Abstract
Background:
Fusobacterium nucleatum activates oncogenic signaling pathways and induces inflammation to promote colorectal carcinogenesis.
Methods:
We characterized F nucleatum and its subspecies in colorectal tumors and examined associations with tumor characteristics and colorectal cancer (CRC) specific survival. We conducted deep sequencing of nusA, nusG, and bacterial 16s rRNA genes in tumors from 1,994 CRC patients and assessed associations between F nucleatum presence and clinical characteristics, CRC-specific mortality, and somatic mutations.
Results:
F nucleatum, which was present in 10.3% of tumors, was detected in a higher proportion of right-sided and advanced-stage tumors—particularly subspecies animalis. Presence of F nucleatum was associated with higher CRC-specific mortality (hazard ratio [HR], 1.97; P=0.0004). This association was restricted to non-hypermutated, microsatellite-stable tumors (HR, 2.13; P=0.0002) and those who received chemotherapy (HR = 1.92, CI: 1.07-3.45, p-value = 0.029). Only F nucleatum subspecies animalis, the main subspecies detected (65.8%), was associated with CRC-specific mortality (HR, 2.16; P=0.0016)—subspecies vincentii and nucleatum were not (HR, 1.07, P=0.86). Additional adjustment for tumor stage suggests that the effect of F nucleatum on mortality is partly driven by a stage shift. Presence of F nucleatum was associated with microsatellite instable tumors, tumors with POLE exonuclease domain mutations, ERBB3 mutations, and suggestively associated with TP53 mutations.
Conclusions:
F nucleatum, and particularly subspecies animalis, was associated with a higher CRC-specific mortality and specific somatic mutated genes.
Impact:
Our findings identify the F nucleatum subspecies animalis as negatively impacting CRC mortality which may occur through a stage shift and its effect on chemoresistance.
INTRODUCTION
The rapid advances in DNA sequencing technologies have allowed unbiased and comprehensive identification of pathogens in human tissues (1,2). It is thought that approximately 15% of all cancers are attributable to microorganisms, including bacteria (3,4). Microbes are not only expected to play a role in susceptibility to some cancers but also in therapeutic response (5). A well-characterized example is gastric cancers, of which 770,000 of all new cancer cases, worldwide, were attributable to Helicobacter pylori infections in 2012 (4,6).
In a subset of colorectal cancer (CRC) cases, Fusobacterium nucleatum, an inhabitant of the oral cavity and gastrointestinal tract, has been found to be enriched in tumor tissues (7-15). Multiple mechanisms have been described by which the F nucleatum may promote CRC. These include activation of WNT/beta-catenin signaling (16), creation of a proinflammatory microenvironment (17), and modulation of the tumor microenvironment to evade the antitumoral immune response (18). The presence of F nucleatum in colorectal (CR) tumors is known to be associated with poor prognosis and resistance to chemotherapy (19). However, less is known about tumor and patient attributes in relation to F nucleatum in CRC. A few studies have shown associations of F nucleatum with somatic mutation burden, tumor site, and tumor stage (7,8,20). However, these studies have relatively small sample sizes and are limited with respect to survival data, somatic mutation information, and epidemiologic data. Moreover, these studies did not investigate F nucleatum composition in CR tumors at the subspecies level. Additionally, as somatic mutations in cancer have been observed to affect the tumor microbiome in other cancer types, it is important to examine associations of F nucleatum with existing mutation data for the same CR tumors (21).
Here, we conducted targeted deep sequencing to assess the presence of F nucleatum across 1,994 CR tumors, which were characterized for somatic mutations in 205 genes (22). In this large dataset from the Genetics and Epidemiology of Colorectal Cancer Consortium (GECCO) and the Colon Cancer Family Registry (CCFR), we characterized F nucleatum and its subspecies, and examined associations with CRC-specific survival, somatically mutated genes, mutational burden, tumor site, and tumor stage. This comprehensive study provides insights into the role of F nucleatum in CRC.
MATERIALS AND METHODS
Study populations
The studies include the Colorectal Cancer Study of Austria (CORSA), Ontario Familial Colorectal Cancer Registry (OFCCR), Seattle-Colon Cancer Family Registry (SCCFR), and Cancer Prevention Study-II (CPS-II). Study descriptions and sample selection are described in the Supplementary Materials and Methods. Institutional review boards approved the study, and all patients (or their proxies) provided written informed consent to allow collection of specimens and data used in this study. Patients or the public WERE NOT involved in the design, or conduct, or reporting, or dissemination plans of our research.
DNA extraction, sequencing, and identification of somatic mutations
Details of materials and F nucleatum sequencing are described in the Supplementary Materials and Methods. Briefly, tumor tissue was macrodissected from formalin-fixed paraffin-embedded (FFPE) tissue sections guided by an H&E stained slide marked for the tumor region, and DNA was extracted using the QIAamp DNA Mini or QIAamp DNA FFPE tissue kits. Paired-end high-depth sequencing was conducted on HighSeq 2500 (Illumina) using a custom-designed AmpliSeq panel. Details of sequencing, identification of somatic mutations, and determination of MSI status have been previously described (22).
Detection and quantification of F nucleatum DNA in CRC tumors
To detect Fusobacterium DNA in CRC tumors, Fusobacterium-specific nusA and nusG genes (NC 003454), and the hypervariable regions, V2, V3, and V6 of the bacterial 16S rRNA genes were sequenced. To identify the Fusobacterium-specific sequence reads and quantify their abundance in each sample, the following steps were taken: (i) Unmapped reads from each BAM file containing sequence reads aligned to the human reference sequence (GRCh37/hg19 assembly) were retrieved; (ii) Sequence reads with low complexity were removed; (iii) Unmapped sequence reads were aligned in single-end mode using the Bowtie2 aligner (23) to a reference sequence database (parameters used: --very-sensitive-local-k 100 --score-min L,0,1.2) composed of five F nucleatum complete genomes (NC003454.1, NC009506.1, NC021277.1, NC021281.1 and NC022196.1) and 17657 16S rRNA genes; (iv) Aligned reads were collapsed using the Picard's MarkDuplicates tool; (v) Reads with ambiguous alignments were reassigned to the most probable F nucleatum genome of origin using a statistical model based on the read alignment scores (24).
To quantify the amount of F nucleatum in tumor samples, we only considered reads that aligned to F nucleatum reference sequences with greater than 95% identity match. The number of F nucleatum mapped reads found in each tumor was normalized to the total number of reads obtained for a given sample so that the abundance of F nucleatum for each sample was measured in parts per million (ppm) of the total number of reads sequenced. To limit the number of potential false positives, we applied a strict threshold of ≥0.5 ppm to define a positive presence of F nucleatum. This stringent cutoff was used to remove nonspecific alignments or sequencing artifacts (Supplementary Materials and Methods, Supplementary Figure S1 and Supplementary Figure S2). F nucleatum next-generation sequencing data was validated using TaqMan qPCR assay (Supplementary Materials and Methods, Supplementary Figure S3).
