Abstract
MAP3K1 is a MAPK family serine-threonine kinase that is frequently mutated in human cancer. The association between mutations in the MAP3K1 gene and the clinicopathological characteristics and prognosis of patients with breast cancer remain unclear in the Chinese population. Thus, the aim of the present retrospective study was to investigate the possible role and function of MAP3K1 in breast cancer. Data obtained from 412 consecutive patients with breast cancer were selected from Guangdong Provincial People's Hospital (GDPH) for analysis in the present study. Mutations were assessed using next-generation sequencing. The association between MAP3K1 mutations and clinicopathological features were analyzed and further compared with the Molecular Taxonomy of Breast Cancer International Consortium (METABRIC) cohort and data from The Cancer Genome Atlas (TCGA). In the GDPH cohort, a total of 45 mutations MAP3K1 were identified in 8.5% (n=35) of the 412 patients, compared with 9.7% (n=244) in METABRIC and 7.9% (n=88) in TCGA. The majority of the mutations identified in the in three cohorts were truncating mutations, followed by mis-sense mutations. Mutations in MAP3K1 were predominant in patients with the luminal A and B breast cancer subtypes in METABRIC datasets (P<0.001), although no significant differences were observed in the GDPH cohort (P=0.227). In the METABRIC cohort, patients with MAP3K1 mutations experienced a improved overall survival (OS) rate than patients without MAP3K1 mutations (P=0.006). In patient with hormone receptor (HR)+ breast cancer, a more significantly higher OS rate was observed in patients with MAP3K1 mutations (P<0.001). MAP3K1 expression was associated with OS in the HR+ subgroup. Moreover, the MAP3K1 methylation levels were reduced in primary breast cancer tissue, compared with normal tissue. Thus, the present findings identified MAP3K1 mutations in Chinese patients with breast cancer, and compared MAP3K1 mutations between the cohorts from Western and Eastern countries.
Keywords: breast cancer, MAP3K1 mutation, next-generation sequencing, DNA methylation
Introduction
Breast cancer is the cancer with the highest incidence worldwide and affects an increasing number of Chinese women, thus representing an important health problem in China (1). In 2017, >25,000 women were newly diagnosed with breast cancer in the United States (2). Systemic therapy has improved the overall survival (OS) and disease-free survival of patients with breast cancer in a manner dependent on the molecular subtype and stage.
Recently, large-scale next-generation sequencing (NGS) has been introduced to characterize multiple cancer types at the genomic level, including breast cancer. These efforts have led to a broad description of the genomic alterations involved in tumorigenesis or tumor progression in breast cancer. Breast cancer is characterized by a large number of genomic alterations, such as ERBB2 amplification (3), PIK3CA mutation (4), FGFR1 amplification (5), CCND1 amplification (6), AKT1 mutation and GATA3 mutation (7,8).
MAPKs are key mediators of evolutionarily conserved signaling networks that play an essential role in multiple aspects of cell physiology. The MAPK pathway is frequently activated in cancerous cells. MAP3K1 or MEKK1 is a 196-kDa serine-threonine kinase in the MAP3K family with functions in cell viability, apoptosis and cell motility/migration in multiple normal and tumor cell types (9). However, the role of MAP3K1 mutations in patient survival in different types of cancer remains poorly understood. In particular, the relationship between MAP3K1 mutations and the clinicopathological characteristics of patients with breast cancer remains an unresolved question.
Thus, the aim of the present study was to extensively describe the relationship between MAP3K1 genomic alterations and breast cancer. A total of 412 Chinese patients with primary breast tumors were enrolled. The prevalence of MAP3K1 mutations and their association with clinicopathological characteristics were assessed. MAP3K1 mutations were also analyzed in data from The Cancer Genome Atlas (TCGA) and from the Molecular Taxonomy of Breast Cancer International Consortium (METABRIC) cohort (10).
Materials and methods
Patients
The study population consisted of three cohorts of patients. The Guangdong Provincial People's Hospital (GDPH) cohort included 412 consecutive patients diagnosed with treatment-naïve breast cancer at Guangdong Provincial People's Hospital from October 2016 to December 2017. The following inclusion criteria were used: i) Patients were diagnosed with invasive breast cancer; ii) complete clinical characterization was available (including age, pathological type, axillary lymph node status, TMN staging, pathological grade, the status of estrogen receptor (ER), the status of progesterone receptor (PR), the status of hormone receptor, human epidermal growth factor receptor 2 (HER2), molecular subtype (11,12); and iii) tumor biopsies were obtained using an Institutional Review Board-approved protocol and analyzed using NGS.
The present study was also based on an analysis of data obtained from TCGA-BRCA and from the METABRIC cohort. Data for the MAP3K1 mutation profiles and clinical characteristics of breast cancer in TCGA and METABRIC cohorts were downloaded from cBioPortal (http://www.cbioportal.org) (11). A total of 1,108 patients with breast cancer were included in TCGA Pan-Cancer analysis project dataset (7). In the METABRIC cohort (10), 2,509 eligible patients with breast cancer for whom MAP3K1 mutation profiles and clinical information were included. Patients with breast cancer with complete MAP3K1 somatic mutation profiles and clinicopathological characteristics were included in this study. For the statistical analysis, individuals with missing variables were excluded. Somatic mutations in the GDPH, TCGA and METABRIC cohorts were analyzed to compare MAP3K1 mutation frequencies. In patient survival analysis, patients with missing MAP3K1 mutation data and missing follow-up were excluded. OncoPrint data was generated from cBioPortal. The expression heatmap of genes in the METABRIC cohort are shown as mRNA expression Z-scores, which were defined as mRNA expression compared to the expression distribution of each gene tumors, that are diploid for this gene.
All experiments described in this study were approved by The Ethics Committee of The Guangdong Provincial People's Hospital (approval no. GDREC2014122H). All patients provided written informed consent to participate in this study.
DNA extraction from the tissues
DNA was extracted from 412 tumor samples using a QIAamp DNA FFPE tissue kit (cat. no. 56404; Qiagen, Inc.) according to the manufacturer's standard protocol. The concentration and purity of DNA were measured using a Qubit dsDNA assay kit (Life Technologies; Thermo Fisher Scientific, Inc.) prior to mutation analysis. The loading concentration of the final library was 4 nmol.
NGS library preparation and sequencing
In the current study, sequencing data were obtained from primary tissue samples. DNA fragmentation was performed using a M220 (Covaris, Inc.), followed by end repair, phosphorylation and adaptor ligation. Fragments 200–400 bp in size were selected using AMPure beads (Agencourt AMPure XP kit; Beckman Coulter, Inc.), followed by hybridization with capture probe baits, hybrid selection with magnetic beads and PCR amplification. A total of 1 to 5 µg PCR products was used for hybridization with NimbleGen SeqCap EZ Hybridization and Wash kits (cat. no. 06953247001; Roche Diagnostics). The captured DNA fragments from the previous step were amplified, and the PCR products were purified with the AxyPrep Mag PCR clean up kit (cat. no. MAG-PCR-CL-250; Corning, Inc.). The quality of the library was then evaluated using the bioanalyzer (Agilent Technologies, Inc.) and Real-Time PCR System (ABI StepOnePlus; 4376600; Thermo Fisher Scientific, Inc.). Indexed samples were sequenced on a NextSeq500 sequencer (Illumina, Inc.) with a NextSeq 500 high output kit V2 (cat. no. FC-404-2002; Illumina, Inc.) with paired-end reads (150 cycles) in a Clinical Laboratory Improvement Amendments/College of American Pathologists-certified laboratory using a panel consisting of 520 cancer-related genes, spanning 1.64 megabases of the human genome (OncoScreen Plus panel; Guangzhou Burning Rock Medical Laboratory Co., Ltd). The panel was designed to capture whole exons of 312 genes and critical exons, introns and promoter regions of the remaining 208 genes.
