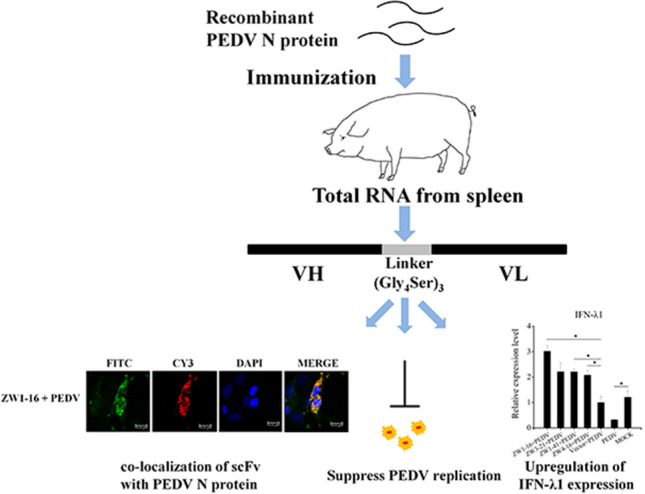
Abstract
Abstract
Porcine epidemic diarrhea virus (PEDV) is the causative agent of porcine epidemic diarrhea, which can cause death in suckling piglets. Vaccines confer only partial protection against new mutant strains, whereas antibodies targeting virus-encoded proteins may be effective prophylactics. In this study, we constructed a recombinant single chain variable fragment (scFv) library from the spleens of two pigs immunized with a recombinant PEDV nucleocapsid (N) protein. Among the positive clones directed against PEDV N protein isolated from the library, four scFvs that showed higher affinity for N were functionally analyzed. These scFvs specifically bound to the PEDV N protein, but not to the transmissible gastroenteritis virus (TGEV) N protein. Their framework regions were highly conserved, whereas their complementarity-determining regions displayed clear diversity. An immunofluorescence assay showed the co-localization of the four scFvs with PEDV N protein in cells. They significantly suppressed PEDV replication, detected with reverse transcription (RT)-quantitative PCR (qPCR; P < 0.01). Two of them significantly reduced the viral titer at 48 hpi and 72 hpi (P < 0.05). In addition, they observably suppressed the production of viral protein at 72 hpi. The expression of interferons, interferon regulatory factor 3 (IRF3), and IRF7 was assessed with RT-qPCR, which indicated that PEDV dramatically suppressed the transcription of interferon-λ1 and IRF7 and that the scFvs significantly upregulated their expression (P < 0.05). These findings facilitated the investigation of the mechanism by which PEDV evaded the host immune response and suggested that these porcine scFvs were potential candidate agents for the prevention and treatment of porcine diarrhea caused by PEDV.
Key points
• Four scFvs targeting PEDV N protein were generated from porcine spleens
• These scFvs co-localized with PEDV N protein and suppressed PEDV replication
• These scFvs significantly upregulated IFN-λ1 expression
Graphical abstract
Supplementary Information
The online version contains supplementary material available at 10.1007/s00253-021-11722-z.
Keywords: Porcine epidemic diarrhea virus, Nucleocapsid protein, Single chain variable fragment (scFv), Replication, Interferon-λ1
Introduction
Porcine epidemic diarrhea (PED), caused by Porcine epidemic diarrhea virus (PEDV), is a highly contagious enteric disease in pigs, which is characterized by anorexia, vomiting, watery diarrhea, dehydration, weight loss, and high mortality in suckling pigs, leading to significant economic losses in the global swine industry (Li et al. 2012; Pensaert and de Bouck 1978; Stevenson et al. 2013).
In recent years, vaccines have been developed to prevent PEDV infection. However, because the virus mutates rapidly to form new variant strains, vaccines frequently show poor efficacy (Sun et al. 2012). Recent studies have demonstrated that the binding specificity and potent neutralizing capacity of antibodies make antibody therapy a promising strategy for the control of PEDV infection (Fu et al. 2017; Gong et al. 2018).
Single chain variable fragments (scFvs) are a kind of genetically engineered antibody, consisting of the variable domains of the heavy (VH) and light (VL) chains of an intact immunoglobulin molecule, linked by a flexible peptide (Bird et al. 1988). Not only does an scFv retain the specific antigen-binding affinity of the antibody, it has better tissue penetration and lower immunogenicity than the intact immunoglobulin because it is smaller (Holliger and Hudson 2005). It is also easy to produce. ScFvs play an important role in the diagnosis and treatment of diseases (Wang et al. 2016b). They can be used as powerful tools that directly target specific antigens to exert strong antitumoral effects and can also be used to visualize tumor masses in vivo (Duranti et al. 2018; Yang et al. 2016). Therefore, passive immunization with scFvs directed against PEDV may be an effective strategy for controlling PEDV infection.
PEDV is an enveloped virus with a single-stranded positive-sense RNA genome of 28 kb, which mainly infects porcine small-intestinal epithelial cells. The genome encodes four structural proteins: the spike (S), envelope (E), membrane (M), and nucleocapsid (N) proteins. As the most abundant viral protein expressed in infected cells (Laude and Masters 1995), N protein binds to the genomic RNA (gRNA) and is involved in several biological activities of the virus (de Haan and Rottier 2005). The interaction between the N and M proteins facilitates the incorporation of the N protein-gRNA ribonucleoprotein complex into the forming viral particle (Kuo et al. 2016). The N protein is also responsible for inducing endoplasmic reticulum (ER) stress, the prolongation of S phase, and the upregulation of interleukin 8 expression; it promotes the recovery of PEDV from infectious clones; and it increases the production of PEDV RNA and virions (Liwnaree et al. 2019; Sungsuwan et al. 2020; Xu et al. 2013).
The innate immune response plays a significant role as the first line of defense against viral infections. During viral infection, pattern recognition receptors in the host cells recognize pathogen-associated molecular patterns, triggering a cascade of signals and the subsequent interferon (IFN) response, which regulates the expression of downstream genes, thus exerting a powerful antiviral effect (Rathinam and Fitzgerald 2011). The IFNs are divided into three different types, type I (IFN-α and IFN-β), type II (IFN-γ), and type III (IFN-λ). Recent studies have shown that the type I and type III IFNs were central to the host’s resistance to viral infection (Mesev et al. 2019) and that IFN-λ1 was more effective against PEDV infection of IPEC-J2 cells than IFN-α (Zhao et al. 2020).
To resist the antiviral effects of IFN and the IFN-induced immune response, many viruses have evolved sophisticated mechanisms to antagonize IFN (Clementz et al. 2010; Liu et al. 2020; Mitra et al. 2019; Totura and Baric 2012). Several PEDV-encoded proteins have been identified as antagonists of the innate immune response: papain-like protease 2 (PLP2), N, endoribonuclease (EndoU), E, nonstructural protein 1 (nsp1), and nsp16 (Deng et al. 2019; Ding et al. 2014; Shi et al. 2019; Xing et al. 2013; Zhang et al. 2016; Zheng et al. 2021). PEDV N protein antagonizes IFN-β and IFN-λ production by targeting various proteins involved in signal transduction (Ding et al. 2014; Shan et al. 2018). Using antibodies to inhibit the function of the target protein is considered to be an effective strategy for the treatment of diseases. However, there have been insufficient studies of the functional effects of antibodies that specifically target the PEDV N protein.
To investigate the effect of scFvs against PEDV N protein during infection, we constructed a porcine recombinant scFv library and selected and identified the scFvs directed against PEDV N protein in this study. The characteristics of the scFvs that bound specifically to PEDV N protein were evaluated. We also examined the effects of the scFvs on PEDV replication in Vero E6 cells and on the expression of IFNs in IPEC-J2 cells. In this study, we demonstrated that scFvs directed against the PEDV N protein may be effective tools for controlling PEDV infection and could allow the development of scFv-based therapeutic agents for the prevention and treatment of porcine diarrhea caused by PEDV.
Materials and methods
Cells and viruses
African green monkey kidney cells (Vero E6) were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Gibco, Carlsbad, CA, USA) supplemented with antibiotics (100 units/mL penicillin and 100 μg/mL streptomycin; Gibco) and 10% fetal bovine serum (FBS; Gibco). The intestinal porcine epithelial cell line J2 (IPEC-J2) was kindly provided by Prof. Jianxiong Xu (Shanghai Jiao Tong University, Shanghai, China) and maintained in DMEM/F12 supplemented with antibiotics (100 units/mL penicillin and 100 μg/mL streptomycin) and 10% FBS. PEDV strain HLJBY was maintained in our laboratory and was passaged in Vero E6 cells.
Construction of recombinant plasmid pCold I-PEDV-N, expression of PEDV N protein, and immunization of pigs
The RNA of PEDV HLJBY was extracted with the E.Z.N.A. Viral RNA Kit (Omega Bio-tek, Norcross, GA, USA). The complementary DNA (cDNA) was synthesized with PrimeScript™ RT Reagent Kit with gDNA Eraser (Perfect Real Time) (Takara Bio, Shiga, Japan). The N gene of PEDV strain HLJBY (GenBank accession number: KP403802) was amplified with an upstream primer containing the XhoI restriction site (underlined) (5′-GTACCACTCGAGATGGCTTCTGTCAGCTTTCAGGA-3′) and a downstream primer containing the HindIII restriction site (underlined) (5′-CGAGGCAAGCTTTT AATTTCCTGTATCGAAGATCTCGT-3′).
