Abstract
SMYD2 is a histone methyltransferase, which methylates both histone H3K4 as well as a number of non-histone proteins. Dysregulation of SMYD2 has been associated with several diseases including cancer. In the present study, we investigated whether and how SMYD2 might contribute to colorectal cancer. Increased expression levels of SMYD2 were detected in human and murine colon tumor tissues compared to tumor-free tissues. SMYD2 deficiency in colonic tumor cells strongly decreased tumor growth in two independent experimental cancer models. On a molecular level, SMYD2 deficiency sensitized colonic tumor cells to TNF-induced apoptosis and necroptosis without affecting cell proliferation. Moreover, we found that SMYD2 targeted RIPK1 and inhibited the phosphorylation of RIPK1. Finally, in a translational approach, pharmacological inhibition of SMYD2 attenuated colonic tumor growth. Collectively, our data show that SMYD2 is crucial for colon tumor growth and inhibits TNF-induced apoptosis and necroptosis.
Subject terms: Colon cancer, Apoptosis
Introduction
Colorectal cancer (CRC) is one of the leading causes of cancer-deaths worldwide and is expected to increase by 60% by the year 2030 [1, 2]. Multiple approaches including surgical removal and chemotherapy have been applied in CRC treatment, but the prognosis of patients with advanced CRC remains poor [3]. In order to improve the clinical management of CRC, it is crucial to identify the molecular mechanisms involved in CRC development and novel targets of anti-tumor therapy.
The SET and MYND domain-containing protein 2 (SMYD2) catalyzes the lysine methylation of histone and non-histone proteins to regulate multiple biological processes in tumor progression [4–6]. SMYD2 has been shown to regulate two opposing processes, cell proliferation and apoptosis [7–9]. The proliferation of tumor cells is crucial for the promotion of the tumor development [10]. Recently, SMYD2 has been implicated in regulating cell proliferation via targeting various molecules [9, 11, 12]. For example, SMYD2 physically interacts with proteins in the TGFβ pathway, including SMAD1–4, SMAD7, and SMURF2 [13]. Moreover, EZH2, a transcriptional repressor of several anti-proliferative genes, has been shown to undergo methylation by SMYD2, leading to EZH2 stability and therefore enhancing cell proliferation in breast cancer [6, 14]. Apoptosis is a form of programmed cell death and its inhibition plays an important role in CRC tumor development [15, 16]. Progressive inhibition of apoptosis is associated with the transformation of colorectal epithelial to carcinoma cells and decreased apoptosis has been observed in CRC tissues [17, 18]. Apoptosis can be triggered via an extrinsic death receptor pathway or by an intrinsic mitochondrial pathway [19]. The extrinsic pathway is triggered upon binding of death receptor ligands to their specific death receptors. TNF, a well-known death ligand, binds to TNFR1 to recruit downstream molecules such as RIPK1, leading to apoptotic pathway activation [20]. On the other hand, the intrinsic pathway is induced by intrinsic signaling such as DNA damage or endoplasmic reticulum stress. Accordingly, SMYD2 was shown to be required to limit the p53-mediated intrinsic apoptotic pathway in cardiac cells [7]. However, it is currently unknown whether SMYD2 may also regulate apoptosis via an extrinsic pathway.
Here we discovered that SMYD2 deficiency in human or murine colon tumor cells compromised tumor growth in orthotopic and non-orthotopic CRC models. Using MC-38 and HT-29 cells, we demonstrated that SMYD2 is dispensable for cell proliferation but crucial for inhibiting TNF-induced apoptosis and necroptosis. Our analysis further revealed that SMYD2 deficiency increased RIPK1 phosphorylation. Finally, we proved that the pharmacological inhibition of SMYD2 using two different SMYD2 inhibitors, AZ505 and BAY-598, attenuates colon tumor growth. Taken together, we identified SMYD2 as an important molecule for colon tumor growth via regulating TNF-induced apoptosis and necroptosis.
Results
SMYD2 is highly expressed in CRC and its deficiency reduces colon tumor growth in vivo
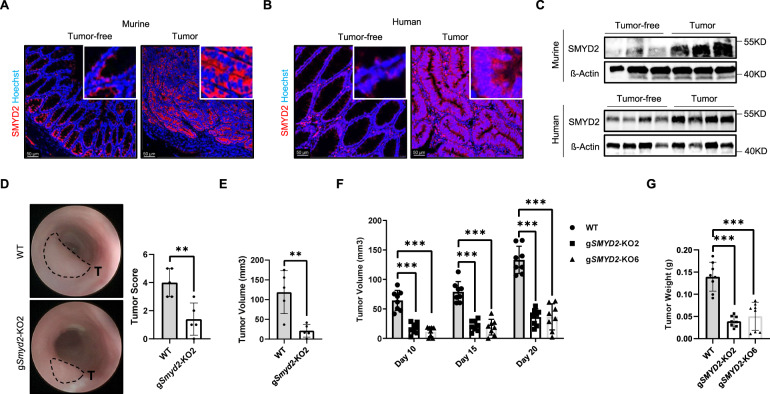
In order to investigate a potential contribution of SMYD2 in colorectal cancer (CRC) development, we initially detected the expression pattern of SMYD2 in colon tumors in murine samples (Fig. 1A). Immunofluorescence staining revealed SMYD2 staining in intestinal epithelial cells, in particular within the crypt region. Interestingly, SMYD2 staining was strongly increased in tumor tissues compared to tumor-free adjacent tissues. A similar increase in SMYD2 levels was observed in tissues derived from human CRC patients (Fig. 1B). Western blot analysis for SMYD2 further confirmed these findings (Fig. 1C). These data show that SMYD2 is highly expressed in human and murine colon tumors.
Fig. 1. SMYD2 is highly expressed in CRC and SMYD2 deficiency reduces colon tumor growth in vivo.
Immunofluorescence staining of murine ApcMin/+ (A) or human (B) colon tumor and tumor-free tissues with anti-SMYD2 antibody. Hoechst was used to stain the nuclei. C Immunoblot analysis of SMYD2 in lysates of human or murine colon tumor and tumor-free tissues. β-Actin served as a loading control. D, E WT and Smyd2 CRISPR/Cas9 knockout MC-38 cells were implanted into mice via submucosal injection. D Representative images of the colonic tumors and miniendoscopy-assisted tumor scoring. E Measurements of colonic tumor volume upon euthanasia. F Tumor volume from mice in which CRISPR/Cas9-directed SMYD2 knockout or WT HT-29 cells were injected subcutaneously and G tumors weight at day 20 from mice shown in (F) above.
To further elucidate a functional contribution of SMYD2 during CRC tumor development, SMYD2 deficient MC-38 cells and HT-29 cells were generated using the CRISPR/Cas9 technology (Fig. S1A). The role of SMYD2 in colorectal carcinogenesis was investigated by subcutaneously injecting the MC-38 Smyd2 deficient and WT (wild type) cells into C57BL/6J mice. Tumor growth was monitored at different time points and tumor weight was determined at the experimental end point. Interestingly, Smyd2 deficient tumors showed a strong reduction in the volume and weight compared with WT tumors (Figs. S1B and S1C). To exclude the possibility that the effects may depend on the nonphysiologic site of injection, we next implanted MC-38 WT and Smyd2 deficient cells directly into the colonic submucosa. Tumor size was monitored and scored via colonoscopy (Fig. 1D). When implanted into the submucosa, the Smyd2 deficient MC-38 cells resulted in tumors that had a significantly reduced tumor size and lower tumor scores (Fig. 1D, E), which was similar to the results obtained from the subcutaneous injection experiment. In summary, our results, from orthotopic and non-orthotopic transplantation of MC-38 WT and Smyd2 deficient cells, suggesting an important role of SMYD2 in murine colon tumor growth.
