Summary
Urinary stem cells (USCs) are a non-invasive, simple, and affordable cell source to study human diseases. Here we show that USCs are a versatile tool for studying Duchenne muscular dystrophy (DMD), since they are able to address RNA signatures and atypical mutation identification. Gene expression profiling of DMD individuals’ USCs revealed a profound deregulation of inflammation, muscle development, and metabolic pathways that mirrors the known transcriptional landscape of DMD muscle and worsens following USCs’ myogenic transformation. This pathogenic transcription signature was reverted by an exon-skipping corrective approach, suggesting the utility of USCs in monitoring DMD antisense therapy. The full DMD transcript profile performed in USCs from three undiagnosed DMD individuals addressed three splicing abnormalities, which were decrypted and confirmed as pathogenic variations by whole-genome sequencing (WGS). This combined genomic approach allowed the identification of three atypical and complex DMD mutations due to a deep intronic variation and two large inversions, respectively. All three mutations affect DMD gene splicing and cause a lack of dystrophin protein production, and one of these also generates unique fusion genes and transcripts. Further characterization of USCs using a novel cell-sorting technology (Celector) highlighted cell-type variability and the representation of cell-specific DMD isoforms. Our comprehensive approach to USCs unraveled RNA, DNA, and cell-specific features and demonstrated that USCs are a robust tool for studying and diagnosing DMD.
We defined urinary stem cell (USC) RNA-seq signatures and provided evidence that they are a non-invasive, affordable cell source to study Duchenne muscular dystrophy (DMD), an X-linked disease, due to dystrophin gene mutations. Based on USCs’ RNA profiling and whole-genome sequencing (WGS), we identified three undetected atypical DMD mutations.
Introduction
Stem cells have been intensively investigated due to their relevance as a cell source for in vitro modeling, drug screening, and regenerative medicine in human diseases.1 The most studied stem cells are both embryonic stem cells (ESCs) and induced pluripotent stem cells (iPSCs). However, major issues, such as low reprogramming and differentiation efficiency, long and expensive procedures, tumorigenicity, and, not lastly, ethical aspects, have limited their usage in a broad range of applications. In addition, when studying human diseases, cells from affected individuals would be greatly preferred over manipulated cell models (e.g., generated by gene editing or transfection methods), since diseased cells better recapitulate the etio-pathogenic disease background. Nevertheless, studies on either cells or tissues from affected individuals often require invasive biopsies, invariably painful and sometimes unachievable (brain biopsies), with very poor patient compliance and with intrinsic ethical concerns.1 This also applies to rare genetic diseases (RDs) where, with a few exceptions like in muscle or skin diseases,2,3 biopsies are rarely an integral part of diagnostic protocols or available for research purposes.
Urine-derived stem cells (USCs) have shown to be a very promising source of mesenchymal stem cells.4 The isolation of USCs from urine is non-invasive, relatively fast, simple, affordable, and readily accepted by patients.5 Moreover, no major ethical issues are applied to urine collection and USC isolation.6 Some recent studies report the use of USCs for tissue engineering,7 gene therapy,8 and as in vitro models for studying several genetic diseases,1 including Duchenne muscular dystrophy (DMD).9
DMD represents one of the most common inherited neuromuscular disorders caused by a variety of mutations in the dystrophin (DMD) gene.10 We have already demonstrated that USCs derived from a DMD individual recapitulate the dystrophin genotype/phenotype in terms of DNA, RNA, and protein profiles of the DMD gene. Moreover, treatment with antisense oligoribonucleotide (AON) in both native and MyoD-transformed DMD USCs induced a corrective transcript-reframing effect leading to restoration of the dystrophin protein (DYS).9
Here, we report detailed studies on the USC transcriptional landscape, also aiming at atypical mutation identification, AON-mediated exon-skipping effects, and characterization of cell subpopulations.
Using RNA sequencing (RNA-seq) in DMD and in control native and myogenic USCs, we identified several deregulated gene circuits, which became more pronounced following myogenic transformation, therefore mirroring circuits already described in DMD muscle, as inflammation, development, and muscle contraction.11 AON-mediated skipping reverted deregulated transcriptomic circuits to normal, suggesting that USCs can be a valuable model for AON in vitro studies and possibly AON therapy monitoring. Full transcript analysis in USCs from three DMD undiagnosed individuals addressed RNA abnormalities in all of them. A very deep intronic variation causing different pseudoexons-including transcripts and two large genomic inversions, one of which causing a DMD transcription mimicry and a fusion mRNA, were identified by whole-genome sequencing (WGS).
Finally, as suggested by gene expression in silico deconvolution, after using Celector (Stem Sel), we confirmed the presence of three cell subpopulations that display DMD-specific features, such as surface markers and DMD-specific isoform expression.
Our results demonstrate that USCs are a robust and versatile cell model in studying DMD by profiling its RNA signature and deciphering atypical genome shapes, supporting their utility in diagnostic, therapeutic, and research applications.
Materials and methods
Individuals enrolled in the study
Table 1 summarizes the information related to each enrolled subject, including the type of study. Informed consent was obtained from all the individuals, both the control individuals and the DMD individuals (UNIFE Ethical Committee approval, no. 161299 [20/05/2020] and no. 66/2020 [23/01/2020]).
Table 1.
DMD individuals and control individuals (C) enrolled for isolation of USCs
| Sample | Disease | Mutation | Gender | Urine processing | USC application |
|---|---|---|---|---|---|
| DMD IG | DMD | del ex 45 | M | within 4 h | RNA sequencing |
| DMD PF | DMD | unknown | M | 24 h after collection | mutation discovery |
| DMD DI | DMD | unknown | M | within 4 h | mutation discovery |
| DMD SE | DMD | unknown | M | within 4 h | mutation discovery |
| DMD IH | DMD | del ex 50–52 | M | within 4 h | Celector separation |
| DMD AF | DMD | del ex 46–47 | M | within 4 h | Celector separation |
| C-1 | healthy control | – | M | within 4 h | RNA sequencing |
| C-2 | healthy control | – | M | within 4 h | RNA sequencing |
| C-3 | healthy control | – | M | within 4 h | RNA sequencing |
| C-4 | healthy control | – | M | within 4 h | Celector separation |
| C-5 | healthy control | – | M | within 4 h | Celector separation |
| C-6 | healthy control | – | M | within 4 h | Celector separation |
The table reports the time of isolation following urine collection (fresh urine: within 4 h; preserved urine: 24 h) and the type of analysis performed on each sample.
USCs from the control individuals (1–6) were isolated as control cell lines. USCs from DMD individuals with known mutations and a confirmed genetic diagnosis (three DMD samples: IG, IH, and AF, carrying the deletion of exons 45, 50–52, and 46–47, respectively) were used for RNA-seq or Celector analysis (Table 1).