Sequencing data analysis
Sequence data were mapped to the human genome (hg19) using Burrows-Wheeler Aligner v.0.7.10 (13). Local alignment optimization, variant calling and annotation were performed using GATK v.3.2 (https://gatk.broadinstitute.org) and VarScan v.2.4.3 (http://varscan.sourceforge.net) (14). Variants were filtered using the VarScan fpfilter pipeline, whereby loci with depth <100 were filtered out. Moreover, ≥5 supporting reads were needed for insertion-deletions (indels); while 8 supporting reads were needed for single nucleotide variations to be called. According to the ExAC (http://http://exac.broadinstitute.org), 1,000 Genomes (https://www.internationalgenome.org), dbSNP (http://www.ncbi.nlm.nih.gov/SNP) and ESP6500SI–V2 (http://www.openbioinformatics.org/annovar/download/hg19_1000g2015aug.zip) databases, variants with population frequency >0.1% were grouped as single nucleotide polymorphism and excluded from further analysis. Remaining variants were annotated with ANNOVAR (15) and SnpEff v.3.6 (16). DNA translocation analysis was performed using Factera v.1.4.3 (17). Data processing methods are also described in previous reports (18–20). All sequencing data are available in the NODE repository under the accession code OEP001295 (http://www.biosino.org/node/project/detail/OEP001295).
Mutation classification
Capture-based targeted sequencing of 412 tumour tissue samples and paired peripheral blood samples was performed using a panel consisting of 520 cancer-related genes. The mutations were classified according to the predicted effect using ANNOVAR (15) and SnoEff v.3.6 (16) on the protein function as follows: i) Mis-sense mutations in the DNA-binding motif (DBM); ii) mis-sense mutations outside of the DBM; and iii) and non-mis-sense mutations (including splice, in-frame, frameshift and nonsense mutations). Samples with no somatic gene alteration were classified as wild-type (WT).
Prognostic analysis of intrinsic molecular subtypes and DNA promoter methylation
The prognostic analysis of MAP3K1 expression in different molecular subtypes of breast cancer was performed using Breast Cancer Gene-Expression Miner v4.4 (http://bcgenex.ico.unicancer.fr) as previously described (21,22). This prognostic analysis tool based on intrinsic molecular subtypes is used for the evaluation of the prognostic factors for a given gene within groups of patients defined by different subtype clustering models. Molecular subtypes were classified into four groups based on three subtype clustering models (SCMOD1, SCMOD2 and SCMGENE). For example, SCMOD models were produced by analyzing the public database cohorts enrolling >2,100 patients based on ER and HER2 module scores (23–28). These groups represented the following intrinsic molecular subtypes: i) ER−/HER2−; ii) ER−/HER2+; iii) ER+/HER2−; and iv) ER+/HER2+, corresponding roughly to the intrinsic basal-like, HER2, combined luminal A/B (HER2-), and luminal B (HER2+) subtypes, respectively (25).
The prognostic effect of MAP3K1 expression was evaluated using univariate Cox proportional hazards model and Kaplan-Meier curves. A univariate Cox analysis table is also provided for robust classification.
The DNA promoter methylation profile from 890 patients in TCGA-BRCA cohort was obtained from UALCAN (http://ualcan.path.uab.edu) (29) to investigate the epigenetic regulation of MAP3K1 expression in breast cancer. In short, BRCA DNA methylation array, mRNA expression datasets and corresponding clinical information from TCGA firehose browser (http://firebrowse.org) was obtained from UALCAN. The DNA methylation array data from tumor tissues from 793 patients with BRCA and 97 adjacent tumor tissues were analyzed.
Statistical analysis
Patient characteristics and sequencing results are summarized using descriptive statistics, including the mean and standard deviations for continuous data. Continuous variables were compared using Student's t-test or Mann-Whitney's U-test. Categorical data were compared using the χ2 test. P-values were two-tailed and P<0.05 was considered to indicate a statistically significant difference. Survival analysis was conducted using the Kaplan-Meier method, and groups were compared using the log-rank test. All statistical analyses were conducted using Statistical Product and Service Solutions 20.0 (IBM Corp.).
Results
Characterization of MAP3K1 mutations
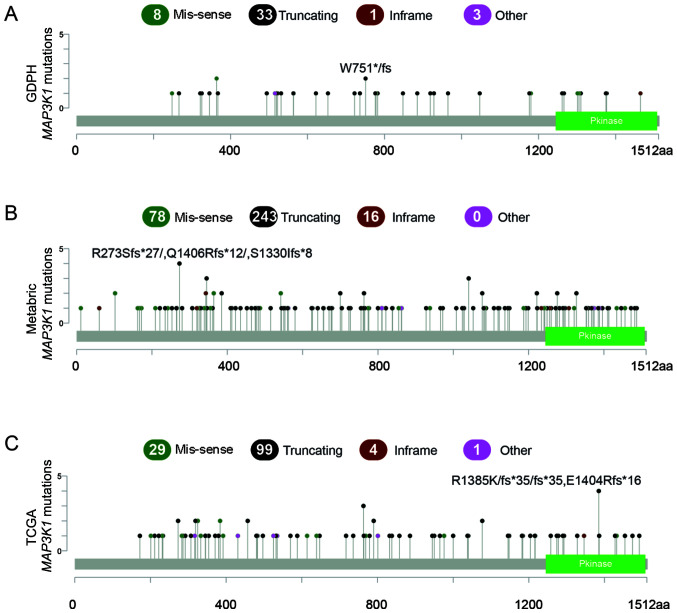
According to the NGS results obtained from the GDPH cohort, 45 MAP3K1 mutations were observed in 35 patients (8.5%). Moreover, 8 of the 35 patients (21.6%) concurrently carried >1 MAP3K1 mutation. Among these eight patients, seven patients had dual MAP3K1 mutations and one patient had a triplet MAP3K1 mutation. The majority of the mutations were truncating mutations (n=33; 74.4%), followed by missense mutations (n=8; 17.8%) (Fig. 1A).
Figure 1.
Somatic mutations in the MAP3K1 gene in patients with breast cancer. (A) MAP3K1 mutations in the GDPH cohort. (B) MAP3K1 mutations in the METABRIC cohort. (C) MAP3K1 mutations in TCGA cohort. Green dots represent missense mutations, while black dots represent truncating mutations. The data of TCGA-BRCA and METABRIC cohorts were obtained from and analyzed using cBioPortal. GDPH, Guangdong Provincial People's Hospital; METABRIC, Molecular Taxonomy of Breast Cancer International Consortium; TCGA, The Cancer Genome Atlas; aa, amino acid; Pkinase, protein kinase domain.
