Summary
Despite a lifetime prevalence of at least 5%, developmental stuttering, characterized by prolongations, blocks, and repetitions of speech sounds, remains a largely idiopathic speech disorder. Family, twin, and segregation studies overwhelmingly support a strong genetic influence on stuttering risk; however, its complex mode of inheritance combined with thus-far underpowered genetic studies contribute to the challenge of identifying and reproducing genes implicated in developmental stuttering susceptibility. We conducted a trans-ancestry genome-wide association study (GWAS) and meta-analysis of developmental stuttering in two primary datasets: The International Stuttering Project comprising 1,345 clinically ascertained cases from multiple global sites and 6,759 matched population controls from the biobank at Vanderbilt University Medical Center (VUMC), and 785 self-reported stuttering cases and 7,572 controls ascertained from The National Longitudinal Study of Adolescent to Adult Health (Add Health). Meta-analysis of these genome-wide association studies identified a genome-wide significant (GWS) signal for clinically reported developmental stuttering in the general population: a protective variant in the intronic or genic upstream region of SSUH2 (rs113284510, protective allele frequency = 7.49%, Z = −5.576, p = 2.46 × 10−8) that acts as an expression quantitative trait locus (eQTL) in esophagus-muscularis tissue by reducing its gene expression. In addition, we identified 15 loci reaching suggestive significance (p < 5 × 10−6). This foundational population-based genetic study of a common speech disorder reports the findings of a clinically ascertained study of developmental stuttering and highlights the need for further research.
Keywords: genome-wide assocation study, stuttering, population-study, complex trait
Despite overwhelming evidence in support of strong genetic influence, the genetic etiology of developmental stuttering has remained elusive. This study reveals genome-wide significant and suggestively significant signals for clinically ascertained stuttering cases in the general population, laying essential groundwork for further research into identification of common stuttering susceptibility variants.
Introduction
Speech and language represents an integral component of the human experience; we unite language (what we say) and speech (how we say it)1 to express our thoughts, feelings, and experiences with one another. Successful verbal communication requires coordination among neurological, cognitive, motor, and linguistic systems; dysregulation across or among any of these systems may result in disordered speech and language.2, 3, 4 Developmental stuttering is a common speech disorder with an onset between 2 and 5 years of age characterized by prolongations, blocks, and repetitions of speech sounds.5 Although most studies report a 5% lifetime prevalence of stuttering,2,6, 7, 8, 9 the true prevalence may be higher due to restrictive reporting criteria and poor subject selection,4,10 particularly since onset and recovery can be transient during early childhood.11 Individuals who are affected by the condition either stutter into adulthood (persistent stuttering) or stutter during early childhood but recover with the assistance of therapy, or spontaneously, typically before age 8 years (recovered stuttering). Persistent developmental stuttering afflicts approximately 1% of the adult population,4,12 which equates to more than 2.5 million adults afflicted with developmental stuttering in the United States.13
This common speech condition impacts the quality of life for many. Persistent stuttering has no known cure, and therapy for affected individuals often results in only a modest reduction in severity.14 Moreover, those who stutter frequently require a lifetime of therapy to manage the speech challenges as well as the psycho-social impact.15, 16, 17 Job performance and employability in adults who stutter can be affected, leading to substantial economic impacts.18, 19, 20 Despite extensive research on the psychological and economic consequences of this speech disorder, the etiology of developmental stuttering remains elusive. Current evidence postulates neurological,21 biological,22 and genetic underpinnings for stuttering,23, 24, 25, 26, 27, 28, 29, 30 though few causal associations have been identified to date. Even though multiple studies in the past few decades12,23,30, 31, 32, 33 evince a genetic predisposition for developmental stuttering, its genetic etiology and architecture largely remain evasive.
Family, twin, and segregation studies overwhelmingly support a strong genetic influence on stuttering risk; many individuals who stutter have a family member who also stutters.12 However, heritability estimates of developmental stuttering have varied widely across studies,7,34, 35, 36, 37, 38 with estimates ranging from 0.42 to 0.84 from the two largest twin studies, each comprising a sample size greater than 20,000 individuals. Although heritability estimates performed in twin studies of developmental stuttering point to genetic causes, such estimates also indicate the presence of environmental factors contributing to developmental stuttering. Monozygotic twin concordance rate estimates range from 38%–62% in these two studies.36,37 Nevertheless, many studies of other complex disorders (e.g., type 2 diabetes39,40 [MIM: 125853], serum lipid levels,41 Parkinson disease,42 and Alzheimer disease43,44 [MIM: 104300]) with similar or smaller heritability estimates have discovered genetic risk factors essential to understanding the molecular basis of the trait, suggesting that similar genetic study designs may offer key insights into the etiology of developmental stuttering.
To date, published literature investigating genetic contributions to developmental stuttering has primarily drawn on family-based analyses and studies of population isolates.23, 24, 25, 26, 27,29, 30, 31,33,45 Linkage and other family-based approaches have been successful at identifying rare and private causal variants with large genetic effects in the absence of genetic heterogeneity. For developmental stuttering, identifying the causal gene(s) within and across families has proven challenging. For example, in 2005 Riaz et al.24 performed linkage analyses in 46 consanguineous Pakistani families where stuttering occurred in at least two generations and diagnosis was confirmed independently by two different clinicians; they discovered a region on 12q23.3 linked with developmental stuttering in a single family without pinpointing an exact causal gene. Five years later in 2010, Kang et al.27 reported the results from a follow-up study of 77 unrelated Pakistani individuals who stutter plus unrelated cases from the same 46 Pakistani families interrogated by Riaz et al. in 2005;24 their investigation pinpointed three causal genes critical for the mannose-6-phosphate lysosomal targeting pathway: GNPTAB (MIM: 607840), GNPTG (MIM: 607838), and NAGPA (MIM: 607985). In 2018, Kazemi et al.46 performed Sanger sequencing and homozygosity mapping for 25 Iranian families afflicted by developmental stuttering and identified an additional 3 variants in GNPTAB and GNPTG that co-segregated with stuttering. Additional studies have revealed several regions across the genome linked with the trait but only identified three candidate risk genes: DRD231 (MIM: 126450), AP4E133 (MIM: 607244), and CYP17A130 (MIM: 609300). Lan et al.31 performed an association study focusing specifically on dopaminergic gene haplotypes and allele frequencies among SNPs in the Han Chinese population and identified risk and protective alleles in DRD2. These results were not replicated in 2011 by Kang et al.32 in a case-control cohort from Brazil and western Europe. In 2015, Raza et al.33 used whole-exome sequencing to identify two heterozygous AP4E1 coding variants that co-segregated with persistent developmental stuttering in a large Cameroonian family (the same polygamous family as published in their earlier work from 201347); they also observed these same two variants in unrelated Cameroonians with persistent stuttering. Although Raza et al.33 also reported 23 additional rare variants (including loss-of-function variants) within AP4E1 among unrelated stuttering individuals from Cameroon, Pakistan, and North America, their findings have yet to be replicated by another group. In 2017, Mohammadi et al.30 performed a case-control study of the Kurdish population aged 3 to 9 years from Western Iran, specifically focusing on the dimorphic nature of stuttering, and identified an allelic polymorphism associated with stuttering susceptibility in CYP17A1, a gene integral for the synthesis of steroid hormones. As reported by Frigerio Domingues et al.48 in 2019, these results were not replicated in an independent case- and population-matched control association study from the United States, Brazil, Pakistan, and Cameroon. Despite these efforts, the molecular pathophysiology of developmental stuttering in general populations remains obscure, in part due to the dearth of studies exploring common genetic risk factors in unrelated individuals and the lack of consensus across studies.
The International Stuttering Project (ISP) was formed to represent global outbred populations of individuals who stutter, specifically to illuminate genetic etiology and broaden investigations of its diverse and variable phenotype (see Web resources). Given the success of investigations for heritable complex diseases, genome-wide association analyses of developmental stuttering are poised to provide insights into its molecular basis. Moreover, prior investigations into the genetics underlying developmental stuttering have comprised samples and study designs ill-equipped to detect common variant effects or reconcile genetic heterogeneity. Our study accommodates both. Here, we accrued a global and multiethnic clinically ascertained developmental stuttering case set through the ISP and report genome-wide significant (GWS) findings in a meta-analysis study of developmental stuttering.
Material and methods
Studies
The multiethnic genome-wide association study (GWAS) meta-analysis included studies with genotype data from clinically ascertained individuals with developmental stuttering from the ISP and their sex- and ancestry-matched control subjects (n = 8,104; n cases = 1,345) and summary statistics (n = 8,357; n cases = 785) from The National Longitudinal Study of Adolescent to Adult Health (Add Health). The ISP comprises 1,345 clinically ascertained developmental stuttering cases collected from the Curtin Stuttering Treatment Clinic in Perth, Australia; the SpeechMatters Clinic and the Irish Stammering Association, in Dublin, Ireland; the National Stuttering Association, USA; online recruitment on reddit.com; and Dr. Shelly Jo Kraft’s research group at Wayne State University (Table 1). Stuttering status was confirmed in all affected individuals by a speech pathologist with expertise in fluency disorders. Up to five ancestry- and sex-matched population-based control subjects per affected individual were drawn from BioVU (n control subjects = 6,759; Table 1), Vanderbilt University Medical Center’s (VUMC’s) electronic health record (EHR)-linked biobank: 49 of the 6,759 control subjects included genotyped unaffected family members of affected individuals. Vanderbilt University Medical Center has recruited and consented individuals to join BioVU since February 2007.49,50 The electronic health record at Vanderbilt University Medical Center offers de-identified demographic data, clinical notes, electronic orders, laboratory measurements, ICD-9 CM/ICD-10 disease diagnosis codes, and CPT codes. Using electronic health records, individuals with diagnoses of developmental, speech, or language disorders as identified via ICD-9 and ICD-10 codes (Table S1) or a phenome risk classifier,51,52 and individuals under age 18 years were excluded as potential control subjects. To select ancestry-matched control subjects, we calculated eigenvectors and eigenvalues through principal component analysis (PCA) run on PLINKv.1.90.53 PCA was performed on the maximally unrelated set of affected individuals and potential control subjects ( < 0.09375, as identified by PRIMUS54,55) using a panel of SNPs in low linkage disequilibrium (LD); additional related affected individuals and potential control subjects were projected along each of the calculated eigenvectors. Pairwise Euclidean distance between each affected individual and potential control subject was calculated using principal components to identify the control subject with the smallest Euclidean pairwise distance for each affected individual.56 This control subject selection method included outlier pruning, which removed any potential control subjects with a pairwise distance more than 2 standard deviations away from the average pairwise distance. Control subject selection also matched according to sex. Age information was not available for 171 samples and therefore was excluded as a covariate during control subject selection.
