Summary
The bone disorder osteogenesis imperfecta (OI) is genetically heterogeneous. Most affected individuals have an autosomal dominant disorder caused by heterozygous variants in either of the type I collagen genes (COL1A1 or COL1A2). To date, two reports have linked Mesoderm Development LRP Chaperone (MESD) to autosomal recessive OI type XX. Four different biallelic pathogenic variants in MESD were shown to cause a progressively deforming phenotype, associated with recurrent fractures and oligodontia in five individuals in five families. Recently, compound heterozygosity for a frameshift predicted to lead to a premature termination codon in exon 2 of the 3-exon gene and a second frameshift in the terminal exon in MESD were detected in three stillbirths in one family with severe OI consistent with the neonatal lethal phenotype. We have identified four additional individuals from four independent families with biallelic variants in MESD: the earlier reported c.632dupA (p.Lys212Glufs∗19) and c.676C>T (p.Arg226∗)—which are associated with a severe form of OI—and one new pathogenic variant, c.603-606delTAAA (p.Asn201Lysfs∗15), which causes a neonatal lethal form of OI. MESD acts in the WNT signaling pathway, where it is thought to play a role in the folding of the WNT co-receptors low-density lipoprotein receptor-related proteins 5 and 6 (LRP5/LRP6) and in chaperoning their transit to the cell surface. Our report broadens the phenotypic and genetic spectrum of MESD-related OI, provides additional insight into the pathogenic pathways, and underscores the necessity of MESD for normal WNT signaling in bone formation.
Keywords: osteogenesis imperfecta, OI, LRP5, LRP6, KDEL receptor, protein recycling from the Golgi
Mesoderm Development LRP Chaperone (MESD) has been linked to autosomal recessive osteogenesis imperfecta type XX. Our report describes four new individuals and broadens the phenotypic and genetic spectrum of MESD-related OI, provides additional insight into the pathogenic pathways, and underscores the necessity of MESD for normal WNT signaling in bone formation.
Main text
Osteogenesis imperfecta (OI [MIM: 166200, 166210, 259420, 166220]; see Table S1 for full list of genes) is a heritable bone dysplasia characterized by low bone mass and fragile bones with fractures—hence, the commonly used description “brittle bone disease.” The vast majority of OI individuals harbor a heterozygous pathogenic variant in one of the two genes that encode the chains of type I procollagen (COL1A1 [MIM: 12050], COL1A2 [MIM: 120160]), the precursor of the major protein of bone.1 In the last 15 years, genetic studies have expanded our understanding of the causative mechanisms that underlie OI. It is now appreciated that rare recessive forms of OI result from variants in almost two dozen genes that encode proteins involved in regulation of collagen production; assembly, transport, chaperoning, and secretion of collagens; extracellular processing of collagen; and regulation of signaling pathways.2 One of these is the Wingless-related integration site (WNT) signaling pathway, whose involvement in bone biology has not been fully elucidated. Variants in WNT1 have been linked to severe OI (MIM: 615220) and to osteoporosis,3, 4, 5, 6 and targets of WNTs that are important for bone development and homeostasis are thought to include alkaline phosphatase (ALPL [MIM: 171760]), which plays a role in bone mineralization, and Specificity Protein 7 (SP7 [MIM: 606633]), which encodes a transcription factor that controls preosteoblast-to-osteoblast transition.7,8 Variants in ALPL cause hypophosphatasia (MIM: 146300 and 241510),9 and two reports associate SP7 with a recessive form of OI (MIM: 613849).10,11
