Abstract
While 9p deletion and duplication syndromes have been studied for several years, small sample sizes and minimal high-resolution data have limited a comprehensive delineation of genotypic and phenotypic characteristics. In this study, we examined genetic data from 719 individuals in the worldwide 9p Network Cohort: a cohort seven to nine times larger than any previous study of 9p. Most breakpoints occur in bands 9p22 and 9p24, accounting for 35% and 38% of all breakpoints, respectively. Bands 9p11 and 9p12 have the fewest breakpoints, with each accounting for 0.6% of all breakpoints. The most common phenotype in 9p deletion and duplication syndromes is developmental delay, and we identified eight known neurodevelopmental disorder genes in 9p22 and 9p24. Since it has been previously reported that some individuals have a secondary structural variant related to the 9p variant, we examined our cohort for these variants and found 97 events. The top secondary variant involved 9q in 14 individuals (1.9%), including ring chromosomes and inversions. We identified a gender bias with significant enrichment for females (p = 0.0006) that may arise from a sex reversal in some individuals with 9p deletions. Genes on 9p were characterized regarding function, constraint metrics, and protein-protein interactions, resulting in a prioritized set of genes for further study. Finally, we achieved precision genomics in one child with a complex 9p structural variation using modern genomic technologies, demonstrating that long-read sequencing will be integral for some cases. Our study is the largest ever on 9p-related syndromes and provides key insights into genetic factors involved in these syndromes.
Keywords: syndrome, 9p, CNV, deletion, duplication, developmental, neurodevelopmental, phenotype
Introduction
In this study, we focus on 9p deletion (MIM: 158170) (also called 9p minus) and duplication syndromes,1,2 which arise from a deletion or duplication involving the p arm of chromosome 9. There are several unresolved features of these syndromes due in part to low incidence and a lack of high-resolution genotype and phenotype data. We present the largest-ever genomic assessment of 9p minus syndrome—comprising of 719 individuals—and identify broad features of this cohort. Through reviewing databases and the literature, we summarize phenotypic features of individuals with 9p syndromes and characterize 9p genes and the proteins they encode. Finally, we present results of a study of one child with a complex 9p structural variation assessed by several modern genomic technologies including short-read whole-genome sequencing (WGS), long-read WGS, and Bionano optical mapping. We compare these methods with previous clinical tests for this individual (karyotype, array, whole-exome sequencing) and show that long-read sequencing is critical to achieving precision genomics. We define precision genomics as “determining all possible relevant genomic variation within an individual to the precise nucleotide.” This term is inspired by “precision medicine,” which is defined by President Barack Obama of the United States of America as “health care tailored to you” with a mission statement “to enable a new era of medicine through research, technology, and policies that empower patients, researchers, and providers to work together toward development of individualized care” (https://obamawhitehouse.archives.gov/precision-medicine). Ultimately, we want to reach precision genomics to strengthen precision medicine in syndromes arising from complex structural variations including 9p deletion and duplication syndromes.
A critical aspect of human genetics and genomics is linking genotype to phenotype. In some diseases, it is clear what gene is underlying the main phenotype (e.g., CFTR [MIM: 602421] in cystic fibrosis [MIM: 219700]3), while in other cases it is not clear. Large, often complex structural variants present a challenge because they can be recurrent with the same breakpoints in all or most individuals (e.g., 22q11.2 [MIM: 192430], 16p11.2 [MIM: 611913],4,5 7q11.23 Williams syndrome region [MIM: 609757])6 or they can show heterogeneity in breakpoints. Further, one gene can underly the majority of the phenotype (e.g., RAI1 [MIM: 607642] in Smith-Magenis syndrome [MIM: 182290]) or several genes can contribute to various phenotypes. 9p deletion and duplication syndromes are particularly challenging because there is heterogeneity in breakpoint locations, they typically encompass several genes, and they have variable phenotypes.
Analysis of different cohorts of individuals with 9p copy-number variants (CNVs) has established that the CNV breakpoint locations are not consistent from patient to patient. This breakpoint variability is found when comparing deletions with duplications as well as when looking at each group independently. For example, one cohort consisting of 65 individuals with 9p deletions found 50 unique breakpoints with only 11 breakpoints shared by at least two individuals.7 Studies comparing the breakpoints of 9p deletions and duplications with the presence of common 9p phenotypes in multiple cohorts have attempted to resolve the critical region for 9p deletion and duplication syndromes.7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17 These studies have suggested a deletion hotspot region within 9p22–9p23,7, 8, 9, 10, 11 although individuals with typical 9p deletion phenotypes and breakpoints outside this region have been described.13,15 When including sex reversal (MIM: 154230) in the deletion syndrome, the proposed critical region extends to 9p24.3.17 The described 9p duplication syndrome critical region occurs at 9p22.314,16; however, individuals with a 9p duplication and less severe phenotypes typically have more proximal duplications occurring between 9p12 and 9p22.1.16 Additionally, in approximately 50% of all cases of 9p minus syndrome, the affected individual also has an associated translocation event, and these translocations have not previously been preferentially linked to any specific chromosome.18 Beyond translocation events, even more complex variations including ring chromosomes2 and mosaicism have also been observed for some rearrangements and CNVs involving 9p19 as well as trisomy 9p mosaic syndrome.20,21 Understanding the exact nature of the variation is essential to identify the genes affected by the variant and to link genotype to phenotype.
The most common phenotype that is seen in nearly every individual with a 9p CNV is developmental delay and intellectual disability (ID).8,18,22,23 Additional shared phenotypes include hypotonia, low-set ears and abnormal ear auricle, high/narrow palate, short/broad neck, broad internipple distance, and the presence of a cardiac murmur or defect.8,18,22 Some phenotypes observed in individuals with 9p deletion and individuals with 9p duplication appear to mirror each other,2 and some phenotypes are variations but not quite mirrors.8,18,22 Generally, the phenotypes in individuals with 9p CNVs are quite variable8,18,22 depending on size and location of the variants.12 An important phenotype to note that often occurs in individuals with a 9p deletion is sex reversal and other differences in sex development (DSDs).8,12,13,17,24, 25, 26, 27, 28, 29, 30 Ambiguous genitalia are estimated to be present in up to 70% of individuals with 9p deletion.27 The 46,XY sex reversal phenotype is more commonly found in individuals with terminal 9p deletions than in those with more proximal deletions.17 Autism spectrum disorder (ASD) is another phenotype that has been associated with 9p deletions and duplications. All ten individuals with 9p deletion described by Hauge et al.12 were reported to display ASD or other behavioral issues, and many additional 9p case reports and cohorts include individuals with ASD.8,26,31, 32, 33 Comparison of 9p CNV individuals with and without ASD and the locations of their CNVs has led to the hypothesis that there is an ASD candidate gene on 9p24.26,31,32
Despite the general genetic and phenotypic variability seen in individuals with 9p deletions and duplications, some progress has been made in associating common 9p phenotypes with genes in the region. These candidates include DMRT1 (MIM: 602424) and DMRT3 (MIM: 614754) in the DSD phenotype;17,27,29 FREM1 (MIM: 608944) implicated for trigonocephaly;19,34 FOXD4 (MIM: 601092) for speech and language deficits;12,19,26 DOCK8 (MIM: 611432) for IDs and seizure disorders that are commonly seen in individuals with 9p CNVs;12,15,19,29 GLDC (MIM: 238300),19 VLDLR (MIM: 192977),19 and ZDHHC21 (MIM: 614605)10 for IDs and/or seizure disorders;19 and CBWD1 (MIM: 611078), which is associated with cobalamin deficiency (feeding difficulties, failure to thrive, hypotonia, seizures, microcephaly, ID, and developmental delay26). KANK1 (MIM: 607704) (previously known as ANKRD15) displays what appears to be a maternal imprinting mechanism in which inherited cerebral palsy can occur when the paternal copy of the gene is disrupted.29,31 Applying precision genomics to 9p deletion and duplication syndromes can further refine these genotype-phenotype associations and presents an opportunity to improve precision medicine in these syndromes.
Materials and methods
Assessment of 9p Network Cohort
De-identified data were accessed through the Chromosome 9p Minus Network for 811 individuals. These data consist of details of the 9p genomic variation, country of origin, and gender. Analyses of these characteristics were conducted using individuals for which the relevant data were available. The genomic variation data are on the level of broad genetic information (e.g., karyotype) and are available for 719 individuals. Bands where the breakpoints occurred for each individual were counted across the cohort. If available, large structural changes on non-9p chromosome bands were also counted in the subset of individuals. Sex chromosomes are included in the broad genetic information for 236 individuals.
9p deletions and duplications from the literature
Genomic data were collected from the literature where approximate breakpoints are known for 53 individuals with 9p deletion and duplication syndromes.12,14, 15, 16, 17,19,27,29,30,33,35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48
Phenotype data from individuals with 9p deletion and duplication syndromes
Phenotype data were collected from three papers8,18,22 assessing individuals with 9p deletions (n = 120 individuals) and one paper22 assessing individuals with 9p duplications (n = 99 individuals). The phenotypes were categorized into the following 13 regions/systems: general, head, ears, nose, mouth, neck, thorax, back, extremities, cardiovascular, respiratory, gastrointestinal, and urogenital. Categories were then further defined into 38 specific phenotypes and aggregated into the percentage of individuals with each phenotype in deletions and duplications, respectively.