Three other individuals with the DMD phenotype but without identified disease-causing mutations following Multiplex ligation-dependent probe amplificationv (MLPA) and DMD gene sequencing12 (DMD PF, DMD DI, and DMD SE; Table 1) were studied by FluiDMD and/or WGS.
PF is a 17-year-old boy who was born in 2003 from non-consanguineous healthy parents after an uneventful pregnancy. Serum creatine kinase (CK) dosing in the mother was normal. The patient had a normal psychomotor development with independent walking at 14 months. At 3 years of age, occasional hyperCKemia was noted with values of 5,000–7,000 U/L. A muscle biopsy was performed in 2011 for western blot (WB) and immunochemistry analysis. At 16 years old, a neurological examination showed Gower’s sign, waddling gait not possible on heels, and lower limb muscle hypotrophy with reduced tibiotarsal (TT) extension. The 6-min walk test (6MWT) was 435 m, North Star Ambulatory assessment (NSAA) was 24/34, height was 156 cm, and body mass index (BMI) was 18.7. There were neither respiratory nor cardiac complications (50% ejection fraction) nor major cognitive defects. The patient is still ambulant and has been on steroid therapy since 5 years of age.
DI is a 36-year-old man who was born in 1984. Serum CK dosing in the mother was normal. His developmental milestones were within normal range. At 3 years of age, he was seen by a neurologist because of frequent falls. Serum CK levels were 10 times normal values. A muscle biopsy was performed for immunochemistry analysis. He was started on prednisone 0.75 mg/kg at age 6 years. He lost ambulation at 14 years of age and steroids were stopped. He was started on non-invasive ventilation (NIV) at age 14 years, initially only at night. He was started on beta blockers at age 9 years. There is no cognitive impairment, and the patient shows a high educational profile, with two university diplomas, and currently works as a computer engineer. On the last neurological examination, the patient is wheelchair-bound and uses NIV for 15 h/day. His cardiac function is within normal range (60% ejection fraction).
SE is an 11-year-old boy who was born in 2009 from non-consanguineous parents after an uneventful pregnancy. Parents were healthy, but the mother was later proven to be an asymptomatic carrier by muscle biopsy and demonstration of a mosaic pattern with dystrophin immunohistochemistry. Serum CK dosing of the mother was 401 U/L (normal value < 145). The patient had a normal psychomotor development in the first year but only walked independently at 18 months. His linguistic development was normal. At the age of 21 months, he presented a tendency to walk on tiptoes and appeared to complain of calf pain. Serum CK was 34,969 U/L. A muscle biopsy was performed for WB and immunochemistry analysis. He was started on deflazacort 0.9 mg/kg/day at the age of 4 years, followed by a rehabilitation protocol with regular physical therapy and use of ankle/foot orthoses. He presented progressive muscle weakness with loss of the ability to rise from the floor autonomously and climb stairs at 9 years old. The patient was 10 years old at the time of this study and still able to walk with a marked waddling gait. There were neither respiratory nor cardiac complications nor major cognitive defects. However, anxiety is present, with a tendency for recurrent, repetitive thoughts and occasional impulsiveness and fits of anger.
Previous MLPA, comparative genomic hybridization (CGH), and sequencing studies failed to identify any pathogenic variants in PF, DI, and SE. Muscle biopsies of these three individuals were utilized for the diagnostic procedures described above without any muscle left for RNA studies, except for a minimum amount from patient SE. We therefore collected urine from these three patients to obtain USCs for RNA studies.
Dystrophin protein analysis
DYS protein was assessed by western blot and densitometry analysis.
Western blot
Fifteen- to 20-μm sections of muscle samples from DMD and normal muscles were solubilized in 100 μL of lysis buffer (50 mM Tris [pH 7.5], 150 mM NaCl, 10 mM MgCl2, 1 mM EDTA, 10% glycerol). Protein concentration was estimated using BCA Protein Assay Reagent (Thermo Scientific, Portsmouth, NH, USA). 40 μg of protein were loaded into each well of 3% to 8% gradient acrylamide gel (NuPAGE Novex Bis-Tris Mini Gels; Invitrogen, Carlsbad, CA, USA) and processed using the following antibodies: anti-Dys rod domain (NCL-DYS1, Novocastra) and anti-carboxyl terminus domain (NCL-DYS2, Novocastra). A secondary antibody was horseradish peroxidase (HRP)-conjugated bovine anti-mouse IgG. The blots were developed using chemiluminescence (ECL System, Amersham) using ImageQuant LAS (GE Healthcare Bio-Sciences). The amount of Dys for each individual was quantitated using ImageJ. To control for muscle protein content in each lane, the actin B signal (47 kDa) was also scanned, and the Dys content was adjusted to actin B. Each patient sample was subjected to electrophoresis adjacent to normal control lanes, and quantitation was done relative to control.
Immunohistochemistry
Eight-micrometer-thick sections were obtained from each muscle biopsy. Cryostat sections were mounted onto Superfrost Plus slides (Thermo Scientific, Portsmouth, NH, USA), hydrated in phosphate-buffered saline (PBS), and incubated with anti-Dys rod domain (NCL-DYS1, Novocastra) (1:100), anti-carboxyl terminus domain (NCL-DYS2, Novocastra) (1:100), and anti-amino terminal domain (NCL-DYS3, Novocastra) (1:100), diluted in PBS. After a triple-wash in PBS, specific labeling was developed by immunofluorescence using anti-mouse Cy-3 conjugated immunoglobulin (1:100) (Caltag, Burlingame, CA, USA). Sections were mounted with anti-fading medium and examined with a video-confocal microscope (ViCo, Nikon Instruments, Melville, NY, USA) or a Nikon Eclipse 80i fluorescence microscope.
Isolation of human USCs
Urine samples were obtained from all individuals listed in Table 1, and USCs were derived and cultured as described by Falzarano et al.9 Briefly, two urine specimens (first morning urine and a second urine sample) were obtained from each subject and processed within 4 h from the collection, except for urine from DMD individual PF, which was collected and preserved in primary medium for 24 h before the processing.
The urine specimens were centrifuged at 400 × g for 10 min at room temperature and washed with PBS (Gibco PBS; Thermo Fisher Scientific, Waltham, MA, USA) supplemented with an antibiotic/antimycotic solution (Sigma-Aldrich, St. Louis, MO, USA). After discarding the supernatant, 1 mL of primary medium was added, and each sample was plated into a coated plate with 0.1% gelatin (Millipore, Billerica, MA, USA). Every 24 h, 1 mL of primary medium was added to each well. The primary medium was removed 96 h after plating, and 1 mL of proliferation medium was added to 1 mL of primary culture medium in each well.