MESD (MIM: 607783), previously called MESDC2, encodes the endoplasmic reticulum (ER) resident chaperone protein MESD (Mesoderm development candidate 2) and consists of a signal sequence (residues 1–33), a chaperone domain (residues 34–164), an escort domain (residues 165–204), and a COOH-terminal KDEL-like sequence (REDL) that distinguishes it from the secreted proteins it chaperones and allows for retrieval from the Golgi.12 The protein facilitates folding of the β-propeller domains of two WNT co-receptors, low-density lipoprotein receptor-related protein 5 and 6 (LRP5/LRP6), and in their localization to the cell surface (Figures 1, 2 A, and 2B).16 LRP5 regulates peak bone mass in vertebrates, and homozygosity for inactivating variants in LRP5 causes osteoporosis-pseudoglioma syndrome (OPPG; MIM: 259770),17 whereas LRP5 gain-of-function variants are implicated in high bone mass (HBM) phenotypes that include endosteal hyperostosis (MIM: 144750), van Buchem disease (MIM: 607636), and osteopetrosis (MIM: 607634).18, 19, 20 In addition, LRP6 loss-of-function variants resulted in altered glycosylation and abrogated activation of the WNT signaling pathway, contributing to the etiology of non-syndromic autosomal dominant oligodontia.21 Homozygous Mesdc2 knockout mice fail to establish a primitive streak and lack a developed mesoderm22 due to a patterning defect in the proximal epiblast.16 This patterning defect is similar to the outcome when another WNT family member, Wnt3, is knocked out in embryos.23
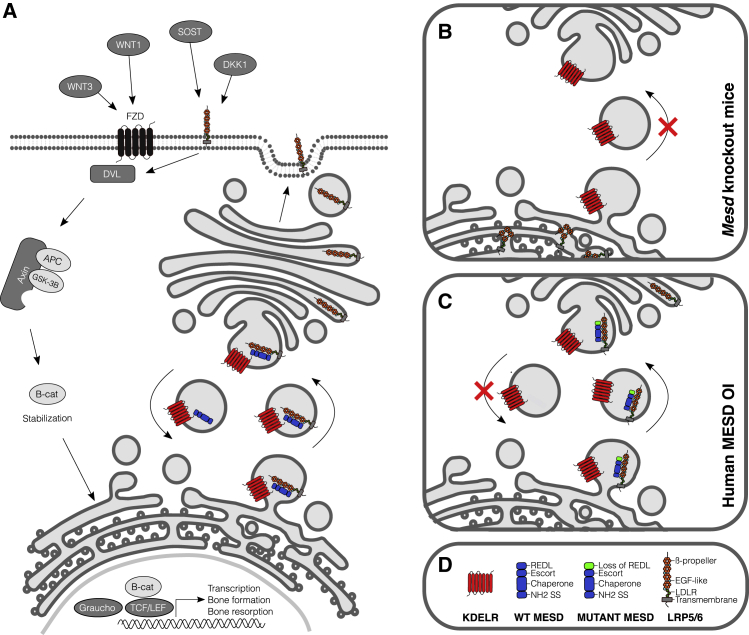
Figure 1.
Function and location of MESD in the WNT signaling pathway (in bone cells)
(A) Canonical WNT signaling involves binding of WNT proteins to the frizzled receptor (FZD) and to the co-receptors LRP5 and LRP6. Formation of this complex leads to the inhibition of glycogen synthase kinase 3β (GSK-3β), which phosphorylates β-catenin. Unphosphorylated β-catenin is safeguarded from proteosomal degradation, accumulates, and moves into the nucleus where it triggers lymphoid enhancer factor (LEF)/T cell factor (TCF)-mediated gene transcription.13 MESD is thought to play a role in the folding of the WNT co-receptors LRP5 and LRP6 and in chaperoning their transit to the cell surface; the C-terminal REDL ER-retention sequence of MESD allows for retrieval from the Golgi.
(B and C) Close-up views of aberrant MESD signaling in mice and humans. LRP5/6 do not fold properly in Mesd knockout mice and appear to aggregate in the RER (B). We hypothesize that the mutant MESD proteins described in this report have (residual) chaperone activity to signal LRP5/6 to the cell surface but fail for retrieval from the Golgi since they lack the REDL sequence (C). This leads to disturbed WNT signaling in bone (LRP5) and tooth development (LRP6).
(D) Legend of most important protein structures in MESD-related WNT signaling: KDELR receptor, red; wild-type (WT) and mutant MESD proteins with/loss of REDL, blue/green; LRP5/6, gray/green/cyan/orange.
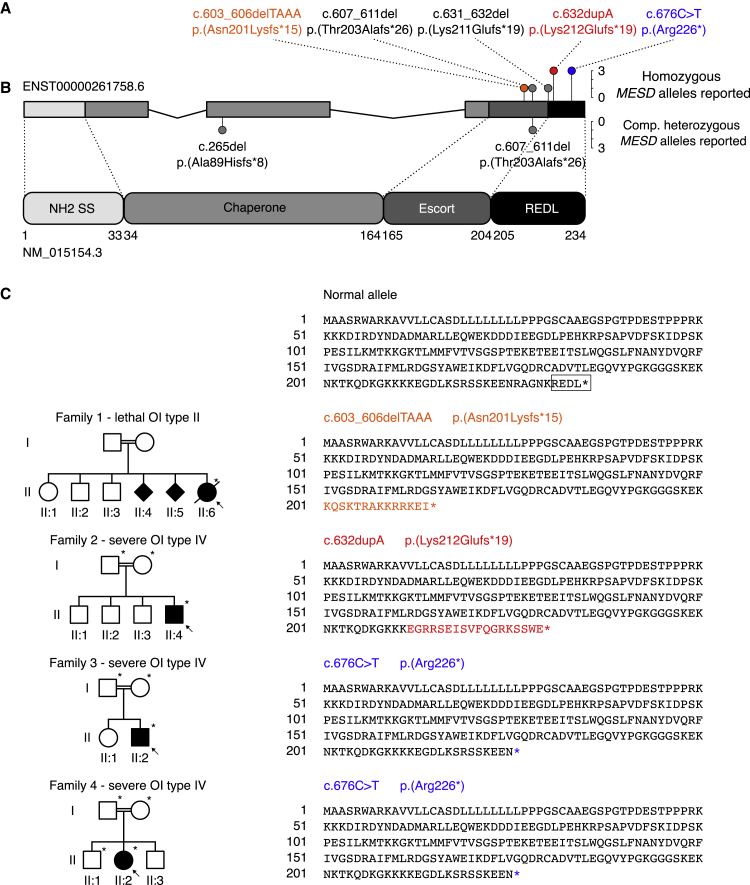
Figure 2.