9p gene constraint and dosage characteristics
For each 9p gene, the pLI score was extracted from gnomAD.49 Dosage characteristics were pulled from a previous publication assessing 29,085 individuals with neurodevelopmental disorders (NDDs) and 19,584 controls.50
Mappability on 9p and dosage of 9p in 1000 Genomes
Mappability tracks for 150-mers on build 38 of the human genome were generated to determine the ability to map short-read Illumina WGS data comprised of 150 base pair reads. The autosome and sex chromosome sequences were extracted from the GRCh38_full_analysis_set_plus_decoy_hla.fa reference file using samtools51 faidx, then the GEMtools52 (https://github.com/Chimera-tools/ChimPipe.git) index was utilized to index the genome, and finally, gem-mappability was used to perform the mappability analysis. The output file was converted from the gem mappability file to a wig and then converted to a bigwig file (https://data.cyverse.org/dav-anon/iplant/home/turnerlabwashu/Turner_Lab_Track_Hubs/genomic_annotations/GRCh38_mappability_150mer.bw). CNV across 9p in the 1000 Genomes Project data was visualized in the UCSC genome browser53 using data from a previous publication54 available at https://github.com/KiddLab/kmer_1KG.
Known 9p gene/phenotype associations
The 435 RefSeq genes on 9p were assessed for their association with known phenotypes by running them through GeneALaCart55 (https://genealacart.genecards.org/). Genes with an elite association were extracted from the file and underwent manual curation via a literature review. The disease associations were then broadly assigned into the following categories: NDD, neurodegenerative, cancer, skeletal, immune, sex reversal, eye, diabetes, obesity, albinism, kidney, premature menopause/ovarian failure, muscle, arthrogryposis, head, mouth, and blood.
Protein-protein interactions on 9p
A STRINGdb56 (https://string-db.org/) analysis was performed using all of the 9p protein-coding genes.
Genomic assessment of 9p.100.p1
Family 9p.100 consists of an unaffected father (9p.100.fa), an unaffected mother (9p.100.mo), and a male child (9p.100.p1) with 9p deletion and duplication syndrome. The child has global developmental delays, hypotonia, joint hypermobility, and immunodeficiency. He has no significant family history. Previous clinical tests include a karyotype, microarray, and whole-exome sequencing. In this study, we assessed individual 9p.100.p1 by Illumina short-read WGS, Bionano optical mapping, and Pacific Biosciences (PacBio) HiFi long-read WGS. Individuals 9p.100.fa and 9p.100.mo were also assessed by PacBio HiFi long-read WGS.
Illumina WGS was performed to a coverage depth of 59.9× for individual 9p.100.p1. Reads were mapped to GRCh38_full_analysis_set_plus_decoy_hla.fa using bwa57 mem v.0.7.10-r789. Single-nucleotide variants (SNVs) and small insertion/deletions (indels) were detected using DeepVariant58 v.1.0.0 using WGS as the model and default settings. CNV was detected using the QuicK-mer254 program with GRCh38 as the reference genome. The steps included running quicKmer2 count followed by quicKmer2 est. An additional QuicK-mer2 analysis using the new Telomere-to-Telomere (T2T) consortium reference genome file59 (https://ftp.ncbi.nlm.nih.gov/genomes/all/GCA/009/914/755/GCA_009914755.3_CHM13_T2T_v1.1/GCA_009914755.3_CHM13_T2T_v1.1_genomic.fna.gz) was also performed on the Illumina data. Bionano optical mapping was carried out as described previously.33 PacBio HiFi long-read sequencing was performed to a coverage depth of 46.12 for individual 9p.100.p1. The CCS fastq files were aligned to build 38 of the human genome (GRCh38_full_analysis_set_plus_decoy_hla.fa) using pbmm2 (https://github.com/PacificBiosciences/pbmm2) v.1.3.0 align. PacBio pbsv (https://github.com/PacificBiosciences/pbsv) v. 2.3.0 was used to call copy-number and structural variants. Read-depth profiles were also generated using mosdepth.60 DeepVariant58 v. 1.0.0, using model PACBIO, was used to generate SNV/indel GVCF files for each individual, and they were joint-genotyped using GLNexus v.1.2.7. De novo assemblies were generated using two different assemblers (HiCanu [Canu v.2.0]61 and Hifiasm62 v.0.13-r307) for each individual.
PCR and Sanger sequencing were performed for the regions on both ends of the rearrangement between chromosome X and chromosome 9 in 9p.100.p1. Primers were designed using Primer3Plus (https://primer3plus.com) to target both rearrangement breakpoint regions, for a total of two amplicons. PCR reactions were performed using the primers, genomic DNA from 9p.100.p1, and Thermo Scientific Phusion High-Fidelity PCR Master Mix with HF Buffer. The two PCR products then underwent PCR cleanup and Sanger sequencing through Genewiz (https://www.genewiz.com). Each product was sequenced in both the forward and reverse directions, for a total of four sequencing products. Sequencing results were obtained from fasta files and aligned to the GRCh38 reference genome using the BLAST-like alignment tool (BLAT) from the UCSC genome browser (https://genome.ucsc.edu). BLAT alignments were further examined to confirm the rearrangement breakpoints.
Results
Insights from the worldwide 9p Network Cohort
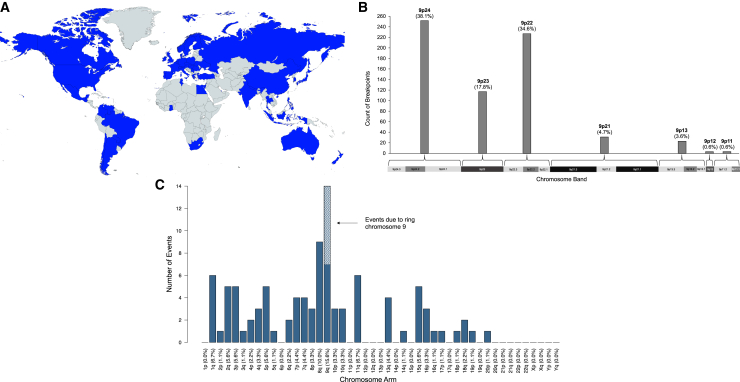
There are 811 individuals (Table S1) in the 9p Network Cohort dataset from 59 different countries representing six continents (Figure 1A). The dataset has low-resolution genetic information for 719 individuals with structural variants involving 9p and is seven to nine times bigger than the largest previously studied cohorts of 9p deletion18 or duplication22 syndromes. Although the genetic information for this cohort is low resolution, the large sample size allows us to investigate broader patterns of structural variants involving 9p.
Figure 1.
Characteristics of 9p network cohort
(A) Global location of individuals in the 9p Network Cohort. Countries represented by at least one individual in the 9p Network Cohort are highlighted in blue. All 811 individuals in the 9p Network Cohort were used to construct the map.
(B) Chromosome band breakpoints of 9p Network Cohort CNVs. The bar plot displays the number and percentage of breakpoints within each chromosome sub-band for CNVs listed in the 9p Network Cohort. Breakpoints are grouped by sub-band to remain consistent with the resolution of breakpoints reported in the 9p Network Cohort.
(C) Other chromosome arms affected in individuals with 9p CNVs. The number and percentage of events involving other chromosome arms in individuals with a 9p CNV are shown. Events include deletions, duplications, translocations, and inversions and seven individuals with ring chromosome 9 (dark blue stripes).
In the 9p Network Cohort, we found that the greatest number of breakpoints are located in the chromosome bands 9p24 and 9p22, with 257 (38.1%) and 233 (34.6%) of the 674 total breakpoints listed in the dataset, respectively (Figure 1B). The least common chromosome bands for 9p breakpoints are 9p12 and 9p11, each with 4 (0.6%) of the total breakpoints in the dataset. These patterns are consistent with the proposed 9p24 and 9p22 critical regions as well as with trends in breakpoint locations in previously published cases (Figure S1). We aggregated data from published cases and the 9p Network Cohort dataset to investigate which chromosome arms are most commonly involved in secondary structural variants in individuals with 9p deletion and duplication syndromes. This analysis revealed that 9q has the highest number of secondary events, due in part to ring chromosome 9 (Figure 1C). Other frequently affected chromosome arms include 1q, 8q, and 11q.
In addition to genetic data, the 9p Network Cohort dataset also lists the gender for all 719 individuals. Of these individuals, 406 individuals are female and 313 are male, indicating a female bias (Binomial test p = 0.0006). This result was surprising considering that no female bias has been previously reported in 9p deletion and duplication syndromes. A possible explanation for the significant bias in the 9p Network Cohort dataset is the XY sex reversal phenotype, which is commonly observed in individuals with 9p deletion syndrome. This phenotype could lead to individuals with XY sex chromosomes being listed in the dataset as having a female gender. To further examine this hypothesis, we subset our dataset to include only the 236 individuals whose sex chromosomes are listed in their genetic information. For this much smaller subset, 125 individuals had female sex chromosomes and 111 had male sex chromosomes, indicating no significant sex bias (Binomial test p = 0.4). We also found no significant gender bias in this group (Binomial test p = 0.2), although we did confirm that four of the individuals with XY sex chromosomes had a gender of female. This comparison suggests that the XY sex reversal phenotype may be responsible for a female gender bias, but not a sex bias, in 9p deletion and duplication syndrome cohorts.
