SUMMARY:
One of the hallmarks of the immune system is a dynamic landscape of cellular communication through the secretion of soluble factors, production of cell-bound ligands, and expression of surface receptors. This communication affects all aspects of immune cell behavior, integrates the responses of immune cells in tissues, and is fundamental to orchestrating effective immunity. Recent pioneering work has shown that the transfer of ribonucleic acids (RNAs) constitutes a novel mode of cellular communication. This communication involves diverse RNA species, with short noncoding RNAs especially enriched in the extracellular space. These RNAs are highly stable and selectively packaged for secretion. Transferred RNAs have functions in target cells that both mirror their cell-intrinsic roles as well as adopt novel mechanisms of action. These extracellular RNAs both impact the behavior of individual immune cells as well as participate in local and systemic immune responses. The impacts of RNA communication on immune cells and disease states have important implications for the development of novel clinical biomarkers and innovative therapeutic designs in immune-related disease. In this review, we will discuss the foundation of knowledge that is establishing RNA communication as an active and functional process in the immune system.
Keywords: extracellular RNA, exRNA, extracellular vesicles, EV, immune, inflammation, T cell, dendritic cell, macrophage, B cell, Argonaute, exomere, exosome, microvesicle, miRNA, tRNA, YRNA, snoRNA, vtRNA
1. INTRODUCTION
A complex ribonucleic acid (RNA) world exists in all cells. Our scientific understanding of this world began with RNAs that participate in protein synthesis through discoveries of messenger RNAs (mRNAs) which template, transfer RNAs (tRNAs) which decode, and ribosomal RNAs (rRNAs) which implement protein synthesis. While essential for cells to function, these RNAs are not the only classes of RNAs present within the cell. There has been an explosion in research which has uncovered multiple classes of noncoding RNAs that perform regulatory functions and partner with other biomolecules in the cell to execute function. Among these are long noncoding RNAs (lncRNAs) which serve diverse roles in regulating transcription, controlling protein function, and modifying the activity of other RNAs. There also exist many classes of <200 nucleotide short noncoding RNAs including small nuclear RNA (snRNAs) which participate in splicing, small nucleolar RNA (snoRNAs) which participate in RNA modification, microRNAs (miRNAs) which serve as post-transcriptional inhibitors of gene expression, vault RNAs (vtRNAs) which are implicated in intracellular transport, and YRNAs which are thought to participate in DNA replication and RNA quality control (Table 1).
Table 1: Summary of different classes of coding and noncoding RNAs.
Size in nucleotides (nt). References (Ref) for review of RNA class included.
| RNA Type | Class | Size | Structure | Function | Ref |
|---|---|---|---|---|---|
| Messenger RNA (mRNA) | Coding | Variable | Single stranded with a guanine 5’ cap and a polyadenylated 3’ tail. | Templates the genetic code and is directly translated into protein. | 100 |
| Transfer RNA (tRNA) | Noncoding, housekeeping | 75–90 nt | Three hairpin loops and stem. Anticodon loop base pairs with mRNA, and stem carries codon-specific amino acid. | Deliver amino acids to the nascent polypeptide chain during translation. | 101 |
| Ribosomal RNA (rRNA) | Noncoding, housekeeping | 120–1900 nt | Complex stem loop configurations form large and small ribosomal subunits. | Combine with proteins to form the ribosome to catalyze protein synthesis. | 102 |
| Long non-coding RNA (lncRNA) | Noncoding, regulatory | >200 nt | Variable (linear, circular, stem loop). | Chromatin remodeling, transcriptional regulation, translational regulation. | 103 |
| Small nuclear RNA (snRNA) | Noncoding, regulatory | ~150 nt | 5’ trimethyl guanosine or monomethylphosphate cap, 3’ stem loop +/− polyuridinylated tail. | Component of small nuclear RNPs (snRNPs) that coordinate RNA splicing. | 104 |
| Small nucleolar RNA (snoRNA) | Noncoding, regulatory | 60–300 nt | Closed loop (Box C/D) or stem loop (box H/ACA). | RNA modification. | 105 |
| MicroRNA (miRNA) | Noncoding, regulatory | ~22 nt | Single stranded with 5’ monophosphate | Post transcriptional inhibition of gene expression through mRNA targeting. | 21 |
| Vault RNA (vtRNA) | Noncoding, regulatory | 80–140 nt | Stem loop. | Regulate signaling pathways and transport of intracellular cargo. | 106 |
| YRNA (YRNA) | Noncoding, regulatory | ~110 nt | Stem loop. | Regulate DNA replication and participate in RNA quality control. | 107 |
We now know that this complex RNA world extends beyond the confines of the cell. Paradigm-shifting observations began 50 years ago when RNAs were first found in extracellular fluids.1 This has sparked much interest in RNAs as biomarkers for disease in plasma and other body fluids (reviewed2). While some have proposed that this extracellular RNA is simply cellular debris, accumulating evidence now strongly points to non-cell autonomous functions for actively secreted RNAs. What then are key features that would support a hypothesis that extracellular RNAs (exRNAs) serve as mediators of intracellular communication within the immune system?
First, we expect that exRNA should be stable outside the cell. Many studies demonstrate that exRNAs are stable, as they fail to degrade with the addition of exogenous RNA degrading enzymes.3–5 exRNAs are also abundant in biofluids which are naturally rich in RNA-degrading enzymes. While biofluids such as blood and airway lining fluid are capable of rapidly degrading naked RNA, endogenous exRNAs remain stable.6,7 Therefore protection, likely through selective packaging of exRNAs, should allow intact RNA to travel between cells.
Second, exRNAs should be secreted from immune cells. Cells of the immune system were among the first cells identified to produce exRNAs. In vitro studies of mast cell lines provided critical first evidence that immune cells can secrete exRNAs including mRNA and miRNA.3 Accumulated evidence now suggests that most, if not all, cells can produce exRNAs. In vitro a diverse population of exRNA are secreted by dendritic cells4,8,9, monocytes/macrophages10,11, mast cells3, T cells12,13, and B cells12. These studies also demonstrate that the exRNA content differs from the total RNA content of the producing immune cell. This especially includes selective secretion of small noncoding RNAs including miRNAs, snRNA, snoRNA, Y RNA, and fragments of tRNAs and rRNAs. Therefore, the RNA secretion into the extracellular space is not a simple dumping of intracellular stores, but the mechanisms of selective packaging by cells are only beginning to be elucidated.
Finally, exRNAs should be bioactive molecules. Early evidence for functional transfer of RNAs was also evident in immune cells. Harvested mast cell ex-mRNAs are competent for in vitro protein translation.3 Cultured dendritic cells are capable of secreting miRNA that can be transferred to target dendritic cells to modify expression of luciferase reporter constructs used to measure sequence-specific gene repression.4 In vivo functional transfer of ex-miRNA resulting in endogenous target gene suppression has been demonstrated using genetic miRNA knockout models.14 In this manuscript, we will review the mounting evidence that functional RNAs transmit essential signals between donor and acceptor immune cells (Figure 1). We will highlight known mechanisms of this RNA transfer, as well as the impact of this novel form of RNA-mediated communication on immunity, inflammation, and disease.
Figure 1: Extracellular RNA communication regulates immune cell function.
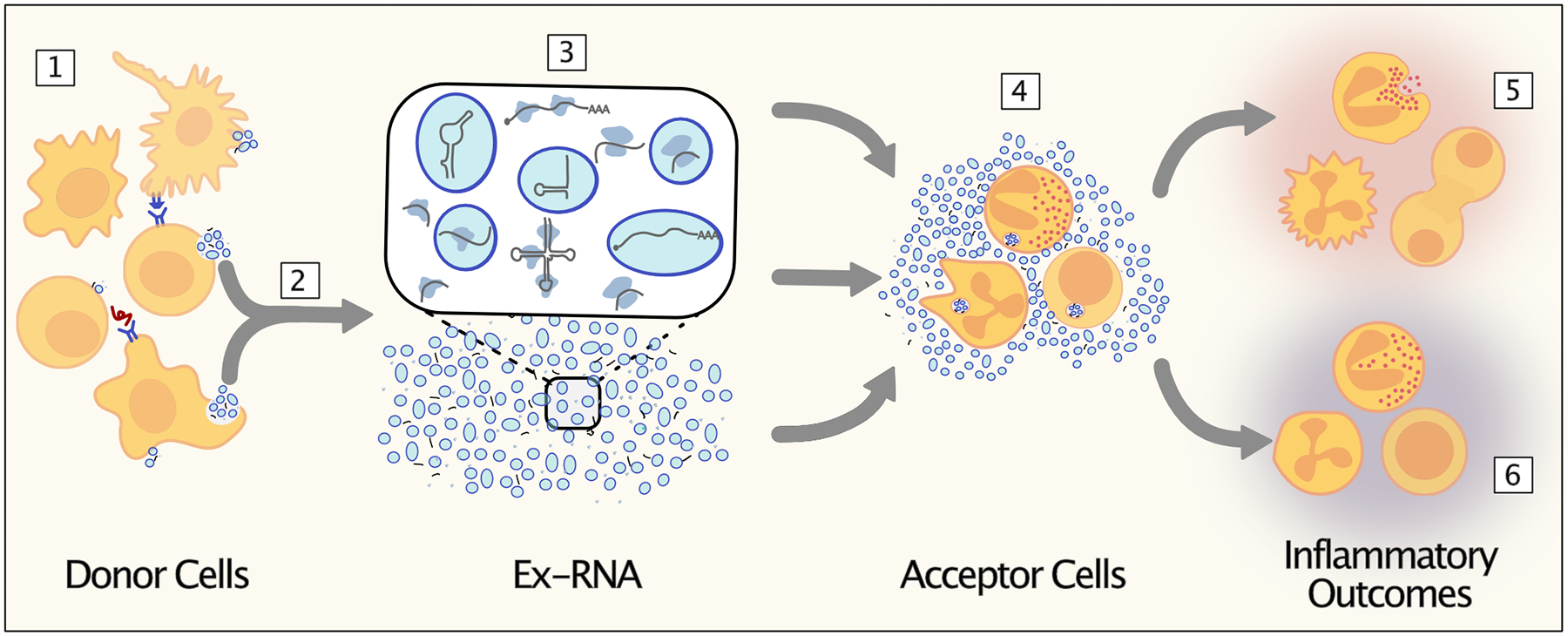
[1] Immune cells receive signals that result in their activation. [2] This stimulates the release of extracellular RNAs (exRNAs) and extracellular vesicles (EVs) from the donor cells. [3] These secreted exRNAs are of various classes and forms, some contained within EVs and some outside of EVs. [4] The exRNAs are delivered to acceptor cells where they are internalized. exRNAs can deliver signals to the acceptor cells to induce a more [5] pro-inflammatory response or [6] more anti-inflammatory response, which depends on the exRNA cell origin, initiating stimulus, form, and class.