All the analyses described in this work were performed using primary USCs, not derived from one single clone. Cultures were on primary medium Dulbecco’s modified Eagle’s medium (DMEM)/high-glucose (EuroClone, Pero, Italy) and GIBCO Ham’s F12 nutrient mix (1:1; Thermo Fisher Scientific), supplemented with 10% (v/v) fetal bovine serum (FBS), antibiotic/antimycotic solution (Sigma-Aldrich), and an REGM (renal epithelial cell growth medium) SingleQuot kit (Lonza, Basel, Switzerland).
Proliferation medium was REGM BulletKit + RE cell basal medium (Lonza), and mesenchymal proliferation medium (DMEM/high glucose, 10% [v/v] FBS, 1% [v/v] Gibco GlutaMAX, 1% [v/v] nonessential amino acids [Gibco NEAA], 1% antibiotic/antimycotic solution, basic fibroblast growth factor [bFGF, 5 ng/mL; ProSpec, Rehovot, Israel], platelet-derived growth factor [PDGF-AB, 5 ng/mL; ProSpec], epidermal growth factor [EGF, 5 ng/mL; Lonza]) was used and mixed at a 1:1 ratio.
Myogenic transformation using MyoD and antisense oligonucleotide cell treatment
We induced myogenesis on USCs obtained from DMD individual IG and control individuals through an infection with adenovirus serotype 5 (Ad5)-derived, EA1-deleted adenoviral vector carrying the MyoD gene, as previously described.13 MyoD-transformed USCs were differentiated into myotubes by serum deprivation. Myotubes obtained from the MyoD-transformed USCs of DMD individual IG were transfected using 2′O-methyl phosphorothioate (2′OMePS) AON to induce DMD exon 44 skipping (IDT Technologies, Coralville, IA, USA) as previously described.9
Transcriptional profiling of USCs by RNA-seq analysis
We analyzed the gene expression levels of (1) native USCs derived from a pool of 3 healthy donors (C-n) and from DMD individual IG (IG-n), (2) MyoD-transformed USCs from control individuals (C-m) and from DMD individual IG (IG-m), and (3) MyoD-transformed USCs from DMD individual IG transfected with AON to skip the exon 44 and reframe the DMD transcript (IG-m 44) (Table 2).
Table 2.
Design of RNA-seq data comparison among USC samples
| Sample abbreviation | Sample description |
|---|---|
| C-n | RNA pool from native USCs of control 1–3 |
| IG-n | RNA from native USCs of DMD IG |
| C-m | RNA pool from MyoD-induced USCs of control 1–3 |
| IG-m | RNA from MyoD-induced USCs of DMD IG |
| IG-m 44 | RNA from DMD IG-m transfected with AON44 |
The table lists the abbreviations used in the text for each sample (column 1) and shows the description of sample composition (column 2).
Total RNA was isolated from native and MyoD-transformed USCs of DMD individual IG and healthy control individuals and from MyoD-transformed USCs of DMD individual IG transfected with AON 44 using the RNeasy-kit (QIAGEN, Chatsworth, CA, USA) according to the manufacturer’s instructions. Libraries were prepared using TruSeq Kit (Illumina) according to the manufacturer’s instructions. The quality and quantity of the RNA library was assessed using the Agilent 2100 Bioanalyzer and the ABI StepOnePlus Real-Time-PCR System. RNA-seq was carried out with the Illumina HiSeq4000 at the Beijing Genomics Institute (BGI, Beijing). Read quality was verified using fastQC (v.0.11.3). Raw reads were trimmed for adapters and for length at 100 bp with Trimmomatic, resulting in about 22 M (range, 14.8–31.7 M) trimmed reads per sample (Table S1). Reads were subsequently aligned to the human reference genome (GRCh38) using STAR (v.2.5.3a14). Raw gene counts were obtained in R-3.4.4 using the featureCounts function of the Rsubread R package (v.1.30.315) and the Gencode gene annotation. Raw counts were normalized to counts per million mapped reads (cpm) using the edgeR package;16 only genes with a cpm greater than 1 in at least 1 sample were further retained for differential analysis, for a total of 20,716 genes. Differential gene expression analysis was performed using the exactTest function of the edgeR package.16 Genes were considered significantly differentially expressed at false discovery rate (FDR) ≤ 0.05. Functional enrichment analysis was performed using the Gene Set Enrichment Analysis (GSEA, v.4.0.3) software and gene sets derived from Hallmark, Reactome, KEGG (Kyoto Encyclopedia of Genes and Genomes), and GO (Gene Ontology) collections of the Molecular Signature Database (MsigDB, v.7.1). The GSEA algorithm was applied using the signal2noise metric and the weighted statistics: gene sets were considered significantly enriched at FDR ≤ 0.05 when using 1,000 permutations of the gene sets.
Deconvolution analysis was performed using the web version of xCell,17 and tissues were considered significantly enriched at p value ≤ 0.05.
Identification and separation of USC subpopulations by Celector technology
Celector analysis was performed on the total native USC population isolated from both control individuals and DMD subjects following the same procedures as described in the paragraph “Isolation of human USCs.” USC cultures at low passage (i.e., p1–p2) from DMD individuals IH and AF and from healthy control individuals 4–6 (Table 1) were sorted and analyzed using a label-free technology named Celector (Stem Sel, Italy) (supplemental methods).18 For each analysis, cells were trypsinized, centrifuged at 1,200 rpm for 5 min, and re-suspended in the appropriate volume of PBS to obtain a concentration of 300,000 cells per 100 μL. This volume was introduced into the system and analyzed at a flow rate of 1 mL/min. Based on the cell profile, samples were divided in subfractions. Cells from each patient sample were run several times to collect the higher number of cells for downstream experiments.
For every analyzed sample, the cell area of eluting cells was quantified using ImageJ software. One representative captured picture for each fraction was used, and the particle analysis plugin was applied. Unpaired t test statistical analyses and plots of results were obtained using GraphPad Prism.
Phenotypic characterization of USC fractions
Cells for each fraction were collected and plated in proliferation medium to obtain the necessary cell number for flow cytometry and RNA analysis. The day after selection, cell images were acquired using a light microscope (Leica) to visualize morphological differences among fractions. When confluent, the cells were trypsinized. 100,000 cells were stained for mesenchymal (CD90-Fitc, CD105-Pe, CD73-Fitc, and CD146-Fitc) and hematopoietic markers (CD34-Fitc and CD45-Pe) and read using the FACS Canto (BD Biosciences). Data were analyzed using FlowJo software and plotted in GraphPad Prism.
Total RNA was extracted from DMD USC fractions using the RNeasy-kit and reverse transcribed into cDNA using random primers and the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). RT-PCR was performed on β-actin to verify cDNA synthesis.
