occasional pathogen. Epidemiol Rev. 1990;12:179–191. doi: 10.1093/oxfordjournals.epirev.a036052. [DOI] [PubMed] [Google Scholar]
- 18.Osorio C R, Klose K E. A region of the transmembrane regulatory protein ToxR that tethers the transcriptional activation domain to the cytoplasmic membrane displays wide divergence among Vibrio species. J Bacteriol. 2000;182:526–528. doi: 10.1128/jb.182.2.526-528.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ottemann K M, Mekalanos J J. Regulation of cholera toxin expression. In: Wachsmuth I K, Blake P A, Olsvik O, editors. Vibrio cholerae and cholera: molecular to global perspectives. Washington, D.C.: ASM Press; 1994. pp. 177–185. [Google Scholar]
- 20.Pilsl H, Smajs D, Braun V. Characterization of colicin S4 and its receptor, OmpW, a minor protein of the Escherichia coli outer membrane. 1999. J Bacteriol. 1999;181:3578–3581. doi: 10.1128/jb.181.11.3578-3581.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Popovic T, Fields P I, Olsvik O. Detection of cholera toxin genes. In: Wachsmuth I K, Blake P A, Olsvik O, editors. Vibrio cholerae and cholera: molecular to global perspectives. Washington, D.C.: ASM Press; 1994. pp. 41–51. [Google Scholar]
- 22.Reich K A, Schoolnik G K. The light organ symbiont Vibrio fisheri possesses a homolog of the Vibrio cholerae transmembrane transcriptional activator ToxR. J Bacteriol. 1994;176:3085–3088. doi: 10.1128/jb.176.10.3085-3088.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rudra S, Mahajan R, Mathur M, Kathuria K, Talwar V. Cluster of cases of clinical cholera due to Vibrio cholerae O10 in east Delhi. Indian J Med Res. 1996;103:71–73. [PubMed] [Google Scholar]
- 24.Ruimy R, Breittmayer V, Elbaze P, Lafay B, Boussemart O, Gauthier M, Christine R. Phylogenetic analysis and assessment of the genera Vibrio, Photobacterium, Aeromonas and Plesiomonas deduced from small subunit rRNA sequences. Int J Syst Bacteriol. 1994;44:416–426. doi: 10.1099/00207713-44-3-416. [DOI] [PubMed] [Google Scholar]
- 25.Sakazaki R. Bacteriology of Vibrio and related organisms. In: Barua D, Greenough III W B, editors. Cholera. New York, N.Y: Plenum Medical Book Company; 1992. pp. 37–55. [Google Scholar]
- 26.Sambrook J, Fritsch E F, Maniatis T. Appendix E: commonly used techniques in molecular cloning. In: Nolan C, editor. Molecular cloning: a laboratory manual. New York, N.Y: CSH Laboratory Press; 1989. pp. E1–E39. [Google Scholar]
- 27.Sengupta T K. Identification and immunological characterisation of cell surface proteins responsible for intestinal adhesion and colonisation of diarrhoeagenic non-O1 Vibrio cholerae. Ph.D. thesis. Calcutta, India: University of Calcutta; 1995. [Google Scholar]
- 28.Sharma C, Thungapathra M, Ghosh A, Mukhopadhyay A K, Basu A, Mitra R, Basu I, Bhattacharya S K, Shimada T, Ramamurthy T, Takeda T, Yamasaki S, Takeda Y, Nair G B. Molecular analysis of non-O1, non-O139 Vibrio cholerae associated with an unusual upsurge in the incidence of cholera-like disease in Calcutta, India. J Clin Microbiol. 1998;36:756–763. doi: 10.1128/jcm.36.3.756-763.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shimada T, Arakawa E, Itoh K, Okitsu T, Matsushima A, Asai Y, Yamai S, Nakazato T, Nair G B, Albert M J, Takeda Y. Extended serotyping scheme for Vibrio cholerae. Curr Microbiol. 1994;28:175–178. [Google Scholar]
- 30.Trucksis M, Michalski J, Deng Y K, Kaper J B. The Vibrio cholerae genome contains two unique circular chromosomes. Proc Natl Acad Sci USA. 1998;95:14464–14469. doi: 10.1073/pnas.95.24.14464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wilson K. Preparation and analysis of DNA. In: Ausubel M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Vol. 1. New York, N.Y: John Willey & Sons, Inc.; 1994. pp. 2.4.1–2.4.2. [Google Scholar]
- 32.Yamai S, Okitsu T, Shimada T, Kaatsube Y. Serogroup of Vibrio cholerae non-O1/non-O139 with specific reference to their ability to produce cholera toxin and addition of novel serogroups. J Jpn Infect Dis. 1997;71:1037–1045. doi: 10.11150/kansenshogakuzasshi1970.71.1037. [DOI] [PubMed] [Google Scholar]