Analysis of mutated genes
We defined a gene as being somatically mutated if it carried any non-silent mutation, which included all nonsynonymous, frameshift and in-frame indels, splicing, and nonsense mutations. We further refined the mutation definition for a subset of genes with more detailed information available for mutational functional relevance. Mutations in APC were restricted to truncating mutations within the first 1600 codons with known functional consequences (25). To specifically examine the effect of mutations in KRAS, BRAF, ERBB2, and ERBB3, only tumors with mutually exclusive mutations in the same signaling pathway were analyzed. For KRAS, codons G12, G13, Q61, K117, and A146 were included, in which mutations are known to confer oncogenic signaling. For BRAF, only the V600E oncogenic mutation was included. For PIK3CA, only nonsynonymous mutations were included. For POLE, only non-synonymous and non-truncating mutations in the exonuclease domain were included. In consideration of statistical power for somatic gene mutation analyses, we restricted analyses to genes on our panel that have been well-established as being mutated in CRC (KRAS, TP53, POLE, BRAF, APC, PIK3CA, and SMAD4) and previously implicated with F nucleatum (KRAS). ERBB2 and ERBB3 genes were also included since they belong to the same tyrosine kinase pathway as KRAS and BRAF.
Statistical analysis
To investigate how different factors such as patients' clinical and tumor molecular features relate to the presence or absence of F nucleatum in tumor tissue logistic regression models were used. To assess associations with receipt of chemotherapy, cancer stage, CpG island methylator phenotype status (CIMP), sex, MSI status, and hypermutation status two different logistic regression models were fitted. The first (simpler) model contained age at diagnosis and sex as covariates, while a second (larger) model further included tumor site, hypermutation status, MSI status and the presence (or absence) of mutations in POLE, TP53, and ERBB3 genes. An analysis of variance (ANOVA) was performed for the age at diagnosis and tumor burden using the aov() function from the R package stats (where age at diagnosis and tumor burden were treated as dependent variables and F nucleatum status (presence/absence) as an independent variable).
We adjusted p-values using the Benjamini-Hochberg method (Supplementary Materials and Methods) to control the false discovery rate. To assess the association between the presence of F nucleatum/subspecies with CRC-specific mortality in 1,320 cases with available survival outcomes data, we used Cox proportional hazards regression analysis, adjusting for sex, age at diagnosis, tumor site, tumor stage, hypermutation status, tumor burden, MSI status, and POLE, TP53 and ERBB3 mutation status. We checked the proportional hazards assumption and used stratification for the Cox model to allow for non-proportionality as necessary. To deal with missing data in the case of univariable and multivariable logistic regressions (results in Table 1), cases with missing data for tumor location were removed before the analysis. For the Cox proportional hazards regression (results in Tables 2 and 3), when fitting the model only the relevant complete cases were used in the analysis. For each F nucleatum subspecies status, we generated adjusted survival curves that were obtained by averaging conditional survival curves based on the Cox model over covariate distribution, as well as the corresponding pointwise confidence intervals (CI) based on 1,000 bootstrapping samples. We performed a series of sensitivity analyses in which different propensity score based methods were used for modeling the association of F nucleatum subspecies with survival (Supplementary Materials and Methods). For mutated genes significantly associated with F nucleatum, we estimated the average treatment effect (ATE) by marginalizing over the covariate distribution to quantify the difference in proportions of F nucleatum presence between mutated and non-mutated.
Table 1.
Patient characteristics, tumor features, and the presence of F nucleatum in colorectal cancer tissue.
| Characteristic | Fuso-negative N (%) |
Fuso-positive N (%) |
OR (95% CI)a | FDR p- valued |
OR (95% CI)b | FDR p-valued |
|---|---|---|---|---|---|---|
| Age at diagnosis c | ||||||
| Mean | 63.3±12.2 | 63.8±12.5 | - | - | ||
| N | 1659 | 192 | - | - | ||
| Sex | ||||||
| Men | 836 (50) | 80 (42) | Ref | Ref | ||
| Women | 824 (50) | 112 (58) | 1.42 (1.05-1.93 p-val=0.023) | 0.031 | 1.31 (0.96-1.79, p-val=0.09) | 0.18 |
| Total | 1660 | 192 | ||||
| Chemotherapy | ||||||
| No | 616 (48.5) | 66 (44.3) | Ref | Ref | ||
| Yes | 653 (51.5) | 83 (55.7) | 1.25 (0.88-1.79, p-val = 0.22) | 0.27 | 1.29 (0.90-1.87, p-val = 0.17) | 0.21 |
| Total | 1269 | 149 | ||||
| Cancer stage | ||||||
| stage I | 462 (35) | 35 (24) | Ref | Ref | ||
| stage II | 356 (27) | 47 (33) | 1.77 (1.11-2.85, p-val = 0.017) | 0.027 | 1.72 (1.06-2.82, p-val = 0.029) | 0.099 |
| stage III | 345 (26) | 48 (33) | 1.84 (1.15-2.98, p-val = 0.012) | 0.022 | 1.88 (1.16-3.10, p-val = 0.011) | 0.061 |
| stage IV | 142 (11) | 14 (10) | 1.36 (0.68-2.58, p-val=0.36) | 0.36 | 1.52 (0.75-2.95, p-val = 0.23) | 0.25 |
| Total | 1305 | 144 | ||||
| Tumor site | ||||||
| rectal | 419 (25.2) | 31 (16.1) | Ref | Ref | ||
| distal colon | 533 (32.1) | 51 (26.6) | 1.27 (0.8-2.04, p-val = 0.31) | 0.34 | 1.22 (0.77-1.97, p-val = 0.40) | 0.4 |
| proximal colon | 708 (42.7) | 110 (57.3) | 2.04 (1.36-3.16, p-val = 8e-4) | 0.0017 | 1.46 (0.94-2.33, p-val = 0.098) | 0.18 |
| left (rectal + distal) | 952 | 82 | Ref | Ref | ||
| right (proximal colon) | 708 | 110 | 1.77 (1.31-2.4, p-val = 0.00026) | 0.00072 | 1.29 (0.92-1.82, p-val=0.14) | 0.19 |
| Total | 1660 | 192 | ||||
| Mutation burden c | ||||||
| Mean | 23.2 ± 47.8 | 34.4 ± 49.02 | ||||
| Total | 1677 | 194 | ||||
| Hypermutation status | ||||||
| non-hypermutated | 1394 (84.0) | 138 (71.9) | Ref | Ref | ||
| hypermutated | 266 (16.0) | 54 (28.1) | 1.98 (1.39-2.78, p-val = 0.0001) | 0.0004 | 0.51 (0.27-0.95, p-val = 0.036) | 0.099 |
| Total | 1660 | 192 | ||||
| Microsatellite instability status | ||||||
| MSS | 1434 (86) | 134 (70) | Ref | Ref | ||
| MSI | 226 (14) | 58 (30) | 2.27 (1.56-3.3, p-val = 1.8e-5) | 9.9e-5 | 3.22 (1.72-5.97, p-val = 0.00023) | 0.0025 |
| Total | 1660 | 192 | ||||
| CpG island methylator phenotype status | ||||||
| CIMP − | 1049 | 107 | Ref | Ref | ||
| CIMP + | 191 | 48 | 1.11 (1.06-1.16, p-val = 3.2e-6) | 3.5e-5 | 1.04 (0.99-1.10, p-val = 0.12) | 0.19 |
| Total | 1240 | 155 |
Adjusted for the age at diagnosis and sex
Adjusted for age at diagnosis, sex, tumor site, hyper mutation status, MSI status and mutations in POLE, TP53, and ERBB3 (for models investigating hypermutation and MSI status, those variables were only included once in the model)
An analysis of variance was performed for age at diagnosis and tumor burden (age at diagnosis p-value = 0.59 and mutation burden p-value = 0.002)
OR = Odds Ratio, CI = Confidence Interval
p-values were adjusted for multiple testing using the Benjamini-Hochberg method.
