Abstract
Bartonella vinsonii subsp. berkhoffii was originally isolated from a dog suffering infectious endocarditis and was recently identified as a zoonotic agent causing human endocarditis. Following the coyote bite of a child who developed clinical signs compatible with Bartonella infection in Santa Clara County, Calif., this epidemiological study was conducted. Among 109 coyotes (Canis latrans) from central coastal California, 31 animals (28%) were found to be bacteremic with B. vinsonii subsp. berkhoffii and 83 animals (76%) had B. vinsonii subsp. berkhoffii antibodies. These findings suggest these animals could be the wildlife reservoir of B. vinsonii subsp. berkhoffii. PCR-restriction fragment length polymorphism (PCR-RFLP) analysis of the gltA and 16S rRNA genes for these 31 isolates yielded similar profiles that were identical to those of B. vinsonii subsp. berkhoffii. Partial sequencing of the gltA and 16S rRNA genes, respectively, indicated 99.5 and 100% homology between the coyote isolate and B. vinsonii subsp. berkhoffii (ATCC 51672). PCR-RFLP analysis of the 16S-23S intergenic spacer region showed the existence of two different strain profiles, as has been reported in dogs. Six (19%) of 31 Bartonella bacteremic coyotes exhibited the strain profile that was identified in the type strain of a canine endocarditis case (B. vinsonii subsp. berkhoffii ATCC 51672). The other 25 bacteremic coyotes were infected with a strain that was similar to the strains isolated from healthy dogs. Based on whole bacterial genome analysis by pulsed-field gel electrophoresis (PFGE) with SmaI restriction endonuclease, there was more diversity in fingerprints for the coyote isolates, which had at least 10 major variants compared to the two variants described for domestic dog isolates from the eastern United States. By PFGE analysis, three Bartonella bacteremic coyotes were infected by a strain identical to the one isolated from three healthy dog carriers. Further studies are necessary to elucidate the mode of transmission of B. vinsonii subsp. berkhoffii, especially to identify potential vectors, and to determine how humans become infected.
Bartonella species are emerging pathogens in human beings and cause severe diseases in immunocompromised patients. At least six Bartonella species are known to be pathogenic for humans: B. bacilliformis, B. quintana, B. henselae, B. elizabethae, B. grahamii, and B. vinsonii subsp. arupensis (2, 27, 50). Among these six species, B. quintana, B. henselae, and B. elizabethae have been identified as causative agents of human endocarditis (2, 18, 19, 48).
Recently, several new Bartonella species have been isolated from rodents (20, 34; R. J. Birtles, E. Fichet-Calvet, D. Raoult, and R. W. Ashford, 13th Sesqui-Annu. Meet. Am. Soc. Rickettsiol. abstr. 34, 1997; R. Heller, M. Kubina, G. Delacour, I. Mahoudeau, F. Lamarque, M. Artois, H. Monteil, B. Jaulhac, and Y. Piemont, Abstr. 97th Gen. Meet. Am. Soc. Microbiol. abstr. B-505, p. 115, 1997), carnivores (32; B. B. Chomel, R. W. Kasten, C. C. Chang, K. Yamamoto, R. Heller, S. Maruyama, H. Ueno, D. Simpson, S. S. Swift, Y. Piemont, and N. C. Pedersen, Abstr. Int. Conf. Emerg. Infect. Dis. vol. 1, p. 21.10, 1998), and wild cervids (12; Chomel et al., Abstr. Int. Conf. Emerg. Infect. Dis., 1998; R. Heller, M. Kubina, G. Delacour, F. Lamarque, G. Van Laere, R. Kasten, B. Chomel, and Y. Piemont, Abstr. Int. Conf. Emerg. Infect. Dis. p. 21.18, 1998). Furthermore, it is likely that other mammals may also serve as reservoirs for zoonotic Bartonella spp., involving various vectors for transmission. B. vinsonii subsp. vinsonii, isolated from a Canadian vole (3), has not yet been identified as a pathogen in either humans or animals. However, B. vinsonii subsp. arupensis was isolated from a cattle rancher with high fever and neurological symptoms (50). Recently, B. vinsonii subsp. berkhoffii was added to the increasing list of zoonotic Bartonella, as a human case of endocarditis was associated with this infectious agent, based on sequencing of the gltA and 16S rRNA genes (43). This agent had been shown to cause endocarditis, arrhythmia, and myocarditis in dogs (9, 10, 32). Kordick and Breitschwerdt (31) further identified two different digestion profiles of B. vinsonii subsp. berkhoffii in dogs, based on the PCR-restriction fragment length polymorphism (PCR-RFLP) analysis of the 16S-23S intergenic spacer (ITS) region using HaeIII restriction endonuclease.
Limited studies have been performed on the epidemiology of this infection in dogs. According to a serosurvey by Pappalardo et al. (38) involving approximately 2,000 sick dogs from North Carolina and Virginia, 3.6% of the dogs tested were seropositive for B. vinsonii subsp. berkhoffii. Because of the low antibody prevalence in domestic dogs, a very small proportion of dogs should be Bartonella bacteremic. Therefore, dogs are not likely to be the main reservoir for B. vinsonii subsp. berkhoffii. The fact that Bartonella organisms are very fastidious bacteria and sometimes are difficult to be isolated by routine laboratory methods (8) may also explain the very small number of B. vinsonii subsp. berkhoffii isolates from dogs. Furthermore, a certain proportion of Bartonella-infected dogs may have been misdiagnosed by blood culture because of sensitive culture techniques or concurrent antibiotic treatment.
It is still unknown how this infectious agent is transmitted. It has been suggested that ticks could be potential vectors for B. vinsonii subsp. berkhoffii transmission in dogs (38). However, which vectors or reservoirs are involved in B. vinsonii subsp. berkhoffii transmission are still unknown and need to be explored.
Because coyotes (Canis latrans) are genetically close to domestic dogs, these wild canids have been shown to be susceptible to or used as sentinels for several viruses (16, 17, 22), bacteria (11, 51), and parasites (39) that infect domestic dogs. Following the coyote bite of a child who developed clinical signs compatible with Bartonella infection in Santa Clara County, Calif., the child and trapped coyotes were serologically tested for possible Bartonella infection (13). The identification of Bartonella-seropositive coyotes prompted us to investigate if coyotes from central coastal California could serve as a potential reservoir of B. vinsonii subsp. berkhoffii. Molecular approaches were applied to determine the characteristics of Bartonella isolates from these animals.
MATERIALS AND METHODS
Sample collection and isolation and identification of B. vinsonii subsp. berkhoffii.
A sample size of 100 coyotes was determined as necessary for a 95% confidence interval (CI) with a 10% error for a 50% prevalence estimate. Between June 1997 and October 1998, a total of 109 coyotes were trapped and euthanized from nine different sites in central coastal California with the help of the Santa Clara County Department of Health Services, Wildlife Unit, Vector Control Section (54 coyotes in 1997 and 55 coyotes in 1998). Blood samples were collected intracardially in plastic 2-ml EDTA tubes (Becton Dickinson, Franklin Lakes, N.J.) and frozen at −70°C until plated. The blood samples were cultured on heart infusion agar containing 5% rabbit blood and incubated in 5% CO2 at 35°C for up to 4 weeks. Identification of the isolates was based on morphological characteristics and growth time on the blood agar plates and then determined by PCR-RFLP analysis of the citrate synthase (gltA), 16S rRNA, and 16S-23S ITS genes. The former two genes were used for comparison because they evolved slowly enough to allow the use of primers to amplify conserved sequences in different strains of Bartonella organisms, yet these genes have regions of diversity that allow for Bartonella species comparison (6, 7, 42). The 16S-23S ITS gene was used for subtyping of B. vinsonii subsp. berkhoffii as previously described (31).