Given the above findings, we studied whether the role of SMYD2 in regulating colon tumor growth is conserved in humans. With this aim, SMYD2-proficient and -deficient HT-29 cells were implanted into Rag1−/− mice via subcutaneous injection, and tumor development was monitored over time. Similar to results obtained from the experiments performed with murine tumor cells, SMYD2 deficient HT-29 tumors showed a significant decrease in tumor size and tumor weight (Fig. 1F, G). Taken together, these data demonstrated that Smyd2 deficiency reduces murine colon tumor growth in vivo and that this process is conserved in human colorectal cancer cells.
SMYD2 is dispensable for CRC proliferation
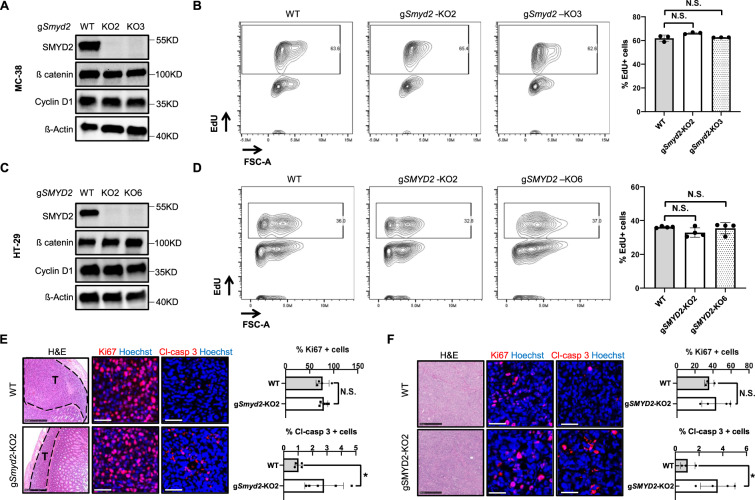
Recently, SMYD2 has been shown to regulate proliferation in multiple cancer cells lines [21]. We, therefore, assessed the effects of SMYD2 deficiency on colon tumor cell proliferation. Interestingly, loss of Smyd2 in MC-38 cells had no effect on the protein levels of ß catenin and Cyclin D1, two important proliferation markers (Fig. 2A). We next analyzed DNA synthesis in cultured MC-38 cells via 5-Ethynyl-2´-deoxyuridine (EdU) assay. In line with the findings above, we detected similar numbers of EdU-positive cells in the Smyd2 deficient group compared with the WT MC-38 group (Fig. 2B), collectively suggesting that SMYD2 is dispensable for regulating cell proliferation in MC-38 cells. In order to investigate whether this observation is a MC-38 cell-specific effect of SMYD2, we further analyzed SMYD2 knockout HT-29 cells. Similar to the results obtained from murine cells, SMYD2 deficiency in human HT-29 cells had no effects on protein levels of ß catenin and Cyclin D1 (Fig. 2C). Comparable levels of EdU-positive cells were observed in WT and SMYD2 deficient HT-29 cells. In summary, our data show that SMYD2 is dispensable for CRC cell proliferation in vitro.
Fig. 2. Proliferation is not impeded in SMYD2 deficient CRC cells.
Immunoblot analysis of SMYD2, Cyclin D1, and ß catenin in lysates of WT and two clones of CRISPR/Cas9-directed SMYD2 knockout MC-38 cells (A) or HT-29 cells (C). β-Actin served as a loading control. Flow cytometry assay of EdU in WT and SMYD2 CRISPR/Cas9 knockout MC-38 cells (B) or HT-29 cells (D). Graph shows quantification of EdU-positive cells. Experiments were performed three times and representative data are shown. Data are presented as mean+SD and student’s t-test was used for statistical calculation. N.S., not significant. Representative images of H&E, Ki67 (red), and cleaved-caspase 3 (red) stained MC-38 tumor tissues (E) or HT-29 tumor tissues (F). Scale bar, 50 µm. Histograms in E and F show quantification of Ki67 or cleaved-caspase 3 positive cells in tumor tissues. n (MC-38 WT) = 4, n (MC-38 gSmyd2-KO2) = 5, n (HT-29 WT) = 3, n (HT-29 gSMYD2-KO2) = 4.
We then studied whether SMYD2 regulates CRC cell proliferation in vivo via detecting Ki67-positive cells in tumor tissues from the orthotopic and xenotopic transplant models. Comparable numbers of Ki67-positive cells were detected in WT and Smyd2 deficient MC-38 tumor implants (Fig. 2E) and also from the WT and SMYD2 deficient HT-29 tumor implants (Fig. 2F), suggesting that SMYD2 is dispensable for regulating cell proliferation in vivo. To our surprise, we found a significant elevation of cleaved-caspase 3 positive cells in the tumor implants of MC-38 and HT-29 SMYD2 deficient cells compared with MC-38 and HT-29 WT cell implants (Fig. 2E, F). Taken together, these data indicate that SMYD2 is dispensable for CRC cell proliferation but restricts apoptosis in vivo.
SMYD2 deficiency sensitizes CRC cells to TNF-induced apoptosis and necroptosis
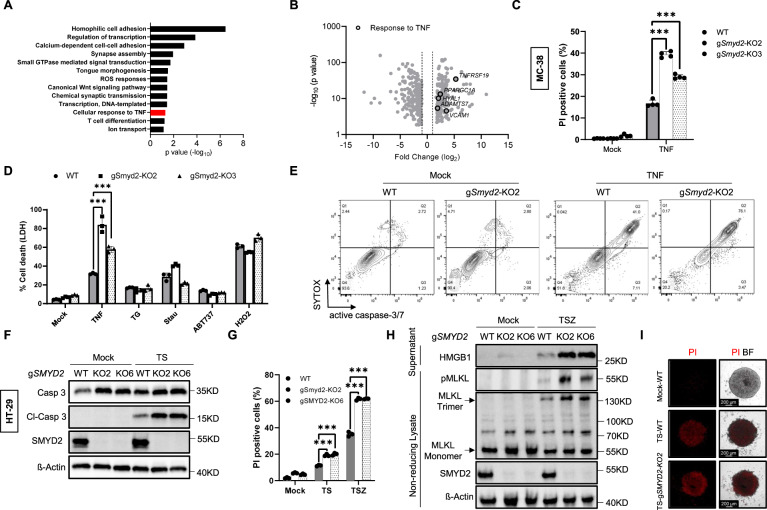
To further narrow down the mechanism of cell death induction upon SMYD2 deficiency, RNA-seq analysis was performed on implanted tumor tissues from two clones of SMYD2 deficient and WT HT-29 cells. We found 163 upregulated and 215 down-regulated genes that were common between the two knockout clones compared with WT (Fig. S2A; full gene list is given in Supplementary Table 1). A significant enrichment was found in the gene ontology for the biological process of cellular response to TNF (Fig. 3A). Furthermore, several TNF-related genes including TNFRSF19, VCAM1, and ADAMTS17 were highly upregulated in SMYD2 deficient cells (Fig. 3B). Based on these data together with the elevated cleaved-caspase 3 positivity in SMYD2 deficient tumors, we hypothesized that SMYD2 deficiency sensitizes colon tumor cells to TNF-induced apoptotic cell death. To evaluate this hypothesis, WT and two different Smyd2 deficient clones of MC-38 cells were stimulated with TNF in vitro and then stained with cell-impermeable propidium iodide (PI) which stains dead cells. Interestingly, the percentage of PI-positive cells was significantly higher in both clones of Smyd2 deficient MC-38 cells compared to WT cells following TNF stimulation (Figs. 3C and S2B), suggesting that SMYD2 plays a role downstream of TNF in regulating cell death. We next confirmed this finding via detecting the release of LDH, a prototypical proxy for cell membrane rupture used to detect cell death. MC-38 cells were stimulated with various inducers of cell death (Fig. 3D). As expected, challenging WT MC-38 cells with TNF, thapsigargin, staurosporine, ABT737, and H2O2 induced significant cell death. Interestingly, Smyd2 deficiency specifically and significantly amplified the TNF-induced cell death but not the cell death induced by other stimuli. In order to confirm that this is apoptotic cell death, we performed a flow cytometry analysis by detecting activated caspase-3/7 and SYTOX, where double positive cells were categorized as apoptotic cells (Figs. 3E and S2C). After TNF stimulation, we observed Smyd2 deficient MC-38 cells showed a highly significant elevation in the number of apoptotic cells compared with WT MC-38 cells (Figs. 3E and S2C). Moreover, the presence of Z-VAD, a pan-caspase inhibitor, reduced TNF-induced apoptosis in WT cells and abolished the increased cell death observed in Smyd2 deficient MC-38 cells (Fig. S2D). These results suggested that SMYD2 regulates TNF-induced apoptosis via a caspase-dependent pathway in MC-38 cells.