PCR thermal cycling was performed with initial denaturation at 95 °C for 5 min, followed by 32 cycles of denaturation at 95 °C for 30 s, annealing at 62 °C for 30 s, and extension at 72 °C for 1 min, with a final extension at 72 °C for 5 min. The PCR products were purified and cloned into the pCold™ I plasmid (Takara Bio). The recombinant plasmid was used to transform Escherichia coli strain BL21(DE3) (TransGen, Beijing, China). The recombinant colonies were cultured in Luria–Bertani (LB) medium containing 100 μg/mL ampicillin until the optical density reached 0.6–0.8. Recombinant protein expression was induced with 1 mM isopropyl β-d-thiogalactoside (IPTG; Sigma-Aldrich, St. Louis, MO, USA) at 16 °C for 30 h. The cells were harvested by centrifugation, resuspended in Ni-Native-0 buffer (50 mM NaH2PO4, 300 mM NaCl, pH 8.0), and disrupted by sonication. After centrifugation at 12,000 × g for 20 min at 4 °C, supernatant was collected. The 6 × His-tagged recombinant protein was purified with Ni–NTA His·Bind® Resin (Merck-Novagen, Darmstadt, Germany), according to the manufacturer’s instructions, concentrated with an Amicon® Ultra-15 Centrifugal Filter (Merck), and analyzed with sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and western blotting, as described below. Imidazole was the competitive agent for the 6 × His-tagged recombinant protein. The protein concentrations were measured with the TaKaRa BCA Protein Assay Kit (Takara Bio).
Two pigs (30–40-days-old) were immunized with the recombinant PEDV N protein (0.5 mg/kg). The protein was homogeneously mixed with Freund’s complete adjuvant and injected intramuscularly into the pigs. Three weeks later, the pigs were immunized twice with the recombinant PEDV N protein homogeneously mixed with Freund’s incomplete adjuvant, with an interval of 3 weeks. Blood was collected and centrifuged to isolate the polyclonal antiserum. After an enzyme-linked immunosorbent assay (ELISA) was performed to measure the antibody titer, the pigs were killed, and their spleens excised, quickly immersed in liquid nitrogen, and stored at − 80 °C.
Construction of the porcine recombinant scFv library
Total RNA was extracted from the porcine spleens with TRIzol™ Reagent (Invitrogen, Carlsbad, CA, USA), and the cDNA was synthesized with PrimeScript™ RT Reagent Kit with gDNA Eraser (Perfect Real Time). The VH and VL genes were amplified with specific primers (Table 1) (Li and Aitken 2004). A linker fragment encoding a short (Gly4Ser)3 peptide was added to the VL genes to generate Linker–VL. Intact scFv genes flanked by the NcoI restriction site and the NotI restriction site were then generated by combining the VH genes with Linker-VL. After the plasmids were digested with the NcoI and NotI restriction enzymes (Thermo Fisher Scientific, Waltham, MA, USA), the scFv genes were ligated into the pOPE101-XP plasmid. Escherichia coli JM109 Chemically Competent Cells (TransGen) were transformed with the recombinant plasmids and incubated on LB-A plates (100 μg/mL ampicillin) at 37 °C for 12 h. Hundreds of transformations were performed to generate the porcine recombinant scFv library. The library size was determined by counting the colonies grown on the LB-A plates. To analyze the diversity of the library, the BstNI restriction enzyme was used to digest randomly selected scFvs at 37 °C for 2 h.
Table 1.
Primers used in the construction of the porcine recombinant scFv library
| Primer | Sequence (5′–3′)a |
|---|---|
| VH-B | GAGGWGAAGCTGGTGGAGTCYGG |
| VH-F | ACGACGACTTCAACGCCTGG |
| VLk-B1 | GCCATYGTGCTGACCCAGASTCC |
| VLk-B2 | GAGCTCGTSATGACCCAGTCTCC |
| VLk-B3 | GAGCTGCGTGATACACAGTCTCC |
| VLk-F | TTTGAKYTCCAGCTTGGTCCC |
| VLλ-B | GATTCTCAGACTGTGATCCAGGAG |
| VLλ-F | GAGGACGGTCAGATGGGTCC |
| Overlap-B1 | CCAGGCGTTGAAGTCGTCGTCGGTGGCGGTGGCTCGGGCGGTGGTGGATCCGGTGGCGGCGGATCTGCCATYGTGCTGACCCAGASTCC |
| Overlap-F1 | GGASTCTGGGTCAGCACRATGGCAGATCCGCCGCCACCGGATCCACCACCGCCCGAGCCACCGCCACCGACGACGACTTCAACGCCTGG |
| Overlap-B2 | CCAGGCGTTGAAGTCGTCGTCGGTGGCGGTGGCTCGGGCGGTGGTGGATCCGGTGGCGGCGGATCTGAGCTCGTSATGACCCAGTCTCC |
| Overlap-F2 | GGAGACTGGGTCATSACGAGCTCAGATCCGCCGCCACCGGATCCACCACCGCCCGAGCCACCGCCACCGACGACGACTTCAACGCCTGG |
| Overlap-B3 | CCAGGCGTTGAAGTCGTCGTCGGTGGCGGTGGCTCGGGCGGTGGTGGATCCGGTGGCGGCGGATCTGAGCTGCGTGATACACAGTCTCC |
| Overlap-F3 | GGAGACTGTGTATCACGCAGCTCAGATCCGCCGCCACCGGATCCACCACCGCCCGAGCCACCGCCACCGACGACGACTTCAACGCCTGG |
| Overlap-λ-B | CCAGGCGTTGAAGTCGTCGTCGGTGGCGGTGGCTCGGGCGGTGGTGGATCCGGTGGCGGCGGATCTGATTCTCAGACTGTGATCCAGGAG |
| Overlap-λ-F | CTCCTGGATCACAGTCTGAGAATCAGATCCGCCGCCACCGGATCCACCACCGCCCGAGCCACCGCCACCGACGACGACTTCAACGCCTGG |
| Overlap-Bprimer | CCAGGCGTTGAAGTCGTCGTC |
| VH-B-NcoI | TCGATCCATGGCTGAGGWGAAGCTGGTGGAGTCYGG |
| VLk-F-NotI | GAACTGCGGCCGCTTTGAKYTCCAGCTTGGTCCC |
| VLλ-F-NotI | TAACTGCGGCCGCGAGGACGGTCAGATGGGTCC |
aThe linker sequence is shown in bold. The restriction sites for NcoI and NotI are underlined
Screening for scFvs directed against PEDV N protein with an ELISA
Recombinant clones randomly selected from the porcine scFv library (n = 768) were cultured in LB medium. The periplasmic expression of the scFv antibody genes in E. coli was induced with 1 mM IPTG at 25 °C for 24 h. The recombinant PEDV N protein (10 μg/mL, 100 µL) was coated onto a 96-well plate and incubated overnight at 4 °C. The plate was washed with phosphate-buffered saline (PBS) containing 0.05% Tween 20 (PBST), blocked at 37 °C for 2 h with 4% bovine serum albumin (BSA) diluted in PBST, and then washed three times with PBST. A periplasmic scFv antibody was added to each well and incubated at 37 °C for 2 h, and the plate was then washed three times with PBST. A mouse monoclonal anti-Myc antibody (diluted 1:3,000) (CMCTAG, Milwaukee, WI, USA) was added to each well and incubated at 37 °C for 2 h, after which the plate was washed four times with PBST. A horseradish peroxidase (HRP)-conjugated Affinipure goat anti-mouse IgG (H + L) antibody (diluted 1:10,000) (Jackson ImmunoResearch, West Grove, PA, USA) was added to each well and incubated at 37 °C for 1 h, after which the plate was washed five times with PBST. 3,3′,5,5′-Tetramethylbenzidine substrate was then added and incubated at room temperature for 10–15 min. Finally, H2SO4 was added to terminate the reaction, and the optical density at a wavelength of 450 nm (OD450) was measured with a Multiskan™ FC Microplate Photometer (Thermo Fisher Scientific).
The N gene of TGEV (GenBank accession number: AF298213) was amplified with an upstream primer containing the XhoI restriction site (underlined) (5′-TACCCTCGAGATGGCCAACCAGGGACAACGTGT-3′) and a downstream primer containing the SalI restriction site (underlined) (5′-GCAGGTCGACTTAGTTCGTTACCTCATCAAT-3′) and cloned into the pCold™ I plasmid. The TGEV N protein was expressed and purified to compare its binding properties to the scFvs with those of the PEDV N protein with an ELISA. The periplasmic expression of the scFvs was detected with western blotting, as described below.
Construction of the pCMV-HA-scFv recombinant plasmid and transient expression of scFvs directed against PEDV N protein
Each complete scFv fragment was amplified with an upstream primer containing the XhoI restriction site and a downstream primer containing the NotI restriction site (Table 2), and then cloned into the pCMV-HA plasmid (Takara Bio). Vero E6 and IPEC-J2 cells were cultured in 6-well plates until 80% confluence. The cells were then transfected with 2.5 μg of pCMV-HA-scFv or pCMV-HA. At 36 h posttransfection (hpt), the cells were harvested and the expression of scFv detected with western blotting, as described below.
Table 2.
