Abstract
Similarity in facial characteristics between relatives suggests a strong genetic component underlies facial variation. While there have been numerous studies of the genetics of facial abnormalities and, more recently, single nucleotide polymorphism (SNP) genome-wide association studies (GWASs) of normal facial variation, little is known about the role of genetic structural variation in determining facial shape. In a sample of Bantu African children, we found that only 9% of common copy number variants (CNVs) and 10-kb CNV analysis windows are well tagged by SNPs (r2 ≥ 0.8), indicating that associations with our internally called CNVs were not captured by previous SNP-based GWASs. Here, we present a GWAS and gene set analysis of the relationship between normal facial variation and CNVs in a sample of Bantu African children. We report the top five regions, which had p values ≤ 9.35 × 10−6 and find nominal evidence of independent CNV association (p < 0.05) in three regions previously identified in SNP-based GWASs. The CNV region with strongest association (p = 1.16 × 10−6, 55 losses and seven gains) contains NFATC1, which has been linked to facial morphogenesis and Cherubism, a syndrome involving abnormal lower facial development. Genomic loss in the region is associated with smaller average lower facial depth. Importantly, new loci identified here were not identified in a SNP-based GWAS, suggesting that CNVs are likely involved in determining facial shape variation. Given the plethora of SNP-based GWASs, calling CNVs from existing data may be a relatively inexpensive way to aid in the study of complex traits.
Keywords: copy number variants, normal facial variation, face shape, CNV, African, Bantu
Introduction
The human face is one of the most visually distinguishable human features. Similarity of facial characteristics within families suggests a strong genetic component in normal facial development. While single nucleotide polymorphism (SNP)-based studies have uncovered many associations with face shape,1, 2, 3 the genetics behind normal facial variation is not well understood. Particularly little is known about the role of copy number variants (CNVs). Previous studies of the sample of Bantu African children studied here have shown that facial shape measures have considerable narrow sense heritability (28%–67% for 32 of 33 facial measurements).4 Cole et al.4 found that much of this heritability is explained by common (minor allele frequency, MAF > 1%) SNPs; nevertheless, for a number of facial phenotype measures, a considerable fraction (26%–64%) of estimated heritability is not explained by common SNPs. We hypothesize that some of this missing heritability may reflect genomic structural variation (SV), especially CNVs that may cause dosage imbalance of genes involved in facial development.
Recent studies in large cohorts have shown that CNVs represent a substantial source of human genetic variation,5 altogether involving 4.8%–9.5% of the genome.6 CNVs have been associated with several common complex diseases and traits, such as schizophrenia7 (MIM: 181500), autism spectrum disorder8 (MIM: 209850), and height9 (MIM: 606255). Additionally, CNVs have been associated with a number of rare diseases, including several in which patients have craniofacial abnormalities, such as 22q11 deletion syndrome10 (MIM: 192430) and Angelman syndrome11 (MIM: 105830).
While recent research has provided insights into SVs, including CNVs, across the genome and in multiple genetic ancestries,12 the role of CNVs in complex traits remains understudied, particularly in non-European ancestral populations.12, 13, 14 Studying CNVs in non-European populations is important, as CNVs and especially rare CNVs may be population specific or differ greatly in frequency.12,15,16 Additionally, the linkage disequilibrium (LD) structure between SNPs and CNVs differs by ancestral population.12
CNV calling and subsequent genome-wide association analysis are both computationally and time intensive; if CNVs are tagged well by SNPs, additional CNV calling and analysis may be unnecessary. The Wellcome Trust Case Control Consortium found that SNP-based analyses captured common CNVs (MAF > 10%) through high LD (r2 > 0.8) with SNPs in a large (n = 16,000) European sample.17 However, only 22% of less common CNVs (MAF < 5%) were well tagged by SNPs. More recently, Collins et al.12 reported lower LD between SNPs and SVs in an African/African American sample compared to other ancestries.
Here, we describe a CNV GWAS and gene set analysis of the relationship between rare and common CNVs and normal facial variation in a sample of 3,388 apparently healthy Bantu African children from Tanzania. We calculated pairwise LD between CNVs and SNPs within 1 megabase (mb) of the CNV to evaluate whether the previous SNP-based GWAS in this sample1 likely captured common and rare CNVs. Within the CNV GWAS, we identify five genomic regions associated with a facial phenotype and nominal CNV associations in three regions that were previously identified by the SNP-based GWAS. Pairwise LD between SNPs and CNVs in these eight regions is low (maximum pairwise r2 < 0.3), further supporting independent and novel associations beyond that of the SNP-based GWAS in African subjects.
Subjects and Methods
Subjects
The study cohort presented here was previously described in detail by Cole et al.1 Briefly, it consisted of 3,631 Bantu African children aged 3–21 from the Mwanza region of Tanzania. Children with abnormal facial features or a relative with known facial abnormalities were excluded. Written informed consent was obtained for all study subjects or their parents, as appropriate. The original SNP GWAS and sample collection was carried out with overall approval and oversight of the Colorado Multiple Institutional Review Board (protocol #09–0731), was additionally approved by the institutional review boards of the University of Calgary, Florida State University, the University of California San Francisco, and the Catholic University of Health and Allied Sciences (Mwanza, Tanzania), and was carried out with the approval of the National Institute for Medical Research (Tanzania). While subjects were apparently unrelated upon data collection, quality control discovered considerable cryptic relatedness within the sample.1 As described below and within Cole et al.,1 this relatedness is accounted for by including a kinship matrix in our model.
Phenotype data
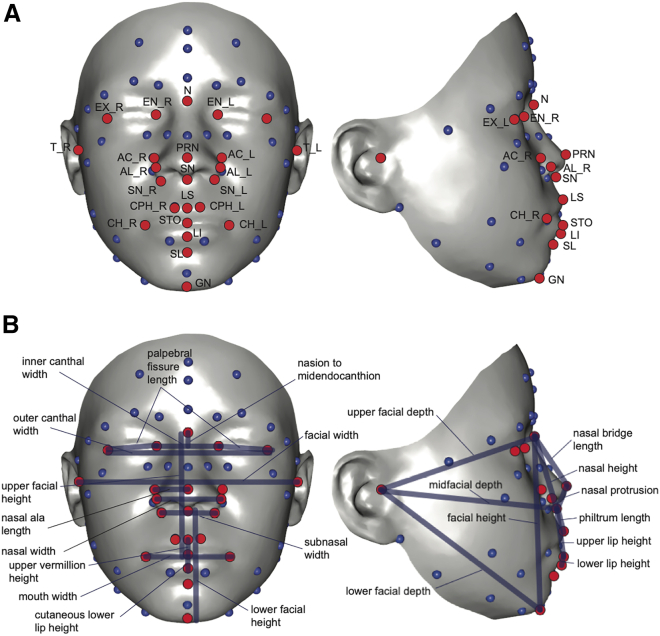
To quantify facial variation, each child was photographed using a 3D camera, as seen in Figure 1. Twenty-nine standard facial morphometric landmarks were extracted, and from these coordinates, twenty-five inter-landmark linear distances, three measures of overall face size (i.e. allometry, centroid size, and head circumference), one summary variable from a principal components analysis (PCA) of the most highly correlated midfacial landmarks (explaining approximately 40% of total midface variation), and five summary variables from a PCA of the whole face (explaining approximately 70% of total facial variation) were derived (Table S1).1 Shape variation associated with size (allometry) was removed by multiple-multivariate regression prior to PCA. Head circumference was measured using a tape measure in 2,686 subjects. This resulted in 34 traits for analysis. See Cole et al.1 for more details.
Figure 1.