Phenotypic characteristics of individuals with 9p deletions and duplications
Since we did not have phenotype information for the 9p Network Cohort, a literature search was performed to characterize common phenotypes in 9p deletion and duplication syndromes (Table 1). From this meta-analysis of 219 individuals, the most frequently observed phenotype is developmental delay (100% in deletions, 99% in duplications). There are nine shared phenotypes between individuals with a deletion or duplication including developmental delays (100% in deletions, 99% in duplications), hypotonia (65.7% in deletions, 61.8% in duplications), low-set ears (85.1% in deletions, 67.1% in duplications), abnormal auricles (51% in deletions, 83.1% in duplications), high/narrow palates (87.7% in deletions, 62.2% in duplications), short/broad necks (93.7% in deletions, 68.8% in duplications), broad internipple distances (92.3% in deletions, 44% in duplications), single palmar crease (69.8% in deletions, 90.6% in duplications), and cardiac murmurs/deficits (48.6% in deletions, 26.7% in duplications). There are also mirrored phenotypes including upward slanting palpebral fissures in deletions and downward slanting palpebral fissures in duplications as well as long philtrum in deletions and short philtrum in duplications.
Table 1.
Summary of 9p deletion and duplication syndrome phenotypic workups with more than 25 individuals
| Region/system affected | Specific phenotype |
Publications studying individuals with deletions (percentage of individuals with phenotype) |
Publication studying individuals with duplications (percent of individuals with phenotype) |
Phenotype comparisons of deletions and duplications |
|---|---|---|---|---|
| Swinkels et al.8Huret et al.18and Young et al.22(n = 120) | Young et al.22(n = 99) | |||
| General | developmental delay | 100.0 | 99.0 | shared |
| speech delay | 100.0 | NA | ||
| motor delay | 100.0 | NA | ||
| hypotonia | 65.7 | 61.8 | shared | |
| Head | trigonocephaly | 84.3 | NA | |
| midface hypoplasia | 82.4 | NA | ||
| upward slanting palpebral fissures | 63.2 | NA | mirror | |
| downward slanting palpebral fissures | 15.0 | 61.2 | mirror | |
| short palpebral fissures | 88.5 | NA | ||
| epicanthal fold | 65.6 | NA | ||
| high, arched eyebrows | 60.0 | NA | ||
| amblyopia | 33.3 | NA | ||
| Ears | low-set | 85.1 | 67.1 | shared |
| abnormal auricle | 51.0 | 83.1 | shared | |
| posteriorly angulated | 45.5 | NA | ||
| small (<p3) | 40.0 | NA | ||
| Nose | short/flat | 85.1 | NA | |
| anteverted nostrils | 88.7 | NA | ||
| Mouth | thin upper lip | 92.3 | NA | |
| long philtrum | 93.1 | NA | mirror | |
| flat philtrum | 46.2 | NA | ||
| high/narrow palate | 87.7 | 62.2 | shared | |
| irregular teeth | 30.0 | NA | ||
| micro/retrognathia | 77.3 | NA | ||
| Neck | short/broad | 93.7 | 68.8 | shared |
| Thorax | broad internipple distance | 92.3 | 44.0 | shared |
| Back | scoliosis | 41.2 | NA | |
| Extremities | tapering fingers | 63.6 | NA | |
| single palmar crease | 69.8 | 90.6 | shared | |
| hyperconvex nails | 66.7 | NA | ||
| flat feet | 72.7 | NA | ||
| hyperlax joints | 50.0 | NA | ||
| Cardiovascular | cardiac murmur/deficit | 48.6 | 26.7 | shared |
| Respiratory | frequent colds/infections | 81.8 | NA | |
| Gastrointestinal | inguinal hernia | 27.7 | NA | |
| omphalocele | 15.4 | NA | ||
| Urogenital | renal abnormalities | 7.7 | NA | |
| abnormal genitals | 36.7 | NA |
NA, not available.
Characteristics of genes on the p arm of chromosome 9
We examined the 435 RefSeq genes on 9p for constraint and dosage features (Table S2). There were 27 constrained genes (BNC2 [MIM: 608669]; CDC37L1 [MIM: 610346]; CLTA [MIM: 118960]; CNTFR [MIM: 118946]; ELAVL2 [MIM: 601673]; MLLT3 [MIM: 159558]; NFIB [MIM: 600728]; NOL6 [MIM: 611532]; PAX5 [MIM: 167414]; PSIP1 [MIM: 603620]; PTPRD [MIM: 601598]; RFX3 [MIM: 601337]; RNF38 [MIM: 612488]; RPS6 [MIM: 180460]; RUSC2 [MIM: 611053]; SHB [MIM: 600314]; SMARCA2 [MIM: 600014]; SMU1 [MIM: 617811]; TAF1L [MIM: 607798]; TEK [MIM: 600221]; TESK1 [MIM: 601782]; TLN1 [MIM: 186745]; TOPORS [MIM: 609507]; UBAP1 [MIM: 609787]; UBE2R2 [MIM: 612506]; UHRF2 [MIM: 615211]; and VCP [MIM: 611745]) with a pLI >0.9, which indicates that dominant disruption of these genes may have phenotypic consequences. We note here that a pLI >0.9 may be too restrictive when considering recessive disruption and that a different pLI cutoff could be considered for recessive genes in the future. This will be possible to explore further with precision genomics in 9p deletion and duplication syndromes. To further understand these genes and potential phenotypic consequences, we looked for enrichment of deletions or duplications in a dataset of 29,085 individuals with NDDs and 19,584 controls.50 Six of the pLI >0.9 genes were enriched for deletions in individuals with NDDs (CDC37L1, NFIB, PTPRD, RFX3, SMARCA2, UHRF2), and all 27 were enriched for duplications in individuals with NDDs (BNC2, CDC37L1, CLTA, CNTFR, ELAVL2, MLLT3, NFIB, NOL6, PAX5, PSIP1, PTPRD, RFX3, RNF38, RPS6, RUSC2, SHB, SMARCA2, SMU1, TAF1L, TEK, TESK1, TLN1, TOPORS, UBAP1, UBE2R2, UHRF2, VCP). This observation suggests that the dosage of these genes may play a role in NDDs. The mappability of most of 9p is quite high for short-read WGS data, indicating that the detection of CNV should be robust (Figure S2). Copy-number assessments generated from short-read WGS data in individuals from the 1000 Genomes Project54 reveal that the copy number of the majority of 9p is not variable in the population (Figure S2).
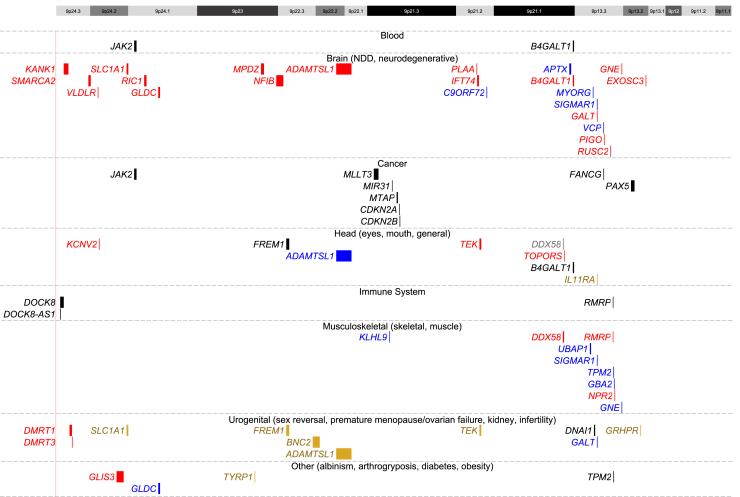
To expand beyond NDDs, a search for other gene/disease associations was carried out (Table S3; Figure 2). This analysis revealed two genes in blood phenotypes (JAK2 [MIM: 147796], B4GALT1 [MIM: 137060]); 17 genes in NDDs (KANK1, SMARCA2, VLDLR, SLC1A1 [MIM: 133550], RIC1 [MIM: 610354], GLDC, MPDZ [MIM: 603785], NFIB, ADAMTSL1 [MIM: 609198], PLAA [MIM: 603873], IFT74 [MIM: 608040], B4GALT1, GALT [MIM: 606999], PIGO [MIM: 614730], RUSC2, GNE [MIM: 603824], EXOSC3 [MIM: 606489]); five genes in neurodegenerative disorders (C9ORF72 [MIM: 614260], APTX [MIM: 606350], MYORG [MIM: 618255], SIGMAR1 [MIM: 601978], VCP); eight genes in cancer (JAK2, MLLT3, MIR31 [MIM: 612155], MTAP [MIM: 156540], CDKN2A [MIM: 600160], CDKN2B [MIM: 600431], FANCG [MIM: 602956], PAX5); eight genes in head-related phenotypes (KCNV2 [MIM: 607604], FREM1, ADAMTSL1, TEK, DDX58 [MIM: 609631], TOPORS, B4GALT1, IL11RA [MIM: 600939]); three genes in immune phenotypes (DOCK8, DOCK8-AS1, RMRP [MIM: 157660]); nine genes in musculoskeletal phenotypes (KLHL9 [MIM: 611201], DDX58, UBAP1, SIGMAR1, TPM2 [MIM: 190990], GBA2 [MIM: 609471], NPR2 [MIM: 607072], GNE, RMRP); ten genes in urogenital phenotypes (DMRT1, DMRT2 [MIM: 604935], SLC1A1, FREM1, BNC2, ADAMTSL1, TEK, DNAI1 [MIM: 604366], GALT, GRHPR [MIM: 604296]); and four genes involved in other phenotypes (GLIS3 [MIM: 610192], a gene known to exhibit imprinting, GLDC, TYRP1 [MIM: 115501], TPM2). Importantly, 29 of these genes are known to be involved in autosomal recessive conditions, including DNAI1 in primary ciliary dyskinesia and GALT in galactosemia. Fourteen of these autosomal recessive genes are associated with neurological phenotypes (e.g., KANK1 in cerebral palsy and MPDZ in congenital hydrocephalus), which may contribute to atypical or severe NDD phenotypes in some patients with 9p CNVs. Disruption of these genes can thus potentially unmask recessive traits and contribute to phenotypic variability and should be explored in patients with complex presentations.