Structure of the MESD gene and MESD protein, pedigrees of the described families, and representation of the mutant allele sequences
(A) The MESD gene consists of 3 exons (4,200 bp) and comprises 14.07 kb on chromosome 15. The cumulative frequency of the six known pathogenic MESD alleles (both this and two previous studies14,15) are denoted with colored lollipop graphs. The pathogenic alleles included in this report are highlighted in orange, red, and blue, respectively.
(B) Full-length MESD protein consists of 234 amino acids (aa) and contains 4 functional domains.
(C) Pedigrees of the 4 families with MESD variants and representation of the WT and mutant alleles. The proband in each family is indicated by an arrow. Filled circles, squares, or diamond structures represent individuals with OI. Individuals who were studied in each family are noted with asterisks. The variants in families 1 and 2 cause frameshifts and introduce premature termination codons 15 and 19 codons downstream of the frameshifts, respectively, and the variant in families 3 and 4 changes an asparagine residue to a stop codon, resulting in premature termination. All three variants cause the loss of the C-terminal REDL ER-retention sequence (marked with a square in the WT allele) needed for retrieval from the Golgi, and their position and mutant AA sequence is highlighted in orange, red and blue, respectively (matching colors in A).
Biallelic pathogenic variants, all located in the final (third) exon, in MESD were recently identified in five individuals from five families who presented with moderately severe, progressively deforming recessive OI, which one individual had with oligodontia.14 With overexpression studies in HEK293T cells, Moosa et al.14 suggested that the OI-associated MESD mutations produced hypomorphic alleles whose failure to remain within the ER was significantly reduced but did not completely eliminate LRP5 and LRP6 trafficking. Recently, infants from one family who harbored compound heterozygous frameshift variants, one that resulted in the premature termination codon in exon two and the other a premature termination in exon three, had a lethal OI picture, similar to OI type II caused by type I collagen gene pathogenic variants. Histological analysis of femoral, calvarial, and spinal bone revealed impaired osseous development with altered osteocyte morphology and reduced canalicular connectivity. Bone mineral density distribution measured by quantitative backscattered electron imaging indicated impaired and more heterogeneous matrix mineralization in the described MESD fetuses than in controls.15 OI that results from pathogenic variants in MESD has been designated as OI type XX (MIM: 618644) in OMIM. Here, we present four new MESD individuals and used fibroblast studies to get additional insight into the pathogenic pathways of the MESD-related OI subtype.
We used whole-exome sequence analysis (WES) to study an infant from a consanguineous family in which we had not previously identified a causative variant in targeted OI-related gene sequences. We identified a homozygous likely pathogenic variant in MESD in the affected proband (c.603-606delTAAA [p.Asn201Lysfs∗15] in family 1, individual 1-II:6, described later). We subsequently identified homozygous variants in three additional independent OI probands, two by means of targeted Sanger sequencing of MESD analysis (c.632dupA [p.Lys212Glufs∗19] and c.676C>T [p.Arg226∗] in families 2 and 3, respectively) and in family 4 by clinical whole exome analysis.
The proband from family 1 (1-II:6; Figure 3A) died in the neonatal period. She had a small chest and multiple fractures. Radiographs showed a thin calvarial mantle, thin deformed ribs with multiple fractures, and short long bones (Figure 3A). The radiographic features were consistent with severe OI (type III).6 No other clinical information was available. Her parents (1-I:1 and 1-I:2) were double first cousins. They had two prior pregnancies that were also affected (1-II:4 and 1-II:5) and had three unaffected children (1-II:1, 1-II:2, and 1-II:3). To identify the causative gene in this family (Figure 2C), we used the same WES dataset previously described by Pyott et al.3 A detailed methodology on the exome filtering is provided in the Supplemental methods (Exome filtering). Of the candidate genes that were identified using this approach, none had known clinical implications relating to skeletal disorders, but one gene, MESD (Figure 2A), was involved in a pathway known to be important to bone development. Individual 1:II-6 was homozygous for a 4-bp deletion in the last exon of MESD (c.603-606delTAAA [p.Asn201Lysfs∗15]) that was predicted to lead to a frameshift and premature termination within the exon (Figure 2C). Sequence analysis of cDNA from the proband’s cultured skin fibroblasts showed that all stable transcripts had the deletion (Figure S1). Analysis of collagen synthesized and secreted by the proband’s cultured dermal fibroblasts revealed no alterations in the electrophoretic mobility of the chains of type I procollagen or in the efficiency of secretion (data not shown). Parental samples were not available to confirm that each carried the variant or to determine if the deletion-bearing transcript was as stable as that encoded by the normal allele.
