Summary
Individuals with orofacial asymmetry due to mucosal overgrowths, ipsilateral bone and dental aberrations with perineurial hyperplasia and/or perineuriomatous pseudo-onion bulb proliferations, comprise a recognizable clinical entity. In this article, we describe three individuals with this clinical entity and mosaic PIK3CA variants c.3140A>G (p. His1047Arg), c.328_330delGAA (p. Glu110del), and c.1353_1364del (p.Glu453_Leu456del). We conclude that the identification of these mosaic variants in individuals with orofacial asymmetry presenting histopathologically perineurial hyperplasia and/or intraneural pseudo-onion bulb perineurial cell proliferations supports the inclusion of this clinical entity in the PIK3CA-related overgrowth spectrum.
Keywords: PIK3CA, overgrowth, oral mucosa, face, peripheral nerve, perineurium, pseudo-onion bulbs
Introduction
Progress in the discovery of molecular signaling pathways and sequencing techniques have allowed the identification of gene variants involved in mosaic or segmental overgrowth conditions. Mosaic variants in PIK3CA have been associated with a group of overgrowth disorders known as PIK3CA-related overgrowth spectrum (PROS; MIM: 171834).1 Nerve enlargement with perineurial hyperplasia (PerHy) has been described in individuals with macrodactyly,2 a form of PROS, and enlarged nerves,3 and mucosal neuromas4 have been reported in biopsies of individuals with facial infiltrating lipomatosis, another form of PROS.
The purpose of this study was to test for the presence of mosaic PIK3CA variants in individuals with orofacial overgrowth characterized by hyperplastic oral soft tissue lesions with PerHy and multiple intraneural pseudo-onion bulb perineurial cell proliferations (POPs).5
Subjects and methods
Three individuals, ages 3–27 years old at the time of first clinical encounter, presented with orofacial asymmetry and oral soft tissue overgrowths. They were clinically evaluated by three of the authors (I.G.K., A.-L.A., and R.A.). Hematoxylin and eosin slides and paraffin tissue blocks of oral lesions were available from all individuals. The peripheral nerve lesions were immunohistochemically studied with S100 (Dako, Carpinteria, CA; 1:1,600, polyclonal), epithelial membrane antigen (EMA) (Dako; 1:50, E29), and neurofilament protein (Dako; 1:800, 2F11) antibodies following a standard protocol.
Participants underwent oral lesional soft tissue biopsies for PIK3CA variant analysis. Genomic analysis of individuals 1 and 2 was reviewed and approved by the National Human Genome Research Institute (NHGRI) institutional review board (IRB). For individual 1, genomic DNA was isolated from two fibroblast cultures established from lingual lesional tissue, and PIK3CA variant allele fraction (VAF) was determined using a custom restriction fragment length polymorphism (RFLP) assay.6
For individual 2, genomic DNA was isolated from biopsy material. A 14-gene custom capture panel was designed to include genes that feature variants that have been associated with mosaic overgrowth (xGen Lockdown Probe Pool, IDT, Coralville, IA). The following genes were included: PIK3CA, AKT1, AKT2, AKT3, FGFR1, GNAQ, GNAS, KRAS, MTOR, PDGFRB, PHLPP1, PIK3R2, PTEN, and RASA1. Analysis was undertaken using next-generation sequencing (MiSeq, Illumina, San Diego, CA). Briefly, ∼1 μg of genomic DNA was fragmented, purified with the QIAGEN MinElute PCR Purification Kit (QIAGEN, Germantown, MD), end-prepped and adaptor-ligated size-selected for 320 bp fragments with AMPure XP beads (Beckman Coulter, Indianapolis, IN), and dual-indexed during seven cycles of PCR enrichment. After library preparation, the library was quantified with Qubit fluorometry (Thermofisher, Rockville, MD) and 100 ng was pooled with other libraries. The capture of DNA fragments was performed using the xGen Lockdown Probe Pool and xGen lockdown reagents according to the manufacturer’s protocols (IDT). Libraries were run on an Illumina MiSeq instrument (Illumina, San Diego, CA) using a v2 nano flow cell at 10 pM loading concentration. After sequencing, demultiplexing was performed on-machine by Illumina’s MiSeq Reporter software version 2.6. Fastq files for each sample were then processed according to Genetic Analysis Toolkit's (GATK) best practices workflow. Reads were filtered for mapping quality >20.