The MAP3K1 mutation profiles were compared between GDPH, METABRIC and TCGA cohorts. MAP3K1 mutation frequencies in the METABRIC and TCGA cohorts were similar to the mutation frequencies in the GDPH cohort, namely, 9.7% in METABRIC (number of patients, 244; number of mutations, 337) and 7.9% in TCGA (number of patients, 88; number of mutations, 133). Similarly, truncating mutations were the major mutation type, accounting for 72.1% in METABRIC and 74.4% in TCGA (Fig. 1B and C).
The mutated domains in the three cohorts are shown in Fig. 1. Mutation hotspots were not uniformly distributed across the three cohorts. No major hotspot was identified in the mutated domains. For example, the most highly recurrent hotspot was Arg364Trp in the METABRIC cohort (n=6), which only occurred in two patients in the GDPH cohort.
Spectrum of MAP3K1 mutations in patients with different molecular subtypes of breast cancer
The relationship between MAP3K1 mutations and clinicopathological characteristics of the three cohorts are shown in Table I. Mutations in MAP3K1 were predominant in the luminal A breast cancer subtype, followed by the luminal B breast cancer subtype, both in TCGA and the METABRIC datasets (P<0.001; Table I). However, there was no significant association in the GDPH cohort (P=0.179). Only four of 35 patients (11.4%) had a hormone receptor (HR)− disease and MAP3K1 mutations were also more dominant in luminal A subtype, compared with luminal B, HER2+ and triple-negative breast cancer (TNBC) (23/35; 65.7%). The non-predominant association with molecular subtype may be due to the small sample size. In addition, a lower pathological grade was observed in the MAP3K1 mutant group in the GDPH and the METABRIC datasets.
Table I.
Association between MAP3K1 mutations and the clinicopathological characteristics of the three cohorts.
| A, GDPH cohort (n=412) | |||
|---|---|---|---|
|
| |||
| Clinicopathological characteristic | Wild-type | Mutant | P-value |
| Mean age ± SD, years | 48.9±10.8 | 51.3±9.7 | 0.207 |
| Type, n (%) | 0.863 | ||
| Ductal | 331 (87.8) | 30 (85.7) | |
| Lobular | - | - | - |
| Other | 44 (11.7) | 5 (14.3) | - |
| Missing | 2 (0.5) | 0 (0) | |
| Node status, n (%) | 0.979 | ||
| pN0 | 184 (48.8) | 17 (48.6) | |
| pN1-pN3 | 193 (51.2) | 18 (51.4) | |
| Missing | 0 (0) | 0 (0) | |
| TNM stage, n (%) | 0.304 | ||
| 1 | 91 (24.1) | 11 (31.4) | |
| 2 | 175 (46.4) | 16 (45.7) | |
| 3 | 85 (22.5) | 6 (17.1) | |
| 4 | 25 (6.6) | 2 (5.7) | |
| Missing | 1 (0.3) | 0 (0) | |
| Pathological grade, n (%) | 0.032 | ||
| 1 | 13 (3.4) | 2 (5.7) | |
| 2 | 169 (44.8) | 22 (62.9) | |
| 3 | 189 (50.1) | 9 (25.7) | |
| Missing | 6 (1.6) | 2 (5.7) | |
| Estrogen receptor, n (%) | 0.116 | ||
| Positive | 265 (70.3) | 29 (82.9) | |
| Negative | 112 (29.7) | 6 (17.1) | |
| Missing | 0 (0) | 0 (0) | |
| Progesterone receptor, n (%) | 0.155 | ||
| Positive | 246 (65.3) | 27 (77.1) | |
| Negative | 131 (34.7) | 8 (22.9) | |
| Missing | 0 (0) | 0 (0) | |
| Hormone receptor, n (%) | 0.056 | ||
| Positive | 279 (74.0) | 31 (88.6) | |
| Negative | 98 (26.0) | 4 (11.4) | |
| Missing | 0 (0) | 0 (0) | |
| HER2, n (%) | 0.889 | ||
| Positive | 108 (28.7) | 9 (25.7) | |
| Negative | 268 (71.1) | 26 (74.3) | |
| Missing | 1 (0.3) | 0 (0) | |
| Subtype, n (%) | 0.179 | ||
| LumA | 76 (20.2) | 11 (31.4) | |
| LumB | 203 (53.8) | 20 (57.1) | |
| HER2+ | 47 (12.5) | 2 (5.7) | |
| TNBC | 51 (13.5) | 2 (5.7) | |
| Claudinlow | - | - | |
| Basal | - | - | |
| Missing | 0 (0) | 0 (0) | |
| HR/HER2 status, n (%) | 0.227 | ||
| HR+/HER2− | 214 (56.8) | 23 (65.7) | |
| HR+/HER2+ | 65 (17.2) | 8 (22.9) | |
| HR−/HER2+ | 47 (12.5) | 2 (5.7) | |
| HR−/HER2− | 51 (13.5) | 2 (5.7) | |
| Missing | 0 (0) | 0 (0) | |
|
| |||
| B, TCGA (n=1,108) | |||
|
| |||
| Clinicopathological characteristic | Wild-type | Mutant | P-value |
|
| |||
| Mean age ± SD, years | 58.1±13.3 | 60.9±12.6 | 0.064 |
| Type, n (%) | 0.054 | ||
| Ductal | 747 (73.2) | 74 (84.1) | |
| Lobular | 195 (19.1) | 12 (13.6) | - |
| Other | 78 (7.6) | 2 (2.3) | - |
| Missing | 0 (0) | 0 (0) | |
| Node status, n (%) | 0.917 | ||
| pN0 | 476 (46.7) | 40 (45.5) | |
| pN1-pN3 | 523 (51.3) | 45 (51.1) | |
| Missing | 21 (2.0) | 3 (3.4) | |
| TNM stage, n (%) | 0.647 | ||
| 1 | 165 (16.2) | 18 (20.5) | |
| 2 | 581 (57.0) | 45 (51.1) | |
| 3 | 233 (22.8) | 18 (20.5) | |
| 4 | 18 (17.6) | 2 (2.3) | |
| Missing | 23 (2.3) | 5 (5.7) | |
| Pathological grade, n (%) | - | ||
| 1 | - | - | - |
| 2 | - | - | - |
| 3 | - | - | - |
| ER, n (%) | 0.043 | ||
| Positive | 740 (72.5) | 74 (84.1) | |
| Negative | 242 (23.7) | 12 (13.6) | |
| Missing | 38 (3.7) | 2 (2.3) | |
