Abstract
Freezing tolerance of triticale is a major trait contributing to its winter hardiness. The identification of genomic regions — quantitative trait loci (QTL) and molecular markers associated with freezing tolerance in winter hexaploid triticale — was the aim of this study. For that purpose, a new genetic linkage map was developed for the population of 92 doubled haploid lines derived from ‘Hewo’ × ‘Magnat’ F1 hybrid. Those lines, together with parents were subjected to freezing tolerance test three times during two winter seasons. Plants were grown and cold-hardened under natural fall/winter conditions and then subjected to freezing in controlled conditions. Freezing tolerance was assessed as the plants recovery (REC), the electrolyte leakage (EL) from leaves and chlorophyll fluorescence parameters (JIP) after freezing. Three consistent QTL for several fluorescence parameters, electrolyte leakage, and the percentage of the survived plants were identified with composite interval mapping (CIM) and single marker analysis (SMA). The first locus Qfr.hm-7A.1 explained 9% of variation of both electrolyte leakage and plants recovery after freezing. Two QTL explaining up to 12% of variation in plants recovery and shared by selected chlorophyll fluorescence parameters were found on 4R and 5R chromosomes. Finally, main locus Qchl.hm-5A.1 was detected for chlorophyll fluorescence parameters that explained up to 19.6% of phenotypic variation. The co-located QTL on chromosomes 7A.1, 4R and 5R, clearly indicated physiological and genetic relationship of the plant survival after freezing with the ability to maintain optimal photochemical activity of the photosystem II and preservation of the cell membranes integrity. The genes located in silico within the identified QTL include those encoding BTR1-like protein, transmembrane helix proteins like potassium channel, and phosphoric ester hydrolase involved in response to osmotic stress as well as proteins involved in the regulation of the gene expression, chloroplast RNA processing, and pyrimidine salvage pathway. Additionally, our results confirm that the JIP test is a valuable tool to evaluate freezing tolerance of triticale under unstable winter environments.
Supplementary Information
The online version contains supplementary material available at 10.1007/s13353-021-00660-1.
Keywords: Freezing tolerance, Plant acclimation, Genetic map, QTL, Chlorophyll fluorescence, Transmembrane, Proteins, Regulation of gene expression, Cereals
Introduction
Freezing is one of the environmental stresses which can constrain agricultural production of winter crops; therefore, increasing freezing tolerance is still important for breeding programs in cold and temperate climates. Many plant species including hexaploid winter triticale (x Triticosecale Wittm.) have developed a specific adaptive response which allows to stand low and freezing temperatures. During cold acclimation process, plants can increase their freezing tolerance after an exposition to low but non-freezing temperatures (Levitt 1980; Winfield et al. 2010; Janeczko et al. 2019). In natural conditions, this process is initiated by the decreasing temperature in late autumn which is complex phenomenon associated with many molecular, biochemical and physiological changes (Salinas 2002; Kaplan et al. 2004; Catala and Salinas 2008; Burbulis et al. 2011).
It has been previously reported that low temperature alters gene expression of a large number of genes encoding proteins could potentially contribute to plant freezing tolerance (Thomashow 1998; Kovi and Ergon, 2016). Certain transcriptional responses are common for most of plant species such as induced expression of vernalization genes, CBF genes, and COR genes (Knox et al. 2008, 2010). Vernalization gene group contain a VRN1, VRN2 and VRN3 genes, whereas CBF gene group includes a set of tandemly duplicated CBF (C-repeat Binding Factors) transcription factors at the FR2 (Frost Resistance 2) locus (Galiba et al. 2009). Furthermore, level of VRN1 transcript increase during exposure to low temperatures (Kobayashi et al. 2005) and it can generate conversion from the vegetative to reproductive plant growth stage (Stockinger et al. 2007). This transition is also associated with the suppressed induction of CBF group of genes in response to cold which results in reduced frost tolerance (Kobayashi et al. 2005; Dhillon et al. 2010; Zhu et al., 2014). Additionally, CBF similarly to COR genes enhance photosynthetic capacity which was reported in A. thaliana and B. napus photosynthetic capacity and freezing tolerance in response to temperature conditions (Kurepin et al. 2013). Furthermore, accumulation of proteins encoded by COR genes can lead to cell membrane stability under freezing conditions (Dong et al. 2002).
The major group of genes associated with cereal freezing tolerance have been reported and identified on the long arms of homeologous group 5 (Roberts 1990; Sutka 1994; Sutka and Snape 1989; Veisz and Sutka 1993; Kocsy et al., 2010). Also, Danyluk et al. (1994) discovered that wCOR410 and wCOR719 genes expression was regulated by factors located on chromosome 5A. Another locus, Fr-A1 on long arm of chromosome 5A was mapped in a close position to the previously described vernalization gene Vrn-A1 (Galiba et al. 1995). The presence of loci controlling expression of cor14b gene was identified on the long arm of 5A in wheat by Vágújfalvi et al. (2000, 2003). Additionally, wheat chromosome 5D was proved to be involved in the regulation of freezing tolerance (Snape et al. 1997).
The measurement of chlorophyll fluorescence parameters proved to be the good method to evaluate the freezing damage of plants reflecting freezing damages of photosynthetic apparatus (Rizza et al. 2001; Rapacz et al. 2011, 2015b). Also the measurements of electrolyte leakage in freeze-damaged leaves are commonly used for estimation of freezing injury as freeze–thaw damages results in disintegration of plasma membranes (Dexter et al. 1932). However, the results of chlorophyll fluorescence measurements, electrolyte leakage test, and plant survival count may give sometimes distinct results depending on environmental conditions (Rapacz et al. 2015a).
Thus, we hypothesized that different effects of freezing on plants may be, at least partially controlled by different genes. In our study we decided to use quantitative trait loci (QTL) mapping technology performed on a newly developed genetic map for triticale to determine the number and to map position of loci of different measures of freezing tolerance such as chlorophyll fluorescence parameters (JIP test) after freezing, membrane integrity affected by freezing, and plant recovery after freezing tests field-laboratory freezing tests. It is known that interactions among loci or between genes/QTL and environment make a substantial contribution to variation in complex traits (Gupta et al. 2007). Additionally, we aimed to identify candidate genes located in genome regions associated with analyzed traits. Doubled haploid mapping population used in our study brought us a unique opportunity to perform all of the experiments to ensure sound quality of the results.
Materials and methods
Plant material
The mapping population consisting of 92 doubled haploid lines was derived from F1 generation of a cross between two triticale cultivars: cv. ‘Hewo’ used as a female parent (Strzelce Plant Breeding Ltd.) and cv. ‘Magnat’ as the pollen parent (Danko Plant Breeding Ltd.). Both parental cultivars differed in tolerance to Microdochium nivale infection (Gołębiowska and Wędzony 2009), as well as in freezing tolerance in pre-tests (unpublished data). The DH ‘Hewo x Magnat’ lines population was developed by the androgenesis in the anther culture according to the method described by Wędzony (2003). The obtained DH lines were numbered from 1 to 92 in relation to the tests results of the degree of Microdochium nivale tolerance. This numbering has been kept for all experiments.
Genetic linkage map
The genomic DNA was isolated from young triticale leaves according to method described by Tyrka et al. (2011). Total DNA purity and integrity were tested on the agarose gels while its quantity was measured using UV–Vis Q500 spectrophotometer (Quawell, San Jose, USA).
Samples were genotyped in the Diversity Arrays Technology Pty Ltd (DArT P/L, Australia, www.diversityarrays.com) to detect different types of DNA variation (single nucleotide polymorphism, indel, and methylation) and to search for diagnostic markers. Using JoinMap 4 (Van Ooijen 2006) software, the segregation data were analyzed to group all markers with LOD value > 3.0. Afterwards, markers within these groups were ordered using the RECORD program (Van Os et al. 2005). The marker order was used to sort all markers within linkage groups and graphical genotypes were examined in Excel 2003. At this step, singletons were replaced by missing values in the dataset and calculations were repeated until no singletons were found (through three rounds). The distance between loci was calculated with the Kosambi function (Kosambi 1944). Finally, the final map has consisted of 680 DArT markers. Nomenclature of markers was synchronized with previously published map for partially overlapping set of lines (Tyrka et al. 2015). Consensus triticale DArT map was used for group identification and orientation (Tyrka et al. 2018).
Simple sequence repeat (SSR) analyses comprised 56 markers that were polymorphic between the two parental lines. The selected 37 mapped SSRs included the following: 20 wms: gwm46, gwm95, gwm126, gwm130, gwm136, gwm146, gwm149, gwm164, gwm169, gwm181, gwm275, gwm332, gwm335, gwm339, gwm368, gwm375, gwm388, gwm495, gwm499, and gwm566 (Röder et al. 1998); 7 scm: scm101, scm120, scm138, scm180, scm268, scm28, and scm304 (Saal and Wricke 1999; Hackauf and Wehling 2002); 7 wmc: wmc168, wmc219, wmc262, wmc289, wmc327, wmc434, and wmc537; and 3 other SSR markers: barc182, gdm109, and gdm147 (Pestsova et al. 2000; Somers et al. 2004). Four primer pairs (barc182, scm304, wmc168, and wmc327) revealed simultaneously two different loci. Redundant markers were shortlisted in final genetic map, and single representative markers with the lowest number of missing data were left to represent the bin with a total number of binned markers given in brackets.
Freezing tolerance assessment with the field-laboratory method
All presented tests took place in Kraków, Poland (N 50.069014, E 19.845528), according to the Koch and Lehman (1966) method with minor modifications, in three independent experiments with three replicates each, in the autumn/winter seasons. Three replicates were performed in a randomized complete block design in order to limit the error resulting from the marginal position of an individual genotype in the planting plastic boxes (30 cm × 38 cm × 9 cm). Kernels were sown in 13 rows of 10 kernels/genotype/box. One row per each parental line was sown in randomized positions in every box as a control. Total of 8 planting boxes/replicate, 24 boxes per each experiment were planted and analyzed.
Plants were grown and cold-hardened under natural fall/winter conditions in the open-air vegetation chamber. The temperature was monitored with an electronic weather station WS-3600–11, Technoline, Berlin, Germany, and the mean values of daily temperatures are presented in Fig. 1. On the days indicated by arrows (Fig. 1), the exact number of growing seedlings was counted in each row, and the boxes with plants were moved to the freezing chamber where they were subjected to the cycle consisting of one day-long growth in − 2 °C, followed by gradual temperature decrease (3 °C/h) down to − 15 °C, and then stable for 6 h. Later, the temperature was increased at the rate 3 °C/h to + 2 °C. After reaching + 2 °C, the boxes were transferred to an unheated glasshouse maintained at 10–15 °C, and the plants were cut 2 cm above the soil level. After 3 weeks, the number of surviving (regrowing) plants was established and the plant survival was expressed as a percentage of the survived plants from initially growing plants.
Fig. 1.