Three dimensional photographs with annotated landmarks. (A) 3D landmarks used for geometric morphometric quantification of facial shape as previously described.1 The full landmark set was used to calculate the shape variables (PCs and allometry) as well as facial size (centroid size). The red landmarks are those used to obtain specific interlandmark distances for analysis. (B) The full set of distances used (Table S1).
Variant calling and quality control
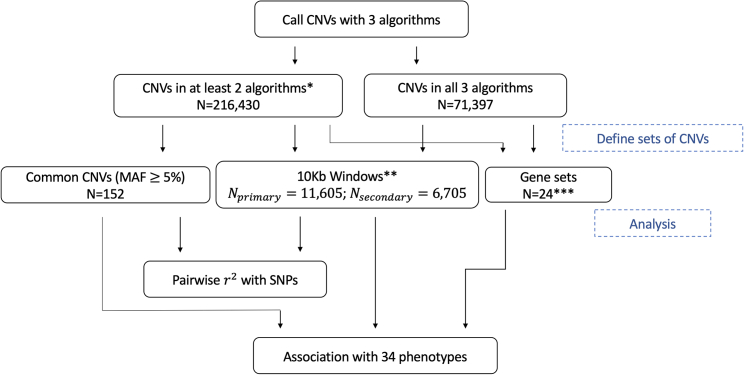
A flowchart of CNV calling and analysis is depicted in Figure 2. Saliva DNA from each subject was genotyped using the HumanOmni2.5Exome array (approximately markers).1 Three CNV calling algorithms were used to call CNVs across autosomes: PennCNV18 (version 1.0.1), DNAcopy19 (version 1.46.0), and VanillaICE20 (version 1.32.2). CNVs were defined as segments of loss or gain >1000 base pair (bp). The scripts used to run the CNV calling algorithms are available (data and code availability). CNV calling results were filtered as described previously.41 In PennCNV, CNV calling from genotype data using high-density SNP arrays can result in artificial splitting of larger CNVs (approximately >500 kilobase [kb]) into multiple smaller CNVs.18 Thus, adjacent CNVs of the same type (i.e., both loss or both gain) and called in at least two algorithms were merged using the approach of Wang et al.18 Briefly, for three adjacent genomic regions A, B, and C, where A and C represent two CNVs of the same type separated by region B, CNVs A and C were merged if .
Figure 2.
Analysis flowchart. Flow chart describing the analysis process. ∗CNVs called in at least two algorithms with ≥10% overlap with CNVs called in all three algorithms; ∗∗10-Kb tiling windows with a 3-Kb overlap; ∗∗∗ 15 gene sets from FaceBase phenotype groups and three gene sets from SNP GWAS of normal facial variation.
After variant calling, we performed subject and CNV quality control (QC) to identify and remove subjects and CNV calls with low confidence. We removed 70 duplicate subjects and 97 subjects with a total CNV count greater than three standard deviations above the cohort mean. CNV QC was then performed to identify and remove low-quality or low-confidence CNV calls. We removed CNVs with fewer than five array probes, any centromere or telomere overlap, more than 50% overlap with a segmental duplication, a PennCNV score ≤10, or a DNAcopy log ratio outside the range (−0.1, 0.1). Restricting to the set of CNVs called in all three algorithms, one additional subject with a total CNV count greater than three standard deviations above the cohort mean was excluded. A high gain-to-loss ratio is indicative of poor quality of CNVs. No subjects had a gain to loss ratio greater than the predefined ratio of four, resulting in no subjects being removed. One subject with no CNVs remaining after CNV QC was removed from the sample. Thus, 3,462 samples passed CNV QC. An additional 74 subjects were removed for not passing phenotype QC, performed by Cole et al.1 The final sample size was 3,388 subjects. The number of subjects with non-missing values for each phenotype is reported in Table S2.
Definition of CNV windows, regions, and common variants
For association analyses, we used internally derived CNVs that were called in at least two algorithms and had at least 10% overlap with a CNV called in all three algorithms (primary; n = 216,430 CNV alleles) or were called in all three algorithms (secondary; n = 71,397 CNV alleles). The intersect command in BEDTools21 was used to identify CNVs that had at least 10% overlap.
CNV tiling windows were defined across the genome as 10-kb regions with a 3-kb overlap. A CNV was included in a window if at least 1 bp of the CNV overlapped the window. Windows with a minimum of ten subjects with a CNV were taken forward for analysis (n = 11,605 primary analysis windows, n = 6,705 secondary analysis windows). Regions were defined as sets of overlapping analysis windows. This resulted in 1,948 primary analysis regions and 988 secondary analysis regions. Common CNVs were defined as a subset of primary analysis CNVs that had the same start and end location and were seen in at least 5% of the sample (170 subjects).
We define CNV sites as called CNVs with the same start and end location, and CNV alleles as the number of observed CNVs in our cohort at each CNV site. A CNV site seen in multiple subjects will have more than one CNV allele, whereas a CNV site only observed in a single subject will have one CNV allele.
Genome-wide association
Subjects were assigned values for each CNV window using two classification functions: (1) absent/present: each subject was assigned a value of 0 or 1 for the absence or presence of a CNV, respectively, and (2) directional: each subject was assigned a value of 0 for a loss, 1 for no CNV, or 2 for a gain. Subjects with both a loss and gain in the same window were coded as 1 (i.e., no CNV) in the directional model and as 1 in the absent/present model. Eighty-two primary analysis windows had at least one subject with both a loss and a gain in non-overlapping portions of the window. Windows with at least ten subjects with phenotype information and a CNV overlapping the window were taken forward for association analysis. Common CNVs were analyzed individually and within the window analysis. For common CNVs, each subject received a 0 or 1 for the absence or presence of the CNV, respectively, in the absent/present model. In the directional model, each subject was assigned a 0, 1, or 2 for a loss, no CNV, or gain for each common CNV of interest.
For all association models, we implemented linear mixed effects regression using Efficient Mixed-Model Association eXpedited22 (EMMAX) via the EPACTS toolbox with default parameters. The kinship matrix was estimated in EMMAX separately for each chromosome using SNPs from all other chromosomes1 (i.e., a leave-one-out strategy) and was used to adjust for relatedness. Additionally, we adjusted for sex, age, and centroid size as described previously.1 Quantile-quantile (QQ) plots were created to assess whether there was bias due to unaccounted for population structure or other confounders. Due to high correlation between overlapping analysis windows within a region, a random window from each region was selected for the QQ plots. To ensure that the top five signals were not driven by small CNVs, which are more likely to be false positives, we performed a sensitivity analysis by restricting CNVs within the region to those >10 kb. Windows with at least 10 subjects with a CNV >10 kb in the window were evaluated.
Gene annotation was performed using GENCODE,23 release 31 (GRCH37). Gene annotation in figures was created using the UCSC Genome Browser.24
Association of CNVs in regions previously identified by SNP-based GWAS
We examined the associated and replicated genetic loci reported in three SNP-based GWASs of normal facial variation: (1) the same sample of Bantu African children studied here from Cole et al.,1 (2) a sample of children and adults with European ancestries from Claes et al.,2 and (3) four cohorts of children and adults with European ancestries from the United States (three cohorts) and United Kingdom (one cohort) examined within a meta-analysis framework by White et al.3 (Table S3). From Cole et al.,1 we investigated the 11 associated and replicated gene regions, as reported by Cole et al. in Table 2 of their manuscript. From Claes et al.,2 we investigated 14 loci (containing 25 genes) that were associated and replicated, as reported by Claes et al. in Table 12 of their manuscript. From White et al.,3 we investigated the 120 loci with consistent genetic effects between the US and UK meta-analyses and passed study-wise significance threshold, as reported in Table S3 of their manuscript. Additional details about the studies, including how replication was defined, are reported in each manuscript.1, 2, 3 We examined windows directly overlapping and within a 50-kb flanking region around the reported genetic region. Here, we report nominally significant CNV windows (i.e., p < 0.05). If the CNV window and SNP were both nominally associated with the same phenotype in our Bantu sample, we performed conditional analysis by including the reported GWAS SNP as a covariate in the CNV window analysis model.