| PR, n (%) | 0.017 | ||
| Positive | 638 (62.5) | 67 (76.1) | |
| Negative | 342 (33.5) | 18 (20.4) | |
| Missing | 40 (3.9) | 3 (3.4) | |
| HR, n (%) | 0.01 | ||
| Positive | 765 (75.0) | 76 (86.4) | |
| Negative | 255 (25.0) | 12 (13.6) | |
| Missing | 0 (0) | 0 (0) | |
| HER2, n (%) | 0.53 | ||
| Positive | 170 (16.7) | 15 (17.0) | |
| Negative | 656 (64.3) | 57 (64.8) | |
| Missing | 194 (19.0) | 16 (18.2) | |
| Subtype, n (%) | - | ||
| LumA | - | - | |
| LumB | - | - | |
| HER2+ | - | - | |
| TNBC | - | - | |
| Claudinlow | - | - | |
| Basal | - | - | |
| Missing | - | - | |
| HR/HER2 status, n (%) | 0.106 | ||
| HR+/HER2− | 514 (50.4) | 51 (58.0) | |
| HR+/HER2+ | 131 (12.8) | 14 (16.0) | |
| HR−/HER2+ | 39 (3.8) | 1 (1.1) | |
| HR−/HER2− | 142 (13.9) | 6 (6.8) | |
| Missing | 194 (19.0) | 16 (18.2) | |
|
| |||
| C, METABRIC (n=2,509) | |||
|
| |||
| Clinicopathological characteristic | Wild-type | Mutant | P-value |
|
| |||
| Mean age ± SD, years | 60.2±13.0 | 62.3±12.8 | 0.018 |
| Type, n (%) | 0.887 | ||
| Ductal | 1,690 (74.6) | 175 (71.7) | |
| Lobular | 172 (7.6) | 20 (8.2) | - |
| Other | 403 (17.8) | 49 (20.1) | - |
| Missing | 0 (0) | 0 (0) | |
| Node status, n (%) | 0.106 | ||
| pN0 | 1,087 (48.0) | 109 (44.7) | |
| pN1-pN3 | 930 (41.0) | 117 (48.0) | |
| Missing | 248 (10.9) | 18 (7.4) | |
| TNM stage, n (%) | 0.933 | ||
| 1 | 566 (25.0) | 64 (26.2) | |
| 2 | 887 (39.2) | 92 (37.7) | |
| 3 | 131 (5.8) | 13 (5.3) | |
| 4 | 10 (0.4) | 1 (0.4) | |
| Missing | 671 (29.6) | 74 (30.3) | |
| Pathological grade, n (%) | <0.001 | ||
| 1 | 173 (7.6) | 41 (16.8) | |
| 2 | 854 (37.7) | 122 (50.0) | |
| 3 | 1,128 (49.8) | 70 (28.7) | |
| Missing | 110 (4.9) | 11 (4.5) | |
| ER, n (%) | <0.001 | ||
| Positive | 1,610 (71.1) | 215 (88.1) | |
| Negative | 617 (27.2) | 27 (11.1) | |
| Missing | 38 (1.7) | 2 (0.8) | |
| PR, n (%) | <0.001 | ||
| Positive | 887 (39.2) | 152 (62.3) | |
| Negative | 887 (39.2) | 54 (22.1) | |
| Missing | 491 (21.7) | 38 (15.6) | |
| HR, n (%) | - | - | - |
| Positive | - | - | - |
| Negative | - | - | - |
| Missing | - | - | - |
| HER2, n (%) | <0.001 | ||
| Positive | 1,022 (45.1) | 153 (62.7) | |
| Negative | 752 (33.2) | 53 (21.7) | |
| Missing | 491 (21.7) | 38 (15.6) | |
| Subtype, n (%) | <0.001 | ||
| LumA | 589 (26.0) | 111 (45.5) | |
| LumB | 433 (19.1) | 42 (17.2) | |
| HER2+ | 208 (9.2) | 16 (6.6) | |
| TNBC | - | - | |
| Claudinlow | 209 (9.2) | 9 (3.7) | |
| Basal | 198 (8.7) | 11 (4.5) | |
| Missing | 628 (27.7) | 55 (22.5) | |
| HR/HER2 status, n (%) | |||
| HR+/HER2− | - | - | - |
| HR+/HER2+ | - | - | - |
| HR−/HER2+ | - | - | - |
| HR−/HER2− | - | - | - |
| Missing | |||
Continuous variables were compared using Student's t-test or Mann-Whitney's U-test. Categorical data were compared using Pearson's χ2 test. ER, estrogen receptor, GDPH, Guangdong Provincial People's Hospital; HER2, human epidermal growth factor receptor 2; Lum, luminal; METABRIC, Molecular Taxonomy of Breast Cancer International Consortium; PR, progesterone receptor; HR, hormone receptor; Pos., possible; TCGA, The Cancer Genome Atlas; TNBC, triple-negative breast cancer; TNM, tumour-node-metastasis.
Prognostic implications of MAP3K1 mutations in breast cancer
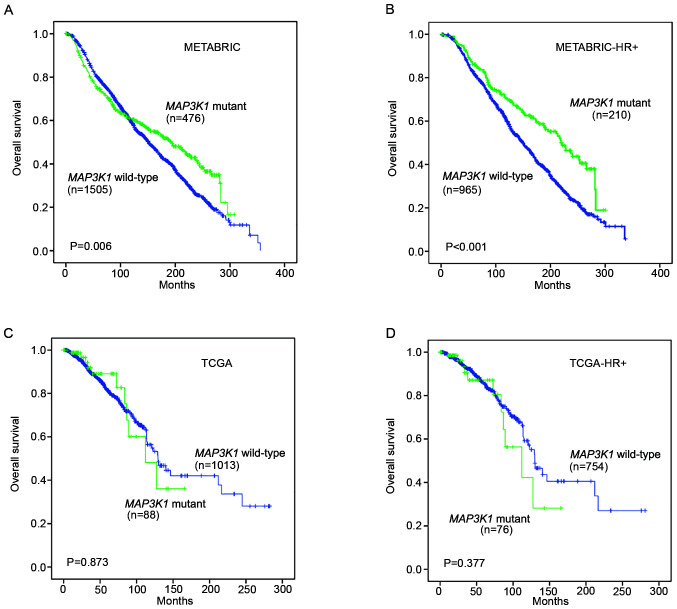
Kaplan-Meier survival analysis was performed on the METABRIC and TCGA datasets to assess the prognostic value of MAP3K1 mutations in the follow-up of patients with breast cancer.
In the METABRIC cohort, patients with MAP3K1 mutations experienced a prolonged OS compared with patients without MAP3K1 mutations (P=0.006; Fig. 2A). In patients with HR+ breast cancer, a significantly longer OS was also observed in the MAP3K1 mutation group (P<0.001; Fig. 2B). Unexpectedly, TCGA cohort and TCGA HR+ mutation subgroup did not display a significant difference in OS (P=0.873 and P=0.377, respectively; Fig. 2C and D).
Figure 2.
Kaplan-Meier analysis of the overall survival of patients stratified according to MAP3K1 mutation status. Kaplan-Meier overall survival curves are shown for (A) 1,981 patients in the METABRIC cohort stratified according to MAP3K1 mutation status, (B) the HR+ subgroup of the METABRIC cohort after stratification according to MAP3K1 mutation status, (C) 1,101 patients in TCGA stratified according to the MAP3K1 mutation status and (D) the HR+ subgroup of TCGA patients stratified according to MAP3K1 mutation status. Cumulative survival rates were analyzed using the Kaplan-Meier method and the log rank test. METABRIC, Molecular Taxonomy of Breast Cancer International Consortium; HR, hormone receptor; TCGA, The Cancer Genome Atlas.
