Abstract
The recent availability of genome-wide sequencing techniques has allowed systematic screening for molecular signatures of adaptation, including in nonmodel organisms. Host–pathogen interactions constitute good models due to the strong selective pressures that they entail. We focused on an adaptive event which affected the poplar rust fungus Melampsora larici-populina when it overcame a resistance gene borne by its host, cultivated poplar. Based on 76 virulent and avirulent isolates framing narrowly the estimated date of the adaptive event, we examined the molecular signatures of selection. Using an array of genome scan methods based on different features of nucleotide diversity, we detected a single locus exhibiting a consistent pattern suggestive of a selective sweep in virulent individuals (excess of differentiation between virulent and avirulent samples, linkage disequilibrium, genotype–phenotype statistical association, and long-range haplotypes). Our study pinpoints a single gene and further a single amino acid replacement which may have allowed the adaptive event. Although our samples are nearly contemporary to the selective sweep, it does not seem to have affected genome diversity further than the immediate vicinity of the causal locus, which can be explained by a soft selective sweep (where selection acts on standing variation) and by the impact of recombination in mitigating the impact of selection. Therefore, it seems that properties of the life cycle of M. larici-populina, which entails both high genetic diversity and outbreeding, has facilitated its adaptation.
Keywords: population genomics, plant–pathogen interactions, coevolution, genome-wide association studies, genome scan
Significance
Fungi are common plant pathogens and can lead to dramatic yield losses in both agriculture and sylviculture. Understanding how they adapt to evade the defenses of their hosts is important in guiding cultural practices and selection programs. In this study, using a combination of population genomic analyses, we identify a single amino acid change potentially responsible for a major adaptive event in the rust fungus Melampsora larici-populina to overcome the defenses of its host. We also find that the selection footprints are consistent with a scenario where the beneficial mutation was already present before the onset of selection (a so-called soft selective sweep), illustrating that the genetic diversity of pathogens can promote fast adaptation.
Introduction
Understanding adaptation in response to natural or anthropic forces is one of the major goals of the study of molecular evolution. In most cases, evolutionary research focuses on events that occur in the past, such as domestication (Haudry et al. 2007; Li et al. 2014; Wang et al. 2014), divergence of populations (Begun et al. 2007; Liti et al. 2009; Pugach and Stoneking 2015), adaptation to environmental conditions (Namroud et al. 2008; Ellison et al. 2011; Burke 2012), or speciation (Kulathinal et al. 2009). In comparison, few biological models allow to address the evolutionary process in contemporary events (but see, e.g., Le Corre et al. 2020). Due to their highly dynamic nature, host–pathogen interactions offer opportunities to study adaptation processes at work. Indeed, host–pathogen interactions evolve constantly and rapidly, due to strong and reciprocal selection pressures, changing environmental conditions and, recently, anthropic interactions (Stukenbrock and Bataillon 2012; Zaman et al. 2014). Microorganisms pathogenic to plants are considered to be capable of extremely fast evolution (Raffaele and Kamoun 2012; Upson et al. 2018; Frantzeskakis et al. 2019).
It has been shown that the fate of an infection depends on a complex interplay of mechanisms involving both partners of the interaction. The pathogen produces molecules manipulating the host to allow entry and co-optation of resources, but also preventing the activation of the host defense machinery (Koenig et al. 2021; Meisrimler et al. 2021). Plants have developed both nonspecific and specific immunity systems to detect pathogens and prevent infection (Jones and Dangl 2006). Plant immunity systems are based on resistance genes which target molecules characteristic of a given pathogen species or even strain. Molecules produced by pathogens and enabling host defense responses (thereby preventing successful infection) are named avirulence factors. Consequently, genes encoding these factors are referred to as avirulence genes. In contrast with nonspecific immunity, such host resistance genes allow complete resistance against the targeted pathogens. Selective pressures on genes controlling these systems can be strong and lead to fast coevolution (Tellier et al. 2014; Ebert and Fields 2020). In particular, genes coding for proteins interacting directly to determine complete immunity, such as pairs constituted of resistance proteins and avirulence factors, are likely to be exposed to the strongest selective pressures (Jones and Dangl 2006; Brown and Tellier 2011).
The strength of selective pressure caused by total host resistance can be extremely high if host resistance results in a substantial reduction of the ecological niche of the pathogen. This is typically the case when disease-resistant hosts are dominant in the environment. Then a mutation restoring virulence is expected to undergo an extremely fast increase in frequency toward fixation, resulting in a strong selective sweep of genomic variation. However, experimental evidence and modeling suggest that relatively mild selective sweeps could be expected (Messer and Petrov 2013), because adaptation based on variants already segregating in the population (soft selective sweep) is expected to be actually faster than adaptation requiring to wait that the appropriate mutation occurs (hard selective sweep).
Identifying genomic regions subjected to selection has long been of interest to evolutionary biologists (Haasl and Payseur 2016). The recent emergence of high-throughput sequencing technologies providing genome-wide coverage (Mardis 2008) raises opportunities to detect loci involved in adaptation. Modern sequencing techniques allow to dramatically increase both the number of loci characterized and the number of individuals used in population samples. The rationale underlying the search for the signatures of adaptation in molecular data has been proposed long before genome sequencing became routine (Lewontin and Krakauer 1973), whereas more recent methodologies have been introduced to leverage the wealth of data that became available thanks to whole-genome sequencing. Essentially it boils down to scanning the genome to pinpoint genes or regions exhibiting characteristic footprints left by selective sweeps (Nielsen et al. 2005; Aguileta et al. 2010). These footprints include a deformation of the allele frequency spectrum, an excess of linkage disequilibrium (LD) and a reduction in genetic diversity around the selected region. It can also include an excess of differentiation in allelic frequencies between individuals from different subpopulations (e.g., if the sweep only affected a single subpopulation). Note that any one of these can be potentially mimicked by demographic effects such as nonequilibrium or undocumented substructure that can result in an inflated rate of false positives, especially for methods that rely on an assumed model (Siol et al. 2010). We can expect however that combining different complementary methods looking for different genomic signatures can increase power, reduce sensitivity to confounding factors (which are unlikely to affect different methods in a similar manner) and increase precision of the detection of selective sweeps (Grossman et al. 2010). Finally if one can show a statistical association between a selected trait and a genotype (like in a QTL or a genome-wide association study [GWAS] analysis), this provides powerful complementary evidence.
We propose to adopt this approach to detect the determinants of a recent adaptation event in a plant pathogenic fungus. We focus on the poplar rust fungus, Melampsora larici-populina, which is a major threat for cultivated poplar in Europe (Pinon and Frey 1997). Melampsora larici-populina needs two hosts from unrelated genera to complete its life cycle: Populus on which it performs several asexual reproduction cycles during summer and autumn and Larix on which it performs a single sexual reproduction cycle once a year in spring. Melampsora larici-populina is particularly damaging on cultivated poplar hybrids, mostly because of their intensive monoclonal cultivation over several decades (Gerard et al. 2006). Many poplar cultivars carrying qualitative resistances were bred, but M. larici-populina eventually overcame all resistances. The most significant resistance breakdown event occurred in 1994 and overcame the RMlp7 resistance, which was at the time carried by most cultivated poplars in Northern France and Belgium. This event has led to the invasion of France by virulent individuals in less than 5 years (Xhaard et al. 2011).
In this article, we focus on this resistance and we denote M. larici-populina isolates that can successfully infect poplar hosts harboring RMlp7 as virulent, as opposed to avirulent isolates.
An analysis of the French population structure of M. larici-populina showed a strong impact of the resistance breakdown (Persoons et al. 2017). The analysis of 594 isolates from a laboratory collection sampled before and after the resistance breakdown identified three genetic groups. These groups have been named “wild” (isolates sampled in a region where cultivated poplars are sparse), “fossil” (isolates sampled before the breakdown and shortly afterward), and “cultivated” (isolates sampled only after the breakdown). All isolates from the two former groups were found to be avirulent and all isolates except one (98AB07) from the latter group were found to be virulent. Furthermore, signatures of population expansion have been detected in the “cultivated” group over time (Persoons et al. 2017).