Gene set analysis
We completed gene set analysis using two primary sources: (1) three gene sets from normal facial variation SNP-based GWAS loci reported in Bantu1 and two European2,3 ancestry groups (Table S3) and (2) 15 gene sets, each corresponding to a phenotype category, from the FaceBase consortium25 (Table S4). The FaceBase gene sets were generated by manual curation of genes associated with abnormal craniofacial phenotypes observed in human subjects. Gene regions from normal facial variation SNP-based GWASs were assessed for each of the three studies. The 11 genes from the Bantu study1 were analyzed as a gene set, as were the 26 genes from the smaller European study.2 The 108 genes from the European meta-analysis gene set3 were divided into seven gene sets. The reported genes were annotated with the region of the face with the strongest association in White et al.3 The regions defining the seven gene sets, seen in Figure 1 of White et al.,3 are as follows: (1) the full face (n = 11), which separates into (2) Segment 2, the midface (n = 14), and (3) Segment 3, the rest of the face (n = 3). Segment 2 was further divided into (4) Quadrant 1, the region of the mouth (n = 14), and (5) Quadrant 2, the region of the nose (n = 29). Likewise, Segment 3 was further divided into (6) Quadrant 3, the lower facial area (n = 21), and (7) Quadrant 4, the upper facial area (n = 27). Eleven of the 108 genes were associated with multiple phenotypic regions and thus in multiple gene sets.
Gene set analysis was completed for both the primary (called in at least two algorithms) and secondary (called in all three algorithms) CNV calling sets using CNVs overlapping a gene (i.e., at least 1 bp) and for CNVs overlapping a ±50-kb flanking region of a gene set. This resulted in four analyses within each of the gene sets described above. Each subject was coded as 1 for at least one CNV in the gene set and 0 for no CNVs in the gene set. Gene sets with at least ten CNVs were analyzed. Association analyses were performed using EMMAX22 as described for the window analysis.
Linkage disequilibrium between SNPs and CNVs
We estimated pairwise r2 between SNPs and common CNVs as well as between SNPs and CNV windows. CNV windows with CNVs called in all three algorithms and the absent/present coding were used. As reported and described in Cole et al.,1 SNPs were imputed using SHAPEIT2 and IMPUTE2 software to 1000 Genomes Project Phase 1 data. We estimated pairwise r2 using the method of Mangin et al.26 that adjusts for relatedness. We calculated r2 between the CNVs called here (i.e., common CNVs and CNV windows that include common and rare CNVs) and each SNP within ±1 mb of the common CNV or window. The SNP with the largest r2 value was reported as the tag SNP for the common CNV, window, or region. A CNV window or common CNV was considered well tagged if the tag SNP had r2 ≥ 0.8.
To evaluate the differences in LD between CNV windows that had more loss versus gain CNVs, we ran a linear mixed effects model using a dummy variable for more gains in the window and the region (as a random effect) to predict r2. Only CNV analysis windows with unequal numbers of loss and gain CNVs were included (n = 6,621).
Significance threshold
For each analysis, we applied a Bonferroni correction for the effective number of independent tests with a family-wise error rate (FWER) significance level of p < 0.05. The Bonferroni correction can be overly conservative, particularly given our correlation in both phenotypes and windows. Using the effective number of tests corrects for multiple testing without being overly conservative. The effective numbers of independent tests and phenotypes were estimated using the method of Gao et al.,27 estimated separately for each chromosome to ensure more subjects than windows. We calculated 23 effectively independent phenotypes, and the effective number of independent tests for each analysis is shown in Table S5. For the primary window analysis, 6,913 effectively independent tests were estimated resulting in an FWER significance threshold .
Results
CNV calling and quality control
We observed 57,142 CNV sites called within our sample with a total of 216,430 CNV alleles across all autosomes (subjects and methods). Of the CNV alleles, 179,729 were losses and 36,701 were gains. About one-third of CNV sites were observed in more than one subject (n = 19,355; 33.9%) and two-thirds were limited to one subject (n = 37,787; 66.1%). The 19,355 CNV sites found in multiple subjects produced the majority of CNV alleles (n = 178,643; 82.5%). The vast majority of the 57,142 CNV sites were rare (MAF ≤5%, n = 56,990, 99.73%) with only 152 common CNV sites (MAF >5%, 0.27%). The subset of CNVs called by all three algorithms used in the secondary analysis included 14,181 CNV sites and a total of 71,397 CNV alleles. A complete list of CNV sites and allele counts is in Table S6. Many of the CNVs are near each other, sometimes sharing either the start and/or end with another CNV site (e.g., Figure 3C). The variability in start and end sites may reflect lack of precision in the CNV calls or true biological sample to sample variability.
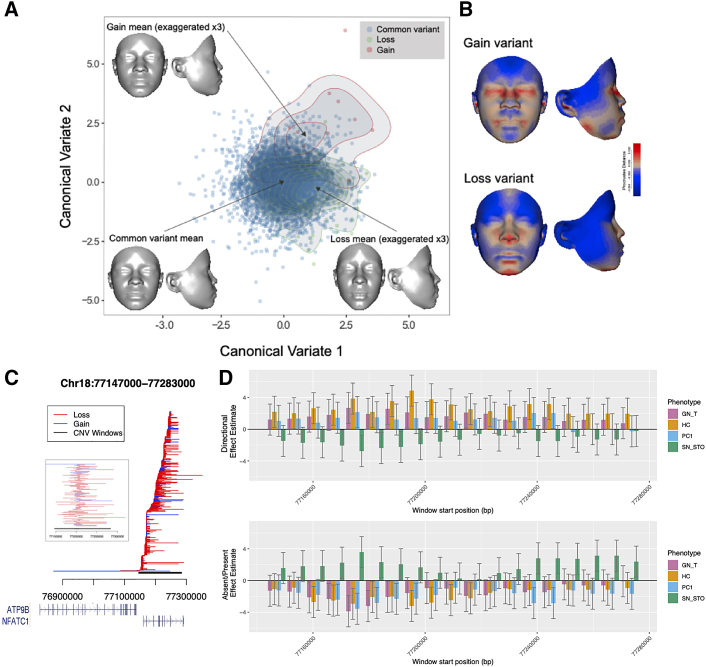
Figure 3.
Region plot, chromosome 18. (A) A canonical variates plot for facial shape variation by CNV variant. The 3D morphs show the mean face for the common variant (no CNV) as well as exaggerated shape contrasts (×3) for the loss and gain variants. (B) Heatmaps for the shape contrasts between the common variant and each of the CNV variants. (C) Loss (red) and gain (blue) CNVs. Each line represents a unique CNV allele from one subject with the genes in the region shown below. A zoom plot of the subset of CNVs overlapping the window with the lowest p value is also shown. The CNV analysis region is shown in black. (D) Test statistic t values (effect estimate/standard error of effect estimate) across the region with 95% confidence intervals in the directional model (top) and absent/present model (bottom). Phenotypes with at least one window with p value < 5 × 10−4 are shown: lower facial depth (GN_T), head circumference (HC), principal component 1: upper facial height and mid facial width (PC1), and upper lip height (SN_STO).