Interactions between MAP3K1, MAP2K1, MAP2K2, MAP2K4 and PIK3CA mutations
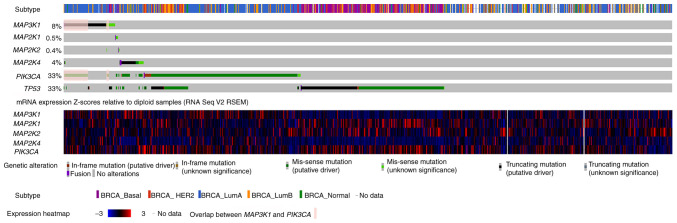
OncoPrint data for MAP3K1, MAP2K4 and PI3K catalytic subunit α (PIK3CA) mutations in the METABRIC cohort are shown in Fig. 3. MAP3K1 mutations in breast cancer were mutually exclusive with respect to mutations in MAP2K4, MAP2K1 and MAP2K2. Indeed, out of 217 patients who had a MAP3K1 mutation, only 3 also had MAP2K4 mutations, none had a MAP2K1 mutation and only one had a MAP2K2 mutation. Nevertheless, 144 PIK3CA mutations were observed in the 217 patients with MAP3K1 mutations, indicating that MAP3K1 alterations were concurrently accompanied by PIK3CA mutations (as indicated by the color overlap in the second and sixth rows; Fig. 3).
Figure 3.
Characterization of MAP3K1, MAP2K1, MAP2K2, MAP4K2 and PIK3CA mutations in patients with breast cancer from the METABRIC cohort. The OncoPrint of MAP3K1, MAP2K4 and PIK3CA genes is shown, in which individual samples and genes in the Molecular Taxonomy of Breast Cancer International Consortium cohort are presented in columns and rows, respectively. Subtype assignments of the patients are shown in the first row. mRNA expression is shown in the next 6 rows. Overlap between MAP3K1 and PIK3CA mutation are shown in pink. The data were obtained from and visualized using the cBioPortal database (accession number, EGAS00001001753) with modifications. RSEM was used for accurate transcript quantification from RNA Seq data. BRCA, breast cancer; HER2, human epidermal growth factor receptor 2; PIK3CA, PI3K catalytic subunit α; TP53, tumor protein 53; RNA Seq, RNA sequencing; RSEM, RNA Seq by Expectation Maximization.
Effect of MAP3K1 expression on the prognosis of patients with different intrinsic molecular subtypes of breast cancer
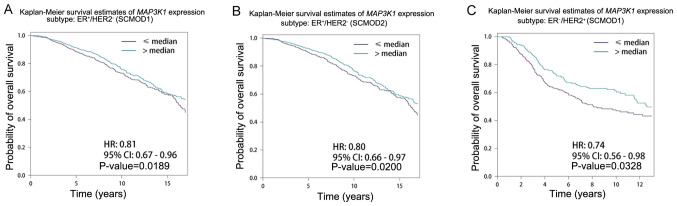
The effect of MAP3K1 expression on patients with different intrinsic molecular subtypes was further examined. Patients were grouped into two groups: High (>median) and low expression (<median) groups. High MAP3K1 expression was associated with longer survival time in the ER+/HER2− and ER−/HER2+ subgroups (Table II). Representative survival curves are shown in Fig. 4. Patients with higher MAP3K1 expression had longer survival time compared with that in patients with lower MAP3K1 expression in the ER+/HER2-subgroup of SCMOD1 (P=0.189; Fig. 4A) and SCMOD2 (P=0.020; Fig. 4B), and ER-/HER2+ subgroup of SCMOD1 (P=0.0328; Fig. 4C).
Table II.
Univariate Cox analysis of MAP3K1 expression level of overall survival in patients with different breast cancer molecular subtypes using three SCMs.
| A, SCMOD1 | ||
|---|---|---|
|
| ||
| Molecular subtype, high vs. low | HR (95% CI) | +P-value |
| ER−/HER2− | 0.87 (0.68-1.11) | 0.2733 |
| ER−/HER2+ | 0.74 (0.56-0.98) | 0.0328 |
| ER+/HER2− | 0.81 (0.67-0.96) | 0.0189 |
| ER+/HER2+ | 0.88 (0.75-1.03) | 0.1027 |
|
| ||
| B, SCMOD2 | ||
|
| ||
| Molecular subtype, high vs. low | HR (95% CI) | P-value |
| ER−/HER2− | 0.91 (0.72-1.14) | 0.3931 |
| ER−/HER2+ | 0.78 (0.60-1.00) | 0.0504 |
| ER+/HER2− | 0.80 (0.66-0.97) | 0.0200 |
| ER+/HER2+ | 0.87 (0.74-1.03) | 0.0991 |
|
| ||
| C, SCMGENE | ||
|
| ||
| Molecular subtype, high vs. low | HR (95% CI) | P-value |
|
| ||
| ER−/HER2− | 0.85 (0.69-1.04) | 0.1192 |
| ER−/HER2+ | 0.87 (0.68-1.12) | 0.2929 |
| ER+/HER2− | 0.91 (0.76-1.09) | 0.3063 |
| ER+/HER2+ | 0.87 (0.74-1.03) | 0.1000 |
The association between MAP3K1 expression and overall survival was analyzed using Univariate Cox analysis. The SCMOD1, SCMOD2 and SCMGENE SCMs were used. All HR and 95% CI values are rounded up or rounded down to 2 decimal places. CI, confidence interval; ER, estrogen receptor, HER2, human epidermal growth factor receptor 2; HR, hazard ratio; SCM, subtype clustering model.
Figure 4.
Kaplan-Meier analysis of the overall survival of patients stratified according to MAP3K1 expression. Kaplan-Meier overall survival curves are shown for the (A) ER+/HER2− subgroup in the SCMOD1 subtype clustering model after stratification according to MAP3K1 expression, (B) the ER+/HER2− subgroup in the SCMOD2 subtype clustering model after stratification according to MAP3K1 expression and (C) the ER−/HER2+ subgroup in the SCMOD1 subtype clustering model after stratification according to MAP3K1 expression. Cumulative survival rates were analyzed using the Kaplan-Meier method and the log rank test. ER, estrogen receptor; HER2, human epidermal growth factor receptor 2; SCMOD, subtype clustering model.
