Abstract
Version 2.0 qualitative and quantitative AMPLICOR reverse transcription-PCR tests for HCV were designed to improve on the performance of first version of the hepatitis C virus (HCV) tests. The new tests were calibrated in international units, the new commonly accepted standard unit of measurement for HCV RNA. The sensitivity of the qualitative tests was enhanced by modifying the specimen processing procedure to achieve a limit of detection 50 IU/ml. The limit of detection for the quantitative tests was 600 IU/ml. Modifications to the amplification reaction mixture and thermal cycling conditions enabled all genotypes to be amplified with similar efficiency. The quantitative tests exhibited a linear range extending from 500 to 500,000 IU/ml and excellent reproducibility, with coefficients of variation ranging from 18 to 39%, within the linear range. These data indicate that the version 2.0 AMPLICOR HCV tests will improve diagnosis of HCV infection and will yield more-accurate titers for prognosis and for monitoring therapeutic efficacy, particularly at low viral loads. Furthermore, it will be possible to compare the performance characteristics and viral load measurements of AMPLICOR tests to those of other tests that adopt the international unit as the standard of measurement.
Both qualitative and quantitative tests for hepatitis C virus (HCV) RNA have contributed to our understanding of the natural history of HCV infection. Quantitative tests are used to measure viral burden for patient identification and stratification (1, 4, 5, 9, 20, 25) and to provide real-time information for modifying treatment (3, 25, 33, 38). Qualitative tests are used to diagnose infection (6, 20, 22, 30) and may serve as a test of cure, where it may suffice to demonstrate that the viral RNA load falls below a critical threshold associated with long-term clinical benefit (14).
The AMPLICOR products have been used successfully for diagnosis (10, 12), prognosis (11, 26, 31, 32), and monitoring therapy (13, 35). Furthermore, a proficiency study found that laboratories using the first version of the qualitative AMPLICOR HCV and quantitative AMPLICOR HCV MONITOR tests obtained more-accurate results than laboratories using either in-house reverse transcription (RT)-PCR HCV tests or other commercially available HCV tests (16). Nevertheless, the AMPLICOR tests had two limitations. First, the limit of detection for the first version of the qualitative tests is only 1,000 copies/ml. Greater sensitivity is required, because some chronic HCV patients have viral loads below this level (11, 31) and because titers fall below this level in a substantial number of patients receiving interferon or combination interferon-ribavirin therapy (25, 36, 38). Also, as has proven true for treatment of human immunodeficiency virus type 1 (HIV-1), aggressive therapies may more effectively suppress viral load and thereby increase the likelihood of achieving a sustained response, making it important to monitor HCV-infected patients with the most-sensitive test available. The second limitation with the first version of the HCV RNA tests is that HCV genotypes 2 and 3 were detected less efficiently than genotype 1 (7, 19, 21). Equivalent detection and quantitation of all genotypes is required to reliably detect and monitor all HCV-positive patients and to determine whether viral load differences explain the relationship between genotype and response to interferon therapy (5, 9, 25).
Version 2.0 (v2.0) AMPLICOR HCV tests were designed to overcome these two limitations. To enhance the sensitivity of the qualitative tests, the specimen processing was modified to increase the specimen volume tested 10-fold, from the equivalent of 5 to 50 μl of plasma. To amplify all genotypes with equivalent efficiency, the composition of the PCR mixture and the thermal cycling parameters were modified.
A limitation for all current HCV RNA assays is that it is impossible to compare assay performance and to determine which assays are most accurate and sensitive. The various assays employ different formats, have different sensitivities, and report results in different units of measurements (e.g., copies and genome equivalents). Additionally, assays that use the same unit of measurement give different results for the same sample; thus, copies measured by one assay format are not equivalent to copies measured by another assay format. Also, the viral load cut-off used to predict response to therapy is assay specific (11, 26, 31, 32). These limitations can be overcome by adopting a commonly accepted standard of measurement, the international unit. By international agreement, 100,000 IU is defined as the amount of virus in 1 ml of the First World Health Organization (WHO) International Standard for Nucleic Acid Amplification Technology Assays for HCV RNA (WHO Standard 96/790), which is distributed in vials containing 50,000 IU per vial (29). Recognizing the importance of a common unit of measurement, the version 2.0 AMPLICOR tests are calibrated with standards containing a known quantity of international units. All results in the quantitative tests and the analytical sensitivity of the qualitative tests are now reported in international units per milliliter. As other test developers adopt the international unit as a unit of measurement, comparisons of assay sensitivity, dynamic range and other performance characteristics will be possible.
Two qualitative version 2.0 AMPLICOR HCV tests are available: the fully automated COBAS AMPLICOR HCV test, v2.0, and the semiautomated AMPLICOR HCV Test, v2.0, which uses a microwell plate format. Similarly, there are two v2.0 quantitative tests: COBAS AMPLICOR HCV MONITOR test, v2.0, and AMPLICOR HCV MONITOR Test, v2.0. Here, we describe the procedure used to calibrate these v2.0 tests to the international unit. We evaluated the sensitivity, specificity and genotype reactivity of all four tests. In addition, we determined the linear range and precision of the quantitative tests.
MATERIALS AND METHODS
Clinical specimens.
The high-titer genotype 1a clinical specimen used to construct the secondary standard and other clinical specimens used to evaluate the performance of the COBAS AMPLICOR and AMPLICOR v2.0 tests were obtained from various sources. For all samples, serum or plasma was separated from whole blood within 6 h of collection, aliquoted, and stored at −70°C or lower.
Standardized, quantitated viral stocks.
The WHO Standard 96/790, which was obtained from the National Institute for Biological Standards and Control (South Mimms, Potters Bar, Hertfordshire, England), served as the primary standard. Each vial of the WHO International Standard was reconstituted in 0.5 ml of distilled water as instructed by the accompanying insert from the National Institute for Biological Standards and Control to give a solution containing 105 IU/ml. Dilutions of the WHO International Standard were made with a pool of human plasma that had tested negative for HCV RNA by RT-PCR. A secondary standard was prepared by diluting a high-titer, HCV-positive (genotype 1a) plasma specimen 48-fold with a pool of human plasma that had tested negative for HCV RNA.
Specimen preparation. (i) Qualitative tests.
HCV RNA was extracted from 200 μl of serum or plasma with 400 μl of working HCV Lysis Buffer containing a known number of internal control (IC) RNA molecules. After a 10-min incubation at 60°C, the RNA was precipitated with isopropanol (600 μl), recovered by microcentrifugation at maximum speed (at least 12,500 × g) for 15 min at room temperature, washed with 1 ml of 70% ethanol, and resuspended in 200 μl of HCV Specimen Diluent. Fifty microliters (50 μl) of the processed specimen was added to Working HCV Master Mix (prepared by adding HCV manganese solution to HCV Master Mix) for the RT-PCR amplification reactions. The amplification reaction contained the HCV RNA recovered from 50 μl of serum or plasma.
(ii) Quantitative tests.
The procedure for the quantitative tests was identical to that used for the qualitative tests with the following exceptions. The Lysis Buffer contained a known number of Quantitation Standard (QS) RNA molecules instead of IC molecules. The input sample volume was reduced to 100 μl, the isopropanol volume was reduced to 500 μl, and the resulting RNA pellet was resuspended in 1 ml of specimen diluent. Consequently, prepared samples for the quantitative tests were 10-fold less concentrated than prepared samples for the qualitative tests. As a result, the amplification reaction mixtures contained the HCV RNA recovered from 5 μl of serum or plasma.