For the 3,388 subjects that passed QC, the mean and median number of CNVs called per person was 63.88 (SE = 0.79) and 50, respectively, with a minimum of 18 and a maximum of 432. For subjects with at least one gain, the mean gain-to-loss ratio was 0.259. All samples had at least one loss; six subjects did not have a gain. The maximum CNV length detected was 2,741 kb, with a mean length of 21.6 kb (SE = 97.9 bp). Complete CNV summary statistics are in Table S7.
CNV analysis windows and regions
Like rare single nucleotide variants (SNVs), rare CNVs cannot be assessed for association individually due to having too few rare alleles. Thus, we use 10-kb tiling windows with a 3-kb overlap to capture CNVs within a genetic region. The window analysis did not contain all CNVS; 11,592 CNV alleles (5.4%) across 7,761 sites (13.6%) were not included in an analysis window due to the minimum requirement of ten CNVs per window. Within the secondary analysis, 6,710 CNV alleles (9.4%) across 4,508 CNV sites (31.8%) were not included in an analysis window.
Of the 11,605 primary analysis windows, 3,525 (30.4%) contained only losses, 1,472 (12.7%) contained only gains, and 6,608 (56.9%) contained both gains and losses. Interestingly, the distribution of the secondary analysis windows was considerably different with an increased proportion of only losses (n = 2,735, 40.8%) and only gains (n = 1,703, 25.4%) and fewer analysis windows containing both gains and losses (n = 2,267, 33.8%). These results show that many of the opposite CNV type (i.e., gains and losses) called near one another are not likely to be called with all three CNV calling algorithms. This may be due to differences in the CNV algorithms' ability to call both losses and gains in the same region or due to biological differences in the likelihood of gains and losses being in the same region.
Genome-wide association
In the window association analysis between the primary CNV windows and the 34 facial measurements, no windows passed the multiple testing corrected threshold of p = 3.14 × 10−7. We report the top five regions, which had p values ≤ 9.35 × 10−6 (Table 1). The QQ plots show well-controlled test statistic distributions for each phenotype (Figures S1, S2, S3, and S4). The median λ values over all phenotypes were 1.002 and 1.028 for the absent/present and directional models, respectively. The most significant CNV association is from a 136-kb CNV region on chromosome 18 (Figure 3). This region contains 19 CNV analysis windows and 466 overlapping CNVs consisting of 412 losses and 54 gains. All but one CNV are between 1 kb and 376 kb in size, with 222 CNVs <10 kb (Figure 3C). However, the window that is most associated within the region contains 73 losses and 12 gains, with 6 loss CNVs <10 kb, as seen within the zoom plot of Figure 3C. Notably, the p value only slightly increased from p = 1.16 × 10−6 to p = 2.41 × 10−6 when restricting to CNVs >10 kb (Table S8). NFATC1 (MIM: 600489) is the only gene to overlap this CNV region, although ATP9B (MIM: 614446) is just outside the CNV region and overlaps two CNVs, including one long CNV (>376 kb). The association appears to be driven by losses, with a loss in the region associated with smaller average lower facial depth (Figures 3A and 3B). The four remaining regions are summarized in Figures S5–S8 and Table S8. Within these four regions, two of the driving windows (i.e., window with the lowest p value) contained no small CNVs (i.e., all CNVs >10 kb), and another region had only one CNV <10 kb in the top window. Only the region on chromosome 10 appeared to be driven by CNVs <10 kb, as ten of the 13 CNVs were <10 kb. Genome-wide results with p values < 0.1 are in Table S9 with complete results available online (data and code availability).
Table 1.
Top CNV regions associated with facial phenotypes
| Region (hg19) | Win (n) | Associated phenotypeb | Minimum p valuea |
Loss (n); gain (n)a | Overlapping genes | r2c | |
|---|---|---|---|---|---|---|---|
| Absent/present | Directional | ||||||
| Chr18: 77,147,000– 77283000 | 19 | head circumference | 1.31 × 10−3 | 1.16 × 10−6 | 73; 12 | NFATC1 | 0.035 |
| lower facial depth (average) | 1.03 × 10−4 | 7.03 × 10−3 | 55; 7 | ||||
| upper lip height | 3.47 × 10−4 | 5.80 × 10−3 | 55; 7 | ||||
| PC1 | 3.71 × 10−4 | 3.05 × 10−2 | 55; 7 | ||||
| Chr10: 111,034,000–111058000 | 3 | upper facial depth (average) | 2.64 × 10−4 | 2.64 × 10−6 | 13; 0 | 0.040 | |
| Chr4: 3,423,000–3538000 | 16 | upper facial depth (average) | 5.20 × 10−6 | 1.51 × 10−1 | 41; 7 | DOK7, LRPAP1, HGFAC, RGS12 | 0.028 |
| midfacial depth (average) | 4.79 × 10−5 | 2.21 × 10−1 | 41; 7 | ||||
| Chr2: 34,230,000–34324000 | 13 | subnasal width | 2.47 × 10−5 | 5.23 × 10−6 | 19; 1 | LINC01317, LINC01318 | 0.063 |
| nasal width | 7.82 × 10−5 | 5.71 × 10−5 | 19; 1 | ||||
| midface PC1 | 6.63 × 10−4 | 4.65 × 10−4 | 19; 1 | ||||
| Chr16: 1,225,000–1508000 | 40 | nasal ala length (average) | 9.35 × 10−6 | 6.26 × 10−5 | 1; 9 | TPSAB1, TPSD1, TPSB2, TPSG1, CACNA1H, UBE2I, BAIAP3, GNPTG, TSR3, UNKL, C16orf91, CCDC154, CLCN7 | 0.100 |
| subnasal width | 5.12 × 10−5 | 1.10 × 10−4 | 1; 9 | ||||
| nasal width | 5.60 × 10−5 | 1.05 × 10−4 | 1; 9 | ||||
| midfacial depth (average) | 2.85 × 10−4 | 3.32 × 10−2 | 12; 7 | ||||
Reported for the window with lowest p value in the region. Details from each window in the region, as well as genes within a 10-kb flanking region are in Table S8.
Associated phenotypes with minimum region p value < 5 × 10−4 in at least one model are reported.
Maximum pairwise r2 between SNP and CNV window in the region is reported.
For the secondary analysis, one window (chr5: 46,130,000–46,140,000, ppresent/absent = 9.70 × 10−7) within an intergenic region passed the multiple testing correction threshold (p < 1.43 × 10−6). This region contains 66 CNV analysis windows and 588 overlapping CNV alleles across 117 CNV sites consisting of 537 losses and 51 gains. There are no genes within 250 kb of the CNV region, and the region is relatively close to a centromere (within 73 kb). Additionally, two of the top five regions in the primary analysis (chromosome 10 and chromosome 2) are also in the top five signals for the secondary analysis. Complete results for the secondary window model are in Table S10.
No common CNVs passed the multiple testing correction of p < 1.31 × 10−5. The most significant association was between PC1 and the common CNV chr5: 17,466,056–17,469,290 (pboth models = 2.04 × 10−4). None of the five most significant regions identified in the primary window analysis contained common CNVs. Complete results for the common CNV analysis are in Table S11.
Association in regions previously identified in SNP-based GWASs
We examined the top reported results from three SNP-based GWASs of normal facial variation for association with CNVs. Of the 145 genes reported by Cole et al.1, Claes et al.,2 and White et al.,3 five gene regions—DPP6 (MIM: 126141), the homeobox D cluster (HOXD@), FGFRL1 (MIM: 605830), ELP1 (MIM: 603722), and SHBG (MIM: 182205)—directly overlapped primary analysis windows with nominal association (p < 0.05). However, ELP1 and SHBG came from phenotypic gene sets that did not contain the phenotype the CNV window was associated with. Table 2 summarizes facial phenotype associations for these three SNP-based GWAS gene regions.