In this study, we selected 76 isolates from the three genetic groups found in Persoons et al. (2017), spread into four samples corresponding each to a given year. We defined two samples in the “cultivated” (virulent) group, one immediately after the resistance breakdown and another 4 years later, following completion of the putative selective sweep (fixation of the virulence allele in the M. larici-populina population). Using over a million single-nucleotide polymorphisms (SNPs), we conducted several genome scan approaches to detect specific regions of the genome potentially involved in virulence. Genome scan methods consistently highlighted a common candidate, which is the first ever described putative avirulence gene in M. larici-populina. However, in spite of the potential strength of selective pressure to overcome resistance, molecular diversity indicates a relatively mild population bottleneck and the signature of selective sweep are rather locally restricted.
Results
Sampling and Polymorphism Detection
We selected 76 M.larici-populina isolates organized in four samples framing the breakdown event, each of them representing a single year of collection: sample Avr1993 (n = 18) from the “fossil” group, samples Vir1994 (n = 18) and Vir1998 (n = 21) from the “cultivated” group, and sample Wild2008 (n = 19) from the “wild” group (supplementary table 1, Supplementary Material online).
The genome of those 76 M. larici-populina isolates was sequenced with the Illumina technology (150-bp paired reads). An average of 68 million reads was generated per isolate (ranging from 26 to 143 million reads). On average, 78% of the reads aligned on the chromosomes of version 2 of the reference genome (Duplessis et al. 2011) generating a sequencing depth of 80× (±24) (supplementary table 1, Supplementary Material online). In total, the site calling procedure identified 81,174,498 fixed sites and 1,125,506 SNPs covering more than 80% of the total length of the 18 chromosomes (supplementary table 2, Supplementary Material online). The remaining 19% of sites were not called due to missing data or, much less frequently, sites with more than two alleles. The amount of variation is higher than previously identified (Persoons et al. 2014), with more than 1% of sites with enough data exhibiting polymorphism.
Genetic Structure
To assess the genetic structure of our data set, we performed a nonparametric structure analysis using a discriminant analysis of principal components (DAPC; Jombart et al. 2010). The Bayesian information criterion (BIC) points to two groups (supplementary fig. 1A, Supplementary Material online), gathering on one hand the Avr1993 and Wild2008 samples and on the other hand the Vir1994 and Vir1998 samples (supplementary fig. 1B, Supplementary Material online). We analyzed the results with three and four groups as well, due to the proximity of BIC values. With three groups, the Avr1993 and Wild2008 samples are separated (supplementary fig. 1C, Supplementary Material online), in a structure identical to Persoons et al. (2017). With four groups, a part of Vir1994 isolates are apart from other Vir1994 isolates who are clustered with the whole Vir1998 sample (supplementary fig. 1D, Supplementary Material online). This group of isolates seems to be only partially related to the region of origin of samples since it includes all five isolates sampled in Belgium, two of the eight isolates sampled in Nord-Pas-de-Calais, one of the three isolates sampled in Picardie and the single isolate sampled in Center. Up to four groups, all assignation coefficients in DAPC analyses are virtually equal to 1 (supplementary fig. 1E–G, Supplementary Material online).
In addition, we reconstructed a neighbor-joining tree based on pairwise distances computed from all SNPs (fig. 1). This tree distinguishes three clades: the Wild2008 sample, the Avr1993 sample and a third clade gathering Vir1994 and Vir1998 samples. These clades exhibit a star-like topology, especially the Wild2008 and Avr1993 samples (with the exception of a pair of Avr1993 samples which exhibit high pairwise similarity). The Vir1994 and Vir1998 samples are interspersed within the third clade that exhibits comparatively longer internal branches. The group of Vir1994 samples that forms a group of its own in the DAPC analysis with four groups forms a subclade, although it does not appear to show marked pairwise divergence with respect to the other isolates.
Fig. 1.
Unrooted neighbor-joining tree of all isolates based on genomic distances. The distances are expressed in number of differences per compared position (due to missing data, the number of compared positions is less than the number of polymorphic sites and varies between comparisons). Colored disks indicate sample membership. Isolates of the Vir1994 sample that are assigned to a specific group by the DAPC analysis are indicated by an additional red circle.
The analysis of genome-wide statistics measuring pairwise differentiation between samples gives results consistent with the tree structure (supplementary table 4I–N, Supplementary Material online). Samples Vir1994 and Vir1998 exhibit strong similarity (Da = 0.0035), as well as Avr1993 and Wild2008 but with a lesser similarity (Da = 0.0109). Between these two pairs of samples, the pairwise comparison results might have been influenced by the presence of substructure within some of the samples with in particular less similarity between samples Avr1993 and Vir1994 than between Avr1993 and Vir1998 (Da = 0.0155 and 0.0142, respectively).
The separation of the Vir1994 and Vir1998 samples on one hand and Avr1993 and Wild2008 on the other hand therefore seems to be the main feature of population structure, but it actually represents a small proportion of the total genetic variance. Weir and Cockerham’s estimation of the fixation index between these two pairs of samples () gives a value of 4% of the total variance (supplementary table 4A, Supplementary Material online). In addition, population substructuration is visible in all but one (Wild2008) sample and especially strong for Vir1994.
Linkage Disequilibrium
In order to monitor the consequences of the assumed population bottleneck and the subsequent fast population growth, we compared the LD decay with physical distance between the four samples (supplementary fig. 2, Supplementary Material online). We found that the average LD drops below r2 = 0.1 before 95–165 kb according to the sample. The Wild2008 sample appears to be the one where LD drops at the fastest rate and Vir1994 at the slowest rate (average r2 below 0.2 before 33 kb for Wild2008 and before 60 kb for Vir1994; average r2 of 0.0955 at 100 kb for Wild2008 and 0.1459 for Vir1994), the Vir1998 and Avr1993 samples having a slightly slower rate of decay than Wild2008 (supplementary table 5, Supplementary Material online).
A survey of the level of pairwise linkage at the chromosome level was performed (supplementary fig. 3, Supplementary Material online). The analysis highlights numerous blocks with extended LD that are shared by several (and often all) samples, such as, for chr01, a region between 5.5 and 6.0 Mb. At a genome-wide scale, it is visible that the overall levels of LD are more elevated in sample Vir1994 and lower in sample Wild2008.
Population Genomics Statistics
At a genome-wide scale, statistics describing nucleotide polymorphism capture the consequences of demographic history. Besides the population similarity statistics addressed in a previous section, we computed statistics characterizing the levels and structure of polymorphism (table 1; see also supplementary table 4, Supplementary Material online) and tested their significance based on coalescent simulations assuming no difference between samples and no change over time (supplementary table 8, Supplementary Material online).
Table 1.
Genome-Wide Diversity Statistics
| Avr1993 | Vir1994 | Vir1998 | Wild2008 | Whole Data Set | |
|---|---|---|---|---|---|
| S | 669,871 | 607,716 | 705,365 | 787,963 | 1,125,506 |
| 0.00199 | 0.00182 | 0.00204 | 0.00231 | 0.00248 | |
| π | 0.00193 | 0.00184 | 0.00192 | 0.00184 | 0.00199 |
| D | −0.12 | 0.05 | −0.22 | −0.77 | −0.65 |
| 0.21 | 0.33 | −0.05 | −0.56 | −0.34 | |
| 0.11 | 0.29 | −0.14 | −0.76 | −0.58 | |
| −0.015 | 0.019 | −0.012 | −0.004 | 0.055 |
Note.—S, number of polymorphic sites; , Watterson’s estimator of θ; π, nucleotide diversity; D, Tajima’s D; , Fu and Li’s D without outgroup; , Fu and Li’s F without outgroup; , deficit of heterozygotes.