Fig. 3. SMYD2 deficiency sensitizes CRC cells to TNF-induced apoptosis and necroptosis.
A Gene ontology analysis for biological processes among the significantly and commonly regulated genes between the two clones of SMYD2 deficient HT-29 implanted tumors compared against WT HT-29 implanted tumors. Cellular response to TNF (highlighted red) among the significantly upregulated biological processes. B Volcano plot showing the expression of significantly downregulated and upregulated genes in SMYD2 deficient tumors compared to WT tumors. Black circles highlight genes related to TNF response. C Flow cytometric quantification of Propidium iodide (PI) stained WT and two different clones of Smyd2 deficient MC-38 cells stimulated with 0.1 ng/ml TNF or vehicle (mock). D LDH release from WT and Smyd2 CRISPR/Cas9 knockout MC-38 cells stimulated with vehicle (Mock), 0.1 ng/ml TNF, 25 µM thapsigargin (TG), 5 µM staurosporine (Stau), 50 µM ABT737, or 0.5 mM H2O2 for 24 h. E Flow cytometric detection of cleaved caspase-3/7 activity and SYTOX from the indicated genotypes of MC-38 cells stimulated with 0.1 ng/ml TNF. F Immunoblot analysis of SMYD2, caspase 3, and Cl-caspase 3 in HT-29 cells treated with or without 40 ng/ml TNF + 4 nM LCL161 (TS). β-Actin served as a loading control. G Flow cytometric quantification of PI stained HT-29 cells stimulated with TS or 10 ng/ml TNF + 1 nM LCL161 + 20 µM Z-VAD (TSZ). H Immunoblot analysis of HMGB1 from the supernatants and pMLKL/MLKL from non-reducing cell lysates. WT and SMYD2 CRISPR/Cas9 knockout HT-29 cells were stimulated with or without TSZ. β-Actin served as a loading control. I Representative images of PI stained HT-29 cells that were cultured in 3D. WT and SMYD2 CRISPR/Cas9 knockout HT-29 cells were stimulated with or without TS. DMSO was used as control. Experiments were performed three times and representative data are shown. Data are presented as mean +SD and student’s t-test was used for statistical calculation. **P < 0.01 and ***P < 0.001. N.S. not significant.
In order to further investigate the effect of SMYD2 in human cells, we also induced apoptosis in SMYD2 proficient and deficient HT-29 cells. The LCL161, a classical second mitochondria-derived activator of caspase (SMAC) mimetic, was used together with TNF to induce RIPK1-dependent apoptosis in HT-29 cells. As expected, TNF plus LCL161 (TS) treatment induced significant caspase 3 activation in HT-29 cells (Fig. 3F). Interestingly, SMYD2 deficiency increased levels of cleaved-caspase 3 upon TS stimulation in both the clones of SMYD2 deficient cells to levels that were higher than those seen in SMYD2 sufficient HT-29 cells. This finding was further confirmed via flow cytometry for PI-positive cells in TS treated HT-29 cells (Fig. 3G). These data indicate that SMYD2 modulates the TNF-RIPK1 signaling arm in HT-29 cells.
A key feature of tumor cells is the ability to evade programs of cell death and one of the strategies employed is the evasion of necroptosis via the activation of initiator caspase modulators such as c-FLIP. We recently showed that forced necroptosis induction in CRC cells downstream of TNF can be a successful strategy to counter such cell death evasion [22]. To address whether SMYD2 is a modulator of the necroptosis arm downstream of TNF, we stimulated HT-29 cells with TS plus Z-VAD (TSZ) (Fig. 3G). As expected, TSZ treatment induced cell death in HT-29 WT cell cultures. Interestingly, we observed a significantly higher number of PI-positive cells in SMYD2 deficient cells, suggesting that SMYD2 inhibits necroptosis. In line with the elevated necroptosis in cells lacking SMYD2, we also observed an increase in the levels of HMGB1 release and MLKL trimerization (Fig. 3H), two biomarkers of necrotic cell death [23, 24]. Furthermore, the presence of necrostatin-1 (Nec-1), a specific inhibitor of RIPK1 kinase activity, prevented TSZ-induced cell death both in WT cells as well as in SMYD2 deficient cells (Fig. S2E). Similarly, RIPK3 inhibition (GSK-872) or MLKL inhibition (NSA: Necrosulfonamide) were both efficient in blocking TSZ-induced cell death in SMYD2-deficient HT29 cells (Fig. S2E). Interestingly, more pMLKL positive cells were detected in SMYD2 deficient HT-29 tumors in vivo (Fig. S2F). Taken together, our data indicated that SMYD2 regulates necroptosis induction in HT-29 cells. Of note, TSZ treatment couldn’t induce cell death in MC-38 cells (Fig. S2G, upper panel). The necroptosis insensitivity of MC-38 cells is likely due to the deficiency of RIPK3 (Fig. S2G, lower panel) [25], a key molecule for necroptosis induction.
The studies described above were performed on 2D-cultured cells and 2D cell culture lacks many aspects of tumors, such as hypoxia and altered cell-cell contacts [26–28]. Therefore we stimulated 3D cultures of HT-29 cells with TS to mimic the in vivo tumor response upon an apoptotic challenge (Figs. 3I and S2H). Similar to the results obtained from the experiments performed in 2D culture of HT-29 cells, SMYD2 deficiency significantly increased cell death induced by TS stimulation. In summary, these data demonstrated that SMYD2 regulates both TNF-induced apoptosis and necroptosis.
SMYD2 regulates RIPK1 recruitment and phosphorylation
The binding of TNF to TNFR1 leads to the recruitment of RIPK1 and a number of other molecules [29]. TNF stimulation induces the expression of pro-survival genes, such as those encoding FLIP or cIAP1/2, which suppress the activation of cell death pathways [29]. FLIP or cIAP1/2 deficiency switches cells from TNF-induced pro-survival pathways to TNF-induced cell death pathways. In the presence of caspase 8, a cytosolic complex named complex IIa can be further formed to induce RIPK1-dependent apoptosis. When caspase-8 activation is blocked, the signaling can be switched to form another complex, named complex IIb, to induce RIPK1-dependent necroptosis [29].