Individual 3 was consented at the local institution (King Saud University), where she underwent a biopsy procedure for molecular diagnostic purposes. The tissue evaluation was performed at the Genetic Diagnostic Laboratory, University of Pennsylvania Perelman School of Medicine, Philadelphia, PA, USA. The test utilized assessed all coding regions of PIK3CA and flanking intron sequences (±20 bp) and specific regions of the AKT1, AKT2, AKT3, GNAQ, GNA11, MTOR, and PIK3R2. The minimum threshold for variant allele detection was set at 2% at 2,000× coverage and ≥10 reads without strand bias. Repeat sequencing of an independent replicate sample was also performed on the same platform to confirm variants. The Ion Reporter Software suite was used for variant filtering. The confidence score was calculated using the Phred quality score.
All sequence variants are numbered using reference sequence NM_006218.4 and Human Genome Variation Society nomenclature.
Results
Individual 1
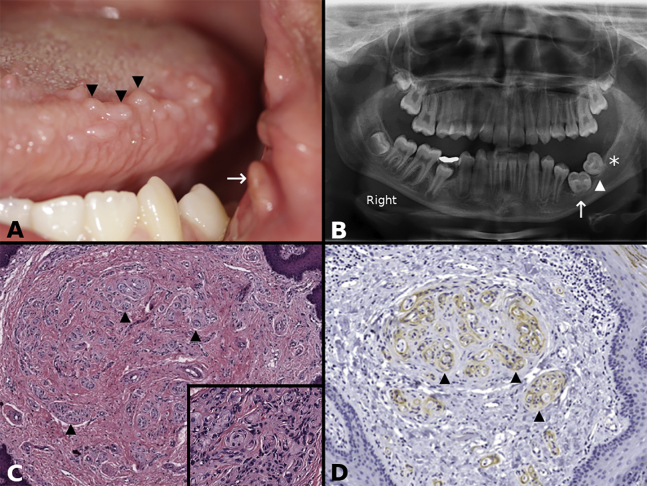
A 13-year-old girl presented with mild left-sided facial overgrowth, multiple small polypoid lesions on the left side of the tongue, and a polyp on the left buccal commissure (Figure 1A). Also, a nodular growth was present in the posterior buccal mucosa. It was unknown if the lesions were congenital; however, the individual had one lesion in the buccal mucosa surgically removed at the age of 6 years with no available information regarding histopathologic evaluation or diagnosis. The family history did not include any similarly affected member. A panoramic radiograph showed arrested development of the roots of the left lower molars and absence of a third lower molar germ (Figure 1B). The lingual lesions were characterized by hyperplastic fibrocollagenous connective tissue with multiple foci of POP (Figure 1C and inset). Immunohistochemically, EMA highlighted perineurial cells (Figure 1D) enveloping Schwann sheath (S100)/neuroaxon units. A somatic PIK3CA c.3140A>G (p. His1047Arg) variant was identified in two fibroblast cultures with a VAF of 0.36 and 0.17 and in DNA isolated directly from excised tissue with a VAF of 0.04. The variant was not detected in a blood sample.
Figure 1.
Individual 1
(A) Clinical photograph of individual 1 showing multiple polypoid lesions on the left side of the tongue (three of many indicated with arrowheads) and a polyp at the oral commissure (arrow).
(B) Orthopantomogram of individual 1 showing arrested root development of the permanent left first (arrow) and second (arrowhead) molars, with a missing third molar germ (asterisk).
(C) Lower-power photomicrograph of individual 1 showing fibrous hyperplasia and multiple pseudo-onion bulb (perineurial cell) intraneural proliferations (three of many indicated with arrowheads) (hematoxylin and eosin stain). (Inset) High-power photomicrograph of individual 1 showing round structures representing pseudo-onion bulb (perineurial cell) intraneural proliferations (hematoxylin and eosin stain).