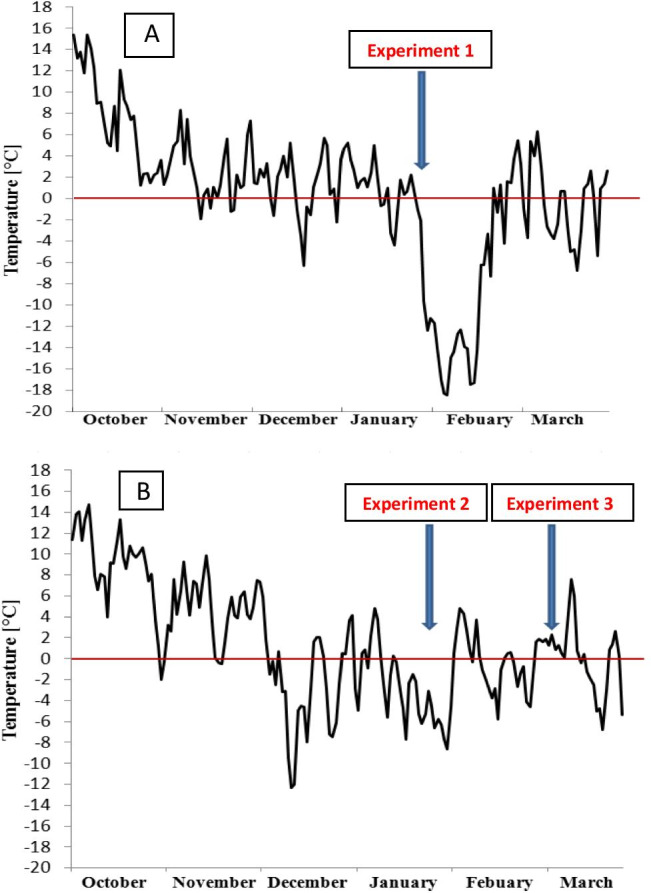
Daily temperatures (means) measured in the open-air vegetation chamber in autumn/winter seasons 2012/2013 (A) and 2013/2014 (B). The dates of plant transfer to the freezing chamber (the start of the freezing test), leaves sampling for chlorophyll fluorescence measurements (JIP test), and electrolyte leakage measurements (on B) are indicated by arrows: 26 January 2012 (A), 21 January 2013, and 4 March 2013 (B). The temperature was monitored with an electronic weather station (Ogimet)
Chlorophyll a fluorescence measurements
Parameters of induction kinetics of chlorophyll a fluorescence in dark-adapted leaves (JIP test) were measured on the middle section of the second fully expanded detached leaves with HandyPEA fluorimeter (Hansatech Kings Lynn, UK). On the days indicated by arrows in Fig. 1 (experiments 1–3), before boxes with plants were put into the freezing chamber to perform freezing tests, 10–12 leaves were randomly collected from each DH line, approximately the same number of 3–4 leaves/DH line from each of 3 replications. Since parental lines were present as a control in every box, parental samples consisted of approximately 26 leaves per each of 3 replications. Leaves were packed into polyethylene bags with a string closure and frozen in − 15 °C for 6 h with the protocol described by Rapacz et al. (2015a). Then, the leaves were brought to room temperature and dark-adapted before measurements for 15 min in a leaf clip (Hansatech, Kings Lynn, UK). Chlorophyll a transient fluorescence was measured according to Rapacz (2007) with a light pulse intensity of 3500 μmol m−2 s−1, and the pulse duration for 0.7 s with the fixed gain (1 ×). The following parameters of JIP test were calculated and described as in Rapacz (2007): absorbed energy flux per leaf cross-section (CS) and the single, active PSII reaction center (RC) (ABS/CS, ABS/RC respectively); trapped energy flux in PSII reaction centers per leaf cross-section and the single, active PSII reaction center (Tr0/CS, Tr0/RC, respectively); the energy flux for electron transport per leaf cross-section and the single, active PSII reaction center (ET0/CS, ET0/RC, respectively); dissipation of energy in PSII reaction centers per leaf cross-section and the single, active PSII reaction center (DI0/CS, DI0/RC, respectively); yield of the energy trapping in PSII (Fv/Fm); performance indexes of PSII (PI) normalized for minimal and maximal densities of active reaction centers per leaf cross-section (PICS0 and PICSm, respectively); minimal and maximal densities of active reaction centers per leaf cross-section (RC/CS0 and RC/CSm, respectively); and the quantum yield of electron transport (φEo) and the efficiency of the electron transfer from QA- to QB (ψo).
Electrolyte leakage measurements
The electrolyte leakage considered as the test of plasma membrane damages was performed in accordance with Flint et al. (1967), two times during winter 2013/2014 (dates indicated by arrows on Fig. 1B, experiments 2 and 3). In each series, measurements were made in 20 biological replicates for each genotype (1 leaf from different plant = 1 replicate). Each leaf was placed separately in a 20-cm3 plastic tube filled with 5 cm3 of deionized water. The material was frozen for 6 h at − 15 °C with the protocol described for chlorophyll fluorescence studies. After removing from the freezing chamber, 10 cm3 of deionized water was added to each tube and then the samples were shaken (ROTH, Linegal Chemicals Sp. z o.o) at room temperature for 24 h. Then, conductivity measurements (EL1) were performed in each tube using a conductivity meter type OK 102/1 (Radelkis). Probes were then frozen in liquid nitrogen for 2 min and again shaken at room temperature for 24 h before the second electrical conductivity measurement (EL2). The EL % was calculated as EL = (EL1/EL2) × 100%, where EL1 = primary electrolyte leakage after − 15 °C treatment and EL2 = total electrolyte leakage after freezing in liquid nitrogen.
Statistical analysis
All the data were analyzed with Statistica 13.0 PL software (Statsoft, Tulsa, OK, USA). Distribution of the data was checked using histograms and accompanied with a Shapiro–Wilk test. For JIP test parameters, one-dimensional variance analysis was performed. Linear correlation coefficients (Pearson’s) were calculated for each of three experiments separately on the basis of mean value of replicates per genotype: (1) between percentage of survived plants and every parameter of fluorescence and (2) between the fluorescence parameters and electrolyte leakage (experiments 2 and 3). The regression line was presented with a 95% coefficient interval.
Quantitative trait loci (QTL) identification
To identify QTL regions associated with the analyzed traits, single marker analysis (SMA) and composite interval mapping (CIM) methods were calculated with the Windows QTLCartographer software version 2.5 (Wang et al. 2012). SMA analysis fits the data to the simple linear regression model while CIM method in turn, determines the linkage between QTL and markers limiting the designated interval on the chromosome map. The threshold logarithms of the odds (LOD) scores were calculated based on 1000 permutations and 1 cM walk speed. QTL was accepted for LOD scores higher than 2.5. The percentage of phenotypic variation was calculated with a single factor regression (R2). Favorable alleles in each QTL region were selected on the basis of the additive effect (Add), where negative additive effect referred to cv. ‘Magnat’ and positive referred to cv. ‘Hewo’. The CIM and SMA analyses were performed separately for each experiment. Results of QTL analysis were visualized using CorelDRAW9 software.
The label of each identified QTL region was created from the short name of each parameter (ex., Qrec for recovery, survival; Qel for electrolyte leakage). For loci explained more than one trait, the general name was given (ex., Qfr for loci co-located for recovery, survival, and electrolyte leakage; Qchl for loci containing analyzed chlorophyll a fluorescence parameters) Hewo × Magnat (hm), chromosome names (wheat A and B group, rye group R), and QTL number on the chromosome (1–4).
The in silico location of genes within the QTL
Within the localized QTL, candidate genes associated with the analyzed traits were identified in silico according to Karbarz et al. (2020) with modifications. Sequences corresponding to the wheat, rye and triticale DArT clones of the flanking and maximal LOD peak markers of the significant QTLs were downloaded from the Diversity Arrays Technology webpage (https://www.diversityarrays.com/technology-and-resources/sequences/). Then, DArT sequences were used to query the IWGSC RefSeq 1.0 wheat genome for physical mapping using the BLAST tool in the Unité de Recherche Génomique URGI database (https://urgi.versailles.inra.fr). Subsequently, DArT sequences were used to query all available wheat and rye genome collections for physical mapping using the BLAST tool of GrainGenes Blast Service beta (10.1093/molbev/msz185). Genes localized on target physical wheat and rye regions were retrieved and annotated with the use of BLAST® (https://blast.ncbi.nlm.nih.gov/Blast.cgi); the sequences producing significant alignments and the highest query cover were selected. Next, the function of candidate genes was deduced from the UniProt database.
Results
Genetic map of the DH ‘Hewo’ x ‘Magnat’ population with SSR and DArT markers
A set of 92 DH lines derived from F1 triticale plants that originated from a cross between cv. ‘Hewo’ and cv. ‘Magnat’ were used to create a new and unique genetic linkage map. Upon the multiple-mapping approaches, a total of 41 SSR and 680 diversity array technology (DArT) markers were ordered into 22 linkage groups assigned to the A, B, and R genomes (Table 1; Fig. S1 A, B, R; Table S1). The mapped markers with common segregation pattern were binned. Markers representing bins were referred as ‘unique’ while number of all markers in a bin was treated as ‘total’ (Table 1). Additionally, three chromosomes, 7A, 2B, and 3B, were represented by double linkage groups. All mapped DArT markers belonged to three groups, rPt, tPt, and wPt, that were developed respectively from rye, triticale, and wheat. Additionally, during the map construction a small number of markers were eliminated, mainly owing to a high percentage of missing data or a lack of linkage with established markers clusters at LOD value of 2.0. Finally, all remaining markers have covered a total of 1367.7 cM with a mean distance between two markers of 4.7 cM. Microsatellite markers allowed assignment of linkage groups to chromosomes of the A and B genomes of wheat and to five chromosomes of rye (1R, 3R, 4R, 5R, and 6R) (Table 1). Comparative analyses with triticale maps (Tyrka et al. 2011, 2015) and additional information on distribution of wheat DArT markers (series wPt) were provided by Diversity Arrays Technology Pty Ltd. and validated the assignment of linkage groups to rye chromosomes. The order of DArT and SSR markers that were used to develop this linkage map are presented in Supplementary Fig. S1 A, B and R and Supplementary Table S1. Details on the distribution of SSR and DArT markers across triticale genomes of Hewo x Magnat population showed the highest saturation of R genome with unique markers, whereas lower densities were identified for the A and B genomes. R genome was covered by 326 markers with total length of 290.4 cM and a mean distance between two unique markers of 2.7 cM, whereas 140 and 255 markers were distributed within genome A and B respectively, with total length of 460.8 and 616.5 cM and density 6.4 and 5.5 cM (Table 1; Table S1).
Table 1.
Genetic linkage map developed for ‘Hewo × Magnat’ DH mapping population. The mapped markers with common segregation pattern were binned. Markers representing bins were referred as ‘unique’ while number of all markers in a bin was treated as ‘total’. The numbers in bold indicate the length of the entire map and the total number of markers
| Genome | Linkage group | Lenght (cM) | Marker nos | ||
|---|---|---|---|---|---|
| Total | Unique | Density | |||
| A | 1A | 54.3 | 24 | 17 | 3.4 |
| 2A | 45.0 | 10 | 6 | 9.0 | |
| 3A | 43.9 | 10 | 3 | 22.0 | |
| 4A | 76.1 | 34 | 13 | 6.3 | |
| 5A | 78.1 | 12 | 8 | 11.2 | |
| 6A | 48.3 | 8 | 4 | 16.1 | |
| 7A.1 | 61.8 | 25 | 11 | 5.6 | |
| 7A.2 | 53.3 | 17 | 10 | 5.9 | |
| Total genome A | 460.8 | 140 | 72 | 6.4 | |
| B | 1B | 93.3 | 55 | 21 | 4.7 |
| 2B.1 | 65.1 | 16 | 10 | 7.2 | |
| 2B.2 | 48.6 | 16 | 8 | 6.9 | |
| 3B.1 | 22.9 | 14 | 8 | 3.3 | |
| 3B.2 | 115.1 | 38 | 18 | 6.8 | |
| 4B | 42.5 | 12 | 7 | 7.1 | |
| 5B | 41.9 | 13 | 8 | 6.0 | |
| 6B | 97.6 | 42 | 16 | 6.5 | |
| 7B | 89.5 | 49 | 17 | 5.6 | |
| Total genome B | 616.5 | 255 | 113 | 5.5 | |
| R | 1R | 17.5 | 20 | 5 | 4.4 |
| 3R | 37.5 | 31 | 7 | 6.3 | |
| 4R | 71.6 | 82 | 17 | 4.2 | |
| 5R | 36.6 | 66 | 19 | 2.0 | |
| 6R | 127.2 | 127 | 58 | 2.2 | |
| Total genome R | 290.4 | 326 | 106 | 2.7 | |
| Total 22 | 1367.7 | 721 | 291 | 4.7 | |
Freezing tolerance of plants cold acclimated under field conditions
From October to January of winter 2012/2013, plants of the experiment 1 grew under an average temperature about + 4 °C, mostly above 0 °C with few days decrease below (minimum in the average daily temperature to − 6.3 °C in December) (Fig. 1A). The winter 2013/2014 was more severe. Plants of experiment 2 grew under large fluctuations of the average temperature which was mostly above 0 °C in October and November, but below 0 °C in December and January, with minimum − 12.3 °C (Fig. 1B). The period with an average temperature of approximately − 2 °C started in December and continued until March, when temperature raised to about + 2 °C just before the experiment 3 measurements were performed (Fig. 1B).
The results of the freezing tolerance testing
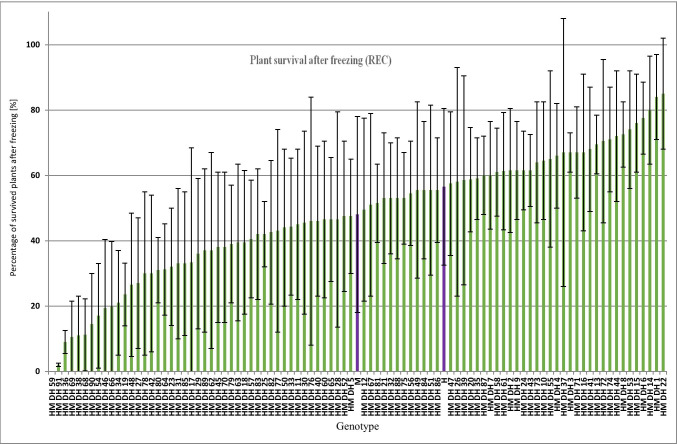
The mean percentage of survived plants from three independent freezing test performed during winter 2012/2013 and 2013/2014 varied from 0 to 85% (Fig. 2). Transgressive segregation of plant recovery was observed in the evaluated population of DH lines. The minimal (0%) percentage of survived plants was observed for HM DH 59 line and the maximal for HM DH 22 (85%) line. Relatively high standard deviation for most DH lines clearly indicates that plants recovery strongly depend on weather conditions. Parent Hewo revealed higher percentage of survived plants after freezing (56%) than parent Magnat (48.5%).
Fig. 2.