Table 1.
Demographic distribution for subjects used in genome-wide association analyses
| ISP GWAS | Add Health GWAS | ||||
|---|---|---|---|---|---|
| Cases | Controls | Cases | Controls | ||
| Total | 1,345 | 6,759 | Total | 785 | 7,572 |
| n (%) | n (%) | ||||
| Male | 965 (71.7) | 4,780 (70.7) | Male | 446 (56.8) | 3,419 (45.2) |
| Female | 380 (28.3) | 1,979 (29.3) | Female | 339 (43.2) | 4,153 (54.8) |
| Ancestry | Ancestry | ||||
| African | 68 (5.1) | 388 (5.7) | Non-Hispanic Black | 182 (23.2) | 1,522 (20.1) |
| Hispanic | 38 (2.8) | 131 (1.9) | Hispanic | 122 (15.5) | 1,055 (13.9) |
| East Asian | 42 (3.1) | 113 (1.7) | Asian | 44 (5.6) | 404 (5.3) |
| European | 1,132 (84.2) | 5,875 (86.9) | Non-Hispanic white | 433 (55.2) | 4,559 (60.2) |
| South Asian | 44 (3.3) | 143 (2.1) | Native American | 4 (0.5) | 32 (0.4) |
| Other/mixed | 21 (1.6) | 109 (1.6) | |||
| Age, years (std) | 28.44 (1.77) | 28.52 (1.81) | |||
For the ISP analysis, ancestry was determined through principal component analysis, and approximately 5 cases were selected for each case, matching on ancestry and sex. For the Add Health GWAS, ancestry was determined through principal component analysis, and affection status was self-reported by each subject.
ISP studies (protocol 0225119MP2E) were approved by Wayne State University’s Committee for the Protection of Human Subjects. The studies were explained to all participants and written informed consent obtained. Genotyping services were provided by Vanderbilt Technologies for Advanced Genomics (VANTAGE). Analysis of deidentified ISP and BioVU data in this study was approved under an institutional review board (IRB) exemption by Vanderbilt University’s Committee for the Projection of Human Subjects (IRB #180583). Use of BioVU data was approved by the Vanderbilt Institute for Clinical and Translational Research (BV247, BV247_A1).
Add Health represents an ongoing, nationally representative, longitudinal study of the social, behavioral, and biological factors influencing health and developmental trajectories from early adolescence into adulthood.57 Add Health collected demographic and health survey data as well as in-home physical and biological data from participants. For our study, 785 self-reported stuttering cases were defined as participants who at one point answered “yes” to the following survey question: “Do you have a problem with stuttering or stammering?” For control subjects, 7,572 participants were included in the analysis. All control individuals answered “no” to the above question and did not mark “delayed speech or other problems with speaking or understanding” or “I don’t know” to the same query across all study visits.
Genotyping, quality control, and imputation
The ISP cases and controls were genotyped using the Illumina Expanded Multi-Ethnic Genotyping Array (MEGAex) at Vanderbilt University Medical Center’s core facility, VANTAGE. Sample and variant filtering for quality control was performed using PLINKv.1.90;53 initial filtering thresholds for the control cohort excluded variants with a call rate less than 98% and samples with a call rate less than 97%. Initial filtering for stuttering cases excluded variants and samples with a call rate less than 90%. Duplicate variants and indels (insertions and deletions) were removed as well as any duplicate samples (the duplicate sample with a lower call rate was removed) in both cases and controls. We applied the methods described by Pluzhnikov et al.58 to identify possible plate or batch effects prior to merging genotype batches. Cases were separately assessed for quality control by ancestry group (European, Admixed American, African, East Asian), assigned using principal components calculated after merging the case samples with HapMap359 reference data for ancestry classification. Each ancestry group was subsequently analyzed, applying a group-specific minor allele filter of 1%, variant missingness filter of 3%, sample missingness filter of 5%, as well as checks for heterozygosity, sex, and variants that deviated strongly from Hardy-Weinberg equilibrium (HWE) (variants with a HWE p < 1 × 10−15 were removed). Cases were then merged with their selected matched controls for imputation according to standard protocols and specifications outlined for the TOPMed Imputation Server, including using William Rayner’s pre-imputation data preparation toolkit (see Web resources).60 Relatedness checks for cases and controls were performed using PRIMUS.54 All autosomal chromosomes were imputed on the TOPMed Imputation Server using EAGLE_v.2.4 phasing,61 Minimac4 imputation,62 and the TOPMed reference.63,64 Post-imputation quality control filtering included removal of variants with a minor allele frequency (MAF) less than 1%, imputation r2 less than 0.4, or with an effective n (neff = 2 × (MAF) × (1 − MAF) × n × r2) less than 30.
In Add Health, data were genotyped on the Illumina Omni Quad 1 and 2.5 and imputed on the Michigan Imputation Server using Minimac262 and the 1000 Genomes (Phase3v.5) reference.63,65 Post-imputation quality control filtering included selecting variants with a MAF above 1%, as well as removing all variants with an imputation r2 less than 0.4 or with an effective n less than 30.
Experimental workflow is depicted in graphical format in Figure S1.
Statistical analysis
In the clinically ascertained developmental stuttering set, ∼9 million imputed variants (Figure S2; Table S2) were analyzed for association with stuttering risk using a frequency-based additive logistic mixed model via SAIGE,66 a method applied and developed for biobank data in order to accommodate imbalanced case-control ratios and sample relatedness. Association analysis corrected for population substructure by using six trait-associated principal components capturing genetic ancestry as covariates.67
In Add Health, ∼9 million imputed variants (Figure S3; Table S3) were analyzed for association with stuttering status using a frequency-based additive logistic model via SUGEN.68 Model covariates included ten ancestry-associated principal components and age.
Meta-analysis
Meta-analysis was performed combining result of the ISP and Add Health studies across 7,275,796 variants imputed in both datasets. Summary statistics (direction of effect and observed p value) from each contributing GWAS were combined in each study to calculate a signed Z-score using METAL.69 The sample size scheme was used in this meta-analysis, since effect size estimates and standard errors were not equivalent between each GWAS. Annotated associations from study-specific and meta-analyses were variants that reached genome-wide significance (p < 5 × 10−8) or were suggestive (p < 5 × 10−6). Variants were aligned to human genome reference build 38. The genome-wide significance threshold of p < 5 × 10−8 was set according to field standards.70 This threshold uses a Bonferroni correction where α = 0.05 and assumes there are approximately 1 million independent (i.e., not in linkage disequilibrium) common signals across the human genome. The suggestive threshold of p < 5 × 10−6 assumes an expectation of one false-positive association per GWAS (i.e., 1/total number of independent SNPs).71,72
Annotation
We annotated top associated variants from study-specific and meta-analyses using ANNOVAR.73 The data accessed from the Genotype Tissue Expression (GTEx)74 portal in July 2021 derive from the following version: dbGaP: phs000424.v8.p2.
Genetic heritability calculation
Genome-wide SNP-based liability scaled heritability within our ISP set was calculated through a genomic-relatedness-based restricted maximum-likelihood (GREML) approach implemented through GCTA software.75,76 Observed variance estimates from the observed scale were transformed to an expected underlying scale with an expected population prevalence being set to 0.01 based conservatively on estimated prevalence among the general adult population.4 Heritability estimates included 485,698 genotyped variants that passed all quality control metrics prior to imputation (see Genotyping, quality control, and imputation) among the PRIMUS54,55-identified maximum unrelated set (up to third degree), which included 7,768 individuals (1,095 cases). We corrected for individual sex and the first six principal components (see Genotyping, quality control, and imputation).
Functional analyses
We performed a Bayesian colocalization analysis between our Add Health-ISP meta-analysis top hits and tissue-specific eQTL signals from GTEx v.8 data74 using fast enrichment aided colocalization analysis (fastENLOC77,78). We looked for colocalization solely for meta-analysis regions with a variant identified as a top hit (Table 2). Evaluated regions in the meta-analysis included all sentinel variants as well as any other variants found in the same LD block in addition to any variants nearby (gene <250 kb upstream or downstream of the sentinel variant) an identified protein-coding gene. LD blocks in the meta-analysis data were defined according to European-based LD calculated from 1000 Genomes phase 1 data by Berisa and Pickrell.79 Colocalization analysis was tissue-specific and included all stuttering-relevant tissues available in GTEx v.8 (skeletal muscle, pituitary, minor salivary gland, all esophageal tissues, and all brain tissues). We reported the results of any colocalization signal with a regional colocalization probability (RCP) (i.e., the probability that one of two SNPs in an LD block is responsible for a genuine association) ≥ 0.05 (Table S6).
Table 2.
Top hits from Add Health and ISP meta-analysis
| rsID |
CHR |
Position |
Effect allele |
Other allele |
EAF |
Z score |
p value |
Nearest gene |
Location |
|---|---|---|---|---|---|---|---|---|---|
| rs113284510a | 3 | 8683501 | T | C | 0.0749 | −5.576 | 2.46E−08 | SSUH2 | intronic or genic upstream transcript |
| rs34919320 | 1 | 217480104 | G | A | 0.2602 | 5.128 | 2.93E−07 | GPATCH2 | intronic |
| rs58528263 | 4 | 17105854 | G | T | 0.249 | 5.102 | 3.37E−07 | LDB2 | 207 kb downstream |
| rs2938894 | 9 | 78480092 | A | T | 0.030 | −5.004 | 5.63E−07 | PSAT1 | 150 kb downstream |
| rs1011275 | 2 | 53611701 | G | T | 0.2393 | 4.804 | 1.55E−06 | ASB3 | 58 kb upstream |
| rs4282275 | 5 | 54451477 | A | G | 0.1347 | 4.783 | 1.72E−06 | HSPB3 | 4 kb upstream |
| rs6547085 | 2 | 76410210 | G | A | 0.0532 | 4.752 | 2.01E−06 | NA | NA |
| rs16855942 | 4 | 43363575 | A | G | 0.3616 | −4.685 | 2.79E−06 | NA | NA |
| rs10994385 | 10 | 46038901 | C | G | 0.3678 | −4.661 | 3.14E−06 | MSMB | intronic |
| rs35612603 | 2 | 98486856 | A | G | 0.1321 | 4.659 | 3.18E−06 | INPP4A | intronic |
| rs16954038 | 15 | 70000732 | G | C | 0.0371 | 4.64 | 3.48E−06 | TLE3 | 47 kb upstream |
| rs11158418 | 14 | 62490392 | T | A | 0.4699 | 4.633 | 3.60E−06 | KCNH5 | 209 kb upstream |
| rs1446110 | 2 | 80290628 | C | A | 0.3264 | −4.622 | 3.79E−06 | CTNNA2 | intronic |
| rs10779884 | 2 | 112130529 | A | G | 0.4826 | 4.62 | 3.83E−06 | FBLN7 | 8 kb upstream |
| rs115327327 | 2 | 61440832 | T | A | 0.2722 | 4.574 | 4.78E−06 | USP34 | intronic |
| rs111962436 | 2 | 223438584 | A | T | 0.0261 | −4.571 | 4.85E−06 | SCG2 | 158 kb upstream |
Genome-wide association summary statistics from Add Health and ISP stuttering studies meta-analyzed using METAL. Sentinel variants from loci with p < 5 × 10−6 reported along with nearest gene annotation. NA (not available) reported for variants where the nearest protein-coding gene was more than 250 kb away (either upstream or downstream according to UCSC reference genome browser). Base-pair positions listed according to human genome reference build 38.