We found substantial levels of diversity, with both and π around 0.002 per site, for the whole data set as well as for the four samples. The Wild2008 sample shows the highest value of but comparatively lower nucleotide diversity. Combined with the results of the tests of neutrality (Tajima’s D and Fu and Li’s and ), this observation suggests that the population from which this sample was collected was not at the mutation–drift equilibrium. The Avr1993 sample exhibits significant values of all three neutrality tests, also suggesting a deviation from the expectations of a nonsubdivised, constant-sized population.
The virulent population shows signature of the expected bottleneck with a marked reduction of diversity and a significantly negative value of Fu and Li’s (indicating a deficit of singleton mutations) in Vir1994 and a recovery of diversity in Vir1998 with an excess of putatively new polymorphisms with unbalanced frequencies (positive Tajima’s D).
values indicate a substantial deficit of heterozygotes in the whole data set, likely due to sample structure. Within samples, deviations are not significantly different from a model without source of inbreeding (such as substructure).
Localization of an Avirulence Gene Candidate
In order to identify the locus or loci involved in the resistance breakdown, we compared the results of several selective sweep detection methods addressing different signatures of a selective sweep event. These signatures fall into three main categories: deformation of the allele frequency spectrum, excess of differentiation between virulent and avirulent isolates and extended LD.
Allele Frequency Spectrum
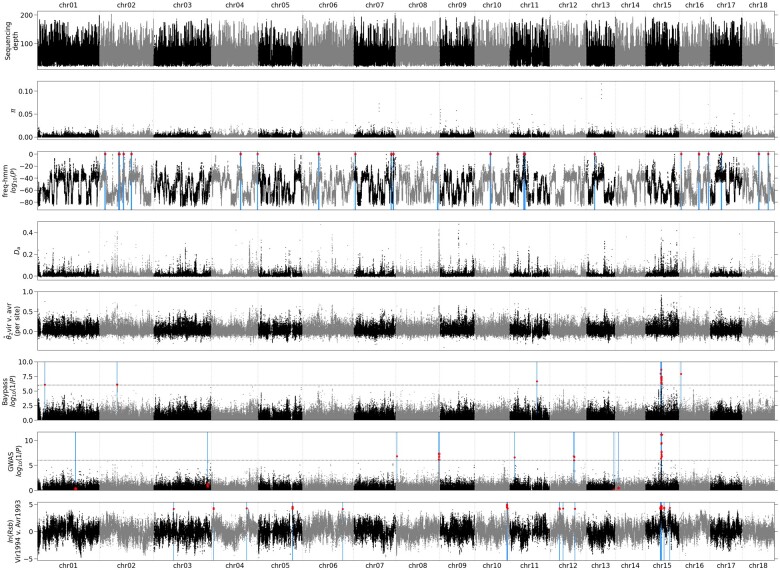
The effect of selection on the allele frequency spectrum should be captured by Tajima’s D. We computed Tajima’s D on a sliding window for all samples separately (supplementary fig. 4, Supplementary Material online) but did not find any clear peak anywhere in the genome. A Bayesian, hidden Markov model method aiming to detect selective sweeps (freq-hmm) was also applied on the Vir1998 sample (when the selective sweep should be completed). We detected 22 potential selective sweeps on 11 out of 18 chromosomes, containing together a total of 614,822 SNPs (fig. 2, third panel, supplementary table 6, Supplementary Material online).
Fig. 2.
Selective sweep signatures in the Melampsora larici-populina genome. π, nucleotide diversity; , net pairwise population divergence; , Weir and Cockerham’s cluster fixation index; ln(Rsb), excess of long-distance haplotype homozygosity. For genome scan methods, the test P value is presented. The average sequencing depth over isolates is given for all considered sites. π and Da are computed over 1,000-bp overlapping windows (step: 250 bp). Each value is placed at the window midpoint. For selective sweep detection methods, the test statistic is given for all SNPs. Significant or outlier values are indicated by larger red disks and their position is outlined by a blue vertical line. For the GWAS results, the SNPs which are significant as cofactors are indicated by a star and the SNPs only passing the threshold in the model without cofactors by red disks.
Differentiation
For addressing signatures of adaptive differentiation between avirulent and virulent isolates, we screened the variation of Weir and Cockerham’s (analogous to ) computed per site. measures the differentiation between Avr1993 and Wild2008 samples on one hand and Vir1994 and Vir1998 on the other hand. varies widely along the genome with clear peaks at different locations, the highest being in chr15 at position 2,198,745 with = 0.91 (fig. 2, fifth panel). A comparison with other differentiation statistics (fig. 2, fourth panel, supplementary fig. 4, Supplementary Material online) shows that the region with the highest differentiation varies according to the statistic, with for example Jost’s D and Hedrick’s pointing to a region near position 2.61 Mb of chr09. It should be noted that is the only statistic that separates the effect of virulent versus avirulent divergence from the effect of divergence between samples.
To screen for potential loci involved in adaptive divergence between virulent and avirulent isolates, we used a Bayesian method aiming to detect adaptive divergence based on allele frequency differentiation between samples (BayPass). The BayPass package (Gautier 2015) provides two models. The core model is based on the sole XtX statistic, which is an analogous to (Güenther and Coop 2013). We identified 17 candidate SNPs, of which 12 are located on chr15 (fig. 2, sixth panel), including the most significant (located at position 2,198,745), and nine of the ten most significant, with positions ranging from 2,103,540 to 2,253,676 (supplementary table 6, Supplementary Material online). The second model of BayPass (covariate model) allows to incorporate phenotypic information. We set the samples Avr1993 and Wild2008 as avirulent and the samples Vir1994 and Vir1998 as virulent. The results are nearly identical, with 16 candidate SNPs of which 15 (including the 12 on chr15) being also detected by the core model.
Association Genetics
We performed a GWAS to identify SNPs significantly associated with virulence. The best model includes seven SNPs as cofactors which, taken together, explain all the phenotypic variance (fig. 2, seventh panel, supplementary fig. 5, Supplementary Material online). The first cofactor included in the model corresponds to the SNP at position 2,237,229 on chr15 which was the most significant in the initial GWAS scan. This SNP alone captures 69% of the phenotypic variance (supplementary fig. 5, Supplementary Material online), and when treated as a cofactor, all the signal in the region drops below significance level, suggesting that there is not more than a single potential causal variant in this region. We consider the seven above-mentioned SNPs as candidates, as well as 53 other SNPs that pass the threshold in the model without cofactors (supplementary table 6, Supplementary Material online).
Extended Haplotype Homozygosity
Finally, we investigated extended haplotype homozygosity (EHH), which identifies long-range tracts of homozygosity as a potential signature of recent positive selection (Sabeti et al. 2007). We computed iEG (integrated EHH) for all sites of the genome separately for the four samples, as well as the ln(Rsb) statistic which characterizes the excess of long-range haplotypes of one population with respect to another (Tang et al. 2007). We compared sample Vir1994 to sample Avr1993, because EHH is meant to detect recent or even ongoing selective sweeps, and applied an arbitrary threshold to pick a proportion of of SNPs. This approach yields 101 candidate SNPs, 61 of them belonging to chr15 and 57 being restricted to the region between 2 and 2.3 Mb of chr15 (fig. 2, eighth panel, supplementary table 6, Supplementary Material online).