The DMD transcript analysis was performed by FluiDMD cards9 that profile all exon-exon junctions of the 79 exons of the DMD transcript, including the DMD isoform promoter and/or first exon unique regions (brain [B], muscle [M], Purkinje [P], Dp260, Dp140, Dp116, Dp71). FluiDMD cards were run on an Applied Biosystems real-time 7900HT appliance (Thermo Fisher Scientific). The cycle threshold (Ct) values obtained for all exon junctions and DMD isoform systems were normalized by using human β-actin as the housekeeping gene (ΔCt = Ct exon junction system − Ct β-actin).
Identification of rare pathogenic variants in the DMD gene by FluiDMD and WGS analysis
Three individuals resulted negative at MLPA (P034/P035 DMD kit, MRC-Holland) followed by Multiplicom NGS (Next-generation sequencing) testing (DMD MASTR assay, Niel, Belgium) and were further studied to identify atypical mutations escaping the diagnostic strategy19 using FluiDMD cards20 and WGS.
RNA analysis
Total RNA was isolated from all DMD USCs and from DMD individual SE’s muscle biopsy using the RNeasy-kit and reverse transcribed into cDNA. FluiDMD cards were used to fully profile the DMD transcript composition. The 2−ΔΔCt method was used to calculate the relative changes in gene expression levels between affected individuals and control samples.
We also analyzed the DMD transcript by RT-PCR in DMD individual PF to amplify the region encompassing exons 10–12 (PCR primer pair sequences will be provided upon request) and in DMD individual SE to amplify the two novel transcript junctions, GRPR exon 1-DMD exon 55 and DMD exon 54-GRPR exon 2, in the two fusion transcripts. PCR products were run on 1.2% agarose gels, and purified PCR products were sequenced by Sanger method on ABI Prism 3130 (Applied Biosystems).
DNA analysis
Genomic DNA was isolated from peripheral blood using QIAsymphony (QIAGEN).
WGS library construction was made by MGIEasy DNALibrary Prep Kit V1 (BGI, cat. no. 85-05533-00) following the manufacturer’s manual. Briefly, 1,000 ng of genomic DNA was sheared with an E 220 Covaris instrument (Covaris) to DNA fragments between 50 bp and ∼800 bp. The DNA fragments were size selected using AMpure XP Beads (Beckman Coulter, Indianapolis, IN, USA) and then underwent end-repairing, phosphorylation, and A-tailing reactions. BGISEQ-500 platform-specific adaptors were ligated to the A-tailed fragments; the ligated fragments were purified and then amplified using PCR. After quantitation and qualification, the libraries were loaded onto a sequencing flow-cell and then processed for 100 bp paired-end sequencing on the BGISEQ-500 platform. Standard analysis of the raw data included sequence alignment and variants (e.g., SNP, insertion or deletion [indel], and copy-number variation [CNV]) calling and annotation. The Integrative Genomics Viewer (IGV) was used to visualize the coverage and the quality of the reads and for the visualization of structural variation (SV).21 IGV was also used to evaluate the aligned reads adjacent to SV breakpoints that mapped to discontinuous parts of the X chromosome.
Patient PF’s DMD mutation was validated by PCR on his genomic DNA. PCR primers were designed to amplify and sequence the entire intron 10 (650 bp long). The breakpoints of the 2 SVs identified by WGS in DMD individuals DI and SE were also validated on genomic DNA. PCR primers were designed to amplify the unique novel junctions that have been generated by the 2 inversions to produce a unique amplicon of the inverted regions. To assess the maternal origin of the rearrangements, the genomic DNA from peripheral blood of the mothers of individuals PF, DI, and SE was also amplified and sequenced as done for the DMD individuals.
All primers were designed using Primer3. Primer sequences and PCR conditions are available upon request. The quality of the amplified products was assessed using agarose gel electrophoresis. Clean PCR products were used for Sanger sequencing on the ABI Prism 3130XL (Applied Biosystems).
RepeatMasker analysis
Interspersed repeats and low-complexity DNA sequences were investigated in the breakpoint regions of the individuals using the RepeatMasker program.
Specifically, sequences of 650 bp overlapping the affected individuals’ breakpoints (325 bp upstream and 325 bp downstream from the breakpoint) were screened using the program’s default conditions: “rmblast” as the search engine and “default” as the speed/sensitivity option. Neither the lineage annotation nor advanced options were selected.
Results
Transcriptional profiling of native and myogenic USCs recapitulates the DMD disease pathways and highlights phenotype reversion by AON treatment
Gene expression profiles of MyoD-induced USCs from control individuals (C-m) and from DMD individual IG (IG-m) revealed many deregulated genes (2,482 and 4,583, respectively) compared to their native counterparts C-n and IG-n (Figure 1A; Figure S1A). This is expected, since it is known that myogenic differentiation causes profound changes in the gene expression profile.22 In particular, several genes representing many different biological functions (e.g., cell adhesion, cell cycle, muscle metabolism, nuclear regulatory factors, receptors/signaling, and structural/cytoskeletal) are deregulated during MyoD-induced myogenesis, as reported by Bergstrom et al.22 Cell myogenic nature was also confirmed by the absence of the MyoD transcript in native USCs and by its upregulation following MyoD transfection (C-n: 0 cpm; C-m: 885 cpm; IG-n: 0.205 cpm; IG-m: 658) (Figure S1B). As a further confirmation of the forced expression of MyoD, we observed deregulation of several representative genes modified by MyoD expression in both the control and DMD USCs (Table S2).
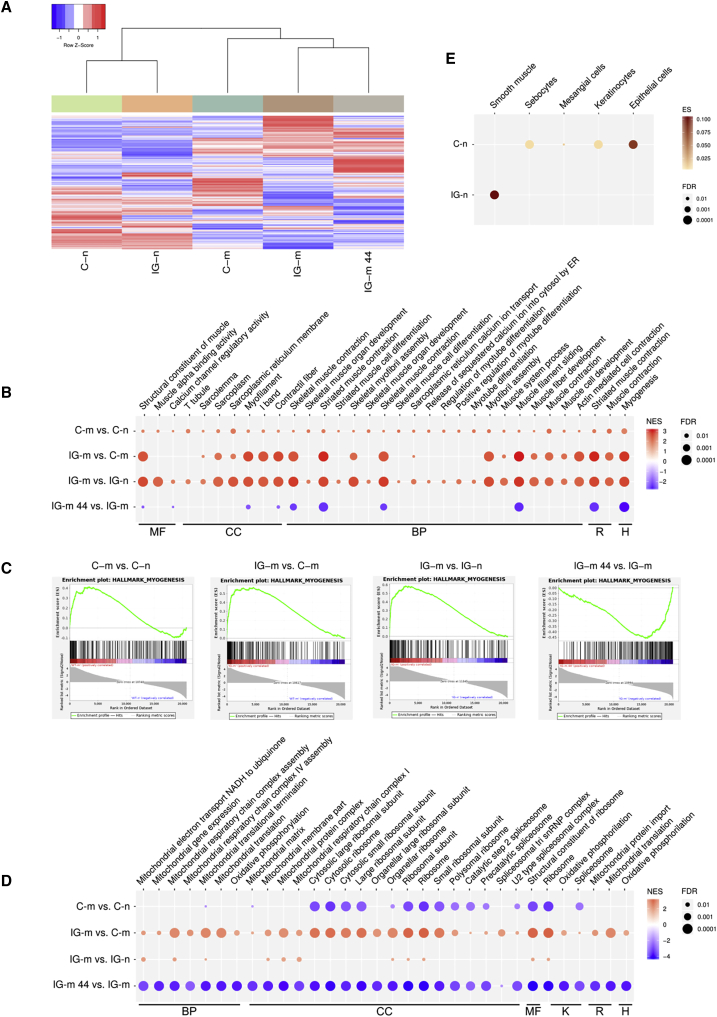
Figure 1.