Primers for PCR cloning and RT–qPCR
| Primer | Sequence (5′–3′)a |
|---|---|
| cmv-ZW1-16-B | TCGATCTCGAGCTGAGGAGAAGCTGGTGGAGTCCGG |
| cmv-ZW1-16-F | TAACTGCGGCCGCTCAGAGCTCGTCCTTTTTGAGTTCCAGCTTGGTCCC |
| cmv-ZW3-21-B | TCGATCTCGAGCTGAGGTGAAGCTGGTGGAGTCTGG |
| cmv-ZW3-21-F | TAACTGCGGCCGCTCAGAGCTCGTCCTTTTTGATCTCCAGCTTGGTCCC |
| cmv-ZW1-41-B | TCGATCTCGAGCTGAGGAGAAGCTGGTGGAGTCTGG |
| cmv-ZW1-41-F | TAACTGCGGCCGCTCAGAGCTCGTCCTTTTTGAGCTCCAGCTTGGTCCC |
| cmv-ZW4-16-B | TCGATCTCGAGCTGAGGTGAAGCTGGTGGAGTCTGG |
| cmv-ZW4-16-F | TAACTGCGGCCGCTCAGAGCTCGTCCTTGAGGACGGTCAGATGGGTCC |
| PEDV-M-B | GGCGTACAGGTAAGTCAATTGCC |
| PEDV-M-F | AGCTGAGTAGTCGCCGTGTT |
| IFN-α-B | AGTTCTGCACTGGACTGGAT |
| IFN-α-F | CTTCTGCCCTGATGATCTCCCA |
| IFN-β-B | TCCTTGTGGAACTTGATGGGC |
| IFN-β-F | CGGAGGTAATCTGTAAGTCTGTT |
| IFN-λ1-B | ACATCCACGTCGAACTTCAGGCTT |
| IFN-λ1-F | AGTCTCCACTGGTAACACTTCTCA |
| IRF3-B | ACGCTACACCCTCTGGTTCTG |
| IRF3-F | GCAGGTCCACCGTGTTCTCC |
| IRF7-B | CCACACTACACCATCTACCTG |
| IRF7-F | CTCGTCATAGAGGCTGTTGGA |
| β-actin-B | GTCACCCACACGGTGCCCATCTA |
| β-actin-F | TCCTTGATGTCCCGCACGATCT |
aThe restriction sites for XhoI and NotI are underlined
Subcellular localization of scFvs directed against PEDV N protein
An immunofluorescence assay (IFA) was used to evaluate the transfection efficiency and the intracellular distribution of the four scFvs. Vero E6 cells were cultured on coverslips placed in the wells of 12-well plates. The cells were transfected with 1 μg of pCMV-HA-scFv or pCMV-HA. At 24 hpt, the culture supernatants were removed, and the cells were infected with PEDV strain HLJBY (multiplicity of infection [MOI] = 0.01). At 36 h postinfection (hpi), the cells were fixed with 4% paraformaldehyde for 30 min and permeabilized with 0.1% Triton-X for 15 min. BSA (5%) was used to block the cells at 37 °C for 1 h. The cells were then incubated with a mouse monoclonal anti-HA antibody (CMCTAG) at 37 °C for 2 h and then with a rabbit polyclonal anti-PEDV N protein antibody at 37 °C for 2 h. A Cy3-conbjugated Affinipure goat anti-mouse IgG (H + L) secondary antibody and fluorescein isothiocyanate (FITC)-conjugated Affinipure goat anti-rabbit IgG (H + L) secondary antibody were added to stain the cells. After the cell nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI), the cells were analyzed with a confocal microscope (Leica, Wetzlar, Germany).
Viral infection
Vero E6 cells were seeded in 12-well plates. Transfection and infection were performed as described above. At 24 hpi, the supernatants were harvested to analyze viral RNA production with reverse transcription (RT)-quantitative PCR (qPCR), as described below. The treated cells were lysed at 36 hpi and 72hpi to measure intracellular PEDV particle content with western blotting. To assess the viral titers, the supernatants were analyzed with a median tissue culture infective dose (TCID50) assay at 24 hpi, 48 hpi, and 72 hpi.
Effects of scFvs directed against PEDV N protein on expression of several IFNs, interferon regulatory factor 3 (IRF3), and IRF7
IPEC-J2 cells were seeded in 24-well plates. The cells were transfected with 0.5 μg of pCMV-HA-scFv or pCMV-HA. At 24 hpt, the culture supernatants were removed, and the cells were infected with PEDV strain HLJBY (MOI = 0.01). At 12 hpi, the treated cells were collected to detect the expression of several IFNs, IRF3, and IRF7 at the mRNA level with RT–qPCR, as described below. The primers are listed in Table 2.
TCID50 assay
Monolayers of Vero E6 cells in 96-well plates were washed twice with PBS. The tenfold serially diluted virus (100 μL) in DMEM was added into each well (eight wells per dilution). After 1 h, 100 μL of DMEM supplemented with tosyl phenylalanyl chloromethyl ketone (TPCK)-treated trypsin (Worthington, Lakewood, NJ, USA) was added into each well. At 3–5 days postinfection, the cells were observed under a microscope. The TCID50 titers were calculated with the Reed–Muench method (Reed and Muench 1938).
RT-qPCR
The viral RNA was extracted from supernatant, and cDNA was synthesized. Virus production was quantified by measuring the expression of the PEDV M gene. The qPCR was performed with a TB Green® Premix Ex Taq™ II Kit (Tli RNaseH Plus) (Takara Bio) in a 20 μL reaction mixture containing the primers for PEDV M gene (Table 2) (Wongthida et al. 2017). The qPCR was performed with initial denaturation at 95 °C for 30 s, followed by 40 cycles at 95 °C for 5 s, and 58 °C for 34 s. To calculate the viral copy number, a recombinant plasmid pMD19-T-PEDV-M containing a truncated M gene fragment was constructed, and tenfold serially diluted pMD19-T-PEDV-M was used to generate a standard curve.
To analyze the expression of several IFNs, IRF3, and IRF7, TRIzol™ reagent was added to each well to lyse and homogenize the treated cells. The total RNA was extracted, and the cDNA was synthesized. The qPCR was performed with the TB Green® Premix Ex Taq™ II Kit (Tli RNaseH Plus) (Takara Bio) in a 20 μL reaction mixture containing the specific primers (Table 1). The two-step qPCR was performed as described above. The cellular housekeeping gene β-actin (ACTB) was used to normalize the individual samples. The relative quantification of gene expression was evaluated with the 2−ΔΔCt method (Livak and Schmittgen 2001).
Western blotting
The target proteins were separated with SDS-PAGE (12.5%) and then transferred onto UltraCruz® Nitrocellulose Pure Transfer Membranes (Santa Cruz Biotechnology, Santa Cruz, CA, USA). The membranes were blocked for 1 h with 5% skim milk diluted in tris buffered saline with tween-20 (TBST) (Sigma-Aldrich) and incubated with the appropriate primary antibodies overnight at 4 °C. Mouse monoclonal anti-His antibody (CMCTAG), mouse monoclonal anti-Myc antibody (CMCTAG), rabbit monoclonal anti-β-tubulin antibody (CMCTAG), mouse monoclonal anti-HA antibody, and rabbit polyclonal anti-PEDV N protein antibody were used as the primary antibodies. The membranes were then incubated with the appropriate HRP-conjugated secondary antibodies: HRP-conjugated Affinipure goat anti-mouse IgG (H + L) antibody or HRP-conjugated Affinipure goat anti-rabbit IgG (H + L) antibody. Finally, Immobilon Western Chemiluminescent HRP Substrate (Merck) was used to detect the signals.
Statistical analysis
The experimental data were analyzed with Student’s t test in the Origin 8.0 software (OriginLab, Northampton, USA). Values are presented as the means ± standard errors of the means (mean ± SEM) of three independent experiments. P < 0.05 was considered to be statistically significant.
Results
Construction and characterization of scFv library
The DNA sequence (1326 bp) of the coding sequence (CDS) region of the PEDV N gene was amplified, purified, and cloned into the pCold™ I plasmid (Fig. 1a). The PEDV N protein was expressed and purified. A protein of approximately 58 kDa was identified with SDS-PAGE and western blotting (Fig. 1b–d).
Fig. 1.
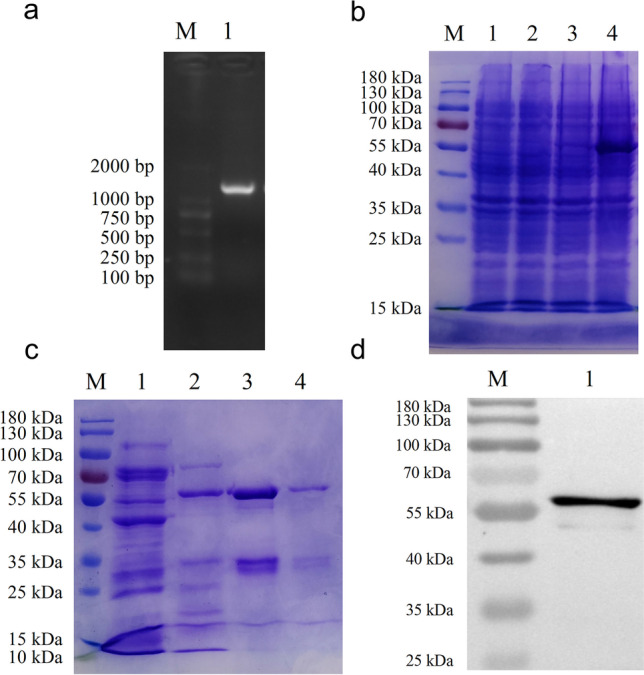
Expression and purification of the recombinant PEDV N protein. a RT-PCR amplification of CDS region of the PEDV N gene. Lane M, 2000-bp DNA ladder; lane 1, PCR product of PEDV N gene. b SDS-PAGE analysis of proteins extracted from bacterial cell lysate. Lane M, protein molecular weight marker; lane 1, total cell lysate of uninduced E. coli containing pCold I; lane 2, total cell lysate of induced E. coli containing pCold I; lane 3, total cell lysate of uninduced E. coli containing pCold I-PEDV-N; lane 4, total cell lysate of induced E. coli containing pCold I-PEDV-N. c SDS-PAGE analysis of the purified protein. Lanes 1–4, purified PEDV N proteins eluted at different imidazole concentration (50 mM, 100 mM, 250 mM, and 500 mM, respectively). d Western blotting analysis of the purified protein. Lane 1, purified PEDV N protein
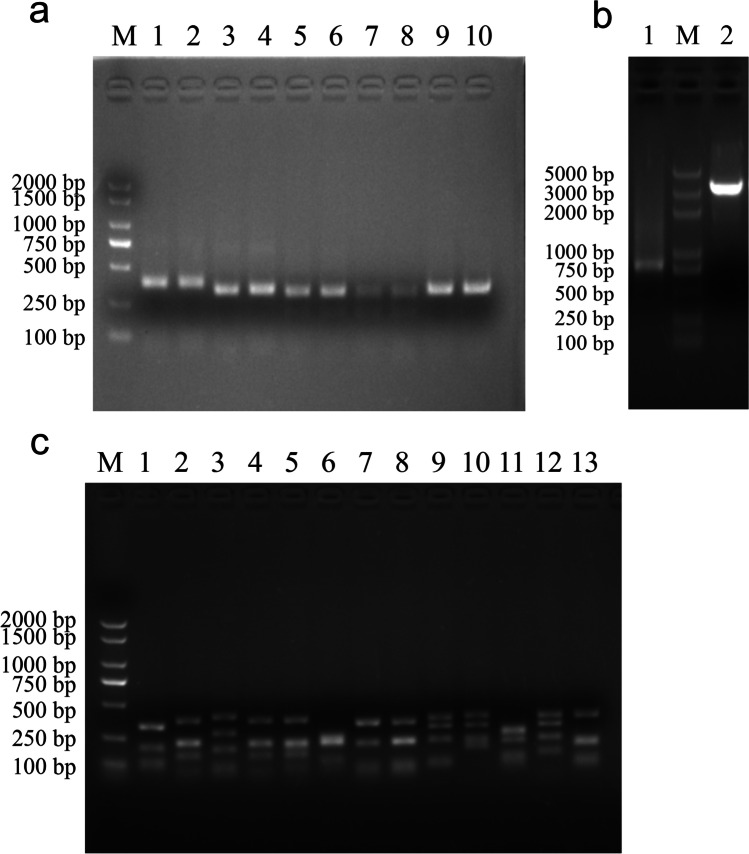
Two pigs were immunized with the recombinant PEDV N protein, and the antibody titers were measured with an ELISA. The total RNA was extracted (Fig. S1) and the cDNA synthesized with reverse transcription. The porcine VH and VL genes (VLk1, VLk2, VLk3, and Vλ) were amplified. Analysis with gel electrophoresis indicated that the fragment lengths were approximately 350 bp (VH) and 330 bp (VL) (Fig. 2a). The scFv genes were cloned into the plasmid pOPE101-XP (Fig. 2b). A porcine scFv library containing 1.63 × 107 clones was constructed. The positivity rate of the library, identified with DNA sequencing, was 85.4%. The diversity of the library was assessed with DNA sequencing and BstNI digestion (Fig. 2c). The digested fragments in the most of the randomly selected scFvs differed from one another.