Congruence between Genome Scan Methods
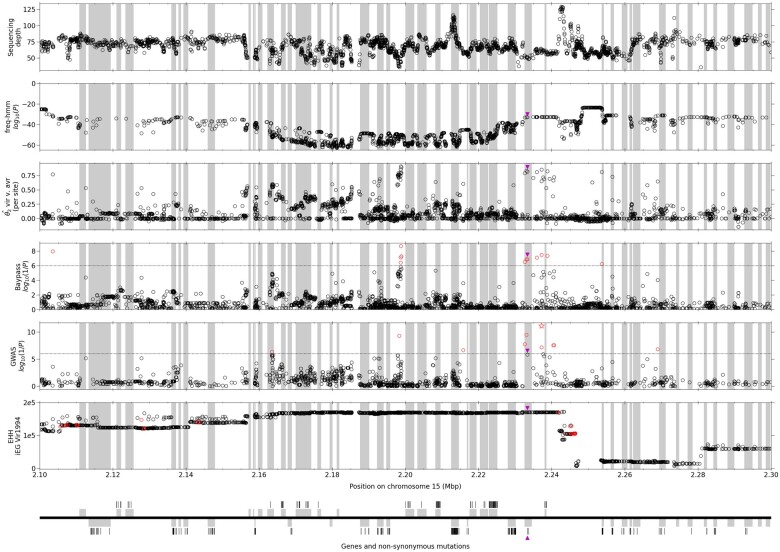
A comparison of the SNPs found using the different methods shows that, of the 15 SNPs which are shared between the two models of BayPass, only three located between 2.2 and 2.4 Mb on chr15 are shared with GWAS, including the top GWAS candidate, at position 2,237,229 of chr15 and none with the other methods. However, this position is within the region (between 2 and 2.3 Mb) containing the majority of EHH candidates. Based on BayPass, GWAS, and EHH results, we focused on the region of chr15 between positions 2.10 and 2.30 Mb (fig. 3). Taken together, BayPass and GWAS candidates are clustered in two regions: in an intergenic region near position 2,198,500 which is upstream of the two flanking genes (jgi.p|Mellp2_3|84583 and jgi.p|Mellp2_3|84584) and in another region spanning three genes (jgi.p|Mellp2_3|1427900, jgi.p|Mellp2_3|71388, and jgi.p|Mellp2_3|104811). Of these SNPs, three fall in gene jgi.p|Mellp2_3|1427900 (positions 2,232,600, 2,233,055 and 2,233,369). The former falls in the 3′-UTR, the second in an intron and the latter causes a nonsynonymous change. The top GWAS candidate is located in an intergenic region at position 2,237,229.
Fig. 3.
Focus on the candidate region. iEG, integrated EHH. Other statistics are like in figure 2. The average sequencing depth over isolates is given for all polymorphic sites. For all statistics, all genes (full genic region) are indicated by gray frames. The significance threshold is indicated by a dotted line for BayPass and the GWAS. GWAS P values from step 1 (without cofactor) are given. Significant SNPs are denoted by red circles (for EHH, significance is assessed by the ln(Rsb) ratio of iEG Vir1994 to iEG Avr1993) and a red star for the top GWAS SNP. The bottom panel replicates the gene localization (upper frames are forward genes and lower frames are reverse genes). Bars show nonsynonymous mutations found in the whole data set. The one nonsynonymous mutation found to be significant in any test is denoted by a purple triangle on all panels.
The EHH analysis shows that the iEG in sample Vir1994 is consistently high from position 2.16 to 2.24 (including both regions mentioned above) and decreases rather progressively on the left side and more abruptly on the right side near position 2.25 Mb. The analysis of iEG for the Vir1998 sample shows a marked decrease of EHH, with a reduced plateau which excludes the jgi.p|Mellp2_3|84583 gene (supplementary fig. 7, Supplementary Material online). However, we note that the bottleneck was probably completed in 1998 and that recombination may already have had an impact at this point, thereby starting to erode long distance homozygosity. In particular, there is a potential recombination hotspot around position 2.25 Mb, where iEG values are consistently lower.
Candidate Gene
Interestingly, the three SNPs falling in gene jgi.p|Mellp2_3|1427900 and the top GWAS candidate are in strong LD (supplementary table 7, Supplementary Material online). Except for missing data, all Vir1994 and Vir1998 samples have a fixed homozygous genotype for one of the alleles at each SNP except 98GC04 which is heterozygous and 98AB07 which is homozygous for the other allele. 98AB07 is the only avirulent of these two samples, justifying its genotype. Conversely, all Avr1993 and Wild2008 samples are either heterozygous or homozygous for the other allele, with the single exception for 08EA95. Therefore, these SNPs, including the one causing a nonsynonymous substitution, show a very good correlation with the genotype pattern expected if the virulence was caused by a recessive mutation.
The candidate gene encodes a 219-amino acid protein (ID: 1427900) which is cysteine-rich (seven cysteine residues) and shows no homology to known proteins in M. larici-populina, as well as in public databases, including in related species. No N-terminal signal peptide is predicted. Nevertheless the protein is predicted to be nonclassically secreted based on SecretomeP 1.0 (Bendtsen et al. 2004) with a NN score of 0.79 (above the threshold of 0.6). No transmembrane domain was identified with TMHMM v.2.0 (Krogh et al. 2001). Finally, machine-learning localization prediction tools point to a nonapoplastic protein with signal of chloroplast and nuclear localizations.
The nonsynonymous SNP at position 2,233,369 causes the substitution of the glycine residue at position 81 of the protein in the reference genome by a serine. Glycine is an aliphatic residue whereas serine is polar. Consistently the substitution is classified as deleterious by the PROVEAN protein prediction tool (Choi et al. 2012) with a score of −6.0, indicating that it is likely to have a functional effect. Note that the reference genome originates from a virulent individual, indicating that serine would actually be the ancestral state of this substitution.
Discussion
Thanks to a sampling design based on collection isolates spanning narrowly the estimated date of the RMlp7 resistance breakdown, we detected consistent molecular signatures pointing to a small region of chr15 and, furthermore, to a specific nonsynonymous polymorphism within the gene jgi.p|Mellp2_3|1427900.
Due to the limited availability of collection isolates, the geographical origins of the four samples are heterogeneous, which might cause substructuration of some of the samples. However, substructuration is expected to cause positive values of within samples, which does not appear to be the case. We assume that the strong dispersal abilities and the low inbreeding reproductive system of M.larici-populina reduce isolation by distance at a regional scale (Barres et al. 2008).
Due to the likely effect of the resistance breakdown on the demography, we cannot assume demographic equilibrium for the Vir1994 and Vir1998 samples which are likely to have experienced a recent bottleneck. For the Avr1993 sample, all three tests of neutrality (D, , and ) show significant deviation from the neutral expectation. However it must be noted that these tests are very sensitive to demographic deviations when computed at the whole-genome scale (Siol et al. 2010). exhibits a larger deviation from zero, whereas D is significantly negative, contrary to the other two test statistics. The three tests are similar in principle, as they are based on the difference between two different estimators of the θ parameter: D is based on the difference between π and and a negative value means that there is an excess of polymorphisms with unbalanced frequencies. is based on the difference between the number of mutations (not different in principle from ) and the number of singleton mutations, whereas is based on the difference between π and the proportion of singleton mutations. Positive values of and can therefore be interpreted as a deficit of singleton mutations. The Avr1993 sample would therefore exhibit at the same time an excess of unbalanced polymorphisms, but comparatively a deficit of singleton mutation. This pattern is not compatible with any simple scenario, requiring to be explained either several population size changes in close succession and/or complex substructure. These hypotheses are difficult to discriminate with a sample of such size.
A very recent or ongoing bottleneck is expected to cause first a strong decrease of diversity more marked for because of the loss of low-frequency variants, and then a recovery of polymorphism causing a relative excess of low-frequency variants corresponding to new mutations. In parallel, levels of long-distance LD should be temporarily inflated due to the rapid expansion of the population from a limited founding population.
The Vir1994 sample shows a pattern of whole-genome statistics compatible with these expectations with values of π and to a larger extent smaller than other samples. Consistently, D is positive, but not significantly so. However, is positive and strongly significant, indicating a deficit of singleton mutations. For Vir1998, the value of is significantly higher than for Vir1994, the signs of D and are reverted and D is now strongly significatively negative whereas is no longer significantly different of zero. The value of D indicates an excess of unbalanced polymorphisms, consistently with the hypothesis of restoration of polymorphism, whereas the value of shows that there is no significant excess of singletons among polymorphisms and shows that there is a moderate excess of singletons compared with the diversity. It is possible that the signature of the bottleneck has already nearly vanished when considering singletons.
Consistently, the Vir1994 sample displays strong signatures of long-range LD and the Vir1998 sample much less so. Such a rapid decay of the effect of the bottleneck on LD can be explained by the low inbreeding of M. larici-populina populations. The Avr1993 sample also displays long-range LD which may be explained by earlier demographic history.