Figure 2.
9p Genes with an associated disease/disorder
The genome browser view shows 9p genes with a manually curated disorder/disease association according to MalaCards. Genes are broadly categorized based on the general region/system affected and are more specifically grouped within each category. Specific groups within each category are indicated by different colors as follows: blood: all = black; brain: NDD = red, neurodegenerative = blue; cancer: all = black; head: general = black, eyes = red, eyes/general = blue, mouth/general = gold, eyes/mouth = gray; immune system: all = black; musculoskeletal: skeletal = red, muscle = blue; urogenital: infertility = black, sex reversal = red, premature menopause/ovarian failure = blue, kidney = gold; other: arthrogryposis = black, diabetes = red, obesity = blue, albinism = gold. NDD, neurodevelopmental disorder.
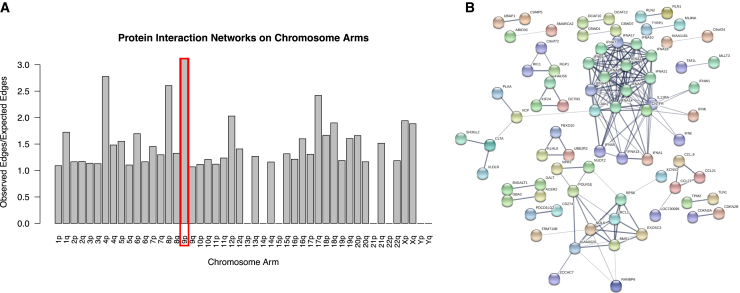
We performed a STRINGdb56 analysis using all of the 9p protein-coding genes (n = 207) to better understand the degree of interaction between the proteins encoded by genes on 9p. There were 57 expected edges (interactions) between the proteins, and we found that there are 177 observed interactions between the 207 proteins from 9p, indicating an observed-versus-expected ratio of 3.11. This represents a significant enrichment of interactions between proteins on 9p (p < 1.0 × 10−16) (Figure 3A). The interaction network driving this enrichment involves the IFNA genes (Figure 3B). These interferon genes are clustered together on 9p and are involved in immune function. We also looked at the observed-versus-expected interactions between proteins on every other chromosome arm. Some chromosome arms were not able to be assessed using this approach due to a lack of gene density on the arm (13p, 14p, 15p, 21p, 22p, Yp, Yq). We observed that the level of interaction enrichment was the highest for 9p (Figure 3A).
Figure 3.
Protein-protein interaction analysis of chromosome arms
(A) The bar plot shows the ratio of observed edges (interactions) versus expected interactions between proteins from genes on each chromosome arm (p < 1.0 × 10−16). Chromosome arms with a ratio of zero did not have enough data to perform the analysis. The p arm of chromosome 9 (boxed in red) shows the highest observed versus expected interaction ratio.
(B) The interaction network for proteins from 9p is shown. Note that there is a cluster of interactions between the IFNA (interferon) proteins, which are involved in immune system function.
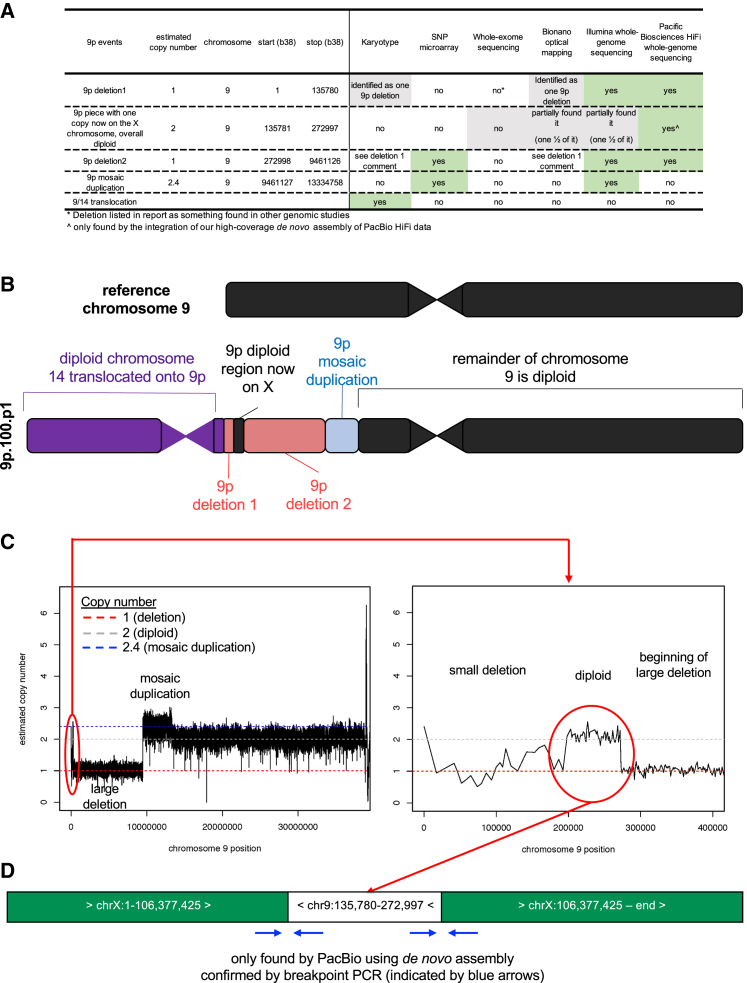
Precision genomics for 9p.100.p1
Several genomic technologies were utilized to determine which could fully resolve the structural variation in an individual (9p.100.p1) with a complex structural variation on 9p (Figure 4). Previous clinical karyotype testing identified a large 9p deletion and a translocation of chromosome 14 on the chromosome 9 containing the deletion (Figure 4A). Prior clinical microarray analysis identified a large 9p deletion and a mosaic 9p duplication (Figure 4A). The final clinical test was whole-exome sequencing, which identified a large 9p deletion (Figure 4A). Three newer genomic technologies were utilized in this study to gather additional data (Illumina short-read WGS, Bionano optical mapping, and PacBio HiFi long-read WGS) (Figure 4A). Bionano optical mapping was the least informative because this technology does not provide actual sequence data, so we instead focused primarily on the final resolution of the complex variation through the use of short-read and long-read WGS (Figures 4A and 4B).
Figure 4.
Precision genomics for 9p.100.p1
(A) Summary of structural variations and resolutions using different genomic technologies.
(B) Schematic of structural variations related to 9p in the individual.
(C) In the left panel, the copy-number estimates are shown for the p arm of chromosome 9 and identify a large deletion followed by a mosaic duplication. In the right panel, a zoom-in of the region near the telomere of chromosome 9p is shown to harbor a small deletion followed by a diploid segment and then a small part of the large deletion.
(D) Shown is the resolved variations, including orientation, for the small diploid segment on the telomeric end of 9p. This was resolved using a de novo assembly built with long-read sequencing.