Mean variability (from three freezing tests) of the percentage of survived plants in Hewo × Magnat DH population. Violet bars represents parental cultivars Magnat (M) and Hewo (H); green bars represents individual DH lines. Error bars represent the value of standard deviation
PSII photochemical efficiency and membrane stability after freezing
Changes in photochemical efficiency under different stresses can be expressed by the chlorophyll a fluorescence parameters (Kurepin et al. 2013; Rapacz 2007). In our study, the photosynthesis efficiency during cold hardening of triticale plants was evaluated based on the maximum quantum yield of photosystem II (PSII) (Fv/Fm), the overall performance indexes of PSII (PI) and particular JIP test parameters which were recommended by Rapacz et al. (2015a) as a good tools to evaluate freezing tolerance (RC/CSm, RC/CS0, TR0/CS, and ET0/CS). For all evaluated chlorophyll fluorescence parameters transgressive segregation was observed in our mapping population. The values of each parameter were different and dependent (Table 2, Figure S2). Relatively high standard deviation of chlorophyll fluorescence parameters for parents may result from the high impact of various weather conditions. In general, the photosynthetic apparatus for Hewo was more active and functioned better than that for Magnat, resulting in significantly higher ABS/CS (characterizing light energy absorption), RC/CSm (number of active reaction centers). Fv/Fm (maximum quantum yield of PSII) and PI (overall performance index of PSII photochemistry) also had trends for higher means in Hewo, though similar amounts of energy used for electron transport and amount of excitation energy trapped in PSII reaction centers (ET0/CS and TR0/CS, respectively) were for both parents. However, the dissipation of energy in PSII reaction centers (DI0/CS and DI0/RC) was higher for Magnat than Hewo.
Table 2.
Phenotypic performance for traits related to chlorophyll a fluorescence parameters and electrolyte leakage (EL) from leaves of doubled haploid lines and their parents
| Traits | Parents (mean ± SD) | DH lines | |||
|---|---|---|---|---|---|
| Hewo | Magnat | Mean ± SD | Min | Max | |
| Fv/Fm | 0.71 ± 0.45 | 0.63 ± 0.36 | 0.66 ± 0.05 | 0.54 | 0.77 |
| PI | 0.89 ± 0.31 | 0.69 ± 0.34 | 0.66 ± 0.13 | 0.32 | 0.95 |
| ψo | 0.39 ± 0.12 | 0.38 ± 0.11 | 0.35 ± 0.03 | 0.29 | 0.44 |
| φEo | 0.28 ± 0.13 | 0.28 ± 0.13 | 0.25 ± 0.03 | 0.19 | 0.34 |
| PICSo | 324.48 ± 143.21 | 300.42 ± 122.62 | 287.98 ± 54.53 | 142.91 | 425.80 |
| PICSm | 1458.53 ± 426.89 | 1384.42 ± 512.49 | 1314.71 ± 319.48 | 525.43 | 2281.35 |
| ABS/CS | 432.54 ± 3.20 | 412.11 ± 16.77* | 444.36 ± 31.73 | 348.74 | 547.45 |
| TRo/CS | 295.09 ± 117.95 | 291.41 ± 59.38 | 293.32 ± 31.66 | 223.70 | 364.88 |
| ETo/CS | 120.06 ± 58.22 | 115.08 ± 55.24 | 113.21 ± 15.14 | 72.19 | 157.70 |
| DIo/CS | 109.82 ± 33.17 | 148.65 ± 70.08* | 151.45 ± 24.27 | 101.80 | 230.45 |
| ABS/RC | 5.20 ± 4.17 | 6.03 ± 5.53 | 6.27 ± 3.19 | 2.78 | 18.05 |
| ETo/RC | 0.87 ± 0.24 | 0.83 ± 0.25 | 0.80 ± 0.06 | 0.58 | 1.00 |
| TRo/RC | 2.31 ± 0.15 | 2.25 ± 0.16 | 2.34 ± 0.13 | 1.88 | 2.61 |
| DIo/RC | 0.77 ± 0.16 | 1.81 ± 0.19* | 1.37 ± 0.52 | 0.68 | 3.26 |
| RC/CSo | 131.85 ± 39.94 | 127.45 ± 38.72 | 129.02 ± 13.61 | 102.28 | 158.47 |
| RC/CSm | 560.25 ± 239.73 | 526.87 ± 240.07* | 521.11 ± 69.45 | 394.86 | 707.95 |
| EL | 69.51 ± 9.61 | 77.05 ± 15.54 | 59.12 ± 21.53 | 18.98 | 90.02 |
*Significance levels between parents ≤ 0.05
The degree of damage to cell membranes under frost was determined by the measurement of electrolyte leakage (EL) from leaf tissues. The statistically significant differences between parents were not observed. The value of the electrolyte leakage for parental cvs.: Hewo and Magnat was 69% and 77%, respectively. The DH lines differed from parental values of electrolyte leakage. The transgression segregation was clearly highlighted: 65% of the DH lines from the population were characterized by a lower statistically significant EL in relation to both Magnat and Hewo parental cultivars. In contrast, 11% of the DH lines showed a higher statistically significant EL value in relation to both parents. The min and max values of EL was 19% and 90%, respectively (Table 2).
Correlation between plant recovery, electrolyte leakage and chlorophyll a fluorescence parameters
A relationship was established between the average percentage of survived plants from three independent experiments and the mean values of the electrolyte leakage and JIP parameters. For that purpose, the Pearson correlation coefficient (r) was calculated between the individual parameters (Table 3). There was no statistically significant correlation between plant recovery and electrolyte leakage. In contrast, a relatively high positive correlation (0.34 to 0.62) was found between plant recovery (REC) and JIP parameters: Fv/Fm, Ψ0, φE0, PI, PICSo, PICSm, ABS/CS, TR0/CS, ET0/CS, ET0/RC, RC/CS0, RC/CSm, and negative statistically significant correlation (− 0.32 to − 0.35) for ABS/RC, DI0/RC, and DI0/CS. The correlation between freezing tolerance and TR0/RC was not statistical significant (Table 3, Fig. 2S).
Table 3.
Correlation matrix using phenotypic mean values for recovery (REC), electrolyte leakage (EL) from leave and chlorophyll a fluorescence parameters. The parameters of JIP test were calculated and described as in Rapacz (2007): absorbed energy flux per leaf cross-section (CS) and the single, active PSII reaction center (RC) (ABS/CS, ABS/RC respectively); trapped energy flux in PSII reaction centers per leaf cross-section and the single, active PSII reaction center (Tr0/CS, Tr0/RC, respectively); the energy flux for electron transport per leaf cross-section and the single, active PSII reaction center (ET0/CS, ET0/RC, respectively); dissipation of energy in PSII reaction centers per leaf cross-section and the single, active PSII reaction center (DI0/CS, DI0/RC, respectively); yield of the energy trapping in PSII (Fv/Fm); performance indexes of PSII (PI) normalized for minimal and maximal densities of active reaction centers per leaf cross-section (PICS0 and PICSm, respectively); minimal and maximal densities of active reaction centers per leaf cross-section (RC/CS0 and RC/CSm, respectively); the quantum yield of electron transport (φEo); and the efficiency of the electron transfer from QA- to QB (ψo)
| REC | EL | Fv/Fm | ψo | φEo | PI | PICS0 | PICSm | ABS/CS | TR0/CS | ET0/CS | DI0/CS | ABC/RC | TR0/RC | ET0/RC | DI0/RC | RC/CS0 | RC/CSm | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| REC | - | 0.52 | 0.47 | 0.56 | 0.45 | 0.51 | 0.52 | 0.34 | 0.58 | 0.62 | − 0.35 | − 0.32 | - | 0.57 | − 0.32 | 0.54 | 0.61 | |
| EL | - | − 0.25 | − 0.28 | − 0.21 | - | - | - | 0.36 | - | - | 0.48 | - | 0.45 | - | 0.40 | - | - |
EL was significantly negatively correlated with the individual chlorophyll a fluorescence parameters: Fv/Fm, Ψ0, and φE0. Positive correlation was statistically significant from 0.36 to 0.48 between EL and the parameters: ABS/CS, DI0/CS, TR0/RC, and DI0/RC (Tqb. 3).
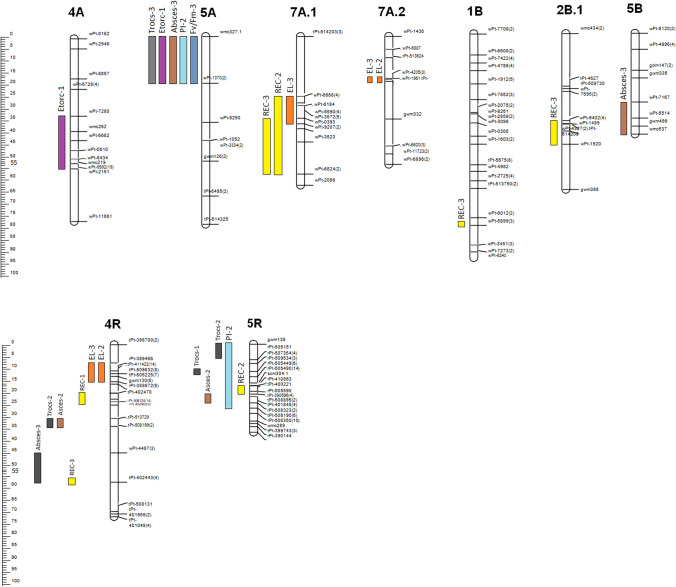
Quantitative trait loci (QTL) for plants recovery
Using CIM analysis, QTL regions for recovery (REC) expressed as percentage of survived plants were identified on chromosomes 7A.1, 1B, 2B.1, 4R, and 5R (Table 4; Fig. 3). Additionally, all QTL regions identified by SMA method are shown in Table S3. The phenotypic variation explained by those QTL ranged from 4.4 to 15.8% depending on winter conditions and method used (Table 4; Table S3). The QTL for percentage of survived plants evaluated in two experiments were co-located on chromosome 7A.1 and 1B and named Qfr.hm-7A.1 and Qrec.hm-1B.1, respectively (Table 4; Fig. 3). Those loci covered 27.5–57.2 cM distance for Qfr.hm-7A.1 and explained up to 9.5% of phenotypic variation. For second locus, Qrec.hm-1B.1 CIM analysis showed only one marker wPt-5899(3) in experiment 3 (Table 4; Fig. 3). Those loci explained up to 8.2% of phenotypic variation with the LOD value 2.5 (Table 4).
Table 4.
Summary of QTL identified using CIM method for plant recovery (REC), percentage of survived plants and electrolyte leakage (EL) from leaves after plant freezing
| QTL name | QTL name | Trait-experiment | Flanking markers | Interval/position (cM) | LOD | R2 (%)a | Addb | Favorable allelec |
|---|---|---|---|---|---|---|---|---|
| Qel.hm-7A-1.4 | Qfr.hm-7A.1 | EL-3 | wPt-6668(4)-wPt9207(2) | 27.5–38.3 | 3.1 | 9.5 | − 5.5 | M |
| Qrec.hm-7A-1.4 | REC-2 | wPt-6668(4)-wPt-6824(2) | 27.5–57.2 | 2.6 | 6.9 | 6.6 | H | |
| REC-3 | wPt-0393-wPt-6824(2) | 36.2–57.2 | 2.5 | 9.2 | 6.5 | H | ||
| Qel.hm-7A-2.1 | Qel.hm-7A.2 | EL-2 | tPt-513624-tPt-2230(2) | 8.9–19.9 | 2.6 | 8.7 | 8.1 | H |
| EL-3 | tPt-513624-wPt-1961 | 8.9–19.3 | 4.1 | 13.0 | 6.4 | H | ||
| Qrec.hm-1B.1 | Qrec.hm.1B.1 | REC-3 | wPt-5899(3) | 79.9 | 2.5 | 8.2 | 6.7 | H |
| Qrec.hm-2B.1 | Qrec.hm-2B.1 | REC-3 | wPt-9402(4)-wPt-1920 | 36.3–46.1 | 2.9 | 10.4 | 7.2 | H |
| Qel.hm-4R.1 | Qel.hm-4R.1 | EL-2 | rPt-389466-rPt-389872(5) | 9.8–16.5 | 6.5 | 22.8 | 18.7 | H |
| EL-3 | rPt-389466-rPt-389872(5) | 9.8–16.5 | 2.5 | 7.7 | 4.9 | H | ||
| Qrec.hm-4R.1 | Qrec.hm-4R.2 | REC-1 | tPt-402470-rPt402563(5) | 22.6–25.2 | 3.5 | 12.9 | -0.8 | M |
| Qrec.hm-4R.2 | Qrec.hm-4R.3 | REC-3 | tPt-402443(4) | 57.2 | 3.0 | 10.8 | − 10.8 | M |
| Qrec.hm-5R.1 | Qrec.hm-5R.1 | REC-2 | rPt-508323(2)-tPt-390596(4) | 22.2–26.5 | 2.5 | 6.3 | − 6.9 | M |
| Qel.hm-5R.1 | Qel.hm-5R.2 | EL-3 | wmc289-rPt-506350(10) | 32.0–32.9 | 2.5 | 5.0 | 6.8 | H |
aR2 (%), the percentage of phenotypic variance explained by the QTL
bAdd, additive effects of QTL expressed in the trait unit
cFavorable allele for each QTL: H, cv. Hewo and M, cv. Magnat
Fig. 3.