RT-amplification.
All HCV tests amplify and detect a 244-base target sequence located in a highly conserved 5′ untranslated region of the HCV genome (37), defined by the primers KY78 and KY80. Only the downstream primer KY78 is biotinylated. The Working HCV master mix is a bicine-buffered solution containing glycerol, dimethyl sulfoxide, potassium acetate, deoxynucleoside triphosphates (dATP, dCTP, dGTP, and dUTP), primer oligonucleotides, thermostable recombinant enzyme Thermus thermophilus DNA polymerase, manganese acetate, AMPERASE, and sodium azide.
Processed specimens were amplified with a GeneAmp PCR system 9600 thermal cycler (Perkin-Elmer Corporation, Norwalk, Conn.) or with the integrated thermal cycler in the COBAS AMPLICOR analyzer. For the qualitative AMPLICOR HCV test, v2.0, the thermal cycling profile consisted of 5 min at 50°C, 30 min at 62°C (for reverse transcription), 37 PCR cycles of 10 s at 95°C and 25 s at 58°C, and 2 h at 91°C. Samples may be removed for denaturation at any time during the final 2-h incubation at 91°C, which provides scheduling flexibility for laboratory personnel. The samples are held at elevated temperature to suppress any residual AMPERASE activity that could otherwise degrade amplification products; to avoid depurination of amplification products, the holding period must not exceed 2 h. The thermal cycling parameters for the quantitative AMPLICOR HCV MONITOR Test, v2.0, were identical except that the number of PCR cycles was reduced from 37 to 32. For the COBAS AMPLICOR HCV test, v2.0, and COBAS AMPLICOR HCV MONITOR test, v2.0, the COBAS AMPLICOR analyzer automatically performed the correct thermal cycling program after the user entered the correct test name.
Hybridization and detection.
The two amplification products, 244-bp sequences generated from HCV and IC (or QS) target RNA, were detected colorimetrically. Immediately upon completion of the amplification reaction, amplification products were denatured by adding 100 μl of Denaturation Solution to each reaction mixture. For both the qualitative and MONITOR tests, this step was performed manually in the AMPLICOR test format or automatically in the COBAS AMPLICOR test format by the COBAS AMPLICOR analyzer.
For the qualitative tests, denatured HCV and IC amplification products were detected by hybridizing 25-μl aliquots of the denatured amplification product to HCV- and IC-specific probes in the presence of 100 μl of Hybridization Buffer. The HCV-specific probe, KY150, is conserved among genotypes; its sequence is 5′ CATAGTGGTCTGCGGAACCGGTGAGT 3′. Probes were coated onto microwell plates and magnetic particles for the AMPLICOR and COBAS AMPLICOR formats, respectively.
For the MONITOR tests, HCV amplification products were quantitatively detected by hybridizing five (AMPLICOR) or four (COBAS AMPLICOR) serial dilutions of each amplification reaction mixture to the HCV-specific oligonucleotide probe. In the AMPLICOR format, these dilutions were prepared by adding 25 μl of the denatured amplification product to 100 μl of Hybridization Buffer and performing four, fivefold serial dilutions using Hybridization Buffer as the diluent. Similarly, QS amplification products were quantitatively detected by hybridizing three (AMPLICOR) or two (COBAS AMPLICOR) serial dilutions of each amplification reaction mixture to the QS-specific oligonucleotide probe. In the AMPLICOR format, these dilutions were prepared by adding 25 μl of the denatured amplification product to 100 μl of Hybridization Buffer and performing two fivefold dilutions using Hybridization Buffer as the diluent. In the AMPLICOR format, the HCV- and QS-specific wells were assembled onto the same microwell plate frame. In the COBAS AMPLICOR format, the COBAS AMPLICOR analyzer automatically performed the required dilutions and added the appropriate dilutions to tubes containing HCV- and QS-specific probe-coated magnetic particles.
The qualitative and MONITOR tests used the same procedures for hybridization and colorimetric detection of the hybridized amplification products. In the AMPLICOR format, these procedures were performed as previously described (17). In the COBAS AMPLICOR format, the COBAS AMPLICOR analyzer automatically performed the hybridization and colorimetric detection steps. The absorbance of the reaction products was measured at 450 and 660 nm (single wavelength) in the AMPLICOR and COBAS AMPLICOR formats, respectively.
Interpretation of results. (i) Qualitative tests.
The results were interpreted by comparing the HCV and IC absorbance values to established cut-offs. The COBAS AMPLICOR analyzer performed this comparison automatically and reported the interpretation along with the actual absorbance values. For the AMPLICOR format, results were interpreted as follows. Specimens with an HCV A450 equal to or greater than 1.0 were interpreted as positive for HCV RNA, regardless of the IC result; specimens with an HCV A450 between 0.3 and 1.0 were interpreted as equivocal (see below). Specimens with an HCV A450 less than 0.3 were interpreted as negative, provided the IC A450 value was equal to or greater than 0.3.
Specimens with an HCV A450 less than 0.3 and an IC A450 less than 0.3 were interpreted as invalid results, and a new aliquot of the specimen was tested.
In the AMPLICOR format, specimens with an HCV A450 equal to or greater than 0.3 but less than 1.0 were interpreted as equivocal for HCV RNA, regardless of the IC result. The COBAS AMPLICOR analyzer automatically interpreted specimens with weak signals (A660 range of 0.15 to 1.0) as equivocal. To obtain an unequivocal result, these specimens were retested in duplicate. The final interpretation was positive if all three tests yielded an A450 equal to or greater than 0.3 for AMPLICOR or an A660 equal to or greater than 0.15 for COBAS AMPLICOR, regardless of the IC results. The final interpretation was negative if any of the three tests had A450 values less than 0.3 for AMPLICOR or A660 values less than 0.15 for COBAS AMPLICOR, provided that the corresponding IC absorbance values for these tests were equal to or greater than 0.3 (A450) for AMPLICOR and 0.15 (A660) for COBAS AMPLICOR.
(ii) Quantitative tests.
For the AMPLICOR HCV MONITOR Test, v2.0, the following calculations were performed manually. For both the target and QS, the dilution that yielded the lowest signal within the linear range of the colorimetric detection assay (i.e., A450 between 0.15 and 2.0) was identified. The absorbance for this dilution was corrected by subtracting the background absorbance (A450, 0.07), and the resulting corrected absorbance was multiplied by the dilution factor to determine the total signal generated. The input HCV RNA concentration was calculated by comparing the total HCV absorbance of the sample to the total QS absorbance for the sample, using the equation RNA = (total target OD/total QS OD) × QS per amplification reaction × 200, where 200 is the conversion factor from international units per amplification reaction to international units/milliliter, RNA is in measured in international units per milliliter, OD is optical density, and QS is measured in international units. For the COBAS AMPLICOR HCV MONITOR test, v2.0, essentially the same calculations were performed automatically with one exception. Instead of choosing the dilution that gave the lowest A450 between 0.15 and 2.0, the COBAS AMPLICOR analyzer identified the dilution with an A660 between 0.10 and 2.0 that yielded the highest total absorbance value when multiplied by its corresponding dilution factor.
