Abstract
Background
Long non-coding RNAs (lncRNAs) play critical roles in the occurrence and progression of various tumors, including ovarian cancer (OC). The lncRNA growth arrest-specific transcript 5 (GAS5) has been shown to be an important modulator in the growth and metastasis of OC cells. Our previous studies confirmed that GAS5 was down-regulated in OC; however, the potential underlying molecular mechanism underlying has not yet been elucidated.
Methods
We screened the Gene Expression Profiling Interactive Analysis (GEPIA) database for the expression of the lncRNA GAS5 in OC. Cell Counting Kit-8 (CCK-8), transwell assay, colony formation assay, flow cytometry analysis, and western blotting were applied to determine the various functions of GAS5 in OC progression. The competing endogenous RNA (ceRNA) mechanism was verified through bioinformatics analysis, dual-spectral luciferase reporter gene assay, and RNA immunoprecipitation assay (RIPA). Finally, the expression interactions between microRNA-96-5p, phosphatase and tensin homolog deleted on chromosome ten (PTEN), and GAS5 were measured.
Results
Our results demonstrated decreased expression levels of GAS5 and PTEN in OC samples and cell lines, while miR-96-5p was up-regulated when compared with the controls. GAS5 overexpression could significantly reduce OC cell proliferation and invasion ability via suppression of miR-96-5p expression. Moreover, GAS5 could influence the PTEN/protein kinase B (AKT)/mammalian target of rapamycin (mTOR) signaling pathway.
Conclusions
Our study identified GAS5 as a ceRNA that can regulate the PTEN/AKT/mTOR axis by sponging miR-96-5p in OC.
Keywords: Long non-coding RNA (lncRNA), growth arrest-specific transcript 5 (GAS5), phosphatase and tensin homolog deleted on chromosome ten (PTEN), miR-96-5p, ovarian cancer (OC)
Introduction
Ovarian cancer (OC) is one of the major malignant tumors of the female reproductive organs, accounting for the highest mortality rate (1). OC is pathologically classified into several subtypes, including epithelial OC, which accounts for 85–90% of malignant ovarian tumors. Statistically, the prognosis of advanced high-grade serous OC is poor, with a 5-year overall survival rate of 27% (2). Despite current therapies, efficient treatment to prevent recurrence remains limited. Therefore, an in-depth understanding of the mechanism of OC progression will contribute to the development of novel and effective therapy strategies.
Long non-coding RNAs (lncRNAs) is a subclass of endogenous, non-protein coding RNAs, which lacks an open reading frame and usually consists of more than 200 nucleotides (3). Increasing evidence has revealed that lncRNAs are involved in various biological processes including gene transcription, genomic imprinting, transcriptional activation, epigenetic modification, and microRNAs (miRNA) regulation (4). LncRNAs have emerged as a new potential cancer treatment strategy (5). Growth arrest-specific transcript 5 (GAS5), an important lncRNA, is transcribed from the non-protein encoded small nucleolar RNA (snoRNA) gene, GAS5 (6). Altered GAS5 expression levels have been demonstrated in multiple cancers. GAS5 can regulate resistance to cisplatin by regulating microRNA-21 (miR-21) in cervical cancer (7). In prostate carcinoma, lncRNA GAS5 can target miR-103 through the protein kinase B (AKT)/mammalian target of rapamycin (mTOR) signaling pathway and inhibit the proliferation and progression of prostate cancer (8). In colorectal cancer, GAS5 snoRNAs were associated with p53 expression and closely related to the DNA damage level (9). One study also confirmed that a significantly reduced level of lncRNA GAS5 in OC is often associated with a poor prognosis (10). However, the biological mechanism of GAS5 in OC remains largely unknown.
MicroRNAs (miRNA) are a group of non-coding RNAs consisting of nearly 22 nucleotides, which can negatively regulate gene expression in a post-transcriptional manner (11,12). MiRNAs play an essential role in various biological behaviors, such as cell proliferation, autophagy, apoptosis, and differentiation (13). Also, previous studies have demonstrated the critical functions exerted by miRNAs in OC (14). Recently, studies have revealed the vital role of miR-96-5p in tumor development. For instance, miR-96-5p influences the radio-sensitivity of squamous cell carcinoma of the head and neck (HNSCC) cell lines by targeting phosphatase and tension homolog deleted on chromosome ten (PTEN) expression (15). In pancreatic cancer, miR-96-5p can inhibit Kirsten rat sarcoma viral oncogene homolog (KRAS) function, acting as a tumor suppressor.
Compared with the previous study, we not only explored the biological functions of lncRNA GAS5 in vitro but also in vivo, which was more convinced. Meanwhile, we explored a new potential molecular mechanism of lncRNA GAS5 as well. In this study, we aimed to elucidate the expression of lncRNA GAS5 in OC patients and its potential function in OC development. Furthermore, our study also revealed the mechanism of lncRNA GAS5 mediation of OC progress via the miR-96a-5p/PTEN axis. We presented the following article in accordance with the ARRIVE reporting checklist (available at https://dx.doi.org/10.21037/atm-21-6134).
Methods
Microarray analysis
Total RNA in the ovarian tissue RNA samples were hybridized to SBC Human(4*180K) ceRNA microarray (Shanghai Biotechnology Co., Shanghai, China). Quantile normalization was applied using GeneSpring GX v11.5.1 software (Agilent Technologies, USA). LncRNAs with differential expression were selected according to the criteria of change fold ≥2 and P<0.05 as previously described (16).
Online website
The online website GEPIA (http://gepia.cancer-pku.cn/detail.php) was used to compare the expression of lncRNA GAS5 between tumor tissues and normal tissues in different types of cancers. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Cell culture
Human OC cell lines HEY and SKOV3 were obtained from China Academia of Sciences (Shanghai, China). Cells were cultured in Dulbecco’s modified Eagle medium (DMEM, Gibco, USA) with 10% fetal bovine serum (FBS, HyClone, USA). All cells were cultured at 37 °C with 5% carbon dioxide (CO2) in a humidified atmosphere. When the cells adhered to the wall bottom, they were digested by 0.25% trypsin for subculturing. Cells in the logarithmic growth phase were collected for further experiments.