The Wild2008 sample, which has been collected in a single location in a distinct region (where cultivated poplars are not dominating the landscape), is assumed not to have been affected by the resistance breakdown. It is also located in the vicinity of large populations of the alternate host of M. larici-populina, Larix decidua, in such a way we assumed large population sizes. Consistently, this sample exhibits the highest value of , but not a high nucleotide diversity (π) compared with other samples, showing a marked unbalance of allelic frequencies (with an excess of singletons and low-frequency variants, as illustrated by the strongly negative values of D, , and as well as the marked star-like structure of the tree (fig. 1). This excess of rare alleles might be explained by recent population expansion, for example, following a population bottleneck, which we assume to be independent from the RMlp7 breakdown.
The molecular signatures of adaptation detected in the current study are of three forms: excess of divergence between virulent and avirulent isolates (tested with BayPass), statistical genotype–phenotype association (tested with GWAS), and presence of long-range homozygosity (tested with EHH). The first two approaches are overlapping since the samples are largely colinear with the virulence status, but use different methodologies. EHH, in contrast, is completely independent and points clearly to the same region. The differentiation and GWAS methods tend to be more precise than EHH and point to one common SNP which is located in an intergenic region. A close survey of the genotypic structure at neighboring SNPs shows that polymorphism within jgi.p|Mellp2_3|1427900 (including a nonsynonymous mutation) is strongly correlated to the top SNP and that it is, furthermore, consistent with a recessive substitution conferring virulence (Flor 1971), with all virulent, and no avirulent, isolates being homozygous (with one exception in each case). The nonsynonymous substitution can be viewed as the best candidate for determining of the RMlp7 breakdown.
We cannot exclude that the true causal mutation is elsewhere in the same genomic region. The nonsynonymous substitution is the best candidate a priori, but silent mutations can have an effect as well, such as mutations in regulatory elements (Romani and Moreno 2021). Furthermore, our analysis is based on the current annotation of the genome of the reference isolate of M. larici-populina (98AG31) which is actually a virulent individual. It is possible that the annotation of jgi.p|Mellp2_3|1427900 is incorrect in the virulent reference isolate because of a frame-shifting mutation or that a large deletion occurred. The availability of genome assemblies for virulent and avirulent isolates, coupled with improvement of gene annotation, will help testing these possibilities.
We should not exclude either that other loci elsewhere in the genome are playing a role in resistance breakdown. Such loci can for example explain the few discrepancies between the genotype at SNPs within jgi.p|Mellp2_3|1427900 and the virulence phenotype while affecting too few isolates to be detectable through the analysis of polymorphism. Therefore the other loci detected by the GWAS should not be excluded completely but, because of the lack of confirmation with other method, the protein 1427900 should at this point viewed as the primary candidate.
Unfortunately, as an obligate biotroph, M. larici-populina exhibits features that hinders functional investigations (in particular, no transformation method is available). Further investigations will be needed in order to validate the avirulent character of the candidate gene. In the short term, a large-scale survey of natural variation may help confirming the role of the nonsynonymous substitution we detected. Other techniques amenable to this system are mutagenesis, controlled crosses, and heterologous systems (Lorrain et al. 2018).
In contrast with the three methods pointing to the candidate region, no selective sweep signatures can be detected in this region through the analysis of the allele frequency spectrum (freq-hmm). It seems unlikely that the failure of detecting the region with this kind of method is purely due to a lack a statistical power, because the selective sweep state of the HMM actually decreases in the region. Besides, several other regions in the genome are highlighted at the level of sensitivity we chose and none of them are confirmed by other methods.
Furthermore, we did not find evidence for a strong bottleneck with virulent isolates, especially in the Vir1994 sample. We expected a strong bottleneck due to the rapidity of the spread of virulence (Xhaard et al. 2011; Persoons et al. 2017). Bottlenecks cause a transient reduction of diversity followed by a recovery whose rate depends on several factors, including mutation rate and population size (Nei et al. 1975). The Vir1994 sample shows a reduction of diversity compared with Vir1998, consistent with a bottleneck that would have recovered already. However the substantial level of diversity in Vir1994 (with values comparable to the other samples), as well as virulence profiles (three different pathotypes in Vir1994) show that the bottleneck was in fact of weak or moderate intensity. Alternatively, the bottleneck could be actually older than what we envision, which would explain why the bottleneck signatures are relatively weak. However, due to active surveillance of rust disease on cultivated poplars (Pinon and Frey 1997) it is rather unlikely that a large population of virulent M. larici-populina existed for a significant amount of time without being detected.
The most likely explanation of the low impact of the bottleneck on the allele frequency spectrum and of the low reduction of diversity in the virulent genetic group is a soft selective sweep. Soft selective sweeps occur when adaptation is mediated by a mutation which was actually segregating in the original population and are thought to be more likely to be involved in fast evolution (Hermisson and Pennings 2017). The extent to which soft selective sweeps contribute to adaptive evolution is debated (Jensen 2014; Harris et al. 2018; Garud et al. 2021). In our study, a soft selective sweep appears as the most likely hypothesis, consistently with the fact that adaptation to human-introduced host resistance was prone to trigger fast adaptation by the pathogen. Consistently with this hypothesis, the putatively causal allele can be found in a heterozygous state in the Avr1993 population which predates the putative breakdown event. In addition to a soft selective sweep, the life history of M. larici-populina reduces inbreeding due to mating types, the need of two hosts to complete the biological cycle and high dispersal abilities (Lorrain et al. 2019). These features likely reduced further the extent of hitch-hiking around the putative avirulence locus. In particular, this can explain why the signatures of the selective sweep do not expand further than 0.2 Mb from the putative selected locus (fig. 3) and the lack of strong signatures on pairwise LD on chr15 (supplementary fig. 3, Supplementary Material online).
Plant pathogen effectors are classically searched on the basis of common features such as secretion signals, small size (<300 amino acids), high cysteine content and lack of homology in other species (Win et al. 2012). The candidate gene found in this study shares some but not all these features. In particular, the lack of a classical secretion signal would have excluded this gene from screening analysis based on common features such as those already performed on M. larici-populina (Hacquard et al. 2012; Lorrain et al. 2019).
Concluding Remarks
Our study identified the best avirulence gene candidate in M.larici-populina so far, based on the survey of genomic diversity. By considering samples framing narrowly the supposed date of the resistance breakdown, we maximized power to identify the causal locus. Our study illustrates the benefit of monitoring the diversity of pathogens through collections of living samples, in order to trace back any significant evolutionary transition that may occur. In complement, we combined a set of complementary genome scan methods to increase accuracy of the screen for adaptation. Our study suggests that features of the pathogen life-cycle such as sexual reproduction, high dispersal, and high levels of diversity foster fast adaptation while resulting in a genetically diverse virulent population which has lost virtually none of its potential for further adaptation.
Materials and Methods
Fungal Material
Four samples were designed based on the population structure previously described in Persoons et al. (2017) (supplementary table 1, Supplementary Material online). All isolates of samples Avr1993 and Wild2008 have been collected in one location, in the North-East of France for Avr1993 and in the South-East for Wild2008. Isolates from samples Vir1994 are spread over the North-East of France and a Belgian location. Isolates of the Vir1998 samples come from two locations, included in the sampling area of Avr1993.
We extracted isolates from a collection available in the laboratory. Note that M. larici-populina isolates are isolated at a stage of their life cycle where they are dikaryotic (two unmerged nuclei per spore cell) and can therefore be treated as diploid. To ensure their purity, genotyping was performed using 25 microsatellite markers with the method used in Persoons et al. (2017). Virulence profiles were characterized on a differential set of nine poplar genotypes each carrying a single qualitative resistance to M.larici-populina (RMlp1: Populus × euramericana “Ogy”; RMlp2: P. × jackii “Aurora”; RMlp3: P. × euramericana “Brabantica”; RMlp4: P. × interamericana “Unal”; RMlp5: P. × interamericana “Rap”; RMlp6: P. × interamericana “84B09”; RMlp7: P. × interamericana “Beaupré”; RMlp8: P. × interamericana “Hoogvorst”; RMlp9: P. deltoides “L270-3”) and on the universal susceptible cultivar P. × euramericana “Robusta” as positive control.