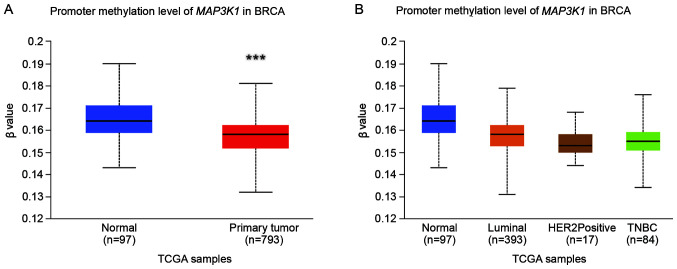
MAP3K1 promoter methylation in breast cancer
The DNA methylation profile was analysed using UALCAN to further explore the epigenetic regulation of MAP3K1 methylation in breast cancer. Significant hypermethylation of MAP3K1 was observed (Fig. 5A; P<0.0001). However, no difference in methylation was observed between the normal, luminal A and B, HER2+ and TNBC subgroups (Fig. 5B).
Figure 5.
MAP3K1 promoter methylation profile in TCGA-BRCA samples. (A) Levels of MAP3K1 promoter methylation in normal tissue and BRCA tissue. Significance of difference was estimated using Mann-Whitney's U-test. (B) Levels of MAP3K1 promoter methylation in normal tissue and tumour tissue from patients with different BRCA molecular subtypes. The β value indicates the levels of DNA methylation and ranges from 0 (unmethylated) to 1 (fully methylated). A β value within the 0.5-0.7 range indicates hypermethylation, whereas a value in the 0.25-0.3 range indicates hypomethylation. ***P<0.001. BRCA, breast cancer; HER2, human epidermal growth factor receptor 2; TCGA, The Cancer Genome Atlas; TNBC, triple-negative breast cancer.
Discussion
The present findings identified MAP3K1 mutations in Chinese patients with breast cancer, as well as in public datasets. Given the differences in cancer biology in patients from different ethnic groups, the MAP3K1 mutation spectrum in Chinese patients was compared with data available in public databases. The current retrospective study is the first comprehensive report to analyse the clinicopathological characteristics of Chinese patients with MAP3K1 mutations in a Chinese cohort from a single centre. In the GDPH cohort, 412 breast cancer samples were analysed using NGS and further compared with data from TCGA and METABRIC cohorts. A similar prevalence in MAP3K1 mutations was observed in these three cohorts, whereas the mutated domains and mutation hotspots did not overlap. A recent study including 122 Chinese patients with breast cancer or ovarian cancer reported a higher frequency of MAP3K1 mutation (14.3%) (30). Although MAP3K1 is among the most frequently mutated genes in cancer, to the best of our knowledge, no study has reported frequent hotspot- or domain-specific alterations associated with MAP3K1 mutations, consistent with the results of the current study. Notably, 21.6% tumors concurrently carried >1 mutation in MAP3K1. Among those tumors, seven tumors contained dual MAP3K1 mutations and one tumor contained triplet mutations. Although MAP3K1 mutations potentially represent a protective marker in breast cancer, it was not possible to determine whether the presence of concurrent dual MAP3K1 mutations had different clinicopathological characteristics from single mutations due to the small cohort size.
Breast cancer is a heterogeneous disease. Distributions of MAP3K1 mutations were analysed. In TCGA and METABRIC cohorts, MAP3K1 mutations were more frequently enriched in patients with HR+ breast cancer than in patients with other subtypes including (HER2+ and TNBC), consistent with previous studies (9,31). Only 4 of the 35 patients who carried the MAP3K1 mutation in the GDPH cohort had HR− disease. The insignificance of the difference in the GDPH cohort may be due to the small cohort size.
Ellis et al (32) conducted whole-genome sequencing and pathway analyses to compare patients who were sensitive or resistant to neoadjuvant treatment with an aromatase inhibitor. Based on the study, mutations in the MAP3K1 gene were found to be related to indolent biological features and low proliferation rates. Therefore, MAP3K1 mutations have been proposed to correlate with favorable outcomes in patients with breast cancer (7). Similarly, a two-gene genomic signature combining with MAP3K1 mutation and FGFR1 amplification was associated with decreased risk of distant metastasis in patients with HR+/HER2− breast cancer (33). MAP3K1-targeting drugs have been developed, including binimetinib (34), fostamatinib (35) and AZD-8330 (36). Binimetinib is used to treat metastatic melanoma and unresectable melanoma, while fostamatinib is used to treat chronic immune thrombocytopenia. To date, Food and Drug Administration-approved MAP3K1-targeting drugs are not available for the treatment of breast cancer.
MAP3K1 activates the JNK pathway by selectively phosphorylating and activating MAP2K4 (37,38). Deficiency in the JNK signaling pathway results in defective apoptosis, leading to unresponsiveness to environmental and genotoxic stresses (39). Ellis et al (7) demonstrated that MAP2K4 mutations were associated with MAP3K1 dysfunction, but mutations in two genes associated with one pathway were rarely observed in a single patient. Tissues with MAP3K1 mutation in the METABRIC cohort did not coincide with mutations in MAP2K4. Considering the differences in samples with MAP3K1 and MAP2K4 mutations, loss of function of either gene in the JNK pathway may result in the deregulation of the other gene and the pathway as a whole.
In addition, MAP3K1 and PIK3CA mutations co-associated in both the METABRIC and TCGA cohorts. In the METABRIC cohort, PIK3CA mutations were accompanied with MAP3K1 mutations in 66.4% of patients. A previous study has identified MAP3K1 and MAP2K2 as powerful predictors of responsiveness to MEK inhibition in patient-derived xenograft models (40). Loss-of-function mutations in MAP3K1 resulted in higher sensitivity to MEK inhibition, demonstrating that MEK inhibitors could represent potential therapeutic targets for tumors with MAP3K1 mutations (40).
In the present study, MAP3K1 expression was confirmed to be associated with OS in different subtype clustering models of breast cancer, supporting the prognostic value of MAP3K1. However, high MAP3K1 was only a protective biomarker in patients with ER+/HER2- and ER-/HER2+ disease.
To the best of our knowledge, MAP3K1 methylation has not yet been reported in breast tumors. In the current study, the methylation of the MAP3K1 gene was decreased in breast cancer tissue samples from TCGA, compared with normal tissue. This result may partially explain the epigenetic regulation of MAP3K1 in breast cancer, although further studies are needed to confirm this finding.
The current study has a number of limitations. Since the present findings were obtained from a retrospective, single-center study, the limited cohort size prevented robust comparison between the Chinese patient cohort and TCGA and METABRIC cohorts. As indicated in our previous report (19), the baseline characteristics of the patients enrolled in the three cohorts, including age, menopausal status, histological grade, ER status, PR status and HER2 status, were significantly different. Moreover, 5.5% patients in TCGA cohort were Asian (60/1084). Ethnicity was not reported for the METABRIC cohort. All patients in the GDPH cohort were Chinese and 98.8% were Han Chinese (407/412). In addition, due to the absence of complete follow-up and prognostic data in the GDPH cohort, the effect of MAP3K1 mutations on the survival of the Chinese patients could not be established. The methylation data for MAP3K1 also lacked from the GDPH cohort. Further multicenter prospective studies with complete follow-up data are needed to verify the present findings.