Cell transfection
Human GAS5 gene (NR_002578) with the BamHI and XhoI restriction site sequences was synthesized (GenePharma, Shanghai, China). After achieving retrovirus-mediated stable expression of GAS5 in SKOV3 and HEY cell lines, real-time quantitative polymerase chain reaction (real-time qPCR) was conducted to validate GAS5 overexpression in the two cell lines, and a mimic-miR-96-5p (sense, 5'-UUUGGCACUAGCACAUUUUUGCU-3'; antisense, 5'-CAAAAAUGUGCUAGUGCCAAAUU-3') was synthesized. The empty vector pcDNA3.1 and scramble small interfering RNA (siRNA) were used as control. Lipofectamine 2000 reagent (Thermo Fisher Scientific, Waltham, MA, USA) was utilized for transfection. After 24 h, cells were harvested for subsequent experiments. Real-time qPCR was used to assess transfection efficiency.
Cell counting kit-8 (CCK-8) assay
Cell proliferation was tested using a CCK-8 assay (Dojindo Molecular Technologies, Kumamoto, Japan). Briefly, OC cells were plated with 5×103 cells per well in 96-well plates. At specific time points (12, 24, 48, and 72 h), the original supernatants were removed from each well and 100 µL culture medium with 10 µL CCK-8 solution was added, after which cells were incubated at 37 °C for 2 h. Cell viability was evaluated by a microplate reader (Multiskan MK3, Thermo Scientific, USA).
Colony formation assay
A total of 1×103 HEY and SKOV3 cells were plated in six-well plates to disperse the cells evenly. Following incubation at 37 °C in 5% CO2 for 10–14 days, the culture was terminated when the colonies were visible by eye. The original medium was removed, and the cells were washed twice using phosphate-buffered saline (PBS). Subsequently, the colonies were fixed by methanol at −20 °C for 15 min and stained with crystal violet for 20 min. The staining fluids were then rinsed off. Images of the stained plates were taken under low magnification, and segments labeled on the plates were identified as scale. Colonies with a diameter larger than scale size were included for calculation.
Wound-healing assay
Cell migration ability was assessed by wound-healing assay. Briefly, 2×105 SKOV3 and HEY cells were cultured in six-well plates. After 24 h, a thin wound was made by scraping with a sterile pipette tip across the center of each well. The cells were then cultured for 24 and 48 h for observation of the wound. Five random views of each well were captured under microscope (Zeiss, Primovert, Axiocam).
Transwell assay
In total, 2×105 cells were transfected after 48 h and resuspended in serum-free DMEM. 200 mL cell suspensions were placed in the upper chambers with an 8 µm pore size polycarbonate membrane Transwell insert (Corning Costar, MD, USA) coated with 50 µL of Matrigel (1:4 dilution in a serum-free medium), and 600 mL DMEM supplemented with 10% serum as a chemotactic factor was added in the lower chambers. Following 24 h of incubation, the cells were stained with crystal violet for those across the porous membrane.
Flow cytometry
HEY and SKOV3 cells were cultured in a six-well plate with 5×106 per well at 37 °C and were classified into Overexpress GAS5 or Normal Contrast (NC) groups. Cells were then digested by 0.25% trypsin without Ethylene Diamine Tetraacetic Acid (EDTA) and resuspended in 100 µL of 1× binding buffer at a density of 1×106/mL staining with Annexin V-fluorescein isothiocyanate (FITC)/propidium iodide (PI). Fluorescence intensity was detected using a FACScan flow cytometer (BD Biosciences, San Jose, CA, USA) to determine the percentage of apoptotic cells.
Dual-luciferase reporter gene assay
Bioinformatics prediction websites were used for the possible binding sites of miR-96-5p and lncRNA GAS5. Several prediction target tools, including miRWalk3 (http://zmf.umm.uni-heidelberg.de/apps/zmf/mirwalk2/) and DIANA Tools (http://carolina.imis.athenainnovation.gr/diana_tools/web/index.php?r=site/tools), were used. The full length of GAS5 and the 3' untranslated regions (3' UTR) of PTEN were amplified separately and cloned into a pmirGLO Dual-Luciferase miRNA Target Expression Vector (Promega, USA). Phosphorylation pGAS5-MUT (mutant type) and pPTEN-MUT vectors were constructed using the pRL-TK vector (Promega, USA). A miR-96-5p mimic and miR-96-5p empty vectors were separately co-transfected with luciferase reporter gene vector into HEY and SKOV3 cells. Luciferase activity was detected using the kit (Promega, USA).
Real-time qPCR
TRIzol reagent (Invitrogen, Thermo Fisher Scientific, USA) and the Universal microRNA Purification Kit (EZBioscience, USA) were used to extract total RNA according to the manufacturers’ protocols. Next, complementary DNAs (cDNAs) were synthesized using PrimeScript RT-PCR kit (TakaraBio, Japan). PCR was conducted on an ABI 7500 Real-Time PCR System (Applied Biosystems, Foster City, USA) using SYBR Premix Ex Taq II kit (Takarabio, Japan) with a miRNA-specific 5' primer (hsa-miR-96-5p: CGGTTTGGCACTAGCACATTTTTG). The primer sequences for GAS5 were as follows: forward, 5'-GGTATGGAGAGTCGGCTTGA-3' and reverse, 5'-GCAAGACCCTTTCAAGCAGT-3'. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an internal control for microRNAs (mRNAs) with primers as follows: forward, 5'-GAAGGTGAAGGTCGGAGTC-3' and reverse, 5'-GAAGATGGTGATGGGATTTC-3'. U6 forward, 5'-CTCGCTTCGG CAGCACA-3', and reverse, 5'-AACGCTTCACGAATTTGCGT-3' was used as an internal control for miRNAs. Gene expression was analyzed using the ΔΔCq method.
RNA immunoprecipitation assays
SKOV3 cells were transfected with lentivirus (LV) LV-GAS5 and LV-NC. RNA immunoprecipitation assay (RIPA) was carried out according to the protocols. The RIPA buffer, which contained magnetic beads conjugated with immunoglobulin G (IgG) (control) and the Eukaryotic Translation Initiation Factor 2C (Ago2) antibody (Abcam, ab32381, UK), was used to obtain the cell lysate. Magna RIPA RNA-Binding Protein Immunoprecipitation Kit (EMD Millipore, USA) was applied for RNA immunoprecipitation. RNAs were then isolated and quantified using Qubit 3.0 (Life Technologies, US). The co-precipitated RNAs were detected by qPCR and were performed to examine the expression levels of lncRNA GAS5 and PTEN.