(D) Epithelial membrane antigen (EMA) immunohistochemical stain decorates the perineurial cells of pseudo-onion bulb (perineurial cell) intraneural proliferations (three of many indicated with arrowheads; EMA with hematoxylin counterstain)
Individual 2
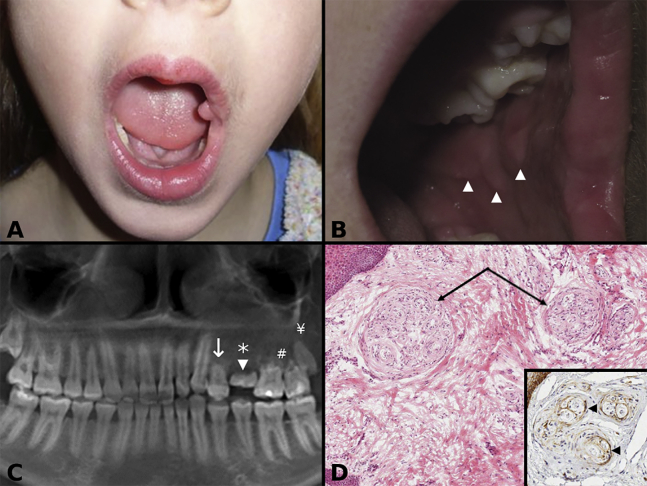
A 3-year-old girl presented with mild left-sided congenital facial overgrowth and a polyp of the left buccal commissure (Figure 2A) that was biopsied and showed an enlarged nerve with POP. Clinical evaluation at age 10 years showed ipsilateral redundant soft tissue on the buccal mucosa (Figure 2B), left maxillary alveolar soft tissue overgrowth, and subtle ipsilateral enlargement of the tongue. Panoramic radiographs at various ages confirmed arrested development of the first maxillary premolar, retained primary second molar, absent second premolar, and a small third molar with root resorption of the left permanent maxillary molars observed at age 16 years (Figure 2C). The dental anatomic variations were associated with left-sided malocclusion. Histopathologic preparations from buccal mucosa reduction tissue at age 10 years showed fibrous hyperplasia and nerves with POP (Figures 2D and inset). A computed tomography (CT) scan, obtained at age 16 years, showed decreased bone density of the left maxilla in the area of premolars and molars, when compared to the right side, and a smaller left maxillary sinus. Magnetic resonance imaging performed at age 16 years showed reduced muscular bulk and fatty overgrowth of the left side of the face. Additional soft tissue reduction for functional and esthetic purposes during left maxillary third molar extraction at age 16 years was necessary and tissue samples were used for variant investigation, which showed in-frame PIK3CA c.1353_1364del (p. Glu453_Leu456del) with a VAF of 0.22. The variant was not detected in the blood sample.
Figure 2.
Individual 2
(A) Clinical photograph of individual 2 at age 3 years showing left side orofacial overgrowth, evident on the left side of the lip and tongue, and an oral commissural polyp.
(B) Clinical photograph of individual 2 at age 10 years showing redundant tissue on the left buccal mucosa tissue (arrowheads).
(C) Panoramic radiograph of individual 2 showing arrested development of the first left maxillary premolar (arrow), a retained primary second molar (arrowhead), an absent second premolar (asterisk), root resorption of the permanent molars (#), and a small third molar (¥).
(D) Photomicrograph of individual 2 showing multiple pseudo-onion bulb (perineurial) intraneural proliferations (arrows) in a fibrous hyperplastic stroma (hematoxylin and eosin). (Inset) Epithelial membrane antigen (EMA) decorates (arrowheads) the perineurial cells of pseudo-onion bulb (perineurial) intraneural proliferations (EMA immunohistochemical stain with hematoxylin counterstain).