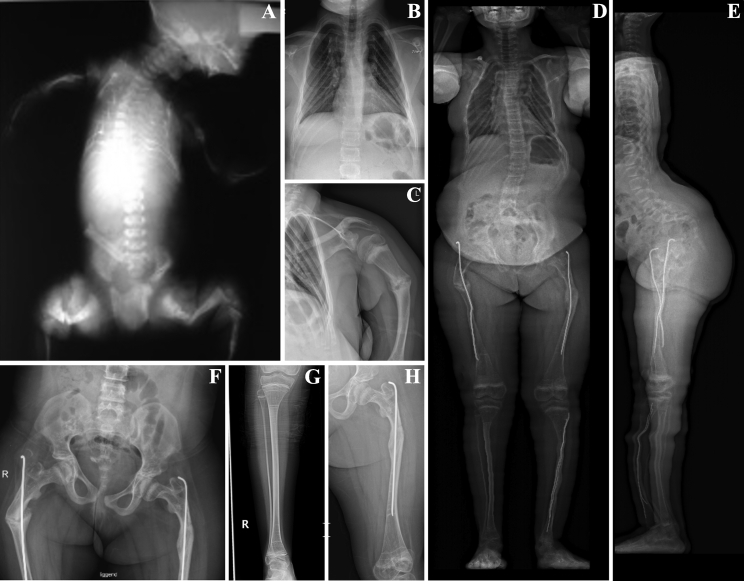
Figure 3.
Clinical spectrum of MESD pathogenic variants
(A) Imaging of individual P1-II:6 shows limited calvarial mineralization, a small chest with thin ribs, fractures in upper and lower extremities, and platyspondyly.
(B–H) Individual P4-II:2 presents with scoliosis (D and E), mild bowing of upper and lower limbs (C–H), fracture of the left humerus (C), and (bilateral) post-surgical rodding of the lower leg (D) and femoral shafts (D, E, F, and H), respectively. The impact of bisphosphonate treatment can be noted in the right lower (G) and left upper leg (H).
Clinical pictures/radiographs taken in the neonatal period (A) or at the age of 11 (B and C) and 12 years (D–H), respectively.
The proband from family 2 (2-II:4) was a 5-year-old boy who had been diagnosed with OI at birth as a result of multiple in utero fractures. When assessed at the age of 5 years, he had a triangular facial shape, lower limb and shoulder deformities, and poor muscle tone. He was non-ambulatory. Most of his teeth had fallen out, but it was unclear whether this was due to dentinogenesis imperfecta (DI), clinical oligodontia (in accordance to previously reported MESD individuals14), or other factors (e.g., poor nutrition, gum disease). His parents (2-I:1 and 2-I:2) were first cousins. Sanger sequencing of MESD revealed a homozygous single base pair duplication in the last exon (c.632dupA [p.Lys212Glufs∗19]) that led to a frameshift and a premature termination in the same exon (Figures 2A and 2C). His parents were each heterozygous for the duplication (Figure S1). Cultured fibroblasts were not available from the proband or from the parents.
The proband in family 3 (3-II:2) was a 5-year-old boy with blue sclerae; multiple fractures in his upper and lower limbs, sternum, and ribs; and deformities of his left femoral and right upper limb following femoral shaft osteotomy and fixation. His parents (3-I:1 and 3-I:2) were first cousins once-removed. The boy was homozygous for a pathogenic nonsense variant in MESD (c.676C>T [p.Arg226∗]) that deletes the last 9 amino acids of the protein (Figures 2A and 2C). His father and mother were each carriers of the pathogenic variant (Figure S1). No fibroblasts were available from the proband.
The 12-year-old proband from family 4 (4-II:2) was the middle child of consanguineous parents of Palestinian origin. She has two healthy brothers and had presented to medical care at age 10 months with a history of multiple low-impact fractures, severe osteoporosis (T- and Z-scores of −7.3 and −5.9), mild bowing of the upper and lower limbs, generalized muscle hypotonia, and severe hyperlaxity of the small joints. She had soft and somewhat translucent skin, had mildly dilated post-operative scars, and did not present with DI. She had severe psychomotor retardation (likely to be caused by the identified variant), polycystic kidney disease (inherited from her mother’s side), and a history of Langerhans cell histiocytosis. Pamidronate treatment was discontinued because of very little improvement, but zoledronate administration led to higher bone mineral content. She had Ilizarov surgery, and at age 3 years, she remained mostly nonambulatory. A detailed overview of the radiological features of this individual is highlighted in Figures 3B–3H. Karyotype of the proband in this family was normal, and whole-genome array comparative genomic hybridization (180k Agilent Array) did not reveal any chromosomal deletions/duplications. No alterations in the electrophoretic migration pattern of the fibrillar collagens type I, III, and V were noted (data not shown). Molecular analysis by means of a clinical WES approach (similar filtering strategy as described in Guillemyn et al.24) revealed the same homozygous pathogenic variant in MESD as described in family 3 (c.676C>T [p.Arg226∗]) (Figures 2A and 2C). Her father and mother were each carriers of the pathogenic variant, and one of her healthy brothers who was available for genetic testing did not carry the pathogenic variant (data not shown).