RESULTS
Calibration to international units.
In the COBAS AMPLICOR HCV MONITOR and AMPLICOR HCV MONITOR tests, the amount of target RNA is calculated by dividing the total optical density [OD] generated from the target by the total OD generated from a known quantity of QS. This dimensionless ratio is then multiplied by the QS concentration and a factor of 200 that has units of reaction per milliliter (this is the inverse of the volume of specimen, 0.005 ml, added to each reaction). Thus, the concentration of the QS must be expressed in international units per amplification reaction to report test results in international units per milliliter.
To calibrate the QS in international units per reaction, we measured the WHO international standard with both the COBAS AMPLICOR and AMPLICOR HCV MONITOR tests. Two dilutions of the WHO international standard, which by definition contained 5,000 and 25,000 IU/ml, were tested. These dilutions were chosen for evaluation because they gave results that were well within the linear range of earlier versions of the COBAS AMPLICOR and AMPLICOR MONITOR tests, which were calibrated to report results in copies per milliliter but otherwise were identical to the tests being used. For each test format, the total OD generated from each diluted WHO standard was divided by the respective total OD generated from the QS. The amount of the WHO standard added to each reaction mixture (5,000 or 25,000 IU/ml × 0.005 ml/amplification reaction) was divided by the total OD ratio to obtain the QS concentration expressed in international units per amplification reaction.
QS concentration values (in international units per amplification reaction mixture) were determined by testing 5,000- and 25,000-IU/ml dilutions of the WHO international standard for HCV RNA with the COBAS AMPLICOR and AMPLICOR HCV MONITOR v2.0 tests. For each dilution, multiple sample extractions and multiple tests were performed using multiple lots of reagents. Within each test format, the results for the two dilutions were similar (geometric mean ± standard deviation = 1.50 ± 0.13 log10 and 1.53 ± 0.14 log10 for 5,000 and 25,000 IU/ml, respectively, in the COBAS AMPLICOR format and 2.04 ± 0.17 log10 and 2.01 ± 0.14 log10 for 5,000 and 25,000 IU/ml, respectively, in the AMPLICOR format). The overall geometric mean for each format was determined by combining the results for both dilutions (n = 223 and 222 for the COBAS AMPLICOR and AMPLICOR formats, respectively), and the anti-log10 was calculated to transform the value to international units per amplification reaction mixture. The value for the AMPLICOR format, 108 IU/amplification reaction mixture, was approximately 3.3 times larger than the value for the COBAS AMPLICOR test format, 33 IU/amplification reaction mixture.
The QS values (international units per amplification reaction for the two formats are different even though the quantity of QS RNA added to each amplification reaction is the same for both formats. The quantity of QS RNA was independently established by end point titration, which measures the number of amplifiable molecules (actually signal-generating units) through the use of end point dilution and Poisson statistics (27). A given number of amplifiable HCV QS RNA molecules generates an approximately fivefold higher total OD in the AMPLICOR HCV format than in the COBAS AMPLICOR format (data not shown). In contrast, the total OD generated from a given number of HCV international units is only 1.5-fold higher in the AMPLICOR HCV format (data not shown). Thus, the HCV target/QS total OD ratio is smaller in the AMPLICOR HCV format, which results in the larger QS international units per amplification reaction factor. By assigning format-specific international unit per amplification reaction values to the QS, we can ensure that a given test sample will yield the same titer (in international units per milliliter) in both the AMPLICOR HCV and COBAS AMPLICOR formats.
Establishment of secondary standard.
The supply of the WHO international standard, however, is limited. Thus, a secondary standard was created for performing routine calibrations. A high-titer, HCV-positive (genotype 1a) plasma specimen was diluted with a pool of negative plasma. Titers of the secondary standard (in international units per milliliter) were determined by testing two dilutions (40- and 200-fold) of the source plasma that were well within the linear range of the versions of the COBAS AMPLICOR and AMPLICOR MONITOR tests that were calibrated to report results in copies per milliliter.
Multiple sample extractions and multiple tests were performed using multiple lots of reagents. For each test format, the 48 replicates at each dilution yielded approximately the same geometric mean for the source plasma. The overall geometric mean for the source plasma was determined in each test format by combining the 96 individual values for both dilutions. The corresponding concentrations of an intermediate dilution (48-fold) of the source plasma that was chosen to be the secondary standard were 26,000 and 28,000 IU/ml in the COBAS AMPLICOR and AMPLICOR formats, respectively. The titers from the COBAS AMPLICOR and AMPLICOR HCV MONITOR tests were averaged to assign a value of 27,000 IU/ml to the secondary standard.
Limit of detection.
The limit of detection was determined using a serial dilution series prepared from the WHO international standard for HCV (96/790). The COBAS AMPLICOR and AMPLICOR HCV v2.0 tests detected all of the samples with concentrations greater than or equal to 50 IU/ml but detected less than 95% of samples at 25 and 15 IU/ml (Table 1). Similarly, the quantitative COBAS AMPLICOR and AMPLICOR HCV MONITOR v2.0 tests detected at least 95% of the samples with concentrations greater than or equal to 600 IU/ml (Table 2). Below 600 IU/ml, the detection rate was less than 95% and decreased as the concentration decreased (Table 2). Because we define the detection limit of the test as the concentration required to yield positive results in at least 95% of replicate reactions, the detection limits for the qualitative and quantitative tests are 50 and 600 IU/ml, respectively.
TABLE 1.
Limit of detection for the COBAS AMPLICOR and AMPLICOR HCV version 2.0 testsa
| Actual input (IU/ml) | % Positive (no. positive/total no. of tests)
|
|
|---|---|---|
| COBAS AMPLICOR format | AMPLICOR format | |
| 150 | 100 (6/6) | 100 (12/12) |
| 125 | 100 (6/6) | 100 (11/11)b |
| 100 | 100 (6/6) | 100 (12/12) |
| 75 | 100 (111/111)c | 100 (117/117)c |
| 50 | 100 (106/106)d | 100 (108/108) |
| 25 | 91 (97/107)b | 90 (96/107)b |
| 15 | 67 (8/12) | 73 (8/11)b |
Limit of detection was determined with dilutions of the WHO international standard for HCV (96/790) as described in Materials and Methods.
One reaction was excluded from the calculation because it gave a negative IC result.
Three reactions were excluded from the calculation because they gave negative IC results.
Two reactions were excluded from the calculation because they gave negative IC results.
TABLE 2.
Limit of detection for the COBAS AMPLICOR and AMPLICOR HCV MONITOR version 2.0 testsa
| Actual input (IU/ml) | % Positive (no. positive/total no. of tests)
|
|
|---|---|---|
| COBAS AMPLICOR format | AMPLICOR format | |
| 1,100 | 100 (48/48) | 100 (48/48) |
| 900 | 95 (91/96) | 99 (94/95)b |
| 700 | 99 (95/96) | 100 (96/96) |
| 600 | 95 (74/78) | 96 (75/78) |
| 500 | 90 (92/102) | 98 (106/108) |
| 400 | 97 (29/30) | 89 (32/36) |
| 300 | 57 (17/30) | 75 (27/36) |
Limit of detection was determined with dilutions of the secondary standard that was calibrated against the WHO international standard for HCV (96/790) as described in Results.
One reaction was excluded from the calculation because it gave a negative IC result.