Table 2.
CNV associations in gene regions previously identified by SNP GWAS
| SNP GWAS | Gene | Association | Phenotype | p valuea | p value (SNP study) | With SNPb | |
|---|---|---|---|---|---|---|---|
| Cole et al.1,4 | DPP6 | reported SNP | rs114189713 | mouth width | 7.26 × 10−8 | 1.87 × 10−7 | 0.067 |
| CNV region | 7: 153,860,000–153,870,000 | lower lip height | 2.93 × 10−3 | ||||
| lower facial depth (average) | 1.78 × 10−2 | ||||||
| Claes et al.2 | HOXD@ | Reported SNP | rs970797 | nose width; mouth and philtrum | 5.47 × 10−1 | 6.17 × 10−11 | 0.254 |
| CNV region | 2: 176,918,000–176,977,000 | head circumference | 1.93 × 10−3 | ||||
| lower facial height | 4.64 × 10−3 | ||||||
| nasal width | 7.58 × 10−3 | ||||||
| inner canthal width | 9.02 × 10−3 | ||||||
| PC5 | 2.45 × 10−2 | ||||||
| White et al.3c | FGFRL1 | Reported SNP | rs74921869 | quadrant 2: region of the nose | 8.95 × 10−1 | 3.51 × 10−11 | 0.0002 |
| CNV region | 4: 931,000–1,060,000 | centroid size | 2.20 × 10−2 | ||||
| PC5 | 3.87 × 10−2 | ||||||
| philtrum length | 2.44 × 10−2 | ||||||
| nasal width | 3.89 × 10−2 |
p value calculated with our sample. Due to CNV QC, our sample is slightly different than that of Cole et al.
Maximum SNP-window pairwise r2 within the region for the reported SNP.
Analysis windows with p values < 0.05 reported. Other two were reported for <2.5 × 10−2.
In the SNP-based GWAS for facial variation in the same sample of Bantu Africans that we use here, Cole et al.1 found that DPP6 was associated with mouth width (rs114189713; Cole et al. replication meta-analysis p = 8.35 × 10−8). In our CNV GWAS, we found the top associated CNV window (chr7: 153,860,000–153,870,000) to be most strongly associated with lower lip height (pboth models = 2.93 × 10−3; 16 losses and 0 gains) (Figure S9). The SNP was not associated with lower lip height in our sample of 3,388 subjects (p = 0.769), nor did we observe a significant association between a CNV window in the DPP6 region and mouth width (minimum p = 0.212). There was low LD between the CNV region and rs114189713 (maximum r2 = 0.067), indicating that the SNP and CNV associations are likely to be independent.
We observed nominally significant associations for CNV windows overlapping the HOXD@ on chromosome 2, previously associated with nose width, and mouth and philtrum by Claes et al.2 (reported from Claes et al. rs970797; discovery p value = 6.17 × 10−11) (Figure S10). For our CNV analysis, the HOXD@ had strongest association with head circumference (pabsent/present = 1.90 × 10−3), lower facial height (pabsent/present = 4.64 × 10−3), and nasal width (pabsent/present = 7.58 × 10−3). In our sample, we observed low LD between the HOXD@ CNV region and the reported SNP from Claes et al. (rs970797, maximum r2 = 0.254), and the SNP identified in Claes et al.2 (rs970797) was not associated with head circumference (p = 0.252), lower facial height (p = 0.875), or nasal width (p = 0.547). The lack of replication for rs970797 may be due to true lack of association or differences in LD between Bantu and European ancestries.
In the SNP-based meta-analysis for normal facial variation White et al.3 found that FGFRL1 was associated with the region of the nose (p = 3.51 × 10−11; US meta-analysis). Our CNV window analysis had nominal associations with PC5 (nose shape, height of mouth, p = 0.039, directional model), and nasal width (p = 0.039, directional model). Within the analysis window most associated with the phenotype, there were 137 losses and 66 gains, and 158 losses and 87 gains for the PC5 and nasal width analysis windows, respectively (Figure S11). Within our cohort, the lead SNP, rs74921869, was not associated with PC5 (p = 0.895) or nasal width (p = 0.303). There was very low LD (max r2 = 0.000177) between the lead SNP and the CNV windows within the region of interest. Unsurprisingly, the conditional analysis incorporating rs74921869 resulted in little change in the CNV window association with FGFRL1 (PC5: minimum punconfitioned = 3.89 × 10−2; minimum ; nasal width: minimum punconfitioned = 3.89 × 10−2; minimum pconfitioned = 3.62 × 10−2). This likely indicates that either the SNP and CNV represent independent signals within the FGFRL1 region, that both are in LD with an as-yet unidentified causal locus, or that there are differences within the two samples, as the SNP was not associated with the nose phenotypes of interest within our Bantu cohort.
Comparison of absent/present and directional CNV classification
By design, CNV windows with all losses or all gains produced identical results. Restricting to windows with both losses and gains, we observed moderate correlation of 0.387 between the for the absent/present and directional models (Figure S12). Using various significance thresholds (1.0 × 10−2, 1.0 × 10−4, 1.0 × 10−3, 1.0 × 10−2, and 5.0 × 10−2), 36.1%–57.1% of the windows with both losses and gains are captured by only one classification. This indicates that both models were likely necessary to capture the relationship between CNVs and normal face shape. The absent/present model had slightly more windows that passed the nominal significance threshold (p < 0.05) than the directional model (n = 9,503 versus n = 7,494, respectively) (Table S12). Out of 77 windows with p values < 1.0 × 10−4, 5, 31, and 41 windows were observed in both, only absent/present, and only directional, respectively.
Gene set analysis
Three normal facial variation gene sets were examined from Cole et al.,1 Claes et al.,2 and White et al.3 with 11, 26, and 108 genes, respectively. The White et al.3 gene set was further divided into seven gene sets based on facial region (methods). Within the Bantu sample, 25 CNV alleles from 25 subjects overlapped 2 genes: DPP6 and EXOC6B (MIM: 607880). When including a ±50-kb flanking region around each gene, 38 CNV alleles from 38 subjects overlapped 5 genes ±50 kb: DPP6, EXOC6B, PDE8A (MIM: 602972), WNK2 (MIM: 606249), and GABRG3 (MIM: 600233). Within the Claes et al.2 gene set, 208 CNV alleles from 207 subjects overlapped one gene region: HOXD@. When including a ±50-kb flanking region around each gene, 332 CNV alleles from 305 subjects overlapped five gene regions: HOXD@ (328 subjects overlapping), ASPM (MIM: 605481), DYNC1I1 (MIM: 603772), RAB7A (MIM: 602298), and RPS12 (MIM: 603660). Notably, 10 gene regions identified in these two SNP-based GWAS did not have an overlapping CNV in our sample even when including the additional 50-kb flanking region, and eight gene regions had fewer than ten individuals with CNVs in the region. This suggests that association signals found in the CNV GWAS may differ from those found in the SNP GWAS simply due lack of detected CNV variation in most of the genes identified from these two GWASs of facial variation.
Within the meta-analysis gene sets from White et al.,3 332 CNV alleles from 292 subjects overlapped 20 genes. When including a ±50-kb flanking region around each gene, 1,745 CNV alleles from 896 subjects overlapped 32 genes ±50 kb.