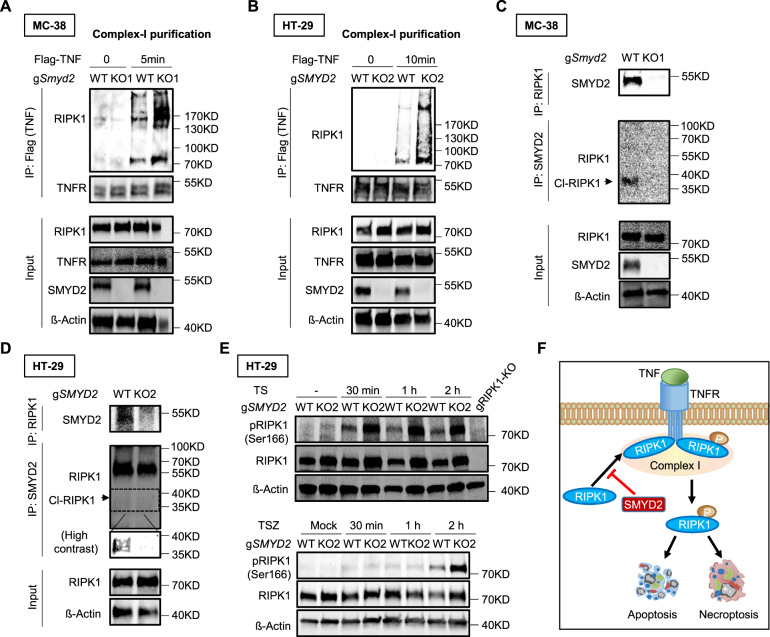
Knowing that SMYD2 regulates TNF-induced apoptosis and necroptosis, we hypothesized that SMYD2 may target the RIPK1 recruitment. To test this hypothesis, we stimulated MC-38 cells with Flag-TNF and pulled-down complex I via Flag immunoprecipitation (Fig. 4A). By doing so, we detected enhanced levels of RIPK1, a key component in the complex I, in the immunoprecipitate of WT cells after TNF treatment (Fig. 4A). Interestingly, in Smyd2 deficient cells, we discovered increased co-precipitation of RIPK1 upon TNF stimulation, suggesting that Smyd2 deficiency promotes RIPK1 recruitment to a TNF/TNF receptor 1 complex. In order to further investigate whether this finding was restricted to MC-38 cells, we also stimulated HT-29 cells with Flag-TNF and purified complex I (Fig. 4B). Similar to the results obtained from experiments performed with murine MC-38 cells, SMYD2 deficient HT-29 cells showed a significant increase of RIPK1 co-precipitation. Collectively, these data suggested that SMYD2 inhibits RIPK1 recruitment after TNF stimulation.
Fig. 4. SMYD2 regulates RIPK1 recruitment and phosphorylation.
Flag TNF-induced complex-I immunoprecipitation. MC-38 cells (A) or HT-29 cells (B) were stimulated with Flag-TNF (1 µg/mL) for the indicated time points, followed by Flag immunoprecipitation and immunoblot analysis. HT-29 cells were pre-treated with 1 nM LCL161 for 1 h before Flag-TNF stimulation cell lysates of MC-38 cells (C) or HT-29 cells (D) stimulated with or without TNF were immunoprecipitated with antibody against RIPK1 or antibody against SMYD2, followed by immunoblot analysis. HT-29 cells were pre-treated with 1 nM LCL161 for 1 h before TNF stimulation. E Immunoblot of phosphorylated or total RIPK1 in lysates of WT and SMYD2 deficient HT-29 cells stimulated with TS (Upper panel) or TSZ (Lower panel) at different time points. β-Actin served as a loading control. F A schematic diagram depicting SMYD2 mediated cell death pathways in CRC cells.
We then tested whether SMYD2 can interact with RIPK1. Cell lysates of MC-38 cells were immunoprecipitated with antibody against RIPK1 and then immunoblotted for SMYD2, or vice versa. Smyd2 deficient cells were used as a negative control. Our results demonstrated an interaction between RIPK1 and SMYD2, as SMYD2 co-immunoprecipitated with RIPK1 (Fig. 4C). Interestingly, SMYD2 specifically interacted with cleaved-RIPK1, not full-length RIPK1, suggesting that SMYD2 regulates the apoptotic pathway via targeting cleaved-RIPK1. We further confirmed this finding in HT-29 cells (Fig. 4D). Since a crucial role of RIPK1 phosphorylation has been demonstrated in its activation [30], we hypothesized that SMYD2 may impact the degree of RIPK1 phosphorylation. Accordingly, we analyzed RIPK1 phosphorylation in HT-29 cells stimulated with TS for activating the apoptotic pathway, or with TSZ for activating the necroptotic pathway (Fig. 4E). As expected, both TS and TSZ stimulation-induced some phosphorylation of RIPK1 in WT cells. However, SMYD2 deficient cells showed increased phosphorylation of RIPK1, suggesting that SMYD2 functions by inhibiting RIPK1 phosphorylation (Fig. 4E). Taken together, our results delineate a signaling pathway in which SMYD2 decreases RIPK1 phosphorylation, which in turn inhibits TNF-induced apoptosis and necroptosis (Fig. 4F).
Pharmacological inhibition of SMYD2 attenuates colon tumor growth
So far, our data revealed a significant impact of SMYD2 in regulating CRC cell death to such an extent that the genetic loss of SMYD2 blocked tumor growth. In order to test whether this function of SMYD2 is therapeutically exploitable against CRC, we inhibited SMYD2 activity using AZ505, a specific SMYD2 inhibitor [8, 31]. The presence of AZ505 significantly increased TNF-induced cell death and RIPK1-SMYD2 interaction (Fig. S3).
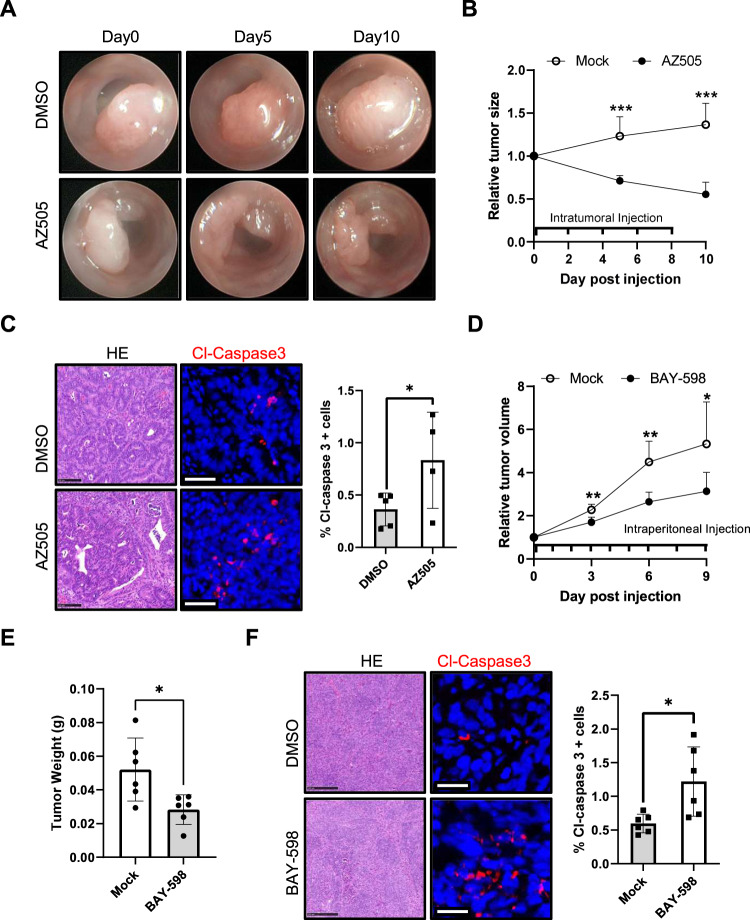
Interestingly, local injection of AZ505 in ApcMin/+ mice via intratumoral injection led to a significant reduction in tumor size (Fig. 5A, B). What is more, elevated levels of cleaved-caspase3 positive cells were detected in AZ505-treated tumors compared with control tumors (Fig. 5C). Taken together, our data indicated that SMYD2 inhibition via AZ505 administration suppresses colon tumor growth.