HCV genotype reactivity.
To verify that the qualitative COBAS AMPLICOR and AMPLICOR HCV v2.0 tests exhibited similar sensitivity for all HCV genotypes, limiting-dilution experiments were performed for RNA transcripts representing genotypes 1a, 1b, 2a, 2b, 3a, 4, and 5. The transcripts were synthesized from cloned HCV DNA and purified by standard methods; the A260 of each preparation was measured to provide an independent measure of the RNA concentration. The samples with 10 copies per reaction yielded positive results at least 95% of the time for all genotypes except genotype 5 (Table 3). The concentration required to yield positive results in at least 95% of replicate reactions was similar for genotypes 1a, 1b, 2a, 2b, 3a, and 4, with the AMPLICOR format exhibiting somewhat greater fluctuation between genotypes (Table 3). In both the COBAS AMPLICOR and AMPLICOR HCV formats, slightly higher concentrations of genotype 5 were required to obtain positive results 95% of the time (Table 3). Thus, the qualitative tests exhibited similar sensitivity for genotypes 1a, 1b, 2a, 2b, 3a, and 4 and were only approximately twofold less sensitive for genotype 5.
TABLE 3.
Genotype reactivity for the COBAS AMPLICOR and AMPLICOR HCV version 2.0 testsa
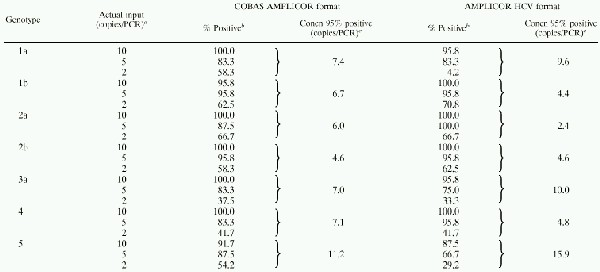 |
Concentrated solutions of transcripts with an A260 of 2.5, which is equivalent to 2.9 × 1014 copies/ml, were diluted with HCV specimen diluent to achieve final concentrations of 40, 100, and 200 copies/ml; 50 μl of each dilution was added to each reaction mixture.
A total of 24 replicate tests were performed at each concentration. Three independent dilutions were prepared for each concentration, and eight replicate reactions were performed for each of the three dilutions.
The concentration that would yield positive results 95% of the time was determined by Probit analysis.
To demonstrate that HCV genotypes 1a, 1b, 2a, 2b, and 3a are quantitated with equal accuracy, sets of serially diluted clinical specimens were tested by both the COBAS AMPLICOR HCV MONITOR and AMPLICOR HCV MONITOR v2.0 tests. The concentrations tested spanned the linear range of the tests. The deviation from the expected concentration was calculated for each dilution of each genotype by subtracting the log10 of the expected titer from the log10 of the measured titer. At each concentration, the measured titers for each genotype were within 0.5 log10 of the expected value; furthermore, the deviations from expected titer were similar for all genotypes (Table 4).
TABLE 4.
Genotype reactivity of the COBAS AMPLICOR and AMPLICOR HCV MONITOR tests
| Format | Genotype | Log10 measured concentration − log10 expected concentrationa (IU/ml)
|
||
|---|---|---|---|---|
| 3.5 log10 | 4.5 log10 | 5.5 log10b | ||
| COBAS AMPLICOR | 1a | 0.2 | 0.1 | −0.1 |
| 1b | 0.3 | 0.2 | 0.1 | |
| 2a | 0.4 | 0.4 | 0.1 | |
| 2b | 0.2 | 0.1 | −0.1 | |
| 3a | 0.0 | 0.0 | −0.2 | |
| AMPLICOR | 1a | 0.2 | 0.0 | −0.2 |
| 1b | 0.3 | 0.2 | 0.1 | |
| 2a | 0.4 | 0.3 | 0.0 | |
| 2b | 0.4 | 0.2 | −0.1 | |
| 3a | −0.1 | −0.2 | −0.3 | |
High-concentration clinical specimens were diluted to the indicated log10 concentration (international units per milliliter). For each sample, results for six replicates were averaged and log10-transformed prior to calculating the difference between the measured and expected log10 titers. The expected concentration was originally measured by performing both the COBAS AMPLICOR and AMPLICOR HCV MONITOR v2.0 tests on a sample of the original specimen that was diluted to achieve a concentration near the middle of the linear range.
For genotype 1b only, the input concentration was 5.2 log10 IU/ml.
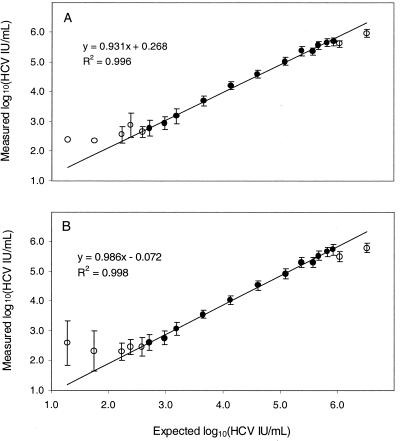
For both the COBAS AMPLICOR and AMPLICOR HCV MONITOR v2.0 tests, the difference between the measured concentration and the expected concentration progressively decreased with increasing expected concentration (Table 4). These data indicate that the measured concentration increased somewhat less rapidly than the expected concentration, an observation consistent with results of experiments that showed the measured concentration increased linearly, but with a slope slightly less than 1.0 (see below and Fig. 1).
FIG. 1.
Linear range for the COBAS AMPLICOR (A) and AMPLICOR HCV MONITOR (B) v2.0 tests. Twenty to 48 replicate tests were performed for each input viral RNA concentration. For expected RNA concentrations below 1,600 IU/mL, some of the replicates gave negative results and were excluded from the data analysis. At each expected RNA concentration, the mean of the log10 measured RNA concentrations (circles) and the standard deviations (error bars) were determined. The closed circles, representing measured RNA concentrations between 500 and 500,000 IU/ml, show the results for concentrations used for linear regression analysis. The open circles show results for concentrations that were not included in the regression analysis. The regression analysis was performed using the averaged result for each expected RNA concentration.
Specificity.
The COBAS AMPLICOR HCV, AMPLICOR HCV, COBAS AMPLICOR HCV MONITOR, and AMPLICOR HCV MONITOR, v2.0, tests utilize the same primers and probe and thus should exhibit identical specificity. Analytical specificity was evaluated for all four v2.0 tests by analyzing microbes and viruses that could potentially coinfect HCV-positive individuals as well as skin flora that could contaminate blood during collection. All of the following organisms gave positive IC (or QS) signals and HCV signals below the negative cut-off: adenovirus types 2, 3, and 7; Chlamydia trachomatis; coxsackievirus B1; cytomegalovirus; echovirus 1; Epstein-Barr virus; hepatitis A virus; hepatitis B virus; herpes simplex virus types 1 and 2; human herpes virus 6; human herpes virus 7; HIV-1, subtypes A through F; human T-cell leukemia virus type 1; human papilloma virus types 16 and 18; Propionibacterium acnes; Staphylococcus aureus; Staphylococcus epidermidis; and varicella-zoster virus (data not shown).