Individual 3
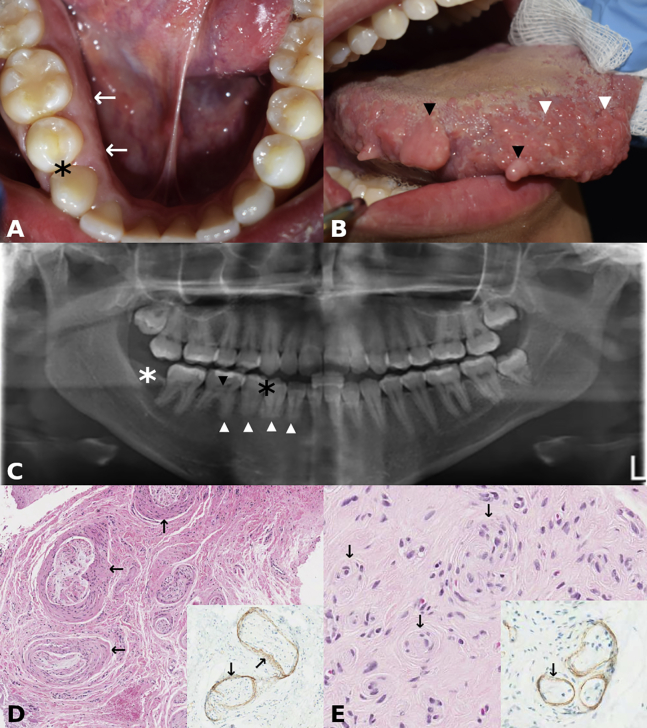
A 27-year-old woman presented with congenital right-sided facial overgrowth, including right side alveolar overgrowth (Figure 3A), multiple ipsilateral polypoid lesions on the tongue, and several enlarged fungiform papillae (Figure 3B). In addition, a single polypoid lesion on the right buccal mucosa commissure was noted. A panoramic radiograph showed absent lower right first premolar, small roots in the lower right teeth, missing right mandibular third molar, and pulpal calcifications in the lower right premolar and molars (Figure 3C). Furthermore, the right body of the mandible was larger with reduced bony trabeculation when compared to the left. A diagnostic biopsy of a polypoid lesion from the tongue showed PerHy (Figure 3D) and POP (Figures 3E and inset). DNA extracted from lesional lingual tissue biopsies showed PIK3CA c.328_330delGAA;p. Glu110del with a VAF of 0.12 and 0.21, respectively, which were not detected in the blood.
Figure 3.
Individual 3
(A) Clinical photograph of individual 3 presenting with enlarged alveolus on the right side when compared to the left side (arrows). Black asterisk indicates missing right first premolar (see also C).
(B) Clinical photograph of individual 3 showing polypoid lesions on the tongue (two of many; black arrowheads) and enlarged fungiform papillae (two of many; white arrowheads).
(C) Panoramic radiograph of individual 3 showing absent right first mandibular premolar (white asterisk), small roots in the lower right teeth (white arrowheads), pulpal calcifications in the lower right premolar and molars (black arrowhead pointing to the pulpal calcification of the first permanent right molar), and missing third molar (black asterisk). The right body of the mandible is larger than the left (L), and there is apparent reduced bony trabeculation of the right side when compared to the left.
(D) Photomicrograph of individual 3 showing perineurial hyperplasia (arrows) and peripheral nerve enlargement (hematoxylin and eosin). (Insert) Hyperplastic perineurium (arrows) decorated by epithelial membrane antigen (EMA; immunohistochemical stain with hematoxylin counterstain).
(E) Photomicrograph of individual 3 showing multiple pseudo-onion bulb (perineurial) intraneural proliferations (arrows) (hematoxylin and eosin). (Inset) Pseudo-onion bulb (perineurial) intraneural proliferations decorated by epithelial membrane antigen (EMA; immunohistochemical stain with hematoxylin counterstain).
Clinicopathologic findings and variants are summarized in Table 1.
Table 1.