Fig. 2.
Construction and diversity analysis of porcine scFv library. a PCR amplification of VH and VL genes. Lanes 1 and 2, PCR product of VH gene; lanes 3 and 4, PCR product of VLκ1 gene; lanes 5 and 6, PCR product of VLκ2 gene; lanes 7 and 8, PCR product of VLκ3 gene; lanes 9 and 10, PCR product of VLλ gene. b Construction of recombinant plasmid pOPE101-XP-scFv. Lane M, 5000-bp DNA ladder; lane 1, PCR product of scFv gene; lane 2, linearized plasmid pOPE101-XP digested with NcoI and NotI restriction enzymes. c Diversity analysis of porcine scFv library. Lanes 1–13, DNA fragments of scFvs digested with BstNI restriction enzyme
Screening and specific binding of scFv directed against PEDV N protein
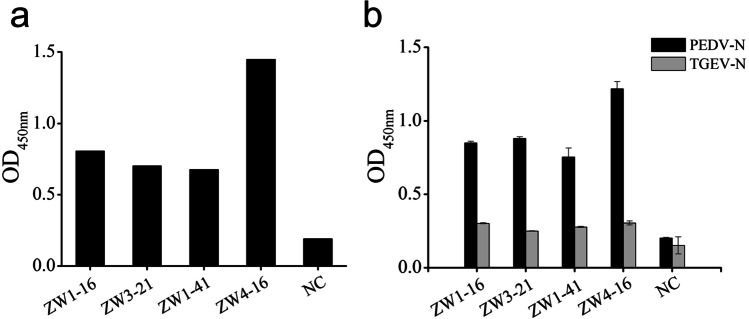
Clones (n = 768) were randomly selected to produce a periplasmic scFv antibody for use in identifying positive clones with an ELISA. When the OD450 values of the clones divided by that of the negative control were > 2.1, they were deemed positive clones. Nine positive clones that showed a binding signal were obtained. Among these, four scFvs showed higher affinity binding to PEDV N protein than the others (Fig. 3a). These four scFvs, designated ZW1-16 (GenBank accession number: MZ424895), ZW3-21 (GenBank accession number: MZ424896), ZW1-41 (GenBank accession number: MZ424897), and ZW4-16 (GenBank accession number: MZ424898), were selected for further study.
Fig. 3.
Binding properties of selected scFvs. a Affinity analysis of selected scFvs for PEDV N protein. b Specific binding analysis of selected scFvs with PEDV N and TGEV N proteins
To evaluate whether the four scFvs cross-reacted with the TGEV N protein, the DNA sequence (1149 bp) encoding the CDS region of the TGEV N gene was cloned into the pCold I plasmid, and the TGEV N protein was expressed and purified (Fig. S2). The results of the ELISA used to examine the binding of the scFvs to the PEDV N and TGEV N proteins are shown in Fig. 3b. All four scFvs reacted strongly with the PEDV N protein, but no reaction with the TGEV N protein was observed. These results indicated that ZW1-16, ZW3-21, ZW1-41, and ZW4-16 reacted specifically with the PEDV N protein.
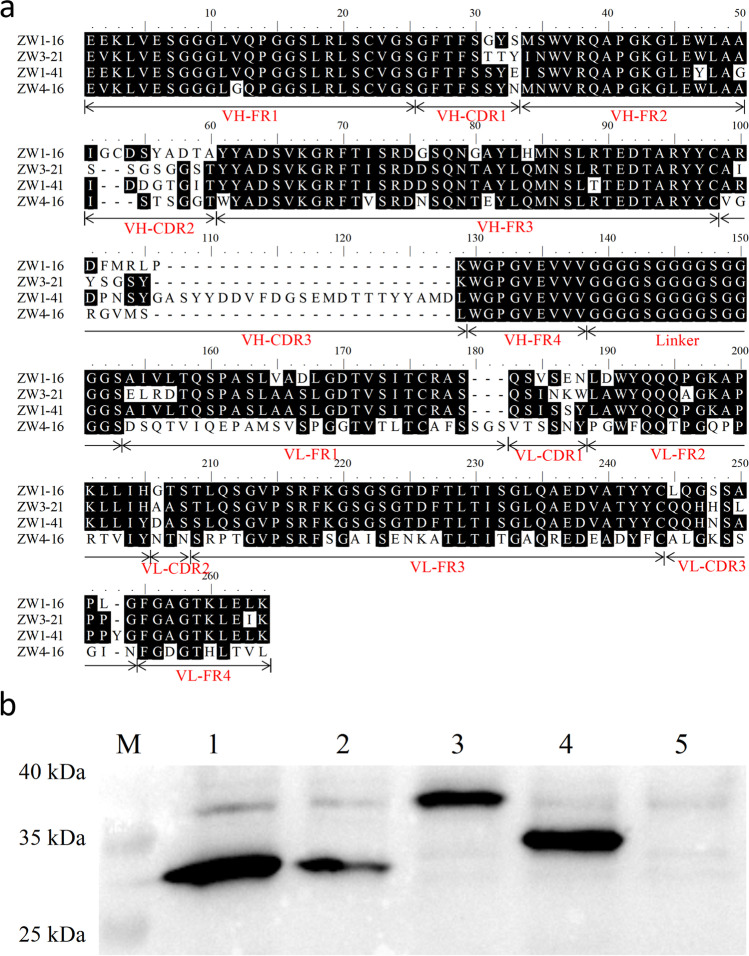
Amino acid sequence analysis and expression of selected scFvs
The DNA sequences of the selected clones containing the scFv fragments were determined. An amino acid sequence analysis of the four scFvs showed that the framework regions (FRs) were highly conserved, whereas the diversity of the complementarity-determining regions (CDRs) was clearly evident, especially CDR3 of VH (Fig. 4a). CDR3 of ZW1-41 was noticeably longer than that of the other three scFvs. The periplasmic proteins extracted from E. coli expressing the Myc-tagged scFvs were subjected to western blotting (Fig. 4b). Prominent bands of approximately 30 kDa were observed, which corresponded to the theoretical molecular weights of the expressed scFvs.
Fig. 4.
Characterization of four selected scFvs. a Amino acid sequences of ZW1-16, ZW3-21, ZW1-41, and ZW4-16. Framework regions (FR) and complementarity-determining regions (CDR) of VH and VL in each scFv and the flexible linker peptide (Linker) are indicated. b Western blotting analysis of the extracted periplasmic proteins. Lanes 1–4, the extracted proteins ZW1-16, ZW3-21, ZW1-41, and ZW4-16, respectively; lane 5, periplasmic protein extracted from E. coli containing pOPE101-XP
ScFvs expressed in Vero E6 cells co-localize with PEDV N protein
To measure the expression of the four scFvs in cells, Vero E6 cells were transfected with pCMV-HA-ZW1-16, pCMV-HA-ZW3-21, pCMV-HA-ZW1-41, pCMV-HA-ZW4-16, or pCMV-HA. At 48 hpt, the treated cells were lysed, and the cell lysates were analyzed with western blotting (Fig. S3). The sizes of the four scFvs expressed in cells were identical when expressed in the periplasm of E. coli.