In addition, cross-reactivity was assessed by testing a minimum of 495 samples obtained from HCV-seronegative blood donors. All of the samples gave positive IC (or QS) signals and HCV signals below the negative cut-off in the COBAS AMPLICOR HCV, AMPLICOR HCV, COBAS AMPLICOR HCV MONITOR, and AMPLICOR HCV MONITOR, v2.0, tests (data not shown).
Linear range.
The linear range for quantitative tests was determined by analysis of a dilution series of an HCV-positive plasma specimen whose titer in international units per milliliter had been determined by the use of the HCV secondary standard. A set of 19 samples with concentrations ranging from 20 to 3,300,000 IU/ml of plasma was generated by diluting the HCV-positive specimen with various amounts of a pool of HCV RNA-negative human plasmas. Three individuals tested the panel in duplicate, for a total of six replicates at each virus concentration. The log10 measured concentration for each replicate was compared to the log10 expected concentration. For both the COBAS AMPLICOR and AMPLICOR HCV test formats, residual analysis showed that the concentration measured by either test was, on average, within 0.3 log10 of the expected concentration for samples with measured titers ranging from 500 to 500,000 IU/ml (data not shown). Both formats yielded similar regression lines for samples within the linear range: the slopes were 0.931 and 0.986, the intercepts were 0.268 and −0.072, and the R2 values were 0.996 and 0.998 for the COBAS AMPLICOR and AMPLICOR HCV formats, respectively (Fig. 1). Because the slopes of the regression lines were slightly less than 1.0 and the intercepts were not 0.0, the log10 of the measured titers over the linear range differed slightly from the log10 of the actual input titers.
Agreement between test formats.
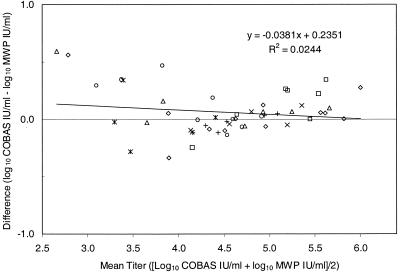
A set of 51 HCV-positive clinical samples with titers ranging from 450 to 1,000,000 IU/ml were tested in parallel with the COBAS AMPLICOR and AMPLICOR HCV MONITOR v2.0 tests. Twenty-two of the 51 samples were tested undiluted, and the remaining 29 high-titer samples were diluted with HCV RNA-negative plasma prior to testing to bring their titers within the linear range of the assay. The set contained at least one sample for each of the following genotypes: 1a, 1b, 2a, 2b, 2c, 2d, 3a, 3b, 4a, 4c, 4f, 4g, 4h, 5a, and 6a (for simplicity, results for the subtypes of genotype 2, 3, and 4 are not distinguished in Fig. 2). To determine whether both tests yielded equivalent titers, we calculated the difference between the two measured titers for each sample in the set. For any one sample, the two titers may differ due to systematic error and test imprecision. If the two formats yield equivalent results, the average of these differences will approach zero (2). Furthermore, the difference between the titers will be independent of the actual titer. To determine if the data satisfied these conditions, we plotted the difference between the two titers versus the actual titer. Since the actual titer of the specimens was not determined by an independent reference test, we estimated the actual titer by averaging the results of the two formats. Because the titers of the samples varied over several orders of magnitude, the measured titers were log transformed before calculating the differences and average titers.
FIG. 2.
Comparison of COBAS AMPLICOR HCV MONITOR test, v2.0, and AMPLICOR HCV MONITOR test (microwell plate [mwp]), v2.0, values for HCV RNA concentration in 51 clinical specimens of genotypes 1a (□), 1b (▵), 2 (◊), 3 (×), 4 (○), 5a (∗), and 6a (+). For each specimen, the difference between the values reported by the COBAS AMPLICOR format and the AMPLICOR HCV (MWP) format is plotted against the average of the two values.
All of the individual differences were less than 0.5 log10, with the exception of two low-titer specimens (Fig. 2). Linear regression analysis indicated that the difference between titers measured by the two formats varied only slightly with concentration; the slope of the regression line was −0.038, indicating that the difference decreased slightly as the viral titer increased (Fig. 2). On average, the difference in titers was small; regression analysis showed that the COBAS AMPLICOR format tended to yield values ranging from 0.02 to 0.13 log10 higher than those obtained in the AMPLICOR format for samples having titers within the linear range of the assays (Fig. 2).
Precision study.
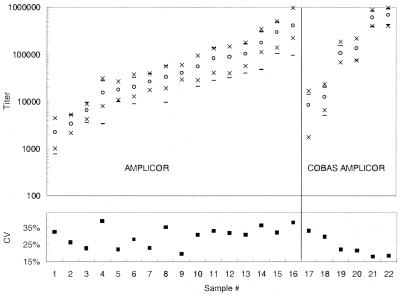
The total imprecision of the COBAS AMPLICOR HCV MONITOR test, v2.0, was assessed by testing each of six samples 40 times over 10 days. Each sample was tested once each day by each of two analysts, each of whom used two different lots of reagents. The samples, which covered most of the linear range of the assay, consisted of serial dilutions prepared from two different clinical specimens. The total imprecision included between-analyst, between-reagent lot, and between-day variation. The estimated total imprecision decreased as the titer increased; the coefficient of variation (CV) ranged from 33% for the lowest-titer sample (8,600 copies/ml) to 18% for a high-titer sample (greater than 600,000 copies/ml [Fig. 3]). For each sample, the upper and lower limits of the 95% confidence interval were within a factor of 3 (0.5 log10) of the mean value (Fig. 3). The minimum and maximum values obtained for each sample were close to the upper and lower limits of the 95% confidence interval (Fig. 3).
FIG. 3.
Precision profile of COBAS AMPLICOR and AMPLICOR HCV MONITOR v2.0 tests. For the COBAS AMPLICOR format, each of the six sets of values plotted represents results from 40 replicates for each of the six samples tested. For the AMPLICOR format, each of the 16 sets of values plotted represents results from 20 replicates for each of the 16 samples tested. The lower portion of the graph shows the CV (■), and the upper portion of the graph shows the mean (○), the minimum and maximum values obtained (×), and the 95% confidence interval (mean ± 2 standard deviations [−]) for a single measurement.
The total imprecision of the AMPLICOR HCV MONITOR test, v2.0, was assessed by testing each of 16 samples 20 times over 10 days. Each sample was tested once each day by each of two analysts. The HCV RNA titers of the 16 samples ranged from 2,200 to 410,000 copies/mL, covering most of the linear range of the assay. The total imprecision included between-analyst and between-day variation. The imprecision for AMPLICOR HCV MONITOR test, v2.0, was similar to that of the COBAS AMPLICOR HCV MONITOR test, v2.0, with CVs ranging from 20 to 39%, but did not vary with HCV RNA titer (Fig. 3). For each sample, the upper and lower limits of the 95% confidence interval were within a factor of 5 (0.7 log10) of the mean value (Fig. 3). The minimum and maximum values obtained for each sample were close to, or well within, the upper and lower limits of the 95% confidence interval (Fig. 3).
DISCUSSION
The version 2.0 qualitative and quantitative AMPLICOR RT-PCR tests for HCV RNA were calibrated in international units, the new commonly accepted standard of measurement for HCV RNA (29). As other test developers adopt the international unit as a unit of measurement, comparisons of assay sensitivity, dynamic range, and other performance characteristics will be possible. For the COBAS AMPLICOR and AMPLICOR HCV qualitative test formats, the limit of detection was calibrated in international units per milliliter by performing the test on dilutions of the WHO international standard, which contains a defined quantity of international units. To calibrate the two quantitative test formats to the international unit, format-specific calibration factors for the QS were calculated by performing both tests on diluted samples of the WHO international standard. The international unit value for the WHO sample was divided by the ratio of the sample total OD/QS total OD to calibrate the QS concentration in international units per amplification reaction.