Clinicopathologic characteristics and PIK3CA mutation analysis
| Individual | Age at the time of initial clinical evaluation | Affected side | Oral findings | Imaging findings | Pathologic findings | PIK3CA variants |
|---|---|---|---|---|---|---|
| 1 | 13 years | left | multiple polypoid lesions; commissural polyp and nodular growth in the posterior buccal mucosa | arrested development of the roots of the left lower molars, absence of a third lower molar germ | POP | PIK3CA c.3140A>G (p.His1047Arg)VAF fibroblast culture 1: 0.36VAF fibroblast culture 2: 0.17 VAF biopsy tissue: 0.04 |
| 2 | 3 years | left | congenital facial overgrowth; polypoid lesion at the commissure; buccal mucosa soft tissue overgrowth; maxillary alveolar soft tissue overgrowth; ipsilateral tongue enlargement | arrested development of the first maxillary premolar, retained primary second molar, absent second premolar and a small third molar; decreased bone density of the left maxilla in the area of premolars and molars, smaller left maxillary sinus; MRI showed reduced muscular bulk and fatty overgrowth of the left side of the face | POP and PerHy | PIK3CA c.328_330delGAA (p.Glu110del)VAF biopsy: 0.22 |
| 3 | 27 years | right | facial overgrowth; polypoid lesions on the tongue and enlarged fungiform papillae; polypoid lesion on right buccal mucosa | absent lower right first premolar, small roots in the lower right teeth, and pulpal calcifications in the lower right premolars and molars; right body of the mandible was larger with reduced bony trabeculation when compared to the left | PerHy and OP | PIK3CA c.1353_1364del (p.Glu453_Leu456del)VAF biopsy #1: 0.12VAF biopsy #2: 0.21 |
Discussion
Phosphatidylinositide 3 kinases (PI3K), members of intracellular lipid kinases, and their downstream mediators, AMP/GMP/protein kinase B (AKT) kinases and mammalian target of rapamycin (mTOR), are key components of the PI3K/AKT/mTOR signaling pathway, which is crucial for cell proliferation, apoptosis, angiogenesis, anabolic reactions, metabolism, therapy resistance, and tumor development.7 The PI3Ks are divided into three classes, with class IA comprising heterodimers of p85 regulatory and p110 catalytic subunits. The p85 and related isoforms are encoded by three genes (PIK3R1, PIK3R2, and PIK3R3), while the catalytic subunits are encoded by PIK3CA, PIK3CB, and PIK3CD.7
Somatic PIK3CA variants have been associated with PROS.1 Strongly activating germline PIK3CA variants are considered incompatible with a viable phenotype, further supporting Happle’s theory that non-hereditary syndromic conditions affecting segment(s) of the body are caused by somatic mosaic variants while complete heterozygosity of the same variants results in an embryonic lethal phenotype.8 Mice with the germline PIK3CAH1047R (p110α) variant, common in PROS and occurring in the kinase region of the gene, die before embryonic day 10 (E10) due to vascular anomalies caused by secondary disruption of vascular endothelial growth factor A (VEGF-A) signaling.9 Constitutional PIK3CA variants can occur if the variant is weakly activating. An example was described in a child with congenital megalencephaly and symmetric macrosomia10 with c.335T>A;p. Ile112Asn identified in peripheral blood, buccal mucosa swab, and skin fibroblasts.
In PROS, the timing and location of postzygotic PIK3CA mutation probably determine the phenotypic variability, thus representing examples of type 1 mosaicism.11 Clinical entities encompassed by PROS include (1) fibroadipose overgrowth (FAO); (2) congenital lipomatous overgrowth, vascular malformations, epidermal nevi, scoliosis/skeletal and spinal (CLOVES) syndrome; (3) hemihyperplasia multiple lipomatosis (HHML), which may overlap with CLOVES syndrome; (4) macrodactyly; (5) capillary malformation of the lower lip, lymphatic malformation of the face and neck, asymmetry, and partial/generalized overgrowth (CLAPO) syndrome; and (6) megalencephaly-capillary malformation (MCAP) syndrome. Recently, there has been a proposal to include Klippel-Trenaunay syndrome (MIM: 149000) in PROS.12
Overgrowth in PROS is typically congenital and static or mildly progressive,13 and manifestations are divided into two groups: A (spectrum; two or more features): (1) adipose, muscle, nerve, or skeletal overgrowth; (2) vascular malformations; and (3) epidermal nevi; and B (isolated features): large lymphatic malformation, macrodactyly, truncal adipose overgrowth, hemimegalencephaly, epidermal nevi, seborrheic keratoses, and benign lichenoid keratoses. A clinical diagnosis of PROS requires identification of the PIK3CA variant and congenital or early childhood onset overgrowth with manifestations from either group A or B.
