Abstract
tRNA intergenic spacer PCR (tDNA-PCR) was evaluated for its usefulness in the differentiation of enterococcal species of human and animal origin. This technique was carried out for 124 strains belonging to 17 enterococcal species and generated DNA fragments, which were separated by capillary electrophoresis. tDNA-PCR enabled us to discriminate for all species tested. Enterococcus faecium showed minor but reproducible differences with Enterococcus durans, while Enterococcus hirae was easily distinguishable. Enterococcus avium, Enterococcus malodoratus, and Enterococcus raffinosus generated highly similar though distinctive patterns.
Enterococci are regarded as one of the leading causes of nosocomial infections (11), and cases of endocarditis, bacteremia, urinary tract infection, and neonatal sepsis have frequently been reported. Several authors have highlighted the need for rapid and accurate identification of enterococcal strains (3, 6, 7, 21). Although Enterococcus faecalis and Enterococcus faecium are responsible for about 95% of all nosocomial infections caused by enterococci, most of the described species have been encountered in human infections (20).
The increasing occurrence of antibiotic resistance, for instance to β-lactam antibiotics and more recently to glycopeptides, has caused great concern (11). Some species, such as E. faecium, are likelier to be more resistant to antimicrobial agents than are others, and Enterococcus gallinarum, Enterococcus casseliflavus, and Enterococcus flavescens show intrinsic low-level resistance to glycopeptide antibiotics. Rapid species identification therefore can be of substantial help in the choice of antibiotic therapy (15).
In the study of outbreaks it is helpful to know that the isolates involved are all the same species of enterococci (1, 5, 23). Fingerprinting techniques acting at infraspecies level, such as restriction analysis in combination with pulsed-field gel electrophoresis, can then be applied (3).
Sequencing of conserved regions, such as the 16S rRNA gene (16) and the sodA gene encoding superoxide dismutase (18), is useful in the identification of enterococci. Several other molecular identification methods have recently been evaluated (3, 6, 14, 16, 19, 24, 29). PCR assays using genes involved in peptidoglycan synthesis (d-alanine:d-alanine ligase genes) and in vancomycin resistance (vanC-1, vanC-2/3) have also been used for identification, classification, or detection of enterococci (7, 9, 17).
tRNA intergenic spacer PCR (tDNA-PCR) (26) has been applied for the species differentiation of streptococci (27), Acinetobacter spp. (8, 28), staphylococci (12), and Listeria spp. (25) and consists of amplification of the tDNAs by use of consensus primers, which are complementary to the highly conserved edges of the flanking tRNA genes and are directed outwardly. The resulting PCR fragments can be separated by capillary electrophoresis. tDNA-PCR makes use of primers complementary to regions conserved throughout the bacteria and should therefore be applicable to a wide range of genera.
MATERIALS AND METHODS
Bacterial strains.
Seventy-one well-characterized enterococcal strains from different origins, identified by whole-cell protein analysis using sodium dodecyl sulfate-polyacrylamide gel electrophoresis, were obtained from the Belgian Coordinated Collection of Microorganisms culture collection (University of Ghent, Ghent, Belgium). This series was extended to include 19 strains isolated from different animal species, which were identified with a biochemical test scheme as described by Devriese et al. (4, 5). E. faecium, E. faecalis, E. gallinarum, and E. casseliflavus strains were confirmed by a specific multiplex PCR using the van and ddl primers as described by Dutka-Malen et al. (7). All reference strains are listed in Table 1.
TABLE 1.