Variant represents a locus that reached genome-wide significance (p < 5 × 10−8).
We performed gene ontology analysis for the top 100 genes associated with variant signals in our meta-analysis. Our top signals were annotated with the Open Targets Genetics “Variant-to-Gene” (V2G) pipeline, which integrates evidence from four main data types (molecular phenotype quantitative trait loci, chromatin interactions, in silico functional predictions from Ensembl, and distance between the variant and each gene’s canonical transcription start site) to assign the most likely functional gene for each variant.80 Next, we used clusterProfiler81,82 to perform a false discovery rate (FDR)-corrected enrichment test for gene ontology terms among our identified top 100 genes. We also performed an enrichment test for gene modules using our identified top 100 genes to determine if any sets of highly correlated genes (gene modules) were associated with stuttering risk. Gene co-expression networks comprised groups of functionally related genes or “modules” Gerring et al.83 identified from GTEx v.7 tissue gene expression data. Module enrichment reported for any gene tissue-specific analysis with a raw p value < 0.05 among stuttering-relevant tissues (skeletal muscle, pituitary, minor salivary gland, all esophageal tissues, and all brain tissues). We performed a competitive gene pathway analysis for reported module enrichments using g:Profiler and subsequently annotated the outputted biological pathways (Table S7).
Power calculation
We calculated our power to detect significant stuttering risk associations across a range of disease allele frequencies for our meta-analysis comprising 2,130 stuttering cases and 14,331 controls. We estimated power assuming a two-sided hypothesis test at p < 5 × 10−8, an additive model, and using a developmental stuttering prevalence of 1%. Calculations were performed using the University of Michigan’s Genetic Association (GAS) Power Calculator.84
Replication for published implicated stuttering genes
We manually reviewed over 200 records on PubMed via the National Center for Biotechnology website for publications in the past 21 years (2000–2021) that mentioned “stuttering” in the title field. Much of the published stuttering literature23, 24, 25, 26,28,29,45,47 implicated large genome regions from linkage studies in families, without determining a specific causal gene. We sought replication for the six genes that have been previously implicated in the stuttering literature27,30,31,33 (Table S5) by evaluating all variants that passed our QC metrics within each gene in our meta-analyzed GWAS. To determine the effective number of tests for each gene, we calculated r2 between each SNP pair within a gene using PLINKv.1.90.53 SNPs that had an r2 > 0.4 were considered to be in linkage disequilibrium. The effective number of tests used for our Bonferroni correction represented the number of independent tag SNPs in each gene with pairwise r2 < 0.4. Results were Bonferroni corrected for the effective number of tests in each gene and the variant with the minimum p value within each gene is reported (Table S5).
Results
Meta-analyzed GWAS
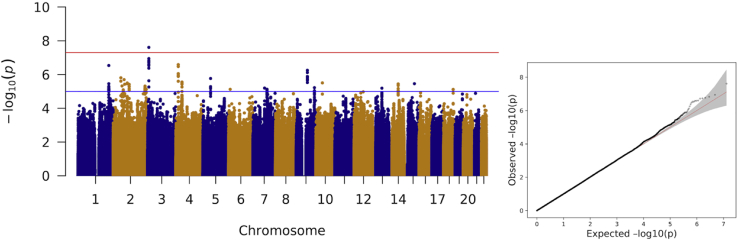
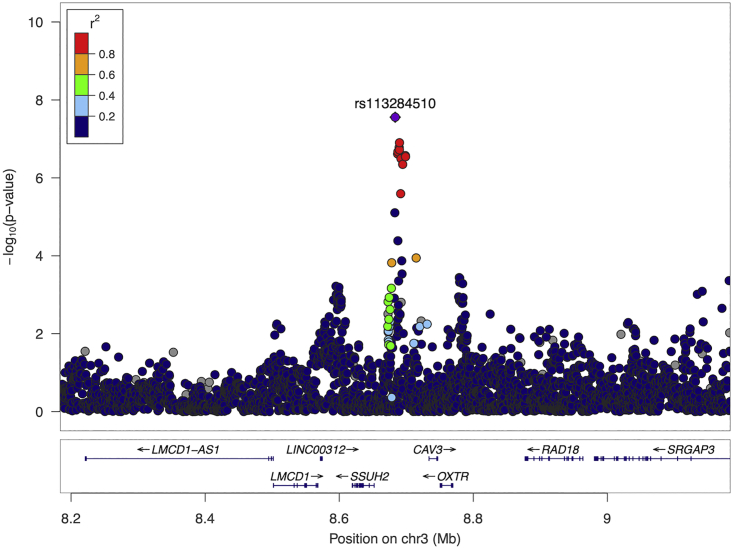
Genome-wide association analyses of stuttering were carried out in 8,104 individuals (1,345 cases) from the ISP study and 8,357 individuals (785 cases) (Table 1) from a self-reported stuttering study, with ∼7.3 million overlapping variants tested. No evidence for residual population stratification or systematic technical artifact was observed in either individual dataset or the meta-analysis. The genomic inflation factor, λ, was 1.0173 (Figure S2) in the ISP GWAS and 1.0161 in the Add Health GWAS (Figure S3). The genomic inflation factor for the meta-analysis was λ = 0.9977 (Figure 1). In the meta-analysis, one genome-wide significant association was observed at rs113284510 (Z = −5.576, p = 2.46 × 10−8). The variant, rs113284510, occurred in either an intronic region or genic upstream region of SSUH2, (MIM: 617479) (Figure 2) depending on the transcript. This variant exhibited consistent direction of effect (p < 5 × 10−6) in the Add Health GWAS (p = 2.23 × 10−7, odds ratio [OR] = 0.455 [0.320–0.591]) and in the ISP GWAS (p = 0.0059, OR = 0.754 [0.617–0.922]) (Table S2). The frequency of the protective effect allele (T) for rs113284510 was 7.49% overall (7.08% in the ISP GWAS and 7.88% in the Add Health GWAS) (Table S2).
Figure 1.
Manhattan and Q-Q plot for meta-analysis of Add Health and ISP stuttering studies
Meta-analysis included 16,461 samples and 7,275,796 variants present in both datasets; variants not present in both datasets were excluded. One locus reached genome-wide significance (red line p < 5 × 10−8); fifteen loci reached suggestive genome-wide significance (blue line p < 5 × 10−6). Q-Q plot x axis represents expected −log10(p) and the y axis represents observed −log10(p).
Figure 2.
Locus zoom plot of rs113284510
Locus zoom plot of meta-analysis stuttering associations with surrounding variants (color coded by r2 bin) and the sentinel variant (denoted by purple diamond) using EUR linkage disequilibrium (LD) generated from 1000 Genomes EUR reference. The x axis represents chromosome position (hg38) with annotated genes found within the region, the y axis represents −log10 (p value) of the association between the genetic variant and stuttering. Sentinel variant is located in either an intronic or genic upstream region of SSUH2.
In the meta-analysis, the index variants for an additional 15 associations reaching a suggestive genome-wide significance threshold of p < 5 × 10−6 are presented in Table 2. No genome-wide significant associations were observed in either the ISP or Add Health GWAS; however, 19 variants reached our suggestive (p < 5 × 10−6) significance threshold for the ISP GWAS (Table S3), and 24 variants reached this same suggestive threshold in the Add Health GWAS (Table S4).
Genetic heritability
We calculated SNP-based liability scaled heritability within our unrelated ISP sample through GCTA.75,76 The proportion of phenotypic variance explained by the genetic factors was reported at 0.791 (SE = 0.043). Through GCTA we also transformed the explained variance estimates from the observed scale to the underlying liability scale, accounting for an expected case prevalence of 0.01. Liability scaled heritability was 0.902 (SE = 0.049).
Functional analyses
Our colocalization analysis identified three regions in our stuttering meta-analysis showing weak association (regional colocalization probability, 0.1 > RCP ≥ 0.05) between cis-eQTLs in GTEx v.8: chr2: 111630529–112630529, chr2: 60940832–6194083, and chr2: 97986856–98986856 (Table S6). In the chr2: 111630529–112630529 region, the lead SNP, rs10779884, was identified as a top hit in our meta-analysis (Table 2) and serves as an eQTL in FBLN7 (MIM: 611551) within muscle skeletal, esophagus mucosa, and brain hypothalamus tissues. The chr2: 60940832–6194083 did not colocalize with any eQTLS for protein coding genes and chr2: 97986856–98986856 region identified rs140321250 as the lead SNP, predicted to act as an eQTL for INPP4A (MIM: 600916) in esophagus mucosa tissue (Table S6).
We did not observe any significant (p < 0.05 after FDR correction) enrichment for gene ontology terms among the top 100 genes identified in our meta-analysis. We observed one significant GTEx tissue-specific enrichment83 for a gene module in the minor salivary gland (FDR-corrected p = 6.63 × 10−3) with biological pathways implicated in processes such as extracellular matrix and structure organization, cell adhesion, anatomical structure development, nervous system development, ossification, neurogenesis, cell migration, and bone morphogenesis (Table S7). The nearest gene to the identified genome-wide significant hit (rs113284510), SSUH2, was found in this gene module as well as the FBLN7 gene near another top variant hit (rs10779884) (Table 2). We did not observe any additional significant GTEx tissue-specific gene module enrichments.