Interval map (cM) for 4A, 5A, 7A.1, 7A.2, 1B, 2B.1, 5B, 4R, and 5R chromosomes of ‘Hewo’ x ‘Magnat’ DH population with QTL identified by CIM method for plant recovery (REC), and electrolyte leakage (EL), Fv/Fm, PI and energy fluxes parameters
Quantitative trait loci (QTL) for electrolyte leakage
Electrolyte leakage (EL) was measured two times during winter 2012/2013. QTL controlling EL after freezing, identified by CIM method were on chromosomes 7A.1, 7A.2, 4R, and 5R (Table 4; Fig. 3). Additionally, all QTL regions identified by SMA method are shown in Table S4. The phenotypic variation explained by those QTL ranged from 3.3 to 22.8% depending on experiment and method used (Table 4; Table S4). The locus Qfr.hm-7A.1, which was identified as associated with EL in experiment 3 was also co-located with QTL identified for plants recovery on chromosome 7A.1. Locus Qfr.hm-7A.1 explained 9.5% of phenotypic variation observed for EL (Table 4) with LOD value 3.1 and was in similar position in cM to loci identified for plants recovery (Table 4). Another locus identified on chromosome 7A, Qel.hm-7A.2 was located in 8.9–19.9 cM distance and explained up to 13% of phenotypic variation; it contained loci found in experiment 2 and 3 (Table 4; Fig. 3). Locus Qel.hm-4R.1 located in 9.8–16.5 cM distance on chromosome 4R was also identified in experiment 2 and 3 (Table 4; Fig. 3). It explained up to 22.8% of phenotypic variation with the LOD value 6.5 in experiment 2 (Table 4). One loci identified in experiment 3 was located on chromosome 5R and covered by wmc289 to rPt-506350(10) markers (Table 4; Fig. 3). This loci explained 6.8% of phenotypic variation (Table 4).
Quantitative trait loci (QTL) for chlorophyll a fluorescence parameters
QTL for chlorophyll a fluorescence parameters identified by CIM method were located on chromosomes 4A, 5A, 5B, 4R, and 5R (Table 5; Fig. 3). Additionally, QTL identified by SMA method are shown in Tables S5–S7.
Table 5.
Summary of QTL for Fv/Fm (the yield of the energy trapping in PSII), PI (performance indexes of PSII) and energy fluxes (RC per single, active PSII reaction center and CS per leaf cross-section) identified using CIM methods. QTL name includes each parameter name (after the letter Q)
| QTL | QTL name | Trait-experiment | Flanking markers | Interval/position (cM) | LOD | R2 (%)a | Addb | Favorable allelec |
|---|---|---|---|---|---|---|---|---|
| Qetorc.hm-4A.1 | Qchl.hm-4A.1 | ET0/RC-1 | wPt-7280-wPt-2151 | 32.3–56.6 | 2.5 | 9.1 | 3.7 | H |
| Qfvfm.hm-5A.2 | Qchl.hm-5A.1 | Fv/Fm-3 | wmc327.1-wPt-1370(2) | 0.0–19.4 | 3.3 | 12.0 | 0.1 | H |
| Qpi.hm-5A.2 | PI-2 | wmc327.1-wPt-1370(2) | 0.0–19.4 | 3.8 | 11.5 | 0.1 | H | |
| Qabscs.hm-5A.2 | ABS/CS-3 | wmc327.1-wPt-1370(2) | 0.0–19.4 | 3.3 | 12.7 | 25.5 | H | |
| Qetocs.hm-5A.2 | ET0/CS-3 | wmc327.1-wPt-1370(2) | 0.0–19.4 | 3.9 | 13.0 | 11.4 | H | |
| Qtrocs.hm-5A.2 | TR0/CS-3 | wmc327.1-wPt-1370(2) | 0.0–19.4 | 5.4 | 19.6 | 36.8 | H | |
| Qabscs.hm-5B.1 | Qchl.hm-5B.1 | ABS/CS-3 | wPt-7167-wmc537 | 28.6–41.9 | 3.9 | 11.6 | 24.8 | H |
| Qabscs.hm-4R.2 | Qchl.hm-4R.1 | ABS/CS-2 | tPt-513729-tPt-508199(2) | 32.9–33.9 | 2.5 | 7.5 | − 11.3 | M |
| Qtrocs.hm-4R.2 | TR0/CS-2 | tPt-513729-tPt-508199(2) | 32.9–33.9 | 2.8 | 8.8 | − 9.0 | M | |
| Qabscs.hm-4R.2 | ABS/CS-3 | wPt-4487(3)-tPt-402443(4) | 45.0–57.2 | 3.1 | 10.3 | − 22.8 | M | |
| Qpi.hm-5R.1 | Qchl.hm-5R.1 | PI-2 | gwm136-tPt-508190(6) | 0.0–28.7 | 3.2 | 9.6 | 0.1 | H |
| Qtrocs.hm-5R.1 | TR0/CS-2 | gwm136-rPt-507354(4) | 0.0–8.0 | 3.1 | 10.8 | − 10.6 | M | |
| Qabscs.hm-5R.1 | ABS/CS-2 | tPt-390596(4)-rPt-508323(2) | 22.2–26.5 | 2.5 | 7.5 | − 11.8 | M | |
| Qtrocs.hm-5R.1 | TR0/CS-1 | rPt-505449(6) | 13.5 | 2.6 | 6.2 | − 6.1 | M |
aR2 (%), the percentage of phenotypic variance explained by the QTL
aAdd, additive effects of QTL expressed in the trait unit
cFavorable allele for each QTL: H, cv. Hewo and M, cv. Magnat
Abbreviations: ABS/CS, absorbed energy flux per leaf cross-section (CS); Tr0/CS, trapped energy flux in PSII reaction centers per leaf cross-section; ET0/CS, the energy flux for electron transport per leaf cross-section; ET0/RC, the energy flux for electron transport per the single, active PSII reaction center; Fv/Fm, yield of the energy trapping in PSII; PI, performance indexes of PSII
Locus Qchl.hm-4A.1 identified on chromosome 4A in a distance between 21.1 cM to 56.6 cM consisted QTL for ET0/RC and explained 9.1%of phenotypic variation with the LOD value 2.5 (Table 5; Fig. 3). Locus Qchl.hm-5A.1 covered by wmc327.1 to wPt-1370(2) markers was identified for Fv/Fm, PI, ABS/CS, ET0/CS and TR0/CS in experiment 2 and 3 (Table 5; Fig. 3). It explained up to 19.6% of phenotypic variation with the LOD value up to 5.4 for TR0/CS (Table 5). On chromosome 5B, locus Qchl.hm-5B.1 was identified for ABS/CS with the LOD value 3.9 and phenotypic variation 11.6% (Table 5; Fig. 3). Locus Qchl.hm-4R.1 was identified on chromosome 4R for TR0/CS measured in experiment 2 and ABS/CS for experiment 2 and 3 (Table 5; Fig. 3). Loci identified for ABS/CS and TR0/CS measured in experiment 2 were located in the same position (32.9–33.9 cM; Table 5). On chromosome 5R, locus Qchl.hm-5R.1 was identified for PI, TR0/CS and ABS/CS measured in experiment 2 and TR0/CS measured in experiment 1 (Table 5; Fig. 3). This QTL explained up to 10.8% of phenotypic variation with LOD value up to 3.2 (Table 5).
Candidate genes for analyzed traits
Seven candidate genes were in silico identified within QTL found for plants recovery (REC) and electrolyte leakage (EL) as well as trapped energy flux in PSII reaction centers per leaf cross-section (Tr0/CS), the energy flux for electron transport per leaf cross-section (ET0/CS) and absorbed energy flux per leaf cross-section (ABS/CS) traits (Table 6). Gene coding PPR protein involved in chloroplast RNA processing, modification, and splicing was identified as associated with plants recovery (Table 6). Genes identified as associated with electrolyte leakage encode mRNA-binding protein BTR1-like and involved in regulation of gene expression as well as transmembrane cyclic nucleotide-gated ion channel (Table 6). In turn, genes associated with chlorophyll a fluorescence parameters were chloroplastic uridine kinase-like protein 1 involved in the pyrimidine salvage pathway, uncharacterized LOC119301557 and phosphoinositide phosphatase SAC9 involved in stress signaling (Table 6).
Table 6.
The characteristics of candidate genes identified in the QTL regions associated with the seedlings traits: electrolytes leakage (EL), plants recovery (REC), trapped energy flux in PSII reaction centers per leaf cross-section (Tr0/CS), the energy flux for electron transport per leaf cross-section (ET0/CS) and absorbed energy flux per leaf cross-section (ABS/CS) in DH ‘Hewo x Magnat’ lines mapping population studied
| QTL name | Trait/experiment | Flanking marker | Sequence query | Position | Sequence ID (mRNA) | Reference organism | Predicted protein | Predicted function |
|---|---|---|---|---|---|---|---|---|
| Qel.hm-7A.2 | EL-2 EL-3 | tPt-513624 | TraesCS7A03G0506500LC (low confidence) | Chr7A:189,793,060..189793458 (+ strand) | XM_037604689.1 | Triticum dicoccoides | Uncharacterized LOC119331524 transcript var. X2 | BTR1-like protein, mRNA binding, regulation of gene expression |
| Qel.hm-5R.2 | EL-3 | rPt-506350 | SECCE5Rv1G0349830 (high confidence) | Chr5R:686,819,682..686824096 (− strand) | XM_037587241.1 | Triticum dicoccoides | Nucleotide-gated ion channel 14 (LOC119311587) | Voltage-gated potassium channel activity, cyclic nucleotide-gated ion channel, integral component of membrane, transmembrane helical protein |
| Qfr.hm-7A.1 | REC-3 | wPt-0393 | TraesCS4B01G034600 (high confidence) | Chr4B:25,384,962..25394077 (− strand) | XM_037576333.1 | Triticum dicoccoides | Pentatricopeptide repeat-containing protein At4g18520, chloroplastic-like (LOC119299015), transcript var. X1 | PPR protein, involved in chloroplast RNA processing, modification and splicing |
| Qchl.hm-4A.1 | ET0/RC-1 | wPt-2151 | TraesCS4A03G1247800 (high confidence) | Chr4A:752,456,325..752459406 (− strand) | XM_020328553.1 | Aegilopstauschii subsp. strangulata | Uncharacterized LOC109769846 | - |
| Qchl.hm-5B.1 | ABS/CS-3 | wPt-7167 | TraesCS5B03G0735400 (high confidence) | Chr5B:473,178,495..473184426 (− strand) | XM_037586621.1 | Triticum dicoccoides | Uridine kinase-like protein 1, chloroplastic (LOC119310993) | Involved in the pyrimidine salvage pathway |
| Qchl.hm-5R.1 | TR0/CS-2 | rPt-507354 | SECCE5Rv1G0327250 (high confidence) | Chr5R:490,868,494..490871516 (− strand) | XM_037578543.1 | Triticum dicoccoides | Uncharacterized LOC119301557 | Gag-Pol polyprotein/retrotransposon, transmembrane helix protein |
| TR0/CS-1 | rPt-505449 | SECCE5Rv1G0298360 (high confidence) | Chr5R:11,157,843..11168368 (+ strand) | XM_037576070.1 | Triticum dicoccoides | Phosphoinositide phosphatase SAC9 (LOC119298775), transcript var. X3 | Probable phosphoinositide phosphatase that could be involved in stress signaling, phosphoric ester hydrolase activity, phosphatidylinositol metabolic process, response to osmotic stress, integral component of membrane |
Discussion
Recently, climate change has been disturbing the natural hardening process (Dalmannsdottir et al. 2017). Global warming caused by the increase in average temperature on Earth is reflected in climate instability and violent local breakdowns of the weather. This leads to the limitation of seamless adjustment of plant physiology to the prevailing conditions, including the course of winters (Rapacz et al. 2014; Dalmannsdottir et al. 2017). Thus, an effective method to evaluate plant freezing tolerance and/or winter hardiness under unstable winter conditions is urgently needed. In our study, we observed differences in plant recovery of DH ‘Hewo x Magnat’ lines population after freezing depending on the experiment condition. Furthermore we observed that differences in plant recovery between those DH lines were closely related to damages of photosynthetic apparatus measured by means of JIP analysis. The relationship between plant survival and photochemical efficiency of PSII under unstable winter conditions was confirmed earlier in wheat (Clement and van Hasselt 1996; Rapacz 2007), oat (Rizza et al. 2001), barley (Francia et al. 2004), and triticale (Rapacz et al. 2011, 2015ab).