(i) PCR-RFLP procedures.
Isolates were analyzed using PCR-RFLP analysis of the gltA gene (37, 40), the 16S rRNA gene (24), and 16S-23S ITS gene (45), as previously described. After approximately 2 cm2 of confluent growth was scraped off and suspended in 100 μl of sterile water, the bacterial suspension was heated at 100°C for 15 min and then centrifuged at 15,000 × g for 10 min at 4°C. Finally, the supernatant diluted 1:10 was used as the DNA template. An approximately 400-bp fragment of the gltA gene, 1,500-bp fragment of the 16S rRNA gene, and 2,900-bp fragment of the 16S-23S ITS gene were amplified and then verified by gel electrophoresis. The amplified product of the gltA gene obtained with the set of primers suggested by Regnery et al. (40) was digested with TaqI (Promega, Madison, Wis.) and HhaI (new England BioLabs, Beverly, Mass.) restriction endonucleases. TaqI and MseI (New England BioLabs) restriction endonucleases were utilized when using the set of primers suggested by Norman et al. (37). The amplified product of the 16S rRNA gene was digested with DdeI (Boehringer GmbH, Mannheim, Germany) and MnlI (New England BioLabs) restriction endonucleases. The digestion conditions used were the ones recommended by the enzymes' manufacturer. Banding patterns were compared with those of a domestic dog isolate (American Type Culture Collection [ATCC] 51672) of B. vinsonii subsp. berkhoffii), B. vinsonii subsp. vinsonii (ATCC VR152), B. henselae (strain U-4; University of California, Davis) and B. clarridgeiae (ATCC 51734); the last two Bartonella species are usually isolated from domestic cats. Finally, the amplified product of the 16S-23S ITS gene was digested with HaeIII restriction endonuclease (Boehringer GmbH) (31).
(ii) DNA sequencing.
The PCR products used for DNA sequencing were purified with Microcon centrifugal filter devices (Millipore Corp., Bedford, Mass.) and sequenced with a fluorescence-based automated sequencing system (Davis Sequencing, Davis, Calif.). Primers BhCS.1137n (5′-AATGCAAAAAGAACAGTAAACA-3′) (37) and Pc1544 (5′-AAGGAGGTGATCCAGCCGCA-3′) (25) were used for partial sequencing of the gltA and 16S rRNA genes, respectively.
IFA test.
B. henselae indirect immunofluorescent-antibody (IFA) test was performed as previously described (14). For IFA slides using B. vinsonii subsp. berkhoffii as the antigen, the reference strain (ATCC 51672) was cultured for 4 days on heart infusion agar containing 5% rabbit blood. The bacteria were harvested into 0.5 ml of sterile saline, washed twice in 0.5 ml of sterile saline, and finally resuspended in 0.5 ml of sterile saline. The bacteria were heat inactivated at 55°C for 30 min and then rewashed twice in 0.5 ml of sterile saline. The final pellet was resuspended in 0.5 ml of sterile saline.
A 90% confluent tissue culture flask (Felis catus whole fetus) was inoculated with the resuspended B. vinsonii subsp. berkhoffii, and the flask was incubated for 3 days at 37°C with 5% CO2. After incubation, the tissue cultures were washed two times with calcium- and magnesium-free phosphate-buffered saline (PBS) (Gibco-BRL, Gaithersburg, Md.) and trypsinized (Gibco-BRL) for 10 min at room temperature. The suspended tissue cultures were combined into one tube and centrifuged at 200 × g for 10 min. The supernatant was discarded, and the cells were resuspended in 30 ml of tissue culture growth medium. Forty microliters of the cell culture were spotted onto HTC supercured glass slides (12-well slides; Cell-Line/Erie Scientific, Co., Newfield, N.J.) and incubated overnight at 37°C with 5% CO2. The slides were then washed twice in PBS (pH 7.4) (Sigma Chemical, St. Louis, Mo.), set for 20 min in acetone at room temperature, air dried, and then stored at −20°C until they were used. Supernatant of the whole blood collected in the EDTA tubes after centrifugation was used for serological testing. Samples added to the test wells were initially screened at 1:32 and 1:64 dilutions in PBS with 5% milk. The slides were then incubated for 35 min at 37°C and were washed for 5 min in PBS twice. Fluorescein-conjugated goat anti-dog immunoglobulin (whole-molecule immunoglobulin G; Organon Teknika Corp., Durham, N.C.) was diluted at 1:1,400 in PBS with 5% milk containing 0.001% Evan's blue, and the mixture was applied to each well. The slides were incubated for 20 min at 37°C and washed again in PBS for 5 min twice prior to being read with a fluorescence microscope (magnification, ×400). The intensity of bacillus-specific fluorescence was scored subjectively from 1 to 4, and a fluorescence score of ≥2 at a dilution of 1:64 was reported as a positive result, as previously described (38). Any sample positive at 1:64 was titrated in serial twofold dilutions to the end point. A double-blind reading of each slide was performed by the same two readers. Negative and positive serum control samples were obtained from two laboratory dogs before and after they were infected with B. vinsonii subsp. berkhoffii.
PFGE.