Fig. 5. Inhibition of SMYD2 with AZ505 or BAY-598 suppressed colon tumor growth.
A−C ApcMin/+ tumors were intratumorally injected with AZ505 or DMSO every two days. A, B Tumor size scored using colonoscopy at indicated time points (n = 5/group). C Representative images of H&E and cleaved-caspase3 (red) stained ApcMin/+ tumor sections (scale bar, 25 µm). The graph shows quantification of cleaved-caspase3 positive cells in tumor tissues. D, E HT-29 cells were injected subcutaneously into Rag1−/− mice. Once tumor size was measureable, mice were intraperitoneal injected with BAY-598 or DMSO daily (n = 6/group). D Relative tumor volume monitored using calipers at three-day intervals. Tumor volume was normalized to the volume at the beginning of treatment. E Tumor weight at day 10 post euthanasia. F Representative images of H&E and cleaved-caspase3 (red) stained tumor sections (scale bar, 25 µm). The graph shows quantification of cleaved-caspase3 positive cells in tumor tissues.
To exclude the possibility of off-target effects of AZ505 or murine cell specific effects, we used BAY-598, an alternative SMYD2 inhibitor that has a different binding mode compared to AZ505 [32, 33], to further investigate the impact of SMYD2 inhibition in human colon tumor growth. HT-29 cells were injected subcutaneously into Rag1−/− mice, and tumor development was monitored over time. BAY-598 treatment significantly inhibited tumor growth (Fig. 5D), reduced tumor weight (Fig. 5E), and increased levels of cleaved-caspase3 positive tumor cells (Fig. 5F). In summary, our results demonstrate that pharmacological inhibition of SMYD2 suppresses colon tumor growth.
Discussion
Although SMYD2 has been shown to play critical roles in tumor development in several types of cancers [6, 32, 34], the functional role of SMYD2 in colorectal cancer is largely unknown. Here, we identified SMYD2 as a key molecule in promoting colon tumor growth. Interestingly, we discovered a yet unknown role of SMYD2 in restricting TNF-induced apoptosis in vivo and in vitro.
Our results confirm previous reports which showed that SMYD2 is highly expressed in CRC [9]. High expression of SMYD2 has been shown to be associated with poor prognosis in several cancers, including CRC [9]. SMYD2 was identified as a methyltransferase that methylates several key cancer-involved proteins, including retinoblastoma, p53, MAPKAPK3, and STAT3, to promote cancer development [5, 8, 21, 32]. SMYD2-regulated cell proliferation is amongst these mechanisms that was reported in various types of cancers, for instance, colon cancer [9, 35]. However, a recent study using small molecule inhibitors and CRISPR/Cas9 technology targeting SMYD2 in hundreds cancer cell lines questioned the functional role of SMYD2 in tumor cell proliferation [36]. Indeed, our data support the findings and indicate that SMYD2 is dispensable for cell proliferation in colon tumor cells, as we detected similar levels of proliferation. Specifically, similar levels of ß catenin and Cyclin D1 as well as of EdU and Ki67 positive cells were detected in SMYD2 sufficient and deficient cells.
TNF is an important cytokine with direct cytotoxic activity on cells. The cytotoxic effects of TNF can be explained by its capacity to activate multiple programmed cell death pathways, such as apoptosis, necroptosis, and pyroptosis [37, 38]. It has been shown that, in mouse renal epithelial cells, TNF stimulation-induced SMYD2 production and Smyd2 deficiency increased the levels of cell death [31]. However, the functional role of SMYD2 in regulating cell death pathways is poorly understood. The present study demonstrates for the first time that SMYD2 specifically regulates TNF-induced apoptosis and necroptosis in colon tumor cells. SMYD2 is known as a lysine methyltransferase that catalyzes the lysine methylation of histone and non-histone proteins [4–6]. Interestingly, we detected an interaction between SMYD2 and cleaved-RIPK1 in murine and human cells, suggesting that SMYD2 may directly target RIPK1. We further found that SMYD2 deficiency increased phosphorylation levels of RIPK1 under apoptotic or necroptotic stimuli. Given the crucial role of RIPK1 in the cell death pathways downstream of the TNFR, our data indicated that SMYD2 inhibits apoptosis and necroptosis, potentially by targeting RIPK1.
The induction of apoptosis and necroptosis has been considered a promising approach in cancer treatment [39]. Along the same line, SMYD2 deficiency in colon tumors showed increased cell death and restricted tumor growth. We confirmed these findings using human and murine cells in orthotopic and non-orthotopic tumor models. Since accumulating evidence suggests that TNF also has the potential of promoting tumor development under certain conditions [40], strategies to induce apoptosis or necroptosis using various downstream inhibitors of TNF without changing proliferation are being tested. We observed that, in vivo, intratumoral administration of AZ505 in the absence of TNF supplementation also induced apoptotic cell death and reduced tumor size. Furthermore, another commercially available SMYD2 inhibitor, BAY-598, also showed similar effects, suggesting that SMYD2 inhibitors may become promising candidates for colon cancer treatment. Our findings, therefore, raise the interesting possibility that SMYD2 inhibition might have therapeutic potential by inducing apoptosis and necroptosis in CRC.
In summary, our study uncovered a novel role of SMYD2 in supporting colon tumor growth. SMYD2 in colon tumor cells inhibits TNF-induced apoptosis and necroptosis. These findings suggest that targeting SMYD2 may be a potential therapeutic strategy for the treatment of colon cancer.
Materials and methods
Mice
ApcMin/+ mice, B6 /J mice and Rag1−/− mice were obtained from the animal facilities of the department of medicine 1, Friedrich-Alexander-Universität Erlangen-Nürnberg (FAU). All mice were maintained under specific pathogen-free conditions and had ad libitum access to normal chow diet and sterile water. Mice of both genders aged 7−15 weeks, were randomly allocated to groups (treatment and control or recipient of control and knockout cells) in the various experiments. For ApcMin/+ mice, tumors were scored (3−5) in a blinded manner from mice randomly allocated to experimental and control groups. Experiments were performed under protocols approved by the Institutional Animal Care and Use Committee of the FAU.
Generation of SMYD2 knockout cells
SMYD2 CRISPR/Cas9 KO Plasmid (m) (sc-432661, Santa Cruz) or SMYD2 CRISPR/Cas9 KO Plasmid (h) (sc-403129, Santa Cruz) was transfected into MC-38 cells or HT-29 cells. The HT-29 cells were purchased from ATCC and MC-38 cells were obtained from Lonza. GFP-positive cells were sorted into 96-well plates (single cell/well) by flow cytometry (FACSAria II; BD). SMYD2 knockout cell lines were established from single-cell colonies. SMYD2 deficiency was verified by western blot (WB) analysis.
Tumor models and treatment
In the xenograft model, 2 × 106 HT-29 cells were implanted into the flanks of Rag1−/− mice via subcutaneous injection (n = 8/group). MC-38 cells were injected subcutaneously (1 × 106 cells) into the flanks of B6/J mice (n = 8/group) or into the colon mucosa (1 × 104 cells) of C57BL/6J mice (n = 5/group). Tumor size was measured at serial time points using a digital caliper and the tumor volume was calculated using V = (length × width × height)/2, or scored as previously described [41, 42]. Each experiments was replicated independently in other experimental models and using other cell lines as described above.