Enterococcal strains used in this study
| Enterococcus species | LMG-BCCM collection type strain no. |
|---|---|
| E. faecalis | 266/5371,a cc193613,a CDC 149-77,a LMG 7937T, LMG 11395, LMG 11636, LMG 11637, LMG 11734, LMG 12161, LMG 13595 |
| E. faecium species group | |
| E. faecium | LMG 11423T, LMG 11397, LMG 11635, LMG 8148, LMG 16198, LMG 16200, LMG 9431, LMG 9448 |
| E. durans | Lab 848, LMG 10741, LMG 12691, LMG 12903, LMG 13604, LMG 14197, LMG 16887, LMG 16888, LMG 16889 |
| E. hirae | LD 13216,a ccm 2423T,a Lab 1342,a LMG 6399T, LMG 11746, LMG 14198, LMG 14200, LMG 14489, LMG 17189, LMG 17190, LMG 17191, LMG 17192, LMG 17285 |
| E. mundtii | LMG 12308, LMG 13044, LMG 10748 |
| E. gallinarum species group | |
| E. gallinarum | LMG 12313, LMG 12904, LMG 13129T, LMG 14166, LMG 14405, LMG 16201, LMG 16202, LMG 16204 |
| E. casseliflavus | LMG 12306, LMG 12307, LMG 12311, LMG 12314, LMG 14406 |
| E. flavescens | LMG 13518T, LMG 13597, LMG 16313, LMG 16314 |
| E. avium species group | |
| E. avium | LMG 10744T, LMG 11394 LMG 12171, LMG 12304, LMG 12305 |
| E. pseudoavium | 266/7909,a 266/7910a |
| E. malodoratus | LMG 12300, LMG 12301, LMG 12905, LMG 15718 |
| E. raffinosus | LMG 12172, LMG 12888T |
| E. cecorum species group | |
| E. cecorum | 2508/122a, 2508/331a, 2508/334a, 98/2539a, LMG 11743, LMG 11744, LMG 12902T, RS15Aa, S566a |
| E. columbae | LMG 11740T, LMG 12295, LMG 12296, LMG 14175, LMG 14595 |
| E. dispar | SS 1295Tb, 266/8667a |
| E. saccharolyticus | CCUG 27643 |
| E. asini | SS 1501Tb |
Provided by L. A. Devriese, University of Ghent, Ghent, Belgium.
Provided by R. R. Facklam, Centers for Disease Control and Prevention, Atlanta, Ga.
Thirty-four strains originating from the intestines of different animal species and from humans were identified by biochemical tests, multiplex PCR according to the method of Dutka-Malen et al. (7), and tDNA-PCR and are shown in Table 2.
TABLE 2.
Strains isolated from intestines of various animals and from humans and used for blind testing in order to evaluate the ability of tDNA-PCR to identify enterococci
| Strain no. | Origin | Species according to biochemical methods | Species according to van-ddl PCR | Species according to tDNA-PCR |
|---|---|---|---|---|
| PAT1084 | Pigeon | E. avium | NDa | E. avium |
| PAT1123 | Horse | E. avium | ND | E. avium |
| KOMA042 | Pig | E. avium | ND | E. avium |
| UHG 98 11 2049 | Human | E. faecium | E. faecium | E. faecium |
| PAT843 | Finch | E. gallinarum group | E. casseliflavus | E. casseliflavus |
| PAT755 | Goat | E. gallinarum group | E. casseliflavus | E. casseliflavus |
| UHG 98 08 5234 | Human | ND | E. casseliflavus | E. casseliflavus |
| PAT999 | Parakeet | E. gallinarum group | E. casseliflavus | E. casseliflavus |
| PAT233 | Pigeon | E. cecorum or E. columbae | ND | E. cecorum |
| PAT664 | Pigeon | E. cecorum or E. columbae | ND | E. cecorum |
| PAT047 | Pigeon | E. cecorum or E. columbae | ND | E. columbae |
| PAT495 | Pigeon | E. cecorum or E. columbae | ND | E. columbae |
| PAT499 | Pigeon | E. cecorum or E. columbae | ND | E. columbae |
| PAT914 | Chicken | E. faecalis | E. faecalis | E. faecalis |
| UHG 98 11 0487 | Human | ND | E. faecalis | E. faecalis |
| PAT397 | Squirrel | E. faecalis | E. faecalis | E. faecalis |
| PAT052 | Turkey | E. faecalis | E. faecalis | E. faecalis |
| UHG 98 06 1902 | Human | ND | E. faecium | E. faecium |
| UHG 98 07 5596 | Human | ND | E. faecium | E. faecium |
| UHG 98 12 2683 | Human | ND | E. faecium | E. faecium |
| UHG 98 12 3779 | Human | ND | E. faecium | E. faecium |
| UHG 99 01 0517 | Human | ND | E. faecium | E. faecium |
| PAT612 | Pig | E. faecium | E. faecium | E. faecium |
| PAT236 | Horse | E. faecium | E. gallinarum | E. gallinarum |
| UHG 98 07 4441 | Human | ND | E. gallinarum | E. gallinarum |
| UHG 98 08 0962 | Human | ND | E. gallinarum | E. gallinarum |
| UHG 98 08 1527 | Human | ND | E. gallinarum | E. gallinarum |
| UHG 98 12 4026 | Human | ND | E. gallinarum | E. gallinarum |
| PAT426 | Rabbit | E. gallinarum group | E. gallinarum | E. gallinarum |
| PAT1100 | Pigeon | E. hirae/durans | ND | E. hirae |
| PAT1105 | Pigeon | E. hirae/durans | ND | E. hirae |
| PAT1236 | Parakeet | E. hirae/durans | ND | E. hirae |
| PAT1238 | Pigeon | E. hirae/durans | ND | E. hirae |
| PAT882 | Rabbit | E. faecium | Negative2 | E. hirae |
ND, not done.