Four canine B. vinsonii subsp. berkhoffii strains isolated at North Carolina State University (including the reference strain, ATCC 51672) were included for comparison with the coyote isolates in the present study. For pulsed-field gel electrophoresis (PFGE), a single colony pick of each Bartonella isolate was subcultured confluently on 5% rabbit agar plate at 35°C for 5 to 7 days in a 5% CO2 incubator. The bacteria grown on the agar plates were scraped off, suspended in sterile saline, and washed twice by centrifugation at 15,000 × g for 5 min at 4°C. The turbidity of the suspension was adjusted to McFarland standard 6. Then, 0.5 ml of the adjusted suspension was mixed gently but thoroughly with the same amount of 2% ultrapure low-melting-point agarose (Gibco-BRL, Life Technologies, Inc., Gaithersburg, Md.), and the mixture was solidified in plug molds at 4°C. The agarose plugs were then transferred into lysozyme solution (10 mM Tris [pH 7.2], 50 mM NaCl, 0.2% sodium deoxycholate, 0.5% sodium lauryl sarcosine, 1 mg of lysozyme per ml) and incubated at 37°C overnight. The plugs were rinsed with sterile water and incubated in proteinase K solution (100 mM EDTA [pH 8.0], 0.2% sodium deoxycholate, 1% sodium lauryl sarcosine, 1 mg of proteinase K per ml) at 50°C overnight. This procedure was repeated a second time. Then, the plugs were washed four times in 10 ml of washing buffer (50 mM EDTA, 20 mM Tris [pH 8.0]) for 1 h at room temperature with gentle agitation. Proteinase K was inactivated by the addition of 1 mM phenylmethylsulfonyl fluoride solution during the second wash. The plugs were stored in wash buffer at 4°C before endonuclease digestion. Before digestion, plugs were transferred to 1.5-ml sterile microcentrifuge tubes with 0.1× washing buffer at 4°C overnight and they were equilibrated in 1× endonuclease-specific reaction buffer for 1 h. SmaI and NotI restriction endonucleases (45) (New England BioLabs) were used for the analysis of the whole Bartonella genome. Bacterial DNA was digested in reaction buffer with SmaI endonuclease at 28°C and with NotI endonuclease at 37°C overnight. After digestion, plugs were equilibrated in 0.5× TBE (45 mM Tris-borate, 1 mM EDTA [pH 8.0]) buffer for 30 min. The chromosomal restriction fragments were separated by PFGE in a CHEF-DRIII system (Bio-Rad, Hercules, Calif.) by a 1.5% (for SmaI digestion) or 1.0% (for NotI digestion) pulsed-field certified agarose (Bio-Rad) gel in 0.5× TBE buffer. The electrophoresis was equilibrated at 14°C for 26 h at a constant voltage of 5.7 V/cm for the SmaI-digested plugs and for 33 h at a constant voltage of 4.5 V/cm for the NotI-digested plugs. Separation of the digested genomic DNA was achieved with pulse times from 3 to 10 s for SmaI-digested plugs and from 5 to 120 s for NotI-digested plugs, respectively. After electrophoresis, the gel was stained with 0.5 μg of ethidium bromide per ml for 30 min, destained with distilled water twice for 15 min each time, and photographed. Lambda ladder pulsed-field gel markers (48.5 to 970 kbp) (Bio-Rad) were used as molecular size standards. Three B. vinsonii subsp. berkhoffii strains isolated from healthy dogs (NC95-C02, NC95-C03, and NC95-C04) and ATCC strain 51672 isolated from a dog with endocarditis were included in PFGE analysis.
(i) Analysis of PFGE profiles.
The PFGE profiles were analyzed by the direct grouping method suggested by Tenover et al. (49). In addition, cluster analysis with Molecular Analyst Software (Fingerprinting version 1.12; Bio-Rad) was performed. The images were processed, and then Jaccard coefficients (SJ) of band-based similarity were calculated as SJ = NAB/(NA + NB − NAB), where NAB is the number of bands common to A and B, NA is the total number of bands in A, and NB is the total number of bands in B. Dendrograms based on results of the matrix of similarity values were created with unweighted-pair group method using average linkage clustering.
(ii) Statistical analysis.
The data were analyzed by Epi-Info version 6.03. The chi-square test for homogeneity was used to evaluate the association between a disease status (bacteremia or seropositivity) and a categorized risk factor, and then P values were calculated using Yates corrected method or two-tailed Fisher's exact test (for analyses with expected numbers of observations of less than five). Mantel extension test for trend was also applied to evaluate for the existence of a seasonal trend for bacteremia and antibody prevalence. The association between seropositivity and bacteremia for Bartonella was evaluated by McNemar's test for paired analysis.
RESULTS
Epidemiological patterns of B. vinsonii subsp. berkhoffii infection in coyotes.
Of 109 coyotes, 31 (28%; 95% CI, 20 to 38%) were positive for Bartonella spp. by blood culture. The seroprevalence for B. vinsonii subsp. berkhoffii was 76% (83 of 109) (95% CI, 67 to 84%). There was no significant difference between the prevalence of bacteremia in 1997 (30% [16 of 54]) and in 1998 (27% [15 of 55]) (P = 0.95). The seroprevalence was also similar in 1997 and 1998 (72 versus 80% [P = 0.47]). Only 10 coyotes less than 1 year old were trapped in this study. The prevalence of Bartonella bacteremia was significantly lower in adult coyotes (25%) than in coyotes less than 1 year old (60%) (Table 1). Conversely, the seroprevalence of Bartonella infection in adult coyotes (91%) was higher than in coyotes less than 1 year old (60%) (Table 1). There was no statistically significant prevalence difference for either bacteremia or antibodies between male and female coyotes.
TABLE 1.
Prevalence of B. vinsonii subsp. berkhoffii bacteremia and seropositivity by age, sex, and collection period for 109 coyotes from central coastal California
| Variable | Bacteremia
|
Seropositivity
|
||
|---|---|---|---|---|
| Prevalencea | P valueb | Prevalence | P value | |
| Age | ||||
| <1 yr old | 60 (6/10) | 0.03 | 60 (6/10) | 0.48 |
| ≥1 yr old | 25 (25/99) | 91 (71/99) | ||
| Sexc | ||||
| Male | 33 (16/49) | 0.42 | 72 (35/49) | 0.44 |
| Female | 24 (14/59) | 80 (47/59) | ||
| Collection period | ||||
| Spring (Mar. to May) | 83 (5/6) | 0.01 | 83 (5/6) | 0.02 |
| Summer (June to Aug.) | 21 (10/47) | 62 (29/47) | ||
| Fall (Sept. to Nov.) | 31 (16/51) | 86 (44/51) | ||
| Winter (Dec. to Feb.) | 0 (0/5) | 100 (5/5) | ||
Prevalence shown as a percentage, with the number of coyotes with bacteremia (or the number of seropositive coyotes) and the total number of coyotes shown in the parentheses.
Chi-square test for homogeneity.
Not available for one coyote.
Using the blood collection date to investigate seasonal differences for the prevalence of Bartonella bacteremia, the lowest prevalence was observed during winter, followed by summer and fall. The highest prevalence was seen during spring. This increasing trend of Bartonella bacteremic prevalence was statistically significant (P = 0.05). The trend of seroprevalence was increased from summer, spring, fall, to winter (P < 0.05). The bacteremia and antibody prevalences varied by collection sites (Table 2). There was no significant association between age and collection periods or between age and collection sites.
TABLE 2.
Bacteremia and antibody prevalences of B. vinsonii subsp. berkhoffii infection in coyotes from central coastal California by capture sites
| Location | Bacteremia prevalencea | Seroprevalence |
|---|---|---|
| Cupertino | 80 (4/5) | 60 (3/5) |
| Gilroy | 36 (9/25) | 68 (17/25) |
| Los Altos | 7 (1/15) | 60 (9/15) |
| Los Gatos | 60 (3/5) | 80 (4/5) |
| Morgan Hill | 22 (5/23) | 70 (16/23) |
| Milpitas | 0 (0/7) | 86 (6/7) |
| San Mateo | 50 (1/2) | 100 (2/2) |
| San Jose | 32 (7/22) | 95 (21/22) |
| Saratoga | 0 (0/4) | 100 (4/4) |
Prevalence shown as a percentage, with the number of coyotes with bacteremia (or the number of seropositive coyotes) and the total number of coyotes shown in the parentheses.
There was no significant association between seropositivity and bacteremia for Bartonella in coyotes. Twenty-four Bartonella bacteremic coyotes had antibody titers ranging from 1:64 (9 coyotes), 1:128 (5 coyotes), to 1:256 (10 coyotes). However, seven of the bacteremic coyotes were seronegative (titer of ≤1:32). Of the 78 nonbacteremic coyotes, 59 (75.6%) had B. vinsonii subsp. berkhoffii antibodies with the following titers: 1:64 (32 coyotes), 1:128 (12 coyotes), 1:256 (13 coyotes), and 1:152 (2 coyotes). Therefore, the positive and negative predictive values of the B. vinsonii subsp. berkhoffii serological assay for bacteremia status were 29% (24 of 83) and 73% (19 of 26), respectively. Sixty-three coyotes that were seropositive for B. vinsonii subsp. berkhoffii were also seropositive for B. henselae (Table 3). However, thirteen coyotes that were seronegative for B. vinsonii subsp. berkhoffii were seropositive for B. henselae (Table 3).