Transcriptional profiling of native and myogenic USCs of control individuals and DMD
(A) Supervised hierarchical clustering using the 9,383 differentially expressed genes in all comparisons. In the heatmap, each column represents a sample and each row a gene. The color scale bar represents the relative gene expression changes normalized by the standard deviation.
(B) Induction of MYOD1 gene expression activates an extensive transcriptional program of gene sets related to muscle differentiation, development, and activity (contraction), both in native and DMD USCs. AON treatment seems to specifically repress contraction-related functions. MF, Gene Ontology, molecular function; CC, Gene Ontology, cellular component; BP, Gene Ontology, biological processes; R, Reactome gene set; H, Hallmark’s gene set; NES, normalized enrichment score. Red, positive NES (i.e., activation of the gene set in the first condition); blue, negative NES (i.e., repression of the gene set in the first condition); FDR, p value after false discovery rate correction. Only significantly (FDR ≤ 0.05) enriched gene sets are shown.
(C) Enrichment plot of the myogenesis gene set from the Hallmark collection (Figure S1): the myogenesis process is activated after MyoD treatment (first 3 plots), while repressed following AON transfection. All genes of the gene set are ranked based on the ratio2noise statistics, with the most upregulated genes (red color) on the left and the most downregulated genes (blue color) on the right. The green curves show the enrichment score and reflect the degree to which each gene (black vertical lines) of the myogenesis gene set is over-represented at the top or at the bottom of the ranked gene list.
(D) MyoD induction promotes the mitochondrial and ribosomal activities, whereas both are repressed by AON treatment. K, KEGG database; further abbreviations as in (B).
(E) Deconvolution analysis indicates that native USCs are significantly enriched in gene sets of specific cell types. In particular, DMD USCs are enriched in gene sets of smooth muscle cells only, whereas USCs from the pool of control individuals are enriched in gene sets characteristic of cells from a different component of the urinary system. ES, enrichment score; FDR, false discovery rate.
A specific “disease-related” transcription signature is observed in DMD USCs, and the myogenic differentiation consistently modifies their transcription profile compared to control cells (Figure 1A; Figure S1A).
To get an exhaustive insight into gene circuits differentially altered in DMD USCs, we performed GSEA using Hallmarks, KEGG, Reactome, and GO collections from MsigDB. Only a few, but focused, gene sets were significantly deregulated (FDR ≤ 0.05) in IG-n compared to C-n. Specifically, we observed the under-representation of gene sets related to the inflammatory response, a process associated with the damage of DMD muscle23 and in skin-related circuits that are known to be altered in the absence of DYS protein24 (Figure S1C). Conversely, gene sets including multiple mechanisms of muscle development and function were activated in C-m and IG-m (Figure 1B). Following forced myogenesis, most signals involved in muscular development, differentiation, and contraction processes were upregulated in IG-m compared to C-m cells, as it occurs in DMD skeletal muscle (Figure 1B).11 Interestingly, several gene sets associated with mitochondrial respiratory functions resulted as activated in IG-m cells (Figure 1D), as identified in skeletal muscle of DMD mouse models and humans.25 In addition, signatures of ribosomal pathways were over-represented in IG-m USCs compared to C-m.
Interestingly, AON treatment of IG-m cells impacts the overall gene expression (Figure 1A), causing the deregulation of 772 genes (Figure S1A), with the reversion of mitochondrial respiratory genes and several muscular-related pathways, such as those involved in inflammation, myogenesis, and muscle contraction (Figures 1B and 1C; Figure S1D). This AON transfection effect suggests a corrective action on exon skipping. Surprisingly, AON induces a global anti-myogenic effect, since most of the core enrichment genes of the Hallmark myogenesis gene set were switched off and most of the upregulated genes in IG-m reverted following the AON treatment (Table S3). Finally, we observed the activation of Myc signaling in IG-m USCs, pointing out the role of dystrophin in mechanisms related to tumor regulation. Interestingly, the activation of Myc signaling was also reverted by AON treatment (Figure S1D).
To elucidate the cellular composition of USCs, we investigated the gene expression profiles of IG-n and IC-n samples with an in silico deconvolution method that suggested different compositions of native cell types. Specifically, gene expression deconvolution indicated a homogeneous cell composition in IG-n USCs, with a significant enrichment only for the smooth muscle cellular component and a higher heterogeneity of control USCs, with enrichment in sebocyte, epithelial, keratinocyte, and mesangial cells (Figure 1E). Interestingly, all these cell types resemble the origin of USCs (i.e., glomerular parietal epithelial cells),26 and this evidence prompted us to further investigate the cellular composition of native USCs by cell sorting.
USC Celector analysis identifies and separates USC subpopulations
To strengthen the evidence from deconvolution analysis of gene expression data, we applied Celector technology to sort the native USCs from the control subjects and DMD individuals and characterize their cellular composition. Sample profiles showed differences between the control and DMD USCs, and some variations were observed even within the two DMD cells.
Control USCs showed a reproducible profile with similar distribution of cells among individuals, having single cells eluting from the 6th until the 14th minute of analysis (Figure 2A) and cell aggregates eluting in fraction F1 (Figure 2D). The main peak was, however, composed by two fractions, F2 and F3, definitively supporting a heterogenous cell type composition (Figure 2A).