Replication analysis of implicated stuttering genes from the literature
To determine whether genetic contributions observed in families and population isolates might replicate in a population-based analysis, we assessed our data for replication of six genes that have previously been implicated in the stuttering literature:27,30,31,33 DRD2, GNTAB, GNPTG, NAGPA, AP4E1, and CYP17A1 (Table S5). We reported the lowest p value observed in our study in imputed variants within the exonic and intronic region for each gene, as well as the Bonferroni corrected p value for each top signal, based on the effective number of tests in that gene. None of the variants measured in our GWAS meta-analysis for these six genes reached statistical significance (p < 0.05) after Bonferroni correction; however, two variants neared statistical significance after Bonferroni correction: rs761057 (intron of GNPTG; p = 0.105; risk allele [T] frequency 9.9%) and rs4919687 (intron of CYP17A1; p = 0.100; protective allele [A] frequency 27%) (Table S5).
Discussion
Our multiethnic GWAS meta-analysis of stuttering in men and women of European, Hispanic, Asian, and African American ancestry led to the identification of one genome-wide significant protective risk locus. The protective T allele for the index variant, rs113284510, occurred within either an intronic or genic upstream region of SSUH2, a gene previously reported to play a major role in odontogenesis. A missense mutation in SSUH2 was shown to disrupt protein structure and production, causing autosomal-dominant dentin dysplasia type I (MIM: 125400) in a large Chinese family.85 Interestingly, a different top meta-analysis locus, rs10779884 (Table 2), is found approximately 8 kb upstream of the FBLN7 transcription start site. FBLN7 encodes a protein that interacts with extracellular matrix molecules in developing teeth and may play important roles in differentiation and maintenance of odontoblasts and dentin formation.86 Moreover, in the GTEx database,74 rs10779884 acts as an eQTL modulating FBLN7 expression across several tissues, including arteries, adipose, tibial nerve, skin, breast, skeletal muscle, heart, esophagus, pancreas, colon, and brain. Interestingly, both these genes (SSUH2 and FBLN7) appeared in a GTEx tissue-specific gene module enriched in the minor salivary gland (Table S7). This enriched gene module also included RELN (MIM: 600514), a gene that encodes a glycoprotein produced within the developing brain. RELN has been implicated in neural traits such as autism spectrum disorder (ASD) (MIM: 209850)87 as well as volumetric brain measures.88 Notably, a recent paper by Peter et al.89 identified likely deleterious variants in RELN inherited by two siblings affected with both ASD and childhood apraxia of speech, suggesting pleiotropic effects for RELN. A pathway analysis showed that genes in this module are involved in a myriad of biological processes such as extracellular matrix organization, nervous system development, neurogenesis, cell migration, and bone morphogenesis. This intriguing analysis provides preliminary support that genes with roles in structural organization and various neural processes might play a role in developmental stuttering risk.
Further investigation of the genome-wide significant sentinel variant, rs113284510, in GTEx74 showed that it acts as an eQTL specifically in esophagus-muscularis tissue by reducing SSUH2 expression in the presence of the protective T allele. This function might in part be explained by its genic upstream position to SSUH2. A review of the GWAS literature also shows suggestive significance (p < 5 × 10−6) for variants located in SSUH2 with ASD.90 ASD is a neurodevelopmental disorder that presents with a gradual or sudden early childhood onset, similar to developmental stuttering. Individuals with ASD exhibit impaired social interaction skills and, in moderate to severe cases, have little to no speech production beyond basic vocalizations. A possible shared genetic liability between ASD and developmental stuttering has not been published; however, disordered speech in ASD was found to be associated with mutations in the FOXP291 (MIM: 605317) and CNTNAP2 (MIM: 604569) genes.92,93 Although not yet specifically implicated in persistent developmental stuttering,94 these genes have known associations with a broad umbrella of speech and language disorders, including developmental verbal dyspraxia and developmental language disorder.95
GTEx74 also shows that the genome-wide significant sentinel variant, rs113284510, acts as an eQTL in tibial artery tissue by increasing CAV3 (MIM: 601253) expression in the presence of the protective T allele. CAV3 encodes instructions for making the caveolin-3 protein, which is found in the membrane surrounding muscle cells; caveolin-3 may also help regulate calcium levels in muscle cells. As such, genetic changes in CAV3 have been implicated in various health conditions with impaired muscle function, such as rippling muscle disease, limb-girdle muscular dystrophy, and hypertrophic cardiomyopathy.96, 97, 98, 99, 100, 101 The rs113284510 variant seems to regulate the expression of two unique genes, CAV3 and SSUH2; investigating pleiotropic effects may help unravel the potential role of these genes in modulating developmental stuttering risk.
Among our meta-analysis top hits (Table 2), we observed nine variants suggestively associated with stuttering risk (p < 5 × 10−6) of obscure functional consequence (rs58528263, rs2938894, rs1011275, rs4282275, rs6547085, rs16855942, rs16954038, rs11158418, and rs111962436). Our gene ontology and GTEx tissue-specific gene module enrichment analysis did not provide any additional illumination for these associations.
For the other six suggestively associated loci, our GTEx,74 Open Targets Genetics Portal,80 and GWAS catalog102 searches uncovered initial clues as to their possible function. For example, our GWAS meta-analysis identified a risk locus that neared genome-wide significance (rs34919320, p = 2.93 × 10−7) in an intronic region of GPATCH2 (MIM: 616836), a gene that encodes a nuclear factor that plays a role in spermatogenesis and tumor growth during breast cancer.103 Investigation of rs34919320 in GTEx74 also supports a possible role for GPATCH2 in spermatogenesis, showing that the risk allele G acts as an eQTL in testis tissue by reducing GPATCH2 expression. Another top hit (rs115327327, p = 4.78 × 10−6) is found in an intronic region of USP34 (MIM: 615295), which encodes a ubiquitin protein. In a large study of testosterone and related sex hormones in 425,097 UK Biobank participants, other intronic variants in USP34 were associated with sex hormone-binding globulin (SHBG).104 SHBG controls how much testosterone, dihydrotestosterone, and estradiol are delivered throughout the body; however, the SHBG blood test is primarily used to determine testosterone levels (see Web resources).105 These findings motivate further exploration into population-based genetic effects that might contribute to the sexually dimorphic nature of stuttering. At stuttering onset, the male-to-female ratio is more even (between 1:1 and 2:18); however, females are more likely to recover from stuttering, changing the male-to-female ratio to 4:1, as observed in adults.4 The mechanisms for this observed sex discrepancy are not well understood, and initial clues into possible causal genetics are unclear. A paper by Mohammadi et al.30 provides preliminary insight: they measured testosterone levels in children who stutter and their control counterparts and found higher levels of testosterone and its metabolites in children who stutter. Observed stuttering susceptibility was also reported in association with CYP17A1,30 a gene that encodes the instructions to make an enzyme involved in steroid hormone synthesis. Interestingly, work by Anthoni et al.106 has also shown that variants in CYP19A1 (MIM: 609300), a gene in the same cytochrome P450 family as CYP17A1, are associated with quantitative measures of language and speech, such as phonological processing and oral motor skills. These results combined with our associations suggest that variants involved in the regulation of sex hormones may contribute to stuttering risk. Follow-up analysis to determine if these associations reach genome-wide significance and replicate in an independent dataset is warranted, particularly in a larger dataset powered to perform sex-stratified analyses.
Another intriguing association with stuttering susceptibility, included the identification of a protective variant in an intronic region of CTNNA2 (MIM: 114025) (rs1446110, p = 3.79 × 10−6), which encodes Catenin alpha-2 protein. Catenin alpha-2 plays a critical role in cortical neuronal migration and neurite growth.107 Another identified protective variant (rs10994385, p = 3.14 × 10−6) occurred in an intronic region of MSMB (MIM: 157145), a gene that encodes a member of the immunoglobulin binding factor and is synthesized by prostate epithelial cells.108,109 Other variants in MSMB have shown association with prostate cancer.110,111 Finally, an identified risk variant, rs35612603, occurred in an intronic region of INPP4A, a gene that encodes a Mg2+ enzyme to hydrolyze the 4-position of the inositol ring.112 In the GWAS Catalog summary statistics repository,102 the top five traits associated with variants within INPP4A included: use of prednisolone medication, time employed in current main job, unspecified personality disorders, hypopituitarism, and brain cancer/tumor. Although these initial associations lay a foundation for possible common developmental stuttering susceptibility variants, future replication analyses to determine if these associations reach genome-wide significance and replicate in independent datasets will be integral to the design of functional validation analyses.
As an initial investigation into possible shared genetic contributions between familial developmental stuttering and the risk of developmental stuttering in the general population, we performed a replication analysis for six published stuttering risk genes (Table S5). Since the observed causal variants in these family-based studies27,30,31,33 were not directly measured on our arrays, were too rare to impute with accuracy, and were too rare to estimate robust effects given our sample size, we instead looked for any potential effects from common variants in and around these genes. After performing locus-based Bonferroni correction for the number of independent tests in each locus, we did not identify any significant common variant effects in these six genes, thus suggesting that the genetic architecture detected to date in families highly enriched with individuals who stutter is largely distinct from the common genetic drivers of stuttering in general populations. However, we did observe common variant signals that neared nominal statistical significance after regional test correction at GNPTG (rs761057; p = 0.105) and CYP17A1 (rs4919687; p = 0.100), suggesting that studies with greater sample size and improved power may identify shared familial and population genetic contributions for these stuttering risk genes. Interestingly, estimation of observed trait heritability in our clinically ascertained subset (ISP dataset) was similar to heritability calculations in several twin studies34,37,38 of developmental stuttering (h2 = 0.791, SE = 0.043), providing evidence that genetic factors for developmental stuttering at a population level exist. Another postulation is that common and rare variation act additively to create risk in developmental stuttering, as was observed in a recent study of the genetics underlying ASD. The authors developed a polygenic transmission disequilibrium test (pTDT) and demonstrated that common and rare variation act additively in ASD.113 As this study is underpowered to detect effects of rare variants (see Figure S4 for power curve), future investigations performing a pTDT, using the data presented here, to create a polygenic risk score capturing common genetic stuttering liability are warranted.