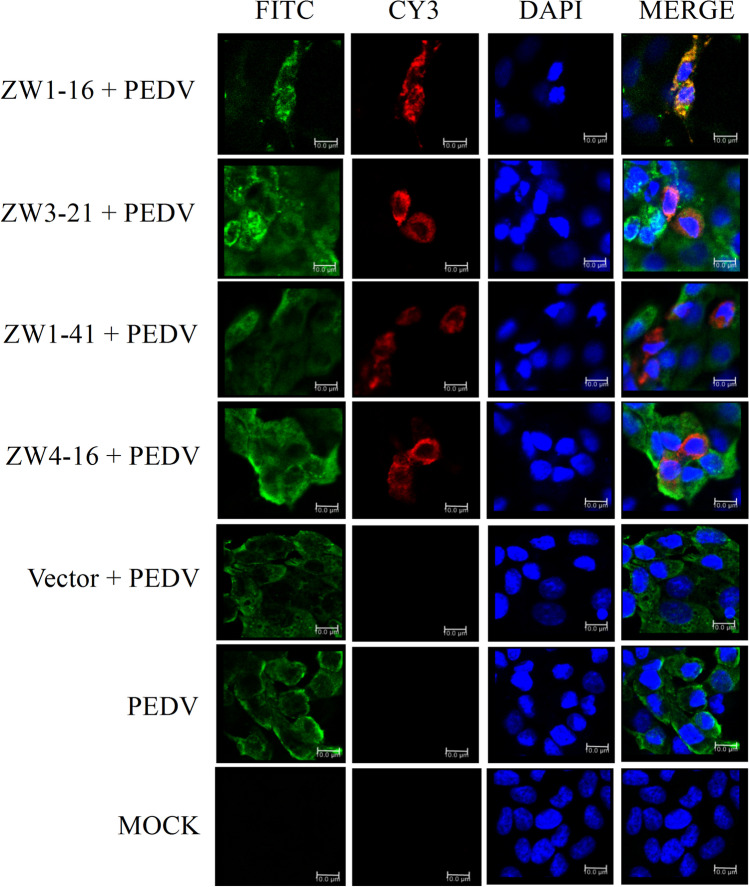
The ability of the four scFvs to co-localize with the intracellular PEDV N protein was analyzed with confocal microscopy. Vero E6 cells were transfected with pCMV-HA-scFv or pCMV-HA before their infection with PEDV. The PEDV N protein was probed with a rabbit anti-PEDV N polyclonal antibody (primary antibody), followed by a FITC-conjugated Affinipure goat anti-rabbit IgG (H + L) secondary antibody. The scFvs were probed with a mouse monoclonal anti-HA antibody and a Cy3-conjugated Affinipure goat anti-mouse IgG (H + L) secondary antibody. The nuclei were stained with DAPI. As shown in Fig. 5, the PEDV N protein and the scFvs were mainly distributed in the cytoplasm, and the four scFvs co-localized with the intracellular PEDV N protein. These results indicated that the four scFvs were successfully expressed in cells and co-localized with the wild-type N protein. These properties of the scFvs suggested that they affected PEDV growth and that they could be used to regulate cellular processes in PEDV-infected cells by blocking the PEDV N protein.
Fig. 5.
Immunofluorescent staining and confocal microscopic analysis of the expression of four scFvs in cells and the scFv binding to intracellular PEDV N protein. Vero E6 cells were transfected with pCMV-HA-scFv or pCMV-HA before PEDV infection. Untransfected infected Vero E6 cells were used as the control. Untransfected uninfected Vero E6 cells were used as the MOCK control. All cells were stained with FITC, CY3, and DAPI. Green color represents the PEDV N protein; red color represents the scFvs; blue color represents nuclei. Merged images show the co-localization of ZW1-16, ZW3-21, ZW1-41, or ZW4-16 with PEDV N protein (orange). Bar = 10 μm for all the figures
ScFvs significantly suppress PEDV replication
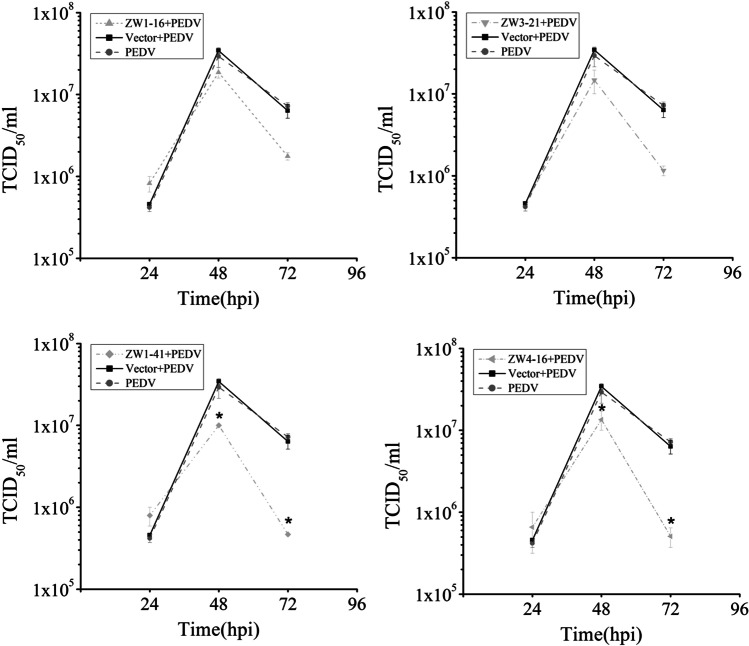
There is evidence that the PEDV N protein contributes to PEDV virion production (Liwnaree et al. 2019). To examine whether the scFvs affected virus production, Vero E6 cells transfected with pCMV-HA-scFv or pCMV-HA were infected with PEDV. At 24 hpi, the cell supernatants were collected to quantify the viral genome with RT-qPCR. The results showed that four scFvs significantly reduced the viral RNA level compared with that in the control group (P < 0.01; Fig. 6a). At 36hpi, the intracellular viral particle content, measured with western blotting, did not differ significantly between the scFv groups and the control group (Fig. 6b). However, based on the results of grayscale analysis, the ratio of the intracellular viral particle content to β-tubulin of the scFv groups decreased about three times compared with that of the control group at 72 hpi (Fig. 6c). The PEDV titers determined with a TCID50 assay at three time points are shown in Fig. 7. At 24 hpi, there was no significant difference in the PEDV titers of the scFv groups and the control group. At 48 hpi, the peak titer in all groups reached or exceeded 107 TCID50/mL. Over time, the viral yields gradually declined. The negative effects of ZW1-41 and ZW4-16 on the viral titer were more marked than those of ZW1-16 and ZW3-21, and they significantly suppressed PEDV production at 48 hpi and 72 hpi (P < 0.05). These results indicated that the scFvs directed against the PEDV N protein significantly suppressed PEDV replication in PEDV-infected cells.
Fig. 6.
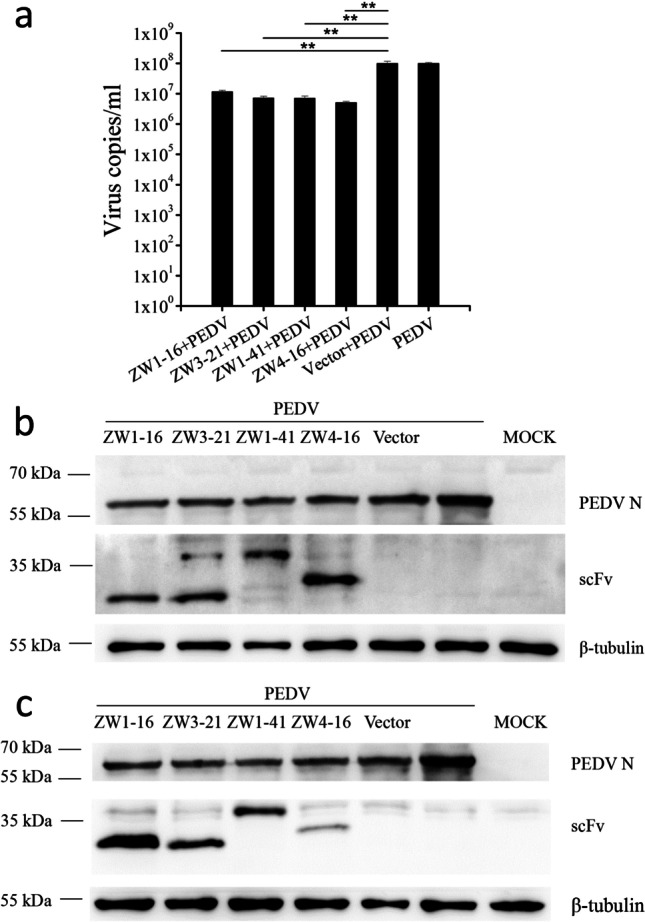
Effects of the four scFvs on PEDV replication in Vero E6 cells. a Quantification of PEDV production with RT–qPCR. Supernatants of treated cells were harvested for quantification of the viral copy number. Values are the means ± standard errors of the means (mean ± SEM) of three independent experiments. b Western blotting analysis of viral production at 36hpi. c Western blotting analysis of viral production at 72hpi. Cell lysates were analyzed with anti-PEDV N antibody, anti-scFv antibody, and anti-β-tubulin antibody. Values are the means ± SEM of three independent experiments. **P < 0.01
Fig. 7.