Negative, no amplification product obtained. Implies that the strain does not belong to the species E. faecalis, E. faecium, E. gallinarum, or E. casseliflavus or E. flavescens.
DNA preparation.
Bacterial cells were grown overnight on Columbia agar (Gibco Technologies, Paisley, Scotland) with 5% sheep blood for 24 h at 37°C in a 5% CO2-enriched environment and were checked for purity. A 1-μl loopful of cells was suspended in 20 μl of lysis buffer (0.25% sodium dodecyl sulfate, 0.05 N NaOH) and heated at 95°C for 5 min. The cell lysate was spun down by brief centrifugation at 16,000 × g and diluted by adding 180 μl of distilled water. The cell debris was removed by centrifugation at 16,000 × g for 5 min. Supernatants were directly used as the template for PCR or were frozen at −20°C until further use.
tDNA-PCR.
PCR was carried out using the outwardly directed tRNA gene consensus primers T5A (5′ AGTCCGGTGCTCTAACCAACTGAG) and T3B (5′ AGGTCGCGGGTTCGAATCC), as described by Welsh and McClelland (26). Reactions were carried out in a 10-μl volume containing 9.1 μl of High Fidelity Mix 1.1× (Gibco Life Technologies). Primers were added at a final concentration of 0.1 μM. Primer T3B consisted of a mixture of 1/5 fluorescent TET-labeled oligonucleotides and 4/5 nonlabeled oligonucleotides (Perkin-Elmer Applied Biosystems, Foster City, Calif.). A volume of 0.7 μl of sample DNA was added (the template was diluted 15 times). After 2 min at 94°C, reaction mixtures were cycled 30 times in a Perkin-Elmer Cetus 9600 thermocycler under the following conditions: 30 s at 94°C, 1 min at 50°C, and 1 min at 72°C. The final extension was 30 min at 72°C. Reaction vials were then cooled to 10°C and kept on ice until used in electrophoresis.
Capillary electrophoresis.
Twelve microliters of deionized formamide was mixed with 0.5 μl of an internal size standard mixture, containing 0.3 μl of the GS-400 High Density size standard and 0.2 μl of the GS-500 size standard, which both have ROX-labeled fragments in the range of 50 to 500 bp (Perkin-Elmer Applied Biosystems). One microliter of tDNA-PCR product was added. The mixtures were denatured by heating at 95°C for 3 min and placed directly on ice for at least 15 min.
Capillary electrophoresis was carried out using an ABI-Prism 310 Genetic Analyzer (Perkin-Elmer Applied Biosystems) at 60°C, a constant voltage of 1.5 kV, and a more or less constant current of approximately 10 mA. Capillaries with a length of 47 cm and a diameter of 50 μm were filled with Performance Optimized Polymer 4. Electropherograms were normalized using Genescan Analysis software, version 2.1 (Perkin-Elmer Applied Biosystems). The fragment lengths were derived from the peak positions after interpolation with the peak positions of the size standard fragments.
Data analysis.
Electropherograms were interpreted visually and with a software program developed at our laboratory (available upon request from the authors).
The software compares samples which are derived from the ABI310 Genescan Analysis program as molecular weight tables. A library, containing one entry per species, was constructed manually as a text file (see Table 3 for an example). Each entry is a list of numerical values which represent those fragment lengths (i.e., peaks) differentiating the species from each other, as established by visual interpretation of superimposed fingerprints obtained with the Genescan software. A positive value indicates that a peak is present in the fingerprint of a particular species, while a negative value penalizes the presence of a peak with this value. After elimination of peaks lower than 50% of the average height of all peaks, the fingerprint of an unknown strain was compared with all entries in the library. The number of fragments in common between the unknown fingerprint and the species entry, divided by the total number of fragments of the species entry in the library, was taken as a measure of similarity. The library does not contain irreproducible peaks or peaks considered irrelevant after visual comparison, which therefore are not taken into consideration by the program when comparing unknown fingerprints with entries in the library. The program enables one to enlarge the peak position tolerance, which corrects for small base pair shifts.