FDR = false discovery rate
Table 2.
Presence of F nucleatum in colorectal cancer tissue and CRC-specific survival.
| F nucleatum | cases (n) | events (n) | aHR (95% CI) | bHR (95% CI) |
|---|---|---|---|---|
| Negative | 1174 | 216 | 1.00 (reference) | 1.00 (reference) |
| Positive | 142 | 34 | 1.97 (1.35-2.86) | 1.68 (1.09-2.59) |
| p-value | 0.0004 | 0.018 |
HR = Hazard Ratio, CI = Confidence Interval
multivariable analysis adjusted for sex, age at diagnosis, tumor site, hypermutation status, tumor burden, mutations in POLE, TP53, and ERBB3, and MSI status
multivariable analysis adjusted for sex, age at diagnosis, tumor site, hypermutation status, tumor burden, mutations in POLE, TP53, and ERBB3, MSI status, and tumor stage
Table 3.
Presence of F nucleatum subspecies in colorectal cancer tissue and CRC-specific survival.
|
F nucleatum subsp animalis |
cases (n) |
events (n) |
aHR (95% CI) | bHR (95% CI) |
|---|---|---|---|---|
| Negative | 1196 | 224 | 1.00 (reference) | 1.00 (reference) |
| animalis | 80 | 20 | 2.16 (1.34-3.47) | 1.75 (0.99- 3.10) |
| p-value | 0.0016 | 0.056 | ||
| vincentii + nucleatum | 40 | 6 | 1.07 (0.48-2.43) | 1.02 (0.42-2.51) |
| p-value | 0.86 | 0.96 |
HR = Hazard Ratio, CI = Confidence Interval
multivariable analysis adjusted for sex, age at diagnosis, tumor site, hypermutation status, tumor burden, mutations in POLE, TP53, and ERBB3, and MSI status
multivariable analysis adjusted for sex, age at diagnosis, tumor site, hypermutation status, tumor burden, mutations in POLE, TP53, and ERBB3, and MSI status, and tumor stage
RESULTS
Characterization of Fusobacterium in CRC cases
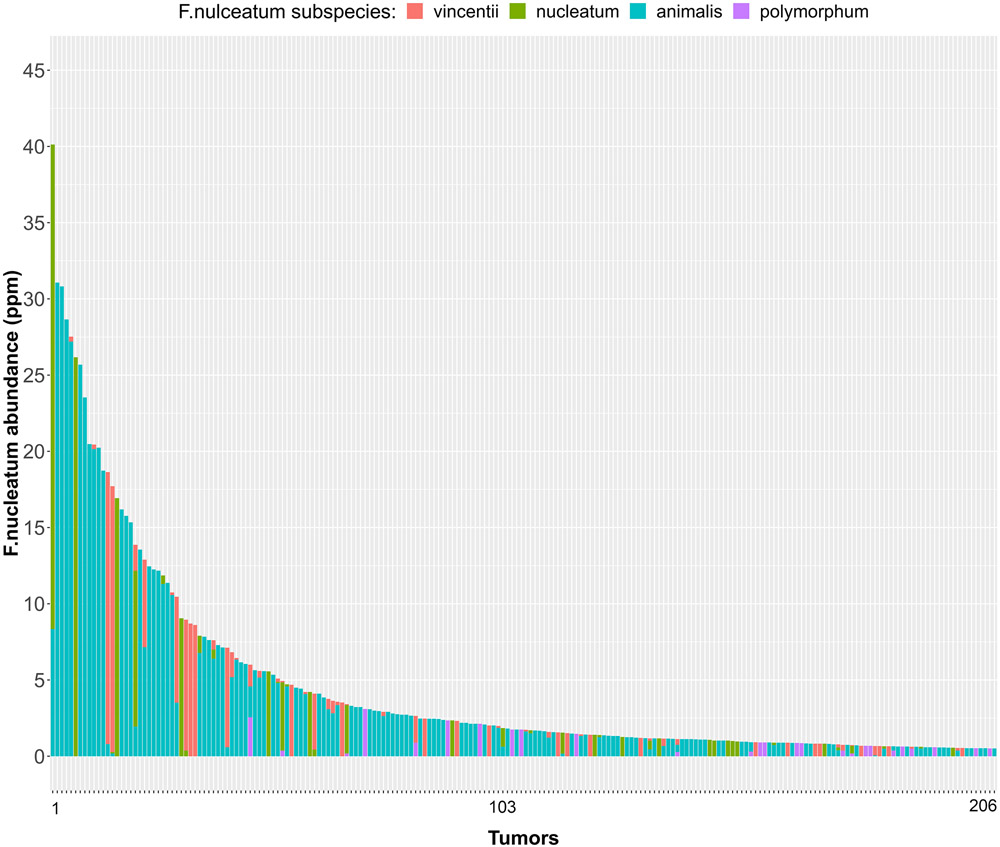
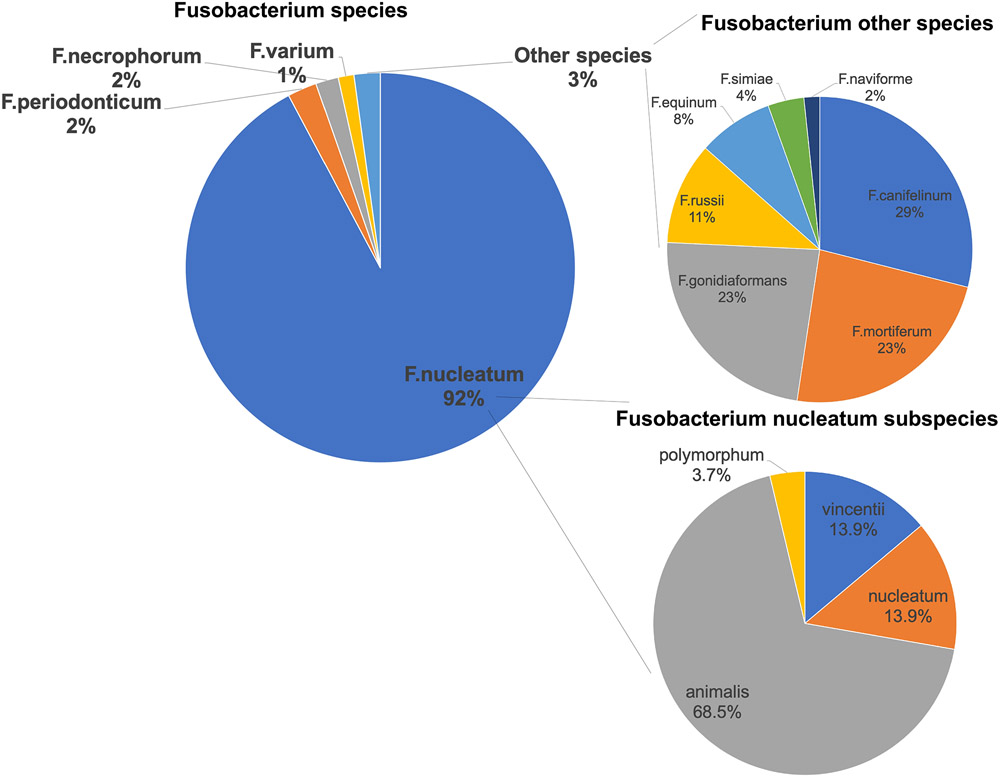
We detected F nucleatum-specific DNA in 10.3% of 1,994 CRC tumors (n = 206) (Figure 1A). Among the sequence reads mapping to Fusobacterium, 92% mapped to F nucleatum. Other species, including F periodonticum, each mapped to ≤ 2% of reads (Figure 2 and Supplementary Table 1). Further characterization of the F nucleatum mapping reads identified subspecies F nucleatum animalis (68.5%), F nucleatum nucleatum (13.9%), F nucleatum vincentii (13.9%), and F nucleatum polymorphum (3.7%) (Supplementary Table 1).