No gene set association passed multiple testing correction of 1.34 × 10−4. However, each gene set derived from SNP GWAS of normal facial variation had at least one nominally significant association (subjects and methods, Table S13). The SNP GWAS gene set from the meta-analysis, quadrant 3 (lower facial area) had the strongest association with philtrum width (p = 2.42 × 10−4), which is in quadrant 1, not quadrant 3. Complete results can be found in Table S13.
The top two associations for the analysis of FaceBase Consortium25 gene sets derived from genes associated with facial abnormalities were between the phenotype upper facial height and FaceBase gene sets for (1) abnormality of the jaws (p = 3.26 × 10−4) and (2) micrognathia (p = 6.02 × 10−4), both with CNVs directly overlapping the gene set (Table S14). The third most significant association had phenotype and gene sets that were related to the nose. The association was between nasal ala length and abnormality of the nose (p = 7.36 × 10−4) with CNVs within the 50-kb flanking region of the gene set. Interestingly, five of the ten most significant results are with the upper facial height phenotype. Although more research in larger sample sizes is needed, these results suggest that regions previously implicated in facial abnormalities may play a role in normal facial variation as well.
Linkage disequilibrium between SNPs and CNVs
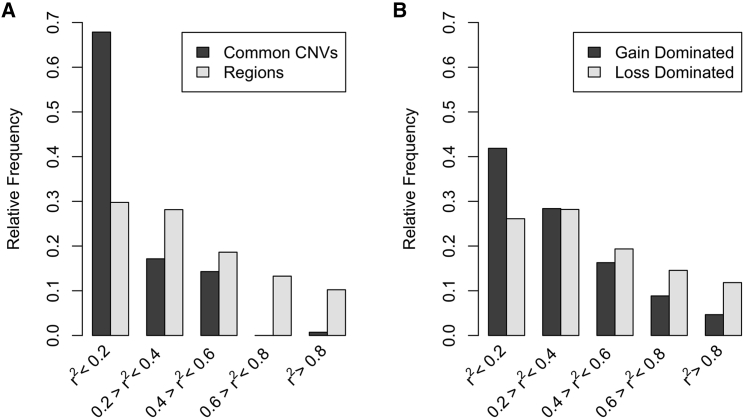
To assess whether GWAS arrays with dense SNP imputation can adequately capture CNVs through LD and thus eliminate the need to call CNVs, we calculated pairwise r2 between SNPs within 1 mb of CNV windows or common CNVs (Tables S15 and S16). Only one common CNV was tagged well (maximum r2 = 0.809). Approximately 36.8% of common CNVs were tagged moderately (n = 56; 0.2 ≤ maximum r2 < 0.8), while the remaining 62.5% were tagged poorly (n = 95, maximum r2 < 0.2) (Figure 4A). Only 10.2% of the 988 CNV window regions were tagged well (n = 101; r2 > 0.8), while 29.8% were tagged poorly by SNPs (n = 296; maximum r2 < 0.2). The low LD supports the need to call and analyze CNVs, as CNV associations are not likely to be captured in SNP GWASs, especially in African ancestry as presented here. After adjusting for region, windows with more gains have lower average maximum pairwise r2 (mean r2 = 0.3147) compared with windows with more losses (mean r2 = 0.3883, p < 2.0 × 10−16) (Figure 4B).
Figure 4.
LD between SNPs and CNVs. (A) Bar chart showing frequency of common CNVs and CNV regions by strength of maximum pairwise r2 with SNPs within 1 Mb of the region. (B) Windows with more than 50% losses (light gray, n = 3,878), windows with more than 50% gains (dark gray, n = 2,743). Eighty-four windows that had an equal number of loss and gains had r2 < 0.2 and are not included.
Discussion
We investigated the role of CNVs in normal facial variation in a sample of Bantu African children. We found that CNVs, both common and rare, are poorly tagged by SNPs, indicating that association signals representing CNVs were likely not captured by previous SNP-based GWASs. We completed a CNV GWAS, identifying five genetic regions with evidence of association with various normal facial phenotypes. The strongest associations were for head circumference, average upper facial depth, subnasal width, and nasal ala length. CNV association windows were poorly tagged by SNPs in all five putative regions (maximum pairwise r2 = 0.100). Four of these five regions were driven by CNVs >10 kb, while the association between face shape and the CNVs in the region on chromosome 10 was driven by relatively small CNVs <10 kb.
The region showing the strongest association (pdirectional = 1.16 × 10−6) is on chromosome 18 and contains the gene NFATC1, a transcription factor that regulates genes in the Wnt signaling pathway, which is known to play an important role in facial morphogenesis.28 NFATC1 was recently identified in a GWAS of facial asymmetry,29 regulates bone mass in mouse models,30 and has been linked to facial morphogenesis28,30,31 and Cherubism,32 a genetic disorder with abnormal bone tissue in the lower face. This is particularly interesting given one of the top associated facial phenotypes is average lower facial depth. We identified 55 losses and seven gains in NFATC1.
The associated region on chromosome 4 (pabsent/present = 5.20 × 10−6) contains four genes, including DOK7, which has been implicated in a disorder characterized by craniofacial abnormalities.33 This region is nested within the microdeletion in 4p16.3 associated with Wolf-Hirschhorn syndrome (MIM: 194190), which includes characteristic dysmorphic facial features.34 Similarly, the associated region on chromosome 16 (pabsent/present = 9.35 × 10−6) is nested within the region associated with the 16p13.3 deletion (MIM: 610543) and duplication (MIM: 613458) syndromes, both of which include dysmorphic facial features.35 Interestingly, the association observed in our study is predominantly due to gain CNVs, which reflect interstitial duplications in 16p13.3.
The other two novel associations, on chromosomes 2 and 10 (Table 1), are localized within regions that are gene-poor and do not have any obvious candidate genes that appear to have roles in facial development. However, the region on chromosome 2 contains two long intervening non-coding RNAs (lincRNAs). Although the downstream targets of these particular lincRNAs are not known, some lincRNAs are known to play a role in gene regulation and other cellular processes.36
Two of 25 gene regions from the two smaller SNP-based GWAS (nCole = 11; nClass = 14) contained nominally significant CNV analysis windows overlapping the gene region (DPP6 and HOXD@, which contains multiple genes). While the gene set analysis supported nominal association between GWAS genes and facial phenotype, this analysis was almost entirely driven by DPP6 and the HOXD@. Three of 108 meta-analysis gene regions contained nominally significant CNV analysis windows overlapping the gene region; one of those three gene regions (FGFRL1) had CNV windows associated with a phenotype similar to the SNP meta-analysis. Using more precise phenotypes within the gene set analysis may eliminate noise to allow for detection of a significant association.
The majority of common CNVs and CNV windows are poorly tagged by SNPs, as measured by pairwise r2. A central assumption is that our CNV calls are true CNVs. An abundance of false CNV calls would also likely result in low LD with SNPs. While we were unable to molecularly verify the CNV calls, the CNVs presented here are optimized for true positive calls based on filtering criteria applied.41 Additionally, SNP haplotypes may result in higher pairwise r2 with CNVs37 relative to the genotype data used here. Thus, the lack of well-tagged CNV windows and common CNVs suggests that SNP-based GWASs are insufficient to capture CNV contributions (and likely other more complex SVs as well), particularly for lower frequency CNVs and in samples of African ancestry. SNP GWASs capturing CNVs are perhaps less problematic in samples of European ancestry where LD is comparatively stronger17 or in studies that include haplotype data.