Altogether, the present findings identified MAP3K1 mutations in Chinese patients with breast cancer, as well as public datasets. MAP3K1 mutations were mainly detected in HR+/HER2− breast cancer and could represent possible prognostic factors. MAP3K1 expression was associated with OS in the HR+ subgroup. Furthermore, a lower level of MAP3K1 methylation was observed in patients with breast cancer. A larger sample size is needed to evaluate the clinical applicability of MAP3K1 mutations in patients with HR+ breast cancer. The results may provide insight into the pathophysiological mechanism of breast cancer and the development of novel therapeutic treatments.
Acknowledgements
Not applicable.
Funding Statement
This study was supported by grants from The Project of Doctoral Scientific Research of Guangdong Provincial People's Hospital (grant. no. 2020bq12), The National Natural Science Foundation of China (grant nos. 81602645, 81702783, 81071851 and 81001189), The Natural Science Foundation of Guangdong Province (grant nos. 2016A030313768, 2017A030310574 and 2018A030313292) and research funds from The Guangzhou Municipal Science and Technology Project (grant nos. 201707010418 and 201804010430).
Availability of data and materials
The datasets generated and/or analyzed in the GDPH cohort are available in the NODE repository (http://www.biosino.org/node) under accession no. OEP001295. Data for the MAP3K1 mutation profiles and clinical characteristics of breast cancer in TCGA and METABRIC cohorts were extracted from cBioPortal (http://www.cbioportal.org).
Authors' contributions
CL and NL conceived the study. YW, GZ, KL, LC, CR, XL collected the data. GZ, YW, BC and NL acquired funding. CL, LW, MJ, HM, JL, WX and XL carried out the experiments. CL, GZ, BC, JL, XL and NL designed the methodology. YW, BC, KL, LC, CR, LW and WX oversaw project administration; YW, KL and LC provided software. CL wrote the original draft; CL, GZ, KL, LC, CR, XL and NL reviewed and edited the manuscript. CL and NL confirm the authenticity of all the raw data. All authors read and approved the final manuscript.
Ethics approval and consent to participate
This study was approved by The Ethics Committee of The Guangdong Provincial People's Hospital (approval no. GDREC2014122H). All patients provided written informed consent to participate in this translational study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Luo CY, Li N, Lu B, Cai J, Lu M, Zhang YH, Chen HD, Dai M. Global and regional trends in incidence and mortality of female breast cancer and associated factors at national level in 2000 to 2019. Chin Med J (Engl) 2021 September 28; doi: 10.1097/CM9.0000000000001814. (Epub ahead of print). doi: 10.1097/CM9.0000000000001814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Waks AG, Winer EP. Breast cancer treatment: A review. JAMA. 2019;321:288–300. doi: 10.1001/jama.2018.19323. [DOI] [PubMed] [Google Scholar]
- 3.Oh DY, Bang YJ. HER2-targeted therapies-a role beyond breast cancer. Nat Rev Clin Oncol. 2020;17:33–48. doi: 10.1038/s41571-019-0268-3. [DOI] [PubMed] [Google Scholar]
- 4.Andre F, Ciruelos E, Rubovszky G, Campone M, Loibl S, Rugo HS, Iwata H, Conte P, Mayer IA, Kaufman B, et al. Alpelisib for PIK3CA-mutated, hormone receptor-positive advanced breast cancer. N Engl J Med. 2019;380:1929–1940. doi: 10.1056/NEJMoa1813904. [DOI] [PubMed] [Google Scholar]
- 5.Hortobagyi GN, Chen D, Piccart M, Rugo HS, Burris HA, III, Pritchard KI, Campone M, Noguchi S, Perez AT, Deleu I, et al. Correlative analysis of genetic alterations and everolimus benefit in hormone receptor-positive, human epidermal growth factor receptor 2-negative advanced breast cancer: Results from BOLERO-2. J Clin Oncol. 2016;34:419–426. doi: 10.1200/JCO.2014.60.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gong X, Litchfield LM, Webster Y, Chio LC, Wong SS, Stewart TR, Dowless M, Dempsey J, Zeng Y, Torres R, et al. Genomic aberrations that activate D-type cyclins are associated with enhanced sensitivity to the CDK4 and CDK6 inhibitor abemaciclib. Cancer Cell. 2017;32:761–776.e6. doi: 10.1016/j.ccell.2017.11.006. [DOI] [PubMed] [Google Scholar]
- 7.Ellis MJ, Ding L, Shen D, Luo J, Suman VJ, Wallis JW, Van Tine BA, Hoog J, Goiffon RJ, Goldstein TC, et al. Whole-genome analysis informs breast cancer response to aromatase inhibition. Nature. 2012;486:353–360. doi: 10.1038/nature11143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pham TT, Angus SP, Johnson GL. MAP3K1: Genomic alterations in cancer and function in promoting cell survival or apoptosis. Genes Cancer. 2013;4:419–426. doi: 10.1177/1947601913513950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pereira B, Chin SF, Rueda OM, Vollan HK, Provenzano E, Bardwell HA, Pugh M, Jones L, Russell R, Sammut SJ, et al. The somatic mutation profiles of 2,433 breast cancers refines their genomic and transcriptomic landscapes. Nat Commun. 2016;7:11479. doi: 10.1038/ncomms11908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hammond ME, Hayes DF, Dowsett M, Allred DC, Hagerty KL, Badve S, Fitzgibbons PL, Francis G, Goldstein NS, Hayes M, et al. American Society of Clinical Oncology/College Of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. J Clin Oncol. 2010;28:2784–2795. doi: 10.1200/JCO.2009.25.6529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wolff AC, Hammond ME, Hicks DG, Dowsett M, McShane LM, Allison KH, Allred DC, Bartlett JM, Bilous M, Fitzgibbons P, et al. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American society of clinical oncology/college of American pathologists clinical practice guideline update. J Clin Oncol. 2013;31:3997–4013. doi: 10.1200/JCO.2013.50.9984. [DOI] [PubMed] [Google Scholar]
- 13.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sahraeian SME, Mohiyuddin M, Sebra R, Tilgner H, Afshar PT, Au KF, Bani Asadi N, Gerstein MB, Wong WH, Snyder MP, et al. Gaining comprehensive biological insight into the transcriptome by performing a broad-spectrum RNA-seq analysis. Nat Commun. 2017;8:59. doi: 10.1038/s41467-017-00050-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang K, Li M, Hakonarson H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cingolani P, Platts A, Wang le L, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Newman AM, Bratman SV, Stehr H, Lee LJ, Liu CL, Diehn M, Alizadeh AA. FACTERA: A practical method for the discovery of genomic rearrangements at breakpoint resolution. Bioinformatics. 2014;30:3390–3393. doi: 10.1093/bioinformatics/btu549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen X, Zhang G, Chen B, Wang Y, Guo L, Cao L, Ren C, Wen L, Liao N. Association between histone lysine methyltransferase KMT2C mutation and clinicopathological factors in breast cancer. Biomed Pharmacother. 2019;116:108997. doi: 10.1016/j.biopha.2019.108997. [DOI] [PubMed] [Google Scholar]