Poplar plants were grown during 2–3 months in a glasshouse at 20–24 °C, with a 16-h photoperiod, as previously described in Pernaci et al. (2014). We excised 12-mm discs on leaves (index 7–14) and placed them in flotation on deionized water in 24-well polystyrene cell culture plates, abaxial surface up. A suspension (approximately spores/ml) of each strain was deposited as 1-µl droplets on each disc. Culture plates were incubated for 13 days at 19 ± 1 °C under continuous illumination before scoring. Isolates resulting in at least ten pathogenic lesions per leaf disk were scored as virulent with respect to the corresponding resistance. Three replications were performed.
The virulence profile of the isolates was determined and showed that all isolates from the Avr1993 and Wild2008 samples were avirulent whereas all isolates from the Vir1994 sample were virulent as well as all but one from the Vir1998 sample (supplementary table 1, Supplementary Material online).
DNA Isolation
The DNA isolation method was performed on 50 mg of urediniospores as previously described in Pernaci et al. (2014) and Persoons et al. (2014). Quality and quantity of recovered high-molecular weight DNA was assessed by electrophoresis on agarose gel, by spectrophotometry (Nanodrop, Saint-Remy-lès-Chevreuse, France) and with the QuBit fluorometric quantitation system (Life Technologies, Villebon-sur-Yvette, France).
Genome Resequencing, Filtering, and Mapping
DNA was used for sequencing by Beckman Coulter Genomics (Grenoble, France) for 22 isolates and at the Joint Genome Institute for the remaining 54 (supplementary table 1, Supplementary Material online). Each library was quantified by qPCR and sequenced on the Illumina HiSeq2000 platform as paired-end 150-bp reads.
The reads were mapped on the M. larici-populina reference genome v2.0, available via the JGI fungal genome portal MycoCosm (Grigoriev et al. 2014). The assembled genome contains 18 chromosomes (101.4 Mb) and 493 unmapped scaffolds (8.4 Mb). The unmapped scaffolds, representing 8% of the total genome length (average length: 17 kb, N50: 36 kb), were not considered. All reads were aligned on the reference genome using the bwa software version 0.7.13 (Li and Durbin 2009). We fixed the maximum number of differences for each read against the reference to 2. All other options were used with default values. We used SAMtools version 1.3 (Li et al. 2009) with default parameters to compute the number of mapped reads, perform genotype calling, and export variant call format (VCF) files.
SNPs were filtered using EggLib version 3 (De Mita and Siol 2012). Genotypes were assigned based on the difference of Phred-scaled likelihoods (PL values in VCF files) between the best genotypes and all others as implemented in EggLib (threshold_PL = 30). Genotypes with a depth below 20 or above 200 reads were called as missing to exclude sites falling potentially in repetitive DNA. After genotype calling for all samples, we excluded sites with more than two alleles overall and less than ten nonmissing isolates in any of the four samples.
Phylogenetic Tree and Population Structure
To build a phylogenetic tree of all isolates, we computed the pairwise distance based on all SNPs. Pairwise distances were computed as the rate of pairwise genotypic differences for a random subset of sites without missing data for the two considered isolates, without correction. One percent of sites was drawn independently for all pairs of individuals. The tree was reconstructed using Phylip v 3.696 (Felsenstein 1981) using the neighbor-joining method. For representation, we used the Interactive Tree of Life online editor (Letunic and Bork 2019). The population structure of the isolates was assessed with DAPC implemented in the Adegenet package in R (Jombart et al. 2010). A random subset of 10% of sites was used. The number of genetic groups was assessed based on decreasing BIC values.
Analysis of Polymorphism
Linkage disequilibrium was computed as r2 using EggLib. Pairwise genotypic LD was computed separately for all four samples and for all pairs of sites of the same chromosome with a distance at most 1 Mb, excluding sites with a genotype at a frequency above 75% or an overall frequency of missing data above 10%. To smooth LD decay curves in the graphical representation, we computed quantiles (including the median) of r2 values grouped in windows of 1,000 bp of pairwise distance. We also monitored the window where the average r2 passed below different thresholds and the average r2 at given distance points. We also computed pairwise genotypic LD between all pairs of SNPs of each chromosome (also separately for the four samples), considering at most one SNP per kb of the reference genome.
We computed summary statistics using EggLib either for individual SNPs, in sliding windows along the chromosomes or for the whole genome (considering the whole set of sites). We defined window boundaries on the reference genome and processed complete windows only. For the sake of symmetry, we shifted window starting points in order to leave equal stretches of unanalyzed sequence at both ends of each chromosome. All per-site statistics were expressed relatively to the number of considered sites (variable or not) within the considered window (or overall), ignoring sites excluded due to an excess of alleles or missing data. Within-population statistics were computed within the four samples and for the whole data set. Differentiation statistics were computed for the whole data sets (four groups) and for pairwise comparisons. We measured the divergence between virulent and avirulent isolates by assessing the differentiation between pairs of samples following Weir and Cockerham (1984) where pairs were defined as the Avr1993 and Wild2008 samples (avirulent) on one hand and the Vir1994 and Vir1998 samples (virulent) on the other hand. Alternatively, we computed differentiation statistics considering two populations with respectively virulent and avirulent isolates. In the latter case, based on phenotyping results, the 98AB07 isolate was treated as avirulent (supplementary table 1, Supplementary Material online).
To test the deviation of whole-genome statistics from the expectations under the standard model, we performed coalescent simulations using EggLib. To avoid complications relative to the implementation of recombination while providing a good sampling of diversity at the genomic scale, we defined 5,000-bp windows with a step of 100,000 bp and neglected recombination within windows and linkage between them (they were simulated independently). Sites with less than ten nonmissing genotypes were discarded, as well as windows with less than 1,000 available sites. We computed for each of sample for each window. For each window, we generated 100,000 simulated samples for the four samples using the average as the value for the θ parameter (the same for the four samples). For all simulated windows, random isolates were removed to match the average number of missing data in each sample. Based on these simulations, both unilateral P values were computed to test the values of statistics, including the difference of some statistics between pairs of samples.
Genome Scan Methods
BayPass (Gautier 2015) version 2.1 was used with default parameters. For the covariate model, virulence status was encoded as an environmental value with a constant value −1 for avirulent populations and +1 for virulent populations. All polymorphic sites were used. The significant threshold was set to .
EHH was estimated for all polymorphic sites with EggLib, separately for all four samples (note that not all sites were polymorphic in all samples). Since our data are unphased, we used a variant based on heterozygosity of isolates (Tang et al. 2007). The main statistic is iEG which integrates the site-wise EHHS statistic. Each site was treated in turn as the core site and iEG was computed by integrating in both directions, until EHHS reached a threshold of 0.2 (or the chromosome limit was reached). To identify candidates, we computed ln(Rsb) as described in Tang et al. (2007) and selected all loci with a ln(Rsb) in the right tail of the distribution (probability density ).
GWAS was performed using a mixed-model approach including multiple loci (Segura et al. 2012), considering the virulence as phenotype and the genotypes as alleles. Since missing data are not supported, they were imputed by assigning the majority genotype to isolates with missing data. Only sites with a minor allele frequency of at least 0.05 were considered. To incorporate the information of genetic structure (including the differentiation between the four samples), we computed a kinship matrix (Korte and Farlow 2013) which was included in the model as the covariance matrix for a random polygenic effect. We used the mBonf and EBIC criteria to select the best model from the forward-backward search (Segura et al. 2012). We report P values from the initial GWAS scan (no cofactor). Candidate loci are determined as those identified as cofactors in the best multilocus model and/or which are significant at a threshold in the initial scan.
The freq-hmm method (Boitard et al. 2009) was applied to all polymorphic sites on allele frequencies using the folded spectrum mode, independently for all four samples. The starting value for θ was set to as computed for the whole genome for each sample. The analysis was performed separately for all chromosomes and the k parameter (which controls the detection rate) was set to .