It is possible that SVs, including CNVs, could be imputed from reference samples containing both structural and SNV data. Given the additional complexity of functional variants with regards to alleles and variant size, reference samples will likely need to be much larger than those needed to impute SNVs, especially for low-frequency CNVs and SVs. Imputation reference panels from large and more diverse whole-genome sequencing studies, such as the Trans-Omics for Precision Medicine consortium,38 have the potential to include structural variation to enable investigation of SVs. Until these reference panels are available and imputation of SVs is assessed, continued study of SVs through direct assay and calling will be necessary.
It is important to consider the ethics surrounding research that aims to understand the genetics behind normal facial variation. While the genetics of face shape have the potential to be used in questionable applications,39 excluding normal facial variation—a highly heritable trait—completely from genetics research would limit the understanding of common and syndromic facial variation. Importantly, we do not predict facial features here, nor do we recommend that these data be used to predict facial features for both ethical reasons as well as poor accuracy to predict facial features for subjects especially for understudied ancestral populations.40
Here, we show that CNVs contribute to the complex phenotype of common facial variation for Bantu African children using CNVs called from GWAS array intensity data. Given the low LD with densely imputed SNPs for the CNV associations identified here, calling CNVs from GWAS array data may identify associations not detectable with solely SNP data. Thus, the large resource of existing SNP GWASs may provide a good resource for calling and assessing the role of CNVs in other complex traits. There is a dearth of studies of CNVs and in people of African ancestry and culture. Here, we add to the understanding of the genetic etiology of common facial variation for both.
Data and code availability
The CNV data presented in this article was previously deposited in the FaceBase Consortium Database (FaceBase: https://doi.org/10.25550/1-7330). The genotype data used for CNV detection were previously deposited in the Database of Genotypes and Phenotypes (dbGaP: http://www.ncbi.nlm.nih.gov/gap; dbGaP study accession: phs000622.v1.p1). Scripts that were used to run the CNV calling algorithms are available at https://github.com/dpastling/facebase_cnv. Complete results from the primary analysis are available at https://github.com/meganmichelle/CNV_FaceShape.
Acknowledgments
This work was supported in part by a grant (DE025363) to T.H.S. by the National Institute of Dental and Craniofacial Research of the National Institutes of Health.
Declaration of interests
F.Y. is now a postdoctoral associate at The Jackson Laboratory. H.Y. is now an employee at Bionano Genomics. J.B.C. has current affiliations with the Programs in Metabolism and Medical & Population Genetics at Broad Institute of Harvard and MIT, the Center for Genomic Medicine at Massachusetts General Hospital, and the Division of Endocrinology and Center for Basic and Translational Obesity Research at Boston Children's Hospital. A.E.H. is on the editorial board for HGG Advances. All other authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xhgg.2021.100082.
Contributor Information
Tamim H. Shaikh, Email: tamim.shaikh@cuanschutz.edu.
Audrey E. Hendricks, Email: audrey.hendricks@ucdenver.edu.
Web resources
BEDTools: https://bedtools.readthedocs.io/en/latest/
DNAcopy: https://bioconductor.org/packages/release/bioc/html/DNAcopy.html.
EPACTS: https://genome.sph.umich.edu/wiki/EPACTS.
GENCODE: https://www.gencodegenes.org/
PennCNV: http://penncnv.openbioinformatics.org/en/latest/
OMIM: http://www.omim.org.
UCSC Genome Browser: https://genome.ucsc.edu/cgi-bin/hgGateway.
VanillaICE: https://www.bioconductor.org/packages/release/bioc/html/VanillaICE.html.
Supplemental information
References
- 1.Cole J.B., Manyama M., Kimwaga E., Mathayo J., Larson J.R., Liberton D.K., Lukowiak K., Ferrara T.M., Riccardi S.L., Li M., et al. Genomewide association study of African children identifies association of SCHIP1 and PDE8A with facial size and shape. PLoS Genet. 2016;12:e1006174. doi: 10.1371/journal.pgen.1006174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Claes P., Roosenboom J., White J.D., Swigut T., Sero D., Li J., Lee M.K., Zaidi A., Mattern B.C., Liebowitz C., et al. Genome-wide mapping of global-to-local genetic effects on human facial shape. Nat Genet. 2018;50:414–423. doi: 10.1038/s41588-018-0057-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.White J.D., Indencleef K., Naqvi S., Eller R.J., Hoskens H., Roosenboom J., Lee M.K., Li J., Mohammed J., Richmond S., et al. Insights into the genetic architecture of the human face. Nat Genet. 2021;53:45–53. doi: 10.1038/s41588-020-00741-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cole J.B., Manyama M., Larson J.R., Liberton D.K., Ferrara T.M., Riccardi S.L., Li M., Mio W., Klein O.D., Santorico S.A., et al. Human facial shape and size heritability and genetic correlations. Genetics. 2017;205:967–978. doi: 10.1534/genetics.116.193185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Karczewski K., Francioli L.C., Tiao G., Cummings B.B., Alföldi J., Wang Q., et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581(7809):434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zarrei M., MacDonald J.R., Merico D., Scherer S.W. A copy number variation map of the human genome. Nat Rev Genet. 2015;16:172–183. doi: 10.1038/nrg3871. [DOI] [PubMed] [Google Scholar]
- 7.Stefansson H., Rujescu D., Cichon S., Pietilainen O.P., Ingason A., Steinberg S., Fossdal R., Sigurdsson E., Sigmundsson T., Buizer-Voskamp J.E., et al. Large recurrent microdeletions associated with schizophrenia. Nature. 2008;455:232–236. doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Glessner J.T., Wang K., Cai G., Korvatska O., Kim C.E., Wood S., Zhang H., Estes A., Brune C.W., Bradfield J.P., et al. Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature. 2009;459:569–573. doi: 10.1038/nature07953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dauber A., Yu Y., Turchin M.C., Chiang C.W., Meng Y.A., Demerath E.W., Patel S.R., Rich S.S., Rotter J.I., Schreiner P.J., et al. Genome-wide association of copy-number variation reveals an association between short stature and the presence of low-frequency genomic deletions. Am J Hum Genet. 2011;89:751–759. doi: 10.1016/j.ajhg.2011.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Butts S.C. The facial phenotype of the velo-cardio-facial syndrome. Int J Pediatr Otorhinolaryngol. 2009;73:343–350. doi: 10.1016/j.ijporl.2008.10.011. [DOI] [PubMed] [Google Scholar]
- 11.Van Buggenhout G., Fryns J.P. Angelman syndrome (AS, MIM 105830) Eur J Hum Genet. 2009;17:1367–1373. doi: 10.1038/ejhg.2009.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collins R.L., Brand H., Karczewski K.J., Zhao X., Alfoldi J., Francioli L.C., Khera A.V., Lowther C., Gauthier L.D., Wang H., et al. A structural variation reference for medical and population genetics. Nature. 2020;581:444–451. doi: 10.1038/s41586-020-2287-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kessler M.D., Yerges-Armstrong L., Taub M.A., Shetty A.C., Maloney K., Jeng L.J.B., Ruczinski I., Levin A.M., Williams L.K., Beaty T.H., et al. Challenges and disparities in the application of personalized genomic medicine to populations with African ancestry. Nat Commun. 2016;7:12521. doi: 10.1038/ncomms12521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lauer S., Gresham D. An evolving view of copy number variants. Curr Genet. 2019;65:1287–1295. doi: 10.1007/s00294-019-00980-0. [DOI] [PubMed] [Google Scholar]