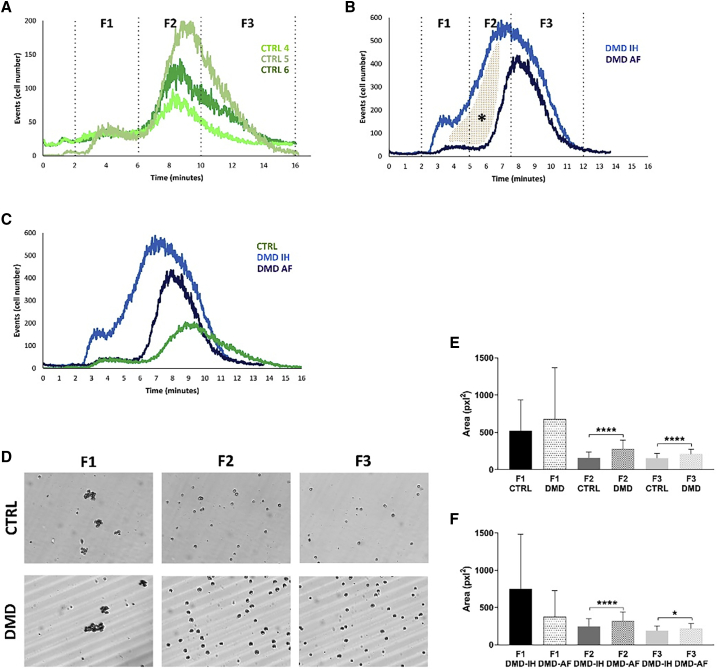
Figure 2.
Celector profile of control individuals and DMD USCs
(A–C) Elution profile of control individuals (4–6) and DMD (IH and AF) samples and cell collection time intervals (fractions F1, F2, and F3). The profile represents the number of eluting cells versus time.
(A) Control cells showed a similar profile, as seen by the same elution time (6 to 15 min). The height of the peak represents the number of cells analyzed. In the control group, cell aggregates eluted in the first minutes are recorded by the first peak.
(B) DMD cells showed a similar profile, but the elution time is shorter compared to control (CTRL) cells. When two DMD samples were analyzed, it was evident that there was a higher distribution of cells in F2 (5 to 7.5 min) in the IH sample compared to the AF sample (colored area, ∗).
(C) The comparison of the three profiles showed a heterogeneity among USCs. DMD-USCs exit earlier from the capillary channel compared to CTRL, meaning that they are bigger and denser.
(D) Based on live images of eluting cells taken by the camera placed at the outlet of the capillary channel, we confirmed the presence of cell aggregates in F1 and then single cells flowed in F2 and F3.
(E) Area of eluting cells confirmed the separation principle. Average of DMD cell area is higher for all three fractions (F1, F2, and F3) compared to CTRL.
(F) Differences in the cell area between the two DMD samples. DMD AF presented bigger cells compared to DMD IH. Graphs show average and standard deviation., Unpaired t test was performed: ∗p < 0.05; ∗∗∗∗p < 0.0001.
Diversely, DMD USC analysis exhibited two distinct profiles. DMD individual IH, carrying an exon 50–52 deletion, showed a larger-base peak with a high proportion of F2 cells, while DMD individual AF carries an exon 46–47 deletion, showing a thin-base peak with the majority of F3 cells (Figure 2B). Notably, the profile of DMD cells is clearly different from those of the control samples (Figure 2C). So, based on the Non-Equilibrium, Earth Gravity-Assisted Dynamic Fractionation (NEEGA-DF) principles that bigger and denser cells elute earlier than smaller ones, the dimensional analysis proved the presence of large-sized DMD USC cells in F2 and F3 compared to control USCs (Figure 2E), suggesting a DMD-related effect on cell size. This is also clearly visible in the capture images of the running analysis (Figure 2D). In addition, both F2 and F3 fractions of DMD individual IH showed smaller cells compared to DMD individual AF (Figure 2F).
Cells collected from every sample were immediately plated in proliferating medium for morphological evaluation and phenotype analysis. In general, no difference was observed among collected fractions in all samples. Cells were quite homogeneous in their morphology, with fibroblastic-like or rounded shapes present in all sorted fractions. The DMD USCs appear larger than the control USCs (Figure 3A), as also shown by quantification of the area of eluting cells.
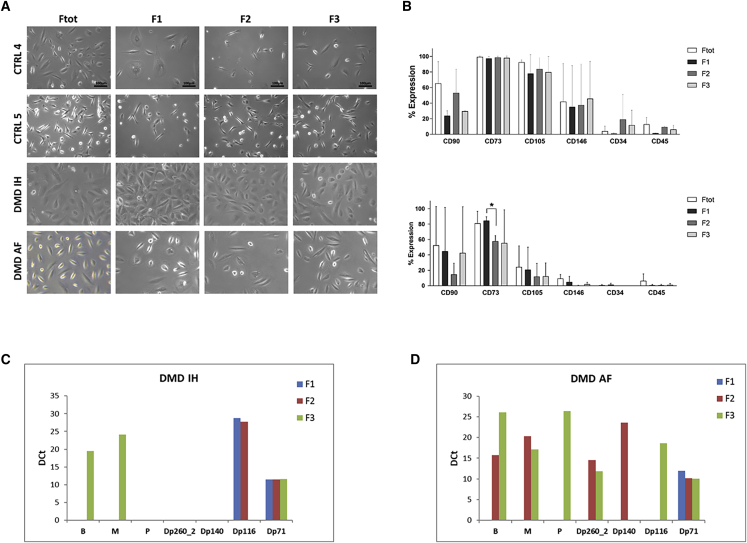
Figure 3.
Characterization of USC fractions from DMD and control individuals
(A) Comparison of cellular morphology among fractions. No differences in the morphology are observed between the total fraction (Ftot) and the sorted fractions F1, F2, and F3: both elongated and rounded cells are present before and after Celector separation. The morphology of the third control sample was like CTRL4.
(B) Evaluation of CD surface markers expressed in the control (upper graph) and DMD (lower graph) USCs by flow cytometry. Bars represent the average of expression between controls (3 samples) and the two DMD samples (unpaired t test: ∗p < 0.05). CD73 and CD105 were highly expressed in all control USC fractions, while significantly decreased in some fractions of the DMD USCs. CD73 is significantly lower both in F1 and F2 of DMD USCs as compared to F1 and F2 of control (p < 0.05 and p < 0.01, respectively) and in F2 compared to F1 in DMD USCs (p < 0.05). CD105 is significantly lower in Ftot, F2, and F3 (p < 0.05) of DMD fractions as compared to controls. CD146 has low expression in USCs of the control and is almost not expressed in DMD USCs.
(C and D) Gene microfluidic card (FluiDMD) profiles of native USCs from DMD IH carrying the deletion of exons 50–52 (C) and DMD AF with the deletion of exons 46–47 (D). FluiDMD shows amplification of the DMD isoforms for the three fractions (F1 blue bar, F2 red bar, and F3 green bar) obtained following the separation by Celector. M and B isoforms are represented both in F2 and F3, while P and Dp140 isoforms are present only in DMD individual AF and in F3. Dp71 is expressed in all the fractions, including F1. Dp260 and Dp116 isoforms are inconsistently expressed across fractions. DCt, delta Ct corresponding to the difference between Ct from DMD isoforms and Ct of the housekeeping gene (β-actin).