Candidate Protein Analysis
The candidate protein 1427900 was evaluated by comparison to proteins of known function in the nonredundant GenBank database. Signal peptide prediction in amino acid sequences was performed using SignalP 5.0 (Armenteros et al. 2019). Nonclassical secretion was predicted using SecretomeP 1.0 (Bendtsen et al. 2004) with the normal threshold of neural network output score of 0.6 as recommended by the authors. Transmembrane domain detection was performed with TMHMM v.2.0 (Krogh et al. 2001) and PHOBIUS (Kall et al. 2007). Potential subcellular localization was predicted in silico using online tools APOPLASTP (Sperschneider et al. 2018) and LOCALIZER (Sperschneider et al. 2017). The PROVEAN online tool (Choi and Chan 2015), based on version 1.1.3 of the software, was used to predict the functional status of the amino acid substitution.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
We are grateful to Jérémy Pétrowski for the production of biological material for our experiments. We thank Pierre Gladieux, Martin Lascoux, Christophe Lemaire, Mathieu Siol, and three anonymous reviewers for comments on earlier versions of the manuscript. This work was supported by the French National Research Agency (ANR-12-ADAP-0009, GANDALF project). The work within the framework of the poplar rust genome project (CSP 1416) conducted by the U.S. Department of Energy Joint Genome Institute, a DOE Office of Science User Facility, is supported by the Office of Science of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231. A.P. was supported by a PhD fellowship from the Region Lorraine and INRAE and a partial post doc fellowship from the Region Grand-Est. A.M. was supported by a PhD fellowship from the Region Lorraine and INRAE. C.L. was supported by a PhD fellowship from the Region Lorraine and the French National Research Agency (ANR-18-CE32-0001, Clonix2D project). The IAM laboratory is part of the Lab of Excellence ARBRE supported by French National Research Agency (ANR-11-LABX-0002-01) which supported A.P. through a partial post doc fellowship.
Data Availability
Sequencing reads for isolates sequenced by Beckman Coulter Genomics have been deposited in GenBank’s Sequence Read Archive as part of BioProject PRJNA702879 (https://www.ncbi.nlm.nih.gov/bioproject/702879, last accessed January 4, 2022). Sequencing reads for isolates sequenced by the JGI are available on the JGI Genome Portal under Proposal ID 1416 (doi: 10.25585/1488093). Accession numbers to the SRA and JGI Project IDs, respectively, are given in supplementary table 1, Supplementary Material online. All scripts used to perform the analysis of polymorphism and processing of genome scan results, including scripts used to generate figures, are available in a dedicated git repository (https://gitlab.com/demita/mlp-genomics-vir7, last accessed January 4, 2022).
Literature Cited
- Aguileta G, et al. 2010. Finding candidate genes under positive selection in non-model species: examples of genes involved in host specialization in pathogens. Mol Ecol. 19(2):292–306. [DOI] [PubMed] [Google Scholar]
- Armenteros JJA, et al. 2019. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat Biotechnol. 37(4):420–423. [DOI] [PubMed] [Google Scholar]
- Barres B, et al. 2008. Genetic structure of the poplar rust fungus Melampsora larici-populina: evidence for isolation by distance in Europe and recent founder effects overseas. Infect Genet Evol. 8(5):577–587. [DOI] [PubMed] [Google Scholar]
- Begun DJ, et al. 2007. Population genomics: whole-genome analysis of polymorphism and divergence in Drosophila simulans. PLoS Biol. 5(11):e310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendtsen JD, Jensen LJ, Blom N, von Heijne G, Brunak S.. 2004. Feature-based prediction of non-classical and leaderless protein secretion. Protein Eng Des Sel. 17(4):349–356. [DOI] [PubMed] [Google Scholar]
- Boitard S, Schloetterer C, Futschik A.. 2009. Detecting selective sweeps: a new approach based on hidden Markov models. Genetics 181(4):1567–1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JKM, Tellier A.. 2011. Plant-parasite coevolution: bridging the gap between genetics and ecology. Annu Rev Phytopathol. 49:345–367. [DOI] [PubMed] [Google Scholar]
- Burke MK. 2012. How does adaptation sweep through the genome? Insights from long-term selection experiments. Proc R Soc B Biol Sci. 279:5029–5038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y, Chan AP.. 2015. PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics 31(16):2745–2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y, Sims GE, Murphy S, Miller JR, Chan AP.. 2012. Predicting the functional effect of amino acid substitutions and indels. PLoS One 7(10):e46688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Mita S, Siol M.. 2012. EggLib: processing, analysis and simulation tools for population genetics and genomics. BMC Genet. 13(1):27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duplessis S, et al. 2011. Obligate biotrophy features unraveled by the genomic analysis of rust fungi. Proc Natl Acad Sci U S A. 108(22):9166–9171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert D, Fields PD.. 2020. Host-parasite co-evolution and its genomic signature. Nat Rev Genet. 21(12):754–768. [DOI] [PubMed] [Google Scholar]
- Ellison CE, et al. 2011. Population genomics and local adaptation in wild isolates of a model microbial eukaryote. Proc Natl Acad Sci U S A. 108(7):2831–2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. 1981. Evolutionary trees from DNA-sequences – a maximum-likelihood approach. J Mol Evol. 17(6):368–376. [DOI] [PubMed] [Google Scholar]
- Flor H. 1971. Current status of gene-for-gene concept. Annu Rev Phytopathol. 9(1):275–296. [Google Scholar]
- Frantzeskakis L, Kusch S, Panstruga R.. 2019. The need for speed: compartmentalized genome evolution in filamentous phytopathogens. Mol Plant Pathol. 20(1):3–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garud NR, Messer PW, Petrov DA.. 2021. Detection of hard and soft selective sweeps from Drosophila melanogaster population genomic data. PLoS Genet. 17(2):e1009373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier M. 2015. Genome-wide scan for adaptive divergence and association with population-specific covariates. Genetics 201(4):1555–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerard PR, Husson C, Pinon J, Frey P.. 2006. Comparison of genetic and virulence diversity of Melampsora larici-populina populations on wild and cultivated poplar and influence of the alternate host. Phytopathology 96(9):1027–1036. [DOI] [PubMed] [Google Scholar]
- Grigoriev IV, et al. 2014. MycoCosm portal: gearing up for 1000 fungal genomes. Nucleic Acids Res. 42(Database issue):D699–D704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman SR, et al. 2010. A composite of multiple signals distinguishes causal variants in regions of positive selection. Science 327(5967):883–886. [DOI] [PubMed] [Google Scholar]
- Günther T, Coop G.. 2013. Robust identification of local adaptation from allele frequencies. Genetics 195(1):205–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haasl RJ, Payseur BA.. 2016. Fifteen years of genomewide scans for selection: trends, lessons and unaddressed genetic sources of complication. Mol Ecol. 25(1):5–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacquard S, et al. 2012. A comprehensive analysis of genes encoding small secreted proteins identifies candidate effectors in Melampsora larici-populina (poplar leaf rust). Mol Plant Microbe Interact. 25(3):279–293. [DOI] [PubMed] [Google Scholar]
- Harris RB, Sackman A, Jensen JD.. 2018. On the unfounded enthusiasm for soft selective sweeps II: examining recent evidence from humans, flies, and viruses. PLoS Genet. 14(12):e1007859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haudry A, et al. 2007. Grinding up wheat: a massive loss of nucleotide diversity since domestication. Mol Biol Evol. 24(7):1506–1517. [DOI] [PubMed] [Google Scholar]
- Hermisson J, Pennings PS.. 2017. Soft sweeps and beyond: understanding the patterns and probabilities of selection footprints under rapid adaptation. Methods Ecol Evol. 8(6):700–716. [Google Scholar]
- Jensen JD. 2014. On the unfounded enthusiasm for soft selective sweeps. Nat Commun. 5:5281. [DOI] [PubMed] [Google Scholar]
- Jombart T, Devillard S, Balloux F.. 2010. Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genet. 11:94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JDG, Dangl JL.. 2006. The plant immune system. Nature 444(7117):323–329. [DOI] [PubMed] [Google Scholar]