This study has a few potential limitations, the most significant of which is the sample size. Our sample of a little over 2,000 cases is not sufficient to identify definitive developmental stuttering susceptibility variants, especially if a myriad of common variants of very small effect prove to impact its liability (Figure S4). Relatedly, our study has insufficient power for stratified analyses examining additional clinical variables of interest such as sex or recovery status (persistence). This study also lacked a sufficient sample size to divide the data into a training and testing set for polygenic risk score development; for example; PRS-CS recommends tens of thousands of cases for their approach.114 Akin to other neurologic polygenic traits such as Tourette syndrome115,116 (MIM: 137580), ASD,117,118 and schizophrenia (MIM: 181500), establishing and independently validating common variant trait liability to stuttering will most likely require studies of tens of thousands of subjects.119 Furthermore, replicating the results herein is critical. Although our study identified rs113284510 as significantly associated with stuttering in our meta-analysis, both the strength of association and quality of imputation (ISP: beta = −0.282 and INFO = 0.860; Add Health: beta = −0.787 and INFO 0.478) for this SNP varied in each contributing study (Table S2). Furthermore, the exact biologic context of this association remains obscure. Our functional analyses implicated the variant, rs113284510, in SSUH2, which our GTEx tissue-specific gene module enrichment test identified as enriched for a gene module in the minor salivary gland (Table S7). Although our pathway analysis showed that genes in this module are involved in processes such as nervous system development and neurogenesis, the module was not significantly enriched within any brain tissues. This lack of observed enrichment in brain tissues might be a model limitation. Furthermore, gene modules were built using adult GTEx data, and it has been hypothesized that genes influencing other speech traits, such as childhood apraxia of speech, are likely expressed during prenatal and early postnatal brains.120 Since our modules relied on adult brain data, we could be missing relevant mechanistic correlations with our genetic findings. Nonetheless, the fact that our stuttering-associated variant and genes appear to have neural functions suggests a role for these genes that warrants future study. Finally, despite the success of this study in identifying both genome-wide significant and suggestively significant signals for stuttering in the general population, we anticipate that the power limitations of our presented study can be resolved by substantial increases in the number of stuttering cases collected for GWAS and inclusion of diverse ancestries.
Most importantly, this study lays necessary groundwork for the identification of additional common developmental stuttering susceptibility variants in larger population-wide cohorts and helps to provide a more complete understanding of the full genetic architecture for this common speech condition, with the potential of uncovering etiology, pathophysiology, and eventual therapeutic targets.
Acknowledgments
This research was supported by National Institutes of Health (NIH) grants from the National Institute on Deafness and Other Communication Disorders (NIDCD) to Wayne State University (1R03DC015329-01) and to Vanderbilt University Medical Center and Wayne State University (R01DC017175). The control samples used for the described clinical GWAS were obtained from Vanderbilt University Medical Center’s BioVU, which is supported by numerous sources: institutional funding, private agencies, and federal grants. These include the NIH-funded Shared Instrumentation Grant S10RR025141 and CTSA grants UL1TR002243, UL1TR000445, and UL1RR024975. Genomic data are also supported by investigator-lead projects that include U01HG004798, R01NS032830, RC2GM092618, P50GM115305, U01HG006378, U19HL065962, and R01HD074711 and additional funding sources listed at the BioVU Funding Acknowledgment (see web resources). H.M.H. was funded by NHLBI training grants T32 HL007055 and T32 HL129982 and ADA grant 1-19-PDF-045. R.M.J. was funded by NIH grant 5R21DC016723-03. We greatly thank every individual who donated DNA to this project in Ireland, Australia, and the United States. This research was made possible due to their participation and support.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xhgg.2021.100073.
Contributor Information
Jennifer E. Below, Email: jennifer.e.below@vanderbilt.edu.
Shelly Jo Kraft, Email: kraft@wayne.edu.
Data and code availability
All summary statistics genome-wide will be available in the GWAS Catalog (Study Accession: GCST90085698 ). ISP GWAS data have not yet been deposited in dbGaP but will be made available to qualified investigators in dbGaP. The Add Health data can be accessed through the Add Health Study. Details are available through the Carolina Population Center (see web resources).
Web resources
1000 Genomes Project, https://www.internationalgenome.org/
BioVU Funding, https://victr.vumc.org/biovu-funding/
Carolina Population Center, HYPERLINK "" Error! Hyperlink reference not valid.https://addhealth.cpc.unc.edu/documentation
g:Profiler, https://biit.cs.ut.ee/gprofiler/gost
GTEx portal, https://gtexportal.org/home/index.html
GWAS Catalog, https://www.ebi.ac.uk/gwas/
OMIM, https://omim.org/
Open Targets Genetics, https://genetics.opentargets.org/
SHBG blood test, https://medlineplus.gov/lab-tests/shbg-blood-test/
-
UCSC Genome Browser, https://genome.ucsc.edu/cgi-bin/hgGateway
International Stuttering Project, https://www.theinternationalstutteringproject.com/
Supplemental information
References
- 1.Levelt W.J. Models of word production. Trends Cogn. Sci. 1999;3:223–232. doi: 10.1016/s1364-6613(99)01319-4. [DOI] [PubMed] [Google Scholar]
- 2.Craig A., Hancock K., Tran Y., Craig M., Peters K. Epidemiology of stuttering in the community across the entire life span. J. Speech Lang. Hear. Res. 2002;45:1097–1105. doi: 10.1044/1092-4388(2002/088). [DOI] [PubMed] [Google Scholar]
- 3.Grigorenko E.L. Speaking genes or genes for speaking? Deciphering the genetics of speech and language. J. Child Psychol. Psychiatry. 2009;50:116–125. doi: 10.1111/j.1469-7610.2008.02006.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yairi E., Ambrose N. Epidemiology of stuttering: 21st century advances. J. Fluency Disord. 2013;38:66–87. doi: 10.1016/j.jfludis.2012.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wingate M.E. A Standard Definition of Stuttering. J. Speech Hear. Disord. 1964;29:484–489. doi: 10.1044/jshd.2904.484. [DOI] [PubMed] [Google Scholar]
- 6.Ambrose N.G., Cox N.J., Yairi E. The genetic basis of persistence and recovery in stuttering. J. Speech Lang. Hear. Res. 1997;40:567–580. doi: 10.1044/jslhr.4003.567. [DOI] [PubMed] [Google Scholar]
- 7.Felsenfeld S., Kirk K.M., Zhu G., Statham D.J., Neale M.C., Martin N.G. A study of the genetic and environmental etiology of stuttering in a selected twin sample. Behav. Genet. 2000;30:359–366. doi: 10.1023/a:1002765620208. [DOI] [PubMed] [Google Scholar]
- 8.Månsson H. Childhood stuttering: Incidence and development. J. Fluency Disord. 2000;25:47–57. [Google Scholar]
- 9.McKinnon D.H., McLeod S., Reilly S. The prevalence of stuttering, voice, and speech-sound disorders in primary school students in Australia. Lang. Speech Hear. Serv. Sch. 2007;38:5–15. doi: 10.1044/0161-1461(2007/002). [DOI] [PubMed] [Google Scholar]
- 10.Bloodstein O., Ratner N.B. New York, NY: Cengage Learning; 2008. A handbook on stuttering, sixth edition. [Google Scholar]
- 11.Ambrose N.G., Yairi E. Normative disfluency data for early childhood stuttering. J. Speech Lang. Hear. Res. 1999;42:895–909. doi: 10.1044/jslhr.4204.895. [DOI] [PubMed] [Google Scholar]
- 12.Frigerio-Domingues C., Drayna D. Genetic contributions to stuttering: the current evidence. Mol. Genet. Genomic Med. 2017;5:95–102. doi: 10.1002/mgg3.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.(2020). Total Population, US Census. https://data.census.gov/cedsci/
- 14.Baxter S., Johnson M., Blank L., Cantrell A., Brumfitt S., Enderby P., Goyder E. The state of the art in non-pharmacological interventions for developmental stuttering. Part 1: a systematic review of effectiveness. Int. J. Lang. Commun. Disord. 2015;50:676–718. doi: 10.1111/1460-6984.12171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Daniels D., Gabel R. The Impact of Stuttering on Identity Construction. Top. Lang. Disord. 2004;24:200–215. [Google Scholar]
- 16.Costa D., Kroll R. Stuttering: an update for physicians. CMAJ. 2000;162:1849–1855. [PMC free article] [PubMed] [Google Scholar]
- 17.Daniels D.E., Gabel R.M., Hughes S. Recounting the K-12 school experiences of adults who stutter: a qualitative analysis. J. Fluency Disord. 2012;37:71–82. doi: 10.1016/j.jfludis.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 18.Klein J.F., Hood S.B. The impact of stuttering on employment opportunities and job performance. J. Fluency Disord. 2004;29:255–273. doi: 10.1016/j.jfludis.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 19.Gabel R.M., Blood G.W., Tellis G.M., Althouse M.T. Measuring role entrapment of people who stutter. J. Fluency Disord. 2004;29:27–49. doi: 10.1016/j.jfludis.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 20.Craig A.R., Calver P. Following up on treated stutterers: studies of perceptions of fluency and job status. J. Speech Hear. Res. 1991;34:279–284. doi: 10.1044/jshr.3402.279. [DOI] [PubMed] [Google Scholar]
- 21.National Academies of Sciences, Engineering, and Medicine; Institute of Medicine; Division of Behavioral and Social Sciences and Education; Board on the Health of Select Populations; Board on Child, Youth, and Families; Committee on the Evaluation of Supplemental Security Income (SSI) Disability Program for Children with Speech Disorders and Language Disorders. Rosenbaum S, Simon P, editors. (2016). Childhood Speech and Language Disorders in the General U.S. Population. In Speech and Language Disorders in Children: Implications for the Social Security Administration’s Supplemental Security Income Program, S. Rosenbaum and P. Simon, eds. (Washington, DC, National Academies Press). [PubMed]