TABLE 3.
Manually constructed tDNA-PCR library, composed of entries that each consists of a list of tDNA spacer fragment lengths (in base pairs)
| Species entry | Lengths of fragments taken into considerationa |
|---|---|
| E. faecalis | 65.8,110.8,266.6 |
| E. faecium | 63.8,95.5,244,269.2 |
| E. hirae | 63.9,93,271.5,303.5 |
| E. durans | 63.8,94.3,−95.5,243.3,269.2 |
| E. mundtii | 63.5,102.5,269.6 |
| E. avium | 65.8,92.9,253.8 |
| E. pseudoavium | 66.2,86.6,220 |
| E. malodoratus | 65.8,91.6,251.5,−256.5 |
| E. raffinosus | 65.8,96,252.5 |
| E. gallinarum | 64.9,95.3,261.5 |
| E. casseliflavus or E. flavescens | 61.3,69.1,97.5,264.6 |
| E. cecorum | 63.8,81.2,−154,−155,240.9,254.7 |
| E. columbae | 66,78.4,237.5,241,255.6 |
| E. dispar | 64,79.2,231,266,284.1 |
| E. saccharolyticus | 64,107.8,231,266 |
| E. asini | 73,98,261.6 |
Values represent peaks that ought to be present (x) or absent (−y) in the fingerprint of an unknown strain in order to be identified as a certain species.
For clustering analysis, the distance matrix was calculated with the in-house software. The similarity between two samples was calculated as described above in a pairwise manner (first considering one sample as the library entry), whereby peaks below a user-defined background threshold were not taken into consideration and the second sample was taken as the library entry. The similarity between the two samples was the average of the two calculated similarity values. Clustering was done with Neighbor software (Phylip) (http://evolution.genetics.washington.edu/phylip.html), employing the algorithm for the unweighted-pair group method using average linkages (UPGMA).
RESULTS
tDNA-PCR.
Capillary electrophoresis of tDNA-PCR amplification products of enterococcal strains generated fingerprint patterns with three to five large, reproducible peaks and several smaller, irreproducible ones. The reproducibility of tDNA-PCR was evaluated by carrying it out for strains LMG 11423 (E. faecium), LMG 13129 (E. gallinarum), and LMG 13595 (E. faecalis) four times, each time using different PCR mixtures, thermal cycling runs, and electrophoresis runs. For each strain, one of these four tDNA-PCR products was run three times in capillary electrophoresis. Similarity was calculated with the in-house software and a background noise level of 50%. Clustering was done using the UPGMA algorithm. The minimal similarity level for the six tDNA-PCR fingerprints obtained for each strain named above was 88.6, 89.3, and 90%, respectively. For all six samples, the standard deviation of the peaks, which are used in the library of the in-house software, ranged from 0.045 bp (for a fragment length of 266.5 bp) to 0.364 bp (for a fragment length of 110.8 bp). Repeated electrophoresis runs of the same PCR product gave a minimum standard deviation of 0.007 bp (for a fragment length of 266.5 bp) and a maximum of 0.418 bp (for a fragment length of 110.8 bp). Repeats of the entire PCR assay (different PCR mixtures, PCR runs, and electrophoresis runs) gave a minimum standard deviation of 0.035 bp (for a fragment length of 269.3 bp) and a maximum of 0.371 bp (for a fragment length of 65.8 bp). The highest reproducibility and the most reliable identifications were obtained by not taking into account those peaks lower than 50% of the average peak height within the range of 60 to 400 bp.