For SMYD2 inhibition in vivo, 100 µg/tumor AZ505 (HY-15226; MedchemExpress) was injected directly into tumors every two days using colonoscopy (n = 5/group). Tumor size was blindly scored at day 5 and day 10. In a different experiment, 50 mg/kg BAY-598 (HY-19546; MedchemExpress) was intraperitoneally injected once daily into mice bearing HT-29 xenografts (n = 6/group). Tumors were measured at serial time points using a digital caliper as mentioned above.
Human material
All human material used in this study was approved by the ethics committee of the University Hospital of Erlangen.
Cell culture
MC-38 cells and HT-29 cells were cultured in complete Dulbecco’s modified Eagle’s medium (DMEM) (31966047, Invitrogen) supplemented with 10% heat-inactivated fetal bovine serum (F7524, Sigma-Aldrich) and 1% penicillin/streptomycin (P4333, Sigma-Aldrich). All cell cultures were free of mycoplasma. Human TNF (11343015; Immunotools) was used to stimulate HT-29 cells while murine TNF (12343016; Immunotools) was used to stimulate MC-38 cells.
For 3D culture of HT-29 cells, cells were cultured with the medium mentioned above supplemented with 3 mg/ml carboxymethyl cellulose (C5013, Sigma-Aldrich).
Cell death assay
For Lactate dehydrogenase (LDH) assay, supernatants were collected from the cultured cell. The release of LDH in supernatant was quantified via Cytotoxicity Detection Kit (LDH) (11644793001; Sigma-Aldrich) according to the manufacturer’s protocol. Supernatants from cells cultured in medium with 0.1% Triton X-100 were used as positive control and culture medium was used as negative control. The cell death was calculated as follows:
For flow cytometry assay, cells were labeled with propidium iodide (PI) (P1304MP; Thermo Fisher) or CellEvent™ Caspase-3/7 Green Flow Cytometry Assay Kit (C10427; Thermo Fisher) and analyzed by flow cytometry (Accuri™ flow cytometers; BD) according to the manufacturer’s protocol. Experiments were performed three times and representative data are shown.
EdU assay
Cells were labeled with EdU Proliferation Kit (ab219801; Abcam) according to the manufacturer’s protocol. Briefly, cells were cultured in medium containing 15 µM EdU for 3 h. After fixation and permeabilization, cells were further stained with iFluor 488 azide. EdU positive cells were analyzed by flow cytometry (Accuri™ flow cytometers; BD). Experiments were performed three times and representative data are shown.
RNA extraction and real-time quantitative PCR
Total RNA was extracted from tissues or cultured cells using peqGOLD RNA Kit (732-2868, Peqlab) according to the manufacturer’s protocol and reverse transcribed into cDNA using the SCRIPT cDNA Synthesis Kit (PCR-511L, Jena Bioscience). Quantitative real-time PCR was carried out using specific QuantiTect Primer assays (Qiagen). The mRNA expression of the reference gene hypoxanthine guanine phosphoribosyl transferase (HPRT) was used to normalize cDNA levels.
RNA-seq analysis
After RNA extraction and quality control, samples were sequenced using an Illumina platform generating paired-end reads. Mapping on the reference genome and quantification were done using STAR (2.7.0d) and featurecounts (v1.6.4) respectively. Differential expression of the groups of samples was performed using deseq2 (1.24.0). Enrichment, clustering, and other analysis were performed using in-house bioinformatic tools.
Histology and immunohistochemistry
Histopathological analyses were performed on formalin-fixed paraffin-embedded tissue after Mayer’s H&E staining. Immunohistochemistry of tissue sections was performed using the TSA system as recommended by the manufacturer (NEL704A001KT, PerkinElmer). Antibodies for immunohistochemistry were as follows: anti-human SMYD2 (HPA029023; Sigma-Aldrich), anti-murine SMYD2 (21290-1-AP; Proteintech), anti-cleaved caspase-3 (9661; Cell Signaling Technology), and anti-rabbit IgG (7074; Dianova). Nuclei were stained with Hoechst 33342 (H3570; Invitrogen). Imaging was carried out using a Leica laser-scanning confocal microscope or Hamamatsu Nanozoomer 2.0 HT (Hamamatsu).
Immunoprecipitation (IP) assay
For immunoprecipitation of complex-I, cells were seeded in 15 cm Petri dishes and treated with 1 µg/ml FLAG-TNF (ALX-522-009-C050 for MC-38 cells and ALX-522-008-C050 for HT-29 cells; Enzo Life Sciences). Whole-cell extracts were prepared in the presence of native lysis buffer (9803; Cell Signaling Technology) supplemented with protease inhibitors (Complete Mini Protease Inhibitor Cocktail, Roche) and phosphatase inhibitors (PhosphoStop Phosphatase Inhibitor Cocktail, Roche). For the control samples, 1 µg/ml FLAG-TNF was added post-lysis. Cell lysates were centrifuged for 20 min at 14,000 rpm and supernatants were collected and incubated with Anti-FLAG® M2 Magnetic Beads (M8823; Sigma-Aldrich) overnight at 4 °C. The beads were washed five times with TBS and co-precipitated proteins were eluted using SDS–PAGE loading buffer.
For immunoprecipitation of RIPK1 and SMYD2, cell lysates were prepared as mentioned above but incubated with anti-RIPK1 antibody (3493; Cell Signaling Technology) or anti-SMYD2 antibody (ab108217; Abcam) overnight at 4 °C. The next day, Protein A Agarose Beads (9863; Cell Signaling Technology) were added into the mixture and incubated in room temperature for 30 min. The beads were washed five times with native lysis buffer and co-precipitated proteins were eluted using SDS–PAGE loading buffer.
Western blot (WB) analysis
For immunoblot analysis of lysates under non-reducing conditions, whole-cell extracts were prepared in NP40 lysis buffer (FNN0021; Thermo Fisher) supplemented with protease inhibitors (Complete Mini Protease Inhibitor Cocktail; Roche) and phosphatase inhibitors (PhosphoStop Phosphatase Inhibitor Cocktail; Roche). Supernatant proteins from cell lysates were quantified and mixed with native sample buffer (161-0738; Bio-Rad). For immunoblot analysis of lysates under reducing conditions, tissues or cell extracts were prepared in RIPA lysis buffer (89900, Thermo Fisher) supplemented with protease inhibitors and phosphatase inhibitors. Supernatant proteins from cell lysates were mixed with LDS Sample Buffer (NP0007; Thermo Fisher) and heated under 95 °C for 5 min.
Proteins were separated by SDS-PAGE using a MiniProtean-TGX gel (4−15% polyacrylamide; BioRad) and blotted on nitrocellulose membrane (Whatman) followed by incubation with the following primary antibodies: anti-SMYD2 (sc-393827; Santa Cruz), anti-caspase-3 (9662; Cell Signaling Technology), anti-cleaved caspase-3 (9661; Cell Signaling Technology), anti-HMGB1 (ab67281; Abcam), anti-MLKL (37705; Cell Signaling Technology), anti-β catenin (8480; Cell Signaling Technology), anti-RIPK1 (3493; Cell Signaling Technology), anti-TNFR (sc-8436; Santa Cruz), anti-murine pRIPK1 (53286; Cell Signaling Technology), anti-human pRIPK1 (44590; Cell Signaling Technology), and HRP-linked β actin (ab49900; Abcam). HRP-linked anti-rabbit IgG (7074; Cell Signaling Technology), anti-Rabbit IgG (Light-Chain Specific) (45262; Cell Signaling Technology), or HRP-linked anti-mouse IgG (7076; Cell Signaling Technology) was used as a secondary antibody.