TABLE 3.
Distribution of anti-B. vinsonii subsp. berkhoffii and anti-B. henselae antibody titers by IFA in coyotes
| Anti-B. henselae antibody titer | Anti-B. vinsonii subsp. berkhoffii antibody titer
|
||||
|---|---|---|---|---|---|
| ≤1:32 | 1:64a | 1:128 | 1:256 | 1:512 | |
| ≤1:32 | 13 | 16 | 3 | 1 | 0 |
| 1:64a | 12 | 21 | 10 | 9 | 1 |
| 1:128 | 1 | 4 | 3 | 6 | 0 |
| 1:256 | 0 | 0 | 0 | 7 | 1 |
| 1:512 | 0 | 0 | 1 | 0 | 0 |
An antibody titer of ≥1:64 was reported as seropositive.
PCR-RFLP-based typing.
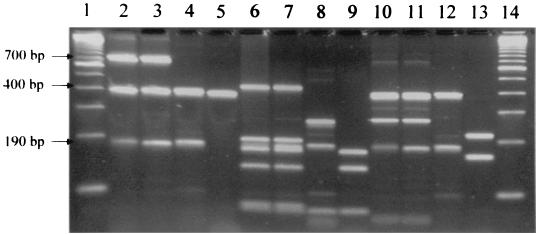
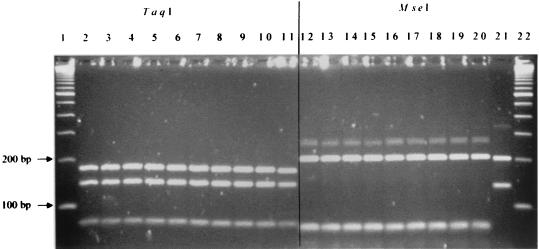
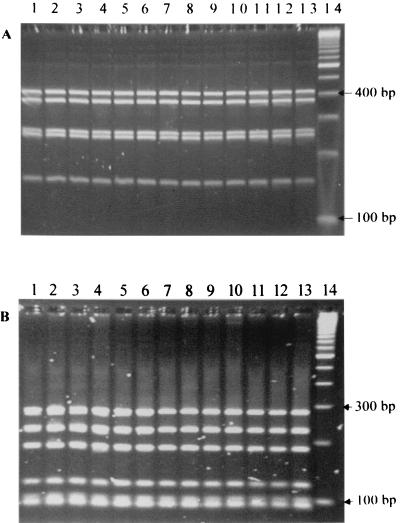
All 31 suspected Bartonella isolates from coyotes were tested by PCR of the gltA gene with two different sets of primers that are specific for Bartonella species (37, 40). Before digestion by the restriction endonucleases, a 400-bp band specific for Bartonella spp. was identified for all 31 coyote isolates by both sets of primers. However, extra 700- and 190-bp bands were also observed for all isolates and B. vinsonii subsp. berkhoffii ATCC 51672 with the primers suggested by Regnery et al. (40) (Fig. 1 and 2). The molecular pattern of all coyote isolates was identical to that of a domestic dog isolate of B. vinsonii subsp. berkhoffii (ATCC 51672), based on PCR-RFLP analysis of the gltA gene with TaqI or HhaI digestion (Fig. 1) and 16S rRNA gene with DdeI and MnlI digestion (Fig. 3). However, these isolates yielded different patterns from that of B. vinsonii subsp. vinsonii (ATCC VR152) when PCR-RFLP analysis of the gltA gene was performed (Fig. 1). The PCR product with the set of primers suggested by Norman et al. (37) could not be digested by HhaI restriction endonuclease; however, the TaqI- and MseI-digested profiles were identical for all isolates (Fig. 2). The partial 16S rRNA gene sequences further showed a 100% homology between the coyote isolate sequenced and a domestic dog isolate (ATCC 51672). By partial sequencing of the gltA gene, there was a 99.5% homology between the coyote isolate and ATCC strain 51672.
FIG. 1.
PCR (lanes 2 to 5) and PCR-RFLP (lanes 6 to 9, TaqI digestion; lanes 10 to 13, HhaI digestion) analyses of the gltA gene of coyote isolates with the set of primers suggested by Regnery et al. (40). Lanes 1 and 14, standard 100-bp molecular ladder; lanes 2, 6, and 10, coyote isolates; lanes 3, 7, and 11, B. vinsonii subsp. berkhoffii ATCC 51672; lanes 4, 8, and 12, B. vinsonii ATCC VR152; lanes 5, 9 and 13, B. henselae (strain U-4, University of California, Davis).
FIG. 2.
PCR-RFLP analysis (lanes 2 to 11, TaqI digestion; lanes 12 to 21, MseI digestion) of the gltA gene of coyote isolates with the set of primers suggested by Norman et al. (37). Lanes 1 and 22, standard 100-bp molecular ladder; lanes 2 to 9 and 12 to 19, coyote isolates; lanes 10 and 20, B. vinsonii subsp. berkhoffii ATCC 51672; lanes 11 and 21, B. henselae (strain U-4; University of California, Davis).
FIG. 3.
PCR-RFLP analysis of the 16S rRNA gene of coyote isolates with DdeI (A) or MnlI (B) restriction endonuclease. Lanes 1 to 12, coyote isolates; lane 13, B. vinsonii subsp. berkhoffii ATCC 51672; lane 14, 100-bp molecular ladder.
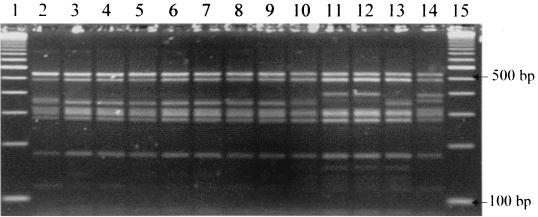
By PCR-RFLP analysis of the 16S-23S interspacer region, 6 (19%) of 31 Bartonella bacteremic coyotes were infected with B. vinsonii subsp. berkhoffii type I strain, similar to the profile of ATCC strain 51672 (Fig. 4). The other 25 Bartonella bacteremic coyotes were all infected with B. vinsonii subsp. berkhoffii type II strain, which has been isolated from healthy dogs. There was no major clustering of these two types by capture sites.
FIG. 4.
PCR-RFLP analysis of the 16S-23S ITS region of coyote isolates with HaeIII restriction endonuclease. Lanes 1 and 15, standard 100-bp molecular ladder; lanes 2 to 10 and 13, coyote isolates (type II); lanes 11 and 12, coyote isolates (type I); lane 14, B. vinsonii subsp. berkhoffii ATCC 51672 (type I).
PFGE-based typing.