The most commonly reported fluorescence parameter for evaluating plant responses to abiotic stress is Fv/Fm which informs about the maximum photochemical PSII activity (Del Rosso et al. 2009). In the studied population, a high positive and statistically significant correlation (0.52) between plant survival and Fv/Fm was obtained. However, there are many reports available showing that different parameters of chlorophyll fluorescence parameters followed by JIP tests could be used as the more effective indicators of freezing tolerance than Fv/Fm, especially when plants are well cold acclimated before the measurements, which was also reported for triticale (Rapacz et al. 2011, 2015ab, 2017). In the present study, we confirmed high statistically significant correlation values of the remaining JIP parameters with plant survival. Similarly to Rapacz et al. (2015a) study, in our results on the JIP test, best correlating with plant survival were ET0/CS and RC/CSm parameters. Moreover, in our study, the dependence of plants ability to recover after freezing on photosynthetic apparatus was also reflected in the colocation of quantitative traits loci on chromosomes 4R and 5R, where many genes related with freezing tolerance were earlier identified.
The location of QTL that control the photochemical efficiency of PSII on chromosome 5 group (5A, 5B, and 5R) may indicate that this trait is partially controlled by the frost-resistance genes. The main QTL associated with photochemical activity of PSII were mapped on chromosome 5A at 58.7–78.1 cM, within the range for which the major frost-resistance loci Fr-A1 and Fr-A2 identified by other authors (Vágújfalvi et al. 2000, 2003). The QTL identified on chromosome 5A for chlorophyll fluorescence parameters may also contain the vrn1 gene (located in close proximity to the Fr-A1 frost-resistance locus). This hypothesis may be supported by the fact that the wPt-1370 marker located on chromosome 5A in DH Hewo x Magnat mapping population used in our study was reported as a marker in close proximity to the gene vrn1 on the genetic map developed for the wheat population Berkut/Krichauff (Genc et al. 2010). On chromosome 5A, the following genes have been also identified: Cor14b, Cbf3 (Vágújfalvi et al. 2000, 2003; Miller et al. 2006), Cbf14, and Cbf15 (Båga et al. 2007). In addition, the Cbf genes: TmCBF12 and HvCbf14 recognized as candidate genes for frost-resistance genes were also located on chromosome 5A at the locus labeled Fr-A2 in wheat and respectively at the locus labeled Fr-H2 in barley at the homeological chromosome 2H (Knox et al. 2008; Fricano et al. 2009). On the 5B wheat chromosome, genes associated with flowering and frost tolerance have been identified and the locus was labeled Fr-B1 (Toth et al. 2003). These results clearly indicate an important role of 5 chromosome groups in the creating of plant freezing tolerance not only in wheat but also in triticale (Alm et al. 2011).
For the purpose of present QTL analysis, new genetic map of DH ‘Hewo’ x ‘Magnat’ lines population was developed. The unique set of SSR and DArT markers enabled to assign a total of 721 markers into 22 linkage groups of triticale. Diversity Arrays Technology (DArT) used in our research is known as a tool used to construct genetic maps of many crop species (Wenzl et al. 2004). That microarray technology can provide the information of several thousand sequence-specific markers without sequence information (Jaccoud et al. 2001; Karbarz et al. 2020). In our research, the distribution of DArT and SSR markers showed the highest saturation of R genome in contrast to the A and B genomes. Similar effect with the best coverage and highest density of R genome in triticale genetic maps were previously described by Tyrka et al. (2011) and Karbarz et al. (2020). Up to now, several genetic maps have been developed and reported QTL analysis in triticale (Alheit et al. 2011; Badea et al. 2011; González et al. 2005; Tyrka et al. 2011, 2015; Krzewska et al. 2012, 2015; Dyda et al. 2019; Karbarz et al. 2020).
Numerous QTL associated with the most commonly analyzed Fv/Fm chlorophyll a fluorescence parameter have been identified in association with various types of stress (Yang et al 2007; Liang et al 2010; Alm et al. 2011; Zhang et al. 2017; Czyczyło-Mysza et al. 2011, 2013; Kumar et al. 2012; Cheng et al. 2012; Bhusal et al. 2018; Dyda et al. 2019). QTL for Fv/Fm under drought stress was identified in wheat on chromosome 5A (Czyczyło-Mysza et al. 2011; Liang et al. 2010; Alm et al. 2011). No correlation was observed in our study between plant recovery after freezing and electrolyte leakage from freezing-damaged leaf tissues of triticale. On the other hand, 10 QTL regions related to the stability of cell membranes measured by electrolyte leakage from leaf tissues after freezing have been identified. Further studies would help assess whether the identified QTLs for the EL could be related to other parameter/s related to plant survival after freezing that was not measured in this work. The QTL identified for freezing tolerance of photosynthetic apparatus was located on chromosomes inherited from wheat, 4A, 5A, 5B, and on rye chromosomes, 4R and 5R. QTL on chromosomes 4A, 5A, and 5B, two loci on chromosome 4R as well as loci on 5R chromosome were identified in both experiments conducted during winter 2012/2013. In our study, loci related to cell membrane stability were identified on chromosome 7A, that is the same chromosome where Morgan and Tan (1996) identified QTL associated with osmoregulation in wheat. In rice (Oryza sativa L.) under drought stress, QTL of the cell membranes stability were identified on chromosomes 1, 3, 7, 9, 11, and 12 (Tripathy et al. 2000) as well as chromosome 8 (Lilley et al. 1996; Tripathy et al. 2000). The literature also provides a lot of information on the genes that are expressed in response to the stress of drought and low temperature and regulate the condition of cell membranes (Danyluk et al. 1998; Nylander et al. 2001; Alm et al. 2011; Kocheva et al. 2014; Janeczko et al. 2019).
In our research, we mainly focused on finding the molecular background of freezing tolerance based on the crosstalk between chlorophyll fluorescence measurements of photosynthetic apparatus freezing tolerance, cell membrane stability, and plant recovery abilities after freezing. For these three components of plant freezing tolerance, we found durable, strong, constant quantitative trait loci regardless on plant growth conditions. A total of 9 genomic regions (QTL on 7A, 1B, 2B, 4R and 5R chromosomes) associated with plant survival after freezing were identified. Four of them were mapped in at least two experiments (7A, 1B and one locus on 4R chromosome). In contrast, QTL identified for one seasons can be considered as potential loci and/or loci specific to particular weather conditions. QTL of plant survival after freezing identified on chromosome 1B confirmed previous results obtained for Norstar × Cappelle-Desprez DH population where locus of frost resistance was mapped (Chodaparambil 2009). Frost-resistance loci in the DH Norstar × Cappelle-Desprez population were identified on chromosome 1B at 90–118 cM. In the present study, QTL of plant recovery was located in a close position on chromosome 1B at position 79.9 cM. Similar to the results obtained for the Hewo × Magnat DH population, also some of the earliest studies have indicated that genes associated with wheat frost resistance are located on chromosomes 7A and 2B (Sutka 1981; 1994; Galiba et al. 2003) and additionally on chromosomes: 4B and 4D (Law and Jenkins 1970; Puchkov and Zhirov 1978; Sutka 1981; Roberts 1986). Chromosome groups 1, 2, and 7 indicated in this study as containing strong QTL of plant survival regardless of environmental conditions, could mainly carry loci responsible for vernalization, flowering and photoperiod regulation, which only may indirectly affect cereal frost resistance (Galiba et al. 1995; Law and Worland 1997; Mahfoozi et al. 2000; Skinner et al. 2004). Genes related to the vernalization process are located, for example, on the wheat group 1 homeological chromosome (vrn3 gene) (Law and Worland 1997) and on barley 7 chromosomes (sh2 gene) (Pan et al. 1994; Galiba et al. 1995) and on the 7 rye chromosome (Plaschke et al. 1993; Galiba et al. 1995). In the presented study, QTL of plant survival after freezing was located exclusively on one of the chromosomes of 5 group, rye chromosome (5R). The reason why the locus responsible for plant survival was not mapped on chromosome 5A in the studied population is probably the lack of variation between parents in this region. Moreover, in triticale the absence of important freezing tolerance loci on wheat chromosome 5D are reported to be compensated by 5R loci.
On chromosome 5R many authors have identified genes of freezing tolerance described as CBF family, closely related to the locus corresponding to the locus Fr2 on homeological chromosomes of the 5 group in barley, diploid, and hexaploid wheat and meadow fescue (Francia et al. 2007; Baga et al. 2007; Knox et al. 2008; Tamura and Yonemaru 2010; Alm et al. 2011; Zhang et al. 2019). Campoli et al. (2009) also identified 12 different Cbf family genes on the long arm of the 5R chromosome. Loci ScCbf (ScCbf2, ScCbf6, ScCbf9b, ScCbf12, ScCbf15, ScIce2, ScDhn3) were mapped on chromosome 5R in similar positions to the frost-resistance loci Fr-H2/Fr-am2 in barley and diploid wheat, respectively (Li et al. 2011a, b). The described Cbf family genes (Campoli et al. 2009; Li et al. 2011a, b) were located on the 5R chromosome in similar position as Qrec.hm-5R.1 identified in our study. Campoli et al. (2009) clearly showed that the expression of Cbf family genes is closely related to the temperature during the acclimation process and the measurement time. This hypothesis could explain why this QTL was identified in our study only in experiment 3.
In the evaluated Hewo × Magnat DH population, QTL regions co-located for plant survival, membrane stability, and photochemical PSII efficiency were identified on rye chromosomes 4R and 5R. QTL for these parameters could indicate the partially general genetic background responsible for the survival of whole plants and the protection of their photosynthetic apparatus after freezing. The colocation of the QTL for photochemical PSII efficiency, membrane stability and plant survival may be due to the fact that co-located genes can control the effectiveness of photosynthesis under stress conditions and affect protection during acclimatization or to aid plant regeneration after stress. The involvement of such mechanisms can be jointly controlled at the genome level, and even the efficiency of the photosynthetic apparatus can directly be responsible for the ability of plant to survive freezing. On the other hand, the incomplete correlation between the chlorophyll fluorescence parameters, membrane stability and the plant survival and the lack of collocation of many QTL obtained between these parameters suggest that not all physiological mechanisms were indicated and plant recovery, membrane stability, and photosynthetic efficiency of PSII under stress conditions in many aspects are controlled by different genes.
Our results of QTL and gene identification strongly indicate physiological and genetic relationship of the plant survival after freezing with the photochemical activity of the photosystem II. For plants recovery after freezing stress, gene coding pentatricopeptide repeat-containing protein At4g18520, involved in chloroplast RNA processing, modification, and splicing, was identified. Such a result indicates a significant role of chloroplast genes in winter triticale seedlings frost survival. Other authors research, conducted in Arabidopsis thaliana showed that protein At4g18520 may be required for proper chloroplast development, the regulation of the plastid gene expression probably through regulation of plastid-encoded polymerase (PEP) dependent chloroplast transcription, for RNA editing of several chloroplastic transcripts (especially accD transcripts), for processing of the chloroplastic rpoA pre-mRNA as well as for the monocistronic rpoA transcript processing (Yin et al. 2012).
In turn, genes identified as associated with chlorophyll a fluorescence parameters were chloroplastic uridine kinase-like protein 1, uncharacterized LOC119301557, and phosphoinositide phosphatase SAC9. Uridine kinase is plasma membrane protein, involved in CTP and UMP salvage, nucleoside metabolic process, protein secretion, regulation of exocytosis, and vesicle docking involved in exocytosis. Phosphoinositide phosphatase could be involved in stress signaling (Williams et al. 2005). At the same time, these genes were identified for the chlorophyll fluorescence parameters, the values of which showed a positive correlation with the plants recovery tests result. Genes found for electrolyte leakage code mRNA-binding protein BTR1-like involved in regulation of gene expression as well as transmembrane cyclic nucleotide-gated voltage-gated potassium channel. Protein-forming potassium channel was identified among abiotic stress responsive proteins of wheat grain determined using proteomics technique by Kamal et al. (2010).
In conclusion, QTL that control the PSII photochemical efficiency were identified on chromosome 5 group (5A, 5B, and 5R). Six genomic regions associated with plant survival after freezing (REC) were identified and two of them were mapped in two experiments (chromosomes 7A.1, 1B, 5R, and 4R). Co-located QTL for plant survival, membrane stability, and photochemical PSII efficiency were identified on chromosomes 4R and 5R. The collinearity of the QTL for PSII photochemical efficiency, membrane stability, and plant survival may be due to the fact that co-located genes can control the effectiveness of photosynthesis under stress conditions and affect protection during acclimatization or to aid plant regeneration after stress. The same QTL for electrolyte leakage and plant recovery Qfr.hm-7A.1 was identified in two experiments indicating that these QTL are responsible for differences in freezing tolerance in our mapping population regardless on strong genotype-environmental interaction observed for freezing tolerance in triticale making this regions particularly interesting for breeders. QTL for percentage of plant survival and electrolyte leakage were both co-located on chromosomes 4R and 5R with QTL for several chlorophyll fluorescence parameters. Such results indicate that the correlation exist between freezing tolerance of photosynthetic apparatus, plasma membranes and plant regrowth after freezing. This conclusion is also supported by chloroplast and membrane genes identified in genome regions associated with these traits.