2. FORMS OF EXTRACELLULAR RNA
The striking stability of RNAs detected in the extracellular space suggested that these RNAs must exist in a form shielded from RNase degrading activity. The most well-studied and widely identified form of packaging for exRNA is within extracellular vesicles (EVs) (Figure 2A). EVs are small lipid-bilayer delimited vesicles secreted from cells which contain cargos of lipids, proteins, and nucleic acids (reviewed15). EVs can either bud directly from the plasma membrane (so called microvesicles or ectosomes), or they can be released when the multivesicular body fuses with the plasma membrane to secrete inwardly budded vesicles that have formed within this organelle (so called exosomes). Although exosomes are typically smaller than microvesicles, they can have overlapping sizes with exosomes typically ranging from 50–150nm in size while microvesicles range from 100–500nm in size. As yet, there are no markers or cargos that universally distinguish EVs arising from these two distinct biogenesis pathways; however, both often carry a common set of EV markers including tetraspanins (i.e. CD9, CD63 and/or CD81) and intracellular cargos important for vesicle biogenesis or cargo loading (i.e. Alix, TSG101, Flotillin, Syntenin, ARF6). Immune cells secrete EVs containing exRNAs. This has been demonstrated through detergent disruption experiments that define exRNAs as cargos loaded within lipid-encapsulated structures secreted from T cells.13 This can also be demonstrated by rigorous EV purification methods which may include ultrafiltration, differential ultracentrifugation, density gradient ultracentrifugation and/or size exclusion chromatography.16 Density gradient ultracentrifugation has been used to separate light EVs from dense protein aggregates to demonstrate the presence of EV-encapsulated exRNAs from both T cells13 and dendritic cells4,8. The functional nature of EV-contained exRNAs has been demonstrated in EV-deficient Rab27a/Rab27b double knockout mice, where defects in systemic responses to lipopolysaccharide (LPS) can only be rescued by EVs carrying miR-155.17
Figure 2: Forms of RNAs in the extracellular space.
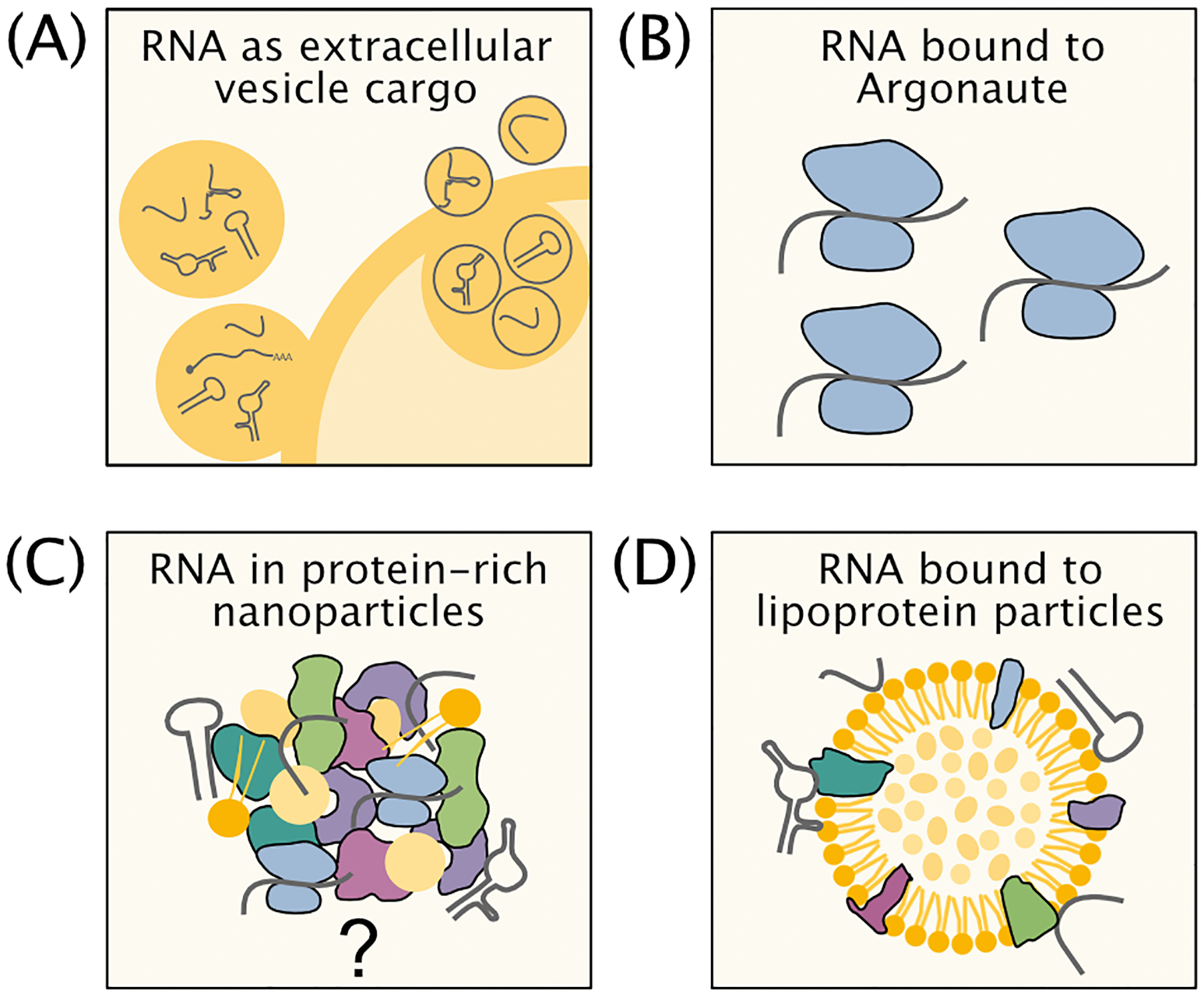
RNAs are packaged and protected from degradation by (A) extracellular vesicles, (B) within Argonaute protein, (C) in protein-rich nanoparticles, and (D) bound to the surface of lipoprotein particles.
While EVs may be common carriers of exRNAs, RNAs can also exist within the extracellular space in non-vesicular forms. In plasma, cerebrospinal fluid, and cell culture supernatants, miRNAs associate with Argonaute (AGO) protein outside of vesicles5,18–20 (Figure 2B). AGO is a central component of the RNA induced silencing complex that binds mature single stranded miRNAs and is required for miRNAs to target and destabilize mRNAs within cells (reviewed21). Identifying miRNAs in association with AGO suggests that loaded and functionally active miRNAs could be available for transfer to acceptor cells. AGO-miRNA complexes isolated from plasma are capable of binding synthetic RNA target sequences, and ≥50% of tested miRNAs can be depleted by this strategy demonstrating that most of these ex-miRNAs retain their targeting capabilities.19 However, their ability to be transferred to and function within target cells remains to be determined. Although we do not yet understand the cellular sources for these complexes and have little insight into their secretion, there is some early evidence that the distribution of ex-miRNAs contained in AGO complexes change in disease. The cerebrospinal fluid of patients with amyotrophic lateral sclerosis (ALS) contains fewer miRNAs in AGO-containing complexes, suggesting a shift or alteration in the form of exRNA present during disease.19 Understanding the function of these complexes is particularly important given that the majority of AGO protein in some extracellular and biofluids is associated with EV-free fractions as shown by density gradient centrifugation and ultrafiltration experiments.5,22,23
Although EVs and Ago-containing protein complexes are the most well-studied forms of exRNAs, there are likely additional packaging partners for secreted RNAs that contribute to their stability and targeting. A wide array of proteins and protein complexes exist within extracellular fluids and biofluids, and some of these are likely to interact with exRNA. exRNAs can be associated with small (<20–40nm) protein-rich nanoparticles, termed exomeres24 (Figure 2C). exRNAs in this non-vesicular fraction appear to be enriched in small noncoding RNAs, especially fragments of tRNAs, fragments of YRNAs, vtRNAs, and a subset of miRNA species.23,25 In circulation, there is evidence that extracellular miRNAs can be carried on lipoprotein particles as well (Figure 2D). Extracellular miRNAs bind to high density lipoprotein (HDL), and the composition of miRNAs carried on HDL changes in hypercholesterolemia, atherosclerosis, and acute coronary syndrome.26,27 HDL-bound miRNAs can be delivered to target cells to change gene expression in target reporter constructs26, and miR-223 delivered by HDL to endothelial cells can change intracellular adhesion molecule 1 (Icam1) expression.28 miR-223 can be exported to HDL from neutrophils and macrophages, and the amount of export to this form changes after cellular activation.29 Therefore, lipoprotein-loaded forms of exRNAs may participate in exRNA communication during inflammation in vivo.
3. STOCHIOMETRY OF PACKAGING RNA INTO EXTRACELLULAR VESICLES
While it is clear that RNAs can be packaged into vesicles in the extracellular space, the distribution of exRNAs within EVs remains an area of active investigation. Conflicting experimental results suggest that we do not yet understand the numbers and distribution of exRNAs in EVs. The stoichiometry of packing RNA into EVs has been most well studied for ex-miRNAs. Comparisons of EV numbers quantified by nanoparticle tracking and miRNA content quantified by quantitative real time PCR and next generation sequencing have suggested that less than 1 miRNA molecule is present on average per EV. These studies have investigated biofluids including plasma/serum, urine, and bronchoalveolar lavage fluid as well as EV-derived RNAs from cell culture supernatants from mast cells, dendritic cells, and tumor cell lines.7,25,30 For the most abundantly expressed miRNAs, ~1 miRNA is present for every 10–100 vesicles. More limited data is available for other exRNA species, but a comprehensive study in glioma cell lines has demonstrated the most abundant exRNAs such as U1/U2 snRNA fragments and RNY1 have ~1 copy per EV present while less abundant miRNA and most mRNAs are present at less than one molecule per ≥ 10,000 vesicles. Our own calibrated small RNA sequencing data in BALF suggests that the most abundant miRNAs are present in ~1 in 100 EVs.7
Although some take these findings as evidence that exRNAs are unlikely to efficiently communicate with target cells, experimental data supports active communication along an exRNA axis (see section 6). At a minimum, this stoichiometry raises critical questions about the distribution of RNAs within EVs. Selective loading of RNA into a minority of vesicles and/or selective targeting of vesicles carrying exRNA cargos to cells could help to explain the dissonance between the exRNA-to-EV ratios and observed biology. Identifying subsets of EVs that carry exRNAs is critical for understanding the function of exRNAs. Indeed, we still do not know whether each EV typically contains a single RNA molecule or a single class of RNA molecule, or whether there are subsets of EVs that are highly abundant in RNA species (Figure 3). The idea that subsets of EVs may be enriched in exRNAs has some supporting experimental evidence within the hematopoietic system. Fluorescence-activated vesicle sorting of EVs carrying either platelet or erythroid markers from blood has shown the most abundant miRNA in each is present at >20 copies per sorted event.31 Continuing to investigate the number of EVs in the body is essential for future exRNA research in the immune system and beyond.
Figure 3: Distribution of extracellular RNAs in extracellular vesicles.