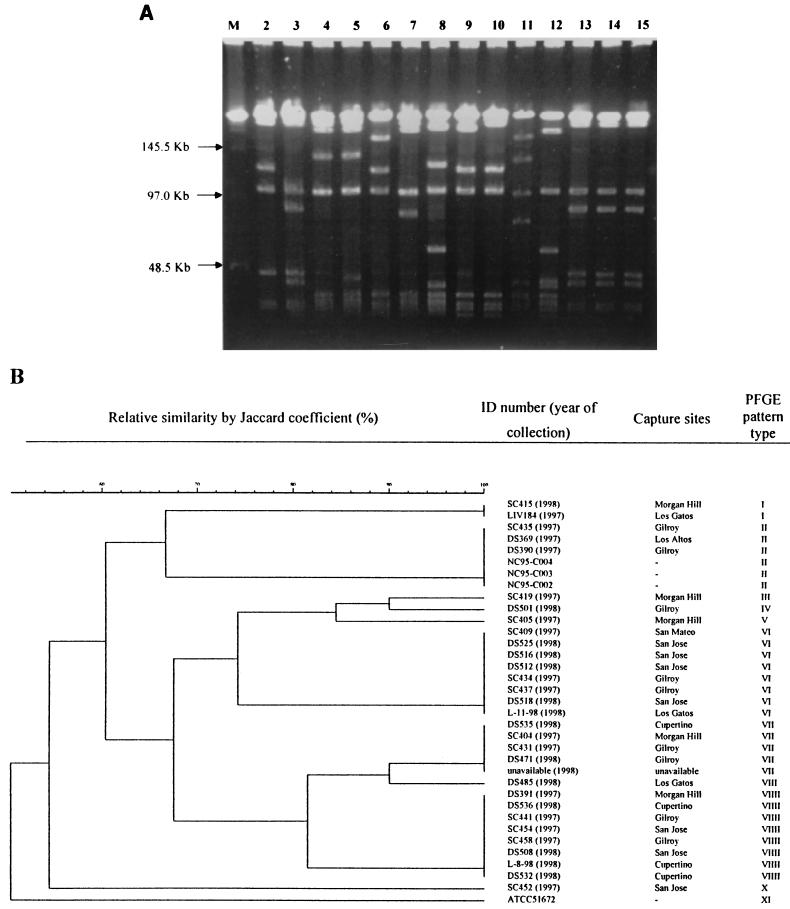
Ten coyote isolates were not digested by NotI restriction enzyme, and only a few high-molecular-weight bands were observed for the remaining 21 isolates after digestion. Therefore, PFGE profiles with SmaI digestion were used for cluster analysis. When a Jaccard coefficient of 70% was applied for grouping the profiles, the fingerprints with SmaI identified 11 major patterns for B. vinsonii subsp. berkhoffii isolates, including ATCC strain 51672, which had a unique pattern (Fig. 5). Based on the fingerprints by SmaI digestion, no clonal distribution according to the place or year of collection was observed for the coyote isolates (Fig. 5). Compared to the 16S-23S ITS fingerprints, five of the six coyotes infected with type I were grouped with the PFGE pattern VII and one type I isolate belonged to the PFGE pattern X.
FIG. 5.
(A) Fingerprints of B. vinsonii subsp. berkhoffii isolates by PFGE with SmaI digestion. Lane M, molecular size markers; lanes 2 to 11, B. vinsonii subsp. berkhoffii patterns I to X of the coyote isolates; lane 12, ATCC strain 51672 (pattern XI); lanes 13 to 15, isolates from three healthy dogs (pattern II). (B) Dendrogram of the fingerprints as determined by unweighted-pair group method using average linkage clustering. NC95-C02, NC95-C03, and NC95-C04 are B. vinsonii subsp. berkhoffii isolates from three healthy dogs. ID, identification.
DISCUSSION
We found 28% of 109 coyotes from central coastal California to be bacteremic with B. vinsonii subsp. berkhoffii, previously isolated from domestic dogs. The prevalence of bacteremia was higher among coyotes <1 year old than among adult coyotes, as previously reported for B. henselae infection in cats (14). The high prevalence (76%) of B. vinsonii subsp. berkhoffii antibodies in these 109 coyotes was also consistent with the previous findings of a serosurvey of California coyotes (13). Seven coyotes that were Bartonella bacteremic were seronegative for Bartonella, possibly because of recently acquired infection; three coyotes were less than 1 year old, and four coyotes were adults. Similarly, cats with Bartonella bacteremia but without detectable antibodies have been previously reported (33).
PCR-RFLP analysis of the gltA and 16S rRNA genes showed that the PCR-RFLP profiles of all 31 coyote isolates were identical to that of a domestic dog isolate of B. vinsonii subsp. berkhoffii (ATCC 51672) but were different from the PCR-RFLP profiles of the other Bartonella strains tested: B. henselae, B. clarridgeiae, B. bacilliformis, B. quintana, B. elizabethae, and B. vinsonii subsp. vinsonii (data not shown). The partial sequencing of the gltA and 16S rRNA genes, respectively, indicated 99.5 and 100% homology between the coyote isolate and B. vinsonii subsp. berkhoffii reference strain (ATCC 51672). There were two different molecular types (designated type I and type II) of B. vinsonii subsp. berkhoffii using PCR-RFLP analysis of the 16S-23S ITS region as previously reported for domestic dogs (31); type I was isolated from a dog with endocarditis, and type II was isolated from three healthy dogs. Unfortunately, PCR-RFLP analysis of the 16S-23S ITS was not conducted in the human endocarditis case caused by B. vinsonii subsp. berkhoffii (43), which did not allow comparison with these canine isolates. In our study, only 19% of 31 Bartonella bacteremic coyotes were infected with the type I strain, i.e., similar to the strain isolated from a dog with endocarditis. Further studies should aimed at determining if the type I strain is specifically associated with canine and human endocarditis cases.
Because of the large number of B. vinsonii subsp. berkhoffii isolates obtained from coyotes, we were able to investigate, for the first time, the molecular diversity of these isolates and domestic dog isolates by analysis of the whole bacterial genome using PFGE. PFGE, using SmaI endonuclease, allowed us to determine 11 variants, including a unique pattern for ATCC 51672 strain. By PFGE, three coyotes were infected with a B. vinsonii subsp. berkhoffii strain similar to the strains isolated from three healthy dogs, but none of the coyotes were infected with strains identical to the ATCC 51672 strain. The wide diversity of B. vinsonii subsp. berkhoffii PFGE profiles could be caused by gene mutation or translocation under natural circumstances that potentially enhance bacterial transmissibility or pathogenicity. In contrast, NotI endonuclease was not an appropriate enzyme for differentiating these isolates because it very infrequently cut B. vinsonii subsp. berkhoffii, as previously reported by Roux and Raoult (45) for B. henselae isolates. As has been suggested in other bacterial studies (49), PFGE analysis appeared to be a more discriminating method in differentiating B. vinsonii subsp. berkhoffii variants in canids than PCR-RFLP analysis. In the future, however, it will be of interest to determine if differences in pathogenicity for canids will be better identified by PCR-RFLP analysis of the 16S-23S ITS region or by PFGE fingerprints.