Supplementary Information
Below is the link to the electronic supplementary material.
Genetic linkage map of A chromosomes with molecular markers. (JPG 3498 KB)
Genetic linkage map of B chromosomes with molecular markers. (JPG 5346 KB)
Genetic linkage map of R chromosomes with molecular markers. (JPG 5349 KB)
Mean values of chlorophyll a fluorescence parameters measured in DH population after cold acclimation in three different field conditions. A, B, C, D, E, F, G, H, I, J charts were made for ABS/RC, TR0/RC, ET0/RC, DI0/RC, ABS/CS, TR0/CS, ET0/CS, DI0/CS, FV/FM and PI, respectively. Plant survival on the x-axis (0-100) is a number of surviving (regrowing) plants examined after three weeks of regrowth in glasshouse (temp. about +15°C) and expressed as a percentage of plant survival. (DOCX 1369 KB)
Genetic map and segregations of markers in population ‘Hewo’ x ‘Magnat’. Number of cosegregating markers was given (in brackets) in the names of representing bin markers. Segregartions of redundant markers were not provided. Physical positions of merkers were obtained afret balstn of marker sequences on ChineseSpring wheat V1.0 (URGI: https://wheat-urgi.versailles.inra.fr/Seq-Repository/BLAST ) and Lo1 rye chromosomes (WheatOmics 1.0: http://202.194.139.32/blast/blast.html). (XLSX 244 KB)
Data on maximum expected number of recombinations per individual (Max_rec) extracted and sum of adjacent recombination frequencies (Mean_rec) from JoinMap. Percentage of lines without crossing-over (c-o) was calculated in Excel. (DOCX 19 KB)
Summary of QTL identified using SMA method for plants recovery REC (percentage of survived plants after freezing) after freezing test performed during winter 2011/2012 (Experiment 1) and 2012/2013 (Experiments 2, 3).Table S4. Summary of QTL for electrolyte leakage (EL) from leaves after freezing identified using SMA method. Table S5. Summary of QTL for Fv/Fm (the yield of the energy trapping in PSII) and PI (performance indexes of PSII) identified using SMA method. QTL name includes each parameter name (after the letter Q). Table S6. Summary of QTL for energy fluxes (per single, active PSII reaction center; RC) identified using SMA method. QTL name includes each parameter name (after the letter Q). Abbreviations: ABS/RC - absorbed energy flux per the single, active PSII reaction center (RC); TR0/RC - trapped energy flux in PSII reaction centers per the single, active PSII reaction center; DI0/RC - dissipation of energy in PSII reaction centers per the single, active PSII reaction center. Table S7. Summary of QTL for phenomenological energy fluxes (per leaf cross-section; CS) identified using SMA method. QTL name includes each parameter name (after the letter Q). Abbreviations: ABS/CS - absorbed energy flux per leaf cross-section (CS); Tr0/CS - trapped energy flux in PSII reaction centers per leaf cross-section; ET0/CS - the energy flux for electron transport per leaf cross-section; DI0/CS – dissipation of energy in PSII reaction centers per leaf cross-section. (DOC 227 KB)
Author contribution
IW — plant culture, REC, EL and JIP analysis, QTL analysis, and manuscript writing. MD — plant culture, REC, EL and JIP analysis, QTL and statistical analysis, and manuscript writing. GG — plant culture, REC, EL and JIP analysis, QTL and statistical analysis, modification of the in silico candidate gene identification method, candidate gene identification, manuscript writing, and corresponding author. MT — developing of the new genetic linkage map, QTL analysis, and manuscript writing. MR — experiment design, procuring funding, and manuscript writing. MS-H — manuscript writing. MW — experiment design, procuring funding, and manuscript writing.
Funding
Research funded by National Science Centre, project N N310 778640 and project GENMARK PBS1/A8/1/2012.
Data availability
Not applicable.
Code availability
Not applicable.
Declarations
Ethics approval
Not applicable.
Consent for publication
Not applicable.
Consent to participate
Not applicable.
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Alheit KV, Reif JC, Maurer HP, Hahn V, Weissmann EA, Miedaner T, Würschum T. Detection of segregation distortion loci in triticale (xTriticosecale Wittmack) based on a high-density DArT marker consensus genetic linkage map. BMC Genomics. 2011;12:380. doi: 10.1186/1471-2164-12-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alm V, Busso CS, Ergon Å, Rudi H, Larsen A, Humphreys MW, Rognli OA. QTL analyses and comparative genetic mapping of frost tolerance, winter survival and drought tolerance in meadow fescue (Festuca pratensis Huds.) Theor Appl Gene. 2011;123(3):369–382. doi: 10.1007/s00122-011-1590-z. [DOI] [PubMed] [Google Scholar]
- Badea A, Eudes F, Salmon D, Tuvesson S, Vrolijk A, Larsson C-T, Caig V, Huttner E, Kilian A, Laroche A. Development and assessment of DArT markers in triticale. Theor Appl Genet. 2011;122:1547–1560. doi: 10.1007/s00122-011-1554-3. [DOI] [PubMed] [Google Scholar]
- Båga M, Chodaparambil SV, Limin AE, Pecar M, Fowler DB, Chibbar RN. Identification of quantitative trait loci and associated candidate genes for low-temperature tolerance in cold-hardy winter wheat. Funct Integr Genomics. 2007;7(1):53–68. doi: 10.1007/s10142-006-0030-7. [DOI] [PubMed] [Google Scholar]
- Bhusal N, Sharma P, Sareen S, Sarial AK. Mapping QTLs for chlorophyll content and chlorophyll fluorescence in wheat under heat stress. Biol Plant. 2018;62(4):721–731. doi: 10.1007/s10535-018-0811-6. [DOI] [Google Scholar]
- Burbulis N, Jonytiene V, Kupriene R, Blinstrubiene A. Changes in proline and soluble sugars content during cold acclimation of winter rapeseed shoots in vitro. J Food Agric Environ. 2011;9:371–374. [Google Scholar]
- Campoli C, Matus-Cádiz MA, Pozniak CJ, Cattivelli L, Fowler DB. Comparative expression of Cbf genes in the Triticeae under different acclimation induction temperatures. Mol Genet Genomics. 2009;282(2):141–152. doi: 10.1007/s00438-009-0451-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catala R, Salinas J. Review. Regulatory mechanisms involved in cold acclimation response. Span J Agric Res. 2008;6:211–220. doi: 10.5424/sjar/200806s1-390. [DOI] [Google Scholar]
- Cheng L, Wang Y, Meng L, Hu X, Cui Y, Sun Y, Ali J, Xu J, Li Z. Identification of salt-tolerant QTLs with strong genetic background effect using two sets of reciprocal introgression lines in rice. Genome. 2012;55(1):45–55. doi: 10.1139/g11-075. [DOI] [PubMed] [Google Scholar]
- Chodaparambil SV (2009). Genetic analysis of low-temperature tolerance in winter wheat (Doctoral dissertation, University of Saskatchewan).
- Clement JMAM, van Hasselt PR. Chlorophyll fluorescence as a parameter for frost hardiness in winter wheat. A comparison with other hardiness parameters. Phyton Ann Rei Bot Austria. 1996;36:29–41. doi: 10.1016/b978-0-12-447602-8.50049-0. [DOI] [Google Scholar]
- Czyczyło-Mysza I, Marcińska I, Skrzypek E, Chrupek M, Grzesiak S, Hura T, Stojałowski S, Myśków B, Milczarski P, Quarrie S. Mapping QTLs for yield components and chlorophyll a fluorescence parameters in wheat under three levels of water availability. Plant Genetic Resources. 2011;9(02):291–295. doi: 10.1017/s1479262111000207. [DOI] [Google Scholar]
- Czyczyło-Mysza I, Tyrka M, Marcińska I, Skrzypek E, Karbarz M, Dziurka M, Hura T, Dziurka K, Quarrie SA. Quantitative trait loci for leaf chlorophyll fluorescence parameters, chlorophyll and carotenoid contents in relation to biomass and yield in bread wheat and their chromosome deletion bin assignments. Mol Breeding. 2013;32:189–210. doi: 10.1007/s11032-013-9862-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalmannsdottir S, Jørgensen M, Rapacz M, Østrem L, Larsen A, Rødven R, Rognli OA. Cold acclimation in warmer extended autumns impairs freezing tolerance of perennial ryegrass (Lolium perenne) and timothy (Phleum pratense) Physiol Plant. 2017;160(3):266–281. doi: 10.1111/ppl.12548. [DOI] [PubMed] [Google Scholar]
- Danyluk J, Houde M, Rassart É, Sarhan F. Differential expression of a gene encoding an acidic dehydrin in chilling sensitive and freezing tolerant Gramineae species. FEBS Lett. 1994;344:20–24. doi: 10.1016/0014-5793(94)00353-x. [DOI] [PubMed] [Google Scholar]
- Danyluk J, Perron A, Houde M, Limin A, Fowler B, Benhamou N, Sarhan F. Accumulation of an acidic dehydrin in the vicinity of the plasma membrane during cold acclimation of wheat. Plant Cell. 1998;10(4):623–638. doi: 10.2307/3870737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Rosso T, Giorgetti E, Cicchi S, Muniz-Miranda M, Margheri G, Giusti A, Rindi A, Ghini G, Sottini S, Marcelli A, Foggi P. Surface-enhanced fluorescence and surface-enhanced Raman scattering of ultrathin layers of bichromophoric antenna systems adsorbed on silver nanoisland films. J Lumin. 2009;129(12):1955–1959. doi: 10.1016/j.jlumin.2008.11.027. [DOI] [Google Scholar]
- Dexter ST, Tottingham WE, Graber LF. Investigations of hardiness of plants by 1008 measurement of electrical conductivity. Plant Physiol. 1932;7:63–78. doi: 10.1104/pp.7.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhillon T, Pearce SP, Stockinger EJ, Distelfeld A, Li C, Knox AK, Vashegyi I, Vagujfalvi A, Galiba G, Dubcovsky J. Regulation of freezing tolerance and flowering in temperate cereals: the VRN-1 connection. Plant Physiol. 2010;153(4):1846–1858. doi: 10.1104/pp.110.159079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyda M, Wąsek I, Tyrka M, Wędzony M, Szechyńska-Hebda M. Local and systemic regulation of PSII efficiency in triticale infected by the hemibiotrophic pathogen Microdochium nivale. Physiol Plant. 2019;165(4):711–727. doi: 10.1111/ppl.12760. [DOI] [PubMed] [Google Scholar]
- Flint HJ, Boyce BR, Brattie DJ. Index of injury, a useful expression of freezing injuries to plant tissues as determined by the electric method. Can J Plant Sci. 1967;47:229–239. doi: 10.4141/cjps67-043. [DOI] [Google Scholar]
- Francia E, Rizza F, Cattivelli L, Stanca AM, Galiba G, Tόth B, Hayes PM, Skinner JS, Pecchioni N. Two loci on chromosome 5H determine low-temperature tolerance in a ‘Nure’ (winter)בTremois’ (spring) barley map. Theor Appl Genet. 2004;108:670–680. doi: 10.1007/s00122-003-1468-9. [DOI] [PubMed] [Google Scholar]
- Francia E, Barabaschi D, Tondelli A, Laidò G, Rizza F, Stanca AM, Busconi M, Fogher C, Stockinger EJ, Pecchioni N. Fine mapping of a HvCBF gene cluster at the frost resistance locus Fr-H2 in barley. Theor Appl Genet. 2007;115:1083–1091. doi: 10.1007/s00122-007-0634-x. [DOI] [PubMed] [Google Scholar]
- Fricano A, Rizza F, Faccioli P, Pagani D, Pavan P, Stella A, Rossini L, Piffanelli P, Cattivelli L. Genetic variants of HvCbf14 are statistically associated with frost tolerance in a European germplasm collection of Hordeum vulgare. Theor Appl Genet. 2009;119:1335–1348. doi: 10.1007/s00122-009-1138-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galiba G, Quarrie SA, Sutka J, Morgounov A. RFLP mapping of the vernalization (Vrn1) and frost resistance (Fr1) genes on chromosome 5A of wheat. Theor Appl Genet. 1995;90:1174–1179. doi: 10.1007/bf00222940. [DOI] [PubMed] [Google Scholar]
- Galiba G, Pecchioni N, Vágújfalvi A, Francia E, Tóth B, Barabaschi D, Stanca MA (2003) Localization of QTLs and candidate genes involved in the regulation of frost resistance in cereals. In Proceedings of the international congress in the wake of the double helix: from the green revolution to the gene revolution (Vol. 27, p. 31).