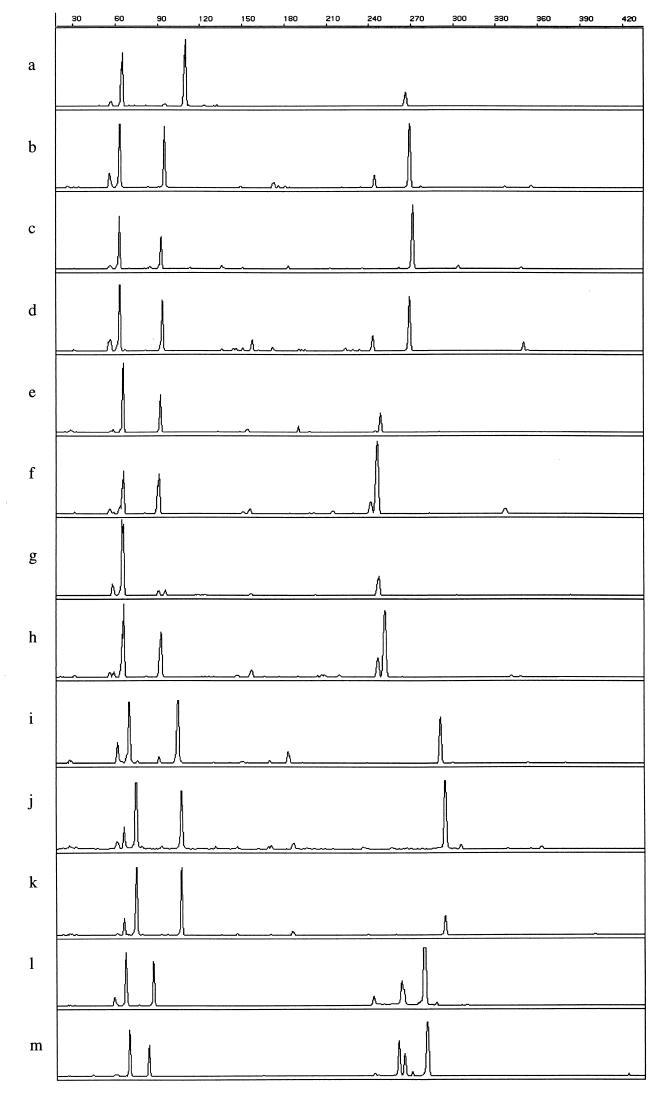
Visual interpretation showed that tDNA-PCR fingerprints of strains belonging to the same species were similar, while strains belonging to different species exhibited different patterns, except for those belonging to the species E. casseliflavus and E. flavescens (Fig. 1). Some species, for instance E. faecium and Enterococcus durans, could be differentiated by the 1-bp length difference of a single tDNA spacer fragment. Enterococcus avium and Enterococcus malodoratus differed in the lengths of two fragments: E. avium strains showed peaks at 65.8, 92.9, and 253.8 bp, whereas the patterns of isolates belonging to E. malodoratus were composed of peaks of 65.8, 91.6, and 251.5 bp.
FIG. 1.
tDNA-PCR fingerprint patterns of enterococcal strains. a, E. faecalis 266/5371; b, E. faecium LMG 11423; c, E. hirae LMG 6399T; d, E. durans LMG 13604; e, E. avium LMG 107444T; f, E. malodoratus LMG 12300; g, E. raffinosus LMG 12888T; h, E. raffinosus LMG 14595; i, E. gallinarum LMG 16204; j, E. casseliflavus LMG 10745; k, E. flavescens LMG 13518; l, E. cecorum LMG 11744; m, E. columbae LMG 11740T. The x axis represents the fragment length in base pairs; the y axis represents the peak intensity.
For use with the in-house software, the tDNA-PCR fingerprint library (Table 3) was constructed using all reference strains (Table 1) from our collection. Each entry contains for each species all reproducible fragment length values that are present in the fingerprints of its different isolates, and some also include negative values to distinguish between highly similar patterns. Using this library, all strains were identified correctly, including those belonging to the species E. durans and E. faecium and those belonging to the species E. avium, Enterococcus raffinosus, and E. malodoratus.
A dendrogram was constructed and is shown in Fig. 2. All species clustered separately except E. casseliflavus and E. flavescens, which clustered together.
FIG. 2.
Dendrogram obtained from tDNA-PCR fingerprints after similarity calculation with the in-house software and clustering with UPGMA using Neighbor software. Bar, distance of 10%.
Identification of unknown strains with tDNA-PCR.
Thirty-four strains isolated from humans and animals were identified in a polyphasic approach using biochemical tests (4, 5) and van-ddl PCR (7) to verify identification results from tDNA-PCR. The results are summarized in Table 2. tDNA fingerprints were analyzed with the in-house software. All 34 strains were identified correctly, regardless of whether peaks lower than 50% of the average peak height were eliminated.
The fingerprints of the enterococcal species were compared with fingerprints obtained from about 400 species belonging to over 40 different genera. All enterococcal strains could be correctly identified.