Assessment of PEDV replication with TCID50 assays at 24, 48, and 72 hpi. Supernatants of treated cells were collected for quantitative analysis of the viral titer. *P < 0.05
Upregulation of IFN-λ1 expression by scFvs
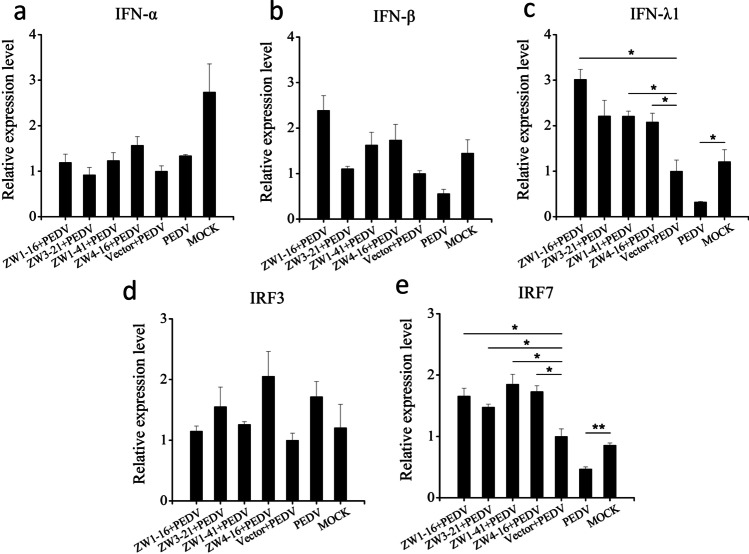
It is well-known that Vero E6 cells are defective in IFN production. Therefore, IPEC-J2 cells were used as the ideal host cells for PEDV infection to examine whether these scFvs affected the expression of IFNs, IRF3, or IRF7 in PEDV-infected cells. The four scFvs were effectively expressed in IPEC-J2 cells, as confirmed with western blotting (Fig. S4). IPEC-J2 cells were transfected with the recombinant plasmid pCMV-HA-ZW1-16, pCMV-HA-ZW3-21, pCMV-HA-ZW1-41, or pCMV-HA-ZW4-16 or the empty vector pCMV-HA. At 24 hpt, the transfected IPEC-J2 cells were infected with PEDV. At 12 hpi, the treated cells were collected to analyze the relative gene expression of IFNs and IFN-responsive genes with RT-qPCR. As shown in Fig. 8, there was no significant change in the expression of IFN-α, IFN-β, or IRF3 after PEDV infection, but the expression of IFN-λ1 and IRF7 was significantly reduced. The scFvs significantly upregulated the expression of IFN-λ1 and IRF7 (P < 0.05), but they had no significant effect on the expression of IFN-α, IFN-β, or IRF3. These results indicated that these scFvs upregulated the expression of IFN-λ1 in PEDV-infected IPEC-J2 cells by blocking PEDV N protein.
Fig. 8.
Analysis of the relative gene expression of IFN-α, IFN-β, IFN-λ1, IRF3, and IRF7 by RT-qPCR. IPEC-J2 cells were transfected with pCMV-HA-scFv or pCMV-HA before PEDV infection. Untransfected infected IPEC-J2 cells were used as the control, and untransfected uninfected IPEC-J2 cells were used as the MOCK control. At 12 hpi, total RNA was isolated from the treated cells, and the transcriptional expression of IFNs, IRF3, and IRF7 was detected with RT–qPCR. The housekeeping gene β-actin was used to normalize the individual samples. Values are the means ± standard errors of the means (mean ± SEM) of three independent experiments. *P < 0.05
Discussion
PEDV infects pigs of all ages, and the mutant strains that emerged in China in 2010–2011 cause up to 100% morbidity (mainly within 7 days of age) and 80–100% mortality in suckling piglets (Sun et al. 2012). At present, vaccines are considered an effective way to prevent and control PEDV. However, commercial inactivated vaccines confer only partial protection against the new mutant strains due to the continuous mutation of the PEDV genome, whereas live attenuated vaccines require a long period of development and are not safe (Li et al. 2020; Sun et al. 2012). Therefore, it is necessary to develop an effective therapy with a short development cycle and excellent safety for the control of PEDV.
Antibodies directed against PEDV N protein have been used for the early detection of PEDV infections because the N protein is highly conserved and induces the production of large numbers of antibodies in the early stage of PEDV infection (Li et al. 2013; Wang et al. 2016a; Yang et al. 2019). The subsequent functions of the N protein during infection make it an ideal target for the prevention and treatment of PEDV infection. The PEDV N protein is one of the essential components of viral particles and promotes viral replication, prolongs the S-phase of the cell cycle, and induces ER stress (Liwnaree et al. 2019; Xu et al. 2013).
The pOPE plasmid derived from the pSEX plasmid was designed for expressing functional antibody fragments in E. coli (Dubel et al. 1992). The pOPE101-XP plasmid, one of the derivatives of the pOPE plasmid, uses the pelB leader sequence to mediate antibody protein secret into the periplasmic space (Dubel et al. 1992; Schmiedl et al. 2000a, 2000b). Schmiedl et al. have used the pOPE101 plasmid to express and purify soluble scFv fragment (Schmiedl et al. 2000b).
In this study, we constructed a recombinant scFv library from pigs immunized with the PEDV N protein, with a capacity of 1.63 × 107. Four porcine scFvs that bound with highest affinity to the PEDV N protein were selected for further study. Previous studies have indicated that the epitopes on the N protein contribute to the cross-reactivity between PEDV and TGEV (Lin et al. 2015; Xie et al. 2019). In this study, the specific binding of the four selected scFvs to PEDV N and TGEV N proteins was analyzed with an ELISA. All the scFvs bound highly specifically to PEDV N protein, but not to TGEV N protein, demonstrating the binding specificity of these scFvs. A multiple sequence alignment of the amino acid sequences of these four scFvs showed that the CDRs of VH and VL displayed clear variations. It should be noted that the diversity in the CDR3 region of VH was significantly higher than that in the other CDRs of VH or VL. The specificity of antigen recognition is predominantly determined by the highly diverse amino acid sequences in the CDR3 region of VH (Xu and Davis 2000). Therefore, these scFvs may target different epitopes on the PEDV N protein, and further studies are required to identify these epitopes.
Previous studies have reported that the PEDV N protein was mainly expressed in the cytoplasm (Shan et al. 2018). In this study, we investigated the distribution of PEDV N protein and the four scFvs with IFA. The results indicated that the four scFvs were uniformly expressed in the cytoplasm and co-localized with the PEDV N protein in PEDV-infected cells. Therefore, these scFvs retained their antigen-binding activity in cells.
Previous studies have shown that the coronavirus N protein, the nucleocapsid protein that binds the gRNA, was crucial for viral RNA synthesis. The interaction between the N protein and nsp3, a component of the replicase-transcriptase complex, plays a key role in the infectivity of the gRNA (Hurst et al. 2013, 2010). The N protein also acts as a bridge involved in the circularization of the coronavirus genome and RNA synthesis (Lo et al. 2019). The PEDV N protein improves the viral titer by promoting the synthesis of viral RNA (Liwnaree et al. 2019). In the present study, the scFvs examined inhibited viral RNA production and reduced the viral titer in Vero E6 cells, when analyzed with RT–qPCR (P < 0.01) and TCID50 assays (P < 0.05), respectively. The production of viral protein, detected with western blotting, was significantly suppressed of the scFv groups compared with that of the control group at 72 hpi. As the Vero E6 cells are defective in IFN production, thus we speculated that these scFvs might reduce the virus titer in Vero E6 cells by impeding the interaction between N protein and nsp3 or blocking the interaction between N protein and the gRNA.
Because PEDV mainly infects porcine small-intestinal epithelial cells and causes severe mucosal atrophy and congestion, the expression of type III IFN, which plays an important role in the mucosal antiviral defenses, should be considered in this context. IFN-λ1 and IFN-λ3 exert more powerful antiviral activity than IFN-α against PEDV in IPEC-J2 cells (Li et al. 2017, 2019; Zhao et al. 2020). To evade the antiviral effects of IFNs, viruses have evolved various sophisticated mechanisms to regulate IFN expression. Previous studies have identified the N proteins of severe acute respiratory syndrome coronavirus (SARS-CoV), severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), Middle East respiratory syndrome coronavirus (MERS-CoV), mouse hepatitis virus (MHV), porcine deltacoronavirus (PDCoV), and PEDV as IFN antagonists (Chang et al. 2020; Chen et al. 2019, 2020; Ding et al. 2014, 2017; Lu et al. 2011). The MERS-CoV N protein suppresses IFN-β and IFN-λ1 production by impeding RIG-I ubiquitination and activation, and the PEDV N protein antagonizes IFN-β and IFN-λ3 production by blocking the interaction between IRF3 and TBK1 and blocking the nuclear translocation of nuclear factor-κB (NF-κB), respectively (Ding et al. 2014; Shan et al. 2018). In the present study, we noted that the expression of IFN-λ1 mRNA was significantly downregulated after PEDV infection (P < 0.05), whereas it was significantly upregulated when cells were transfected with the scFvs before PEDV infection (P < 0.05). There was no significant difference in the expression of IFN-α or IFN-β mRNAs among any of the treatment groups. Therefore, we speculated that the scFvs counteracted the antagonistic effect of the PEDV N protein on IFN-λ1 production in PEDV-infected IPEC-J2 cells by blocking the N protein.
Recent studies have demonstrated that the induction of IFN-λ1 correlated with the activation of IRF3, IRF7, and NF-κB (Cao et al. 2014; Osterlund et al. 2007). The nuclear translocation of IRF3, IRF7, or NF-κB activated the transcription of IFN-λ1. However, the detailed mechanism by which IRF7 regulates IFN-λ1 production in swine remains unclear. In the present study, no significant difference in IRF3 mRNA expression was detected among any of the treatment groups, whereas IRF7 mRNA expression was significantly downregulated after PEDV infection (P < 0.01). When cells were transfected with the scFvs before PEDV infection, the expression of IRF7 was significantly upregulated (P < 0.05). As the expression changes of IFN-λ1 and IRF3 were unexpected, the effects of scFvs alone on the expression of IFN-λ1 and IRF3 in IPEC-J2 cells were evaluated (Fig. S5). As shown in Figure S5, the expression of IFN-λ1 or IRF3 was not observably changed by the scFvs. Since different scFvs can recognize different epitopes, it may be attribute that these scFvs recognized the epitopes of PEDV N protein which are responsible for regulating the expression of IFN-λ1 and IRF7. Furthermore, the scFvs may eliminate the inhibitory effect of PEDV on the transcription of IRF7 by blocking the PEDV N protein, causing increased activation of IRF7 and IFN-λ1 expression in PEDV-infected IPEC-J2 cells.