Western blot
Total protein was extracted and separated using 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and transferred to polyvinylidene fluoride (PVDF) membranes (Millipore, USA). Subsequently, the membranes were then blocked in 5% skimmed milk, and incubated with primary antibodies at 4 °C overnight. After blocking with 5% bovine serum albumin (BSA) (Sigma-Aldrich, USA) at 37 °C for 1 h, the membrane was incubated with primary antibodies of PTEN (1:1,000; cat. no. 9188T; Cell Signaling Technology, USA), AKT (1:1,000; cat. no. 4691T; Cell Signaling Technology, USA), p-AKT (1:2,000; cat. no. 4060T; Cell Signaling Technology, USA), N-Cadherin (1:1,000; cat. no. 13116T; Cell Signaling Technology, USA), Phosphatidylinositol 3-kinase (PI3K) PI3K-p85 (1:1,000; cat. no. 173665; Cell Signaling Technology, USA), Cleaved Caspase-3 (1:1,000; cat. no. 9664T; Cell Signaling Technology, USA), Snail (1:1,000; cat. no. 3897T; Cell Signaling Technology; USA), p-mTOR (1:1,000; cat. no. 5536T; Cell Signaling Technology, USA), and β-actin (1:500; cat. no. F3002; Sigma-Aldrich, USA) at 4 °C overnight. Following incubation with a horseradish peroxidase (HRP)-conjugated goat anti-rabbit (1:5,000; cat. no. ab205718; Abcam, USA) or goat anti-mouse (1:5,000; cat. no. ab205719; Abcam) secondary antibody at 4 °C for 2 h, densitometric analysis was performed using ImageJ v2.4.1 (National Institutes of Health, Shanghai, China), with the protein expression levels normalized to β-actin.
Tumorigenicity assays in nude mice
Experiments were performed under a project license (No. [2018]114) granted by the Medical Research Ethics Committee of Renji Hospital, School of Medicine, Shanghai Jiao Tong University, in compliance with Institutional Animal Care and Use Committee(IACUC) guidelines for the care and use of animals. Briefly, HEY cells were stably transfected with different vectors (Lenti-GAS5 overexpression or Lenti-control) for 72 h. The cells were then harvested for the injection. Four to six weeks old nude mice were randomly divided into two groups [NC group and GAS5 group (n=6 in each group)] and injected intraperitoneally with approximately 6×106 cells diluted in 200 µL PBS at the right flanks. Tumors were evaluated every week for 5 weeks. The observation was then ceased and the mice were executed at 35 days, and tumor development was monitored. The difference in the number of visible tumors and the weight between the two groups and tumor sizes were measured. The protein expression levels in the tumors of both groups were also detected though immunohistochemistry.
Immunohistochemistry
Sections were prepared after tissues from nude mice were embedded in paraffin. PTEN expression was detected by staining. Slides were observed under the microscope (400×) (Olympus, Japan), and the results were graded by the percentage of positive cells.
RNA-fluorescence in situ hybridization (FISH)
Cy3-labeled GAS5 and diamidino-2-phenylindole (DAPI)-labeled U6 probes were customized from RiboBio (Guangzhou, China). RNA FISH were conducted using a fluorescent in situ hybridization kit according to the manufacturer’s instructions.
Statistical analysis
Statistical analysis was performed using GraphPad Prism 8.0 software (GraphPad Software, San Diego, CA, USA). Quantitative results were expressed as the mean ± standard deviation (SD) and were compared using Student’s t-test in two groups. One-way analysis of variance (ANOVA) was used to compare in three or more groups. P values less than 0.05 were considered statistically significant.
Results
Overexpression of GAS5 inhibited the proliferation of OC cells in vivo
The microarray results [three epithelial ovarian cancer (EOC) samples vs. three normal samples] demonstrated that the lncRNA GAS5 expression level was significantly lower in cancer samples. Real-time qPCR also revealed that GAS5 had a markedly lower expression in 53 EOC samples compared to 10 normal ovarian samples. Similarly, in SKOV3 cell lines, GAS5 expression was obviously reduced between OC and normal human ovarian epithelial cell lines. Conversely, GAS5 expression was positively associated with the International Federation of Gynecology and Obstetrics (FIGO) stage and histological type (data not shown) (16). The present results are consistent with those of the Gene Expression Profiling Interactive Analysis (GEPIA) database. Herein, GAS5 was down-regulated in most tumors, including OC (Figure 1A). Following transfection with the GAS5 expression vector, real-time qPCR exhibited overexpression of GAS5 in SKOV3 and HEY cell lines (Figure 1B,1C). Figure 1D displays GAS5 overexpression in cells transfected with the GAS5 expression vector rather than in those transfected with the empty vector. Moreover, in a mouse xenograft model, we investigated the growth inhibition effects of GAS5 in vivo. As shown in Figure 1E-1G, the number and weight of tumors were significantly decreased in the GAS5 overexpression group compared with control group 5 weeks after injection. Nevertheless, there was no difference in weight between these two groups (Figure 1H).
Figure 1.
The lncRNA GAS5 is down-regulated in OC and inhibits OC tumor growth in mouse xenograft models. (A) GAS5 expression in different cancer types from the GEPIA database. (B,C) LncRNA GAS5 expression after transfection of LV-GAS5 or empty LV vector plasmids into HEY or SKOV3 cells by real-time qPCR. (D) Stable transfection of mutation type GAS5 (GAS5 Mut) plasmid (magnification: 100×) GFP fluorescence of LV-GAS5 or empty LV vector plasmids transfected into HEY or SKOV3 cells was observed by fluorescence microscope. (E) Images of disseminated tumors in intraperitoneal OC xenograft model treated with vehicle NC buffer or LV-GAS5. (F,G) Whole tumor number and weight in intraperitoneal OC xenograft model treated with vehicle NC buffer (n=6) or LV-GAS5 (n=6). (H) Body weight was monitored during the detectable period in nude mice. **, P<0.01. lncRNA, long non-coding RNA; GAS5, growth arrest-specific transcript 5; OC, ovarian cancer.
GAS5 suppressed the proliferation and migration ability of OC cells in vitro
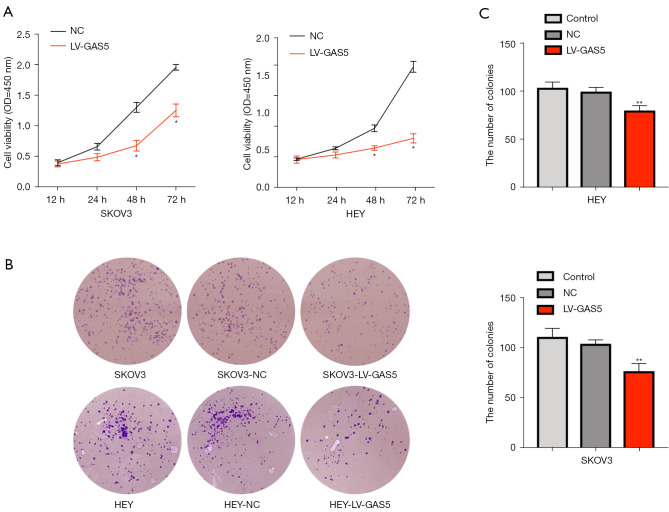
Viability of HEY and SKOV3 cell lines were assessed after LV-GAS5 transduction (P<0.05) by CCK-8 assay (Figure 2A). The colony formation assay revealed that GAS5 overexpression significantly reduced the proliferation of HEY and SKOV3 cells in vitro (P<0.01) (Figure 2B,2C) The wound-healing assay also showed that overexpression of GAS5 could decrease the cell migratory capacity both in HEY and SKOV3 cells (Figure 3A,3B). The results of the transwell migration assay suggested that GAS5 overexpression decreased the migration ability of OC cells in vitro (Figure 3C,3D).