Figure 1. F nucleatum DNA abundance detected in 1994 colorectal cancer tumors.
F nucleatum DNA abundance in units of parts per million (ppm) detected in 206 colorectal tumor DNA samples with minimum abundance ≥ 0.5 ppm. The F nucleatum subspecies are shown in different colors.
Figure 2. Percent of sequencing reads mapping to Fusobacterium species and subspecies.
A total of 206 colorectal cancer tumors positive for F nucleatum were used in the plots.
As survival, tumor feature, and chemotherapy data were not available for all tumors (see Table 1 and Supplementary Materials and Methods), the total number of samples used in analyses varied.
Association of F nucleatum with clinical features
F nucleatum was found more often in right-sided tumors compared to left-sided tumors (Odds Ratio (OR) = 1.77, p-value = 0.00072, Table 1). Within the proximal region, tumors in the cecum were most likely to be positive for F nucleatum (OR =1.5, 95% CI: 1.0-2.3, p-value = 0.035). Tumors diagnosed at stage II (OR = 1.77, p-value = 0.027) or stage III (OR = 1.84, p-value = 0.022) were more likely to be positive for F nucleatum compared to stage I tumors. Furthermore, F nucleatum was more prevalent among women than men (OR = 1.42, p-value = 0.031). However, none of these associations remained significant in the multivariate adjusted model and after accounting for multiple testing. Age at diagnosis was not associated with presence of F nucleatum in tumors (p = 0.59).
We performed subsequent analyses to examine associations at the subspecies level. Consistent with our overall findings, presence of the predominant subspecies, F nucleatum subspecies animalis, was associated with right-sided versus left-sided tumors (OR = 2.47, p-value = 2.5e-5), female versus male sex (OR = 1.52, p-value = 0.04), and stage II (OR = 2.3, p-value = 0.02), stage III (OR = 2.55, p-value = 0.009) versus stage I tumors, and microsatelitte instable vs tumors not microsatellite instable (OR = 2.84, P-value = 4.86e-06) (Supplementary Table 2). Except for sex, these associations remained significant in the multivariate adjusted model and accounting for multiple testing. F nucleatum subspecies vincentii, and F nucleatum subspecies nucleatum were not associated with the above features (Supplementary Table 2).
Impact of F nucleatum on survival
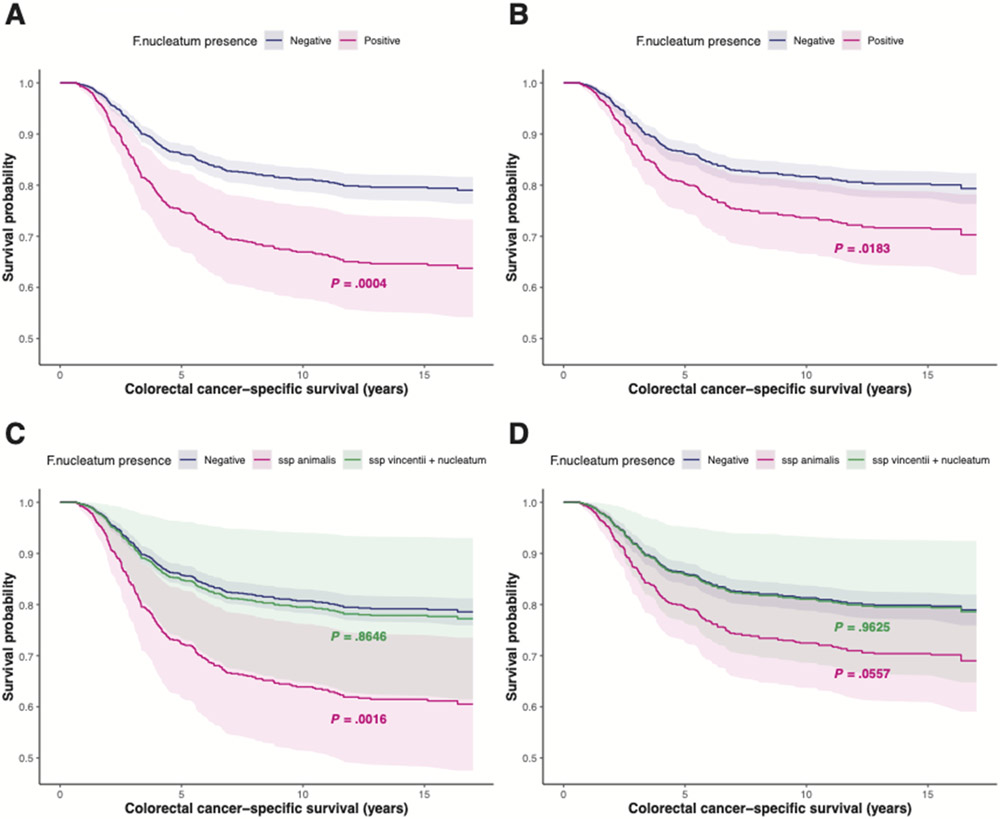
We observed that patients with F nucleatum present in tumors were more likely to die of CRC than those without F nucleatum present (Hazard Ratio (HR) = 1.97, CI: 1.35-2.86, p-value = 0.0004). This association remains statistically significant, even after adjustment for tumor stage (p-value = 0.018, Figure 3A and 3B, Table 2). A sub-analysis restricted to non-hypermutated, MSS tumors was consistent with this overall finding (HR = 2.13, 95% CI: 1.44-3.15, p-value = 0.0002). In contrast, among cases with hypermutated tumors with MSI, the presence of F nucleatum was not significantly associated with survival (HR = 0.84, 95% CI: 0.21-3.34, p-value = 0.81). As F nucleatum has been linked to chemoresistance (19), we stratified the association between F nucleatum and CRC-specific mortality by chemotherapy. We observed that F nucleatum was associated with increased CRC-specific mortality in CRC cases receiving chemotherapy (HR = 1.92, CI: 1.07-3.45, p-value = 0.029), but not in those without chemotherapy (HR = 0.42, CI: 0.06-3.16, p-value = 0.40).
Figure 3. Adjusted survival curves for colorectal cancer-specific survival based on Cox models.
(A) According to the presence of F nucleatum DNA in colorectal cancer tissues adjusted for sex, age at diagnosis, tumor site, hypermutation status, tumor burden, mutations in POLE, TP53, and ERBB3 and MSI. (B) According to the presence of F nucleatum DNA in colorectal cancer tissues adjusted for sex, age at diagnosis, tumor site, hypermutation status, tumor burden, mutations in POLE, TP53, and ERBB3, MSI, and tumor stage. (C) According to the presence of F nucleatum subspecies DNA in colorectal cancer tissues adjusted for sex, age at diagnosis, tumor site, hypermutation status, tumor burden, mutations in POLE, TP53, and ERBB3, and MSI. (D) According to the presence of F nucleatum subspecies DNA in colorectal cancer tissues adjusted for sex, age at diagnosis, tumor site, hypermutation status, tumor burden, mutations in POLE, TP53, and ERBB3, MSI, and tumor stage. P-values resulting from the multivariate analyses are shown on the plots. Pointwise confidence intervals are based on 1000 bootstrapping samples.