The WHO international standard is available in limited quantities. For routine use, a secondary standard was established by measuring the titer of an HCV-positive specimen using the QS that was calibrated against the WHO international standard. Once established, the secondary standard was then used to calibrate subsequent QS preparations in international units per amplification reaction. This value is supplied in each amplification kit, enabling the user to calculate the specimen concentration in international units per milliliter directly from the target and QS absorbance values. The assignment of format-specific values for the QS in international units per amplification reaction enables a given sample to yield similar HCV RNA titers in both test formats. Format-specific values are required because the QS yields a stronger signal (relative to the HCV signal) in the AMPLICOR HCV format than in the COBAS AMPLICOR format. Differences in hybridization kinetics for the HCV and QS in the two detection formats probably account for this phenomenon.
Unlike the first version of the AMPLICOR HCV tests, the v2.0 tests amplified all genotypes with similar efficiency. End point dilution assays with RNA transcripts of each genotype demonstrated that the qualitative tests exhibit similar sensitivity for genotypes 1a, 1b, 2a, 2b, 3a, and 4 and slightly lower (twofold) sensitivity for genotype 5. This improved sensitivity for non-1 genotypes was confirmed in an independent end point dilution study (7). At concentrations throughout the linear range, the quantitative tests yielded titers within 0.5 log10 of the actual titer for genotype 1a, genotype 1b, genotype 2a, genotype 2b, and genotype 3a clinical specimens. This improved genotype reactivity is attributed to a reformulated PCR mixture and modified thermal cycling parameters, which most likely promoted unfolding of secondary structures that could form at the relatively low annealing and extension temperatures employed in the first version of the tests.
A recent independent evaluation of the AMPLICOR HCV MONITOR test, v2.0, also demonstrated that RNA transcripts of each genotype and a genotype 1 clinical specimen were accurately quantitated; the difference between the measured titer and the expected titer was nearly constant throughout the test's linear range (19). A genotype 2 clinical specimen and a genotype 3 clinical specimen, however, gave titers that were much lower than the expected titer when tested at concentrations near the top of the linear range (19). The difference between the measured titer and the expected titer decreased progressively and dramatically as the samples were diluted (19). The reason for this discrepancy is not clear. One possibility is that the efficiency of RT may be different for viral RNA and transcripts (19). The genotype 2 and genotype 3 specimens tested by Mellor et al. may have contained factors that reduced the efficiency of RT of the viral RNA without similarly reducing RT of the QS RNA transcript, which would cause the titer to be underestimated. This degree of underestimation would decrease as the samples were diluted, because the concentration of the hypothesized interfering factors would be reduced. Additional genotype 2 and genotype 3 specimens need to be studied to determine whether inaccurate quantitation is a specimen-specific phenomenon.
The qualitative COBAS AMPLICOR and AMPLICOR HCV v2.0 tests were shown to be more sensitive than the first versions of the tests, exhibiting a limit of detection of 50 IU/ml. While these v2.0 tests were also able to detect samples with lower titers, a single result cannot reliably detect the presence of viral RNA when the titer is below 50 IU/ml; by performing replicate tests, we demonstrated that such specimens do not always yield positive results. The sensitivity of the first version of the tests is approximately 1,000 copies/ml (these tests were not evaluated using a standard calibrated in international units). Since an international unit is equivalent to approximately 0.93 to 3.1 copies as measured in the AMPLICOR HCV and COBAS AMPLICOR formats, respectively (data not shown), the v2.0 tests were approximately 6- to 22-fold more sensitive than the first versions of the tests. This is consistent with the results of a recent independent study (7) and agrees with the expected 10-fold difference given the modifications made to the specimen processing procedure: the input sample volume was increased by a factor of two and the final resuspension volume was decreased by a factor of five.
The clinical performance of the qualitative version 2.0 tests has been evaluated in a recent study. Compared to enzyme immunoassay, sensitivity was 94 to 95% and specificity was 97 to 100% (Shiffer et al., unpublished data). The failure to detect HCV RNA in some serologically positive patients is expected; some patients may be infected but not viremic, and others may have resolved earlier infections. The IC showed that less than 3% of specimens were inhibitory when initially tested (Shiffer et al., unpublished data).
The COBAS AMPLICOR and AMPLICOR HCV MONITOR v2.0 tests exhibited good reproducibility. The overall CVs ranged from 18 to 39%. For the COBAS AMPLICOR HCV MONITOR test, v2.0, lower titer specimens exhibited higher CVs, but for AMPLICOR HCV MONITOR test, v2.0, there was no apparent relationship between titer and CV. Given this level of imprecision, any one result will be within a factor of 3 to 5 (0.5 to 0.7 log10) of the actual titer and a greater-than-three (0.5 log10, COBAS AMPLICOR)- or fivefold (0.7 log10, AMPLICOR) change between successive samples can be considered significant.
The COBAS AMPLICOR and AMPLICOR HCV MONITOR v2.0 tests produced a linear response for specimens with measured titers ranging from 500 to 500,000 IU of HCV RNA/ml of plasma. Measured titers at the upper end of the range were somewhat lower than, but within 0.3 log10 of, the actual titer. Also, the measured titers leveled off rapidly above the upper end of the linear range, which increases the chance that out-of-range, high-titer specimens will be assigned a value within the linear range. Consequently, if precise titers need to be determined for samples yielding results near the top of range, the sample should be tested after it is diluted to give a titer that is well within the linear range. The two formats yielded virtually identical regression lines for specimens having titers within the linear range. This implies that the two tests can be used interchangeably, a prediction that was borne out by demonstrating that the tests gave very similar titers when individual specimens were tested in parallel. For any one specimen, the difference between the two results was generally small and within the limit predicted by the precision data; furthermore, the average difference was negligible (approximately 0.06 log10, with COBAS AMPLICOR giving slightly higher values) for the group of specimens tested.
The broad linear range of the v2.0 MONITOR tests makes them well-suited for predicting and monitoring the response to therapy. A large body of data has demonstrated that patients with lower baseline viral loads exhibit higher response rates when treated with interferon, although the positive and negative predictive values are not high enough for deciding whether to administer or withhold interferon treatment from an individual patient (5, 9, 20). Current European and U.S. guidelines recommend a cut-off of 2 × 106 copies/ml, as determined by an in-house RT-PCR assay (8, 20). Other studies, however, have shown that there is little difference in prognosis for viral RNA titers above 105 to 106 copies/ml (11, 15, 18, 28, 31, 34). Thus, the fact that the HCV MONITOR tests underestimate viral loads above 5.0 × 105 IU/ml may not be a limitation. Indeed, several studies have shown that the first version of the AMPLICOR HCV MONITOR test and the Quantiplex HCV test, which does quantitate viral loads above 106 copies/ml, are equally accurate for predicting response to therapy from baseline viral load (26, 32). Future studies that use international units per milliliter as a standardized reporting format should identify a prognostic cut-off. It should be noted, however, that the prognostic value of pretreatment viral load may diminish with the availability of more potent antiviral treatment regimens since reasonably high sustained response rates can be achieved in patients with high viral loads (25).