Segmental arrested development of the roots of teeth was observed in all three individuals. In facial infiltrating lipomatosis, however, macrodontia has been observed.4 Given that the PIK3/AKT pathway is an important regulator of the sonic hedgehog pathway,14 and, as such, is essential for craniofacial development including dentition,15 PIK3CA variants may affect the development of teeth. A PIK3CA variant was identified in the pulp of an extracted tooth of an individual with MCAP.16
There are reports of individuals with asymmetric facial overgrowth who have features similar to those described herein who we hypothesize to have mosaic PIK3CA variants. Vargo et al.17 reported an individual with asymmetric mandibular enlargement, including enlargement of the mandibular canal, ipsilateral mandibular condyle enlargement, exostoses, and multiple fibrotic lesions on the tongue, shown to be POP on histologic evaluation. Siponen et al.18 described an individual with hemifacial overgrowth and characteristics of PROS. In their report, these authors concentrated on peripheral nerve lesions observed microscopically in the ipsilateral buccal mucosa and vestibule and referred to them as orofacial intraneural perineuriomas, but they are likely to be POP.5 As well, the individual described by Pujol et al.19 as having multiple idiopathic mucosal neuromas may instead have PROS.
Maclellan et al.3 and Couto et al.4 reported individuals with facial infiltrating lipomatosis that shared features with CLOVES, including abnormal nerves3 and mucosal neuromas,4 respectively. However, neither of these publications included detailed histopathologic descriptions. In the past, some individuals with apparent PROS and oral neuromatous lesions have been classified as having partial MEN2B (pure mucosal neuroma syndrome; MIM: 162300), although RET variants were not identified in lesional tissue or blood.20 At that time, the concept of PROS was not established. Re-evaluation of those reports in light of current knowledge leads us to suggest that at least some of these individuals had PROS.
Most identified PIK3CA pathogenic variants to date are missense variants or small inframe deletions and are located in the kinase and helical domains.21 Individual 2 reported here had PIK3CA c.1353_1364del, while individual 3 had p.(Glu110del), mapping to the C2 and P85BD domains, respectively. Although additional studies are needed to confirm that these variants represent functional mutations, we are of the opinion that they relate to the phenotype observed. Because this disorder has a gain-of-function pathogenetic mechanism, it is unsurprising that there are no nonsense or splice site variants involving PIK3CA identified to date. Commonly identified PIK3CA somatic variants in PROS include (1) c.1258T>C (p. Cys420Arg), (2) c.1624G>A (p.Glu542Lys), (3) c.1633G>A (p. Glu545Lys), (4) c.3140A>G (p. His1047Arg), and (5) c.3140A>T (p. His1047Leu), with three of them being recurrent cancer-associated variants: p.Glu542Lys, p.Glu545Lys, and p.His1047Arg.22 Failure to detect PIK3CA variants does not exclude the clinical diagnosis of PROS in individuals with suggestive features as undetected variants may be the result of low-level mosaicism.
In summary, we presented here the clinicopathologic findings of individuals with orofacial overgrowth that is clinically distinct from other variants of PROS with histopathologic evidence of POP and PerHy in lesional tissue and somatic mosaic PIK3CA variants. PTEN-related overgrowth syndromes (MIM: 158350)23 and Proteus syndrome (MIM: 176920), which is caused by AKT124 variants, do not manifest, to the best of our knowledge, such neuropathologic findings. Also, individuals with MEN2B, although presenting with multiple mucosal neuromas, representing enlarged nerves, have distinct clinical manifestations and RET germline mutations. Thus, the identification of orofacial overgrowth with multifocal POP and abnormal nerves with PerHy should raise the diagnostic possibility of PROS, which can be confirmed by molecular testing available from a number of genetic testing laboratories.
Data and code availability
This study did not generate/analyze new datasets or code.
Acknowledgments
The research was partially funded by the Oral Pathology Research Fund, University of Minnesota, School of Dentistry. The authors are grateful to Mr. Brian Dunnette of the University of Minnesota for his assistance with the illustrations.
Declaration of interests
The authors declare no competing interests.