- 19.Zhang G, Wang Y, Chen B, Guo L, Cao L, Ren C, Wen L, Li K, Jia M, Li C, et al. Characterization of frequently mutated cancer genes in Chinese breast tumors: A comparison of Chinese and TCGA cohorts. Ann Transl Med. 2019;7:179. doi: 10.21037/atm.2019.04.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stephens PJ, Tarpey PS, Davies H, Van Loo P, Greenman C, Wedge DC, Nik-Zainal S, Martin S, Varela I, Bignell GR, et al. The landscape of cancer genes and mutational processes in breast cancer. Nature. 2012;486:400–404. doi: 10.1038/nature11017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jezequel P, Campone M, Gouraud W, Guerin-Charbonnel C, Leux C, Ricolleau G, Campion L. bc-GenExMiner: An easy-to-use online platform for gene prognostic analyses in breast cancer. Breast Cancer Res Treat. 2012;131:765–775. doi: 10.1007/s10549-011-1457-7. [DOI] [PubMed] [Google Scholar]
- 22.Jezequel P, Frenel JS, Campion L, Guerin-Charbonnel C, Gouraud W, Ricolleau G, Campone M. bc-GenExMiner 3.0: New mining module computes breast cancer gene expression correlation analyses. Database (Oxford) 2013;2013:bas060. doi: 10.1093/database/bas060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sorlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel A, Deng S, Johnsen H, Pesich R, Geisler S, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci USA. 2003;100:8418–8423. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu Z, Fan C, Oh DS, Marron JS, He X, Qaqish BF, Livasy C, Carey LA, Reynolds E, Dressler L, et al. The molecular portraits of breast tumors are conserved across microarray platforms. BMC Genomics. 2006;7:96. doi: 10.1186/1471-2164-7-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Desmedt C, Haibe-Kains B, Wirapati P, Buyse M, Larsimont D, Bontempi G, Delorenzi M, Piccart M, Sotiriou C. Biological processes associated with breast cancer clinical outcome depend on the molecular subtypes. Clin Cancer Res. 2008;14:5158–5165. doi: 10.1158/1078-0432.CCR-07-4756. [DOI] [PubMed] [Google Scholar]
- 26.Wirapati P, Sotiriou C, Kunkel S, Farmer P, Pradervand S, Haibe-Kains B, Desmedt C, Ignatiadis M, Sengstag T, Schütz F, et al. Meta-analysis of gene expression profiles in breast cancer: Toward a unified understanding of breast cancer subtyping and prognosis signatures. Breast Cancer Res. 2008;10:R65. doi: 10.1186/bcr2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parker JS, Mullins M, Cheang MC, Leung S, Voduc D, Vickery T, Davies S, Fauron C, He X, Hu Z, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27:1160–1167. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haibe-Kains B, Desmedt C, Rothe F, Piccart M, Sotiriou C, Bontempi G. A fuzzy gene expression-based computational approach improves breast cancer prognostication. Genome Biol. 2010;11:R18. doi: 10.1186/gb-2010-11-2-r18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK, Varambally S. UALCAN: A portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19:649–658. doi: 10.1016/j.neo.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kwong A, Cheuk IW, Shin VY, Ho CY, Au CH, Ho DN, Wong EY, Yu SW, Chen J, Chan KK, et al. Somatic mutation profiling in BRCA-negative breast and ovarian cancer patients by multigene panel sequencing. Am J Cancer Res. 2020;10:2919–2932. [PMC free article] [PubMed] [Google Scholar]
- 31.Griffith OL, Spies NC, Anurag M, Griffith M, Luo J, Tu D, Yeo B, Kunisaki J, Miller CA, Krysiak K, et al. The prognostic effects of somatic mutations in ER-positive breast cancer. Nat Commun. 2018;9:3476. doi: 10.1038/s41467-018-07407-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ellis MJ, Suman VJ, Hoog J, Goncalves R, Sanati S, Creighton CJ, DeSchryver K, Crouch E, Brink A, Watson M, et al. Ki67 proliferation index as a tool for chemotherapy decisions during and after neoadjuvant aromatase inhibitor treatment of breast cancer: Results from the American college of surgeons oncology group Z1031 trial (Alliance) J Clin Oncol. 2017;35:1061–1069. doi: 10.1200/JCO.2016.69.4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carene D, Tran-Dien A, Lemonnier J, Dalenc F, Levy C, Pierga JY, Jacot W, Canon JL, Richon C, Lacroix L, et al. Association between FGFR1 copy numbers, MAP3K1 mutations, and survival in axillary node-positive, hormone receptor-positive, and HER2-negative early breast cancer in the PACS04 and METABRIC studies. Breast Cancer Res Treat. 2020;179:387–401. doi: 10.1007/s10549-019-05462-y. [DOI] [PubMed] [Google Scholar]
- 34.Koelblinger P, Dornbierer J, Dummer R. A review of binimetinib for the treatment of mutant cutaneous melanoma. Future Oncol. 2017;13:1755–1766. doi: 10.2217/fon-2017-0170. [DOI] [PubMed] [Google Scholar]
- 35.Sweeny DJ, Li W, Clough J, Bhamidipati S, Singh R, Park G, Baluom M, Grossbard E, Lau DT. Metabolism of fostamatinib, the oral methylene phosphate prodrug of the spleen tyrosine kinase inhibitor R406 in humans: Contribution of hepatic and gut bacterial processes to the overall biotransformation. Drug Metab Dispos. 2010;38:1166–1176. doi: 10.1124/dmd.110.032151. [DOI] [PubMed] [Google Scholar]
- 36.Haasbach E, Hartmayer C, Planz O. Combination of MEK inhibitors and oseltamivir leads to synergistic antiviral effects after influenza A virus infection in vitro. Antiviral Res. 2013;98:319–324. doi: 10.1016/j.antiviral.2013.03.006. [DOI] [PubMed] [Google Scholar]
- 37.Yan M, Dai T, Deak JC, Kyriakis JM, Zon LI, Woodgett JR, Templeton DJ. Activation of stress-activated protein kinase by MEKK1 phosphorylation of its activator SEK1. Nature. 1994;372:798–800. doi: 10.1038/372798a0. [DOI] [PubMed] [Google Scholar]
- 38.Fanger GR, Johnson NL, Johnson GL. MEK kinases are regulated by EGF and selectively interact with Rac/Cdc42. EMBO J. 1997;16:4961–4972. doi: 10.1093/emboj/16.16.4961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wagner EF, Nebreda AR. Signal integration by JNK and p38 MAPK pathways in cancer development. Nat Rev Cancer. 2009;9:537–549. doi: 10.1038/nrc2694. [DOI] [PubMed] [Google Scholar]
- 40.Xue Z, Vis DJ, Bruna A, Sustic T, van Wageningen S, Batra AS, Rueda OM, Bosdriesz E, Caldas C, Wessels LFA, Bernards R. MAP3K1 and MAP2K4 mutations are associated with sensitivity to MEK inhibitors in multiple cancer models. Cell Res. 2018;28:719–729. doi: 10.1038/s41422-018-0044-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and/or analyzed in the GDPH cohort are available in the NODE repository (http://www.biosino.org/node) under accession no. OEP001295. Data for the MAP3K1 mutation profiles and clinical characteristics of breast cancer in TCGA and METABRIC cohorts were extracted from cBioPortal (http://www.cbioportal.org).