Detection and identification of Bartonella organisms have been done mainly by PCR-RFLP analysis and/or sequencing targeting the gltA gene (37, 40), 16S rRNA gene (5, 18, 29, 35, 40, 42), and/or 16S-23S ITS region (31, 36, 44; G. M. Matar, Letter, J. Clin. Microbiol. 33:3370, 1995). The low rate of evolutionary sequence divergence of the 16S rRNA gene is useful for designing primers targeting conserved sequences for broad-range PCR. Currently, sequencing data of the 16S rRNA gene for more than 2,000 bacterial species is available in GenBank. However, it could be difficult to design the genus-specific primers that are able to specifically amplify one bacterial genus, e.g., Bartonella, for rapid diagnosis. It has been shown that the sequence of the gltA gene is less conserved than that of the 16S rRNA gene within the genus Bartonella (7), facilitating the design of Bartonella-specific primers for PCR amplification. We suggest using PCR-RFLP analysis and/or sequencing of the gltA gene as a first step in confirmation and identification of Bartonella species and to generate phylogenic trees for the Bartonellaceae family, as done by Kosoy et al. (34). PCR-RFLP and/or sequencing of the 16S rRNA gene could be used in a second step for analysis of phylogenic relationships with the other closely related bacterial genera (41, 42). Finally, PCR-RFLP and/or sequencing of the 16S-23S ITS gene or PFGE could be applied for subspecies identification (45, 47).
When preparing IFA slides, it was observed that B. vinsonii subsp. berkhoffii-infected F. catus whole fetus cells were prone to clump together, making the preparation of IFA slides difficult. We found that heat inactivation during antigen preparation could prevent cell clumping and improve the quality of IFA tests for detection of B. vinsonii subsp. berkhoffii antibodies.
Sixty-three coyotes which were seropositive for B. vinsonii subsp. berkhoffii were also seropositive for B. henselae, but thirteen coyotes that were seronegative for B. vinsonii subsp. berkhoffii were seropositive for B. henselae. These results may be related to cross-reactivity between various Bartonella species, which has been reported in humans and animals (4, 21, 26). Moreover, it is still unknown if canids can be naturally infected with B. henselae, isolated at present only from felids and humans, and become seropositive. However, all 31 Bartonella bacteremic coyotes were found to be infected only with B. vinsonii subsp. berkhoffii, not with B. henselae or any other Bartonella species.
Compared to B. vinsonii subsp. berkhoffii infection in domestic dogs (38), the significantly higher bacteremia and antibody prevalences in coyotes implies that coyotes might be a wildlife reservoir. The ability to maintain a particular infectious agent for a long period of time and the high prevalence of infection with this infectious agent in a given animal species are the characteristics of animal reservoirs, as illustrated for cats and B. henselae. The high prevalence of B. vinsonii subsp. berkhoffii infection found in coyotes also suggests that these animals could serve as a potential reservoir for B. vinsonii subsp. berkhoffii. Repeated isolation of the infectious agent from captured wild coyotes provides presumptive evidence of reservoir competency, although this capacity was not fully demonstrated by the present study.
In our laboratory, experimental inoculation of two domestic dogs by the intradermal route with a coyote isolate led to a prolonged bacteremia (at least 8 weeks) (B. B. Chomel et al., unpublished data). As seen for B. henselae infection in cats, which can remain bacteremic for periods ranging from several months to years (28, 30, 46), both dogs had a high level of Bartonella bacteremia for a few weeks after infection but without fever and clinical signs. These data suggest that, in addition to the capacity to be infected, canids can maintain levels of bacteremia for periods long enough to allow possible arthropod transmission. However, this cross-sectional study did not allow us to determine the duration of B. vinsonii subsp. berkhoffii bacteremia in wild coyotes. Coyotes could be a source of infection for domestic dogs, especially when Bartonella enzootic coyote populations have an overlapping home range with domestic dogs.
Modes of transmission between coyotes and domestic dogs could be either by mechanical means (biting and scratching) or through arthropod vectors. However, Bartonella spp. are usually transmitted by arthropod vectors (2). B. bacilliformis, the agent of Carrión's disease, mainly found in the Andes mountains, is transmitted by sand flies. B. quintana, which is transmitted by the human body louse, causes trench fever. Cats are the main reservoir for B. henselae (28), and cat fleas are a competent vector for the transmission between cats (15). No direct transmission of B. henselae from cat to cat has been documented in experimental settings (1, 23). The seasonal fluctuation of Bartonella bacteremic prevalence in coyotes from central coastal California observed in this study could possibly be related to arthropod activity. Based on our previous study, the clustered distribution of higher Bartonella seroprevalence in coyotes from coastal California compared to coyotes from the inland regions also suggested that the geographical distribution of this Bartonella infection in coyotes could be associated with the presence of certain arthropod species, such as ticks or sand flies (13). We therefore hypothesize that B. vinsonii subsp. berkhoffii transmission within coyotes and between coyotes and dogs could be arthropodborne. Until now, investigations of which vectors are competent for B. vinsonii subsp. berkhoffii transmission among dogs have been very limited. Tickborne infection has been suggested for dogs from the eastern United States (38). To confirm this hypothesis, one needs to show vector competency by allowing ticks to feed on experimentally infected dogs and/or coyotes to determine if ticks are able to acquire B. vinsonii subsp. berkhoffii infection and further transmit this agent to noninfected canids. Furthermore, testing of field-collected tick samples should be performed.
Dogs and possibly humans could become infected during recreational activities through bites of infected arthropods that may have fed on Bartonella bacteremic coyotes. The recent identification of B. vinsonii subsp. berkhoffii in a human endocarditis case (43) warrants further investigation to elucidate the mode of transmission of B. vinsonii subsp. berkhoffii, especially to identify potential vectors, and to determine how humans become infected.
ACKNOWLEDGMENTS
We thank Didier Raoult for kindly sharing unpublished data on the human case of Bartonella vinsonii subsp. berkhoffii endocarditis. We also thank Ian Gardner (Department of Medicine and Epidemiology, School of Veterinary Medicine, University of California, Davis) and Robert S. Lane (Department of Environmental Science, Policy and Management, College of Natural Resources, University of California, Berkeley) for suggestions and help in preparing this manuscript.