References
- 1.Keppler-Noreuil K.M., Parker V.E., Darling T.N., Martinez-Agosto J.A. Somatic overgrowth disorders of the PI3K/AKT/mTOR pathway & therapeutic strategies. Am. J. Med. Genet. C. Semin. Med. Genet. 2016;172:402–421. doi: 10.1002/ajmg.c.31531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rios J.J., Paria N., Burns D.K., Israel B.A., Cornelia R., Wise C.A., Ezaki M. Somatic gain-of-function mutations in PIK3CA in patients with macrodactyly. Hum. Mol. Genet. 2013;22:444–451. doi: 10.1093/hmg/dds440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maclellan R.A., Luks V.L., Vivero M.P., Mulliken J.B., Zurakowski D., Padwa B.L., Warman M.L., Greene A.K., Kurek K.C. PIK3CA activating mutations in facial infiltrating lipomatosis. Plast. Reconstr. Surg. 2014;133:12e–19e. doi: 10.1097/01.prs.0000436822.26709.7c. [DOI] [PubMed] [Google Scholar]
- 4.Couto J.A., Konczyk D.J., Vivero M.P., Kozakewich H.P.W., Upton J., Fu X., Padwa B.L., Mulliken J.B., Warman M.L., Greene A.K. Somatic PIK3CA mutations are present in multiple tissues of facial infiltrating lipomatosis. Pediatr. Res. 2017;82:850–854. doi: 10.1038/pr.2017.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koutlas I.G., Scheithauer B.W. On pseudo-onion bulb intraneural proliferations of the non-major nerves of the oral mucosa. Head Neck Pathol. 2013;7:334–343. doi: 10.1007/s12105-013-0446-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lindhurst M.J., Parker V.E., Payne F., Sapp J.C., Rudge S., Harris J., Witkowski A.M., Zhang Q., Groeneveld M.P., Scott C.E., et al. Mosaic overgrowth with fibroadipose hyperplasia is caused by somatic activating mutations in PIK3CA. Nat. Genet. 2012;44:928–933. doi: 10.1038/ng.2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Engelman J.A., Luo J., Cantley L.C. The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat. Rev. Genet. 2006;7:606–619. doi: 10.1038/nrg1879. [DOI] [PubMed] [Google Scholar]
- 8.Happle R. Lethal genes surviving by mosaicism: a possible explanation for sporadic birth defects involving the skin. J. Am. Acad. Dermatol. 1987;16:899–906. doi: 10.1016/s0190-9622(87)80249-9. [DOI] [PubMed] [Google Scholar]
- 9.Hare L.M., Schwarz Q., Wiszniak S., Gurung R., Montgomery K.G., Mitchell C.A., Phillips W.A. Heterozygous expression of the oncogenic Pik3ca(H1047R) mutation during murine development results in fatal embryonic and extraembryonic defects. Dev. Biol. 2015;404:14–26. doi: 10.1016/j.ydbio.2015.04.022. [DOI] [PubMed] [Google Scholar]
- 10.Di Donato N., Rump A., Mirzaa G.M., Alcantara D., Oliver A., Schrock E., Dobyns W.B., O’Driscoll M. Identification and characterization of a novel constitutional PIK3CA mutation in a child lacking the typical segmental overgrowth of “PIK3CA-related overgrowth spectrum”. Hum. Mutat. 2016;37:242–245. doi: 10.1002/humu.22933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Happle R. The categories of cutaneous mosaicism: A proposed classification. Am. J. Med. Genet. A. 2016;170A:452–459. doi: 10.1002/ajmg.a.37439. [DOI] [PubMed] [Google Scholar]
- 12.Vahidnezhad H., Youssefian L., Uitto J. Klippel-Trenaunay syndrome belongs to the PIK3CA-related overgrowth spectrum (PROS) Exp. Dermatol. 2016;25:17–19. doi: 10.1111/exd.12826. [DOI] [PubMed] [Google Scholar]