The estimated copy number was calculated across the genome using short-read WGS, which revealed a large deletion and a mosaic duplication on chromosome 9 (Figure 4C). Attempts at finding the expected translocation breakpoint involving chromosomes 9 and 14, known from karyotype analysis, instead revealed a breakpoint involving chromosomes 9 and X. A deeper examination of the estimated copy number near the telomere on chromosome 9 found a small deletion followed by a segment with copy number 2 followed by the large deletion (Figure 4C). The exact base pairs at the border of this diploid region could not be determined using short-read WGS. To explore this variation further, the de novo assembly built from long-read WGS was queried to look for the sequences in the diploid segment (see Figure S3 for details on assembly comparisons). A contig was identified that revealed that one copy of this segment was on the X chromosome (Figure 4D), and Sanger sequencing confirmed the rearrangement breakpoints. The expected structure for the chromosomes 9 and X rearrangement in 9p.100.p1 involves an insertion of the region chr9:135,780–272,997 at chrX:106,377,425. Importantly, the chromosome 9 region is inverted relative to the chromosome X sequence. The BLAT alignment for the first PCR product covers chrX:106,377,216–106,377,425 and chr9:272,759–272,998, and the alignment of the reverse product for this region covers chrX:106,377,193–106,377,425 and chr9:272,797–272,998. The BLAT alignment for the forward product of the second rearrangement breakpoint covers chrX:106,377,430–106,377,786 and chr9:135,779–136,055, and the reverse product alignment covers chrX:106,377,430–106,377,744 and chr9:135,779–136,090. These alignments support the expected rearrangement coordinates and directionality. To summarize, the minimal technologies needed to resolve all variations in this individual (i.e., precision genomics) were a karyotype to find the 9/14 translocation since chromosome 14 had no large dosage changes, either microarray or short-read WGS to find the mosaic 9p duplication, and long-read WGS to find the complex variation near the telomere on chromosome 9.
Since the field is moving toward using newer reference genomes (i.e., T2T genome59), copy-number estimates from short-read WGS were also assessed using T2T reference, and the overall results were the same (Figure S4). A query of the sequence at breakpoints derived by long-read WGS was also compared with the T2T reference genome, and there was a slight shift of coordinates, as expected, when comparing any two genome builds (Figure S4). Overall, the comparison with T2T may be useful for resolving variations in some individuals in the future but did not change the overall interpretation for this individual.
With the precise variation determined for 9p.100.p1, the genes located within each variant were identified (Table 2). Important genes within the variant regions include DOCK8, implicated in immune phenotypes, and DOCK8, GLDC, KANK1, VLDLR, and MPDZ, implicated in NDDs. Other genes of interest include CDC37L1, PTPRD, RFX3, SMARCA2, and UHRF2, which all have a pLI >0.9 and are enriched for deletions/duplications in individuals with NDDs (Table 2). As a research study, we are also working on the process of reporting research results back to participants who would like access to the detailed genomic information. A concise one-page report was determined to be the best strategy for relaying the precision genomics research results. This report (Figure S5) clearly notes that this is a research report and has three main features: a schematic of the variation, a table of the precise breakpoints, and a table of the genes affected in each of the variant regions on chromosome 9. This approach could be a template for other research studies involving complex structural variations.
Table 2.
Genes involved in the structural variation identified in 9p.100.p1
| Variant | Gene names | Variation notes | Genes implicated as having a possible 9p phenotype | Genes with deletion nominal enrichment in NDDs | Genes with duplication nominal enrichment in NDDs | Genes with pLI > 0.9 | Genes with known phenotype (category) |
|---|---|---|---|---|---|---|---|
| 9p- deletion1 | CBWD1, DDX11L5, FAM138C, FOXD4, MIR1302-9, PGM5P3-AS1, WASHC1 | FOXD4 (speech and language development) | None | none | none | none | |
| 9p piece with one copy now on the X chromosome, overall diploid | CBWD1, DOCK8, DOCK8-AS1 | both the CBWD1 and DOCK8 genes are broken even though this full piece of DNA is diploid and moved to the X chromosome | DOCK8 (ID, seizures, autism) | DOCK8 | none | none | DOCK8 (immune) |
| 9p- deletion2 | AK3, CD274, CDC37L1, CDC37L1-DT, DMAC1, DMRT1, DMRT2, DMRT3, DOCK8, ERMP1, GLDC, GLIS3, GLIS3-AS1, IL33, INSL4, INSL6, JAK2, KANK1, KCNV2, KDM4C, KIAA2026, LINC01230, LINC01231, MIR101-2, MIR4665, MLANA, PDCD1LG2, PLGRKT, PLPP6, PTPRD, PTPRD-AS1, PUM3, RANBP6, RCL1, RFX3, RFX3-AS1, RIC1, RLN1, RLN2, SLC1A1, SMARCA2, SPATA6L, TPD52L3, UHRF2, VLDLR, VLDLR-AS1 | KANK1 (cerebral palsy), DMRT3 (disorders of sex development), DMRT1 (disorders of sex development), GLDC (intellectual disability, seizures), DOCK8 (intellectual disability, seizures, autism), VLDLR (intellectual disability, seizures, cerebellar hypoplasia/ataxia) | AK3, CD274, CDC37L1, CDC37L1-DT, DMAC1, DMRT1, DMRT2, DMRT3, DOCK8, DOCK8-AS1, ERMP1, GLDC, GLIS3, GLIS3-AS1, IL33, INSL4, INSL6, JAK2, KANK1, KCNV2, KDM4C, KIAA2026, LINC01230, LINC01231, MIR101-2, MIR4665, MLANA, PDCD1LG2, PLGRKT, PLPP6, PTPRD, PTPRD-AS1, PTPRD-AS2, PUM3, RANBP6, RCL1, RFX3, RFX3-AS1, RIC1, RLN1, RLN2, SLC1A1, SMARCA2, SPATA6L, TPD52L3, UHRF2, VLDLR, VLDLR-AS1 | AK3, CD274, CDC37L1, CDC37L1-DT, DMAC1, DMRT1, DMRT2, DMRT3, ERMP1, GLDC, GLIS3, GLIS3-AS1, IL33, INSL4, INSL6, JAK2, KANK1, KCNV2, KDM4C, KIAA2026, LINC01230, LINC01231, MIR101-2, MIR4665, MLANA, PDCD1LG2, PLGRKT, PLPP6, PTPRD, PTPRD-AS1, PTPRD-AS2, PUM3, RANBP6, RCL1, RFX3, RFX3-AS1, RIC1, RLN1, RLN2, SLC1A1, SMARCA2, SPATA6L, TPD52L3, UHRF2, VLDLR, VLDLR-AS1 | CDC37L1, PTPRD, RFX3, SMARCA2, UHRF2 | DMRT1 (sex reversal), DMRT3 (sex reversal), DOCK8 (immune), DOCK8-AS1 (immune), GLDC (NDD, obesity), GLIS3 (diabetes), JAK2 (cancer), KANK1 (NDD), KCNV2 (eye), SLC1A1 (kidney, NDD), SMARCA2 (NDD), VLDLR (NDD) | |
| 9p- mosaic duplication | PTPRD, LOC105375972, PTPRD-AS2, TYRP1, LURAP1L-AS1, LURAP1L, SNORD137, MPDZ | of the genes in the region, LOC105375972, PTPRD-AS2, TYRP1, LURAP1L-AS1, LURAP1L, SNORD137, and MPDZ are fully duplicated | PTPRD, LOC105375972, PTPRD-AS2, TYRP1, LURAP1L, LURAP1L-AS1, SNORD137, MPDZ | PTPRD, LOC105375972, PTPRD-AS2, TYRP1, LURAP1L, LURAP1L-AS1, SNORD137, MPDZ | PTPRD | TYRP1 (albinism), MPDZ (NDD) |
Discussion
In this study, we present an analysis of the largest cohort of individuals with 9p deletions and duplications studied to date. We first assessed the genomic variation in this cohort to determine if there are any trends in the 9p breakpoint region and confirmed 9p22 and 9p24 as the regions with the most breakpoints, as previously described in studies seven to nine times smaller than the present study. We then assessed the genomic variations to determine if there were any trends in the chromosome arms involved in secondary structural variations. A similar investigation of structural variation patterns in other phenotypes and syndromes has proven crucial to improving clinical management and developing therapeutic applications. For example, many cases of chronic myeloid leukemia (CML) are driven by a fusion protein derived from a translocation between chromosomes 9 and 22 (MIM: 608232). Importantly, identification of this variation pattern, known as “The Philadelphia Chromosome,”63 has enabled successful targeting by clinical therapeutics (MIM: 608232). In contrast to The Philadelphia Chromosome and CML, we found that the pattern of breakpoints and chromosome arms affected by secondary structural variants in 9p deletion and duplication syndromes is more heterogeneous. This reinforces the genetic and clinical complexities of these syndromes and the need for a precision genomics approach.
While DSDs have been reported in 9p deletion and duplication syndromes, we identify for the first time a significant gender bias in the full cohort with an enrichment for females. Among those with available sex chromosome information, we found individuals in the cohort with a gender of female and a sex chromosome complement of XY as expected in some DSDs. To make this a comprehensive study of phenotypes and genes in 9p deletion and duplication syndromes, we performed a meta-analysis of phenotypes observed in 9p deletion and duplication syndromes and found shared, similar, mirrored, and differing phenotypes. Several gene features were also considered for prioritization including constraint, enrichment for deletions/duplications in NDDs, and prior established disease associations. These are useful resources for the assessment of 9p-related structural variations. Recently developed genomic technologies are revolutionizing the way we assess syndromes with complex structural variations. We applied several of these technologies in this study to an individual with a complex 9p deletion, duplication, and associated translocation. We found that the classical karyotype is essential, that either a microarray or short-read WGS is critical to identify the mosaic duplication, and that long-read sequencing is the only technology able to resolve the intricate complexities of this variation.