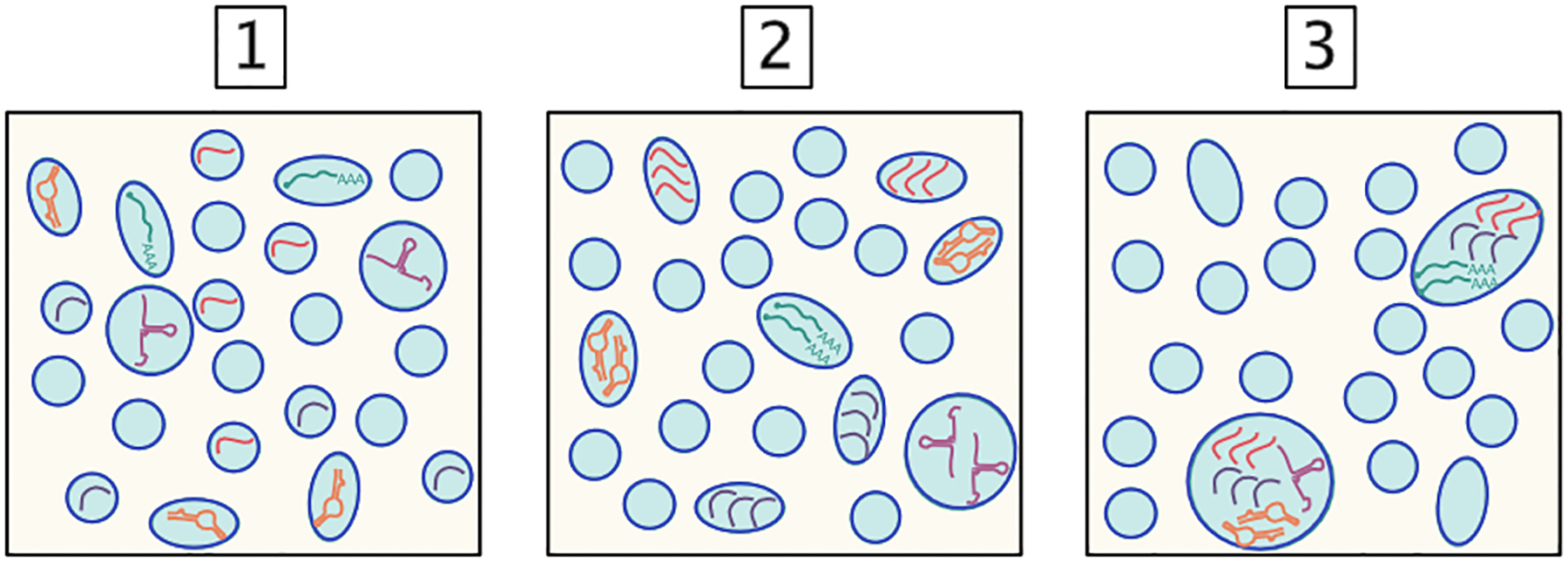
The distribution of RNAs within populations of extracellular vesicles (EVs) is not known. [1] There could be one RNA macromolecule per EV, [2] multiple RNAs of identical sequences or classes could be packaged together, or [3] there could be a subset of EVs that contain many RNAs packaged together. These possibilities are not mutually exclusive and may depend on the identity and activation state of the donor cell.
4. PACKAGING EXTRACELLULAR RNAs FOR EXPORT
Although there is little data about the origin and mechanisms of RNA secretion found outside of vesicles in the extracellular space, there is a body of emerging research about the sources and mechanisms of export of exRNAs within EVs. First, sorting into EVs is intimately linked to endogenous cellular pathways for RNA function. This is most evident to date in emerging associations between pathways of canonical miRNA function with EV secretion. This includes both spatial relationships within the cell as well as molecular interactions (Figure 4A). Key proteins that are required for miRNA-mediated gene silencing, including AGO and the AGO-binding protein GW182, have been found in close spatial association with endosomes and multivesicular bodies in monocytes by cellular fractionation and immunofluorescent studies.32 Depletion of select proteins of endosomal sorting complexes including HRS and Alix compromise miRNA-mediated gene silencing, suggesting this may be an active site for post transcriptional inhibition of gene expression in the cell.32 AGO shuttling to the multivesicular body is also regulated, and shuttling is inhibited by phosphorylation at serine 387 downstream of KRAS-driven MAPK activation.33 Given that expression of mutant AGO protein incapable of being phosphorylated at this site is sufficient to drive increased secretion of a subset of miRNAs within colon cancer cell lines33, packaging of some miRNAs into EVs may require shuttling to multivesicular bodies by AGO protein. In addition, competitive binding of miRNA away from AGO by the AU-rich binding protein HuR (derived from the Elavl1 gene) is also required to deliver some miRNAs to EVs for export, as has been demonstrated in liver cells under starvation conditions.34 Together this work supports an intimate and complex association between miRNA endogenous function and secretion. Activation-induced changes in mRNA target transcripts also can indirectly alter loading of miRNAs into EVs in macrophages. High levels of endogenous targets or synthetic miRNA binding sponge sequences can reduce miRNA secretion into EVs in these cells after activation, shifting the miRNA out of the multi vesicular bodies (sites of exosome EV production) and into P bodies (sites of miRNA-mediated target transcript repression).11
Figure 4: Extracellular RNA export is mediated by RBPs and lipids.
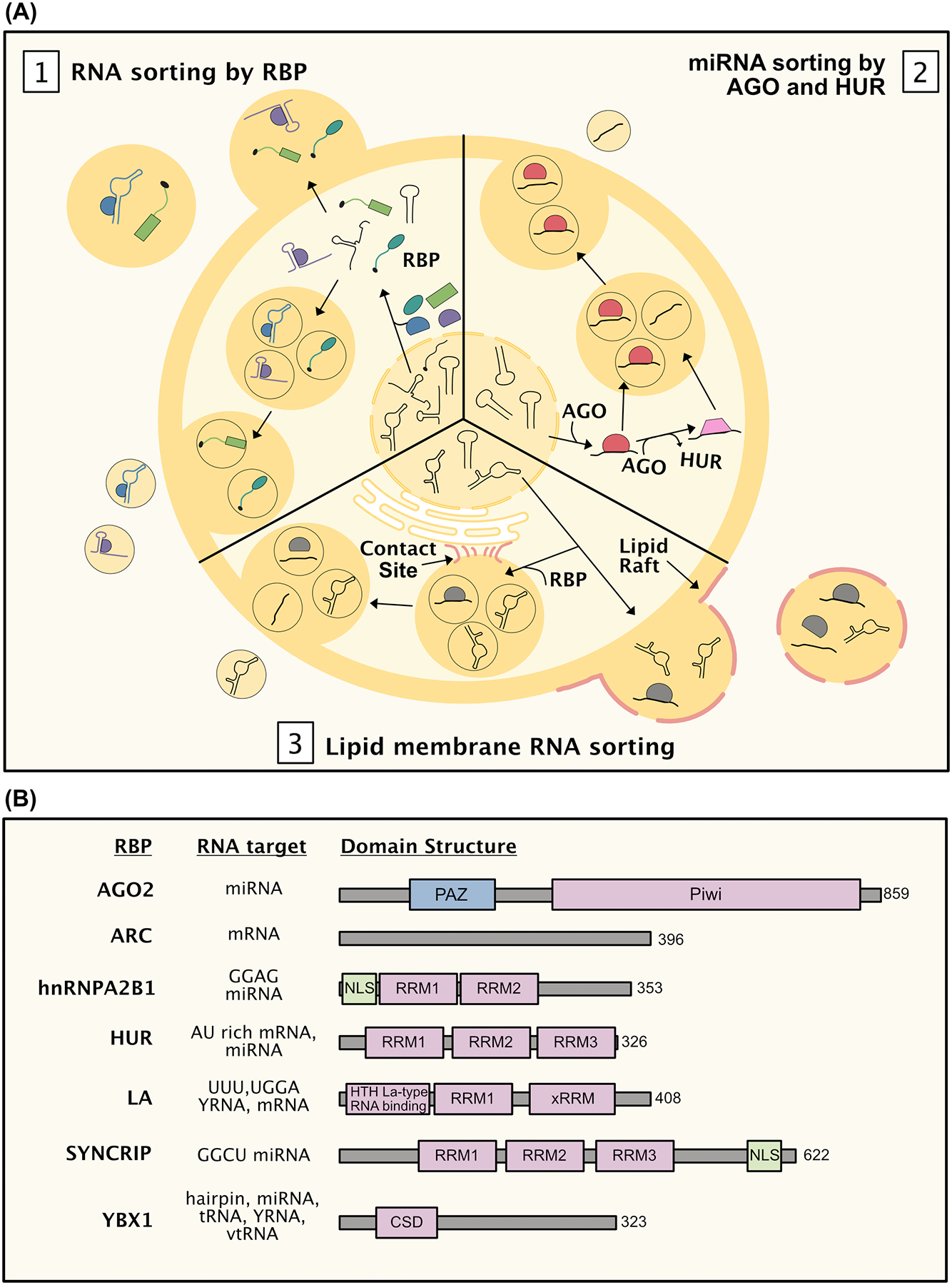
A, Various proteins and lipids are involved in RNA packaging into extracellular vesicles and release from the cell. [1] Upon leaving the nucleus, RNA binding proteins (RBPs) bind to and sort RNA into vesicles based on primary sequences and secondary structure. [2] Mature miRNAs packaging into vesicles can depend on Ago2 and/or HuR. [3] Membrane contact sites connecting the multivesicular body with the endoplasmic reticulum are important for RNA and lipid loading into vesicles. Additionally, interaction between RNA lipid raft domains may drive RNA packaging into EVs. B, Summary of RBPs involved in RNA packaging/export. Domain abbreviations in include RNA recognition motif (RRM), nuclear localization signal (NLS), helix-turn-helix (HTH) La-type RNA binding, cold shock domain (CSD), and Piwi Argonaut Zwille (PAZ) domains. Nucleic acid binding domains are highlighted in purple.
RNA binding proteins (RBP) are common cargos of EVs and have been implicated in sorting RNA into EVs for secretion. RBP cargos have been identified from EV proteomic studies from dendritic cells35, neutrophils36, T cells37, and B cells38. Efforts to compile the EV proteome have shown that a diversity of RBPs are secreted into the extracellular space across multiple cell types (Vesicleopedia39, microvesicle.org; Exocarta40, exocarta.org). Primary RNA sequence motifs within noncoding exRNAs help to dictate sorting into EVs. Sequence-specific sorting of ex-miRNAs depend on the RBPs hnRNPA2B1 in T cells, SYNCRIP (also known as hnRNPQ) in hepatocytes, and the lupus LA protein in tumor cell lines. In all these cases, short sequences are targets for RBP binding and control loading (Figure 4A). However, the sequences targeted by each are unique, with the most common EV-enriched RNA motifs being ‘GGAG’ for hnRNPA2B141, ‘GGCU’ for SYNCRIP42,43, and both ‘UUU’ and ‘UGGA’ for LA protein44 (Figure 4B). This results in differential sorting of miRNAs by each RBP. Sorting specificity therefore will depend on the paired activity of both the RBP and miRNA expression within a cell. In addition, both hnRNPA2B1 and SYNCRIP can be found in a sumoylated form within EVs.41,42 Sumoylation controls miRNAs binding to hnRNPA2B141, providing evidence for additional post-translational control to RBP-mediated ex-miRNA secretion.
mRNAs can also contain primary RNA sequence motifs, termed “zip codes”, which allow for sorting into vesicles for secretion. A 25-nucleotide sequence in the 3’UTR of transcripts containing a ‘CUGCC’ core on a stem loop structure is sufficient to drive reporter constructs into vesicles for secretion in HEK293T cells.45 Interestingly, miRNA binding enhances secretion of mRNA carrying this zip code, suggesting that multiple sequence-based interaction can cooperate to regulate delivery of mRNAs into secretion pathways. mRNAs can also be exported via interaction with the GAG-like protein ARC, which is derived from ancient retrotransposons.46 Within neuronal cells, ARC proteins form capsid-like structures that shuttle functional mRNAs to target cells, though whether or how specificity is imparted in this interaction is not yet known. Whether these mechanisms for packaging of ex-mRNAs exist in immune cells remains to be determined.
In addition to primary RNA motifs, exRNAs are also sorted and packaged for export based on structural RNA motifs. Given the diversity of structures present among small noncoding RNAs and their relative enrichment in the extracellular space, this may be a critical feature for their sorting. The RBP YBX1 is capable of binding to and sorting into vesicles diverse small noncoding RNAs, including miRNAs, tRNAs, YRNAs and vtRNAs.47,48 The binding between YBX1 and RNA has been shown to depend on hairpin structural motifs in target RNAs as opposed to primary sequence motifs.49,50 Finally, there are many RBPs present in EVs whose sorting capability and specificity remain to be investigated. For example, the RBP major vault protein (MVP) participates in miRNA sorting into EVs for secretion51,52, although the molecular mechanisms or motifs by which MVP interact with these extracellular-bound miRNAs are not known.