DISCUSSION
For the identification of the most important enterococci, a simple, conventional biochemical test scheme exists (10) and a more complex, phylogenetically based differential identification scheme has been described (4). However, the results of these tests are sometimes unreliable or ambiguous, and some species are too closely related to show biochemical differences. This is especially the case for Enterococcus hirae and E. durans, E. gallinarum and E. casseliflavus, and Enterococcus cecorum and Enterococcus columbae (4). In this study, we evaluated a universally applicable genotypic method for the identification of enterococci.
tDNA-PCR enabled us to differentiate between all species tested, except E. casseliflavus and E. flavescens. However, these species are most probably synonymous, as is also apparent from several other studies (3, 7, 19, 22). Closely related organisms showed peak shifts of no more than one or a few base pairs. Therefore, the high resolution of capillary electrophoresis was needed to separate fragments differing by 1 bp in length.
Values for similarity between fingerprints of the same strain obtained in different PCR and electrophoresis runs were very high, around 90%. In the reproducibility test, standard deviations of the peak positions in base pairs were not higher than 0.364 bp, which indicates that the peak positions in the fingerprints are highly reproducible.
To be able to compare large numbers of unidentified strains to a database of well-characterized strains, a suitable software package was necessary. When comparing complete fingerprints, as is done when using the Dice coefficient, calculation of the similarity between visually highly identical fingerprints can still give low values (as a consequence of small peak shifts and variable presence of minor peaks). Software which enabled a different approach was developed. A library in which only reproducible peaks were included was manually constructed. Peaks in new fingerprints were regarded to be identical to a peak of a reference pattern when their positions lay within a range of −0.7 bp to +0.7 bp of the reference peak. This is twice the maximum standard deviation obtained in the reproducibility tests. Because the peak position of each fragment is Gauss distributed, 95% of the electrophoretic profiles of the same sample should have a peak within this range. The use of a manually constructed library also prevents nonreproducible peaks from influencing the calculation of similarity values, which in turn leads to a higher discriminatory power.
Analysis of the molecular weight tables with the in-house software permitted discrimination of the highly similar tDNA patterns of E. faecium and E. durans strains. Blind testing of enterococcal strains which were previously identified with biochemical tests or multiplex PCR showed that this software enabled us to process tDNA-PCR fingerprints originating from an ABI Prism 310 Genetic Analyzer. One of the most important advantages of tDNA-PCR is the use of universal primers. In theory, this technique can be used for species identification for a wide range of genera. It requires little time and manual labor. tDNA-PCR takes about 3 h; the GeneAmp 9600 PCR cycler permits testing of 96 strains at once. Capillary electrophoresis requires half an hour per run; one run can include up to three samples if primers are labeled with different fluorescent dyes. Its high reproducibility and satisfactory discriminatory power make it possible to develop an identification tool which can be used by different laboratories. Normalization of the fingerprints is done automatically by the Genescan Analysis program, and quality control of the different steps in the protocol takes between 2 and 10 min for approximately 50 strains. Using our software, a list of identifications for up to 50 strains at once is available within 5 min of exportation in table format of the normalized fingerprints as obtained on ABI310.
Taken together, the whole procedure as described above, starting from a pure culture to a final identification, can be completed in 24 h for 45 strains, requiring only 4 to 5 h hands-on time. The cost per strain, comprising DNA preparation, tDNA-PCR, and capillary electrophoresis reagents (capillary, buffer, size marker, POP4 gel, and laser wear, excluding PCR and electrophoresis equipment), was calculated to add up to $2.50 (U.S.) (labor not included). tDNA-PCR fingerprints can be obtained within 8 h after colony picking for the first five electrophoresis samples, which can contain PCR products of up to three strains.
From these results, we conclude that tDNA-PCR is very suitable for rapid, discriminatory, and reliable identification of all currently described enterococcal species and can be extended to include newly described ones.
In addition to the high discriminatory power of tDNA-PCR for other groups, like Acinetobacter (8, 28), Listeria (25), staphylococci (12), and streptococci (2, 13), one can start to consider the applicability of this technique for the species identification of cultured organisms in the average laboratory.
ACKNOWLEDGMENTS
M.V. is indebted to FWO Flanders for an appointment as research associate.
We are grateful to A. Vandekerckhove, L. Van Simaey, and F. Grillaert for excellent technical assistance.