REFERENCES
- 1.Abbott R C, Chomel B B, Kasten R W, FloydHawkins K A, Kikuchi Y, Koehler J E, Pedersen N C. Experimental and natural infection with Bartonella henselae in domestic cats. Comp Immunol Microbiol Infect Dis. 1997;20:41–51. doi: 10.1016/s0147-9571(96)00025-2. [DOI] [PubMed] [Google Scholar]
- 2.Anderson B E, Neuman M A. Bartonella spp. as emerging human pathogens. Clin Microbiol Rev. 1997;10:203–219. doi: 10.1128/cmr.10.2.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baker J A. A rickettsial infection in Canadian voles. J Exp Med. 1946;84:37–50. [PubMed] [Google Scholar]
- 4.Baneth G, Kordick D L, Hegarty B C, Breitschwerdt E B. Comparative seroreactivity to Bartonella henselae and Bartonella quintana among cats from Israel and North Carolina. Vet Microbiol. 1996;50:95–103. doi: 10.1016/0378-1135(96)00006-5. [DOI] [PubMed] [Google Scholar]
- 5.Bergmans A M, Schellekens J F, van Embden J D, Schouls L M. Predominance of two Bartonella henselae variants among cat-scratch disease patients in The Netherlands. J Clin Microbiol. 1996;34:254–260. doi: 10.1128/jcm.34.2.254-260.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Birtles R J. Differentiation of Bartonella species using restriction endonuclease analysis of PCR-amplified 16S rRNA genes. FEMS Microbiol Lett. 1995;129:261–265. doi: 10.1111/j.1574-6968.1995.tb07590.x. [DOI] [PubMed] [Google Scholar]
- 7.Birtles R J, Raoult D. Comparison of partial citrate synthase gene (gltA) sequences for phylogenetic analysis of Bartonella species. Int J Syst Bacteriol. 1996;46:891–897. doi: 10.1099/00207713-46-4-891. [DOI] [PubMed] [Google Scholar]
- 8.Breathnach A S, Hoare J M, Eykyn S J. Culture-negative endocarditis: contribution of Bartonella infections. Heart. 1997;77:474–476. doi: 10.1136/hrt.77.5.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Breitschwerdt E B, Atkins C E, Brown T T, Kordick D L, Snyder P S. Bartonella vinsonii subsp. berkhoffii and related members of the alpha subdivision of the Proteobacteria in dogs with cardiac arrhythmias, endocarditis, or myocarditis. J Clin Microbiol. 1999;37:3618–3626. doi: 10.1128/jcm.37.11.3618-3626.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Breitschwerdt E B, Kordick D L, Malarkey D E, Keene B, Hadfield T L, Wilson K. Endocarditis in a dog due to infection with a novel Bartonella subspecies. J Clin Microbiol. 1995;33:154–160. doi: 10.1128/jcm.33.1.154-160.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burgess E C, Windberg L A. Borrelia sp. infection in coyotes, black-tailed jack rabbits and desert cottontails in southern Texas. J Wildl Dis. 1989;25:47–51. doi: 10.7589/0090-3558-25.1.47. [DOI] [PubMed] [Google Scholar]
- 12.Chang C C, Chomel B B, Kasten R W, Heller R, Kocan K M, Ueno H, Yamamoto K, Bleich V C, Pierce B M, Gonzales B J, Swift P K, Boyce W M, Jang S S, Boulouis H, Piemont Y. Bartonella spp. isolated from wild and domestic ruminants in North America. Emerg Infect Dis. 2000;6:306–311. doi: 10.3201/eid0603.000313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang C C, Yamamoto K, Chomel B B, Kasten R W, Simpson D C, Smith C R, Kramer V L. Seroepidemiology of Bartonella vinsonii subsp. berkhoffii infection in California coyotes, 1994–1998. Emerg Infect Dis. 1999;5:711–715. doi: 10.3201/eid0505.990514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chomel B B, Abbott R C, Kasten R W, Floyd-Hawkins K A, Kass P H, Glaser C A, Pedersen N C, Koehler J E. Bartonella henselae prevalence in domestic cats in California: risk factors and association between bacteremia and antibody titers. J Clin Microbiol. 1995;33:2445–2450. doi: 10.1128/jcm.33.9.2445-2450.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chomel B B, Kasten R W, Floyd-Hawkins K, Chi B, Yamamoto K, Roberts-Wilson J, Gurfield A N, Abbott R C, Pedersen N C, Koehler J E. Experimental transmission of Bartonella henselae by the cat flea. J Clin Microbiol. 1996;34:1952–1956. doi: 10.1128/jcm.34.8.1952-1956.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clark K A, Neill S U, Smith J S, Wilson P J, Whadford V W, McKirahan G W. Epizootic canine rabies transmitted by coyotes in south Texas. J Am Vet Med Assoc. 1994;204:536–540. [PubMed] [Google Scholar]
- 17.Cypher B L, Scrivner J H, Hammer K L, O'Farrell T P. Viral antibodies in coyotes from California. J Wildl Dis. 1998;34:259–264. doi: 10.7589/0090-3558-34.2.259. [DOI] [PubMed] [Google Scholar]
- 18.Daly J S, Worthington M G, Brenner D J, Moss C W, Hollis D G, Weyant R S, Steigerwalt A G, Weaver R E, Daneshvar M I, O'Connor S P. Rochalimaea elizabethae sp. nov. isolated from a patient with endocarditis. J Clin Microbiol. 1993;31:872–881. doi: 10.1128/jcm.31.4.872-881.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Drancourt M, Mainardi J L, Brouqui P, Vandenesch F, Carta A, Lehnert F, Etienne J, Goldstein F, Acar J, Raoult D. Bartonella (Rochalimaea) quintana endocarditis in three homeless men. N Engl J Med. 1995;332:419–423. doi: 10.1056/NEJM199502163320702. [DOI] [PubMed] [Google Scholar]
- 20.Ellis B A, Regnery R L, Beati L, Bacellar F, Rood M, Glass G G, Marston E, Ksiazek T G, Jones D, Childs J E. Rats of the genus Rattus are reservoir hosts for pathogenic Bartonella species: an old world origin for a new world disease? J Infect Dis. 1999;180:220–224. doi: 10.1086/314824. [DOI] [PubMed] [Google Scholar]
- 21.Engbaek K, Koch C. Immunoelectrophoretic characterization and cross-reactivity of Rochalimaea henselae, Rochalimaea quintana and Afipia felis. APMIS. 1994;102:931–942. [PubMed] [Google Scholar]
- 22.Foreyt W J, Evermann J F. Serologic survey of canine coronavirus in wild coyotes in the western United States. J Wildl Dis. 1985;21:428–430. doi: 10.7589/0090-3558-21.4.428. [DOI] [PubMed] [Google Scholar]
- 23.Guptill L, Slater L, Wu C C, Lin T L, Glickman L T, Welch D F, HogenEsch H. Experimental infection of young specific pathogen-free cats with Bartonella henselae. J Infect Dis. 1997;176:206–216. doi: 10.1086/514026. [DOI] [PubMed] [Google Scholar]
- 24.Gurfield A N, Boulouis H J, Chomel B B, Heller R, Kasten R W, Yamamoto K, Piemont Y. Coinfection with Bartonella clarridgeiae and Bartonella henselae and with different Bartonella henselae strains in domestic cats. J Clin Microbiol. 1997;35:2120–2123. doi: 10.1128/jcm.35.8.2120-2123.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heller R, Artois M, Xemar V, De Briel D, Gehin H, Jaulhac B, Monteil H, Piemont Y. Prevalence of Bartonella henselae and Bartonella clarridgeiae in stray cats. J Clin Microbiol. 1997;35:1327–1331. doi: 10.1128/jcm.35.6.1327-1331.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jackson L A, Spach D H, Kippen D A, Sugg N K, Regnery R L, Sayers M H, Stamm W E. Seroprevalence to Bartonella quintana among patients at a community clinic in downtown Seattle. J Infect Dis. 1996;173:1023–1026. doi: 10.1093/infdis/173.4.1023. [DOI] [PubMed] [Google Scholar]