Since USCs have been shown to express mesenchymal markers,9,27, 28, 29 we evaluated if there were some differences in the expression of these markers among the fractions (Figure 3B). Mesenchymal markers CD73 and CD105 were highly expressed in all control USC fractions, while they significantly decreased in some fractions of the DMD USCs. CD73 was significantly lower in both F1 and F2 of DMD USCs as compared to the same fractions of the control (p < 0.05 and p < 0.01, respectively) and in F2 compared to F1 in DMD USCs (p < 0.05). CD105 was lower in all DMD fractions compared to controls, with statistically significant differences in Ftot, F2, and F3 (p < 0.05), while pericyte marker CD146 was almost not expressed in DMD USCs. The hematopoietic markers CD34 and CD45 were not expressed in both groups. These results confirmed the elevated heterogeneity of USCs.
We also analyzed the DMD transcript in the DMD USC fractions. FluiDMD card showed that the DMD isoforms are differentially represented in the two DMD individuals’ fractions (Figures 3C and 3D). Full-length M and B isoforms are represented both in F2 and F3, while full-length P and the short Dp140 isoforms are present only in DMD individual AF and in F3.30
The shortest and ubiquitous Dp71 isoform, normally present in many adult tissues and with higher levels in the central nervous system, is expressed in all the tested fractions, including F1, where it represents the only isoform detected. Dp260 and Dp116 isoforms are inconsistently expressed across fractions. Dp140, Dp260, and Dp116 are tissue-specific isoforms normally present only in the brain, retina, and peripheral nerves, respectively.30,31
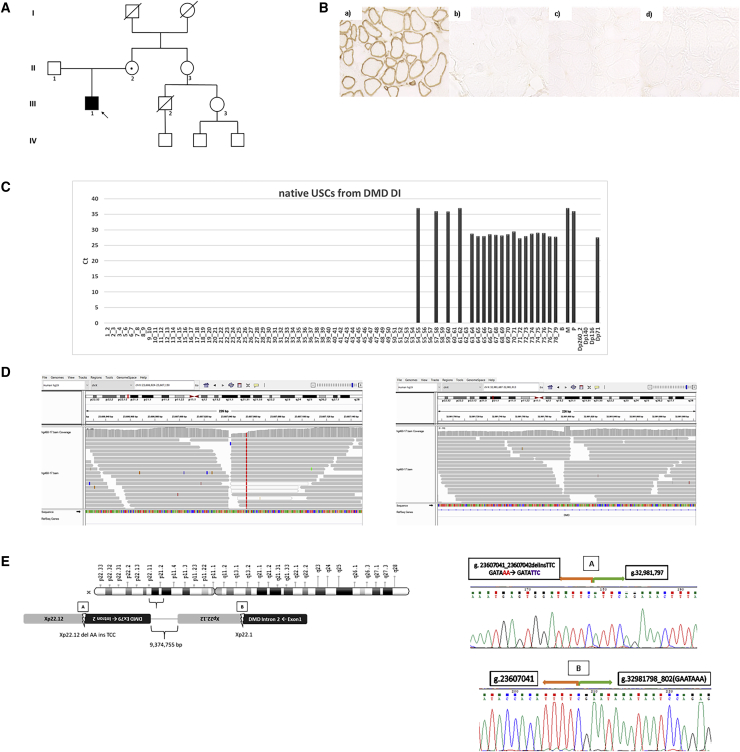
USCs represent a robust and effective model to diagnose atypical DMD mutations in undiagnosed individuals
We used USCs to profile the DMD transcript using FluiDMD in three individuals (PF, DI, and SE), where no DMD mutations were identified by MLPA or DMD gene full sequencing.
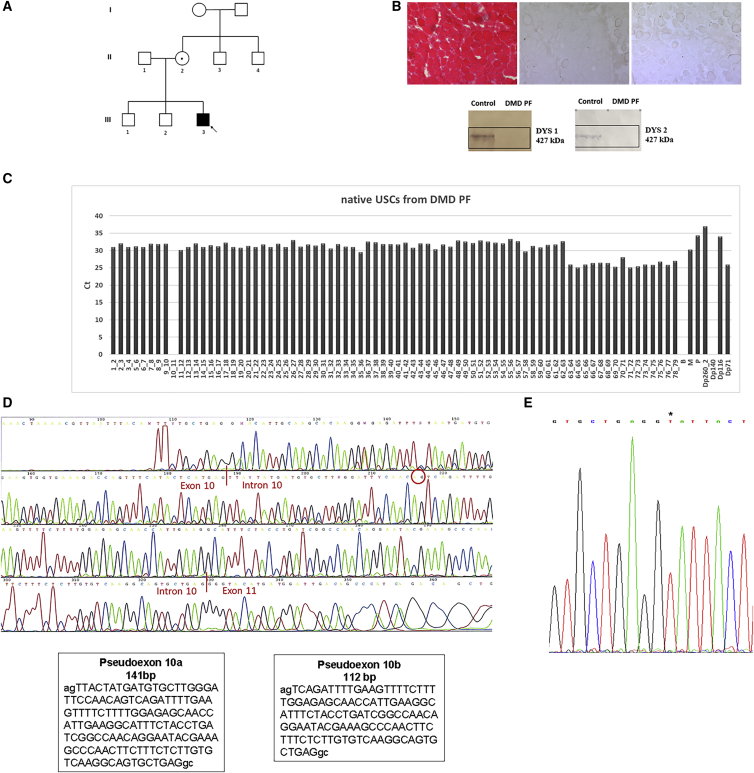
PF has no family history of any neuromuscular disorders (Figure 4A). Muscle biopsy showed dystrophic changes and a complete absence of DYS protein on WB (Figure 4B). An absence of amplification of the exon-exon junction 10–11 (Figure 4C) was observed in his USCs by FluiDMD analysis. RT-PCR amplification followed by sequencing revealed the presence of two PCR products between exons 10 and 11, one with the inclusion of an additional 141 bp and the second one with the inclusion of 112 bp (Figure 4D). Sequencing of DMD intron 10 identified a previously unreported SNP c.1149+250C>T, which generates a novel donor splice site (Figure 4E). This donor splice site activates two cryptic acceptor splice sites (c.1149+106_107 and c.1149+135_136) creating two novel alternative pseudo-exons, defined as c.1149+108_248 and 1149+137_248, incorporated into the DMD transcript and both leading to early premature stop codons. The sequence of the mother’s genomic DNA confirmed the maternal inheritance of the mutation, being that she is a carrier (data not shown).
Figure 4.
Clinical features, transcriptional studies, and identification of mutation in DMD PF
(A) Pedigree of DMD individual PF’s family. The index individual (III3, arrow) has a DMD diagnosis and carries the c.1149+250C>T mutation in intron 10 of the DMD gene (D, right). The individual’s mother (II2) is a carrier of the DMD mutation.