- 22.Fitzpatrick E. Neurocognitive development in congenitally deaf children. Handb. Clin. Neurol. 2015;129:335–356. doi: 10.1016/B978-0-444-62630-1.00019-6. [DOI] [PubMed] [Google Scholar]
- 23.Shugart Y.Y., Mundorff J., Kilshaw J., Doheny K., Doan B., Wanyee J., Green E.D., Drayna D. Results of a genome-wide linkage scan for stuttering. Am. J. Med. Genet. A. 2004;124A:133–135. doi: 10.1002/ajmg.a.20347. [DOI] [PubMed] [Google Scholar]
- 24.Riaz N., Steinberg S., Ahmad J., Pluzhnikov A., Riazuddin S., Cox N.J., Drayna D. Genomewide significant linkage to stuttering on chromosome 12. Am. J. Hum. Genet. 2005;76:647–651. doi: 10.1086/429226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suresh R., Ambrose N., Roe C., Pluzhnikov A., Wittke-Thompson J.K., Ng M.C., Wu X., Cook E.H., Lundstrom C., Garsten M., et al. New complexities in the genetics of stuttering: significant sex-specific linkage signals. Am. J. Hum. Genet. 2006;78:554–563. doi: 10.1086/501370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wittke-Thompson J.K., Ambrose N., Yairi E., Roe C., Cook E.H., Ober C., Cox N.J. Genetic studies of stuttering in a founder population. J. Fluency Disord. 2007;32:33–50. doi: 10.1016/j.jfludis.2006.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kang C., Riazuddin S., Mundorff J., Krasnewich D., Friedman P., Mullikin J.C., Drayna D. Mutations in the lysosomal enzyme-targeting pathway and persistent stuttering. N. Engl. J. Med. 2010;362:677–685. doi: 10.1056/NEJMoa0902630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Raza M.H., Riazuddin S., Drayna D. Identification of an autosomal recessive stuttering locus on chromosome 3q13.2-3q13.33. Hum. Genet. 2010;128:461–463. doi: 10.1007/s00439-010-0871-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Domingues C.E., Olivera C.M., Oliveira B.V., Juste F.S., Andrade C.R., Giacheti C.M., Moretti-Ferreira D., Drayna D. A genetic linkage study in Brazil identifies a new locus for persistent developmental stuttering on chromosome 10. Genet. Mol. Res. 2014;13:2094–2101. doi: 10.4238/2014.March.24.13. [DOI] [PubMed] [Google Scholar]
- 30.Mohammadi H., Joghataei M.T., Rahimi Z., Faghihi F., Khazaie H., Farhangdoost H., Mehrpour M. Sex steroid hormones and sex hormone binding globulin levels, CYP17 MSP AI (-34T:C) and CYP19 codon 39 (Trp:Arg) variants in children with developmental stuttering. Brain Lang. 2017;175:47–56. doi: 10.1016/j.bandl.2017.09.004. [DOI] [PubMed] [Google Scholar]
- 31.Lan J., Song M., Pan C., Zhuang G., Wang Y., Ma W., Chu Q., Lai Q., Xu F., Li Y., et al. Association between dopaminergic genes (SLC6A3 and DRD2) and stuttering among Han Chinese. J. Hum. Genet. 2009;54:457–460. doi: 10.1038/jhg.2009.60. [DOI] [PubMed] [Google Scholar]
- 32.Kang C., Domingues B.S., Sainz E., Domingues C.E.F., Drayna D., Moretti-Ferreira D. Evaluation of the association between polymorphisms at the DRD2 locus and stuttering. J. Hum. Genet. 2011;56:472–473. doi: 10.1038/jhg.2011.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Raza M.H., Mattera R., Morell R., Sainz E., Rahn R., Gutierrez J., Paris E., Root J., Solomon B., Brewer C., et al. Association between Rare Variants in AP4E1, a Component of Intracellular Trafficking, and Persistent Stuttering. Am. J. Hum. Genet. 2015;97:715–725. doi: 10.1016/j.ajhg.2015.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ooki S. Genetic and environmental influences on stuttering and tics in Japanese twin children. Twin Res. Hum. Genet. 2005;8:69–75. doi: 10.1375/1832427053435409. [DOI] [PubMed] [Google Scholar]
- 35.Dworzynski K., Remington A., Rijsdijk F., Howell P., Plomin R. Genetic etiology in cases of recovered and persistent stuttering in an unselected, longitudinal sample of young twins. Am. J. Speech Lang. Pathol. 2007;16:169–178. doi: 10.1044/1058-0360(2007/021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Beijsterveldt C.E., Felsenfeld S., Boomsma D.I. Bivariate genetic analyses of stuttering and nonfluency in a large sample of 5-year-old twins. J. Speech Lang. Hear. Res. 2010;53:609–619. doi: 10.1044/1092-4388(2009/08-0202). [DOI] [PubMed] [Google Scholar]
- 37.Fagnani C., Fibiger S., Skytthe A., Hjelmborg J.V. Heritability and environmental effects for self-reported periods with stuttering: a twin study from Denmark. Logoped. Phoniatr. Vocol. 2011;36:114–120. doi: 10.3109/14015439.2010.534503. [DOI] [PubMed] [Google Scholar]
- 38.Rautakoski P., Hannus T., Simberg S., Sandnabba N.K., Santtila P. Genetic and environmental effects on stuttering: a twin study from Finland. J. Fluency Disord. 2012;37:202–210. doi: 10.1016/j.jfludis.2011.12.003. [DOI] [PubMed] [Google Scholar]
- 39.Mahajan A., Go M.J., Zhang W., Below J.E., Gaulton K.J., Ferreira T., Horikoshi M., Johnson A.D., Ng M.C.Y., Prokopenko I., et al. DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium. Asian Genetic Epidemiology Network Type 2 Diabetes (AGEN-T2D) Consortium. South Asian Type 2 Diabetes (SAT2D) Consortium. Mexican American Type 2 Diabetes (MAT2D) Consortium. Type 2 Diabetes Genetic Exploration by Nex-generation sequencing in muylti-Ethnic Samples (T2D-GENES) Consortium Genome-wide trans-ancestry meta-analysis provides insight into the genetic architecture of type 2 diabetes susceptibility. Nat. Genet. 2014;46:234–244. doi: 10.1038/ng.2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Palmer N.D., Goodarzi M.O., Langefeld C.D., Wang N., Guo X., Taylor K.D., Fingerlin T.E., Norris J.M., Buchanan T.A., Xiang A.H., et al. Genetic Variants Associated With Quantitative Glucose Homeostasis Traits Translate to Type 2 Diabetes in Mexican Americans: The GUARDIAN (Genetics Underlying Diabetes in Hispanics) Consortium. Diabetes. 2015;64:1853–1866. doi: 10.2337/db14-0732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Willer C.J., Schmidt E.M., Sengupta S., Peloso G.M., Gustafsson S., Kanoni S., Ganna A., Chen J., Buchkovich M.L., Mora S., et al. Global Lipids Genetics Consortium Discovery and refinement of loci associated with lipid levels. Nat. Genet. 2013;45:1274–1283. doi: 10.1038/ng.2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nalls M.A., Pankratz N., Lill C.M., Do C.B., Hernandez D.G., Saad M., DeStefano A.L., Kara E., Bras J., Sharma M., et al. International Parkinson’s Disease Genomics Consortium (IPDGC) Parkinson’s Study Group (PSG) Parkinson’s Research: The Organized GENetics Initiative (PROGENI) 23andMe. GenePD. NeuroGenetics Research Consortium (NGRC) Hussman Institute of Human Genomics (HIHG) Ashkenazi Jewish Dataset Investigator. Cohorts for Health and Aging Research in Genetic Epidemiology (CHARGE) North American Brain Expression Consortium (NABEC) United Kingdom Brain Expression Consortium (UKBEC) Greek Parkinson’s Disease Consortium. Alzheimer Genetic Analysis Group Large-scale meta-analysis of genome-wide association data identifies six new risk loci for Parkinson’s disease. Nat. Genet. 2014;46:989–993. doi: 10.1038/ng.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lambert J.-C., Ibrahim-Verbaas C.A., Harold D., Naj A.C., Sims R., Bellenguez C., DeStafano A.L., Bis J.C., Beecham G.W., Grenier-Boley B., et al. European Alzheimer’s Disease Initiative (EADI) Genetic and Environmental Risk in Alzheimer’s Disease. Alzheimer’s Disease Genetic Consortium. Cohorts for Heart and Aging Research in Genomic Epidemiology Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat. Genet. 2013;45:1452–1458. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Escott-Price V., Bellenguez C., Wang L.-S., Choi S.-H., Harold D., Jones L., Holmans P., Gerrish A., Vedernikov A., Richards A., et al. United Kingdom Brain Expression Consortium. Cardiovascular Health Study (CHS) Gene-wide analysis detects two new susceptibility genes for Alzheimer’s disease. PLoS ONE. 2014;9:e94661. doi: 10.1371/journal.pone.0094661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Raza M.H., Amjad R., Riazuddin S., Drayna D. Studies in a consanguineous family reveal a novel locus for stuttering on chromosome 16q. Hum. Genet. 2012;131:311–313. doi: 10.1007/s00439-011-1134-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kazemi N., Estiar M.A., Fazilaty H., Sakhinia E. Variants in GNPTAB, GNPTG and NAGPA genes are associated with stutterers. Gene. 2018;647:93–100. doi: 10.1016/j.gene.2017.12.054. [DOI] [PubMed] [Google Scholar]
- 47.Raza M.H., Gertz E.M., Mundorff J., Lukong J., Kuster J., Schäffer A.A., Drayna D. Linkage analysis of a large African family segregating stuttering suggests polygenic inheritance. Hum. Genet. 2013;132:385–396. doi: 10.1007/s00439-012-1252-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Frigerio Domingues C.E., Grainger K., Cheng H., Moretti-Ferreira D., Riazuddin S., Drayna D. Are variants in sex hormone metabolizing genes associated with stuttering? Brain Lang. 2019;191:28–30. doi: 10.1016/j.bandl.2019.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roden D.M., Pulley J.M., Basford M.A., Bernard G.R., Clayton E.W., Balser J.R., Masys D.R. Development of a large-scale de-identified DNA biobank to enable personalized medicine. Clin. Pharmacol. Ther. 2008;84:362–369. doi: 10.1038/clpt.2008.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McGregor T.L., Van Driest S.L., Brothers K.B., Bowton E.A., Muglia L.J., Roden D.M. Inclusion of pediatric samples in an opt-out biorepository linking DNA to de-identified medical records: pediatric BioVU. Clin. Pharmacol. Ther. 2013;93:204–211. doi: 10.1038/clpt.2012.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pruett D.G., Shaw D.M., Chen H.-H., Petty L.E., Polikowsky H.G., Kraft S.J., Jones R.M., Below J.E. Identifying developmental stuttering and associated comorbidities in electronic health records and creating a phenome risk classifier. J. Fluency Disord. 2021;68:105847. doi: 10.1016/j.jfludis.2021.105847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shaw D.M., Polikowsky H.P., Pruett D.G., Chen H.