The four individuals reported here had OI phenotypes that ranged from moderately severe to lethal in the perinatal period and are similar in clinical presentation to the MESD individuals that were described by Moosa et al.14 A cumulative overview of the molecular and salient clinical and radiographic findings of all 12 autosomal recessive MESD individuals reported to date is provided in Table 1: all individuals presented with a history of fractures (7/12 confirmed prenatal fractures), and homozygous or compound heterozygous frameshift variants were identified in all 12. Nine had a consanguineous background, five had blue sclerae, three presented with clinical oligodontia, only one had hearing impairment, and a delayed gross motor function was noted in half of the individuals reported to date (6/12).
Table 1.
Cumulative overview of molecular, clinical, and radiographic findings of the autosomal recessive MESD individuals reported to date
| Findings | Cumulative numbers (12 individuals) |
|---|---|
| Location of pathogenic MESD alleles | homozygous in last exon (3): 9/12; compound heterozygous in exon 2 and 3: 3/12 |
| Gender | 4 females, 8 males |
| Consanguinity | 9/12 |
| Bisphosphonate treatment | 5 have a history of bisphosphonate treatment |
| Confirmed prenatal fractures | 7/12 |
| Color of sclera | 5 bluish, 3 white, 4 N/A |
| Disorganized dentition/clinical oligodontia | 3 yes, 1 no, 1?, 7 N/A |
| Hearing impairment | 3 no, 1 yes, 8 N/A |
| History of fractures | 12/12 |
| Vertebral/thoracic cage/rib fractures | 11 yes, 1 no |
| Retarded gross motor function | 6 yes |
The explanation for the variation in severity of the phenotypes is not yet apparent. The variants that we identified are not noted in the NHLBI Exome Variant Server (EVS), the ExAC database, gnomAD, or in dbSNP. In the gnomAD database, there are 8 heterozygous variants (12 individuals) that produce premature termination codons in the last exon that might have similar effects to those seen here. This represents a carrier frequency of about 1/10,000 individuals, meaning homozygosity or compound heterozygosity would lead to a very low frequency of the disorder in the population represented in those collections. MESD variants accounted for about 5% of the approximately 100 unsolved OI families we tested but represent only 4 of more than 3,500 OI individuals in whom we have identified the causative variants.
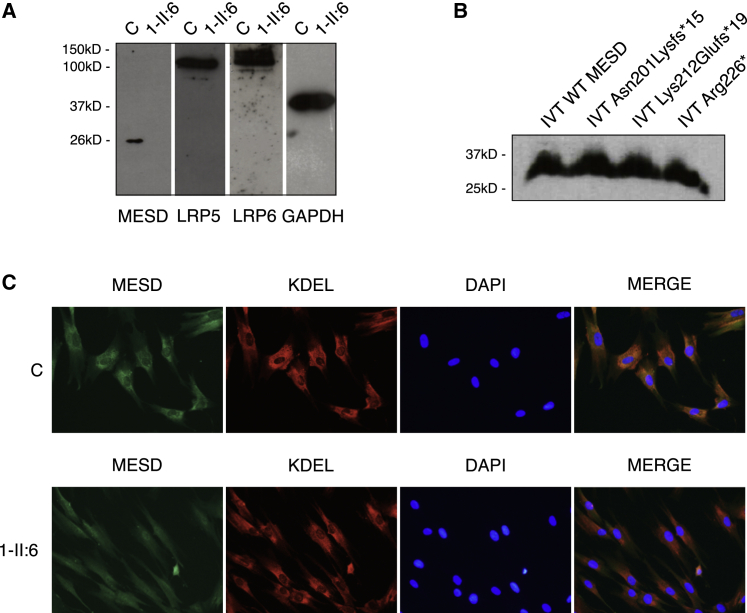
As also noted by Moosa et al.,14 the three pathogenic variants that we identified were found in the last exon of MESD, as were the resulting premature termination codons. We could isolate and sequence cDNA from the cultured cells available from individual 1-II:6, thereby complementing the earlier observations of stable mRNA transcripts. Despite these, the MESD protein was not detected in the cell lysate from 1-II:6 fibroblasts, while it was detected in control cells (Figure 4A). To assess the relative stability of the mutant MESD proteins, we performed coupled in vitro transcription/translation (IVTT) for each of the variant sequences and found that the wild-type sequence and all the variant constructs produced stable MESD proteins (Figure 4B). Nonetheless, immunocytochemical studies of MESD in primary fibroblasts from 1-II:6 showed weak and diffuse staining compared to the strong ER-localized staining observed in control fibroblasts (Figure 4C). Cultured fibroblasts from 1-II:6 (family 1) had intracellular levels of LRP5 and LRP6 that were similar to control (Figure 4A), but we were unable to determine if those proteins tracked to the cell surface.
Figure 4.
Loss of the REDL sequence in MESD is pathogenic
(A) Protein studies on cultured fibroblasts from the proband in family 1 (1-II:6) demonstrate the absence of MESD protein in the cell lysate. Levels of LRP5 and LRP6 appear normal. Images are of a single blot that was probed, washed, and re-probed for each of the targets (MESD, LRP5, LRP6, GAPDH). Western blot.
(B) IVTT of the variant MESD sequences results in production of stable MESD protein in all cases.
(C) Variant MESD lacking the REDL sequence is weakly staining and diffuse throughout the cytoplasm in 1-II:6 primary fibroblasts, while it is properly ER-localized in healthy control cells. Immunocytochemical.