Statistical analysis and sample sizes
Significances were determined using the two-tailed students t-test for experiments with two groups. Comparisons of experiments with more than two groups were done with ANOVA. Differences were considered significant at *p < 0.05; **p < 0.01; ***p < 0.001; N.S. = not significant (p ≥ 0.05). Normal distribution and equal variance were assumed for all endpoints subjected to statistical testing. For cell line-based experiments, samples sizes were chosen based on power calculation and using the standard deviations calculated from preliminary experiments [43]. For these experiments, the estimated sample sizes were exceeded in order to ensure enough power. In addition, we also replicated the same experiments in another, independent knockout cell line to increase the power and the confidence in the outcomes. For studies involving mice, power and sample sizes were estimated using empirical data from several years of experience in the laboratory for the expected standard deviations involved in xenograft and transplant-mouse models of cancer and supported by the available sample size calculators. For e.g., sample size estimations for mouse tumor models were calculated to be n = 4, at an alpha error of 0.05 and a beta error of 0.05, at a power of 95%, and Cohens d. In the actual experiments, we budgeted one extra mouse for unforeseen events and used an n of 5 for the actual experiment. Also for all in vivo experiments, we also replicated the findings using independent mouse strains and cell lines to increase the power and confidence in experimental outcomes.
Supplementary information
Author contributions
YY and CB designed the research. FBE, MS, and MW provided material that made the study possible. YY, MFN, MG, EL, and CB analyzed data and wrote the paper. YY, VT, JVP, OT, MZ, LB, and SW performed experiments. All authors reviewed the manuscript.
Funding
This work received funding from the DFG projects FOR2438 (TP02, TP05), BE3686/9-1, SFB1181 (C05), Wilhelm Sander-Stiftung (2019.143.1 to FBE) and TRR241 (A02, A03, A06, B05, C04). Further support came from the China Scholarship Council and the Polish Ministry of Science and Higher Education (Mobility plus 5, 1658/1/MOB/17/2018/0). Open Access funding enabled and organized by Projekt DEAL.
Data availability
Sequencing data have been deposited at ArrayExpress under the accession number E-MTAB-11053. Full size blots have been included in the supplemental Fig. 4.
competing interests
The authors declare no competing interests.
Footnotes
Edited by Professor Jianke Zhang
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41419-021-04483-0.
References
- 1.Global Burden of Disease Cancer C. Fitzmaurice C, Allen C, Barber RM, Barregard L, Bhutta ZA, et al. Global, regional, and national cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life-years for 32 cancer groups, 1990 to 2015: A systematic analysis for the global burden of disease study. JAMA Oncol. 2017;3:524–48. doi: 10.1001/jamaoncol.2016.5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arnold M, Sierra MS, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global patterns and trends in colorectal cancer incidence and mortality. Gut. 2017;66:683–91. doi: 10.1136/gutjnl-2015-310912. [DOI] [PubMed] [Google Scholar]
- 3.Marmol I, Sanchez-de-Diego C, Pradilla Dieste A, Cerrada E, Rodriguez Yoldi MJ. Colorectal carcinoma: A general overview and future perspectives in colorectal cancer. Int J Mol Sci. 2017;18:197. [DOI] [PMC free article] [PubMed]
- 4.Brown MA, Sims RJ, 3rd, Gottlieb PD, Tucker PW. Identification and characterization of Smyd2: a split SET/MYND domain-containing histone H3 lysine 36-specific methyltransferase that interacts with the Sin3 histone deacetylase complex. Mol Cancer. 2006;5:26. doi: 10.1186/1476-4598-5-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang J, Perez-Burgos L, Placek BJ, Sengupta R, Richter M, Dorsey JA, et al. Repression of p53 activity by Smyd2-mediated methylation. Nature. 2006;444:629–32. doi: 10.1038/nature05287. [DOI] [PubMed] [Google Scholar]
- 6.Zeng Y, Qiu R, Yang Y, Gao T, Zheng Y, Huang W, et al. Regulation of EZH2 by SMYD2-mediated lysine methylation is implicated in tumorigenesis. Cell Rep. 2019;29:1482–98. doi: 10.1016/j.celrep.2019.10.004. [DOI] [PubMed] [Google Scholar]
- 7.Sajjad A, Novoyatleva T, Vergarajauregui S, Troidl C, Schermuly RT, Tucker HO, et al. Lysine methyltransferase Smyd2 suppresses p53-dependent cardiomyocyte apoptosis. Biochim Biophys Acta. 2014;1843:2556–62. doi: 10.1016/j.bbamcr.2014.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li LX, Zhou JX, Calvet JP, Godwin AK, Jensen RA, Li X. Lysine methyltransferase SMYD2 promotes triple negative breast cancer progression. Cell Death Dis. 2018;9:326. doi: 10.1038/s41419-018-0347-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meng F, Liu X, Lin C, Xu L, Liu J, Zhang P, et al. SMYD2 suppresses APC2 expression to activate the Wnt/beta-catenin pathway and promotes epithelial-mesenchymal transition in colorectal cancer. Am J Cancer Res. 2020;10:997–1011. [PMC free article] [PubMed] [Google Scholar]
- 10.Koliaraki V, Pallangyo CK, Greten FR, Kollias G. Mesenchymal cells in colon cancer. Gastroenterology. 2017;152:964–79. doi: 10.1053/j.gastro.2016.11.049. [DOI] [PubMed] [Google Scholar]
- 11.She P, Zhang H, Peng X, Sun J, Gao B, Zhou Y, et al. The Gridlock transcriptional repressor impedes vertebrate heart regeneration by restricting expression of lysine methyltransferase. Development. 2020;147:dev190678. [DOI] [PMC free article] [PubMed]
- 12.Lai Y, Yang Y. SMYD2 facilitates cancer cell malignancy and xenograft tumor development through ERBB2-mediated FUT4 expression in colon cancer. Mol Cell Biochem. 2020. 10.1007/s11010-020-03738-2. [DOI] [PubMed]
- 13.Gao S, Wang Z, Wang W, Hu X, Chen P, Li J, et al. The lysine methyltransferase SMYD2 methylates the kinase domain of type II receptor BMPR2 and stimulates bone morphogenetic protein signaling. J Biol Chem. 2017;292:12702–12. doi: 10.1074/jbc.M117.776278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li S, Wang C, Liu X, Hua S, Liu X. The roles of AXIN2 in tumorigenesis and epigenetic regulation. Fam Cancer. 2015;14:325–31. doi: 10.1007/s10689-014-9775-7. [DOI] [PubMed] [Google Scholar]
- 15.Ismail NI, Othman I, Abas F, Lajis NH, Naidu R. Mechanism of apoptosis induced by curcumin in colorectal cancer. Int J Mol Sci. 2019;20:2454. [DOI] [PMC free article] [PubMed]
- 16.Schmidt S, Gay D, Uthe FW, Denk S, Paauwe M, Matthes N, et al. A MYC-GCN2-eIF2alpha negative feedback loop limits protein synthesis to prevent MYC-dependent apoptosis in colorectal cancer. Nat Cell Biol. 2019;21:1413–24. doi: 10.1038/s41556-019-0408-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bedi A, Pasricha PJ, Akhtar AJ, Barber JP, Bedi GC, Giardiello FM, et al. Inhibition of apoptosis during development of colorectal cancer. Cancer Res. 1995;55:1811–6. [PubMed] [Google Scholar]