We further examined CRC-specific survival within F nucleatum subspecies. The presence of F nucleatum subspecies animalis was associated with higher mortality (HR = 2.16, 95% CI: 1.34-3.47, p-value = 0.0016, Table 3); however, the presence of F nucleatum subspecies vincentii and F nucleatum subspecies nucleatum were not (HR = 1.07, 95% CI: 0.48-2.43, p-value = 0.86) (Figure 3C). When further adjusting for tumor stage in the subspecies analysis, F nucleatum subspecies animalis exhibited the same survival trend, but did not remain statistically significant. This may be due to a limited sample size (Table 3) or may suggest that the effect of F nucleatum and the subspecies animalis on CRC-specific mortality is partly driven by a shift in stage.
Importantly, all results from the Cox proportional hazards regression analyses described above were consistent with results from our sensitivity analyses using different propensity score-based methods (Supplementary Tables 3, 4, and 5).
F nucleatum, MSI, and tumor hypermutation features
F nucleatum was more often present in hypermutated tumors compared to non-hypermutated tumors (OR = 1.98, p-value = 0.0004, Table 1). Mutations in DNA mismatch repair genes (MLH1, MLH3, MSH2, MSH6, PM2) and in the exonuclease domain of POLE contribute to hypermutation in CRC (26). DNA mismatch repair-deficient, MSI tumors were more likely to have F nucleatum present compared to the MSS tumors (OR = 2.27, p-value = 9.9e-5, Table 1). Additionally, F nucleatum was associated with CIMP status (OR = 1.11, p-value = 3.5e-5, Table 1). We found that tumors with POLE exonuclease domain mutations were also more likely to have F nucleatum present (OR = 4.14, p-value = 0.029, Table 4). The association for MSI status and POLE mutation status remained significant in the multivariate adjusted model and accounting for multiple testing.
Table 4.
Associations between somatically mutated genes and the presence of F nucleatum in colorectal tumors.
| Mutation status mutant vs non-mutated |
Univariate OR (95% CI) |
FDR p-valueb | Multivariable OR (95% CI)a |
FDR p- valueb |
|---|---|---|---|---|
| KRAS c | 1.35 (0.94-1.94, p-val = 0.099) | 0.16 | 1.42 (0.97 2.07, p-val = 0.067) | 0.15 |
| TP53 | 0.58 (0.43-0.79 p-val = 0.00044) | 0.002 | 0.64 (0.44- 0.94, p-val = 0.021) | 0.064 |
| POLE d | 2.82 (1.1-6.4 p-val = 0.019) | 0.057 | 4.14 (1.42- 11.19, p-val = 0.0064) | 0.029 |
| ERBB2 | 0.95 (0.28-2.43, p-val = 0.93) | 0.93 | 0.65 (0.18-1.77, p-val = 0.44) | 0.665 |
| ERBB3 | 6.22 (2.87-12.89, p-val = 1.49e-6) | 1.34e-5 | 4.33 (1.84-9.85, p-val = 0.00056) | 5.0e-3 |
| APC | 0.77 (0.57-1.05), p-val = 0.098) | 0.16 | 1.02 (0.73-1.42, p-val = 0.93) | 0.93 |
| BRAF | 1.58 (0.89-2.70, p-val = 0.12) | 0.16 | 0.88 (0.43-1.73, p-val = 0.72) | 0.81 |
| PIK3CA | 1.37 (0.9-2.02, p-val = 0.12) | 0.16 | 1.22 (0.80-1.83, p-val = 0.33) | 0.60 |
| SMAD4 | 1.08 (0.64-1.72, p-val = 0.75) | 0.85 | 1.11 (0.65-1.7, p-val = 0.68) | 0.81 |
In the regression analysis, F nucleatum was used as a binary outcome variable (negative = 0 vs positive = 1) and gene mutation status as the independent variable.
Details of genes with mutations are presented in the Methods section.
logistic regression model adjusted for sex, age at diagnosis, tumor site, MSI, and hypermutation status
p-values were adjusted for multiple testing using the Benjamini-Hochberg method.
FDR = False Discovery Rate, OR = Odd Ratio, CI = Confidence Interval
Codons G12/13, Q61, K117, and A146
Exonuclease domain mutations
At the subspecies level, F nucleatum subspecies animalis was more often present in hypermutated tumors compared to non-hypermutated tumors (OR = 2.59, p-value = 2.5e-5, Supplementary Table 2). Tumors with MSI were also more likely to have F nucleatum subspecies animalis present than the MSS tumors (OR = 2.84, p-value = 2.5e-5).
F nucleatum and tumor gene mutations
When testing the association of F nucleatum with mutation status of somatic genes frequently mutated in CRC, while accounting for multiple testing, we observed that the presence of F nucleatum was associated with mutations in ERBB3 (OR = 4.33, p-value = 0.005, Table 4). The average treatment effect of F nucleatum on ERBB3 was 18.8% (p-value 0.007, Supplementary Table 6) with 8.2% for ERBB3 non-mutated and 27.0% for ERBB3 mutant. Mutations in TP53 appeared to be negatively associated with the presence of F nucleatum, but results were not statistically significant after accounting for multiple comparisons (OR = 0.64, p-value = 0.064, Table 4). Mutation status of APC, PIK3CA, KRAS, BRAF, ERBB2, and SMAD4 was not associated with F nucleatum prevalence.
At the subspecies level, the presence of F nucleatum subspecies animalis was associated with mutations in ERBB3 (OR = 3.76, p-value = 0.043, Supplementary Table 7). No significant associations were found between presence of F nucleatum subspecies animalis and mutation status for APC, TP53, KRAS, PIK3CA, KRAS, BRAF, ERBB2, or SMAD4.
DISCUSSION
We used next-generation sequencing to comprehensively characterize the presence of F nucleatum in 1,994 CRC cases. Presence of F nucleatum was associated with tumor site, tumor stage, sex, microsatilite instability and mutation status of a subset of genes frequently mutated in CRC. CRC-specific mortality increased with the presence of F nucleatum, an association that was restricted to non-hypermutated, microsatellite-stable tumors and to patients receiving chemotherapy. F nucleatum subspecies animalis is the most common subspecies in CRC, and is most significantly associated with patient tumor characteristics, CRC-specific mortality, and somatic mutations.
We used eighteen primer pairs to amplify F nucleatum specific nusA, nusG genes, and 16S rRNA, and generated consistent results across our studies at the species and subspecies levels. The percent of cases we observed as having F nucleatum present (10.3%) is in line with most previous studies, which identified F nucleatum in 9-13% of CRC cases (7,9,19,27,28).
Tumors containing F nucleatum were more likely to present in the right side of the colon, at advanced stages, and in women. The higher prevalence of F nucleatum bearing tumors in the cecum, the far right of the colon, is consistent with at least one previous publication (8). More tumors in the cecum with F nucleatum may have occurred due to its direct connection to the appendix, which is known to harbor Fusobacteria (29). We discovered that right-sided tumor location, and advanced tumor stages (particularly stage II and III), were significantly associated with the presence of F nucleatum subspecies animalis, but not with other abundant F nucleatum subspecies. These findings are relevant for CRC prognosis and treatment.