Finally, exRNA secretion is partially dependent on lipids. Lipids including cholesterol, ceramide, and phosphatidyl serine are enriched in EVs and play important roles in their biogenesis pathway (reviewed53). Lipids in the EV membrane also contribute to loading of miRNA into EVs for secretion (Figure 4A). Pharmacologic inhibition of Neutral sphingomyelinase 2 with the chemical inhibitor GW4869 or by gene knockdown reduces ex-miRNAs and EV secretion of exRNAs.54 Neutral sphingomyelinase 2 is a rate-limiting enzyme for ceramide biosynthesis, implicating this lipid in EV-mediated exRNA secretion. Given the role of ceramide in coalescing lipid raft domains, it is likely that interactions with these microdomains drive loading functions.55 In the immune system, transfer of ex-miRNAs from T cells to antigen presenting cell lines is dependent on Neutral sphingomyelinase 2.9 Some RNA-lipid interactions may be mediated by proteins. For example, knockdown of the calcium dependent phospholipid binding protein Annexin A2 results in reduced ex-miRNA secretion without changes in total EV numbers.56 Annexin A2 may act as a bridge between lipids and RNAs, as this protein interacts with miRNAs as shown by immunoprecipitation experiments.56 However, it is intriguing to propose that direct RNA-lipid binding may help package RNA for secretion as well. RNA structure and lipid membrane composition can affect direct interactions between RNAs and vesicles.57,58 Lipid interactions may also be important within the cell for exRNA secretion. Regions of close apposition of the endoplasmic reticulum and other organelles, termed membrane contact sites, are important for loading of cholesterol, ceramide, RBPs, and miRNAs into a dense fraction of small EVs dependent on the expression of a tether protein VAPA.59 Together it is clear that multiple and diverse mechanisms contribute to RNA loading into EVs for extracellular communication.
5. EXTRACELLULAR RNAs CHANGE WITH INFLAMMATION
Secretion of soluble factors is a defining feature of both the classification and function of cells of the immune system. It is widely known that the secretion of soluble molecules not only is critical for immune cell recruitment and activation, but also helps define unique types and subsets of immune cells. exRNA secretion is emerging as a unique and defining property of immune cells. Dendritic cells4,8,9, monocytes/macrophage10,11, mast cells3, T cells12,13, and B cells12 each produce a distinct exRNAome. The complement of RNAs secreted by each cell type partially reflects the unique content of RNAs present within the parental cell, giving cell-type specificity to the secreted products. However, exRNAs produced by these cells are also distinct from their parental cells with clear evidence of selective secretion of RNA subsets. How shared and unique secreted products define related immune cell function will provide a useful tool to understand the role of exRNAs in immune responses.
exRNAs secreted by immune cells contribute unique and measurable RNA content to extracellular fluids in vivo. Analysis of blood ex-miRNAs of lymphocyte knockout mice or mice with a lymphocyte-specific deletion of miRNA biogenesis machinery demonstrate up to an ~80% reduction of select ex-miRNAs in the blood of mice, including miR-128, miR-142a, miR-150, miR-297a, and miR-706.60 T cells, B cells and NKT cells also make unique contributions to the pool of ex-miRNA. This is particularly striking for NKT cells which comprise ≤ 1% of circulating cells but contribute much of the miR-181c found in blood61, a miRNA that is essential in their development60. Fluorescence activated vesicle sorting of EVs from plasma has directly demonstrated unique exRNA content from different hematopoietic cells in circulation. Sorted CD41+ platelet EVs contain high levels of miR-223 and miR-199a, while sorted CD235a+ erythroid EVs contain high levels of miR-451a.31 It will be interesting to directly assess EVs from immune cells to determine preferential cargos carried into biofluids in vivo.
exRNA cargos depend not only on their immune cell origin, but also on the activation state of that immune cell as part of a local or systemic immune response. Some of these changes reflect alterations in the relative abundance of miRNAs in the parental cell in response to preferential activation, while others reflect new differential secretion into exosomes. Activating LPS stimulation of dendritic cells enhances ex-miRNA production of multiple miRNA species such as miR-9, miR-155, and miR-146.4,8 LPS stimulation in dendritic cells also reduces secretion of several other species of noncoding RNAs, especially snoRNAs and YRNAs.8 In macrophages, stimulation with the cytokine IL-4, which induces an alternative activation program, results in changes in ex-miRNA secretion. Some of the increased ex-miRNAs reflect upregulation in cellular expression, including miR-138 and miR-149, while other ex-miRNAs show new selective secretion, including miR-22, miR-99a, miR-125a and miR-130b.11 Pro-inflammatory LPS also leads to changes in exRNA production by macrophages, with rapid secretion of EV-contained snoRNAs within 1–2 hours after stimulation.62 And macrophages treated with other activating signals including oxidized low-density lipoprotein and palmitic acid secrete more YRNA fragments than unstimulated cells.63
exRNA cargos are also changed by activation of adaptive immune cells. In CD4+ T cells activated by the mitogen phytohemagglutinin (PHA), several miRNAs are both increased within the cell as well as within the extracellular space including miR-155 and members of the miR-17~92 family such as miR-17, miR-19b, and miR-20a.12 However, another set of miRNAs decrease within the cell and increase within the extracellular space, especially miR-150 and miR-342, suggesting a rapid activation-dependent selective secretion of miRNAs. T cell activation in vitro with CD3 and CD28 also increases secretion of select tRNA fragments, among other noncoding RNAs.13 And B cells stimulated with CpG, anti-CD40 and anti-IgM show enhanced secretion of miRNAs including miR-150 and miR-223.12
Activation-dependent changes in immune ex-miRNA secretion have also been documented with systemic inflammation in vivo. Correlating with in vitro secretion of miR-150 after lymphocyte activation, both humans and mice show enhanced miR-150 levels in circulation after immunization.12 Our own work assessing lavage fluid in the lungs of mice strongly suggest that immune cells also contribute unique exRNA content to tissues with local inflammation. miRNAs preferentially expressed by immune cells including miR-142a, miR-150, and miR-223 are increased in extracellular fluid as well as small EVs during allergic lung inflammation.64 The in vivo dynamics, cellular targets, and immunologic functions of increased ex-miRNAs during inflammation remain to be determined. However, these findings support exRNA secretion as a new and fundamental aspect to coordinate systemic and local immune responses.
6. EXTRACELLULAR miRNA COMMUNICATION FOR TARGET GENE SUPRESSION
Communication among cells of the immune system is critical for both effective immune responses and pathologic inflammation. Functional ex-miRNA transfer provides a novel form of immune cell to immune cell communication. In some cases, this transfer of ex-miRNAs occurs among cells of the same cell type. Transfer of miR-451 and miR-148a from bone marrow derived dendritic cell donors to an acceptor dendritic cell line results in suppression of target luciferase reporter constructs within acceptor cells.4 Transfer of miR-155 and miR-146 within EVs also occurs in vitro among bone marrow derived dendritic cells and is sufficient to repress known endogenous mRNA targets of these miRNAs.14 EV-mediated transfer of these miRNAs programs the response of dendritic cells to LPS in vitro and sepsis in vivo. Ex-miR-155 enhances pro-inflammatory IL-6 and TNFα production, while ex-miR-146 enhances anti-inflammatory IL-10 and limits pro-inflammatory cytokine production14 (Figure 5A). These functions are consistent with the known cell-intrinsic functions of both miR-155 and miR-14665–68, demonstrating that transferred ex-miRNAs can function equivalently to endogenously expressed miRNAs. RNA communication between cells of the same immune cell subset may help propagate signals through immune cell populations as well as coordinate robust immune responses.
Figure 5: Extracellular miRNAs as part of immune cell signaling to suppress target genes and change cell function.
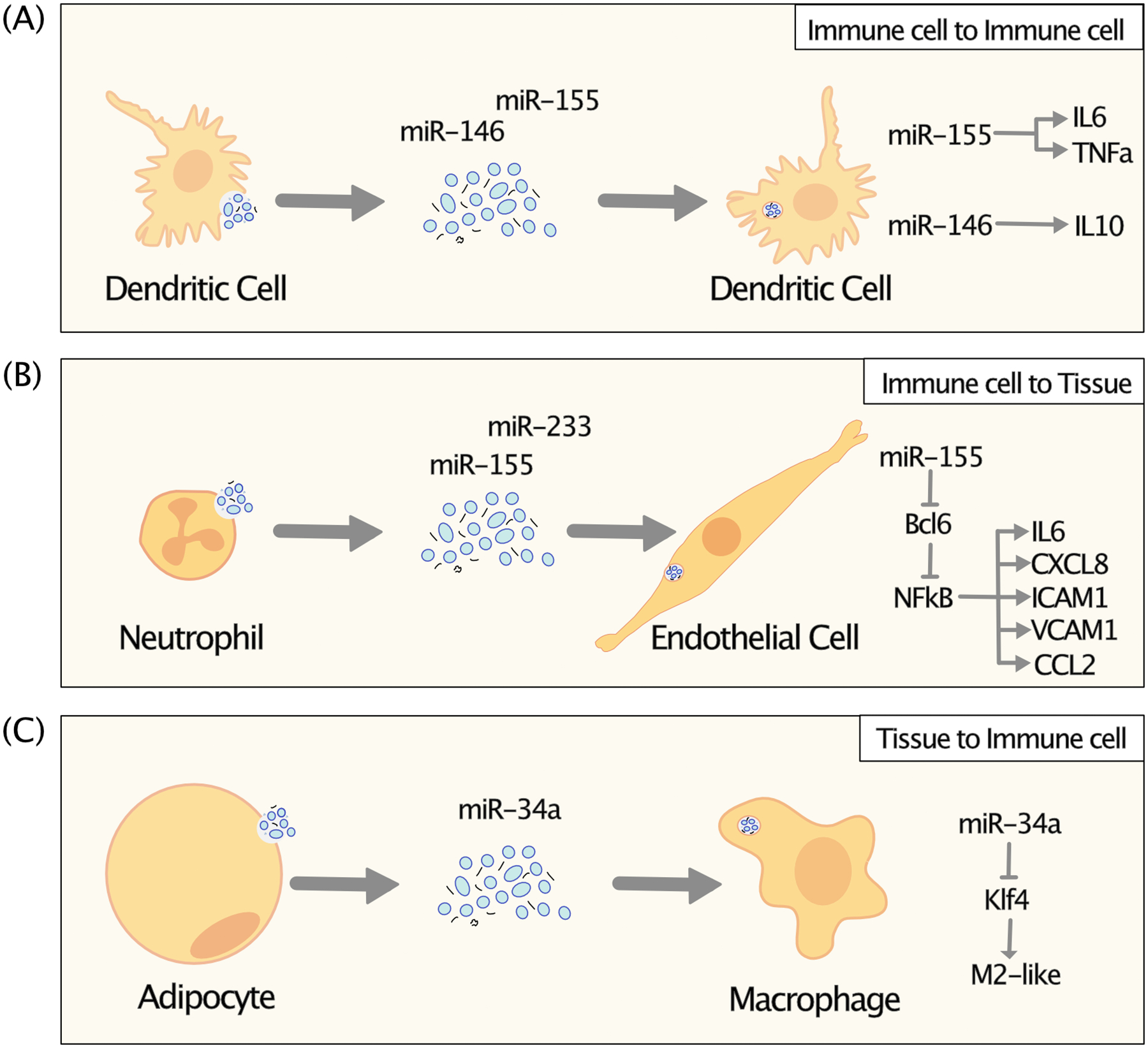
Extracellular miRNAs can be communicated between immune cells (A) or between immune cells and tissues (C,D). Upon transfer, they can serve as post transcriptional inhibitors of gene expression of direct mRNA targets and affect signaling pathways within cells.