REFERENCES
- 1.Alonso-Echanove J, Robles B, Jarvis W R. Proficiency of clinical laboratories in Spain in detecting vancomycin-resistant Enterococcus spp. J Clin Microbiol. 1999;37:2148–2152. doi: 10.1128/jcm.37.7.2148-2152.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Degheldre Y, Vandamme P, Goossens H, Struelens M. Identification of clinically relevant viridans streptococci by analysis of transfer DNA intergenic spacer length polymorphism. Int J Syst Bacteriol. 1999;49:1591–1598. doi: 10.1099/00207713-49-4-1591. [DOI] [PubMed] [Google Scholar]
- 3.Descheemaeker P, Lammens C, Pot B, Vandamme P, Goossens H. Evaluation of arbitrarily primed PCR analysis and pulsed-field gel electrophoresis of large genomic DNA fragments for identification of enterococci important in human medicine. Int J Syst Bacteriol. 1997;47:555–561. doi: 10.1099/00207713-47-2-555. [DOI] [PubMed] [Google Scholar]
- 4.Devriese L A, Pot B, Collins M D. Phenotypic identification of the genus Enterococcus and differentiation of phylogenetically distinct enterococcal species and species groups. J Appl Bacteriol. 1993;75:399–408. doi: 10.1111/j.1365-2672.1993.tb02794.x. [DOI] [PubMed] [Google Scholar]
- 5.Devriese L A, Pot B, Kersters K, Lauwers S, Haesebrouck F. Acidification of methyl-α-d-glucopyranoside: a useful test to differentiate Enterococcus casseliflavus and Enterococcus gallinarum from Enterococcus faecium species group and from Enterococcus faecalis. J Clin Microbiol. 1996;34:2607–2608. doi: 10.1128/jcm.34.10.2607-2608.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Donabedian S, Chow J W, Shlaes D M, Green M, Zervos M J. DNA hybridization and contour-clamped homogeneous electric field electrophoresis for identification of enterococci to the species level. J Clin Microbiol. 1995;33:141–145. doi: 10.1128/jcm.33.1.141-145.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dutka-Malen S, Evers S, Courvalin P. Detection of glycopeptide resistance genotypes and identification to the species level of clinically relevant enterococci by PCR. J Clin Microbiol. 1995;33:24–27. doi: 10.1128/jcm.33.1.24-27.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ehrenstein B, Bernards A T, Dijkshoorn L, Gerner-Smidt P, Towner K J, Bouvet P J M, Daschner F D, Grundmann H. Acinetobacter species identification by using tRNA spacer fingerprinting. J Clin Microbiol. 1996;34:2414–2420. doi: 10.1128/jcm.34.10.2414-2420.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Evers S, Casadewall B, Charles M, Dutka-Malen S, Galimand M, Courvalin P. Evolution of structure and substrate specificity in d-alanine:d-alanine ligases and related enzymes. J Mol Evol. 1996;42:706–712. doi: 10.1007/BF02338803. [DOI] [PubMed] [Google Scholar]
- 10.Facklam R, Collins M D. Identification of Enterococcus species isolated from human infections by a conventional test scheme. J Clin Microbiol. 1989;27:731–734. doi: 10.1128/jcm.27.4.731-734.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huycke M M, Sahm D F, Gilmore M S. Multiple-drug resistant enterococci: the nature of the problem and an agenda for the future. Emerg Infect Dis. 1998;4:239–249. doi: 10.3201/eid0402.980211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maes N, De Gheldre Y, De Ryck R, Vaneechoutte M, Meugnier H, Etienne J, Struelens M J. Rapid and accurate identification of Staphylococcus species by tRNA intergenic spacer length polymorphism analysis. J Clin Microbiol. 1997;35:2477–2481. doi: 10.1128/jcm.35.10.2477-2481.1997. . (Erratum, 36:1468, 1998.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McClelland M, Welsh J. Length polymorphisms in tRNA intergenic spacers detected by using the polymerase chain reaction can distinguish streptococcal strains and species. J Clin Microbiol. 1992;30:1499–1504. doi: 10.1128/jcm.30.6.1499-1504.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Monstein H J, Quednau M, Samuelsson A, Ahrne S, Isaksson B, Jonasson J. Division of the genus Enterococcus into species groups using PCR-based molecular typing methods. Microbiology. 1998;144:1171–1179. doi: 10.1099/00221287-144-5-1171. [DOI] [PubMed] [Google Scholar]