- Kall L, Krogh A, Sonnhammer ELL.. 2007. Advantages of combined transmembrane topology and signal peptide prediction – the Phobius web server. Nucl Acids Res. 35(Web Server):W429–W432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenig A, Mueller R, Mogavero S, Hube B.. 2021. Fungal factors involved in host immune evasion, modulation and exploitation during infection. Cell Microbiol. 23:e13272. [DOI] [PubMed] [Google Scholar]
- Korte A, Farlow A.. 2013. The advantages and limitations of trait analysis with GWAS: a review. Plant Methods. 9(1):29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer ELL.. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 305(3):567–580. [DOI] [PubMed] [Google Scholar]
- Kulathinal RJ, Stevison LS, Noor MAF.. 2009. The genomics of speciation in Drosophila: diversity, divergence, and introgression estimated using low-coverage genome sequencing. PLoS Genet. 5(7):e1000550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Corre V, Siol M, Vigouroux Y, Tenaillon MI, Délye C.. 2020. Adaptive introgression from maize has facilitated the establishment of teosinte as a noxious weed in Europe. Proc Natl Acad Sci U S A. 117(41):25618–25627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Bork P.. 2019. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 47(W1):W256–W259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewontin R, Krakauer J.. 1973. Distribution of gene frequency as a test of theory of selective neutrality of polymorphisms. Genetics 74(1):175–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R.. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25(14):1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, et al. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25(16):2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, et al. 2014. Whole-genome sequencing of Berkshire (European native pig) provides insights into its origin and domestication. Sci Rep. 4:4678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liti G, et al. 2009. Population genomics of domestic and wild yeasts. Nature 458(7236):337–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorrain C, Petre B, Duplessis S.. 2018. Show me the way: rust effector targets in heterologous plant systems. Curr Opin Microbiol. 46:19–25. [DOI] [PubMed] [Google Scholar]
- Lorrain C, dos Santos KCG, Germain H, Hecker A, Duplessis S.. 2019. Advances in understanding obligate biotrophy in rust fungi. New Phytol. 222(3):1190–1206. [DOI] [PubMed] [Google Scholar]
- Mardis ER. 2008. Next-generation DNA sequencing methods. Annu Rev Genom Hum Genet. 9(1):387–402. [DOI] [PubMed] [Google Scholar]
- Meisrimler C-N, Allan C, Eccersall S, Morris RJ.. 2021. Interior design: how plant pathogens optimize their living conditions. New Phytol. 229(5):2514–2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messer PW, Petrov DA.. 2013. Population genomics of rapid adaptation by soft selective sweeps. Trends Ecol Evol. 28(11):659–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namroud M-C, Beaulieu J, Juge N, Laroche J, Bousquet J.. 2008. Scanning the genome for gene single nucleotide polymorphisms involved in adaptive population differentiation in white spruce. Mol Ecol. 17(16):3599–3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Maruyama T, Chakraborty R.. 1975. Bottleneck effect and genetic variability in populations. Evolution 29(1):1–10. [DOI] [PubMed] [Google Scholar]
- Nielsen R, et al. 2005. Genomic scans for selective sweeps using SNP data. Genome Res. 15(11):1566–1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pernaci M, et al. 2014. Genome-wide patterns of segregation and linkage disequilibrium: the construction of a linkage genetic map of the poplar rust fungus Melampsora larici-populina. Front Plant Sci. 5(454):454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persoons A, et al. 2014. Patterns of genomic variation in the poplar rust fungus Melampsora larici-populina identify pathogenesis-related factors. Front Plant Sci. 5:450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persoons A, et al. 2017. The escalatory Red Queen: population extinction and replacement following arms race dynamics in poplar rust. Mol Ecol. 26(7):1902–1918. [DOI] [PubMed] [Google Scholar]
- Pinon J, Frey P.. 1997. Structure of Melampsora larici-populina populations on wild and cultivated poplar. Eur J Plant Pathol. 103(2):159–173. [Google Scholar]
- Pugach I, Stoneking M.. 2015. Genome-wide insights into the genetic history of human populations. Invest Genet. 6:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffaele S, Kamoun S.. 2012. Genome evolution in filamentous plant pathogens: why bigger can be better. Nat Rev Microbiol. 10(6):417–430. [DOI] [PubMed] [Google Scholar]
- Romani F, Moreno JE.. 2021. Molecular mechanisms involved in functional macroevolution of plant transcription factors. New Phytol. 230(4):1345–1353. [DOI] [PubMed] [Google Scholar]
- Sabeti PC, et al. 2007. Genome-wide detection and characterization of positive selection in human populations. Nature 449(7164):913–U12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segura V, et al. 2012. An efficient multi-locus mixed-model approach for genome-wide association studies in structured populations. Nat Genet. 44(7):825–U144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siol M, Wright SI, Barrett SCH.. 2010. The population genomics of plant adaptation. New Phytol. 188(2):313–332. [DOI] [PubMed] [Google Scholar]
- Sperschneider J, et al. 2017. LOCALIZER: subcellular localization prediction of both plant and effector proteins in the plant cell. Sci Rep. 7:44598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperschneider J, Dodds PN, Singh KB, Taylor JM.. 2018. APOPLASTP: prediction of effectors and plant proteins in the apoplast using machine learning. New Phytol. 217(4):1764–1778. [DOI] [PubMed] [Google Scholar]
- Stukenbrock EH, Bataillon T.. 2012. A population genomics perspective on the emergence and adaptation of new plant pathogens in agro-ecosystems. PLoS Pathog. 8(9):e1002893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang K, Thornton KR, Stoneking M.. 2007. A new approach for using genome scans to detect recent positive selection in the human genome. PLoS Biol. 5(7):e171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tellier A, Moreno-Gamez S, Stephan W.. 2014. Speed of adaptation and genomic footprints of host-parasite coevolution under arms race and trench warfare dynamics. Evolution 68(8):2211–2224. [DOI] [PubMed] [Google Scholar]
- Upson JL, Zess EK, Białas A, Wu C-H, Kamoun S.. 2018. The coming of age of EvoMPMI: evolutionary molecular plant-microbe interactions across multiple timescales. Curr Opin Plant Biol. 44:108–116. [DOI] [PubMed] [Google Scholar]
- Wang M, et al. 2014. The genome sequence of African rice (Oryza glaberrima) and evidence for independent domestication. Nat Genet. 46(9):982–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir B, Cockerham C.. 1984. Estimating F-statistics for the analysis of population structure. Evolution 38(6):1358–1370. [DOI] [PubMed] [Google Scholar]
- Win J, et al. 2012. Effector biology of plant-associated organisms: concepts and perspectives. Cold Spring Harb Sym Quant Biol. 77(0):235–247. [DOI] [PubMed] [Google Scholar]
- Xhaard C, et al. 2011. The genetic structure of the plant pathogenic fungus Melampsora larici-populina on its wild host is extensively impacted by host domestication. Mol Ecol. 20(13):2739–2755. [DOI] [PubMed] [Google Scholar]
- Zaman L, et al. 2014. Coevolution drives the emergence of complex traits and promotes evolvability. PLoS Biol. 12(12):e1002023. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing reads for isolates sequenced by Beckman Coulter Genomics have been deposited in GenBank’s Sequence Read Archive as part of BioProject PRJNA702879 (https://www.ncbi.nlm.nih.gov/bioproject/702879, last accessed January 4, 2022). Sequencing reads for isolates sequenced by the JGI are available on the JGI Genome Portal under Proposal ID 1416 (doi: 10.25585/1488093). Accession numbers to the SRA and JGI Project IDs, respectively, are given in supplementary table 1, Supplementary Material online. All scripts used to perform the analysis of polymorphism and processing of genome scan results, including scripts used to generate figures, are available in a dedicated git repository (https://gitlab.com/demita/mlp-genomics-vir7, last accessed January 4, 2022).