Immune cells also use ex-miRNAs to communicate with other cell types within the immune system. In mixed hematopoietic chimera experiments, wild type immune cells can transfer miR-155 to T cells, B cells, and CD11b+ myeloid linage miR-155−/− cells in vivo.14 This data suggests that a complex network of cell communication is likely to occur throughout the immune system. Regulatory T cells have also been shown to transfer both transfected fluorescently tagged miRNAs as well as endogenous miRNAs including miR-155, Let7b and Let7d to target effector T cells.69 EVs from regulatory T cells inhibited Th1 cell proliferation in vitro, and this effect was lost in EVs derived from Dicer−/− regulatory T cells which have impaired global miRNA biogenesis. While this result is consistent with transferred miRNA content being a critical functional cargo, Dicer−/− regulatory T cells have severely impaired function70–72, and therefore other aspects of EV biogenesis and function may be changed to affect suppression. T cell lines are also capable of transferring miR-335 to antigen presenting cells for target gene suppression.9 Interestingly, this transfer was dependent on formation of an immunologic synapse and correlates with alignment of multivesicular bodies with the synapse.
Multiple tissue types have also been shown to be direct targets of ex-miRNA communication from immune cells. Neutrophil-derived large EVs can deliver ex-miR-155 to endothelial cells in vitro and the aorta in vivo, reducing expression of the known miR-155 target mRNA Bcl6 to induce NFκB-mediated inflammation, adhesion, and chemokine secretion by acceptor endothelial cells73 (Figure 5B). These neutrophil large EVs worsen lesion size, and this effect is lost in EVs derived from miR-155−/− cells. Monocyte-derived large EVs transfer functional miR-223 to lung epithelial cell lines as measured by luciferase reporter constructs, increasing expression of those miRNAs in target cells 10–100x.10 Macrophage cell lines can also transfer miR-150 to acceptor endothelial cell lines to enhance migration through the direct inhibition of target mRNAs74. Adipose tissue macrophage derived EVs can transfer miRNAs including miR-155 and/or miR-223 to adipocyte cell lines in vitro and to primary adipocytes in bone marrow chimeric mice. This transfer has been proposed to promote insulin resistance in this target tissue in part through the suppression of the miR-155 target gene Pparg.75 This activity of immune cell exRNAs may be critical for effector cell function in diverse types of tissue inflammation.
Immune cells can additionally be the targets of ex-miRNA transfer from other cell types, allowing tissues to instruct and modify local inflammation. This transfer participates in pathologic, disease associated inflammation. Ex-miRNA communication may sensitize monocytes and macrophages to adopt a pro-inflammatory phenotype in diseases characterized by chronic inflammation. Hepatocyte cell lines treated with alcohol upregulate miR-122 and transfer this miRNA to macrophage cell lines.76 Transfection of this miRNA is sufficient to enhance IL-1β and TNF-α production in these macrophages, raising the possibility that delivery of ex-miR-122 is sufficient to exacerbate pro-inflammatory phenotypes. Adipocytes secrete miR-34a in EVs which can be transferred to macrophages in vitro to suppress the mRNA of a critical transcription factor Klf4 required for M2 polarization77 (Figure 5C). Suggesting a critical role for this paracrine transfer in obesity, loss of miR-34a in adipocytes is sufficient to shift macrophages from away from a pro-inflammatory M1-like and toward a protective M2-like phenotype ameliorating high fat diet induced metabolic dysfunction in mice. Tumor cells also are capable of transferring miRNAs to macrophages. Dying tumor cells transfer miR-375 to macrophages within the tumor microenvironment, targeting genes including Tensin 3 and Paxillin to enhance macrophage migration.78 In addition, glioma tumor cells transfer miR-21 via EVs to target microglial cells, inhibiting the expression of Btg2 to increase microglial proliferation.79 In each of these studies, macrophages are identified as a major acceptor for ex-miRNA transfer. As cells designed to both take-up material and sense cues from the local tissue microenvironment, macrophages may be particularly apt acceptors of exRNAs from non-immune cells.
7. OTHER RNA TRANSFER IN THE IMMUNE SYSTEM
Although ex-miRNA communication is best studied, there is emerging evidence that multiple classes of both long and short RNAs communicate signals via RNA transfer to and from immune cells. Mast cells of human and murine origin can secrete and receive mRNA containing EVs, and mRNA harvested from these EVs can be in vitro translated3. These data show that functional mRNAs are packaged in EVs by immune cells and raises the possibility that they drive new protein expression in a target cell. mRNA transfer events have been captured in vivo using Cre recombinase mRNA transfer to floxed reporter systems in acceptor cells.80,81 Immune cell Cre recombinase driven by the pan-hematopoietic cell promoter for the Vav1 gene was found to deliver Cre mRNA to neurons, and this delivery increased with the induction of systemic inflammation.80 In this study, injection of EVs carrying Cre mRNA into the blood was sufficient for transfer, suggesting that immune cell RNA communication can act at a distance in endocrine-type signaling. However, the functional impact of endogenous mRNA transfer on immune cell function and inflammation is not yet known. Communication via long RNA transfer also is likely to occur via lncRNA transfer. The myeloid cell specific HIF-1α stabilizing lncRNA HISLA is packaged in tumor associated macrophage EVs, and treatment of breast cancer cells or tumors with HISLA-containing EVs enhances their aerobic glycolysis and apoptosis resistance.82 siRNA knockdown of HISLA in tumor associated macrophages is sufficient to abrogate this effect, supporting a direct role for lncRNA in macrophage-to-tumor cell intercellular communication.82
Additional short RNAs have been implicated in cell-to-cell communication in the immune system. Defined fragments from the 5’ end and 3’ internal regions of tRNAs are enriched in EVs released by T cells in an activation-dependent manner.13 Antisense oligonucleotide knockdown of these tRNA fragments inhibits T cell activation.13 It is therefore interesting to hypothesize that both removal of tRNA fragments from donor cells as well as their transfer to acceptor cells could modulate T cell activation, possibly serving to equilibrate activation levels among a population of responding cells. Transwell co-culture assays have shown that snoRNAs can be transferred from wild type to snoRNA-deficient macrophages, with collection into nuclear punctae consistent with snoRNA activity.62 Following parabiont surgery with a wildtype mouse, snoRNA knockout mice show increased levels of 2’-O-methylation in tissues with high levels of new rRNA synthesis due to rapid cell proliferation, a canonical sign for snoRNA activity.62 Whether this transfer occurs via snoRNA carried through the blood to distant site or through local secretion by migratory immune cells to those sites was not established. Together these data show that transfer of endogenous RNA within the immune system is a novel communication axis by which a donor cell may regulate the function and fate of target cells.
8. NONCANNONICAL FUNCTIONS OF EXTRACELLULAR RNA
exRNA communication can occur when an RNA is transferred and takes on its canonical cellular function within an acceptor cell. However, RNAs secreted into the extracellular space may also take on new or additional roles during or after transfer. Accumulating evidence suggests that exRNAs are capable of ligating receptors on target immune cells, and that this interaction may communicate signals that modify immune cell function. The most well-studied interaction of this type is the interaction of exRNAs with pattern recognition receptors (PRRs), which classically recognize exogenous pathogen-associated molecular patterns (PAMPs) and endogenous damage-associated molecular patterns (DAMPs) (reviewed83). Ex-YRNA fragments as well as ex-miRNA have been shown to interact with the endosomal PRR Toll-like receptor 7 (TLR7) in mice and TLR8 in humans (Figure 6). These receptors bind GU-rich single stranded RNAs of both viral and endogenous origin.84,85. They can recognize very short RNAs down to ~10 nucleotides in length84 with GU stretches of ≥ 4 nucleotides86 The role of mouse TLR7 and human TLR8 in recognizing these communicating endogenous exRNAs has been demonstrated through inhibition of endosome acidification using chloroquine63, specific chemical antagonists63, co-immunoprecipitation experiments87,88 and knockout of TLR7or its signaling adaptor MyD8887–90.
Figure 6: Extracellular RNAs can function as pattern recognition receptor ligands.
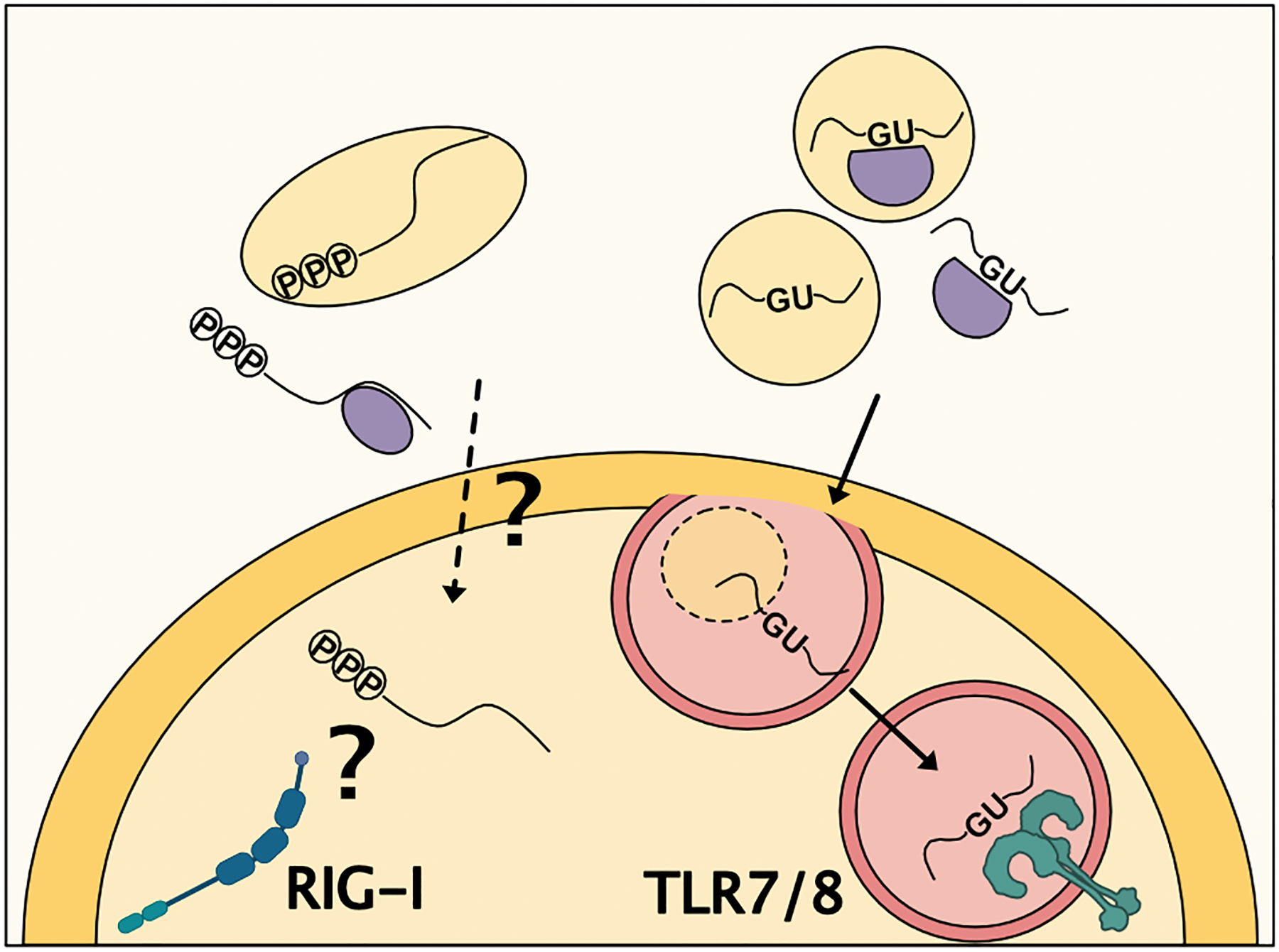
RNA from donor cells has been shown to activate Toll-like receptor 7/8 (TLR 7/8) and PRR Retinoic Acid-Inducible Gene I (RIG-I). miRNA binding to TLR7/8 is dependent on GU-rich sequences. Activation of RIG-I by exRNA requires the RNA to have an unshielded 5’ tri-phosphate. Evidence of RIG-I activation by extracellular RNA is in tumor cells, so its role in immune cell signaling and inflammation requires further study.