- 15.Armengol L., Villatoro S., Gonzalez J.R., Pantano L., Garcia-Aragones M., Rabionet R., Caceres M., Estivill X. Identification of copy number variants defining genomic differences among major human groups. PLoS One. 2009;4:e7230. doi: 10.1371/journal.pone.0007230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sudmant P.H., Rausch T., Gardner E.J., Handsaker R.E., Abyzov A., Huddleston J., Zhang Y., Ye K., Jun G., Fritz M.H., et al. An integrated map of structural variation in 2,504 human genomes. Nature. 2015;526:75–81. doi: 10.1038/nature15394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wellcome Trust Case Control Consortium. Craddock N., Hurles M.E., Cardin N., Pearson R.D., Plagnol V., Robson S., Vukcevic D., Barnes C., Conrad D.F., Giannoulatou E., et al. Genome-wide association study of CNVs in 16,000 cases of eight common diseases and 3,000 shared controls. Nature. 2010;464:713–720. doi: 10.1038/nature08979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang K., Li M., Hadley D., Liu R., Glessner J., Grant S.F., Hakonarson H., Bucan M. PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res. 2007;17:1665–1674. doi: 10.1101/gr.6861907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Venkatraman E.S., Olshen A.B. A faster circular binary segmentation algorithm for the analysis of array CGH data. Bioinformatics. 2007;23:657–663. doi: 10.1093/bioinformatics/btl646. [DOI] [PubMed] [Google Scholar]
- 20.Scharpf R.B., Parmigiani G., Pevsner J., Ruczinski I. Hidden Markov models for the assessment of chromosomal alterations using high-throughput SNP arrays. Ann Appl Stat. 2008;2:687–713. doi: 10.1214/07-AOAS155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kang H.M., Sul J.H., Service S.K., Zaitlen N.A., Kong S.Y., Freimer N.B., Sabatti C., Eskin E. Variance component model to account for sample structure in genome-wide association studies. Nat Genet. 2010;42:348–354. doi: 10.1038/ng.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frankish A., Diekhans M., Ferreira A.M., Johnson R., Jungreis I., Loveland J., Mudge J.M., Sisu C., Wright J., Armstrong J., et al. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. 2019;47:D766–D773. doi: 10.1093/nar/gky955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brinkley J.F., Fisher S., Harris M.P., Holmes G., Hooper J.E., Jabs E.W., Jones K.L., Kesselman C., Klein O.D., Maas R.L., et al. The FaceBase Consortium: a comprehensive resource for craniofacial researchers. Development. 2016;143:2677–2688. doi: 10.1242/dev.135434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mangin B., Siberchicot A., Nicolas S., Doligez A., This P., Cierco-Ayrolles C. Novel measures of linkage disequilibrium that correct the bias due to population structure and relatedness. Heredity (Edinb) 2012;108:285–291. doi: 10.1038/hdy.2011.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao X., Starmer J., Martin E.R. A multiple testing correction method for genetic association studies using correlated single nucleotide polymorphisms. Genet Epidemiol. 2008;32:361–369. doi: 10.1002/gepi.20310. [DOI] [PubMed] [Google Scholar]
- 28.Brugmann S.A., Goodnough L.H., Gregorieff A., Leucht P., ten Berge D., Fuerer C., Clevers H., Nusse R., Helms J.A. Wnt signaling mediates regional specification in the vertebrate face. Development. 2007;134:3283–3295. doi: 10.1242/dev.005132. [DOI] [PubMed] [Google Scholar]
- 29.Rolfe S., Lee S.I., Shapiro L. Associations between genetic data and quantitative assessment of normal facial asymmetry. Front Genet. 2018;9:659. doi: 10.3389/fgene.2018.00659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Winslow M.M., Pan M., Starbuck M., Gallo E.M., Deng L., Karsenty G., Crabtree G.R. Calcineurin/NFAT signaling in osteoblasts regulates bone mass. Dev Cell. 2006;10:771–782. doi: 10.1016/j.devcel.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 31.Doraczynska-Kowalik A., Nelke K.H., Pawlak W., Sasiadek M.M., Gerber H. Genetic factors involved in mandibular prognathism. J Craniofac Surg. 2017;28:e422–e431. doi: 10.1097/SCS.0000000000003627. [DOI] [PubMed] [Google Scholar]
- 32.Kadlub N., Sessiecq Q., Dainese L., Joly A., Lehalle D., Marlin S., Badoual C., Galmiche L., Majoufre-Lefebvre C., Berdal A., et al. Defining a new aggressiveness classification and using NFATc1 localization as a prognostic factor in cherubism. Hum Pathol. 2016;58:62–71. doi: 10.1016/j.humpath.2016.07.019. [DOI] [PubMed] [Google Scholar]
- 33.Vogt J., Morgan N.V., Marton T., Maxwell S., Harrison B.J., Beeson D., Maher E.R. Germline mutation in DOK7 associated with fetal akinesia deformation sequence. J Med Genet. 2009;46:338–340. doi: 10.1136/jmg.2008.065425. [DOI] [PubMed] [Google Scholar]
- 34.Battaglia A., Filippi T., Carey J.C. Update on the clinical features and natural history of Wolf-Hirschhorn (4p-) syndrome: experience with 87 patients and recommendations for routine health supervision. Am J Med Genet C Semin Med Genet. 2008;148C:246–251. doi: 10.1002/ajmg.c.30187. [DOI] [PubMed] [Google Scholar]
- 35.Thienpont B., Bena F., Breckpot J., Philip N., Menten B., Van Esch H., Scalais E., Salamone J.M., Fong C.T., Kussmann J.L., et al. Duplications of the critical Rubinstein-Taybi deletion region on chromosome 16p13.3 cause a novel recognisable syndrome. J Med Genet. 2010;47:155–161. doi: 10.1136/jmg.2009.070573. [DOI] [PubMed] [Google Scholar]
- 36.Ulitsky I., Bartel D.P. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154:26–46. doi: 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nyangiri O.A., Noyes H., Mulindwa J., Ilboudo H., Kabore J.W., Ahouty B., Koffi M., Asina O.F., Mumba D., Ofon E., et al. Copy number variation in human genomes from three major ethno-linguistic groups in Africa. BMC Genomics. 2020;21:289. doi: 10.1186/s12864-020-6669-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Taliun D., Harris D.N., Kessler M.D., Carlson J., Szpiech Z.A., Torres R., Taliun S.A.G., Corvelo A., Gogarten S.M., Kang H.M., et al. Sequencing of 53,831 diverse genomes from the NHLBI TOPMed Program. Nature. 2021;590:290–299. doi: 10.1038/s41586-021-03205-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Arnold C. The controversial company using DNA to sketch the faces of criminals. Nature. 2020;585:178–181. doi: 10.1038/d41586-020-02545-5. [DOI] [PubMed] [Google Scholar]
- 40.Sirugo G., Williams S.M., Tishkoff S.A. The missing diversity in human genetic studies. Cell. 2019;177:1080. doi: 10.1016/j.cell.2019.04.032. [DOI] [PubMed] [Google Scholar]
- 41.Yilmaz Feyza, Null Megan, Astling David, Yu Hung-Chun, Cole Joanne, Santorico Stephanie A Santorico, et al. Genome-wide copy number variations in a large cohort of bantu African children. BMC Med Genomics. 2021;14(1) doi: 10.1186/s12920-021-00978-z. https://pubmed.ncbi.nlm.nih.gov/34001112/ [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The CNV data presented in this article was previously deposited in the FaceBase Consortium Database (FaceBase: https://doi.org/10.25550/1-7330). The genotype data used for CNV detection were previously deposited in the Database of Genotypes and Phenotypes (dbGaP: http://www.ncbi.nlm.nih.gov/gap; dbGaP study accession: phs000622.v1.p1). Scripts that were used to run the CNV calling algorithms are available at https://github.com/dpastling/facebase_cnv. Complete results from the primary analysis are available at https://github.com/meganmichelle/CNV_FaceShape.