The decrease in viral load early after treatment initiation appears to have more prognostic value than baseline viral load (11, 13, 25, 35, 36). Thus, measuring the early viral load response may prove useful for guiding changes in therapeutic regimens in individual patients (3, 25, 33, 38) and for comparing the efficacy of new treatment regimens. During interferon-treatment, viral load often drops rapidly to low levels (25, 38). The ability of the HCV MONITOR tests to quantitate viral loads as low as 600 IU/ml make them ideal for measuring this early response. Less-sensitive tests such as the Quantiplex HCV test have limited value for monitoring the response to therapy, since the viral load quickly drops below their limit of detection (11, 23, 24).
In summary, the v2.0 AMPLICOR HCV tests offer several advantages over their predecessors. The new qualitative COBAS AMPLICOR and AMPLICOR HCV v2.0 tests are approximately 10-fold more sensitive, with a detection limit of 50 IU/ml. The quantitative COBAS AMPLICOR and AMPLICOR HCV MONITOR v2.0 tests accurately measure titers of all genotypes. Because they are calibrated in international units, the performance characteristics and viral load measurements of AMPLICOR tests can be compared to those of other tests that adopt the international unit as the standard of measurement. Given these features, the v2.0 AMPLICOR HCV tests will improve diagnosis of HCV infection and will yield more-accurate titers for prognosis and for monitoring therapeutic efficacy.
ACKNOWLEDGMENTS
We thank Minh Nguyen, Karen Mabee, and Kathy Mohr for their excellent technical assistance in the preparation and characterization of the secondary standard.
REFERENCES
- 1.Berenguer M, Wright T L. Hepatitis C and liver transplantation. Gut. 1999;45:159–163. doi: 10.1136/gut.45.2.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bland J M, Altman D J. Regression analysis. Lancet. 1986;i:908–909. doi: 10.1016/s0140-6736(86)91008-1. [DOI] [PubMed] [Google Scholar]
- 3.Bonetti P, Chemello L, Antona C, Breda A, Brosolo P, Casarin P, Crivellaro C, Dona G, Martinelli S, Rinaldi R, Zennaro V, Santonastaso M, Urban F, Pontisso P, Alberti A. Treatment of chronic hepatitis C with interferon-alpha by monitoring the response according to viraemia. J Viral Hepat. 1997;4:107–112. doi: 10.1111/j.1365-2893.1997.tb00212.x. [DOI] [PubMed] [Google Scholar]
- 4.Charlton M, Seaberg E, Wiesner R, Everhart J, Zetterman R, Lake J, Detre K, Hoofnagle J. Predictors of patient and graft survival following liver transplantation for hepatitis C. Hepatology. 1998;28:823–830. doi: 10.1002/hep.510280333. [DOI] [PubMed] [Google Scholar]
- 5.Davis G L, Lau J Y N. Factors predictive of a beneficial response to therapy of hepatitis C. Hepatology. 1997;26(Suppl. 1):122S–127S. doi: 10.1002/hep.510260721. [DOI] [PubMed] [Google Scholar]
- 6.Dhumeaux D, Doffoel M, Galmiche J P. A French consensus conference on hepatitis C: screening and treatment. J Hepatol. 1997;27:941–944. doi: 10.1016/s0168-8278(97)80337-6. [DOI] [PubMed] [Google Scholar]
- 7.Doglio A, Laffont C, Caroli-Bosc F X, Rochet P, Lefebvre J. Second generation of the automated Cobas Amplicor HCV assay improves sensitivity of hepatitis C virus RNA detection and yields results that are more clinically relevant. J Clin Microbiol. 1999;37:1567–1569. doi: 10.1128/jcm.37.5.1567-1569.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.EASL International Consensus Conference on Hepatitis C. Consensus statement. J Hepatol. 1999;30:956–961. [PubMed] [Google Scholar]
- 9.Fried M W. Clinical applications of hepatitis C virus genotyping and quantitation. Clin Liver Dis. 1997;1:631–645. doi: 10.1016/s1089-3261(05)70326-3. [DOI] [PubMed] [Google Scholar]
- 10.Gane E J, Maertens G, Ducatteeuw A, Qian K P, Lau J Y, Jones H, Davies E, Naoumov N V, Williams R. Antibodies to hepatitis C virus envelope proteins correlate with hepatitis C viraemia after liver transplantation. Transplantation. 1999;67:78–84. doi: 10.1097/00007890-199901150-00013. [DOI] [PubMed] [Google Scholar]
- 11.Ichijo T, Matsumoto A, Kobayashi M, Furihata K, Tanaka E. Quantitative measurement of HCV RNA in the serum: a comparison of three assays based on different principles. J Gastroenterol Hepatol. 1997;12:500–506. doi: 10.1111/j.1440-1746.1997.tb00473.x. [DOI] [PubMed] [Google Scholar]
- 12.John M, Flexman J, French M A. Hepatitis C virus-associated hepatitis following treatment of HIV-infected patients with HIV protease inhibitors: an immune restoration disease? AIDS. 1998;12:2289–2293. doi: 10.1097/00002030-199817000-00010. [DOI] [PubMed] [Google Scholar]
- 13.Kakumu S, Aiyama T, Okumura A, Iwata K, Ishikawa T, Yoshioka K. Earlier loss of hepatitis C virus RNA in interferon therapy can predict a long-term response in chronic hepatitis C. J Gastroenterol Hepatol. 1997;12:468–472. doi: 10.1111/j.1440-1746.1997.tb00468.x. [DOI] [PubMed] [Google Scholar]
- 14.Lau D T Y, Kleiner D E, Ghany M G, Park Y, Schmid P, Hoofnagle J H. 10-year follow-up after interferon-alpha therapy for chronic hepatitis C. Hepatology. 1998;28:1121–1127. doi: 10.1002/hep.510280430. [DOI] [PubMed] [Google Scholar]
- 15.Lau J Y, Davis G L, Kniffen J, Qian K P, Urdea M S, Chan C S, Mizokami M, Neuwald P D, Wilber J C. Significance of serum hepatitis C virus RNA levels in chronic hepatitis C. Lancet. 1993;341:1501–1504. doi: 10.1016/0140-6736(93)90635-t. [DOI] [PubMed] [Google Scholar]
- 16.Lelie P N, Cuypers H T M, van Drimmelen A A J, Quint W G V. Quality assessment of hepatitis C virus nucleic acid amplification methods. Infusionsther Transfusionsmed. 1998;25:102–110. [Google Scholar]
- 17.Loeffelholz M J, Lewinski C A, Silver S R, Purohit A P, Herman S A, Buonagurio D A, Dragon E A. Detection of Chlamydia trachomatis in endocervical specimens by polymerase chain reaction. J Clin Microbiol. 1992;30:2847–2851. doi: 10.1128/jcm.30.11.2847-2851.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Manesis E K, Papaioannou C, Gioustozi A, Kafiri G, Koskinas J, Hadziyannis S J. Biochemical and virological outcome of patients with chronic hepatitis C treated with interferon alfa-2b for 6 or 12 months: a 4-year follow-up of 211 patients. Hepatology. 1997;26:734–739. doi: 10.1002/hep.510260327. [DOI] [PubMed] [Google Scholar]