In all four families, the frameshift or stop-gain variants result in loss of the REDL sequence but retention of the chaperone and escort domains. It is possible that loss of the REDL signal results in MESD secretion rather than being recycled to the ER. Previous studies demonstrated that in HEK293T cells engineered to overexpress a mutant MESD protein that lacked the REDL sequence, MESD was not retrieved from the Golgi, and some traveled through the secretory pathway into the medium.16 One limitation of those studies is that the gene was overexpressed, and saturation of available KDEL receptors would mean either that there was degradation or that some of the protein was contained in vesicles that proceeded from the rough endoplasmic reticulum (RER) to the Golgi and then to the cell surface. Alternatively, loss of the REDL sequence may destabilize the protein, a possibility suggested by study of other OI-causing genes involved in the collagen synthetic pathway. In the Prolyl-3 Hydroxylase (P3H1)-Cartilage-Associated Protein (CRTAP)-Cyclophilin B (CypB) molecular complex, responsible for the post-translational modification of type I collagen proα chains, if either the KDEL-containing P3H1 or the KDEL non-containing CRTAP is null, both proteins are unstable and absent from the cell.25 It was later reported that loss of the P3H1 KDEL retention sequence alone was sufficient for loss of both P3H1 and CRTAP and the cause of OI in one family, though only intracellular P3H1 and CRTAP were analyzed, leaving open the question of whether stable secreted protein from these cells would be detectable.26 While we were able to detect LRP5 and LRP6 protein in cell lysates from affected individuals’ cells, we were not able to determine their localization using immunocytochemistry. This could have been due to poor cell quality. It is also unclear whether the expression and activity of MESD, LRP5, LRP6, and the appropriately interacting WNTs in fibroblasts are representative of osteogenic cells.
The range of the clinical presentations in individuals with biallelic pathogenic variants in MESD is striking, as it extends from a very severe peri- or prenatal lethal phenotype to one that fits into the more affected end of the original OI type IV range of Sillence (Moosa et al.,14 Stürznickel et al.,15 and this report). From these reports it is clear that heterozygosity, even for a likely null allele, is tolerated without clinical effect. Compound heterozygosity for a null and a frameshift premature termination codon in the last exon produces a very severe and lethal form of OI, similar to the severe OI type II picture. The mildest phenotype appears to result from a late premature termination codon that deletes the last 8 amino acids of the MESD protein, including the REDL sequence that permits recycling of the protein from the Golgi but retains the domains of the protein involved in the stabilization of the propeller motifs of the LRP proteins.
In the absence of MESD in knockout mice, LRP5 and LRP6 are retained in the ER as high-molecular-weight aggregates.16 This appears to reflect the loss of proper folding in the propeller domains of the LRP proteins that is contributed by the chaperone function of MESD. MESD is vectorially inserted into the RER as a consequence of its amino terminal signal sequence. The carboxy-terminal RDEL sequence would then permit attachment to a KDEL receptor protein in the RER membrane. LRP5 and LRP6, both of which interact with MESD, are transmembrane proteins with the large, ultimately external facing domain in the RER lumen. These portions of the LRP proteins contain both the propeller domains and the LDL-like receptor domains that interact with frizzled proteins and WNT proteins on the cell surface. It seems likely that the loss of the REDL anchor would limit interaction of MESD with the membrane-anchored LRP proteins and so limit the correct folding of the propeller domains. The normal life cycle of MESD would be to facilitate incorporation of the two LRP proteins into RER vesicles that are then transported to the Golgi. Once there, they should dissociate and return to the RER, while the LRP proteins would continue to their cell surface localization. In the absence of MESD, both the chaperone and guidance to the Golgi functions would be lost. It appears, however, that all the mutations that we have encountered must facilitate LRP folding, after which at least some of the folded LRP proteins could negotiate transport to the Golgi and then on to the cell surface without the MESD fellow traveler. The fate of the RDEL-lacking MESD remains uncertain. Overexpression studies show secretion into the culture medium, but this level of expression is likely to overwhelm the capture of the protein by the KDEL-receptor mechanisms. It is also not clear how LRP5 and/or LRP6 would be secreted, given their status as transmembrane-anchored proteins. In-depth investigation of fibroblast or osteogenic cells of MESD individuals, as well as animal models deficient for Mesd, Lrp5, and/or Lrp6, will be key to (1) study the exact role and fate of mutant MESD proteins, (2) shed light on the direct consequences on LRP5 and/or LRP6 secretion, and (3) define the respective roles of LRP5/6 in the development of bone and teeth phenotypes, respectively. In addition to these studies, the identification of more individuals with MESD defects, potentially including structural variants that disrupt different functional domains of the MESD protein, will further increase our understanding of the pathophysiological mechanism underlying this condition.