-H., Petty L.E., Viljoen K.Z., et al. Phenome risk classification enables phenotypic imputation and gene discovery in developmental stuttering. Am. J. Hum. Genet. 2021;108:2271–2283. doi: 10.1016/j.ajhg.2021.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A.R., Bender D., Maller J., Sklar P., de Bakker P.I.W., Daly M.J., Sham P.C. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Staples J., Nickerson D.A., Below J.E. Utilizing graph theory to select the largest set of unrelated individuals for genetic analysis. Genet. Epidemiol. 2013;37:136–141. doi: 10.1002/gepi.21684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Staples J., Qiao D., Cho M.H., Silverman E.K., Nickerson D.A., Below J.E., University of Washington Center for Mendelian Genomics PRIMUS: rapid reconstruction of pedigrees from genome-wide estimates of identity by descent. Am. J. Hum. Genet. 2014;95:553–564. doi: 10.1016/j.ajhg.2014.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Luca D., Ringquist S., Klei L., Lee A.B., Gieger C., Wichmann H.E., Schreiber S., Krawczak M., Lu Y., Styche A., et al. On the use of general control samples for genome-wide association studies: genetic matching highlights causal variants. Am. J. Hum. Genet. 2008;82:453–463. doi: 10.1016/j.ajhg.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Harris K.M., Halpern C.T., Whitsel E.A., Hussey J.M., Killeya-Jones L.A., Tabor J., Dean S.C. Cohort Profile: The National Longitudinal Study of Adolescent to Adult Health (Add Health) Int. J. Epidemiol. 2019;48 doi: 10.1093/ije/dyz115. 1415–1415k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pluzhnikov A., Below J.E., Konkashbaev A., Tikhomirov A., Kistner-Griffin E., Roe C.A., Nicolae D.L., Cox N.J. Spoiling the whole bunch: quality control aimed at preserving the integrity of high-throughput genotyping. Am. J. Hum. Genet. 2010;87:123–128. doi: 10.1016/j.ajhg.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Altshuler D.M., Gibbs R.A., Peltonen L., Altshuler D.M., Gibbs R.A., Peltonen L., Dermitzakis E., Schaffner S.F., Yu F., Peltonen L., et al. International HapMap 3 Consortium Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rayner N.W., Robertson N., McCarthy M.I. 2004. McCarthy Group Tools. www.well.ox.ac.uk/∼wrayner/tools/ [Google Scholar]
- 61.Loh P.-R., Danecek P., Palamara P.F., Fuchsberger C., A Reshef Y., K Finucane H., Schoenherr S., Forer L., McCarthy S., Abecasis G.R., et al. Reference-based phasing using the Haplotype Reference Consortium panel. Nat. Genet. 2016;48:1443–1448. doi: 10.1038/ng.3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fuchsberger C., Abecasis G.R., Hinds D.A. minimac2: faster genotype imputation. Bioinformatics. 2015;31:782–784. doi: 10.1093/bioinformatics/btu704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Das S., Forer L., Schönherr S., Sidore C., Locke A.E., Kwong A., Vrieze S.I., Chew E.Y., Levy S., McGue M., et al. Next-generation genotype imputation service and methods. Nat. Genet. 2016;48:1284–1287. doi: 10.1038/ng.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Taliun D., Harris D.N., Kessler M.D., Carlson J., Szpiech Z.A., Torres R., Taliun S.A.G., Corvelo A., Gogarten S.M., Kang H.M., et al. NHLBI Trans-Omics for Precision Medicine (TOPMed) Consortium Sequencing of 53,831 diverse genomes from the NHLBI TOPMed Program. Nature. 2021;590:290–299. doi: 10.1038/s41586-021-03205-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Auton A., Brooks L.D., Durbin R.M., Garrison E.P., Kang H.M., Korbel J.O., Marchini J.L., McCarthy S., McVean G.A., Abecasis G.R., 1000 Genomes Project Consortium A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhou W., Nielsen J.B., Fritsche L.G., Dey R., Gabrielsen M.E., Wolford B.N., LeFaive J., VandeHaar P., Gagliano S.A., Gifford A., et al. Efficiently controlling for case-control imbalance and sample relatedness in large-scale genetic association studies. Nat. Genet. 2018;50:1335–1341. doi: 10.1038/s41588-018-0184-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Price A.L., Patterson N.J., Plenge R.M., Weinblatt M.E., Shadick N.A., Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 68.Lin D.Y., Tao R., Kalsbeek W.D., Zeng D., Gonzalez F., 2nd, Fernández-Rhodes L., Graff M., Koch G.G., North K.E., Heiss G. Genetic association analysis under complex survey sampling: the Hispanic Community Health Study/Study of Latinos. Am. J. Hum. Genet. 2014;95:675–688. doi: 10.1016/j.ajhg.2014.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Willer C.J., Li Y., Abecasis G.R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Risch N., Merikangas K. The future of genetic studies of complex human diseases. Science. 1996;273:1516–1517. doi: 10.1126/science.273.5281.1516. [DOI] [PubMed] [Google Scholar]
- 71.Duggal P., Gillanders E.M., Holmes T.N., Bailey-Wilson J.E. Establishing an adjusted p-value threshold to control the family-wide type 1 error in genome wide association studies. BMC Genomics. 2008;9:516. doi: 10.1186/1471-2164-9-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Xu C., Tachmazidou I., Walter K., Ciampi A., Zeggini E., Greenwood C.M.T., UK10K Consortium Estimating genome-wide significance for whole-genome sequencing studies. Genet. Epidemiol. 2014;38:281–290. doi: 10.1002/gepi.21797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang K., Li M., Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.The G.C., GTEx Consortium The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science. 2020;369:1318–1330. doi: 10.1126/science.aaz1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yang J., Benyamin B., McEvoy B.P., Gordon S., Henders A.K., Nyholt D.R., Madden P.A., Heath A.C., Martin N.G., Montgomery G.W., et al. Common SNPs explain a large proportion of the heritability for human height. Nat. Genet. 2010;42:565–569. doi: 10.1038/ng.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lee S.H., Wray N.R., Goddard M.E., Visscher P.M. Estimating missing heritability for disease from genome-wide association studies. Am. J. Hum. Genet. 2011;88:294–305. doi: 10.1016/j.ajhg.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wen X., Pique-Regi R., Luca F. Integrating molecular QTL data into genome-wide genetic association analysis: Probabilistic assessment of enrichment and colocalization. PLoS Genet. 2017;13:e1006646. doi: 10.1371/journal.pgen.1006646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pividori M., Rajagopal P.S., Barbeira A., Liang Y., Melia O., Bastarache L., Park Y., Consortium G., Wen X., Im H.K. PhenomeXcan: Mapping the genome to the phenome through the transcriptome. Sci. Adv. 2020;6:eaba2083. doi: 10.1126/sciadv.aba2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Berisa T., Pickrell J.K. Approximately independent linkage disequilibrium blocks in human populations. Bioinformatics. 2016;32:283–285. doi: 10.1093/bioinformatics/btv546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ghoussaini M., Mountjoy E., Carmona M., Peat G., Schmidt E.M., Hercules A., Fumis L., Miranda A., Carvalho-Silva D., Buniello A., et al. Open Targets Genetics: systematic identification of trait-associated genes using large-scale genetics and functional genomics. Nucleic Acids Res. 2021;49(D1):D1311–D1320. doi: 10.1093/nar/gkaa840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wu T., Hu E., Xu S., Chen M., Guo P., Dai Z., Feng T., Zhou L., Tang W., Zhan L., et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation (N Y) 2021;2:100141. doi: 10.1016/j.xinn.2021.100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yu G., Wang L.G., Han Y., He Q.Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gerring Z.F., Gamazon E.R., Derks E.M., Major Depressive Disorder Working Group of the Psychiatric Genomics Consortium A gene co-expression network-based analysis of multiple brain tissues reveals novel genes and molecular pathways underlying major depression. PLoS Genet. 2019;15:e1008245. doi: 10.1371/journal.pgen.1008245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Skol A.D., Scott L.J., Abecasis G.R., Boehnke M. Joint analysis is more efficient than replication-based analysis for two-stage genome-wide association studies. Nat. Genet. 2006;38:209–213. doi: 10.1038/ng1706. [DOI] [PubMed] [Google Scholar]
- 85.Xiong F., Ji Z., Liu Y., Zhang Y., Hu L., Yang Q., Qiu Q., Zhao L., Chen D., Tian Z., et al. Mutation in SSUH2 Causes Autosomal-Dominant Dentin Dysplasia Type I. Hum. Mutat. 2017;38:95–104. doi: 10.1002/humu.23130. [DOI] [PubMed] [Google Scholar]
- 86.Consortium T.U., UniProt Consortium UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res. 2021;49(D1):D480–D489. doi: 10.1093/nar/gkaa1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.De Rubeis S., He X., Goldberg A.P., Poultney C.S., Samocha K., Cicek A.E., Kou Y., Liu L., Fromer M., Walker S., et al. DDD Study. Homozygosity Mapping Collaborative for Autism. UK10K Consortium Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515:209–215. doi: 10.1038/nature13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhao B., Luo T., Li T., Li Y., Zhang J., Shan Y., Wang X., Yang L., Zhou F., Zhu Z., Zhu H., Alzheimer’s Disease Neuroimaging Initiative. Pediatric Imaging, Neurocognition and Genetics Genome-wide association analysis of 19,629 individuals identifies variants influencing regional brain volumes and refines their genetic co-architecture with cognitive and mental health traits. Nat. Genet. 2019;51:1637–1644. doi: 10.1038/s41588-019-0516-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Peter B., Dinu V., Liu L., Huentelman M., Naymik M., Lancaster H., Vose C., Schrauwen I. Exome Sequencing of Two Siblings with Sporadic Autism Spectrum Disorder and Severe Speech Sound Disorder Suggests Pleiotropic and Complex Effects. Behav. Genet. 2019;49:399–414. doi: 10.1007/s10519-019-09957-8. [DOI] [PubMed] [Google Scholar]
- 90.Matoba N., Liang D., Sun H., Aygün N., McAfee J.C., Davis J.E., Raffield L.M., Qian H., Piven J., Li Y., et al. Common genetic risk variants identified in the SPARK cohort support DDHD2 as a candidate risk gene for autism. Transl. Psychiatry. 2020;10:265. doi: 10.1038/s41398-020-00953-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lai C.S., Fisher S.E., Hurst J.A., Vargha-Khadem F., Monaco A.P. A forkhead-domain gene is mutated in a severe speech and language disorder. Nature. 2001;413:519–523. doi: 10.1038/35097076. [DOI] [PubMed] [Google Scholar]