- 19.Mellor J, Hawkins A, Simmonds P. Genotype dependence of hepatitis C virus load measurement in commercially available quantitative assays. J Clin Microbiol. 1999;37:2525–2532. doi: 10.1128/jcm.37.8.2525-2532.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.National Institutes of Health. Management of hepatitis C. NIH Consensus Statement Online. 1997;15(3):1–41. http://odp.od.nih.gov/consensus/cons/105/105_statement.htm . [Online.] http://odp.od.nih.gov/consensus/cons/105/105_statement.htm. . [PubMed] [Google Scholar]
- 21.Olmedo E, Costa J, Lopez-Labrador F X, Forns X, Ampurdanes S, Maluenda M D, Guilera M, Sanchez-Tapias J M, Rodes J, Jimenez de Anta M T. Comparative study of a modified competitive RT-PCR and Amplicor HCV monitor assays for quantitation of hepatitis C virus RNA in serum. J Med Virol. 1999;58:35–43. doi: 10.1002/(sici)1096-9071(199905)58:1<35::aid-jmv5>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 22.Pawlotsky J M, Lonjon I, Hezode C, Raynard B, Darthuy F, Remire J, Soussy C J, Dhumeaux D. What strategy should be used for diagnosis of hepatitis C virus infection in clinical laboratories? Hepatology. 1998;27:1700–1702. doi: 10.1002/hep.510270632. [DOI] [PubMed] [Google Scholar]
- 23.Pawlotsky J M, Martinot-Peignoux M, Poveda J D, Bastie A, Le Breton V, Darthuy F, Remire J, Erlinger S, Dhumeaux D, Marcellin P. Quantification of hepatitis C virus RNA in serum by branched DNA-based signal amplification assays. J Virol Methods. 1999;79:227–235. doi: 10.1016/s0166-0934(99)00024-5. [DOI] [PubMed] [Google Scholar]
- 24.Pockros P J, Bain V G, Hunter E B, Conrad A, Balart A, Hollinger F B, Albert D the Consensus Interferon Study Group. A comparison of reverse transcription-polymerase chain reaction and branched-chain DNA assays for hepatitis C virus RNA in patients receiving interferon treatment. J Viral Hepat. 1999;6:145–150. doi: 10.1046/j.1365-2893.1999.00147.x. [DOI] [PubMed] [Google Scholar]
- 25.Poynard T, McHutchison J, Goodman Z, Ling M-H, Albrecht J for the Algovirc Project Group. Is an “à la carte” combination interferon alfa-2b plus ribavirin regimen possible for the first line treatment in patients with chronic hepatitis C? Hepatology. 2000;31:211–218. doi: 10.1002/hep.510310131. [DOI] [PubMed] [Google Scholar]
- 26.Reichard O, Norkrans G, Fryden A, Braconier J H, Sonnerborg A, Weiland O. Comparison of 3 quantitative HCV RNA assays—accuracy of baseline viral load to predict treatment outcome in chronic hepatitis C. Scand J Infect Dis. 1998;30:441–446. doi: 10.1080/00365549850161395. [DOI] [PubMed] [Google Scholar]
- 27.Rosenstraus M, Wang Z, Chang S Y, DeBonville D, Spadoro J P. An internal control for routine diagnostic PCR: design, properties and effect on clinical performance. J Clin Microbiol. 1998;36:191–197. doi: 10.1128/jcm.36.1.191-197.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rumi M, Del Ninno E, Parravicini M L, Romeo R, Soffredini R, Donato M F, Wilber J, Russo A, Colombo M. A prospective, randomized trial comparing lymphoblastoid to recombinant interferon alfa 2a as therapy for chronic hepatitis C. Hepatology. 1996;24:1366–1370. doi: 10.1002/hep.510240609. [DOI] [PubMed] [Google Scholar]
- 29.Saldanha J, Lelie N, Heath A. Establishment of the first international standard for nucleic acid amplification technology (NAT) assays for HCV RNA. WHO Collaborative Study Group. Vox Sang. 1999;76:149–158. doi: 10.1159/000031040. [DOI] [PubMed] [Google Scholar]
- 30.Schneeberger P M, Keur I, van der Vliet W, van Hoek K, Boswijk H, van Loon A M, van Dijk W C, Kauffmann R H, Quint W, van Doorn L J. Hepatitis C virus infections in dialysis centers in The Netherlands: a national survey by serological and molecular methods. J Clin Microbiol. 1998;36:1711–1715. doi: 10.1128/jcm.36.6.1711-1715.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shiratori Y, Kato N, Yokosuka O, Hashimoto E, Hayashi N, Nakamura A, Asada M, Kuroda H, Ohkubo H, Arakawa Y, Iwama A, Omata M. Quantitative assays for hepatitis C virus in serum as predictors of the long-term response to interferon. J Hepatol. 1997;27:437–444. doi: 10.1016/s0168-8278(97)80346-7. [DOI] [PubMed] [Google Scholar]
- 32.Soffredini R, Rumi M G, Del Ninno E, Parravicini M L, Russo A, Colombo M. Serum levels of hepatitis C virus RNA predict non-response to interferon therapy: comparison of two commercial assays. J Viral Hepat. 1999;6:65–71. doi: 10.1046/j.1365-2893.1999.6120130.x. [DOI] [PubMed] [Google Scholar]
- 33.Tong M J, Blatt L M, Tong L T, Sayadzadeh K, Conrad A. Long-term retreatment in chronic hepatitis C patients who were non-responders to an initial course of interferon-alpha 2b. J Viral Hepat. 1998;5:323–331. doi: 10.1046/j.1365-2893.1998.00120.x. [DOI] [PubMed] [Google Scholar]
- 34.Yamada G, Takatani M, Kishi F, Takahashi M, Doi T, Tsuji T, Shin S, Tanno M, Urdea M S, Kolberg J A. Efficacy of interferon alfa therapy in chronic hepatitis C patients depends primarily on hepatitis C virus RNA level. Hepatology. 1995;22:1351–1354. [PubMed] [Google Scholar]
- 35.Yamakawa Y, Sata M, Suzuki H, Tanaka K, Tanaka E, Noguchi S, Ono K, Tanikawa K. Monitoring of serum levels of HCV RNA in early phase of IFN therapy; as a predictive marker of subsequent response. Hepatogastroenterology. 1998;45:133–136. [PubMed] [Google Scholar]
- 36.Yasui K, Okanoue T, Murakami Y, Itoh Y, Minami M, Sakamoto S, Sakamoto M, Nishioji K. Dynamics of hepatitis C viremia following interferon-alpha administration. J Infect Dis. 1998;177:1475–1479. doi: 10.1086/515309. [DOI] [PubMed] [Google Scholar]
- 37.Young K, Resnick R, Myers T. Detection of hepatitis C virus RNA by a combined reverse-transcriptase-polymerase chain reaction assay. J Clin Microbiol. 1993;31:882–886. doi: 10.1128/jcm.31.4.882-886.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zeuzem S, Lee J H, Franke A, Ruster B, Prummer O, Herrmann G, Roth W K. Quantification of the initial decline of serum hepatitis C virus RNA and response to interferon alfa. Hepatology. 1998;27:1149–1156. doi: 10.1002/hep.510270433. [DOI] [PubMed] [Google Scholar]