- Galiba G, Vágújfalvi A, Li Ch, Soltész A, Dubcovsky J. Regulatory genes involved in the determination of frost tolerance in temperate cereals. Plant Sci. 2009;176:12–19. doi: 10.1016/j.plantsci.2008.09.016. [DOI] [Google Scholar]
- Genc Y, Oldach K, Verbyla AP, Lott G, Hassan M, Tester M, Wallwork H, McDonald GK. Sodium exclusion QTL associated with improved seedling growth in bread wheat under salinity stress. Theor Appl Genet. 2010;121:877–894. doi: 10.1007/s00122-010-1357-y. [DOI] [PubMed] [Google Scholar]
- Gołębiowska G, Wędzony M. Cold-hardening of winter triticale (x Triticosecale Wittm.) results in increased resistance to pink snow mould Microdochium nivale (Fr., Samuels & Hallett) and genotype-dependent chlorophyll fluorescence modulations. Acta Physiol Plant. 2009;31:1219–1227. doi: 10.1007/s11738-009-0357-5. [DOI] [Google Scholar]
- González JM, Muñiz LM, Jouve N. Mapping of QTLs for androgenetic response based on a molecular genetic map of ×Triticosecale Wittmack. Genome. 2005;48(6):999–1009. doi: 10.1139/g05-064. [DOI] [PubMed] [Google Scholar]
- Gupta PK, Balyan HS, Kulwal PL, Kumar N, Kumar A, Mir RR, Mohan A, Kumar J (2007) QTL analysis for some quantitative traits in bread wheat. Journal of Zhejiang University SCIENCE B pp. 807–814. 10.1631/jzus.2007.b0807 [DOI] [PMC free article] [PubMed]
- Hackauf B, Wehling P. Identification of microsatellite polymorphisms in an expressed portion of the rye genome. Plant Breed. 2002;121:17–25. doi: 10.1046/j.1439-0523.2002.00649.x. [DOI] [Google Scholar]
- Jaccoud D, Peng K, Feinstein D, Kilian A. Diversity arrays: a solid state technology for sequence information independent genotyping. Nucleic Acids Res. 2001;29:25. doi: 10.1093/nar/29.4.e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janeczko A, Pociecha E, Dziurka M, Jurczyk B, Libik-Konieczny M, Oklestkova J, Novak O, Pilarska M, Filek M, Rudolphi-Skórska E, Sadura I, Siwek A. Changes in content of steroid regulators during cold hardening of winter wheat - steroid physiological/biochemical activity and impact on frost tolerance. Plant Physiol Biochem. 2019;139:215–228. doi: 10.1016/j.plaphy.2019.03.020. [DOI] [PubMed] [Google Scholar]
- Kamal AHM, Kim KH, Shin KH, Choi JS, Baik BK, Tsujimoto H, Woo SH. Abiotic stress responsive proteins of wheat grain determined using proteomics technique. Aust J Crop Sci. 2010;4(3):196. [Google Scholar]
- Kaplan F, Kopka J, Haskell DW, Zhao W, Schiller KC, Gatzke N, Sung DY, Guy CL. Exploring the temperature-stress metabolome of Arabidopsis. Plant Physiol. 2004;136:4159–4168. doi: 10.1104/pp.104.052142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karbarz M, Tyrka M, Woźniak-Strzembicka A, Czajowski G, Wardyńska A, Tyrka D, Pojmaj M, Wędzony M. Quantitative trait loci mapping of adult-plant resistance to powdery mildew in triticale. Annals of Applied Biology. 2020;177(2):223–231. doi: 10.1111/aab.12613. [DOI] [Google Scholar]
- Knox AK, Li C, Vágújfalvi A, Galiba G, Stockinger EJ, Dubcovsky J. Identification of candidate CBF genes for the frost tolerance locus Fr-A m 2 in Triticum monococcum. Plant Mol Biol. 2008;67(3):257–270. doi: 10.1007/s11103-008-9316-6. [DOI] [PubMed] [Google Scholar]
- Knox AK, Dhillon T, Cheng H, Tondelli A, Pecchioni N, Stockinger EJ. CBF gene copy number variation at Frost Resistance-2 is associated with levels of freezing tolerance in temperate-climate cereals. Theor Appl Genet. 2010;121(1):21–35. doi: 10.1007/s00122-010-1288-7. [DOI] [PubMed] [Google Scholar]
- Kobayashi F, Takumi S, Kume S, Ishibashi M, Ohno R, Murai K, Nakamura C. Regulation by Vrn-1/Fr-1 chromosomal intervals of CBF-mediated Cor/Lea gene expression and freezing tolerance in common wheat. J Exp Bot. 2005;56(413):887–895. doi: 10.1093/jxb/eri081. [DOI] [PubMed] [Google Scholar]
- Koch MD, Lehman EO. Resistenzeigeschatten im Gärsten und Weizensortiment Gatersleben, 7 Prüfung der Frostresistenzpflanze. DAI. 1966;14:263–282. doi: 10.1007/bf02095288. [DOI] [Google Scholar]
- Kocheva K, Nenova V, Karceva T, Petrov P, Georgiev GI, Börner A, Landjeva S. Changes in water status, membrane stability and antioxidant capacity of wheat seedlings carrying different Rht-B1 dwarfing alleles under drought stress. J Agron Crop Sci. 2014;200(2):83–91. doi: 10.1111/jac.12047. [DOI] [Google Scholar]
- Kocsy G, Athmer B, Perovic D, Himmelbach A, Szücs A, Vashegyi I, Schweizer P, Galiba G, Stein N. Regulation of gene expression by chromosome 5A during cold hardening in wheat. Mol Genet Genomics. 2010;283:351–363. doi: 10.1007/s00438-010-0520-0. [DOI] [PubMed] [Google Scholar]
- Kosambi DD. The estimation of map distance from recombination value. Ann Eug. 1944;12:172–175. doi: 10.1111/j.1469-1809.1943.tb02321.x. [DOI] [Google Scholar]
- Kovi MR, Ergon Å. Rognli OA (2016) Freezing tolerance revisited - effects of variable temperatures on gene regulation in temperate grasses and legumes. Curr Opin Plant Biol. 2016;33:140–146. doi: 10.1016/j.pbi.2016.07.006. [DOI] [PubMed] [Google Scholar]
- Krzewska M, Czyczyło-Mysza I, Dubas E, Gołębiowska-Pikania G, Golemiec E, Stojałowski S, Chrupek M, Żur I. Quantitative trait loci associated with androgenic responsiveness in triticale (× Triticosecale Wittm.) anther culture. Plant Cell Rep. 2012;31:2099–2108. doi: 10.1007/s00299-012-1320-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzewska M, Czyczyło-Mysza I, Dubas E, Gołębiowska-Pikania G, Żur I. Identification of QTLs associated with albino plant formation and some new facts concerning green versus albino ratio determinants in triticale (× Triticosecale Wittm.) anther culture. Euphytica. 2015;206:263–278. doi: 10.1007/s10681-015-1509-x. [DOI] [Google Scholar]
- Kumar S, Sehgal SK, Kumar U, Prasad PV, Joshi AK, Gill BS. Genomic characterization of drought tolerance-related traits in spring wheat. Euphytica. 2012;186(1):265–276. doi: 10.1007/s10681-012-0675-3. [DOI] [Google Scholar]
- Kurepin LV, Dahal KP, Savitch LV, Singh J, Bode R, Ivanov AG, Hurry V, Kurepin LV, Dahal KP, Savitch LV, Singh J, Bode R, Ivanov AG, Hurry V, Huener N. Role of CBFs as integrators of chloroplast redox, phytochrome and plant hormone signaling during cold acclimation. Int J Mol Sci. 2013;14:12729–12763. doi: 10.3390/ijms140612729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law CN, Jenkins G. A genetic study of cold resistance in wheat. Genet Res. 1970;15:197–208. doi: 10.1017/s001667230000152x. [DOI] [Google Scholar]
- Law CN, Worland AJ. Genetic analysis of some flowering time and adaptive traits in wheat. New Phytol. 1997;137(1):19–28. doi: 10.1046/j.1469-8137.1997.00814.x. [DOI] [Google Scholar]
- Levitt J (1980) Responses of plants to environmental stresses. Chilling, freezing, and high temperature stresses. Ed. 2. Academic Press, New York, NY. 10.1016/b978-0-12-445501-6.50011-7
- Li Y, Haseneyer G, Schön CC, Ankerst D, Korzun V, Wilde P, Bauer E. High levels of nucleotide diversity and fast decline of linkage disequilibrium in rye (Secale cereale L.) genes involved in frost response. BMC Plant Biology. 2011;11(1):1–14. doi: 10.1186/1471-2229-11-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Böck A, Haseneyer G, Korzun V, Wilde P, Schön CC, Ankerst DP, Bauer E. Association analysis of frost tolerance in rye using candidate genes and phenotypic data from controlled, semi-controlled, and field phenotyping platforms. BMC Plant Biol. 2011;11(1):1–14. doi: 10.1186/1471-2229-11-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Q, Cheng X, Mei M, Yan X, Liao H. QTL analysis of root traits as related to phosphorus efficiency in soybean. Ann Bot. 2010;106(1):223–234. doi: 10.1093/aob/mcq097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley JM, Ludlow MM, McCouch SR, O’toole JC, Locating QTL for osmotic adjustment and dehydration tolerance in rice. J Exp Bot. 1996;47(9):1427–1436. doi: 10.1093/jxb/47.9.1427. [DOI] [Google Scholar]
- Mahfoozi S, Limin AE, Hayes PM, Hucl P, Fowler DB. Influence of photoperiod response on the expression of cold hardiness in wheat and barley. Can J Plant Sci. 2000;80(4):721–724. doi: 10.4141/p00-031. [DOI] [Google Scholar]
- Miller AK, Galiba G, Dubcovsky J. A cluster of 11 CBF transcription factors is located at the frost tolerance locus Fr-A m 2 in Triticum monococcum. Mol Genet Genomics. 2006;275(2):193–203. doi: 10.1007/s00438-005-0076-6. [DOI] [PubMed] [Google Scholar]
- Morgan JM, Tan MK. Chromosomal location of wheat osmoregulation gene using RFLP analysis. Aust J Plant Physiol. 1996;23:803–806. doi: 10.1071/pp9960803. [DOI] [Google Scholar]
- NDong Ch, Danyluk J, Wilson KE, Pocock T, Huner NPA, Sarhan F, Cold-regulated cereal chloroplast late embryogenesis abundant-like proteins. Molecular Characterization and Functional Analyses. Plant Physiol. 2002;129:1368–1381. doi: 10.1104/pp.001925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nylander M, Svensson J, Palva ET, Welin BV. Stress-induced accumulation and tissue-specific localization of dehydrins in Arabidopsis thaliana. Plant Mol Biol. 2001;45(3):263–279. doi: 10.1023/a:1006469128280. [DOI] [PubMed] [Google Scholar]
- Pan A, Hayes PM, Chen F, Chen THH, Blake T, Wright S, Karsai I, Bedö Z. Genetic analysis of the components of winterhardiness in barley (Hordeum vulgare L.) Theor Appl Genet. 1994;89:900–910. doi: 10.1007/bf00224516. [DOI] [PubMed] [Google Scholar]
- Plaschke J, Bӧrner A, Xie DX, Koebner RMD, Schlegel R, Gale MD. RFLP mapping of genes affecting plant height and growth habit in rye. Theor Appl Genet. 1993;85:1049–1054. doi: 10.1007/bf00215046. [DOI] [PubMed] [Google Scholar]
- Pestsova E, Ganal MW, Röder MS. Isolation and mapping of microsatellite markers specific for the D genome of bread wheat. Genome. 2000;43:689–697. doi: 10.1139/g00-042. [DOI] [PubMed] [Google Scholar]
- Puchkov YM, Zhirov EG. Breeding of common wheat varieties with a high frost resistance and genetic aspects of it. World Sci News. 1978;15:17–22. [Google Scholar]
- Pugsley AT. A genetic analysis of the spring-winter habit in wheat. Aust J Agric Res. 1971;22:21–31. doi: 10.1071/ar9710021. [DOI] [Google Scholar]
- Rapacz M. Chlorophyll a fluorescence transient during freezing and recovery in winter wheat. Photosynthetica. 2007;45:409–418. doi: 10.1007/s11099-007-0069-2. [DOI] [Google Scholar]
- Rapacz M, Sasal M, Gut M. Chlorophyll fluorescence-based studies of frost damage and the tolerance for cold-induced photoinhibition in freezing tolerance analysis of triticale (×Triticosecale Wittmack) J Agron Crop Sci. 2011;197:378–389. doi: 10.1111/j.1439-037x.2011.00472.x. [DOI] [Google Scholar]
- Rapacz M, Ergon Å, Höglind M, Jørgensen M, Jurczyk B, Østrem L, Rognli OA, Tronsmo AM. Overwintering of herbaceous plants in a changing climate. Still more questions than answers. Plant Sci. 2014;225:34–44. doi: 10.1016/j.plantsci.2014.05.009. [DOI] [PubMed] [Google Scholar]
- Rapacz M, Sasal M, Wójcik-Jagła M. Direct and indirect measurements of freezing tolerance: advantages and limitations. Acta Physiol Plant. 2015;37:157. doi: 10.1007/s11738-015-1907-7. [DOI] [Google Scholar]
- Rapacz M, Sasal M, Kalaji HM, Kościelniak J. Is the OJIP test a reliable indicator of winter hardiness and freezing tolerance of common wheat and triticale under variable winter environments? PLoS ONE. 2015;10(7):e0134820. doi: 10.1371/journal.pone.0134820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapacz M, Jurczyk B, Sasal M. Deacclimation may be crucial for winter survival of cereals under warming climate. Plant Sci. 2017;256:5–15. doi: 10.1016/j.plantsci.2016.11.007. [DOI] [PubMed] [Google Scholar]
- Rizza F, Pagani D, Stanca AM, Cattivelli L. Use of chlorophyll fluorescence to evaluate the cold acclimation and freezing tolerance of winter and spring oats. Plant Breed. 2001;120:389–396. doi: 10.1046/j.1439-0523.2001.00635.x. [DOI] [Google Scholar]
- Roberts DWA. Chromosomes in ‘Cadet’ and ‘Rescue’ wheats carrying loci for cold hardiness and vernalization response. Can J Genet Cytol. 1986;28:991–997. doi: 10.1139/g86-137. [DOI] [Google Scholar]
- Roberts DWA. Identification of loci on chromosome-5A of wheat involved in control of cold hardiness, vernalization, leaf length, rosette growth habit, and height of hardened plants. Genome. 1990;33:247–259. doi: 10.1139/g90-039. [DOI] [Google Scholar]
- Röder MS, Korzun V, Wendehake K, Plaschke J, Tixier MH, Leroy P, Ganal MW. A microsatellite map of wheat. Genetics. 1998;149:2007–2023. doi: 10.1007/s001220051185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saal B, Wricke G. Development of simple sequence repeat markers in rye (Secale cereale L.) Genome. 1999;42:964–972. doi: 10.1139/g99-052. [DOI] [PubMed] [Google Scholar]
- Salinas J (2002) Molecular mechanism of signal transduction in cold acclimation. Frontiers in Molecular Biology (Hames B.D., Glover D.M, eds). Oxford University Press, pp. 116–139.