- 92.Nisar S., Hashem S., Bhat A.A., Syed N., Yadav S., Azeem M.W., Uddin S., Bagga P., Reddy R., Haris M. Association of genes with phenotype in autism spectrum disorder. Aging (Albany NY) 2019;11:10742–10770. doi: 10.18632/aging.102473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Newbury D.F., Fisher S.E., Monaco A.P. Recent advances in the genetics of language impairment. Genome Med. 2010;2:6. doi: 10.1186/gm127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Han T.U., Park J., Domingues C.F., Moretti-Ferreira D., Paris E., Sainz E., Gutierrez J., Drayna D. A study of the role of the FOXP2 and CNTNAP2 genes in persistent developmental stuttering. Neurobiol. Dis. 2014;69:23–31. doi: 10.1016/j.nbd.2014.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Vernes S.C., Newbury D.F., Abrahams B.S., Winchester L., Nicod J., Groszer M., Alarcón M., Oliver P.L., Davies K.E., Geschwind D.H., et al. A functional genetic link between distinct developmental language disorders. N. Engl. J. Med. 2008;359:2337–2345. doi: 10.1056/NEJMoa0802828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Aboumousa A., Hoogendijk J., Charlton R., Barresi R., Herrmann R., Voit T., Hudson J., Roberts M., Hilton-Jones D., Eagle M., et al. Caveolinopathy--new mutations and additional symptoms. Neuromuscul. Disord. 2008;18:572–578. doi: 10.1016/j.nmd.2008.05.003. [DOI] [PubMed] [Google Scholar]
- 97.Traverso M., Bruno C., Broccolini A., Sotgia F., Donati M.A., Assereto S., Gazzerro E., Lo Monaco M., Modoni A., D’Amico A., et al. Truncation of Caveolin-3 causes autosomal-recessive Rippling Muscle Disease. J. Neurol. Neurosurg. Psychiatry. 2008;79:735–737. doi: 10.1136/jnnp.2007.133207. [DOI] [PubMed] [Google Scholar]
- 98.Catteruccia M., Sanna T., Santorelli F.M., Tessa A., Di Giacopo R., Sauchelli D., Verbo A., Lo Monaco M., Servidei S. Rippling muscle disease and cardiomyopathy associated with a mutation in the CAV3 gene. Neuromuscul. Disord. 2009;19:779–783. doi: 10.1016/j.nmd.2009.08.015. [DOI] [PubMed] [Google Scholar]
- 99.Gazzerro E., Sotgia F., Bruno C., Lisanti M.P., Minetti C. Caveolinopathies: from the biology of caveolin-3 to human diseases. Eur. J. Hum. Genet. 2010;18:137–145. doi: 10.1038/ejhg.2009.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gazzerro E., Bonetto A., Minetti C. Caveolinopathies: translational implications of caveolin-3 in skeletal and cardiac muscle disorders. Handb. Clin. Neurol. 2011;101:135–142. doi: 10.1016/B978-0-08-045031-5.00010-4. [DOI] [PubMed] [Google Scholar]
- 101.Ullrich N.D., Fischer D., Kornblum C., Walter M.C., Niggli E., Zorzato F., Treves S. Alterations of excitation-contraction coupling and excitation coupled Ca(2+) entry in human myotubes carrying CAV3 mutations linked to rippling muscle. Hum. Mutat. 2011;32:309–317. doi: 10.1002/humu.21431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Buniello A., MacArthur J.A.L., Cerezo M., Harris L.W., Hayhurst J., Malangone C., McMahon A., Morales J., Mountjoy E., Sollis E., et al. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 2019;47(D1):D1005–D1012. doi: 10.1093/nar/gky1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lin M.L., Fukukawa C., Park J.H., Naito K., Kijima K., Shimo A., Ajiro M., Nishidate T., Nakamura Y., Katagiri T. Involvement of G-patch domain containing 2 overexpression in breast carcinogenesis. Cancer Sci. 2009;100:1443–1450. doi: 10.1111/j.1349-7006.2009.01185.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ruth K.S., Day F.R., Tyrrell J., Thompson D.J., Wood A.R., Mahajan A., Beaumont R.N., Wittemans L., Martin S., Busch A.S., et al. Endometrial Cancer Association Consortium Using human genetics to understand the disease impacts of testosterone in men and women. Nat. Med. 2020;26:252–258. doi: 10.1038/s41591-020-0751-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Medline Plus . National Library of Medicine; 2020. SHBG Blood Test.https://medlineplus.gov/lab-tests/shbg-blood-test/ [Google Scholar]
- 106.Anthoni H., Sucheston L.E., Lewis B.A., Tapia-Páez I., Fan X., Zucchelli M., Taipale M., Stein C.M., Hokkanen M.E., Castrén E., et al. The aromatase gene CYP19A1: several genetic and functional lines of evidence supporting a role in reading, speech and language. Behav. Genet. 2012;42:509–527. doi: 10.1007/s10519-012-9532-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Schaffer A.E., Breuss M.W., Caglayan A.O., Al-Sanaa N., Al-Abdulwahed H.Y., Kaymakçalan H., Yılmaz C., Zaki M.S., Rosti R.O., Copeland B., et al. Biallelic loss of human CTNNA2, encoding αN-catenin, leads to ARP2/3 complex overactivity and disordered cortical neuronal migration. Nat. Genet. 2018;50:1093–1101. doi: 10.1038/s41588-018-0166-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Mbikay M., Nolet S., Fournier S., Benjannet S., Chapdelaine P., Paradis G., Dubé J.Y., Tremblay R., Lazure C., Seidah N.G., et al. Molecular cloning and sequence of the cDNA for a 94-amino-acid seminal plasma protein secreted by the human prostate. DNA. 1987;6:23–29. doi: 10.1089/dna.1987.6.23. [DOI] [PubMed] [Google Scholar]
- 109.Ulvsbäck M., Lindström C., Weiber H., Abrahamsson P.-A., Lilja H., Lundwall A. Molecular cloning of a small prostate protein, known as β-microsemenoprotein, PSP94 or β-inhibin, and demonstration of transcripts in non-genital tissues. Biochem. Biophys. Res. Commun. 1989;164:1310–1315. doi: 10.1016/0006-291x(89)91812-3. [DOI] [PubMed] [Google Scholar]
- 110.Thomas G., Jacobs K.B., Yeager M., Kraft P., Wacholder S., Orr N., Yu K., Chatterjee N., Welch R., Hutchinson A., et al. Multiple loci identified in a genome-wide association study of prostate cancer. Nat. Genet. 2008;40:310–315. doi: 10.1038/ng.91. [DOI] [PubMed] [Google Scholar]
- 111.Eeles R.A., Kote-Jarai Z., Giles G.G., Olama A.A., Guy M., Jugurnauth S.K., Mulholland S., Leongamornlert D.A., Edwards S.M., Morrison J., et al. UK Genetic Prostate Cancer Study Collaborators. British Association of Urological Surgeons’ Section of Oncology. UK ProtecT Study Collaborators Multiple newly identified loci associated with prostate cancer susceptibility. Nat. Genet. 2008;40:316–321. doi: 10.1038/ng.90. [DOI] [PubMed] [Google Scholar]
- 112.Joseph R.E., Walker J., Norris F.A. Assignment of the inositol polyphosphate 4-phosphatase type I gene (INPP4A) to human chromosome band 2q11.2 by in situ hybridization. Cytogenet. Cell Genet. 1999;87:276–277. doi: 10.1159/000015448. [DOI] [PubMed] [Google Scholar]
- 113.Weiner D.J., Wigdor E.M., Ripke S., Walters R.K., Kosmicki J.A., Grove J., Samocha K.E., Goldstein J.I., Okbay A., Bybjerg-Grauholm J., et al. iPSYCH-Broad Autism Group. Psychiatric Genomics Consortium Autism Group Polygenic transmission disequilibrium confirms that common and rare variation act additively to create risk for autism spectrum disorders. Nat. Genet. 2017;49:978–985. doi: 10.1038/ng.3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ge T., Chen C.-Y., Ni Y., Feng Y.A., Smoller J.W. Polygenic prediction via Bayesian regression and continuous shrinkage priors. Nat. Commun. 2019;10:1776. doi: 10.1038/s41467-019-09718-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Yu D., Sul J.H., Tsetsos F., Nawaz M.S., Huang A.Y., Zelaya I., Illmann C., Osiecki L., Darrow S.M., Hirschtritt M.E., et al. Tourette Association of America International Consortium for Genetics, the Gilles de la Tourette GWAS Replication Initiative, the Tourette International Collaborative Genetics Study, and the Psychiatric Genomics Consortium Tourette Syndrome Working Group Interrogating the Genetic Determinants of Tourette’s Syndrome and Other Tic Disorders Through Genome-Wide Association Studies. Am. J. Psychiatry. 2019;176:217–227. doi: 10.1176/appi.ajp.2018.18070857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Scharf J.M., Yu D., Mathews C.A., Neale B.M., Stewart S.E., Fagerness J.A., Evans P., Gamazon E., Edlund C.K., Service S.K., et al. North American Brain Expression Consortium. UK Human Brain Expression Database Genome-wide association study of Tourette’s syndrome. Mol. Psychiatry. 2013;18:721–728. doi: 10.1038/mp.2012.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Gaugler T., Klei L., Sanders S.J., Bodea C.A., Goldberg A.P., Lee A.B., Mahajan M., Manaa D., Pawitan Y., Reichert J., et al. Most genetic risk for autism resides with common variation. Nat. Genet. 2014;46:881–885. doi: 10.1038/ng.3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Sanders S.J., He X., Willsey A.J., Ercan-Sencicek A.G., Samocha K.E., Cicek A.E., Murtha M.T., Bal V.H., Bishop S.L., Dong S., et al. Autism Sequencing Consortium Insights into Autism Spectrum Disorder Genomic Architecture and Biology from 71 Risk Loci. Neuron. 2015;87:1215–1233. doi: 10.1016/j.neuron.2015.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lee S.H., Ripke S., Neale B.M., Faraone S.V., Purcell S.M., Perlis R.H., Mowry B.J., Thapar A., Goddard M.E., Witte J.S., et al. Cross-Disorder Group of the Psychiatric Genomics Consortium. International Inflammatory Bowel Disease Genetics Consortium (IIBDGC) Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nat. Genet. 2013;45:984–994. doi: 10.1038/ng.2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Eising E., Carrion-Castillo A., Vino A., Strand E.A., Jakielski K.J., Scerri T.S., Hildebrand M.S., Webster R., Ma A., Mazoyer B., et al. A set of regulatory genes co-expressed in embryonic human brain is implicated in disrupted speech development. Mol. Psychiatry. 2019;24:1065–1078. doi: 10.1038/s41380-018-0020-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All summary statistics genome-wide will be available in the GWAS Catalog (Study Accession: GCST90085698 ). ISP GWAS data have not yet been deposited in dbGaP but will be made available to qualified investigators in dbGaP. The Add Health data can be accessed through the Add Health Study. Details are available through the Carolina Population Center (see web resources).