In conclusion, four scFvs specifically targeting the PEDV N protein with greatest affinity, but not targeting the TGEV N protein, were isolated from the spleens of pigs immunized with a recombinant PEDV N protein. IFA showed that these scFvs were effectively expressed in the cytoplasm and co-localized with the wild-type N protein in PEDV-infected cells. These scFvs significantly suppressed PEDV replication and positively regulated the transcription of the host IFN-λ1 and IRF7 genes compared with that in cells transfected with the empty vector, suggesting that PEDV circumvented the host immune response through other mechanisms. Our findings demonstrated the potential utility of scFvs directed against PEDV N protein as novel therapeutic agents and should facilitate the development of scFv-based therapeutic agents for the prevention and treatment of porcine diarrhea caused by PEDV.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
Grateful thanks for the Analysis and Testing Platform of Shanghai Jiao Tong University, School of Agriculture and Biology for providing laser confocal imaging analysis.
Author contribution
F.W. conceived and designed research. J.Z. applied for funding and supervised research. F.W. conducted experiments. M.W. and L.Z. contributed analytical tools. M.C. and M.L. contributed new reagents. F.W. analyzed data. M.W validated data. F.W. wrote the manuscript. All authors read and approved the manuscript.
Funding
This study was supported by the Key Project of Science and Technology for Agriculture of Shanghai (Hu Nong Ke Gong Zi (2015) No. 1–8) (To Jianguo Zhu).
Data availability
All data and material are presented in the manuscript and its supplementary information file, and the authors promise the availability of supporting data.
Code availability
Not applicable
Declarations
Ethics approval
All animal care and procedures were performed in accordance with the recommendations of the Guidelines for the Use of Laboratory Animals provided by the Science and Technology Commission of Shanghai Municipality (STCSM) (permit number: SYXK-2018–0006). All animal experiments underwent approval by the Animal Ethics Committee of Shanghai Jiao Tong University, School of Agriculture and Biology.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Bird RE, Hardman KD, Jacobson JW, Johnson S, Kaufman BM, Lee SM, Lee T, Pope SH, Riordan GS, Whitlow M. Single-chain antigen-binding proteins. Science. 1988;242(4877):423–426. doi: 10.1126/science.3140379. [DOI] [PubMed] [Google Scholar]
- Cao Y, Zhang R, Zhang W, Zhu C, Yu Y, Song Y, Wang Q, Bai L, Liu Y, Wu K, Wu J. IL-27, a cytokine, and IFN-lambda1, a type III IFN, are coordinated to regulate virus replication through type I IFN. J Immunol. 2014;192(2):691–703. doi: 10.4049/jimmunol.1300252. [DOI] [PubMed] [Google Scholar]
- Chang CY, Liu HM, Chang MF, Chang SC (2020) Middle East respiratory syndrome coronavirus nucleocapsid protein suppresses type I and type III interferon induction by targeting RIG-I signaling. J Virol 94(13):e00099-20. 10.1128/JVI.00099-20 [DOI] [PMC free article] [PubMed]
- Chen J, Fang P, Wang M, Peng Q, Ren J, Wang D, Peng G, Fang L, Xiao S, Ding Z. Porcine deltacoronavirus nucleocapsid protein antagonizes IFN-beta production by impairing dsRNA and PACT binding to RIG-I. Virus Genes. 2019;55(4):520–531. doi: 10.1007/s11262-019-01673-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Xiao F, Hu D, Ge W, Tian M, Wang W, Pan P, Wu K, Wu J (2020) SARS-CoV-2 nucleocapsid protein interacts with RIG-I and represses RIG-mediated IFN-beta production. Viruses 13(1):47. 10.3390/v13010047 [DOI] [PMC free article] [PubMed]
- Clementz MA, Chen Z, Banach BS, Wang Y, Sun L, Ratia K, Baez-Santos YM, Wang J, Takayama J, Ghosh AK, Li K, Mesecar AD, Baker SC. Deubiquitinating and interferon antagonism activities of coronavirus papain-like proteases. J Virol. 2010;84(9):4619–4629. doi: 10.1128/JVI.02406-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haan CA, Rottier PJ. Molecular interactions in the assembly of coronaviruses. Adv Virus Res. 2005;64:165–230. doi: 10.1016/S0065-3527(05)64006-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X, van Geelen A, Buckley AC, O'Brien A, Pillatzki A, Lager KM, Faaberg KS, Baker SC (2019) Coronavirus endoribonuclease activity in porcine epidemic diarrhea virus suppresses type I and type III interferon responses. J Virol 93(8):e02000-18. 10.1128/JVI.02000-18 [DOI] [PMC free article] [PubMed]
- Ding Z, Fang L, Jing H, Zeng S, Wang D, Liu L, Zhang H, Luo R, Chen H, Xiao S. Porcine epidemic diarrhea virus nucleocapsid protein antagonizes beta interferon production by sequestering the interaction between IRF3 and TBK1. J Virol. 2014;88(16):8936–8945. doi: 10.1128/JVI.00700-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Z, Fang L, Yuan S, Zhao L, Wang X, Long S, Wang M, Wang D, Foda MF, Xiao S. The nucleocapsid proteins of mouse hepatitis virus and severe acute respiratory syndrome coronavirus share the same IFN-beta antagonizing mechanism: attenuation of PACT-mediated RIG-I/ MDA5 activation. Oncotarget. 2017;8(30):49655–49670. doi: 10.18632/oncotarget.17912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubel S, Breitling F, Klewinghaus I, Little M. Regulated secretion and purification of recombinant antibodies in E. coli. Cell Biophys. 1992;21(1–3):69–79. doi: 10.1007/BF02789479. [DOI] [PubMed] [Google Scholar]
- Duranti C, Carraresi L, Sette A, Stefanini M, Lottini T, Crescioli S, Crociani O, Iamele L, De Jonge H, Gherardi E, Arcangeli A. Generation and characterization of novel recombinant anti-hERG1 scFv antibodies for cancer molecular imaging. Oncotarget. 2018;9(79):34972–34989. doi: 10.18632/oncotarget.26200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu F, Li L, Shan L, Yang B, Shi H, Zhang J, Wang H, Feng L, Liu P. A spike-specific whole-porcine antibody isolated from a porcine B cell that neutralizes both genogroup 1 and 2 PEDV strains. Vet Microbiol. 2017;205:99–105. doi: 10.1016/j.vetmic.2017.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong L, Lin Y, Qin J, Li Q, Xue C, Cao Y. Neutralizing antibodies against porcine epidemic diarrhea virus block virus attachment and internalization. Virol J. 2018;15(1):133. doi: 10.1186/s12985-018-1042-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holliger P, Hudson PJ. Engineered antibody fragments and the rise of single domains. Nat Biotechnol. 2005;23(9):1126–1136. doi: 10.1038/nbt1142. [DOI] [PubMed] [Google Scholar]
- Hurst KR, Ye R, Goebel SJ, Jayaraman P, Masters PS. An interaction between the nucleocapsid protein and a component of the replicase-transcriptase complex is crucial for the infectivity of coronavirus genomic RNA. J Virol. 2010;84(19):10276–10288. doi: 10.1128/JVI.01287-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst KR, Koetzner CA, Masters PS. Characterization of a critical interaction between the coronavirus nucleocapsid protein and nonstructural protein 3 of the viral replicase-transcriptase complex. J Virol. 2013;87(16):9159–9172. doi: 10.1128/JVI.01275-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo L, Hurst-Hess KR, Koetzner CA, Masters PS. Analyses of coronavirus assembly interactions with interspecies membrane and nucleocapsid protein chimeras. J Virol. 2016;90(9):4357–4368. doi: 10.1128/JVI.03212-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laude H, Masters PS. The coronavirus nucleocapsid protein. In: Siddell SG, editor. The Coronaviridae. Boston; 1995. pp. 141–163. [Google Scholar]
- Li F, Aitken R. Cloning of porcine scFv antibodies by phage display and expression in Escherichia coli. Vet Immunol Immunopathol. 2004;97(1–2):39–51. doi: 10.1016/j.vetimm.2003.08.011. [DOI] [PubMed] [Google Scholar]