The early studies of 9p deletion and duplication syndromes relied on the use of karyotyping,1 which does not have the resolution to define CNV breakpoints beyond the chromosome band and can fail to detect microdeletions and microduplications.26 The absence of high-resolution alignments and precise breakpoint analysis is one factor that has contributed to the difficulty in establishing genotype-phenotype correlations with 9p CNVs.9,64 The advancements of modern sequencing technologies provide an opportunity to precisely resolve breakpoints to the exact base,33 thus allowing for a better characterization of 9p CNVs both in terms of genomic variation and phenotypes. The power of long-read sequencing technologies (e.g., Oxford Nanopore Technologies and PacBio) is enabling complete genomic variant resolution within individuals,65,66 as shown in the present study. In addition, advancing the bioinformatic assessment of long-read sequencing data is also providing insight into methylation and will be useful for examining imprinted genes on 9p.67 These types of technologies will be critical to the growing understanding of 9p CNVs, especially when many affected individuals present with complex rearrangements. Another recent use of modern high-resolution sequencing technologies by Ng et al.33 for a patient with a complex rearrangement involving a 9p deletion and a 13q duplication that translocated onto the chromosome 9 containing the deletion allowed for the resolution of breakpoints to the single-nucleotide level.33 Analysis of single-nucleotide resolution breakpoints provides the ability to precisely resolve the genomic region associated with patient phenotypes and ultimately identify genes affected by the genomic variation.
Another area for future development is the linking of genotypes with phenotypes in individuals with 9p deletion and duplication syndromes. Many approaches can be utilized including critical region delineation. However, we also highlight the application of machine learning to complex biological problems and suggest it as a strategy to combine all of the genomic and phenotypic data. The application of novel genomic technologies with deep phenotyping on a large cohort of individuals would be the ideal input to these types of models.
Considerable progress has been made in the assessment of 9p deletion and duplication syndromes. However, it is still challenging to predict an affected individual's phenotypes with the currently available data. Part of this phenotypic unpredictability is attributable to the low genomic resolution possible with older genotyping methods. Current technological advances in genomics are providing strategies to detect all forms of variations in the genome at a large scale and a reasonable cost. For example, in 9p deletion syndrome, these technologies can identify the precise breakpoints of the event, detect potentially relevant variations on the remaining allele, and look at the remainder of the genome for other relevant events (e.g., a second hit). Utilizing this information across many individuals with 9p deletion and duplication syndromes and combining it with leading edge analyses of phenotypic data (e.g., the parsing of electronic health records) will enable the delineation of complete genotype-phenotype correlations. This combined work will bring the dream of precision genomics to reality in 9p deletion and duplication syndromes.
Data and code availability
Data are consented for sharing in a controlled-access database and are available at dbGaP (https://www.ncbi.nlm.nih.gov/gap/) phs002054.v1.p1.
Acknowledgments
This work was supported by grants from the National Institutes of Health (R00MH117165 and P50HD103525), funds from the Department of Genetics and Pediatrics at Washington University School of Medicine, and a donation to the McDonnell Genome Institute to support work on Chromosome 9p Minus syndrome. This study was approved by the IRB (IRB ID #201102181 and ID #201706062) at the Washington University School of Medicine, and written informed consent for this study was obtained from study participants. We would also like to acknowledge and thank Mr. Kyle Greig and the Chromosome 9p Minus Network Board for generously providing us with the 9p Network Cohort data.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xhgg.2021.100081.
Web resources
GEMtools, https://github.com/Chimera-tools/ChimPipe.git
Mappability 150-mer for b38, https://data.cyverse.org/dav-anon/iplant/home/turnerlabwashu/Turner_Lab_Track_Hubs/genomic_annotations/GRCh38_mappability_150mer.bw
1000 Genomes copy-number tracks, https://github.com/KiddLab/kmer_1KG
GeneALaCart, https://genealacart.genecards.org/
STRINGdb, https://string-db.org/
T2T reference genome, https://ftp.ncbi.nlm.nih.gov/genomes/all/GCA/009/914/755/GCA_009914755.3_CHM13_T2T_v1.1/GCA_009914755.3_CHM13_T2T_v1.1_genomic.fna.gz
pbmm2, https://github.com/PacificBiosciences/pbmm2
pbsv, https://github.com/PacificBiosciences/pbsv.
Primer3Plus, https://primer3plus.com.
Genewiz, https://www.genewiz.com.
UCSC genome browser, https://genome.ucsc.edu
dbGaP, https://www.ncbi.nlm.nih.gov/gap/
OMIM, https://omim.org/
Supplemental information
References
- 1.Alfi O., Donnell G.N., Crandall B.F., Derencsenyi A., Menon R. Deletion of the short arm of chromosome no.9 (46,9p-): a new deletion syndrome. Ann Genet. 1973;16:17–22. [PubMed] [Google Scholar]
- 2.Alfi O.S., Donnell G.N., Allderdice P.W., Derencsenyi A. The 9p- syndrome. Ann Genet. 1976;19:11–16. [PubMed] [Google Scholar]
- 3.Kerem B., Rommens J.M., Buchanan J.A., Markiewicz D., Cox T.K., Chakravarti A., Buchwald M., Tsui L.C. Identification of the cystic fibrosis gene: genetic analysis. Science. 1989;245:1073–1080. doi: 10.1126/science.2570460. [DOI] [PubMed] [Google Scholar]
- 4.Marshall C.R., Noor A., Vincent J.B., Lionel A.C., Feuk L., Skaug J., Shago M., Moessner R., Pinto D., Ren Y., et al. Structural variation of chromosomes in autism spectrum disorder. Am J Hum Genet. 2008;82:477–488. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weiss L.A., Shen Y., Korn J.M., Arking D.E., Miller D.T., Fossdal R., Saemundsen E., Stefansson H., Ferreira M.A., Green T., et al. Association between microdeletion and microduplication at 16p11.2 and autism. N Engl J Med. 2008;358:667–675. doi: 10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]
- 6.Sanders S.J., Ercan-Sencicek A.G., Hus V., Luo R., Murtha M.T., Moreno-De-Luca D., Chu S.H., Moreau M.P., Gupta A.R., Thomson S.A., et al. Multiple recurrent de novo CNVs, including duplications of the 7q11.23 Williams syndrome region, are strongly associated with autism. Neuron. 2011;70:863–885. doi: 10.1016/j.neuron.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schwartz S.B.S., Christ L. , Eichenmiller M., Graf M., Vance H., Crowe C. (2005). Delineation of Chromosome 9p Deletions: A Model of Phenotype and Chromosomal Mechanisms for Terminal Deletions, 209. https://www.ashg.org/wp-content/uploads/2019/10/2005-all-abstracts.pdf
- 8.Swinkels M.E., Simons A., Smeets D.F., Vissers L.E., Veltman J.A., Pfundt R., de Vries B.B., Faas B.H., Schrander-Stumpel C.T., McCann E., et al. Clinical and cytogenetic characterization of 13 Dutch patients with deletion 9p syndrome: delineation of the critical region for a consensus phenotype. Am J Med Genet A. 2008;146A:1430–1438. doi: 10.1002/ajmg.a.32310. [DOI] [PubMed] [Google Scholar]
- 9.Christ L.A., Crowe C.A., Micale M.A., Conroy J.M., Schwartz S. Chromosome breakage hotspots and delineation of the critical region for the 9p-deletion syndrome. Am J Hum Genet. 1999;65:1387–1395. doi: 10.1086/302606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kawara H., Yamamoto T., Harada N., Yoshiura K., Niikawa N., Nishimura A., Mizuguchi T., Matsumoto N. Narrowing candidate region for monosomy 9p syndrome to a 4.7-Mb segment at 9p22.2-p23. Am J Med Genet A. 2006;140:373–377. doi: 10.1002/ajmg.a.31094. [DOI] [PubMed] [Google Scholar]
- 11.Faas B.H., de Leeuw N., Mieloo H., Bruinenberg J., de Vries B.B. Further refinement of the candidate region for monosomy 9p syndrome. Am J Med Genet A. 2007;143A:2353–2356. doi: 10.1002/ajmg.a.31961. [DOI] [PubMed] [Google Scholar]