- 27.Kerkhoff F T, Bergmans A M C, Van Der Zee A, Rothova A. Demonstration of Bartonella grahamii DNA in ocular fluids of a patient with neuroretinitis. J Clin Microbiol. 1999;37:4034–4038. doi: 10.1128/jcm.37.12.4034-4038.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koehler J E, Glaser C A, Tappero J W. Rochalimaea henselae infection—a new zoonosis with the domestic cat as reservoir. J Am Med Assoc. 1994;271:531–535. doi: 10.1001/jama.271.7.531. [DOI] [PubMed] [Google Scholar]
- 29.Koehler J E, Quinn F D, Berger T G, Le Boit P E, Tappero J W. Isolation of Rochlimaea species from cutaneous and osseous lesions of bacillary angiomatosis. N Engl J Med. 1992;327:1625–1631. doi: 10.1056/NEJM199212033272303. [DOI] [PubMed] [Google Scholar]
- 30.Kordick D L, Breitschwerdt E B. Intraerythrocytic presence of Bartonella henselae. J Clin Microbiol. 1995;33:1655–1656. doi: 10.1128/jcm.33.6.1655-1656.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kordick D L, Breitschwerdt E B. Persistent infection of pets within a household with three Bartonella species. Emerg Infect Dis. 1998;4:325–328. doi: 10.3201/eid0402.980225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kordick D L, Swaminathan B, Greene C E, Wilson K H, Whitney A M, O'Connor S, Hollis D G, Matar G M, Steigerwalt A G, Malcolm G B, Hayes P S, Hadfield T L, Breitschwerdt E B, Brenner D J. Bartonella vinsonii subsp. berkhoffii subsp. nov., isolated from dogs; Bartonella vinsonii subsp. vinsonii; and emended description of Bartonella vinsonii. Int J Syst Bacteriol. 1996;46:704–709. doi: 10.1099/00207713-46-3-704. [DOI] [PubMed] [Google Scholar]
- 33.Kordick D L, Wilson K H, Sexton D J, Hadfield T L, Berkhoff H A, Breitschwerdt E B. Prolonged Bartonella bacteremia in cats associated with cat-scratch disease patients. J Clin Microbiol. 1995;33:3245–3251. doi: 10.1128/jcm.33.12.3245-3251.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kosoy M Y, Regnery R L, Tzianabos T, Marston E L, Jones D C, Green D, Maupin G O, Olson J G, Childs J E. Distribution, diversity, and host specificity of Bartonella in rodents from the southeastern United States. Am J Trop Med Hyg. 1997;57:578–588. doi: 10.4269/ajtmh.1997.57.578. [DOI] [PubMed] [Google Scholar]
- 35.Lucey D, Dolan M J, Moss C W, Garcia M, Hollis D G, Wegner S. Relapsing illness due to Rochalimaea henselae in immunocompetent host: implication for therapy and new epidemiological associations. Lin Infect Dis. 1992;14:683–688. doi: 10.1093/clinids/14.3.683. [DOI] [PubMed] [Google Scholar]
- 36.Matar G M, Swaminathan B, Hunter S B, Slater L N, Welch D F. Polymerase chain reaction-based restriction fragment length polymorphism analysis of a fragment of the ribosomal operon from Rochalimaea species for subtyping. J Clin Microbiol. 1993;31:1730–1734. doi: 10.1128/jcm.31.7.1730-1734.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Norman A F, Regnery R, Jameson P, Greene C, Krause D C. Differentiation of Bartonella-like isolates at the species level by PCR-restriction fragment length polymorphism in the citrate synthase gene. J Clin Microbiol. 1995;33:1797–1803. doi: 10.1128/jcm.33.7.1797-1803.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pappalardo B L, Correa M T, York C C, Peat C Y, Breitschwerdt E B. Epidemiologic evaluation of the risk factors associated with exposure and seroreactivity to Bartonella vinsonii in dogs. Am J Vet Res. 1997;58:467–471. [PubMed] [Google Scholar]
- 39.Pappas L G, Lunzman A T. Canine heartworm in the domestic and wild canids of southeastern Nebraska. J Parasitol. 1985;71:828–830. [PubMed] [Google Scholar]
- 40.Regnery R L, Anderson B E, Clarridge III J E, Rodriguez-Barradas M C, Jones D C, Carr J H. Characterization of a novel Rochalimaea species, R. henselae sp. nov., isolated from blood of a febrile, human immunodeficiency virus-positive patient. J Clin Microbiol. 1992;30:265–274. doi: 10.1128/jcm.30.2.265-274.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Relman D A, Lepp P W, Sadler K N, Schmidt T M. Phylogenetic relationships among the agent of bacillary angiomatosis, Bartonella bacilliformis, and other alpha-proteobacteria. Mol Microbiol. 1992;6:1801–1807. doi: 10.1111/j.1365-2958.1992.tb01352.x. [DOI] [PubMed] [Google Scholar]
- 42.Relman D A, Loutit J S, Schmidt T M, Falkow S, Tompkins L S. The agent of bacillary angiomatosis. An approach to the identification of uncultured pathogens. N Engl J Med. 1990;323:1573–1580. doi: 10.1056/NEJM199012063232301. [DOI] [PubMed] [Google Scholar]
- 43.Roux V, Eykyn S J, Wyllie S, Raoult D. Bartonella vinsonii subsp. berkhoffii as an agent of afebrile blood culture-negative endocarditis in a human. J Clin Microbiol. 2000;38:1698–1700. doi: 10.1128/jcm.38.4.1698-1700.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roux V, Raoult D. The 16S–23S rRNA intergenic spacer region of Bartonella (Rochalimaea) species is longer than usually described in other bacteria. Gene. 1995;156:107–111. doi: 10.1016/0378-1119(94)00919-j. [DOI] [PubMed] [Google Scholar]
- 45.Roux V, Raoult D. Inter- and intraspecies identification of Bartonella (Rochalimaea) species. J Clin Microbiol. 1995;33:1573–1579. doi: 10.1128/jcm.33.6.1573-1579.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sander A, Buhler C, Pelz K, von Cramm E, Bredt W. Detection and identification of two Bartonella henselae variants in domestic cats in Germany. J Clin Microbiol. 1997;35:584–587. doi: 10.1128/jcm.35.3.584-587.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sander A, Ruess M, Bereswill S, Schuppler M, Steinbrueckner B. Comparison of different DNA fingerprinting techniques for molecular typing of Bartonella henselae isolates. J Clin Microbiol. 1998;36:2973–2981. doi: 10.1128/jcm.36.10.2973-2981.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Spach D H, Callis K P, Paauw D S, Houze Y B, Schoenknecht F D, Welch D F, Rosen H, Brenner D J. Endocarditis caused by Rochalimaea quintana in a patient infected with human immunodeficiency virus. J Clin Microbiol. 1993;31:692–694. doi: 10.1128/jcm.31.3.692-694.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tenover F C, Arbeit R D, Goering R V, Mickelsen P A, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Welch D F, Carroll K C, Hofmeister E K, Persing D H, Robison D A, Steigerwalt A G, Brenner D J. Isolation of a new subspecies, Bartonella vinsonii subsp. arupensis, from a cattle rancher: identity with isolates found in conjunction with Borrelia burgdorferi and Babesia microti among naturally infected mice. J Clin Microbiol. 1999;37:2598–2601. doi: 10.1128/jcm.37.8.2598-2601.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Willeberg P W, Ruppanner R, Behymer D E, Higa H H, Franti C E, Thompson R A. Epidemiologic survey of sylvatic plague by serotesting coyote sentinels with enzyme immunoassay. Am J Epidemiol. 1979;110:328–334. doi: 10.1093/oxfordjournals.aje.a112818. [DOI] [PubMed] [Google Scholar]