- Snape JW, Semikhodskii A, Fish L, Sarma RN, Quarrie SA, Galiba G, Sutka J. Mapping frost resistance loci in wheat and comparative mapping with other cereals. Acta Agr Hungarica. 1997;45:265–270. [Google Scholar]
- Somers DJ, Isaac P, Edwards K. A high-density microsatellite consensus map for bread wheat (Triticum aestivum L.) Theor Appl Genet. 2004;109(6):1105–1114. doi: 10.1007/s00122-004-1740-7. [DOI] [PubMed] [Google Scholar]
- Stockinger EJ, Skinner JS, Gardner KG, Francia E, Pecchioni N. Expression levels of barley Cbf genes at the Frost resistance-H2 locus are dependent upon alleles at Fr-H1 and Fr-H2. Plant J. 2007;51(2):308–321. doi: 10.1111/j.1365-313X.2007.0141.x. [DOI] [PubMed] [Google Scholar]
- Sutka J. Genetic studies of frost resistance in wheat. Theor Appl Genet. 1981;59:145–152. doi: 10.1007/bf00264968. [DOI] [PubMed] [Google Scholar]
- Sutka J. Genetic control of frost tolerance in wheat (Triticum aestivum L.) Euphytica. 1994;77:277–282. doi: 10.1007/bf02262642. [DOI] [Google Scholar]
- Sutka J, Snape JW. Location of a gene for frost resistance on chromosome 5A of wheat. Euphytica. 1989;42:41–44. doi: 10.1007/bf00042613. [DOI] [Google Scholar]
- Tamura KI, Yonemaru JI. Next‐generation sequencing for comparative transcriptomics of perennial ryegrass (Lolium perenne L.) and meadow fescue (Festuca pratensis Huds.) during cold acclimation. Grassland Science. 2010;56(4):230–239. doi: 10.1111/j.1744-697X.2010.00199.x. [DOI] [Google Scholar]
- Thomashow MF. Role of cold-responsive genes in plant freezing tolerance. Plant Physiol. 1998;118:1–7. doi: 10.1104/pp.118.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth B, Galiba G, Feher E, Sutka J, Snape JW. Mapping genes affecting flowering time and frost resistance on chromosome 5B in wheat. Theor Appl Genet. 2003;107:509–514. doi: 10.1007/s00122-003-1275-3. [DOI] [PubMed] [Google Scholar]
- Tripathy JN, Zhang J, Robin S, Nguyen TT, Nguyen HT. QTLs for cell-membrane stability mapped in rice (Oryza sativa L.) under drought stress. Theor Appl Genet. 2000;100(8):1197–1202. doi: 10.1007/s001220051424. [DOI] [Google Scholar]
- Tyrka M, Bednarek PT, Kilian A, Wędzony M, Hura T, Bauer E. Genetic map of triticale compiling DArT. SSR and AFLP Markers Genome. 2011;54(5):391–401. doi: 10.1139/g11-009. [DOI] [PubMed] [Google Scholar]
- Tyrka M, Tyrka D, Wędzony M. Genetic map of triticale integrating microsatellite. DArT and SNP Markers Plos One. 2015;10(12):e0145714. doi: 10.1371/journal.pone.0145714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyrka M, Oleszczuk S, Rabiza-Swider J, Woś H, Wędzony M, Zimny J, Ponitka A, Ślusarkiewicz-Jarzina A, Metzger RJ, Baenziger PS, Łukaszewski AJ. Populations of doubled haploids for genetic mapping in hexaploid winter triticale. Mol. Breeding. 2018;38(4):46. doi: 10.1007/s11032-018-0804-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Ooijen JW (2006) JoinMap® 4, Software for the calculation of genetic linkage maps in experimental populations. Kyazma BV, Wageningen, 33(10.1371).
- Van Os H, Stam P, Visser RG, Van Eck HJ. RECORD: a novel method for ordering loci on a genetic linkage map. Theor Appl Genet. 2005;112(1):30–40. doi: 10.1007/s00122-005-0097-x. [DOI] [PubMed] [Google Scholar]
- Vágújfalvi A, Crosatti C, Galiba G, Dubcovsky J, Cattivelli L. Two loci on wheat chromosome 5A regulate the differential cold-dependent expression of the cor14b gene in frost tolerant and sensitive genotypes. Mol Gen Genet. 2000;263:194–200. doi: 10.1007/s004380051160. [DOI] [PubMed] [Google Scholar]
- Vágújfalvi A, Galiba G, Cattivelli L, Dubcovsky J. The cold-regulated transcriptional activator Cbf3 is linked to the frost-tolerance locus Fr-A2 on wheat chromosome 5A. Mol Genet Genomics. 2003;269:60–67. doi: 10.1007/s00438-003-0806-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veisz O, Sutka J (1993) Ditelosomic analysis of frost resistance in wheat (cv Chinese Spring). Cereal Res Comm 21: 263–267. https://www.jstor.org/stable/23783977
- Wang S, Basten CJ, Zeng BZ (2012) Windows QTL cartographer 2.5. Raleigh: Department of Statistics, North Carolina State University.
- Wenzl P, Carling J, Kudrna D, Jaccoud D, Huttner E, Kleinhofs A, Kilian A. Diversity arrays technology (DArT) for whole-genome profiling of barley. Proc Natl Acad Sci USA. 2004;101:9915–9920. doi: 10.1073/pnas.0401076101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wędzony M (2003) Protocol for doubled haploid production in hexaploid triticale (×Triticosecale Wittm.) by crosses with maize. In: Maluszynski M, Kasha KJ, Forster BP, Szarejko I (eds) Doubled haploid production in crop plants. A manual. Kluwer Academic Publishers, Dordrecht, pp. 135–140. 10.1007/978-94-017-1293-4_21
- Williams ME, Torabinejad J, Cohick E, Parker K, Drake EJ, Thompson JE, DeWald DB. Mutations in the Arabidopsis phosphoinositide phosphatase gene SAC9 lead to overaccumulation of PtdIns (4, 5) P2 and constitutive expression of the stress-response pathway. Plant Physiol. 2005;138(2):686–700. doi: 10.1104/pp.105.061317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winfield MO, Lu C, Wilson ID, Coghill JA, Edwards KJ. Plant responses to cold: transcriptome analysis of wheat. Plant Biotechnol J. 2010;8(7):749–771. doi: 10.1111/j.1467-7652.2010.00536.x. [DOI] [PubMed] [Google Scholar]
- Yang DL, Jing RL, Chang XP, Li W. Quantitative trait loci mapping for chlorophyll fluorescence and associated traits in wheat (Triticum aestivum) J Integr Plant Biol. 2007;49(5):646–654. doi: 10.1111/j.1744-7909.2007.00443.x. [DOI] [Google Scholar]
- Yin Q, Cui Y, Zhang G. The Arabidopsis pentatricopeptide repeat protein PDM1 is associated with the intergenic sequence of S11-rpoA for rpoA monocistronic RNA cleavage. Chin Sci Bull. 2012;57:3452–3459. doi: 10.1007/s11434-012-5278-9. [DOI] [Google Scholar]
- Zhang X, Shabala S, Koutoulis A, Shabala L, Zhou M. Meta-analysis of major QTL for abiotic stress tolerance in barley and implications for barley breeding. Planta. 2017;245(2):283–295. doi: 10.1007/s00425-016-2605-4. [DOI] [PubMed] [Google Scholar]
- Zhang H, Wang Y, Bao M, Chan Z. Physiological changes and DREB1s expression profiles of tall fescue in response to freezing stress. Sci Hortic. 2019;245:116–124. doi: 10.1016/j.scienta.2018.09.052. [DOI] [Google Scholar]
- Zhu J, Pearce S, Burke A, See DR, Skinner DZ, Dubcovsky J, Garland-Campbell K. Copy number and haplotype variation at the VRN-A1 and central FR-A2 loci are associated with frost tolerance in hexaploid wheat. Theor Appl Genet. 2014;127(5):1183–1197. doi: 10.1007/s00122-014-2290-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Genetic linkage map of A chromosomes with molecular markers. (JPG 3498 KB)
Genetic linkage map of B chromosomes with molecular markers. (JPG 5346 KB)
Genetic linkage map of R chromosomes with molecular markers. (JPG 5349 KB)
Mean values of chlorophyll a fluorescence parameters measured in DH population after cold acclimation in three different field conditions. A, B, C, D, E, F, G, H, I, J charts were made for ABS/RC, TR0/RC, ET0/RC, DI0/RC, ABS/CS, TR0/CS, ET0/CS, DI0/CS, FV/FM and PI, respectively. Plant survival on the x-axis (0-100) is a number of surviving (regrowing) plants examined after three weeks of regrowth in glasshouse (temp. about +15°C) and expressed as a percentage of plant survival. (DOCX 1369 KB)
Genetic map and segregations of markers in population ‘Hewo’ x ‘Magnat’. Number of cosegregating markers was given (in brackets) in the names of representing bin markers. Segregartions of redundant markers were not provided. Physical positions of merkers were obtained afret balstn of marker sequences on ChineseSpring wheat V1.0 (URGI: https://wheat-urgi.versailles.inra.fr/Seq-Repository/BLAST ) and Lo1 rye chromosomes (WheatOmics 1.0: http://202.194.139.32/blast/blast.html). (XLSX 244 KB)
Data on maximum expected number of recombinations per individual (Max_rec) extracted and sum of adjacent recombination frequencies (Mean_rec) from JoinMap. Percentage of lines without crossing-over (c-o) was calculated in Excel. (DOCX 19 KB)
Summary of QTL identified using SMA method for plants recovery REC (percentage of survived plants after freezing) after freezing test performed during winter 2011/2012 (Experiment 1) and 2012/2013 (Experiments 2, 3).Table S4. Summary of QTL for electrolyte leakage (EL) from leaves after freezing identified using SMA method. Table S5. Summary of QTL for Fv/Fm (the yield of the energy trapping in PSII) and PI (performance indexes of PSII) identified using SMA method. QTL name includes each parameter name (after the letter Q). Table S6. Summary of QTL for energy fluxes (per single, active PSII reaction center; RC) identified using SMA method. QTL name includes each parameter name (after the letter Q). Abbreviations: ABS/RC - absorbed energy flux per the single, active PSII reaction center (RC); TR0/RC - trapped energy flux in PSII reaction centers per the single, active PSII reaction center; DI0/RC - dissipation of energy in PSII reaction centers per the single, active PSII reaction center. Table S7. Summary of QTL for phenomenological energy fluxes (per leaf cross-section; CS) identified using SMA method. QTL name includes each parameter name (after the letter Q). Abbreviations: ABS/CS - absorbed energy flux per leaf cross-section (CS); Tr0/CS - trapped energy flux in PSII reaction centers per leaf cross-section; ET0/CS - the energy flux for electron transport per leaf cross-section; DI0/CS – dissipation of energy in PSII reaction centers per leaf cross-section. (DOC 227 KB)
Data Availability Statement
Not applicable.
Not applicable.