- 12.Hauge X., Raca G., Cooper S., May K., Spiro R., Adam M., Martin C.L. Detailed characterization of, and clinical correlations in, 10 patients with distal deletions of chromosome 9p. Genet Med. 2008;10:599–611. doi: 10.1097/gim.0b013e31817e2bde. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barbaro M., Balsamo A., Anderlid B.M., Myhre A.G., Gennari M., Nicoletti A., Pittalis M.C., Oscarson M., Wedell A. Characterization of deletions at 9p affecting the candidate regions for sex reversal and deletion 9p syndrome by MLPA. Eur J Hum Genet. 2009;17:1439–1447. doi: 10.1038/ejhg.2009.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Recalcati M.P., Bellini M., Norsa L., Ballarati L., Caselli R., Russo S., Larizza L., Giardino D. Complex rearrangement involving 9p deletion and duplication in a syndromic patient: genotype/phenotype correlation and review of the literature. Gene. 2012;502:40–45. doi: 10.1016/j.gene.2012.04.030. [DOI] [PubMed] [Google Scholar]
- 15.Di Bartolo D.L., El Naggar M., Owen R., Sahoo T., Gilbert F., Pulijaal V.R., Mathew S. Characterization of a complex rearrangement involving duplication and deletion of 9p in an infant with craniofacial dysmorphism and cardiac anomalies. Mol Cytogenet. 2012;5:31. doi: 10.1186/1755-8166-5-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kowalczyk M., Tomaszewska A., Podbioł-Palenta A., Constantinou M., Wawrzkiewicz-Witkowska A., Kowalski J., Kałużewski B., Zajączek S., Srebniak M.I. Another rare case of a child with de novo terminal 9p deletion and co-existing interstitial 9p duplication: clinical findings and molecular cytogenetic study by array-CGH. Cytogenet Genome Res. 2013;139:9–16. doi: 10.1159/000342165. [DOI] [PubMed] [Google Scholar]
- 17.Onesimo R., Orteschi D., Scalzone M., Rossodivita A., Nanni L., Zannoni G.F., Marrocco G., Battaglia D., Fundarò C., Neri G. Chromosome 9p deletion syndrome and sex reversal: novel findings and redefinition of the critically deleted regions. Am J Med Genet A. 2012;158A:2266–2271. doi: 10.1002/ajmg.a.35489. [DOI] [PubMed] [Google Scholar]
- 18.Huret J.L., Leonard C., Forestier B., Rethore M.O., Lejeune J. Eleven new cases of del(9p) and features from 80 cases. J Med Genet. 1988;25:741–749. doi: 10.1136/jmg.25.11.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sivasankaran A., Kanakavalli M.K., Anuradha D., Samuel C.R., Kandukuri L.R. Ring chromosome 9 and chromosome 9p deletion syndrome in a patient Associated with developmental delay: a case report and review of the literature. Cytogenet Genome Res. 2016;148:165–173. doi: 10.1159/000445862. [DOI] [PubMed] [Google Scholar]
- 20.Beaudry S.M., Shchelochkov O., Trapane P., Darbro B., Nagy J.M.W. Case report of a pseudo-isodicentric chromosome 9 resulting in mosaic trisomy 9. Clin Case Rep. 2021;9:2340–2344. doi: 10.1002/ccr3.4031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li M., Glass J., Du X., Dubbs H., Harr M.H., Falk M., Smolarek T., Hopkin R.J., Zackai E., Sheppard S.E. Trisomy 9 mosaic syndrome: sixteen additional patients with new and/or less commonly reported features, literature review, and suggested clinical guidelines. Am J Med Genet A. 2021;185:2374–2383. doi: 10.1002/ajmg.a.62251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Young R.S., Reed T., Hodes M.E., Palmer C.G. The dermatoglyphic and clinical features of the 9p trisomy and partial 9p monosomy syndromes. Human Genet. 1982;62:31–39. doi: 10.1007/bf00295601. [DOI] [PubMed] [Google Scholar]
- 23.Mohamed A.M., Kamel A.K., Eid M.M., Eid O.M., Mekkawy M., Hussein S.H., Zaki M.S., Esmail S., Afifi H.H., El-Kamah G.Y., et al. Chromosome 9p terminal deletion in nine Egyptian patients and narrowing of the critical region for trigonocephaly. Mol Genet Genomic Med. 2021:e1829. doi: 10.1002/mgg3.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bennett C.P., Docherty Z., Robb S.A., Ramani P., Hawkins J.R., Grant D. Deletion 9p and sex reversal. J Med Genet. 1993;30:518–520. doi: 10.1136/jmg.30.6.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ogata T., Muroya K., Matsuo N., Hata J., Fukushima Y., Suzuki Y. Impaired male sex development in an infant with molecularly defined partial 9p monosomy: implication for a testis forming gene(s) on 9p. J Med Genet. 1997;34:331–334. doi: 10.1136/jmg.34.4.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vinci G., Chantot-Bastaraud S., El Houate B., Lortat-Jacob S., Brauner R., McElreavey K. Association of deletion 9p, 46,XY gonadal dysgenesis and autistic spectrum disorder. Mol Hum Reprod. 2007;13:685–689. doi: 10.1093/molehr/gam045. [DOI] [PubMed] [Google Scholar]
- 27.Quinonez S.C., Park J.M., Rabah R., Owens K.M., Yashar B.M., Glover T.W., Keegan C.E. 9p partial monosomy and disorders of sex development: review and postulation of a pathogenetic mechanism. Am J Med Genet A. 2013;161A:1882–1896. doi: 10.1002/ajmg.a.36018. [DOI] [PubMed] [Google Scholar]
- 28.Mitsui N., Shimizu K., Nishimoto H., Mochizuki H., Iida M., Ohashi H. Patient with terminal 9 Mb deletion of chromosome 9p: refining the critical region for 9p monosomy syndrome with trigonocephaly. Congenit Anom (Kyoto) 2013;53:49–53. doi: 10.1111/j.1741-4520.2012.00362.x. [DOI] [PubMed] [Google Scholar]
- 29.Chen C.P., Su Y.N., Chen C.Y., Chern S.R., Wu P.S., Su J.W., Lee C.C., Chen L.F., Wang W. Prenatal diagnosis and molecular cytogenetic characterization of a de novo pure distal 9p deletion and literature review. Genomics. 2013;102:265–269. doi: 10.1016/j.ygeno.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 30.Bruni V., Roppa K., Scionti F., Apa R., Sestito S., Di Martino M.T., Pensabene L., Concolino D. A 46,XY female with a 9p24.3p24.1 deletion and a 8q24.11q24.3 duplication: a case report and review of the literature. Cytogenet Genome Res. 2019;158:74–82. doi: 10.1159/000500619. [DOI] [PubMed] [Google Scholar]
- 31.Yang Y., Wang C., Wang F., Zhu L., Liu H., He X. Novel chromosomal translocation t(11;9)(p15;p23) involving deletion and duplication of 9p in a girl associated with autism and mental retardation. Gene. 2012;502:154–158. doi: 10.1016/j.gene.2012.04.036. [DOI] [PubMed] [Google Scholar]
- 32.Güneş S., Ekinci Ö., Ekinci N., Toros F. Coexistence of 9p deletion syndrome and autism spectrum disorder. J Autism Dev Disord. 2017;47:520–521. doi: 10.1007/s10803-016-2943-x. [DOI] [PubMed] [Google Scholar]
- 33.Ng J., Sams E., Baldridge D., Kremitzki M., Wegner D.J., Lindsay T., Fulton R., Cole F.S., Turner T.N. Precise breakpoint detection in a patient with 9p- syndrome. Cold Spring Harb Mol Case Stud. 2020;6 doi: 10.1101/mcs.a005348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vissers L.E., Cox T.C., Maga A.M., Short K.M., Wiradjaja F., Janssen I.M., Jehee F., Bertola D., Liu J., Yagnik G., et al. Heterozygous mutations of FREM1 are associated with an increased risk of isolated metopic craniosynostosis in humans and mice. PLoS Genet. 2011;7:e1002278. doi: 10.1371/journal.pgen.1002278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cordes Selby S., Iwata-Otsubo A., Delk P., Nebesio T.D., Gohil A., Matlock P., Torres-Martinez W., Vance G.H. A brother and sister with the same karyotype: case report of two siblings with partial 3p duplication and partial 9p deletion and sex reversal. Clin Case Rep. 2021;9:e04141. doi: 10.1002/ccr3.4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Banerjee I., Senniappan S., Laver T.W., Caswell R., Zenker M., Mohnike K., Cheetham T., Wakeling M.N., Ismail D., Lennerz B., et al. Refinement of the critical genomic region for congenital hyperinsulinism in the Chromosome 9p deletion syndrome. Wellcome Open Res. 2019;4:149. doi: 10.12688/wellcomeopenres.15465.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.D'Angelo C.S., Varela M.C., de Castro C.I.E., Otto P.A., Perez A.B.A., Lourenço C.M., Kim C.A., Bertola D.R., Kok F., Garcia-Alonso L., Koiffmann C.P. Chromosomal microarray analysis in the genetic evaluation of 279 patients with syndromic obesity. Mol Cytogenet. 2018;11:14. doi: 10.1186/s13039-018-0363-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hou Q.F., Wu D., Chu Y., Liao S.X. Clinical findings and molecular cytogenetic study of de novo pure chromosome 9p deletion: pre- and postnatal diagnosis. Taiwan J Obstet Gynecol. 2016;55:867–870. doi: 10.1016/j.tjog.2016.11.001. [DOI] [PubMed] [Google Scholar]