- 18.Kikuchi Y, Dinjens WN, Bosman FT. Proliferation and apoptosis in proliferative lesions of the colon and rectum. Virchows Arch. 1997;431:111–7. doi: 10.1007/s004280050076. [DOI] [PubMed] [Google Scholar]
- 19.Carneiro BA, El-Deiry WS. Targeting apoptosis in cancer therapy. Nat Rev Clin Oncol. 2020;17:395–417. doi: 10.1038/s41571-020-0341-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seyrek K, Ivanisenko NV, Richter M, Hillert LK, Konig C, Lavrik IN. Controlling cell death through post-translational modifications of DED proteins. Trends Cell Biol. 2020;30:354–69. doi: 10.1016/j.tcb.2020.02.006. [DOI] [PubMed] [Google Scholar]
- 21.Yi X, Jiang XJ, Fang ZM. Histone methyltransferase SMYD2: ubiquitous regulator of disease. Clin Epigenetics. 2019;11:112. doi: 10.1186/s13148-019-0711-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He GW, Gunther C, Thonn V, Yu YQ, Martini E, Buchen B, et al. Regression of apoptosis-resistant colorectal tumors by induction of necroptosis in mice. J Exp Med. 2017;214:1655–62. doi: 10.1084/jem.20160442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cai Z, Jitkaew S, Zhao J, Chiang HC, Choksi S, Liu J, et al. Plasma membrane translocation of trimerized MLKL protein is required for TNF-induced necroptosis. Nat Cell Biol. 2014;16:55–65. doi: 10.1038/ncb2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Christofferson DE, Yuan J. Cyclophilin A release as a biomarker of necrotic cell death. Cell Death Differ. 2010;17:1942–3. doi: 10.1038/cdd.2010.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takemura R, Takaki H, Okada S, Shime H, Akazawa T, Oshiumi H, et al. PolyI:C-induced, TLR3/RIP3-dependent necroptosis backs up immune effector-mediated tumor elimination in vivo. Cancer Immunol Res. 2015;3:902–14. doi: 10.1158/2326-6066.CIR-14-0219. [DOI] [PubMed] [Google Scholar]
- 26.Kim JW, Ho WJ, Wu BM. The role of the 3D environment in hypoxia-induced drug and apoptosis resistance. Anticancer Res. 2011;31:3237–45. [PubMed] [Google Scholar]
- 27.Loessner D, Stok KS, Lutolf MP, Hutmacher DW, Clements JA, Rizzi SC. Bioengineered 3D platform to explore cell-ECM interactions and drug resistance of epithelial ovarian cancer cells. Biomaterials. 2010;31:8494–506. doi: 10.1016/j.biomaterials.2010.07.064. [DOI] [PubMed] [Google Scholar]
- 28.Han K, Pierce SE, Li A, Spees K, Anderson GR, Seoane JA, et al. CRISPR screens in cancer spheroids identify 3D growth-specific vulnerabilities. Nature. 2020;580:136–41. doi: 10.1038/s41586-020-2099-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yuan J, Amin P, Ofengeim D. Necroptosis and RIPK1-mediated neuroinflammation in CNS diseases. Nat Rev Neurosci. 2019;20:19–33. doi: 10.1038/s41583-018-0093-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Delanghe T, Dondelinger Y, Bertrand MJM. RIPK1 kinase-dependent death: A symphony of phosphorylation events. Trends Cell Biol. 2020;30:189–200. doi: 10.1016/j.tcb.2019.12.009. [DOI] [PubMed] [Google Scholar]
- 31.Li LX, Fan LX, Zhou JX, Grantham JJ, Calvet JP, Sage J, et al. Lysine methyltransferase SMYD2 promotes cyst growth in autosomal dominant polycystic kidney disease. J Clin Invest. 2017;127:2751–64. doi: 10.1172/JCI90921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reynoird N, Mazur PK, Stellfeld T, Flores NM, Lofgren SM, Carlson SM, et al. Coordination of stress signals by the lysine methyltransferase SMYD2 promotes pancreatic cancer. Genes Dev. 2016;30:772–85. doi: 10.1101/gad.275529.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eggert E, Hillig RC, Koehr S, Stockigt D, Weiske J, Barak N, et al. Discovery and characterization of a highly potent and selective aminopyrazoline-based in vivo probe (BAY-598) for the protein lysine methyltransferase SMYD2. J Med Chem. 2016;59:4578–4600. doi: 10.1021/acs.jmedchem.5b01890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ohtomo-Oda R, Komatsu S, Mori T, Sekine S, Hirajima S, Yoshimoto S, et al. SMYD2 overexpression is associated with tumor cell proliferation and a worse outcome in human papillomavirus-unrelated nonmultiple head and neck carcinomas. Hum Pathol. 2016;49:145–55. doi: 10.1016/j.humpath.2015.08.025. [DOI] [PubMed] [Google Scholar]
- 35.Ren H, Wang Z, Chen Y, Liu Y, Zhang S, Zhang T, et al. SMYD2-OE promotes oxaliplatin resistance in colon cancer through MDR1/P-glycoprotein via MEK/ERK/AP1 pathway. Onco Targets Ther. 2019;12:2585–94. doi: 10.2147/OTT.S186806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thomenius MJ, Totman J, Harvey D, Mitchell LH, Riera TV, Cosmopoulos K, et al. Small molecule inhibitors and CRISPR/Cas9 mutagenesis demonstrate that SMYD2 and SMYD3 activity are dispensable for autonomous cancer cell proliferation. PLoS One. 2018;13:e0197372. doi: 10.1371/journal.pone.0197372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hou J, Zhao R, Xia W, Chang CW, You Y, Hsu JM, et al. PD-L1-mediated gasdermin C expression switches apoptosis to pyroptosis in cancer cells and facilitates tumour necrosis. Nat Cell Biol. 2020;22:1264–1275. [DOI] [PMC free article] [PubMed]
- 38.Wilson CA, Browning JL. Death of HT29 adenocarcinoma cells induced by TNF family receptor activation is caspase-independent and displays features of both apoptosis and necrosis. Cell Death Differ. 2002;9:1321–33. doi: 10.1038/sj.cdd.4401107. [DOI] [PubMed] [Google Scholar]
- 39.Gong Y, Fan Z, Luo G, Yang C, Huang Q, Fan K, et al. The role of necroptosis in cancer biology and therapy. Mol Cancer. 2019;18:100. doi: 10.1186/s12943-019-1029-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Elinav E, Nowarski R, Thaiss CA, Hu B, Jin C, Flavell RA. Inflammation-induced cancer: Crosstalk between tumours, immune cells, and microorganisms. Nat Rev Cancer. 2013;13:759–71. doi: 10.1038/nrc3611. [DOI] [PubMed] [Google Scholar]
- 41.Becker C, Fantini MC, Wirtz S, Nikolaev A, Kiesslich R, Lehr HA, et al. In vivo imaging of colitis and colon cancer development in mice using high resolution chromoendoscopy. Gut. 2005;54:950–4. doi: 10.1136/gut.2004.061283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Neurath MF, Wittkopf N, Wlodarski A, Waldner M, Neufert C, Wirtz S, et al. Assessment of tumor development and wound healing using endoscopic techniques in mice. Gastroenterology. 2010;139:1837–43. doi: 10.1053/j.gastro.2010.10.007. [DOI] [PubMed] [Google Scholar]
- 43.Krzywinski M, Altman N. Power and sample size. Nat Methods. 2013;10:1139–40. doi: 10.1038/nmeth.2738. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data have been deposited at ArrayExpress under the accession number E-MTAB-11053. Full size blots have been included in the supplemental Fig. 4.