- 13.Keppler-Noreuil K.M., Rios J.J., Parker V.E.R., Semple R.K., Lindhurst M.J., Sapp J.C., Alomari A., Ezaki M., Dobyns W., Biesecker L.G. PIK3CA-related overgrowth spectrum (PROS): diagnostic and testing eligibility criteria, differential diagnosis, and evaluation. Am. J. Med. Genet. A. 2015;167A:287–295. doi: 10.1002/ajmg.a.36836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Riobó N.A., Lu K., Ai X., Haines G.M., Emerson C.P., Jr. Phosphoinositide 3-kinase and Akt are essential for Sonic Hedgehog signaling. Proc. Natl. Acad. Sci. USA. 2006;103:4505–4510. doi: 10.1073/pnas.0504337103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seppala M., Fraser G.J., Birjandi A.A., Xavier G.M., Cobourne M.T. Sonic Hedgehog signaling and development of dentition. J. Dev. Biol. 2017;5:E6. doi: 10.3390/jdb5020006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McDermott J.H., Byers H., Clayton-Smith J. Detection of a mosaic PIK3CA mutation in dental DNA from a child with megalencephaly capillary malformation syndrome. Clin. Dysmorphol. 2016;25:16–18. doi: 10.1097/MCD.0000000000000099. [DOI] [PubMed] [Google Scholar]
- 17.Vargo R.J., Potluri A., Bauer R.E., Seethala R.R., Bilodeau E.A. Intraoral pseudo-onion bulb intraneural prolferations in a patient with hemimandibular hyperplasia: a case report and review of the literature. Head Neck Pathol. 2016;10:475–480. doi: 10.1007/s12105-016-0725-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Siponen M., Sándor G.K., Ylikontiola L., Salo T., Tuominen H. Multiple orofacial intraneural perineuriomas in a patient with hemifacial hyperplasia. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endod. 2007;104:e38–e44. doi: 10.1016/j.tripleo.2006.12.030. [DOI] [PubMed] [Google Scholar]
- 19.Pujol R.M., Matias-Guiu X., Miralles J., Colomer A., de Moragas J.M. Multiple idiopathic mucosal neuromas: a minor form of multiple endocrine neoplasia type 2B or a new entity? J. Am. Acad. Dermatol. 1997;37:349–352. [PubMed] [Google Scholar]
- 20.Gordon C.m., Majzoub J.A., Marsh D.J., Mulliken J.B., Ponder B.A., Robinson B.G., Eng C. Four cases of mucosal neuroma syndrome: multiple endocrine neoplasm 2B or not 2B? J. Clin. Endocrinol. Metab. 1998;83:17–20. doi: 10.1210/jcem.83.1.4504. [DOI] [PubMed] [Google Scholar]
- 21.Rivière J.B., Mirzaa G.M., O’Roak B.J., Beddaoui M., Alcantara D., Conway R.L., St-Onge J., Schwartzentruber J.A., Gripp K.W., Nikkel S.M., et al. Finding of Rare Disease Genes (FORGE) Canada Consortium De novo germline and postzygotic mutations in AKT3, PIK3R2 and PIK3CA cause a spectrum of related megalencephaly syndromes. Nat. Genet. 2012;44:934–940. doi: 10.1038/ng.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mirzaa G., Timms A.E., Conti V., Boyle E.A., Girisha K.M., Martin B., Kircher M., Olds C., Juusola J., Collins S., et al. PIK3CA-associated developmental disorders exhibit distinct classes of mutations with variable expression and tissue distribution. JCI Insight. 2016;1:e87623. doi: 10.1172/jci.insight.87623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lachlan K.L., Lucassen A.M., Bunyan D., Temple I.K. Cowden syndrome and Bannayan Riley Ruvalcaba syndrome represent one condition with variable expression and age-related penetrance: results of a clinical study of PTEN mutation carriers. J. Med. Genet. 2007;44:579–585. doi: 10.1136/jmg.2007.049981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lindhurst M.J., Sapp J.C., Teer J.K., Johnston J.J., Finn E.M., Peters K., Turner J., Cannons J.L., Bick D., Blakemore L., et al. A mosaic activating mutation in AKT1 associated with the Proteus syndrome. N. Engl. J. Med. 2011;365:611–619. doi: 10.1056/NEJMoa1104017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This study did not generate/analyze new datasets or code.