Figure 2.
The lncRNA GAS5 suppresses the proliferation ability of HEY and SKOV3 cells. (A) The viability of HEY and SKOV3 cell lines in the LV-GAS5 and NC groups was assessed by CCK-8. (B,C) Proliferation of HEY and SKOV3 cell lines were compared between LV-GAS5 and NC groups (magnification: 100×, crystal violet staining). The experiments were repeated three times. *, P<0.05; **, P<0.01. lncRNA, long non-coding RNA; GAS5, growth arrest-specific transcript 5.
Figure 3.
The lncRNA GAS5 suppresses HEY and SKOV3 cell migration and invasion ability. (A) The migration ability of HEY and SKOV3 cells was detected via the wound-healing test after transfection between LV-GAS5 and NC groups (magnification: 100×, crystal violet staining). (B) The wound-healing rate in HEY and SKOV3 cells was evaluated. (C) The cell invasion ability of HEY and SKOV3 cells was measured in the GAS5 and NC groups (magnification: 200×, crystal violet staining). (D) The number of invasive HEY and SKOV3 cells was evaluated following transfection in the LV-GAS5 and NC groups. The data were presented as the mean ± SD and analyzed using one-way ANOVA. The experiment was independently repeated three times. **, P<0.01; ***, P<0.001. lncRNA, long non-coding RNA; GAS5, growth arrest-specific transcript 5; SD, standard deviation; ANOVA, analysis of variance.
LncRNA GAS5 influenced PTEN expression by competitively binding to miR-96-5p
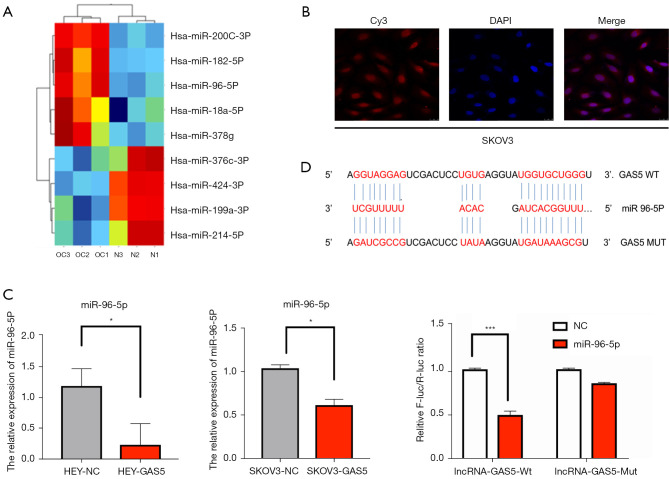
To explore the potential molecular mechanism of GAS5 in OC progression, we investigated the related recruitment of miRNAs. The microarray results (three EOC samples vs. three normal samples) demonstrated that the lncRNA GAS5 expression was markedly decreased in cancer samples in our previous research. At the same time, miRNAs were involved in the occurrence of OC cells, as shown in our results of Agilent ceRNA in heatmaps, which showed the top nine miRNAs (Figure 4A).
Figure 4.
The lncRNA GAS5 sponges miR-96-5p by direct targeting. (A) Heatmap showing the hierarchical clustering of differentially-expressed miRNAs in patients and normal controls. Red: up-regulated genes; green: down-regulated genes. (B) The FISH assay revealed that lnc-GAS5 was localized in the cytoplasm and the nucleus of SKOV3 cells. (C) MiR-96-5p expression was detected by real-time qPCR in HEY and SKOV3 cell lines transfected with LV-GAS5 and NC. (D) The miR-96-5p binding site in lnc-GAS5 3' UTR. The binding relationship between GAS5 and miR-96-5p was detected using the luciferase reporter assay with NC or Mut luciferase reporter plasmids of GAS5. *, P<0.05; ***, P<0.001. lncRNA, long non-coding RNA; GAS5, growth arrest-specific transcript 5; FISH, fluorescence in situ hybridization; qPCR, quantitative reverse transcription polymerase chain reaction.
Bioinformatics analysis indicated that GAS5 possesses the conserved target site of miR-96-5p, with a high affinity score (http://carolina.imis.athenainnovation.gr/diana_tools/web/index.php?r=site/tools). FISH images indicated that the lncRNA GAS5 was located in the cytoplasm and the nucleus of the SKOV3 (Figure 4B). Furthermore, real-time qPCR showed the expression level of miR-96-5p in the empty vector and GAS5 groups. We found that the expression of miR-96-5p was significantly lower in GAS5 overexpression cell lines than those in the normal OC cell line (P<0.05, Figure 4C).
Subsequently, a dual-luciferase reporter gene assay was applied to confirm the relationship between lncRNA GAS5 and miR-96-5p. Compared with the empty vector group, the luciferase activity of the GAS5 WT-binding site was inhibited by miR-96-5p (P<0.001), while the GAS5 MUT-binding site was not affected (P>0.05). MiR-96-5p could specifically bind to the lncRNA GAS5 (Figure 4D). These results indicated that GAS5 functioned as a ceRNA of miR-96-5p.
GAS5 and miR-96-5p co-regulated the proliferation and migration of OC cells
MiR-96-5p mimics were generated to determine whether miR-96-5p or lncRNA GAS5 influences OC cell proliferation and migration. QPCR results showed that the expression of miR-96-5p was enhanced under the influence of the miR-96-5p mimic (P<0.001, Figure S1). CCK-8 assay was performed to analyze the effects exerted by GAS5 and miR-96-5p on the proliferation of OC cells. Compared with the NC group, GAS5 up-regulation and miR-96-5p down-regulation deterred the proliferation of OC cells (P<0.05, Figure 5A,5B). The scratch test revealed that compared to the control group in HEY cell lines, the cellular migration ability was enhanced in the miR-96-5p mimic groups (P<0.01), while that of the miR-96-5p inhibitor groups declined (P<0.01). It was concluded that down-regulated GAS5 expression and up-regulated miR-96-5P expression could both induce strong migration ability of OC cells (P<0.001). Furthermore, up-regulated GAS5 expression and down-regulated miR-96-5P expression retrained the migration ability (P<0.001, Figure 5C). Similar results were obtained in the SKOV3 cell line (Figure 5D). The colony formation assay was applied to analyze the effects exerted by GAS5 and miR-96-5p on the proliferation of OC cells. GAS5 up-regulation and miR-96-5p down-regulation also deterred the proliferation of OC cells compared with the NC group (P<0.01; P<0.001) (Figure 6A,6B).