We found the presence of F nucleatum associated with increased CRC-specific mortality. This observation is consistent with previous publications (7,20,30-32). Our finding of an association of F nucleatum with higher CRC-specific mortality in patients receiving chemotherapy further supports its role in chemoresistance (19). Moreover, we identified the F nucleatum subspecies F nucleatum subspecies animalis, but not F nucleatum subspecies vincentii or F nucleatum subspecies nucleatum, to be associated with CRC-specific mortality. This difference is likely not driven by statistical power as the association with CRC-specific survival is qualitatively different (HR 2.16 vs 1.07). Our approach was capable of detecting all four F nucleatum subspecies, of these F nucleatum subspecies polymorphum was the least abundant and present in fewer patients. These findings strongly suggest a pathogenic role of F nucleatum animalis in CRC.
Comparing stage-unadjusted and stage-adjusted results suggests that the effect of F nucleatum and its subspecies animalis on survival is mediated, in part, through a shift towards a higher tumor stage given that the stage-adjusted association is weaker. We consider the stage-adjusted analyses as secondary analyses that provide mechanistic insights into the association. While studies that aim to identify predictors for survival tend to default to stage-adjusted analyses to explore if the marker adds information above the commonly used stage information, our study is focused on establishing and characterizing the association between F nucleatum status and survival.
The availability of tumor sequencing data allowed us to examine associations between the presence of F nucleatum and select tumor molecular features. F nucleatum, and its subspecies F nucleatum subspecies animalis, were more commonly found in tumors with MSI. Furthermore, we found that F nucleatum was associated with the presence of mutations in the proofreading exonuclease domain of POLE, independent of MSI status. Hypermutated tumors are known to exhibit abundant neo-antigens and thereby elicit an immune response; however, mechanisms of immunosuppression by F nucleatum in CRC also have been described (18). As F nucleatum was not associated with CRC-specific mortality in hypermutated tumors, it is unlikely that F nucleatum impacts improved survival in this tumor subtype.
We further examined the association of F nucleatum with frequently mutated genes belonging to the following pathways implicated in CRC: WNT, p53, receptor-tyrosine kinase, TGF-beta, and PI-3-kinase pathways (33). Among these genes, only an association between KRAS and F nucleatum was described in a previous study (32). Consistent with this prior study, we found suggestive evidence (although not surpassing the multiple comparison test) that the presence of F nucleatum is associated with mutations in KRAS (32). As KRAS belongs to the receptor tyrosine kinase pathway, we further examined the association of F nucleatum with mutually exclusive mutations in BRAF, NRAS, ERBB2, and ERBB3, all belonging to the same pathway. Interestingly, the presence of F nucleatum, and its subspecies F nucleatum subspecies animalis, was significantly associated with mutations in ERBB3, after adjusting for covariates. Nonsynonymous oncogenic mutations in ERBB3 have been described in CRC and other cancers (34-36). Confirmed oncogenic mutations, V104M/L, A232V, P262S, G284R, and Q809R (34,35) as well as all known and predicted oncogenic mutations described in OncoKB curated dataset (37) were present in our CRC tumors. Among these mutations, the V104M/L substitutions in the extracellular domain of the ERBB3 receptor were the most common alterations. Expression of ERBB family of receptors on the surface of epithelial cells exposes these receptors directly to gut bacteria. Several pathogenic bacteria are known to exploit host cell signaling pathways to promote their adherence and internalization. For example, for entry of N meningitidis and N gonorrhoeae into epithelial cells, translocation of ERBB receptors to the apical surface and their recruitment have been demonstrated (38). F nucleatum has also been shown to internalize in the epithelial and endothelial cells of colon and other tissues (11,39-41). Internalization of F nucleatum into epithelial cells and recruitment of the ERBB receptors by other bacteria for host cell entry may suggest a similar mechanism for F nucleatum. Additional studies are required to test this hypothesis and potential blockade of ERBB receptors.
The second most commonly mutated gene in CRC is TP53, having mutations in approximately 53% of tumors (33),(22). Although our result is slightly above the significance threshold after adjusting for multiple testing, our analysis suggests that F nucleatum is more likely present in tumors with wildtype TP53, in line with the results from a previous study (15). Consistent with our findings, TP53 deletion and inhibition in the host were shown to enhance the clearance of extracellular bacteria during pneumonia by increasing the function of microbicidal neutrophils (42). Some pathogenic bacteria are also known to inhibit p53 by inducing its degradation resulting in the alteration of the p53 stress response (43). This phenomenon was first described in gastric cells co-cultured with Helicobacter pylori (44). By interfering with the wildtype p53 function, bacterial pathogens extend the life of host cells to access sufficient nutrients during intracellular replication (45). Although the exact role of F nucleatum in the p53 pathway remains unknown, similar mechanisms of rapid clearance of F nucleatum from tumors with mutated TP53 and the requirement of wildtype p53 for successful infection could be operating in CRC tumors.
In CRC, F nucleatum has been shown to activate WNT/beta-catenin signaling, generate a pro-inflammatory microenvironment, and modulate the tumor microenvironment to evade the antitumoral immune response. Interaction of the FadA protein of F nucleatum to E-cadherin mediates F nucleatum attachment and invasion into the epithelial cells leading to the activation of beta-catenin signaling, increased expression of transcription factors, oncogenes, WNT genes, and inflammatory genes, as well as growth stimulation of CRC cells (16). F nucleatum has also been shown to generate a pro-inflammatory microenvironment by the expansion of myeloid-derived immune cells that is conducive for CRC neoplasia progression (17). Enrichment of the F nucleatum in colonic adenomas indicates its involvement in the early development of CRC (16,17). Interaction of the Fap2 protein of F nucleatum with the TIGIT receptor on natural killer cells, T-cells, and tumor-infiltrating lymphocytes inhibits anti-tumor immune activities of these cells (18). Collectively, modulation of the tumor microenvironment, oncogenic signaling in tumor cells, and immune evasion by F nucleatum exacerbate tumor progression affecting survival.
Our findings advance our understanding of the potential contributions of the gut microbiome to CRC outcomes. Potential interventions that could stem from this work include the use of antibiotics, pre- and pro-biotics, vaccines, fecal transplants, and dietary/exercise interventions. Previous work in CRC patient-derived xenograft mice, has shown that treatment with a broad-spectrum antibiotic reduces F nucleatum load, cancer cell proliferation, and overall tumor growth (13). These findings demonstrate the potential therapeutic value of antimicrobial interventions in patients with F nucleatum-associated CRC. However, as the use of broad-spectrum antibiotics in CRC patients may have negative consequences via impact on the commensal microbial community, further studies to identify a narrow-spectrum antibiotic targeting F nucleatum are warranted. Other alternative approaches include the use of “predatory” bacteria such as Bdellovibrio bacteriovorus to combat F nucleatum (46) or other oncogenic bacteria and dietary interventions (9,47,48) that alter the gut microbiome.
The enrichment of F nucleatum in CRC, as well as its role in CRC tumorigenesis, have been described in several studies (7-13,30). One primary strength of the current study is the use of next-generation sequencing of archival FFPE tumor tissues, which enabled identification of F nucleatum at the subspecies level. We adjusted for multiple comparisons, allowing for careful evaluation of findings from prior studies that lacked this more stringent approach. Another advantage of this study, which had robust findings across different propensity score-based methods, is the analysis of well-characterized cohorts with long follow-up periods and data on CRC-specific survival, tumor characteristics, somatic mutations. Because F nucleatum was found in only 10% of tumors, some analyses of subspecies and less frequently mutated genes were limited due to sample size, even in the context of this large study. Thus, additional tumor sequencing is required to further improve statistical power.