Acknowledgments
We thank the families for participating in this study. This work was supported by the Freudmann Research Fund at the University of Washington, the National Institute of Arthritis and Musculoskeletal and Skin Diseases (F31AR069971), the Cell & Molecular Biology Training Grant (5T32GM007270), the Molecular Medicine Training Grant (5T32GM095421), the UW Department of Pathology, the Collagen Diagnostic Laboratory, Ghent University (Methusalem grant 08/01M01108), and Research Foundation Flanders (1842318N to F.M.). Analysis assistance was provided by the University of Washington Center for Mendelian Genomics (UW-CMG) and was funded by the National Human Genome Research Institute and the National Heart, Lung, and Blood Institute grant HG006493 to D.A.N., M.J.B., and Suzanne Leal. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Declaration of interests
M.J.B. is the Editor of Human Genetics and Genomics Advances. All other authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xhgg.2021.100051.
Web resources
OMIM, https://www.omim.org
ExAC, http://exac.broadinstitute.org
Supplemental information
References
- 1.Marini J.C., Forlino A., Cabral W.A., Barnes A.M., San Antonio J.D., Milgrom S., Hyland J.C., Körkkö J., Prockop D.J., De Paepe A., et al. Consortium for osteogenesis imperfecta mutations in the helical domain of type I collagen: regions rich in lethal mutations align with collagen binding sites for integrins and proteoglycans. Hum. Mutat. 2007;28:209–221. doi: 10.1002/humu.20429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marini J.C., Forlino A., Bächinger H.P., Bishop N.J., Byers P.H., Paepe A., Fassier F., Fratzl-Zelman N., Kozloff K.M., Krakow D., et al. Osteogenesis imperfecta. Nat. Rev. Dis. Primers. 2017;3:17052. doi: 10.1038/nrdp.2017.52. [DOI] [PubMed] [Google Scholar]
- 3.Pyott S.M., Tran T.T., Leistritz D.F., Pepin M.G., Mendelsohn N.J., Temme R.T., Fernandez B.A., Elsayed S.M., Elsobky E., Verma I., et al. WNT1 mutations in families affected by moderately severe and progressive recessive osteogenesis imperfecta. Am. J. Hum. Genet. 2013;92:590–597. doi: 10.1016/j.ajhg.2013.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Keupp K., Beleggia F., Kayserili H., Barnes A.M., Steiner M., Semler O., Fischer B., Yigit G., Janda C.Y., Becker J., et al. Mutations in WNT1 cause different forms of bone fragility. Am. J. Hum. Genet. 2013;92:565–574. doi: 10.1016/j.ajhg.2013.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Laine C.M., Joeng K.S., Campeau P.M., Kiviranta R., Tarkkonen K., Grover M., Lu J.T., Pekkinen M., Wessman M., Heino T.J., et al. WNT1 mutations in early-onset osteoporosis and osteogenesis imperfecta. N. Engl. J. Med. 2013;368:1809–1816. doi: 10.1056/NEJMoa1215458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fahiminiya S., Majewski J., Mort J., Moffatt P., Glorieux F.H., Rauch F. Mutations in WNT1 are a cause of osteogenesis imperfecta. J. Med. Genet. 2013;50:345–348. doi: 10.1136/jmedgenet-2013-101567. [DOI] [PubMed] [Google Scholar]
- 7.Fujita K., Janz S. Attenuation of WNT signaling by DKK-1 and -2 regulates BMP2-induced osteoblast differentiation and expression of OPG, RANKL and M-CSF. Mol. Cancer. 2007;6:71. doi: 10.1186/1476-4598-6-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heo J.S., Lee S.-Y., Lee J.-C. Wnt/β-catenin signaling enhances osteoblastogenic differentiation from human periodontal ligament fibroblasts. Mol. Cells. 2010;30:449–454. doi: 10.1007/s10059-010-0139-3. [DOI] [PubMed] [Google Scholar]
- 9.Weiss M.J., Cole D.E., Ray K., Whyte M.P., Lafferty M.A., Mulivor R.A., Harris H. A missense mutation in the human liver/bone/kidney alkaline phosphatase gene causing a lethal form of hypophosphatasia. Proc. Natl. Acad. Sci. USA. 1988;85:7666–7669. doi: 10.1073/pnas.85.20.7666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lapunzina P., Aglan M., Temtamy S., Caparrós-Martín J.A., Valencia M., Letón R., Martínez-Glez V., Elhossini R., Amr K., Vilaboa N., Ruiz-Perez V.L. Identification of a frameshift mutation in Osterix in a patient with recessive osteogenesis imperfecta. Am. J. Hum. Genet. 2010;87:110–114. doi: 10.1016/j.ajhg.2010.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fiscaletti M., Biggin A., Bennetts B., Wong K., Briody J., Pacey V., Birman C., Munns C.F. Novel variant in Sp7/Osx associated with recessive osteogenesis imperfecta with bone fragility and hearing impairment. Bone. 2018;110:66–75. doi: 10.1016/j.bone.2018.01.031. [DOI] [PubMed] [Google Scholar]