Figure 5.
The lncRNA GAS5 inhibits HEY and SKOV3 cell viability and migration ability via modulation of miR-96-5p. (A,B) The viability of HEY and SKOV3 cell lines was assessed with in the LV-GAS5, miR-96-5p mimic, miR-96-5p inhibitor, and NC groups. (C,D) A wound-healing test was conducted to examine the migration of HEY and SKOV3 cells in the three groups at 24 and 48 h following transfection (magnification: 100×, crystal violet staining). The data were presented as the mean ± SD and analyzed using one-way ANOVA. The experiment was independently repeated three times. *, P<0.05; **, P<0.01; ***, P<0.001. lncRNA, long non-coding RNA; GAS5, growth arrest-specific transcript 5; SD, standard deviation; ANOVA, analysis of variance.
Figure 6.
lncRNA GAS5 inhibits HEY and SKOV3 cell proliferation ability via modulation of miR-96-5p. (A,B) The proliferation of HEY and SKOV3 cell lines were assessed in the LV-GAS5, miR-96-5p mimic, miR-96-5p inhibitor, and NC groups (magnification: 100×, crystal violet staining). The data were presented as the mean ± SD and analyzed using one-way ANOVA. The experiment was independently repeated three times. *, P<0.05; **, P<0.01; ***, P<0.001. lncRNA, long non-coding RNA; GAS5, growth arrest-specific transcript 5; SD, standard deviation; ANOVA, analysis of variance.
LncRNA GAS5 and miR-96-5p repressed the invasion ability of OC cells
Using the transwell assay, we observed the influence of OC cell invasion by miR-96-5p or the lncRNA GAS5. As shown in Figure 7A-7D, the invasion ability of HEY cells in the control group was not significantly influenced (P>0.05), while in the oe-lncRNA GAS5 and miR-96-5p inhibitor groups, the invasion ability was inhibited (P<0.05). Compared with oe-lncRNA GAS5 and miR-96-5p inhibitor groups, the invasion ability increased in the oe-lncRNA GAS5 + miR-96-5p mimic group and miR-96-5p mimic group (all P<0.05). Similar results were also observed in the SKOV3 cell line (Figure 7A-7D).
Figure 7.
The lncRNA GAS5 inhibits HEY and SKOV3 cells invasion ability via modulation of miR-96-5p. (A) Transwell assay of HEY and SKOV3 cells in the LV-GAS5 and miR-96-5p mimic groups after transfection (magnification: 200×, crystal violet staining). (B) The number of invasive HEY and SKOV3 cells in the LV-GAS5 and miR-96-5p mimic groups after transfection. (C) Transwell assay of HEY and SKOV3 cells in the miR-96-5p inhibitor group after transfection (magnification: 100×, crystal violet staining). (D) The number of invasive HEY and SKOV3 cells in the LV-GAS5 and miR-96-5p inhibitor groups following transfection. The data were presented as the mean ± SD and analyzed using one-way ANOVA. The experiment was independently repeated three times. *, P<0.05. lncRNA, long non-coding RNA; GAS5, growth arrest-specific transcript 5; SD, standard deviation; ANOVA, analysis of variance.
Up-regulation of the lncRNA GAS5 increased the PTEN protein level but inhibited the phosphorylation of AKT
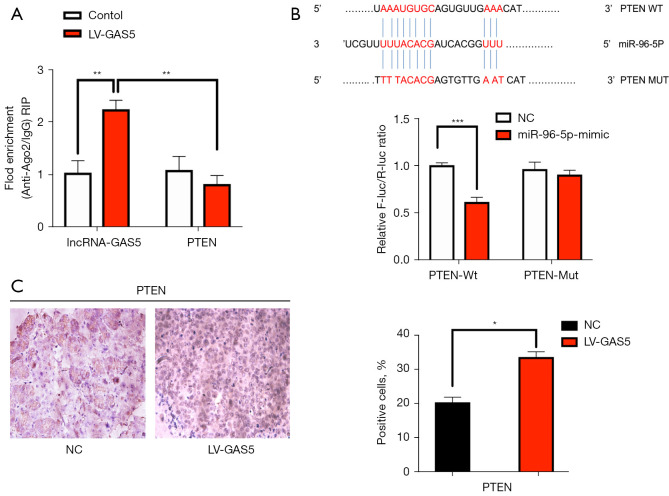
First, we performed RIPA based on Ago2. We overexpressed the lncRNA GAS5 in SKOV3 cells then pulled down Ago2 using an anti-Ago2 antibody. Overexpression of GAS5 can result in a significant decrease in the enrichment of PTEN transcripts pulled down by Ago2 (P<0.01, Figure 8A), indicating the existence of miRNA-bound PTEN transcripts. As the PTEN/AKT/mTOR pathway is essential for cell proliferation and invasion (17), we investigated the correlations of GAS5, miR-96-5p, and the PTEN/AKT/mTOR pathway in OC cells.
Figure 8.
PTEN expression level was correlated with GAS5 expression. (A) qPCR results of the RIPA showed lncRNA GAS5 could compete with PTEN by binding to the Ago2 protein. (B) The binding relationship between PTEN and miR-96-5p was detected by luciferase reporter assay with WT or Mut luciferase reporter plasmids of PTEN. (C) PTEN expression was detected in tissues with and without GAS5 overexpression by immunohistochemical staining (magnification: 100×). PTEN, phosphatase and tensin homolog deleted on chromosome ten; GAS5, growth arrest-specific transcript 5; qPCR, quantitative reverse transcription polymerase chain reaction; RIPA, RNA immunoprecipitation assay; lncRNA, long non-coding RNA. *, P<0.05; **, P<0.01; ***, P<0.001.