- 39.Vásquez-Velásquez A.I., García-Castillo H.A., González-Mercado M.G., Dávalos I.P., Raca G., Xu X., Dwyer E., Rivera H. Duplication 5q and deletion 9p due to a t(5;9)(q34;p23) in 2 cousins with features of Hunter-McAlpine syndrome and hypothyroidism. Cytogenet Genome Res. 2011;132:233–238. doi: 10.1159/000321647. [DOI] [PubMed] [Google Scholar]
- 40.Sgardioli I.C., de Mello Copelli M., Monteiro F.P., Dos Santos A.P., Lustosa Mendes E., Paiva Vieira T., Gil-da-Silva-Lopes V.L. Diagnostic approach to microdeletion syndromes based on 22q11.2 investigation: challenges in four cases. Mol Syndromol. 2017;8:244–252. doi: 10.1159/000477598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Durmaz C.D., Yararbaş K., Kutlay N.Y., Türedi Ö., Akın İ., Gürbüz C., Karataş G., Tükün A. Unusual chromosomal rearrangement resulted in interstitial monosomy 9p: case report. Cytogenet Genome Res. 2016;148:19–24. doi: 10.1159/000444872. [DOI] [PubMed] [Google Scholar]
- 42.Hulick P.J., Noonan K.M., Kulkarni S., Donovan D.J., Listewnik M., Ihm C., Stoler J.M., Weremowicz S. Cytogenetic and array-CGH characterization of a complex de novo rearrangement involving duplication and deletion of 9p and clinical findings in a 4-month-old female. Cytogenet Genome Res. 2009;126:305–312. doi: 10.1159/000251966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schlade-Bartusiak K., Tucker T., Safavi H., Livingston J., van Allen M.I., Eydoux P., Armstrong L. Independent post-zygotic breaks of a dicentric chromosome result in mosaicism for an inverted duplication deletion 9p and terminal deletion 9p. Eur J Med Genet. 2013;56:229–235. doi: 10.1016/j.ejmg.2013.01.013. [DOI] [PubMed] [Google Scholar]
- 44.Nakayama T., Nabatame S., Saito Y., Nakagawa E., Shimojima K., Yamamoto T., Kaneko Y., Okumura K., Fujie H., Uematsu M., et al. 8p deletion and 9p duplication in two children with electrical status epilepticus in sleep syndrome. Seizure. 2012;21:295–299. doi: 10.1016/j.seizure.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 45.Martín-De Saro M.D., Valdés-Miranda J.M., Plaza-Benhumea L., Pérez-Cabrera A., Gonzalez-Huerta L.M., Guevara-Yañez R., Cuevas-Covarrubias S.A. Characterization of a complex chromosomal rearrangement involving a de novo duplication of 9p and 9q and a deletion of 9q. Cytogenet Genome Res. 2015;147:124–129. doi: 10.1159/000444138. [DOI] [PubMed] [Google Scholar]
- 46.Yokoyama E., Del Castillo V., Sánchez S., Ramos S., Molina B., Torres L., Navarro M.J., Avila S., Castrillo J.L., García-De Teresa B., et al. Derivative chromosomes involving 5p large rearranged segments went unnoticed with the use of conventional cytogenetics. Mol Cytogenet. 2018;11:30. doi: 10.1186/s13039-018-0374-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chehimi S.N., Zanardo É A., Ceroni J.R.M., Nascimento A.M., Madia F.A.R., Dias A.T., Filho G.M.N., Montenegro M.M., Damasceno J., Costa T., et al. Breakpoint delineation in 5p- patients leads to new insights about microcephaly and the typical high-pitched cry. Mol Genet Genomic Med. 2020;8:e957. doi: 10.1002/mgg3.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tang Z., Berlin D.S., Toji L., Toruner G.A., Beiswanger C., Kulkarni S., Martin C.L., Emanuel B.S., Christman M., Gerry N.P. A dynamic database of microarray-characterized cell lines with various cytogenetic and genomic backgrounds. G3 (Bethesda) 2013;3:1143–1149. doi: 10.1534/g3.113.006577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Karczewski K.J., Francioli L.C., Tiao G., Cummings B.B., Alföldi J., Wang Q., Collins R.L., Laricchia K.M., Ganna A., Birnbaum D.P., et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Coe B.P., Witherspoon K., Rosenfeld J.A., van Bon B.W., Vulto-van Silfhout A.T., Bosco P., Friend K.L., Baker C., Buono S., Vissers L.E., et al. Refining analyses of copy number variation identifies specific genes associated with developmental delay. Nature Genet. 2014;46:1063–1071. doi: 10.1038/ng.3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rodríguez-Martín B., Palumbo E., Marco-Sola S., Griebel T., Ribeca P., Alonso G., Rastrojo A., Aguado B., Guigó R., Djebali S. ChimPipe: accurate detection of fusion genes and transcription-induced chimeras from RNA-seq data. BMC Genomics. 2017;18:7. doi: 10.1186/s12864-016-3404-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shen F., Kidd J.M. Rapid, paralog-sensitive CNV analysis of 2457 human genomes using QuicK-mer2. Genes (Basel) 2020;11 doi: 10.3390/genes11020141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stelzer G., Rosen N., Plaschkes I., Zimmerman S., Twik M., Fishilevich S., Stein T.I., Nudel R., Lieder I., Mazor Y., et al. The GeneCards suite: from gene data mining to disease genome sequence analyses. Curr Protoc Bioinformatics. 2016;54:1.30.31–31.30.33. doi: 10.1002/cpbi.5. [DOI] [PubMed] [Google Scholar]
- 56.Szklarczyk D., Gable A.L., Lyon D., Junge A., Wyder S., Huerta-Cepas J., Simonovic M., Doncheva N.T., Morris J.H., Bork P., et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47 doi: 10.1093/nar/gky1131. D607–d613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv. 2013 https://arxiv.org/abs/1303.3997 [Google Scholar]
- 58.Poplin R., Chang P.C., Alexander D., Schwartz S., Colthurst T., Ku A., Newburger D., Dijamco J., Nguyen N., Afshar P.T., et al. A universal SNP and small-indel variant caller using deep neural networks. Nature Biotechnol. 2018;36:983–987. doi: 10.1038/nbt.4235. [DOI] [PubMed] [Google Scholar]
- 59.Nurk S., Koren S., Rhie A., Rautiainen M., Bzikadze A.V., Mikheenko A., Vollger M.R., Altemose N., Uralsky L., Gershman A., et al. The complete sequence of a human genome. bioRxiv. 2021 doi: 10.1101/2021.05.26.445798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pedersen B.S., Quinlan A.R. Mosdepth: quick coverage calculation for genomes and exomes. Bioinformatics. 2018;34:867–868. doi: 10.1093/bioinformatics/btx699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nurk S., Walenz B.P., Rhie A., Vollger M.R., Logsdon G.A., Grothe R., Miga K.H., Eichler E.E., Phillippy A.M., Koren S. HiCanu: accurate assembly of segmental duplications, satellites, and allelic variants from high-fidelity long reads. Genome Res. 2020;30:1291–1305. doi: 10.1101/gr.263566.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cheng H., Concepcion G.T., Feng X., Zhang H., Li H. Haplotype-resolved de novo assembly using phased assembly graphs with hifiasm. Nature Methods. 2021 doi: 10.1038/s41592-020-01056-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.de Klein A., van Kessel A.G., Grosveld G., Bartram C.R., Hagemeijer A., Bootsma D., Spurr N.K., Heisterkamp N., Groffen J., Stephenson J.R. A cellular oncogene is translocated to the Philadelphia chromosome in chronic myelocytic leukaemia. Nature. 1982;300:765–767. doi: 10.1038/300765a0. [DOI] [PubMed] [Google Scholar]
- 64.Sirisena N.D., Wijetunge U.K., de Silva R., Dissanayake V.H. Child with deletion 9p syndrome presenting with craniofacial dysmorphism, developmental delay, and multiple congenital malformations. Case Rep Genet. 2013;2013:785830. doi: 10.1155/2013/785830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wenger A.M., Peluso P., Rowell W.J., Chang P.C., Hall R.J., Concepcion G.T., Ebler J., Fungtammasan A., Kolesnikov A., Olson N.D., et al. Accurate circular consensus long-read sequencing improves variant detection and assembly of a human genome. Nature Biotechnol. 2019;37:1155–1162. doi: 10.1038/s41587-019-0217-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jain M., Koren S., Miga K.H., Quick J., Rand A.C., Sasani T.A., Tyson J.R., Beggs A.D., Dilthey A.T., Fiddes I.T., et al. Nanopore sequencing and assembly of a human genome with ultra-long reads. Nature Biotechnol. 2018;36:338–345. doi: 10.1038/nbt.4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tse O.Y.O., Jiang P., Cheng S.H., Peng W., Shang H., Wong J., Chan S.L., Poon L.C.Y., Leung T.Y., Chan K.C.A., et al. Genome-wide detection of cytosine methylation by single molecule real-time sequencing. Proc Natl Acad Sci U S A. 2021;118 doi: 10.1073/pnas.2019768118. e2019768118. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are consented for sharing in a controlled-access database and are available at dbGaP (https://www.ncbi.nlm.nih.gov/gap/) phs002054.v1.p1.