- 12.Chen J., Liu C.-C., Li Q., Nowak C., Bu G., Wang J. Two structural and functional domains of MESD required for proper folding and trafficking of LRP5/6. Structure. 2011;19:313–323. doi: 10.1016/j.str.2011.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kubota T., Michigami T., Ozono K. Wnt signaling in bone metabolism. J. Bone Miner. Metab. 2009;27:265–271. doi: 10.1007/s00774-009-0064-8. [DOI] [PubMed] [Google Scholar]
- 14.Moosa S., Yamamoto G.L., Garbes L., Keupp K., Beleza-Meireles A., Moreno C.A., Valadares E.R., de Sousa S.B., Maia S., Saraiva J., et al. Autosomal-Recessive Mutations in MESD Cause Osteogenesis Imperfecta. Am. J. Hum. Genet. 2019;105:836–843. doi: 10.1016/j.ajhg.2019.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stürznickel J., Jähn-Rickert K., Zustin J., Hennig F., Delsmann M.M., Schoner K., Rehder H., Kreczy A., Schinke T., Amling M., et al. Compound heterozygous frameshift mutations in MESD cause a lethal syndrome suggestive of osteogenesis imperfecta type XX. J. Bone Miner. Res. 2021;36:1077–1087. doi: 10.1002/jbmr.4277. [DOI] [PubMed] [Google Scholar]
- 16.Hsieh J.-C., Lee L., Zhang L., Wefer S., Brown K., DeRossi C., Wines M.E., Rosenquist T., Holdener B.C. Mesd encodes an LRP5/6 chaperone essential for specification of mouse embryonic polarity. Cell. 2003;112:355–367. doi: 10.1016/s0092-8674(03)00045-x. [DOI] [PubMed] [Google Scholar]
- 17.Gong Y., Slee R.B., Fukai N., Rawadi G., Roman-Roman S., Reginato A.M., Wang H., Cundy T., Glorieux F.H., Lev D., et al. Osteoporosis-Pseudoglioma Syndrome Collaborative Group LDL receptor-related protein 5 (LRP5) affects bone accrual and eye development. Cell. 2001;107:513–523. doi: 10.1016/s0092-8674(01)00571-2. [DOI] [PubMed] [Google Scholar]
- 18.Boyden L.M., Mao J., Belsky J., Mitzner L., Farhi A., Mitnick M.A., Wu D., Insogna K., Lifton R.P. High bone density due to a mutation in LDL-receptor-related protein 5. N. Engl. J. Med. 2002;346:1513–1521. doi: 10.1056/NEJMoa013444. [DOI] [PubMed] [Google Scholar]
- 19.Little R.D., Carulli J.P., Del Mastro R.G., Dupuis J., Osborne M., Folz C., Manning S.P., Swain P.M., Zhao S.C., Eustace B., et al. A mutation in the LDL receptor-related protein 5 gene results in the autosomal dominant high-bone-mass trait. Am. J. Hum. Genet. 2002;70:11–19. doi: 10.1086/338450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Van Wesenbeeck L., Cleiren E., Gram J., Beals R.K., Bénichou O., Scopelliti D., Key L., Renton T., Bartels C., Gong Y., et al. Six novel missense mutations in the LDL receptor-related protein 5 (LRP5) gene in different conditions with an increased bone density. Am. J. Hum. Genet. 2003;72:763–771. doi: 10.1086/368277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Massink M.P.G., Créton M.A., Spanevello F., Fennis W.M.M., Cune M.S., Savelberg S.M.C., Nijman I.J., Maurice M.M., van den Boogaard M.-J.H., van Haaften G. Loss-of-Function Mutations in the WNT Co-receptor LRP6 Cause Autosomal-Dominant Oligodontia. Am. J. Hum. Genet. 2015;97:621–626. doi: 10.1016/j.ajhg.2015.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holdener B.C., Faust C., Rosenthal N.S., Magnuson T. msd is required for mesoderm induction in mice. Development. 1994;120:1335–1346. doi: 10.1242/dev.120.5.1335. [DOI] [PubMed] [Google Scholar]
- 23.Liu P., Wakamiya M., Shea M.J., Albrecht U., Behringer R.R., Bradley A. Requirement for Wnt3 in vertebrate axis formation. Nat. Genet. 1999;22:361–365. doi: 10.1038/11932. [DOI] [PubMed] [Google Scholar]
- 24.Guillemyn B., Nampoothiri S., Syx D., Malfait F., Symoens S. Loss of TANGO1 Leads to Absence of Bone Mineralization. JBMR Plus. 2021;5:e10451. doi: 10.1002/jbm4.10451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chang W., Barnes A.M., Cabral W.A., Bodurtha J.N., Marini J.C. Prolyl 3-hydroxylase 1 and CRTAP are mutually stabilizing in the endoplasmic reticulum collagen prolyl 3-hydroxylation complex. Hum. Mol. Genet. 2010;19:223–234. doi: 10.1093/hmg/ddp481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takagi M., Ishii T., Barnes A.M., Weis M., Amano N., Tanaka M., Fukuzawa R., Nishimura G., Eyre D.R., Marini J.C., Hasegawa T. A novel mutation in LEPRE1 that eliminates only the KDEL ER- retrieval sequence causes non-lethal osteogenesis imperfecta. PLoS ONE. 2012;7:e36809. doi: 10.1371/journal.pone.0036809. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.