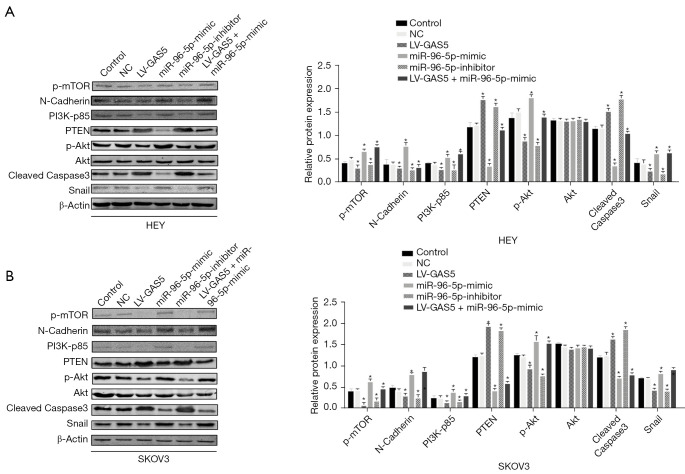
Secondly, bioinformatics analysis showed that there were specific binding sites between the sequences of miR-96-5p and 3' UTR of PTEN. We also found that the luciferase activity of the PTEN-3' UTR (WT) was significantly inhibited by miR-96-5p (P<0.05), whereas the activity of PTEN-3' UTR (MUT) was not affected (P>0.05), indicating that miR-96-5p specifically bound to PTEN-3' UTR and reduced the expression of PTEN after transcription (Figure 8B). Next, we compared the PTEN expression in tumor tissues of nude mice by immunohistochemical staining. The results showed that the proportion of positive cells in the GAS5 overexpression group were lower than that of the NC group in Figure 8C (P<0.05). Western blot examined the protein expression of the PTEN pathway in the HEY and SKOV3 cell lines. Compared with the control group, the protein expression of PTEN did not show notable differences in the empty vector group (all P>0.05). In contrast to the control group, the oe-lncRNA GAS5 group and miR-96-5p inhibitor group showed increased protein expression of PTEN, but decreased phosphorylation of AKT, N-cadherin, Snail, PI3K-p85, and P-mTOR (all P<0.05). Compared to the oe-lncRNA GAS5 group and miR-96-5p inhibitor groups, the protein expression of PTEN was lower, while the phosphorylation of AKT, N-cadherin, Snail, PI3K-p85, and mTOR were higher in the oe-lncRNA GAS5 + miR-96-5p mimic group (all P<0.05). However, the protein expression of PTEN did not differ significantly between the oe-lncRNA GAS5 group and miR-96-5p inhibitor group (all P>0.05). The protein expression of AKT remained nearly identical between all groups in the HEY and SKOV3 cell lines (Figure 9A,9B).
Figure 9.
The lncRNA GAS5 increases the protein levels of PTEN and cleaved caspase-3, but decreases the phosphorylation of AKT, mTOR, N-cadherin, and Snail. (A) The protein bands of P-mTOR, N-cadherin, PI3K-p85, PTEN, AKT, p-AKT, cleaved caspase-3, Snail, and β-actin in HEY cells were detected by western blot assay. (B) The protein bands of P-mTOR, N-cadherin, PI3K-p85, PTEN, AKT, P-AKT, cleaved caspase-3, Snail, and β-actin in SKOV3 cells were detected by western blot. The data were presented as the mean ± SD and analyzed using one-way ANOVA. The experiment was independently repeated three times. *, P<0.05. lncRNA, long non-coding RNA; GAS5, growth arrest-specific transcript 5; PTEN, phosphatase and tensin homolog deleted on chromosome ten; SD, standard deviation; ANOVA, analysis of variance.
Discussion
An increasing number of studies have confirmed the important role of lncRNAs in numerous biological processes. Aberrantly expressed lncRNAs has been discovered and investigated in the pathogenesis and development of OC. Our study showed that the lncRNA GAS5 can inhibit OC cell proliferation and promote apoptosis by negatively targeting the miR-96-5p/PTEN axis.
GAS5 expression has been examined in various cancers including breast, ovarian, and cervical carcinomas (16,18-20). A previous study confirmed that GAS5 expression in human tumor cells is relatively lower than that in the normal tissues (21), which is consistent with the speculation that GAS5 exerts an anti-proliferative, tumor-suppressing function. The lncRNA GAS5 is involved in cell proliferation, drug resistance, tumor cell invasion, apoptosis, and autophagy. However, the underlying mechanism remains unclear. This study focused on the biological role of the lncRNA GAS5 in OC.
First, we observed that GAS5 overexpression could suppress OC cell proliferation and increase cell apoptosis, which is consistent with the previously reported function of GAS5 in other carcinomas, such as pancreatic and breast cancers (22). CeRNA, including lncRNA, binds to miRNA and serves as a “sponge” to absorb miRNA in cells. It is well-known that miRNA can bind to the 3' UTRs of the mRNAs of target gene, thereby regulating their expressions. As an effective miRNA sponge, lncRNAs are primarily located in the cytoplasm (23,24). We isolated RNA from the nuclei and cytoplasms of SKOV3 cells. The FISH assay revealed lncRNA GAS5 up-regulation in the cytoplasm, suggesting that the lncRNA GAS5 could play a regulatory role in a ceRNA-dependent manner. Therefore, we speculated that GAS5 could bind to miRNAs to exert its function. Previous reports have also confirmed this hypothesis; for example, GAS5 can positively regulate miR-222 in colorectal cancers (25).
Other lncRNA, like PTAR, can also bind to miR-101-3p to regulate zinc finger E-box-binding homeobox 1 (ZEB1) expression, thus promoting epithelial-mesenchymal transition (EMT) and invasion in OC (26). So, we investigated the related recruitment of miRNAs in OC. From the top miRNAs listed here from our own data, we observed that GAS5 possesses the conserved target site of miR-96-5p with a high affinity score after bioinformatics analysis. MiR-96-5P is a vital miRNA in cancer development. The overexpression of miR-96-5P has been discovered in HNSCC (15), and the lncRNA GAS5 acts as a competing endogenous RNA of miR-96-5p in renal tubular epithelial fibrosis (27). Next, from the results of the luciferase reporter assay, we confirmed the interaction between lncRNA GAS5 and miR-96-5p. The results showed that the binding of the lncRNA GAS5 to miR-96-5p was sequence-specific. MiRNA regulates the stability and translation of mRNA at the post-transcriptional level (28). Overall, our results suggested that GAS5 could exert its biological functions by directly binding to miR-96-5p.