- 15.Murray B E. The life and times of the Enterococcus. Clin Microbiol Rev. 1990;3:46–65. doi: 10.1128/cmr.3.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Patel R, Piper K E, Rouse M S, Steckelberg J M, Uhl J R, Kohner P, Hopkins M K, Cockerill F R, Kline B C. Determination of 16S rRNA sequences of enterococci and application to species identification of nonmotile Enterococcus gallinarum isolates. J Clin Microbiol. 1998;36:3399–3407. doi: 10.1128/jcm.36.11.3399-3407.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Patel R, Uhl J R, Kohner P, Hopkins M K, Cockerill F R., III Multiplex PCR detection of vanA, vanB, vanC-1, and vanC-2/3 genes in enterococci. J Clin Microbiol. 1997;35:703–707. doi: 10.1128/jcm.35.3.703-707.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Poyart C, Quesnes G, Trieu-Cuot P. Sequencing the gene encoding manganese-dependent superoxide dismutase for rapid species identification of enterococci. J Clin Microbiol. 2000;38:415–418. doi: 10.1128/jcm.38.1.415-418.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Quednau M, Ahrne S, Petersson A C, Molin G. Identification of clinically important species of Enterococcus within 1 day with randomly amplified polymorphic DNA (RAPD) Curr Microbiol. 1998;36:332–336. doi: 10.1007/s002849900318. [DOI] [PubMed] [Google Scholar]
- 20.Ruoff K L, de la Maza L, Murtagh M J, Spargo J D, Ferraro M J. Species identities of enterococci isolated from clinical specimens. J Clin Microbiol. 1990;28:435–437. doi: 10.1128/jcm.28.3.435-437.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schaberg D R, Culver D H, Gaynes R P. Major trends in the microbial etiology of nosocomial infection. Am J Med. 1991;91:72S–75S. doi: 10.1016/0002-9343(91)90346-y. [DOI] [PubMed] [Google Scholar]
- 22.Teixeira L M, Carvalho M G, Merquior V L, Steigerwalt A G, Teixeira M G, Brenner D J, Facklam R R. Recent approaches on the taxonomy of the enterococci and some related microorganisms. Adv Exp Med Biol. 1997;418:397–400. doi: 10.1007/978-1-4899-1825-3_95. [DOI] [PubMed] [Google Scholar]
- 23.Toye B, Shymanski J, Bobrowska M, Woods W, Ramotar K. Clinical and epidemiologic significance of enterococci intrinsically resistant to vancomycin (possessing the vanC genotype) J Clin Microbiol. 1997;35:3166–3170. doi: 10.1128/jcm.35.12.3166-3170.1997. . (Erratum, 36:1469, 1998.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tyrrell G J, Bethune R N, Willey B, Low D E. Species identification of enterococci via intergenic ribosomal PCR. J Clin Microbiol. 1997;35:1054–1060. doi: 10.1128/jcm.35.5.1054-1060.1997. . (Erratum, 35:2434.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vaneechoutte M, Boerlin P, Tichy H V, Bannerman E, Jäger B, Bille J. Comparison of PCR-based DNA fingerprinting techniques for the identification of Listeria species and their use for atypical Listeria isolates. Int J Syst Bacteriol. 1998;48:127–139. doi: 10.1099/00207713-48-1-127. [DOI] [PubMed] [Google Scholar]
- 26.Welsh J, McClelland M. Genomic fingerprints produced by PCR with consensus tRNA gene primers. Nucleic Acids Res. 1991;19:861–866. doi: 10.1093/nar/19.4.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Welsh J, McClelland M. PCR-amplified length polymorphisms in tRNA intergenic spacers for categorizing staphylococci. Mol Microbiol. 1992;6:1673–1680. doi: 10.1111/j.1365-2958.1992.tb00892.x. [DOI] [PubMed] [Google Scholar]
- 28.Wiedmann-Al-Ahmad M, Tichy H V, Schön G. Characterization of Acinetobacter type strains and isolates obtained from wastewater treatment plants by PCR fingerprinting. Appl Environ Microbiol. 1994;60:4066–4071. doi: 10.1128/aem.60.11.4066-4071.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Williams A M, Rodrigues U M, Collins M D. Intrageneric relationships of enterococci as determined by reverse transcriptase sequencing of small-subunit rRNA. Res Microbiol. 1991;142:67–74. doi: 10.1016/0923-2508(91)90098-u. [DOI] [PubMed] [Google Scholar]