This TLR7-mediated exRNA communication contributes to pro-inflammatory signals. It leads to monocyte/macrophage NFκB pathway and caspase 3 activation63,87,88, as well as microglia and macrophage TNFα and IL-6 cytokine production87,88,90. In animal models, introduction of stabilized extracellular let-7b is sufficient to induce neuronal cell loss in a TLR7 dependent fashion, and this may have relevance for pathologic neurodegeneration as patients with Alzheimer’s disease have increased levels of this ex-miRNA in the CSF.90 In addition, EV-independent ex-miR-122 released from the liver after damage can be taken up by macrophages in the lung and drive pulmonary inflammation.88 Activation of TLR7 has been documented for miR-21, miR-29a, miR-147 and let-7 family members87,90, and in the case of let-7b the activation was shown to depend on GU-rich sequences90. To date, these noncanonical functions of secreted RNAs triggering TLR7 have been predominantly associated with cell death-associated changes in exRNA secretion. It will be important to determine whether this pathway is utilized in immune cell signaling in the absence of cell death.
exRNAs have also been shown to interact with additional PRRs, though this work has largely investigated roles of exRNAs in tumor cell communication rather than immune cell biology. The cytoplasmic PRR Retinoic acid-inducible gene I (RIG-I) has been implicated in exRNA communication (Figure 6). RIG-I typically recognizes short (20–300 nucleotide) dsRNA sequences that are 5’ tri-phosphorylated, and this receptor is capable of recognizing both viral RNAs as well as endogenous RNAs, typically derived from polymerase III transcription (reviewed91). The signal recognition particle RNA RN7SL1 can be transferred from stromal to breast cancer cells, bind to RIG-I receptor in target cells, and enhance tumor growth.92 This activity requires unshielding of the 5’ triphosphate of this RNA in EVs, as the endogenous cytoplasmic form of RN7SL1 is bound to RBPs to prevent RIG-I recognition. This EV encapsulated unshielded 5’ tri-phosphorylated RNA is also capable of inducing macrophage and dendritic cell maturation and activation92, suggesting it could be important for shaping the immune response in the tumor microenvironment. However, this possibility remains to be directly tested. This may expand the repertoire of ligands and scope of endogenous RNA recognition
9. CONCLUSIONS
In this review, we have presented emerging evidence that exRNAs constitute a novel communication axis within the immune system. The concept that RNAs may be readily transferred between cells is paradigm-shifting, challenging what has previously defined a single cell as an autonomous unit. exRNAs are enriched in small noncoding RNAs, are packaged into RNase-resistant forms, and are actively secreted from cells. They rely on interactions with RBPs and lipids to organize and select RNA cargos for secretion. In many instances, their functions in target cells mirror their endogenous cell-autonomous function. However, through the ligation of PRRs they can also take on new signaling functions once secreted into the extracellular space. Although there is good empirical evidence that exRNAs are delivered to the cytoplasm of target cells with functional effects, the mechanisms of this delivery remain largely unknown. Many processes have been suggested to participate in the delivery of exRNAs to target cells including micropinocytosis, phagocytosis, endocytosis, and back fusion (reviewed53). However, much work remains to understand the molecular mechanism by which active RNAs can be delivered to an acceptor cell, and this is one of the most pressing questions in the field.
Within the immune system, exRNA communication has broad functions to regulate immune cells and inflammation. Likely we have only begun to understand their contributions to fundamental physiologic and pathologic processes. From the work to date, several themes and principles have emerged. First, a subset of exRNAs is both selectively secreted and has been repeatedly implicated in potent immune exRNA communication functions, especially ex-miRNAs miR-150, miR-155, and miR-223. Understanding the mechanisms by which these critical regulators act will help define principles for effective exRNA communication. Second, although most immune cells can participate in exRNA communication, the mechanism and efficiencies of this communication may vary significantly, depending on both the cell and its activation state. For example, determining how macrophages function as major recipients of exRNA communication from tissues will uncover critical features of exRNA communication between non-immune and immune cells. Finally, immune cell exRNAs may have a particularly broad reach within the body, both through delivery of immune exRNAs through the circulation as well as the cells’ widespread infiltration of tissues.
The study of exRNAs is not only of profound biological relevance, but the investigation of exRNA in the immune system has major implications for clinical medicine. Broad efforts are underway to characterize the exRNA profiles of biofluids including plasma, serum, urine, and cerebrospinal fluid2,20,93,94. This research will establish which exRNA are present in different biofluid, the steady-state levels of exRNAs in healthy individuals, and methodologies to facilitate and standardize the use of exRNAs as novel biomarkers for disease. While this new class of biomarkers offers important tools for the diagnosis and monitoring of disease, perhaps even more exciting are the therapeutic opportunities afforded through understanding endogenous exRNA transfer. In 2018 the FDA approved the first siRNA therapeutic delivered by lipid nanoparticles for the treatment of hereditary transthyretin-mediated amyloidosis95. Furthermore, the remarkable mRNA vaccines which addressed the COIVD-19 global pandemic used extracellular RNA-mediated delivery of viral proteins to induce potent immunity.96–99 In development are numerous other RNA-based therapies, some of which are loading siRNAs (NCT03608631) and miRNAs (NCT03384433) into EVs to leverage the biology of exRNA communication. exRNA communication is likely to both guide design of novel drug classes and afford new therapeutic targets. Together, strong partnerships between basic, translational, and clinical science will help propel this field forward and lead to exciting discoveries in this new domain of RNA biology.
Acknowledgements:
This work was supported by DP2HL152426, R01CA249424, and funds from the Department of Pathology, Microbiology, and Immunology at Vanderbilt University Medical Center to HHP. Parts of some images were made with BioRender.
Abbreviations:
- AGO
Argonaute
- DAMP
damage associated molecular pattern
- exRNAs
extracellular RNAs
- EV
extracellular vesicle
- LPS
lipopolysaccharide
- mRNA
messenger RNA
- miRNA
microRNA
- NKT
natural killer T cell
- PAMP
pathogen associated molecular pattern
- PRR
pattern recognition receptor
- PHA
phytohemagglutinin
- RIG-I
Retinoic acid-inducible gene I
- RNA
ribonucleic acid
- rRNA
ribosomal RNA
- RBP
RNA binding protein
- snRNA
small nuclear RNA
- snoRNA
small nucleolar RNA
- TLR
Toll like receptor
- tRNA
transfer RNA
- vtRNA
vault RNA
Footnotes
Conflict of Interest: HHP has been a consultant for Janssen Research & Development.
REFERENCES
- 1.Kolodny GM, Culp LA, Rosenthal LJ. Secretion of RNA by normal and transformed cells. Experimental Cell Research. 1972;73(1):65–72. doi: 10.1016/0014-4827(72)90102-4 [DOI] [PubMed] [Google Scholar]
- 2.Das S, Abdel-Mageed AB, Adamidi C, et al. The Extracellular RNA Communication Consortium: Establishing Foundational Knowledge and Technologies for Extracellular RNA Research. Cell. 2019;177(2):231–242. doi: 10.1016/j.cell.2019.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nature Cell Biology. 2007;9(6):654–659. doi: 10.1038/ncb1596 [DOI] [PubMed] [Google Scholar]
- 4.Montecalvo A, Larregina AT, Shufesky WJ, et al. Mechanism of transfer of functional microRNAs between mouse dendritic cells via exosomes. Blood. 2012;119(3):756–766. doi: 10.1182/blood-2011-02-338004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Turchinovich A, Weiz L, Langheinz A, Burwinkel B. Characterization of extracellular circulating microRNA. Nucleic Acids Research. 2011;39(16):7223–7233. doi: 10.1093/nar/gkr254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arroyo JD, Chevillet JR, Kroh EM, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proceedings of the National Academy of Sciences. 2011;108(12):5003–5008. doi: 10.1073/pnas.1019055108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pua HH, Happ HC, Gray CJ, et al. Increased Hematopoietic Extracellular RNAs and Vesicles in the Lung during Allergic Airway Responses. Cell Reports. 2019;26(4):933–944.e4. doi: 10.1016/j.celrep.2019.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Driedonks TAP, van der Grein SG, Ariyurek Y, et al. Immune stimuli shape the small non-coding transcriptome of extracellular vesicles released by dendritic cells. Cellular and Molecular Life Sciences. 2018;75(20):3857–3875. doi: 10.1007/s00018-018-2842-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mittelbrunn M, Gutiérrez-Vázquez C, Villarroya-Beltri C, et al. Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nature Communications. 2011;2(1):1–10. doi: 10.1038/ncomms1285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ismail N, Wang Y, Dakhlallah D, et al. Macrophage microvesicles induce macrophage differentiation and miR-223 transfer. Blood. 2013;121(6):984–995. doi: 10.1182/blood-2011-08-374793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Squadrito ML, Baer C, Burdet F, et al. Endogenous RNAs Modulate MicroRNA Sorting to Exosomes and Transfer to Acceptor Cells. Cell Reports. 2014;8(5):1432–1446. doi: 10.1016/j.celrep.2014.07.035 [DOI] [PubMed] [Google Scholar]
- 12.de Candia P, Torri A, Gorletta T, et al. Intracellular Modulation, Extracellular Disposal and Serum Increase of MiR-150 Mark Lymphocyte Activation. PLoS ONE. 2013;8(9):1–13. doi: 10.1371/journal.pone.0075348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chiou NT, Kageyama R, Ansel KM. Selective Export into Extracellular Vesicles and Function of tRNA Fragments during T Cell Activation. Cell Reports. 2018;25(12):3356–3370.e4. doi: 10.1016/j.celrep.2018.11.073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alexander M, Hu R, Runtsch MC, et al. Exosome-delivered microRNAs modulate the inflammatory response to endotoxin. Nature Communications. 2015;6(1):7321. doi: 10.1038/ncomms8321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Niel G, D’Angelo G, Raposo G. Shedding light on the cell biology of extracellular vesicles. Nature Reviews Molecular Cell Biology. 2018;19(4):213–228. doi: 10.1038/nrm.2017.125 [DOI] [PubMed] [Google Scholar]