PTEN is a tumor-suppressor gene that is frequently deleted or mutated in various human cancers (29), and can be regulated by the PI3K/AKT axis. Up-regulation of PTEN can reduce the generation of reactive oxygen species in cells (30). Furthermore, PTEN might be a potential target of miR-96-5p, as predicted by bioinformatics analysis. PTEN can also influence the progression of OC (31,32). Increasing evidence indicates that miRNA is vital during the pathogenesis of tumors, and can affect the invasion and proliferation of tumors by regulating the PI3K/AKT signing pathway (33). For example, the lamin A/C protein can promote prostate cancer cell growth, migration, and invasion through the PI3K/AKT/mTOR pathway (34). Moreover, in traumatic brain injury, the PI3K/AKT/mTOR pathway contributes to neuroprotection through PPAR (35). Inactivation of PTEN is positively correlated with the aggressive capacity of OC, whereas PTEN plays a tumor-suppressive role in OC (36,37). Our study initially identified an inverse relationship between miR-96-5p and PTEN expression levels in OC lines through bioinformatics analysis. Dual-luciferase reporter assay also confirmed the interaction between miR-96-5p and PTEN. Therefore, we can infer that miR-96-5p is at least partially responsible for oncogenic role of PTEN. Moreover, miR-96-5p could inhibit proliferation proteins by targeting PTEN.
The relationship between lncRNA GAS5 and PTEN was further testified by western blot assay. The rescuing experiment results showed that the lncRNA GAS5 up-regulated PTEN expression and suppressed cell proliferation and migration-related proteins by acting as a miR-96-5p sponge. Mice models overexpressing the lncRNA GAS5 were established by intraperitoneal injection of transfected GAS5 OC. We found that overexpression of lncRNA GAS5 significantly inhibited tumor metastasis and growth, as well as PTEN expression in tumor tissues. It has been reported that GAS5 inhibits the proliferation, migration, invasion, and epithelial stromal transformation of oral squamous cell carcinoma (OSCC) by regulating the miR21/PTEN axis (38). In the present study, the activation of PTEN/AKT/mTOR was enhanced when GAS5 was overexpressed. These results demonstrate that GAS5 can regulate the PTEN/AKT/mTOR pathway in OC.
From our study, we conclude that the lncRNA GAS5 can regulate the PTEN/AKT/mTOR axis by sponging miR-96-5p in OC progression as a ceRNA. Although we demonstrated the role of the AKT/PTEN signaling pathway in OC cells, the mechanism underlying the activation of the AKT/PTEN pathway by the lncRNA GAS5 and miR-96-5p has not been completely elucidated; therefore, further investigations are needed.
Supplementary
The article’s supplementary files as
Acknowledgments
We thank the Department of Obstetrics and Gynecology, Ren Ji Hospital, School of Medicine, Shanghai Jiao Tong University for their support. The authors are grateful to all staff members who contributed to this study.
Funding: This work was supported by the National Key Research and Development Program (No. 863) of China (No. 2016YFC1302900), the National Natural Science Foundation of China (No. 81772770), the Science and Technology Commission of Shanghai Municipality (No. 18441904800), the Shanghai Municipal Commission of Health and Family Planning (No. 2017ZZ02016 and ZY (2018-2020)-FWTX-3006), and the Shanghai Municipal Key Clinical Specialty and the Program of Shanghai Hospital Development Center (No. 16CR2001A).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Experiments were performed under a project license (No. [2018]114) granted by the Medical Research Ethics Committee of Renji Hospital, School of Medicine, Shanghai Jiao Tong University, in compliance with Institutional Animal Care and Use Committee (IACUC) guidelines for the care and use of animals.
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at https://dx.doi.org/10.21037/atm-21-6134
Data Sharing Statement: Available at https://dx.doi.org/10.21037/atm-21-6134
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/atm-21-6134). The authors have no conflicts of interest to declare.
(English Language Editor: A. Kassem)
References
- 1.Jayson GC, Kohn EC, Kitchener HC, et al. Ovarian cancer. Lancet 2014;384:1376-88. 10.1016/S0140-6736(13)62146-7 [DOI] [PubMed] [Google Scholar]
- 2.Torre LA, Trabert B, DeSantis CE, et al. Ovarian cancer statistics, 2018. CA Cancer J Clin 2018;68:284-96. 10.3322/caac.21456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet 2014;15:7-21. 10.1038/nrg3606 [DOI] [PubMed] [Google Scholar]
- 4.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet 2009;10:155-9. 10.1038/nrg2521 [DOI] [PubMed] [Google Scholar]
- 5.Chen G, Wang Z, Wang D, et al. LncRNADisease: a database for long-non-coding RNA-associated diseases. Nucleic Acids Res 2013;41:D983-6. 10.1093/nar/gks1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith CM, Steitz JA. Classification of gas5 as a multi-small-nucleolar-RNA (snoRNA) host gene and a member of the 5'-terminal oligopyrimidine gene family reveals common features of snoRNA host genes. Mol Cell Biol 1998;18:6897-909. 10.1128/MCB.18.12.6897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wen Q, Liu Y, Lyu H, et al. Long Noncoding RNA GAS5, Which Acts as a Tumor Suppressor via microRNA 21, Regulates Cisplatin Resistance Expression in Cervical Cancer. Int J Gynecol Cancer 2017;27:1096-108. 10.1097/IGC.0000000000001028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xue D, Zhou C, Lu H, et al. LncRNA GAS5 inhibits proliferation and progression of prostate cancer by targeting miR-103 through AKT/mTOR signaling pathway. Tumour Biol 2016. [Epub ahead of print]. 10.1007/s13277-016-5429-8 [DOI] [PubMed] [Google Scholar]
- 9.Krell J, Frampton AE, Mirnezami R, et al. Growth arrest-specific transcript 5 associated snoRNA levels are related to p53 expression and DNA damage in colorectal cancer. PLoS One 2014;9:e98561. 10.1371/journal.pone.0098561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gao J, Liu M, Zou Y, et al. Long non-coding RNA growth arrest-specific transcript 5 is involved in ovarian cancer cell apoptosis through the mitochondria-mediated apoptosis pathway. Oncol Rep 2015;34:3212-21. 10.3892/or.2015.4318 [DOI] [PubMed] [Google Scholar]
- 11.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004;116:281-97. 10.1016/S0092-8674(04)00045-5 [DOI] [PubMed] [Google Scholar]