Our study produced important and significant results, including that right-sided tumors are more likely to contain F nucleatum, and that presence of F nucleatum is significantly associated with advanced stage tumors, CRC-specific mortality, and specific tumor molecular features. F nucleatum was also associated with increased CRC-specific mortality in chemotherapy-receiving CRC cases. Additionally, we found that F nucleatum was more prevalent in tumors with KRAS and ERBB3 mutated genes and wild-type TP53. Although multiple mechanisms have been described for F nucleatum’s role in CRC, its connection to TP53 and ERBB3 pathways remains unknown. Particularly intriguing is the novel finding pointing toward F nucleatum subspecies animalis as the most likely pathogenic subspecies of F nucleatum. In this regard, our results may shed new light on the role of F nucleatum in CRC and aid in designing better approaches for the diagnosis and treatment of CRC patients harboring F nucleatum in their tumors.
Supplementary Material
Acknowledgments
GECCO is supported in part by NCI/NIH awards U01 CA137088 (NCI, U. Peters), U01 CA164930 (NCI, U. Peters), R01 CA176272 (NCI, P.A. Newcomb and A.T. Chan). The CCFR is supported in part by NIH/NCI awards U01 CA167551 (NCI, M.A. Jenkins), U01 CA074794 (NCI, P.A. Newcomb), U24 CA074794 (NCI, P.A. Newcomb), and R01 CA076366 (NCI, P.A. Newcomb). OFCCR is supported by U01 CA074783 (S. Gallinger), U24 CA074783 (S. Gallinger), and the Ontario Research Fund GL201-043 (B.W. Zanke). American Cancer Society funds the creation, maintenance, and update of the CPS-II cohort. CORSA: “Österreichische Nationalbank Jubiläumsfondsprojekt” 12511 (A. Gsur) and Austrian Research Funding Agency (FFG) grant 829675 (A. Gsur). S. Ogino was supported by NCI (R35 CA197735), a Nodal Award from Dana-Farber Harvard Cancer Center, and Cancer Research UK Grand Challenge Award (through the OPTIMISTICC Team). D.D. Buchanan is supported by an NHMRC R.D. Wright Career Development Fellowship. Additional funding was provided by the Ontario Institute for Cancer Research. We thank all those who agreed to participate in the CORSA study, including the patients and the control persons, as well as all the physicians and students. The authors thank the CPS-II participants and Study Management Group for their invaluable contributions to this research. The authors would also like to acknowledge the contribution to this study from central cancer registries supported through the Centers for Disease Control and Prevention National Program of Cancer Registries, and cancer registries supported by the National Cancer Institute Surveillance Epidemiology and End Results program. The authors would like to thank all those at the GECCO Coordinating Center for helping bring together the data and people that made this project possible.
Dr. Giannakis was supported by the Cancer Research UK Grand Challenge Award and a Stand Up to Cancer Colorectal Cancer Dream Team Translational Research Grant (SU2C-AACR-DT22-17). Dr. Buchanan is supported by a NHMRC R.D. Wright Career Development Fellowship. Additional funding was provided by the Ontario Institute for Cancer Research. Stand Up To Cancer is a division of the Entertainment Industry Foundation. The indicated SU2C grant is administered by the American Association for Cancer Research, the scientific partner of SU2C.
Footnotes
Conflict of interests:
Daniel Buchanan served as a consultant on the Tumour Agnostic (dMMR) Advisory Board of Merck Sharp and Dohme in 2017 and 2018 for Pembrolizumab. Reiko Nishihara’s current employer is Pfizer, Inc. Her contribution to this work was prior to employment at Pfizer, Inc. M. Giannakis receives research funding from Bristol-Myers Squibb and Merck. Nickolas Papadopoulos is a co-founder of, holds equity in, and is a consultant to Thrive Earlier Detection and Personal Genome Diagnostics. He is on the Thrive Earlier Detection Board of Directors and is a consultant to and holds equity in NeoPhore. Sysmex, Qiagen, Invitae, Personal Genome Diagnostics, PapGene, Thrive Earlier Detection, Horizon Discovery, Thermo FIsher, and other companies, have licensed previously described technologies from Johns Hopkins University. Nickolas Papadopoulos is an inventor on some of these technologies. Licenses to these technologies are or will be associated with equity or royalty payments to the inventors, as well as to Johns Hopkins University. The terms of all these arrangements are being managed by Johns Hopkins University in accordance with its conflict-of-interest policies.
Data availability:
All data relevant to the study are included in the article or uploaded as supplementary information.
REFERENCES
- 1.Gwinn M, MacCannell D, Armstrong GL. Next-Generation Sequencing of Infectious Pathogens. JAMA. 2019;321:893–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Forbes JD, Knox NC, Ronholm J, Pagotto F, Reimer A. Metagenomics: The Next Culture-Independent Game Changer. Front Microbiol. 2017;8:1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parkin DM. The global health burden of infection-associated cancers in the year 2002. Int J Cancer. 2006;118:3030–44. [DOI] [PubMed] [Google Scholar]
- 4.Plummer M, de Martel C, Vignat J, Ferlay J, Bray F, Franceschi S. Global burden of cancers attributable to infections in 2012: a synthetic analysis. Lancet Glob Health. 2016;4:e609–16. [DOI] [PubMed] [Google Scholar]
- 5.Helmink BA, Khan MAW, Hermann A, Gopalakrishnan V, Wargo JA. The microbiome, cancer, and cancer therapy. Nat Med. 2019;25:377–88. [DOI] [PubMed] [Google Scholar]
- 6.Bouvard V, Baan R, Straif K, Grosse Y, Secretan B, El Ghissassi F, et al. A review of human carcinogens--Part B: biological agents. Lancet Oncol. 2009;10:321–2. [DOI] [PubMed] [Google Scholar]
- 7.Mima K, Nishihara R, Qian ZR, Cao Y, Sukawa Y, Nowak JA, et al. Fusobacterium nucleatum in colorectal carcinoma tissue and patient prognosis. Gut. 2016;65:1973–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mima K, Cao Y, Chan AT, Qian ZR, Nowak JA, Masugi Y, et al. Fusobacterium nucleatum in Colorectal Carcinoma Tissue According to Tumor Location. Clin Transl Gastroenterol. 2016;7:e200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mehta RS, Nishihara R, Cao Y, Song M, Mima K, Qian ZR, et al. Association of Dietary Patterns With Risk of Colorectal Cancer Subtypes Classified by Fusobacterium nucleatum in Tumor Tissue. JAMA Oncol. 2017;3:921–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brennan CA, Garrett WS. Fusobacterium nucleatum - symbiont, opportunist and oncobacterium. Nat Rev Microbiol. 2019;17:156–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Castellarin M, Warren RL, Freeman JD, Dreolini L, Krzywinski M, Strauss J, et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012;22:299–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kostic AD, Gevers D, Pedamallu CS, Michaud M, Duke F, Earl AM, et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 2012;22:292–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bullman S, Pedamallu CS, Sicinska E, Clancy TE, Zhang X, Cai D, et al. Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science. 2017;358:1443–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Repass J, Reproducibility Project: Cancer Biology, Iorns E, Denis A, Williams SR, Perfito N, et al. Replication Study: Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. elife. 