- 16.Théry C, Witwer KW, Aikawa E, et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. Journal of Extracellular Vesicles. 2018;7(1). doi: 10.1080/20013078.2018.1535750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alexander M, Ramstead AG, Bauer KM, et al. Rab27-Dependent Exosome Production Inhibits Chronic Inflammation and Enables Acute Responses to Inflammatory Stimuli. The Journal of Immunology. 2017;199(10):3559–3570. doi: 10.4049/jimmunol.1700904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arroyo JD, Chevillet JR, Kroh EM, et al. SUPPLEMENT Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proceedings of the National Academy of Sciences. doi: 10.1107/S1600536809047072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Geekiyanage H, Rayatpisheh S, Wohlschlegel JA, Brown R, Ambros V. Extracellular microRNAs in human circulation are associated with miRISC complexes that are accessible to anti-AGO2 antibody and can bind target mimic oligonucleotides. Proceedings of the National Academy of Sciences of the United States of America. 2020;117(39):24213–24223. doi: 10.1073/pnas.2008323117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murillo OD, Thistlethwaite W, Rozowsky J, et al. exRNA Atlas Analysis Reveals Distinct Extracellular RNA Cargo Types and Their Carriers Present across Human Biofluids. Cell. 2019;177(2):463–477.e15. doi: 10.1016/j.cell.2019.02.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bartel DP. Metazoan MicroRNAs. Cell. 2018;173(1):20–51. doi: 10.1016/j.cell.2018.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van Deun J, Mestdagh P, Sormunen R, et al. The impact of disparate isolation methods for extracellular vesicles on downstream RNA profiling. Journal of Extracellular Vesicles. 2014;3(1):24858. doi: 10.3402/jev.v3.24858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jeppesen DK, Fenix AM, Franklin JL, et al. Reassessment of Exosome Composition. Cell. 2019;177(2):428–445.e18. doi: 10.1016/j.cell.2019.02.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang H, Freitas D, Kim HS, et al. Identification of distinct nanoparticles and subsets of extracellular vesicles by asymmetric flow field-flow fractionation. Nature Cell Biology. 2018;20(3):332–343. doi: 10.1038/s41556-018-0040-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen CC, Carter B, Balaj L, et al. Coding and noncoding landscape of extracellular RNA released by human glioma stem cells. Nature Communications. 2017;8(1):1145. doi: 10.1038/s41467-017-01196-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, Remaley AT. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nature Cell Biology. 2011;13(4):423–433. doi: 10.1038/ncb2210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wagner J, Riwanto M, Besler C, et al. Characterization of levels and cellular transfer of circulating lipoprotein-bound microRNAs. Arteriosclerosis, Thrombosis, and Vascular Biology. 2013;33(6):1392–1400. doi: 10.1161/ATVBAHA.112.300741 [DOI] [PubMed] [Google Scholar]
- 28.Tabet F, Vickers KC, Cuesta Torres LF, et al. HDL-transferred microRNA-223 regulates ICAM-1 expression in endothelial cells. Nature Communications. 2014;5. doi: 10.1038/ncomms4292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cuesta Torres LF, Zhu W, Öhrling G, et al. High-density lipoproteins induce miR-223–3p biogenesis and export from myeloid cells: Role of scavenger receptor BI-mediated lipid transfer. Atherosclerosis. 2019;286:20–29. doi: 10.1016/j.atherosclerosis.2019.04.227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chevillet JR, Kang Q, Ruf IK, et al. Quantitative and stoichiometric analysis of the microRNA content of exosomes. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(41):14888–14893. doi: 10.1073/pnas.1408301111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kondratov K, Nikitin Y, Fedorov A, et al. Heterogeneity of the nucleic acid repertoire of plasma extracellular vesicles demonstrated using high-sensitivity fluorescence-activated sorting. Journal of Extracellular Vesicles. 2020;9(1):1743139. doi: 10.1080/20013078.2020.1743139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gibbings DJ, Ciaudo C, Erhardt M, Voinnet O. Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity. Nature Cell Biology. 2009;11(9):1143–1149. doi: 10.1038/ncb1929 [DOI] [PubMed] [Google Scholar]
- 33.McKenzie AJ, Hoshino D, Hong NH, et al. KRAS-MEK Signaling Controls Ago2 Sorting into Exosomes. Cell Reports. 2016;15(5):978–987. doi: 10.1016/j.celrep.2016.03.085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mukherjee K, Ghoshal B, Ghosh S, et al. Reversible HuR-micro RNA binding controls extracellular export of miR-122 and augments stress response. EMBO reports. 2016;17(8):1184–1203. doi: 10.15252/embr.201541930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kowal J, Arras G, Colombo M, et al. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proceedings of the National Academy of Sciences. 2016;113(8):E968–E977. doi: 10.1073/pnas.1521230113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dalli J, Montero-Melendez T, Norling L v., et al. Heterogeneity in neutrophil microparticles reveals distinct proteome and functional properties. Molecular and Cellular Proteomics. 2013;12(8):2205–2219. doi: 10.1074/mcp.M113.028589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perez-Hernandez D, Gutiérrez-Vázquez C, Jorge I, et al. The intracellular interactome of tetraspanin-enriched microdomains reveals their function as sorting machineries toward exosomes. Journal of Biological Chemistry. 2013;288(17):11649–11661. doi: 10.1074/jbc.M112.445304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Buschow SI, van Balkom BWM, Aalberts M, Heck AJR, Wauben M, Stoorvogel W. MHC class II-associated proteins in B-cell exosomes and potential functional implications for exosome biogenesis. Immunology and Cell Biology. 2010;88(8):851–856. doi: 10.1038/icb.2010.64 [DOI] [PubMed] [Google Scholar]
- 39.Kalra H, Simpson RJ, Ji H, et al. Vesiclepedia: A Compendium for Extracellular Vesicles with Continuous Community Annotation. PLoS Biology. 2012;10(12):e1001450. doi: 10.1371/journal.pbio.1001450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Keerthikumar S, Chisanga D, Ariyaratne D, et al. ExoCarta: A Web-Based Compendium of Exosomal Cargo. Journal of Molecular Biology. 2016;428(4):688–692. doi: 10.1016/j.jmb.2015.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Villarroya-Beltri C, Gutiérrez-Vázquez C, Sánchez-Cabo F, et al. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nature Communications. 2013;4:1–10. doi: 10.1038/ncomms3980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Santangelo L, Giurato G, Cicchini C, et al. The RNA-Binding Protein SYNCRIP Is a Component of the Hepatocyte Exosomal Machinery Controlling MicroRNA Sorting. Cell Reports. 2016;17(3):799–808. doi: 10.1016/j.celrep.2016.09.031 [DOI] [PubMed] [Google Scholar]
- 43.Hobor F, Dallmann A, Ball NJ, et al. A cryptic RNA-binding domain mediates Syncrip recognition and exosomal partitioning of miRNA targets. Nature Communications. 2018;9(1). doi: 10.1038/s41467-018-03182-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Temoche-Diaz MM, Shurtleff MJ, Nottingham RM, et al. Distinct mechanisms of microrna sorting into cancer cell-derived extracellular vesicle subtypes. eLife. 2019;8. doi: 10.7554/eLife.47544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bolukbasi MF, Mizrak A, Ozdener GB, et al. MiR-1289 and “zipcode”-like sequence enrich mRNAs in microvesicles. Molecular Therapy - Nucleic Acids. 2012;1(2):e10. doi: 10.1038/mtna.2011.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ashley J, Cordy B, Lucia D, Fradkin LG, Budnik V, Thomson T. Retrovirus-like Gag Protein Arc1 Binds RNA and Traffics across Synaptic Boutons. Cell. 2018;172(1–2):262–274.e11. doi: 10.1016/j.cell.2017.12.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shurtleff MJ, Temoche-Diaz MM, Karfilis K v., Ri S, Schekman R. Y-box protein 1 is required to sort microRNAs into exosomes in cells and in a cell-free reaction. eLife. 2016;5(AUGUST):1–23. doi: 10.7554/eLife.19276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shurtleff MJ, Yao J, Qin Y, et al. Broad role for YBX1 in defining the small noncoding RNA composition of exosomes. Proceedings of the National Academy of Sciences. 2017;114(43):E8987–E8995. doi: 10.1073/pnas.1712108114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kossinova OA, Gopanenko A v., Tamkovich SN, et al. Cytosolic YB-1 and NSUN2 are the only proteins recognizing specific motifs present in mRNAs enriched in exosomes. Biochimica et Biophysica Acta - Proteins and Proteomics. 2017;1865(6):664–673. doi: 10.1016/j.bbapap.2017.03.010 [DOI] [PubMed] [Google Scholar]
- 50.Yanshina DD, Kossinova OA, Gopanenko A v., et al. Structural features of the interaction of the 3′-untranslated region of mRNA containing exosomal RNA-specific motifs with YB-1, a potential mediator of mRNA sorting. Biochimie. 2018;144:134–143. doi: 10.1016/j.biochi.2017.11.007 [DOI] [PubMed] [Google Scholar]
- 51.Teng Y, Ren Y, Hu X, et al. MVP-mediated exosomal sorting of miR-193a promotes colon cancer progression. Nature Communications. 2017;8(1):1–16. doi: 10.1038/ncomms14448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Statello L, Maugeri M, Garre E, et al. Identification of RNA-binding proteins in exosomes capable of interacting with different types of RNA: RBP-facilitated transport of RNAs into exosomes. PLoS ONE. 2018;13(4):e0195969. doi: 10.1371/journal.pone.0195969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van Niel G, D’Angelo G, Raposo G. Shedding light on the cell biology of extracellular vesicles. Nature Reviews Molecular Cell Biology. 2018;19(4):213–228. doi: 10.1038/nrm.2017.125 [DOI] [PubMed] [Google Scholar]
- 54.Kosaka N, Iguchi H, Yoshioka Y, Takeshita F, Matsuki Y, Ochiya T. Secretory mechanisms and intercellular transfer of microRNAs in living cells. Journal of Biological Chemistry. 2010;285(23):17442–17452. doi: 10.1074/jbc.M110.107821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Janas T, Janas MM, Sapoń K, Janas T. Mechanisms of RNA loading into exosomes. FEBS Letters. 2015;589(13):1391–1398. doi: 10.1016/j.febslet.2015.04.036 [DOI] [PubMed] [Google Scholar]
- 56.Hagiwara K, Katsuda T, Gailhouste L, Kosaka N, Ochiya T. Commitment of Annexin A2 in recruitment of microRNAs into extracellular vesicles. FEBS Letters. 2015;589(24):4071–4078. doi: 10.1016/j.febslet.2015.11.036 [DOI] [PubMed] [Google Scholar]
- 57.Janas T, Janas T, Yarus M. Specific RNA binding to ordered phospholipid bilayers. Nucleic Acids Research. 2006;34(7):2128–2136. doi: 10.1093/nar/gkl220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Khvorova A, Kwak YG, Tamkun M, Majerfeld I, Yarus M. RNAs that bind and change the permeability of phospholipid membranes. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(19):10649–10654. doi: 10.1073/pnas.96.19.10649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Barman1 B, Ping2 J, Krystofiak3 E, Allen4 R. Biogenesis of RNA-containing extracellular vesicles at endoplasmic reticulum membrane contact sites. bioRxiv. Published online December 5, 2020:2020.12.04.412379. doi: 10.1101/2020.12.04.412379 [DOI] [Google Scholar]