- 12.Lim LP, Lau NC, Garrett-Engele P, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 2005;433:769-73. 10.1038/nature03315 [DOI] [PubMed] [Google Scholar]
- 13.Farazi TA, Hoell JI, Morozov P, et al. MicroRNAs in human cancer. Adv Exp Med Biol 2013;774:1-20. 10.1007/978-94-007-5590-1_1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deb B, Uddin A, Chakraborty S. miRNAs and ovarian cancer: An overview. J Cell Physiol 2018;233:3846-54. 10.1002/jcp.26095 [DOI] [PubMed] [Google Scholar]
- 15.Vahabi M, Pulito C, Sacconi A, et al. miR-96-5p targets PTEN expression affecting radio-chemosensitivity of HNSCC cells. J Exp Clin Cancer Res 2019;38:141. 10.1186/s13046-019-1119-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Long X, Song K, Hu H, et al. Long non-coding RNA GAS5 inhibits DDP-resistance and tumor progression of epithelial ovarian cancer via GAS5-E2F4-PARP1-MAPK axis. J Exp Clin Cancer Res 2019;38:345. 10.1186/s13046-019-1329-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carnero A, Blanco-Aparicio C, Renner O, et al. The PTEN/PI3K/AKT signalling pathway in cancer, therapeutic implications. Curr Cancer Drug Targets 2008;8:187-98. 10.2174/156800908784293659 [DOI] [PubMed] [Google Scholar]
- 18.Du Z, Fei T, Verhaak RG, et al. Integrative genomic analyses reveal clinically relevant long noncoding RNAs in human cancer. Nat Struct Mol Biol 2013;20:908-13. 10.1038/nsmb.2591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pickard MR, Williams GT. Regulation of apoptosis by long non-coding RNA GAS5 in breast cancer cells: implications for chemotherapy. Breast Cancer Res Treat 2014;145:359-70. 10.1007/s10549-014-2974-y [DOI] [PubMed] [Google Scholar]
- 20.Li J, Huang H, Li Y, et al. Decreased expression of long non-coding RNA GAS5 promotes cell proliferation, migration and invasion, and indicates a poor prognosis in ovarian cancer. Oncol Rep 2016;36:3241-50. 10.3892/or.2016.5200 [DOI] [PubMed] [Google Scholar]
- 21.Pickard MR, Williams GT. Molecular and Cellular Mechanisms of Action of Tumour Suppressor GAS5 LncRNA. Genes (Basel) 2015;6:484-99. 10.3390/genes6030484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu B, Wu S, Ma J, et al. lncRNA GAS5 Reverses EMT and Tumor Stem Cell-Mediated Gemcitabine Resistance and Metastasis by Targeting miR-221/SOCS3 in Pancreatic Cancer. Mol Ther Nucleic Acids 2018;13:472-82. 10.1016/j.omtn.2018.09.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu C, Zhang Y, Wang Q, et al. Long non-coding RNA GAS5 controls human embryonic stem cell self-renewal by maintaining NODAL signalling. Nat Commun 2016;7:13287. 10.1038/ncomms13287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dong H, Hu J, Zou K, et al. Activation of LncRNA TINCR by H3K27 acetylation promotes Trastuzumab resistance and epithelial-mesenchymal transition by targeting MicroRNA-125b in breast Cancer. Mol Cancer 2019;18:3. 10.1186/s12943-018-0931-9 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 25.Zhao X, Wang P, Liu J, et al. Gas5 Exerts Tumor-suppressive Functions in Human Glioma Cells by Targeting miR-222. Mol Ther 2015;23:1899-911. 10.1038/mt.2015.170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liang H, Yu T, Han Y, et al. LncRNA PTAR promotes EMT and invasion-metastasis in serous ovarian cancer by competitively binding miR-101-3p to regulate ZEB1 expression. Mol Cancer 2018;17:119. 10.1186/s12943-018-0870-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang W, Jia YJ, Yang YL, et al. LncRNA GAS5 exacerbates renal tubular epithelial fibrosis by acting as a competing endogenous RNA of miR-96-5p. Biomed Pharmacother 2020;121:109411. 10.1016/j.biopha.2019.109411 [DOI] [PubMed] [Google Scholar]
- 28.Thivierge C, Tseng HW, Mayya VK, et al. Alternative polyadenylation confers Pten mRNAs stability and resistance to microRNAs. Nucleic Acids Res 2018;46:10340-52. 10.1093/nar/gky666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lei L, He L, Chen K, et al. The expression of SOX9, Tiam1, and PTEN is correlated with angiogenesis and prognosis in gastric cancer. Transl Cancer Res 2020;9:3998-4004. 10.21037/tcr-20-2071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu J, Tian W, Ma X, et al. The molecular mechanism underlying morphine-induced Akt activation: roles of protein phosphatases and reactive oxygen species. Cell Biochem Biophys 2011;61:303-11. 10.1007/s12013-011-9213-5 [DOI] [PubMed] [Google Scholar]
- 31.Jang H, Lee OH, Lee Y, et al. Melatonin prevents cisplatin-induced primordial follicle loss via suppression of PTEN/AKT/FOXO3a pathway activation in the mouse ovary. J Pineal Res 2016;60:336-47. 10.1111/jpi.12316 [DOI] [PubMed] [Google Scholar]
- 32.Martins FC, Santiago Id, Trinh A, et al. Combined image and genomic analysis of high-grade serous ovarian cancer reveals PTEN loss as a common driver event and prognostic classifier. Genome Biol 2014;15:526. 10.1186/s13059-014-0526-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shao S, Wang C, Wang S, et al. LncRNA STXBP5-AS1 suppressed cervical cancer progression via targeting miR-96-5p/PTEN axis. Biomed Pharmacother 2019;117:109082. 10.1016/j.biopha.2019.109082 [DOI] [PubMed] [Google Scholar]
- 34.Kong L, Schäfer G, Bu H, et al. Lamin A/C protein is overexpressed in tissue-invading prostate cancer and promotes prostate cancer cell growth, migration and invasion through the PI3K/AKT/PTEN pathway. Carcinogenesis 2012;33:751-9. 10.1093/carcin/bgs022 [DOI] [PubMed] [Google Scholar]
- 35.Kitagishi Y, Matsuda S. Diets involved in PPAR and PI3K/AKT/PTEN pathway may contribute to neuroprotection in a traumatic brain injury. Alzheimers Res Ther 2013;5:42. 10.1186/alzrt208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Patch AM, Christie EL, Etemadmoghadam D, et al. Whole-genome characterization of chemoresistant ovarian cancer. Nature 2015;521:489-94. 10.1038/nature14410 [DOI] [PubMed] [Google Scholar]
- 37.Russo A, Czarnecki AA, Dean M, et al. PTEN loss in the fallopian tube induces hyperplasia and ovarian tumor formation. Oncogene 2018;37:1976-90. 10.1038/s41388-017-0097-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zeng B, Li Y, Jiang F, et al. LncRNA GAS5 suppresses proliferation, migration, invasion, and epithelial-mesenchymal transition in oral squamous cell carcinoma by regulating the miR-21/PTEN axis. Exp Cell Res 2019;374:365-73. 10.1016/j.yexcr.2018.12.014 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as