- Li W, Li H, Liu Y, Pan Y, Deng F, Song Y, Tang X. He Q (2012) New variants of porcine epidemic diarrhea virus, China. Emerg Infect Dis. 2011;18(8):1350–1353. doi: 10.3201/eid1808.120002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Chen F, Yuan Y, Zeng X, Wei Z, Zhu L, Sun B, Xie Q, Cao Y, Xue C, Ma J, Bee Y. Sequence and phylogenetic analysis of nucleocapsid genes of porcine epidemic diarrhea virus (PEDV) strains in China. Arch Virol. 2013;158(6):1267–1273. doi: 10.1007/s00705-012-1592-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Fu F, Xue M, Chen W, Liu J, Shi H, Chen J, Bu Z, Feng L, Liu P. IFN-lambda preferably inhibits PEDV infection of porcine intestinal epithelial cells compared with IFN-alpha. Antiviral Res. 2017;140:76–82. doi: 10.1016/j.antiviral.2017.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Xue M, Fu F, Yin L, Feng L, Liu P. IFN-lambda 3 mediates antiviral protection against porcine epidemic diarrhea virus by inducing a distinct antiviral transcript profile in porcine intestinal epithelia. Front Immunol. 2019;10:2394. doi: 10.3389/fimmu.2019.02394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Ma Z, Li Y, Gao S, Xiao S. Porcine epidemic diarrhea virus: molecular mechanisms of attenuation and vaccines. Microb Pathog. 2020;149:104553. doi: 10.1016/j.micpath.2020.104553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CM, Gao X, Oka T, Vlasova AN, Esseili MA, Wang Q, Saif LJ. Antigenic relationships among porcine epidemic diarrhea virus and transmissible gastroenteritis virus strains. J Virol. 2015;89(6):3332–3342. doi: 10.1128/JVI.03196-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Fang P, Ke W, Wang J, Wang X, Xiao S, Fang L. Porcine deltacoronavirus (PDCoV) infection antagonizes interferon-lambda1 production. Vet Microbiol. 2020;247:108785. doi: 10.1016/j.vetmic.2020.108785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Liwnaree B, Narkpuk J, Sungsuwan S, Jongkaewwattana A, Jaru-Ampornpan P. Growth enhancement of porcine epidemic diarrhea virus (PEDV) in Vero E6 cells expressing PEDV nucleocapsid protein. PLoS ONE. 2019;14(3):e0212632. doi: 10.1371/journal.pone.0212632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo CY, Tsai TL, Lin CN, Lin CH, Wu HY. Interaction of coronavirus nucleocapsid protein with the 5′- and 3′-ends of the coronavirus genome is involved in genome circularization and negative-strand RNA synthesis. FEBS J. 2019;286(16):3222–3239. doi: 10.1111/febs.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Pan J, Tao J, Guo D. SARS-CoV nucleocapsid protein antagonizes IFN-beta response by targeting initial step of IFN-beta induction pathway, and its C-terminal region is critical for the antagonism. Virus Genes. 2011;42(1):37–45. doi: 10.1007/s11262-010-0544-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesev EV, LeDesma RA, Ploss A. Decoding type I and III interferon signalling during viral infection. Nat Microbiol. 2019;4(6):914–924. doi: 10.1038/s41564-019-0421-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra B, Wang J, Kim ES, Mao R, Dong M, Liu Y, Zhang J, Guo H (2019) Hepatitis B virus precore protein p22 inhibits alpha interferon signaling by blocking STAT nuclear translocation. J Virol 93(13):e00196-19. 10.1128/JVI.00196-19 [DOI] [PMC free article] [PubMed]
- Osterlund PI, Pietila TE, Veckman V, Kotenko SV, Julkunen I. IFN regulatory factor family members differentially regulate the expression of type III IFN (IFN-lambda) genes. J Immunol. 2007;179(6):3434–3442. doi: 10.4049/jimmunol.179.6.3434. [DOI] [PubMed] [Google Scholar]
- Pensaert MB, de Bouck P. A new coronavirus-like particle associated with diarrhea in swine. Arch Virol. 1978;58(3):243–247. doi: 10.1007/BF01317606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathinam VA, Fitzgerald KA. Cytosolic surveillance and antiviral immunity. Curr Opin Virol. 2011;1(6):455–462. doi: 10.1016/j.coviro.2011.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed LJ, Muench H. A simple method of estimating fifty percent endpoints. Am J Epidemiol. 1938;3:493–497. doi: 10.1093/oxfordjournals.aje.a118408. [DOI] [Google Scholar]
- Schmiedl A, Breitling F, Dubel S. Expression of a bispecific dsFv-dsFv' antibody fragment in Escherichia coli. Protein Eng. 2000;13(10):725–734. doi: 10.1093/protein/13.10.725. [DOI] [PubMed] [Google Scholar]
- Schmiedl A, Breitling F, Winter CH, Queitsch I, Dubel S. Effects of unpaired cysteines on yield, solubility and activity of different recombinant antibody constructs expressed in E. coli. J Immunol Methods. 2000;242(1–2):101–14. doi: 10.1016/s0022-1759(00)00243-x. [DOI] [PubMed] [Google Scholar]
- Shan Y, Liu ZQ, Li GW, Chen C, Luo H, Liu YJ, Zhuo XH, Shi XF, Fang WH, Li XL. Nucleocapsid protein from porcine epidemic diarrhea virus isolates can antagonize interferon-lambda production by blocking the nuclear factor-kappaB nuclear translocation. J Zhejiang Univ Sci B. 2018;19(7):570–580. doi: 10.1631/jzus.B1700283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi P, Su Y, Li R, Liang Z, Dong S, Huang J. PEDV nsp16 negatively regulates innate immunity to promote viral proliferation. Virus Res. 2019;265:57–66. doi: 10.1016/j.virusres.2019.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson GW, Hoang H, Schwartz KJ, Burrough ER, Sun D, Madson D, Cooper VL, Pillatzki A, Gauger P, Schmitt BJ, Koster LG, Killian ML, Yoon KJ. Emergence of porcine epidemic diarrhea virus in the United States: clinical signs, lesions, and viral genomic sequences. J Vet Diagn Invest. 2013;25(5):649–654. doi: 10.1177/1040638713501675. [DOI] [PubMed] [Google Scholar]
- Sun RQ, Cai RJ, Chen YQ, Liang PS, Chen DK, Song CX. Outbreak of porcine epidemic diarrhea in suckling piglets, China. Emerg Infect Dis. 2012;18(1):161–163. doi: 10.3201/eid1801.111259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sungsuwan S, Jongkaewwattana A, Jaru-Ampornpan P. Nucleocapsid proteins from other swine enteric coronaviruses differentially modulate PEDV replication. Virology. 2020;540:45–56. doi: 10.1016/j.virol.2019.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Totura AL, Baric RS. SARS coronavirus pathogenesis: host innate immune responses and viral antagonism of interferon. Curr Opin Virol. 2012;2(3):264–275. doi: 10.1016/j.coviro.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Xie C, Zhang J, Zhang W, Yang D, Yu L, Jiang Y, Yang S, Gao F, Yang Z, Zhou Y, Tong G. The identification and characterization of two novel epitopes on the nucleocapsid protein of the porcine epidemic diarrhea virus. Sci Rep. 2016;6:39010. doi: 10.1038/srep39010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Zhang Y, Zhu J. Anti-Staphylococcus aureus single-chain variable region fragments provide protection against mastitis in mice. Appl Microbiol Biotechnol. 2016;100(5):2153–2162. doi: 10.1007/s00253-015-7045-8. [DOI] [PubMed] [Google Scholar]
- Wongthida P, Liwnaree B, Wanasen N, Narkpuk J, Jongkaewwattana A. The role of ORF3 accessory protein in replication of cell-adapted porcine epidemic diarrhea virus (PEDV) Arch Virol. 2017;162(9):2553–2563. doi: 10.1007/s00705-017-3390-5. [DOI] [PubMed] [Google Scholar]
- Xie W, Ao C, Yang Y, Liu Y, Liang R, Zeng Z, Ye G, Xiao S, Fu ZF, Dong W, Peng G. Two critical N-terminal epitopes of the nucleocapsid protein contribute to the cross-reactivity between porcine epidemic diarrhea virus and porcine transmissible gastroenteritis virus. J Gen Virol. 2019;100(2):206–216. doi: 10.1099/jgv.0.001216. [DOI] [PubMed] [Google Scholar]
- Xing Y, Chen J, Tu J, Zhang B, Chen X, Shi H, Baker SC, Feng L, Chen Z. The papain-like protease of porcine epidemic diarrhea virus negatively regulates type I interferon pathway by acting as a viral deubiquitinase. J Gen Virol. 2013;94(Pt 7):1554–1567. doi: 10.1099/vir.0.051169-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu JL, Davis MM. Diversity in the CDR3 region of V(H) is sufficient for most antibody specificities. Immunity. 2000;13(1):37–45. doi: 10.1016/s1074-7613(00)00006-6. [DOI] [PubMed] [Google Scholar]
- Xu X, Zhang H, Zhang Q, Huang Y, Dong J, Liang Y, Liu HJ, Tong D. Porcine epidemic diarrhea virus N protein prolongs S-phase cell cycle, induces endoplasmic reticulum stress, and up-regulates interleukin-8 expression. Vet Microbiol. 2013;164(3–4):212–221. doi: 10.1016/j.vetmic.2013.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JL, Pan XY, Zhao WX, Hu QC, Ding F, Feng Q, Li GY, Luo Y. The antitumor efficacy of a novel adenovirus-mediated anti-p21Ras single chain fragment variable antibody on human cancers in vitro and in vivo. Int J Oncol. 2016;48(3):1218–1228. doi: 10.3892/ijo.2016.3334. [DOI] [PubMed] [Google Scholar]
- Yang W, Chen W, Huang J, Jin L, Zhou Y, Chen J, Zhang N, Wu D, Sun E, Liu G. Generation, identification, and functional analysis of monoclonal antibodies against porcine epidemic diarrhea virus nucleocapsid. Appl Microbiol Biotechnol. 2019;103(9):3705–3714. doi: 10.1007/s00253-019-09702-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Shi K, Yoo D. Suppression of type I interferon production by porcine epidemic diarrhea virus and degradation of CREB-binding protein by nsp1. Virology. 2016;489:252–268. doi: 10.1016/j.virol.2015.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M, Li L, Zhai L, Yue Q, Liu H, Ren S, Jiang X, Gao F, Bai S, Li H, Zhang Y, Xu H, Zhang L, Liu P, Tan M, Yu Q. Comparative transcriptomic and proteomic analyses prove that IFN-lambda1 is a more potent inducer of ISGs than IFN-alpha against porcine epidemic diarrhea virus in porcine intestinal epithelial cells. J Proteome Res. 2020;19(9):3697–3707. doi: 10.1021/acs.jproteome.0c00164. [DOI] [PubMed] [Google Scholar]
- Zheng L, Wang X, Guo D, Cao J, Cheng L, Li X, Zou D, Zhang Y, Xu J, Wu X, Shen Y, Wang H, Yu W, Li L, Xiao L, Song B, Ma J, Liu X, Li P, Xu S, Xu X, Zhang H, Wu Z, Cao H. Porcine epidemic diarrhea virus E protein suppresses RIG-I signaling-mediated interferon-beta production. Vet Microbiol. 2021;254:108994. doi: 10.1016/j.vetmic.2021.108994. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data and material are presented in the manuscript and its supplementary information file, and the authors promise the availability of supporting data.
Not applicable