- 60.Henao-Mejia J, Williams A, Goff LA, et al. The MicroRNA miR-181 Is a Critical Cellular Metabolic Rheostat Essential for NKT Cell Ontogenesis and Lymphocyte Development and Homeostasis. Immunity. 2013;38(5):984–997. doi: 10.1016/j.immuni.2013.02.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.de Candia P, Torri A, Fedeli M, et al. The circulating microRNome demonstrates distinct lymphocyte subset-dependent signatures. European Journal of Immunology. 2016;46(3):725–731. doi: 10.1002/eji.201545787 [DOI] [PubMed] [Google Scholar]
- 62.Rimer JM, Lee J, Holley CL, et al. Long-range function of secreted small nucleolar RNAs that direct 2-O-methylation. Journal of Biological Chemistry. 2018;293(34):13284–13296. doi: 10.1074/jbc.RA118.003410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hizir Z, Bottini S, Grandjean V, Trabucchi M, Repetto E. RNY (YRNA)-derived small RNAs regulate cell death and inflammation in monocytes/macrophages. Cell Death and Disease. 2017;8(1):e2530–e2530. doi: 10.1038/cddis.2016.429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pua HH, Happ HC, Gray CJ, et al. Increased Hematopoietic Extracellular RNAs and Vesicles in the Lung during Allergic Airway Responses. Cell Reports. 2019;26(4). doi: 10.1016/j.celrep.2019.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.O’Connell RM, Chaudhuri AA, Rao DS, Baltimore D. Inositol phosphatase SHIP1 is a primary target of miR-155. Proceedings of the National Academy of Sciences. 2009;106(17):7113–7118. doi: 10.1073/pnas.0902636106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Androulidaki A, Iliopoulos D, Arranz A, et al. The Kinase Akt1 Controls Macrophage Response to Lipopolysaccharide by Regulating MicroRNAs. Immunity. 2009;31(2):220–231. doi: 10.1016/j.immuni.2009.06.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-κB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(33):12481–12486. doi: 10.1073/pnas.0605298103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hou J, Wang P, Lin L, et al. MicroRNA-146a Feedback Inhibits RIG-I-Dependent Type I IFN Production in Macrophages by Targeting TRAF6, IRAK1, and IRAK2. The Journal of Immunology. 2009;183(3):2150–2158. doi: 10.4049/jimmunol.0900707 [DOI] [PubMed] [Google Scholar]
- 69.Okoye IS, Coomes SM, Pelly VS, et al. MicroRNA-Containing T-Regulatory-Cell-Derived Exosomes Suppress Pathogenic T Helper 1 Cells. Immunity. 2014;41(1):89–103. doi: 10.1016/j.immuni.2014.05.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cobb BS, Hertweck A, Smith J, et al. A role for Dicer in immune regulation. Journal of Experimental Medicine. 2006;203(11):2519–2527. doi: 10.1084/JEM.20061692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhou X, Jeker LT, Fife BT, et al. Selective miRNA disruption in T reg cells leads to uncontrolled autoimmunity. Journal of Experimental Medicine. 2008;205(9):1983–1991. doi: 10.1084/JEM.20080707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liston A, Lu L-F, O’Carroll D, Tarakhovsky A, Rudensky AY. Dicer-dependent microRNA pathway safeguards regulatory T cell function. Journal of Experimental Medicine. 2008;205(9):1993–2004. doi: 10.1084/JEM.20081062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gomez I, Ward B, Souilhol C, et al. Neutrophil microvesicles drive atherosclerosis by delivering miR-155 to atheroprone endothelium. Nature Communications. 2020;11(1):1–18. doi: 10.1038/s41467-019-14043-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang Y, Liu D, Chen X, et al. Secreted Monocytic miR-150 Enhances Targeted Endothelial Cell Migration. Molecular Cell. 2010;39(1):133–144. doi: 10.1016/j.molcel.2010.06.010 [DOI] [PubMed] [Google Scholar]
- 75.Ying W, Riopel M, Bandyopadhyay G, et al. Adipose Tissue Macrophage-Derived Exosomal miRNAs Can Modulate In Vivo and In Vitro Insulin Sensitivity. Cell. 2017;171(2):372–384.e12. doi: 10.1016/j.cell.2017.08.035 [DOI] [PubMed] [Google Scholar]
- 76.Momen-Heravi F, Bala S, Kodys K, Szabo G. Exosomes derived from alcohol-treated hepatocytes horizontally transfer liver specific miRNA-122 and sensitize monocytes to LPS. Scientific Reports. 2015;5(1):9991. doi: 10.1038/srep09991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pan Y, Hui X, Chong Hoo RL, et al. Adipocyte-secreted exosomal microRNA-34a inhibits M2 macrophage polarization to promote obesity-induced adipose inflammation. Journal of Clinical Investigation. 2019;129(2):834–849. doi: 10.1172/JCI123069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Frank AC, Ebersberger S, Fink AF, et al. Apoptotic tumor cell-derived microRNA-375 uses CD36 to alter the tumor-associated macrophage phenotype. Nature Communications. 2019;10(1):1–18. doi: 10.1038/s41467-019-08989-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Abels ER, Maas SLN, Nieland L, et al. Glioblastoma-Associated Microglia Reprogramming Is Mediated by Functional Transfer of Extracellular miR-21. Cell Reports. 2019;28(12):3105–3119.e7. doi: 10.1016/j.celrep.2019.08.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ridder K, Keller S, Dams M, et al. Extracellular Vesicle-Mediated Transfer of Genetic Information between the Hematopoietic System and the Brain in Response to Inflammation. PLoS Biology. 2014;12(6). doi: 10.1371/journal.pbio.1001874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zomer A, Maynard C, Verweij FJ, et al. In vivo imaging reveals extracellular vesicle-mediated phenocopying of metastatic behavior. Cell. 2015;161(5):1046–1057. doi: 10.1016/j.cell.2015.04.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chen F, Chen J, Yang L, et al. Extracellular vesicle-packaged HIF-1α-stabilizing lncRNA from tumour-associated macrophages regulates aerobic glycolysis of breast cancer cells. Nature Cell Biology. 2019;21(4):498–510. doi: 10.1038/s41556-019-0299-0 [DOI] [PubMed] [Google Scholar]
- 83.Fitzgerald KA, Kagan JC. Toll-like Receptors and the Control of Immunity. Cell. 2020;180(6):1044–1066. doi: 10.1016/j.cell.2020.02.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Heil F, Hemmi H, Hochrein H, et al. Species-Specific Recognition of Single-Stranded RNA via Till-like Receptor 7 and 8. Science. 2004;303(5663):1526–1529. doi: 10.1126/science.1093620 [DOI] [PubMed] [Google Scholar]
- 85.Diebold SS, Kaisho T, Hemmi H, Akira S, Reis E Sousa C. Innate Antiviral Responses by Means of TLR7-Mediated Recognition of Single-Stranded RNA. Science. 2004;303(5663):1529–1531. doi: 10.1126/science.1093616 [DOI] [PubMed] [Google Scholar]
- 86.Forsbach A, Nemorin J-G, Montino C, et al. Identification of RNA Sequence Motifs Stimulating Sequence-Specific TLR8-Dependent Immune Responses. The Journal of Immunology. 2008;180(6):3729–3738. doi: 10.4049/jimmunol.180.6.3729 [DOI] [PubMed] [Google Scholar]
- 87.Fabbri M, Paone A, Calore F, et al. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(31):E2110–E2116. doi: 10.1073/pnas.1209414109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang Y, Liang H, Jin F, et al. Injured liver-released miRNA-122 elicits acute pulmonary inflammation via activating alveolar macrophage TLR7 signaling pathway. Proceedings of the National Academy of Sciences of the United States of America. 2019;116(13):6162–6171. doi: 10.1073/pnas.1814139116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Feng Y, Chen H, Cai J, et al. Cardiac RNA induces inflammatory responses in cardiomyocytes and immune cells via Toll-like receptor 7 signaling. Journal of Biological Chemistry. 2015;290(44):26688–26698. doi: 10.1074/jbc.M115.661835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lehmann SM, Krüger C, Park B, et al. An unconventional role for miRNA: Let-7 activates Toll-like receptor 7 and causes neurodegeneration. Nature Neuroscience. 2012;15(6):827–835. doi: 10.1038/nn.3113 [DOI] [PubMed] [Google Scholar]
- 91.Schlee M, Hartmann G. Discriminating self from non-self in nucleic acid sensing. Nature Reviews Immunology. 2016;16(9):566–580. doi: 10.1038/nri.2016.78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Nabet BY, Qiu Y, Shabason JE, et al. Exosome RNA Unshielding Couples Stromal Activation to Pattern Recognition Receptor Signaling in Cancer. Cell. 2017;170(2):352–366.e13. doi: 10.1016/j.cell.2017.06.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Srinivasan S, Yeri A, Cheah PS, et al. Small RNA Sequencing across Diverse Biofluids Identifies Optimal Methods for exRNA Isolation. Cell. 2019;177(2):446–462.e16. doi: 10.1016/j.cell.2019.03.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rozowsky J, Kitchen RR, Park JJ, et al. exceRpt: A Comprehensive Analytic Platform for Extracellular RNA Profiling. Cell Systems. 2019;8(4):352–357.e3. doi: 10.1016/j.cels.2019.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Adams D, Gonzalez-Duarte A, O’Riordan WD, et al. Patisiran, an RNAi Therapeutic, for Hereditary Transthyretin Amyloidosis. New England Journal of Medicine. 2018;379(1):11–21. doi: 10.1056/nejmoa1716153 [DOI] [PubMed] [Google Scholar]
- 96.Jackson LA, Anderson EJ, Rouphael NG, et al. An mRNA Vaccine against SARS-CoV-2 — Preliminary Report. New England Journal of Medicine. 2020;383(20):1920–1931. doi: 10.1056/nejmoa2022483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Baden LR, el Sahly HM, Essink B, et al. Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine. New England Journal of Medicine. 2021;384(5):403–416. doi: 10.1056/nejmoa2035389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mulligan MJ, Lyke KE, Kitchin N, et al. Phase I/II study of COVID-19 RNA vaccine BNT162b1 in adults. Nature. 2020;586(7830):589–593. doi: 10.1038/s41586-020-2639-4 [DOI] [PubMed] [Google Scholar]
- 99.Polack FP, Thomas SJ, Kitchin N, et al. Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine. New England Journal of Medicine. 2020;383(27):2603–2615. doi: 10.1056/nejmoa2034577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhao BS, Roundtree IA, He C. Post-transcriptional gene regulation by mRNA modifications. Nature Reviews Molecular Cell Biology. 2016;18(1):31–42. doi: 10.1038/nrm.2016.132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Suzuki T The expanding world of tRNA modifications and their disease relevance. Nature Reviews Molecular Cell Biology. Published online 2021. doi: 10.1038/s41580-021-00342-0 [DOI] [PubMed] [Google Scholar]
- 102.Steitz TA. A structural understanding of the dynamic ribosome machine. Nature Reviews Molecular Cell Biology. 2008;9(3):242–253. doi: 10.1038/nrm2352 [DOI] [PubMed] [Google Scholar]
- 103.Yao RW, Wang Y, Chen LL. Cellular functions of long noncoding RNAs. Nature Cell Biology. 2019;21(5):542–551. doi: 10.1038/s41556-019-0311-8 [DOI] [PubMed] [Google Scholar]
- 104.Matera AG, Wang Z. A day in the life of the spliceosome. Nature Reviews Molecular Cell Biology. 2014;15(2):108–121. doi: 10.1038/nrm3742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kufel J, Grzechnik P. Small Nucleolar RNAs Tell a Different Tale. Trends in Genetics. 2019;35(2):104–117. doi: 10.1016/j.tig.2018.11.005 [DOI] [PubMed] [Google Scholar]
- 106.Hahne JC, Lampis A, Valeri N. Vault RNAs: hidden gems in RNA and protein regulation. Cellular and Molecular Life Sciences. 2021;78(4):1487–1499. doi: 10.1007/s00018-020-03675-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kowalski MP, Krude T. Functional roles of non-coding Y RNAs. International Journal of Biochemistry and Cell Biology. 2015;66:20–29. doi: 10.1016/j.biocel.2015.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]