2018;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tahara T, Yamamoto E, Suzuki H, Maruyama R, Chung W, Garriga J, et al. Fusobacterium in colonic flora and molecular features of colorectal carcinoma. Cancer Res. 2014;74:1311–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rubinstein MR, Wang X, Liu W, Hao Y, Cai G, Han YW. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/β-catenin signaling via its FadA adhesin. Cell Host Microbe. 2013;14:195–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kostic AD, Chun E, Robertson L, Glickman JN, Gallini CA, Michaud M, et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe. 2013;14:207–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gur C, Ibrahim Y, Isaacson B, Yamin R, Abed J, Gamliel M, et al. Binding of the Fap2 protein of Fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity. 2015;42:344–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu T, Guo F, Yu Y, Sun T, Ma D, Han J, et al. Fusobacterium nucleatum Promotes Chemoresistance to Colorectal Cancer by Modulating Autophagy. Cell. 2017;170:548–563.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen Y, Lu Y, Ke Y, Li Y. Prognostic impact of the Fusobacterium nucleatum status in colorectal cancers. Medicine (Baltimore). 2019;98:e17221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Greathouse KL, White JR, Vargas AJ, Bliskovsky VV, Beck JA, von Muhlinen N, et al. Interaction between the microbiome and TP53 in human lung cancer. Genome Biol. 2018;19:123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zaidi SH, Harrison TA, Phipps AI, Steinfelder R, Trinh QM, Qu C, et al. Landscape of somatic single nucleotide variants and indels in colorectal cancer and impact on survival. Nat Commun. 2020;11:3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hong C, Manimaran S, Shen Y, Perez-Rogers JF, Byrd AL, Castro-Nallar E, et al. PathoScope 2.0: a complete computational framework for strain identification in environmental or clinical sequencing samples. Microbiome. 2014;2:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Kinzler KW. Cancer genome landscapes. Science. 2013;339:1546–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hino H, Shiomi A, Kusuhara M, Kagawa H, Yamakawa Y, Hatakeyama K, et al. Clinicopathological and mutational analyses of colorectal cancer with mutations in the POLE gene. Cancer Med. 2019;8:4587–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mima K, Sukawa Y, Nishihara R, Qian ZR, Yamauchi M, Inamura K, et al. Fusobacterium nucleatum and T Cells in Colorectal Carcinoma. JAMA Oncol. 2015;1:653–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nosho K, Sukawa Y, Adachi Y, Ito M, Mitsuhashi K, Kurihara H, et al. Association of Fusobacterium nucleatum with immunity and molecular alterations in colorectal cancer. World J Gastroenterol. 2016;22:557–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rogers MB, Brower-Sinning R, Firek B, Zhong D, Morowitz MJ. Acute appendicitis in children is associated with a local expansion of fusobacteria. Clin Infect Dis. 2016;63:71–8. [DOI] [PubMed] [Google Scholar]
- 30.Lee D-W, Han S-W, Kang J-K, Bae JM, Kim H-P, Won J-K, et al. Association Between Fusobacterium nucleatum, Pathway Mutation, and Patient Prognosis in Colorectal Cancer. Ann Surg Oncol. 2018;25:3389–95. [DOI] [PubMed] [Google Scholar]
- 31.Flanagan L, Schmid J, Ebert M, Soucek P, Kunicka T, Liska V, et al. Fusobacterium nucleatum associates with stages of colorectal neoplasia development, colorectal cancer and disease outcome. Eur J Clin Microbiol Infect Dis. 2014;33:1381–90. [DOI] [PubMed] [Google Scholar]
- 32.Yamaoka Y, Suehiro Y, Hashimoto S, Hoshida T, Fujimoto M, Watanabe M, et al. Fusobacterium nucleatum as a prognostic marker of colorectal cancer in a Japanese population. J Gastroenterol. 2018;53:517–24. [DOI] [PubMed] [Google Scholar]
- 33.Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jaiswal BS, Kljavin NM, Stawiski EW, Chan E, Parikh C, Durinck S, et al. Oncogenic ERBB3 mutations in human cancers. Cancer Cell. 2013;23:603–17. [DOI] [PubMed] [Google Scholar]
- 35.Loree JM, Bailey AM, Johnson AM, Yu Y, Wu W, Bristow CA, et al. Molecular landscape of ERBB2/ERBB3 mutated colorectal cancer. J Natl Cancer Inst. 2018;110:1409–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ross JS, Fakih M, Ali SM, Elvin JA, Schrock AB, Suh J, et al. Targeting HER2 in colorectal cancer: The landscape of amplification and short variant mutations in ERBB2 and ERBB3. Cancer. 2018;124:1358–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chakravarty D, Gao J, Phillips SM, Kundra R, Zhang H, Wang J, et al. Oncokb: A precision oncology knowledge base. JCO Precis Oncol. 2017;2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ho J, Moyes DL, Tavassoli M, Naglik JR. The role of erbb receptors in infection. Trends Microbiol. 2017;25:942–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Biedermann A, Kriebel K, Kreikemeyer B, Lang H. Interactions of anaerobic bacteria with dental stem cells: an in vitro study. PLoS ONE. 2014;9:e110616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kriebel K, Biedermann A, Kreikemeyer B, Lang H. Anaerobic co-culture of mesenchymal stem cells and anaerobic pathogens - a new in vitro model system. PLoS ONE. 2013;8:e78226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu M, Yamada M, Li M, Liu H, Chen SG, Han YW. FadA from Fusobacterium nucleatum utilizes both secreted and nonsecreted forms for functional oligomerization for attachment and invasion of host cells. J Biol Chem. 2007;282:25000–9. [DOI] [PubMed] [Google Scholar]
- 42.Madenspacher JH, Azzam KM, Gowdy KM, Malcolm KC, Nick JA, Dixon D, et al. p53 Integrates host defense and cell fate during bacterial pneumonia. J Exp Med. 2013;210:891–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zaika AI, Wei J, Noto JM, Peek RM. Microbial regulation of p53 tumor suppressor. PLoS Pathog. 2015;11:e1005099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wei J, Nagy TA, Vilgelm A, Zaika E, Ogden SR, Romero-Gallo J, et al. Regulation of p53 tumor suppressor by Helicobacter pylori in gastric epithelial cells. Gastroenterology. 2010;139:1333–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Siegl C, Rudel T. Modulation of p53 during bacterial infections. Nat Rev Microbiol. 2015;13:741–8. [DOI] [PubMed] [Google Scholar]
- 46.Negus D, Moore C, Baker M, Raghunathan D, Tyson J, Sockett RE. Predator Versus Pathogen: How Does Predatory Bdellovibrio bacteriovorus Interface with the Challenges of Killing Gram-Negative Pathogens in a Host Setting? Annu Rev Microbiol. 2017;71:441–57. [DOI] [PubMed] [Google Scholar]
- 47.Liu Y, Ajami NJ, El-Serag HB, Hair C, Graham DY, White DL, et al. Dietary quality and the colonic mucosa-associated gut microbiome in humans. Am J Clin Nutr. 2019;110:701–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang P, Liu Y. A reasonable diet promotes balance of intestinal microbiota: prevention of precolorectal cancer. Biomed Res Int. 2019;2019:3405278. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data relevant to the study are included in the article or uploaded as supplementary information.