(B) Histological analysis with hematoxylin and eosin (H&E) (left part) and immunostaining with the dystrophin antibody to rod-domain and C-terminal regions (right part) of muscle sections from DMD PF. Dystrophic changes (variation in fiber size, fibrosis, inflammatory infiltrate) are observed with H&E and only few revertant fibers are dystrophin positive. Western blot analysis (lower part) shows the complete absence of the dystrophin protein in the DMD PF muscle.
(C) Expression of dystrophin transcript in native USCs from DMD individual PF. FluiDMD shows the absence of amplification of the exon-exon junction 10–11 of the dystrophin transcript. Ct, cycle threshold.
(D) Sequencing of the dystrophin transcript of the DMD individual. The region spanning exons 10–12 was amplified, and PCR products were sequenced. Two transcripts spliced between exon 10 and exon 11 were found, the first one of 141 bp (pseudo-exon 10a, left box) and the second one of 112 bp (pseudo-exon 10a, right box); the red circle highlights the acceptor site of the shorter pseudo-exon 10b.
(E) Sequencing of intron 10 identified the pathogenic mutation c.1149+250C>T (∗) generating a donor splice site.
DI has no family history of any neuromuscular disorders (Figure 5A). The immunochemistry analysis of his muscle biopsy showed complete absence of DYS (Figure 5B). Complete absence of the transcript region spanning exons 1–53 was observed in his USCs by FluiDMD analysis (Figure 5C), suggesting the presence of a large genomic rearrangement. FluiDMD profile shows that Dp71 is the only isoform expressed in DI’s USCs. Indeed, the Dp71 promoter, located in intron 62, is spared by the inversion. Exon-exon junctions 54–55, 57–58, 59–60, and 61–62 are observed, though with extremely high Ct, and could be due to intermediate splicing products related to some residual activity of shorter isoform promoters (Dp260, Dp140, and Dp116), which might have some inverse-oriented, inefficient transcription. M and P isoform exon 1 junctions are detected, since their promoter regions are not involved in the inversion (Figure 5E). The absence of amplification of the expected M 1–2 exon-exon junction is probably due to a non-sequential splicing, which has been previously described in the DMD transcript.32 WGS analysis (Figure 5D) identified a large genomic inversion of about 10 Mb encompassing the Xp region g.23607043_32981797inv. The two breakpoints of the inversion were localized in an Xp22.12 non-coding region (g.23607041_23607042) and in Xp21.1 (g.32981798_802) corresponding to intron 2 of the DMD gene, respectively. The genomic inversion, therefore, includes almost all DMD intron 2 and exons 3–79 (Figure 5E, left panel). Amplification and sequencing of the 5′ and 3′ unique genome fusion junctions confirmed the two breakpoint sequences (Figure 5E, left panel). Sanger sequencing also highlighted the occurrence of a small deletion/insertion due to a 2 bp deletion (delAA) at position g.23607041_042 and an insertion of 3 bp (insTTC) at the breakpoint in Xp22.12. A short 5 bp GAATA-like motif localized at the Xp21.1 breakpoint (g.32981798_802) could be possibly implicated in causing the large inversion through a microhomology-mediated recombination mechanism. The mother resulted as being a carrier of the inversion (data not shown). RepeatMasker analysis showed a 201 bp LINE1 element in intron 2 of the DMD gene, which is involved in the inversion breakpoint.
Figure 5.
Clinical features, transcriptional studies, and identification of mutation in DMD DI
(A) Pedigree of DMD individual DI’s family. The index individual (III1, arrow) has a DMD diagnosis and carries the genomic inversion of about 10Mb encompassing the Xp region g.23607043_32981797inv. The individual’s mother (II2) is a carrier of the DMD mutation. She is asymptomatic with a normal serum CK level.
(B) Immunohistochemistry of dystrophin. Immunohistochemistry of spectrin (Leica) (a) revealed normal sarcolemma integrity. Dystrophin (b–d) immunohistochemistry (Leica) showed no labeling of antibodies directed to N-terminal (b), rod-domain (c), and C-terminal (d) of dystrophin (20× magnification).
(C) FluiDMD of RNA from native USCs of DMD patient DI shows the absence of amplification of the region spanning exon 1–53 of dystrophin transcript with only Dp71 fully transcribed, since maintaining their promoter and first exon. Sporadic amplification of some upstream exons is due to mimicry, since no adjacent exons are present in the DMD transcript. Ct, cycle threshold.
(D) Genomic inversion of about 10 Mb encompassing the region Xp g.23607043_32981797inv revealed by WGS analysis. The breakpoints are localized in a non-coding region of Xp22.12, position g.23607041_23607042, and in Xp21.1, position g.32981798_802, corresponding to intron 2 of the DMD gene.
(E) Left panel: genomic inversion including part of intron 2 and the exons from 3 to 79 of the dystrophin gene. Right panel: the unique junctions generated by the inversion (A and B) were amplified and sequenced. A small deletion/insertion was found at breakpoint 3′ (A) due to 2 bp deletion (delAA) and the insertion of 3 bp (insTTC). At breakpoint 5′ (B), a GAATA-like motif was observed.
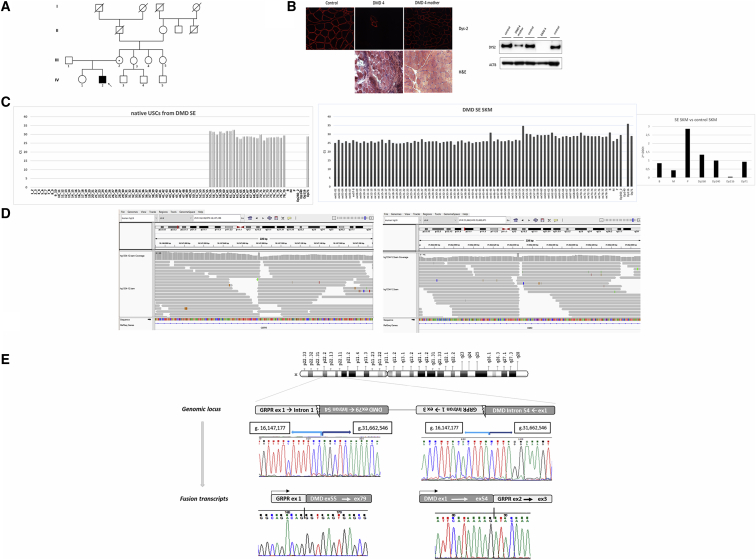
Patient SE’s pedigree does not show any family history of neuromuscular disorders (Figure 6A). His muscle biopsy showed a dystrophic pattern and an absence of DYS protein (Figure 6B, left panel). The mother also has a muscle biopsy showing a mosaic of DYS staining, therefore being a possible carrier (Figure 6B). The FluiDMD analysis of the USCs from patient SE showed the absence of exons 1–54 and Dp71 as the only isoform expressed (Figure 6C, left panel). Interestingly, FluiDMD on muscle biopsy RNA showed a different profile, apparently with all DMD exons represented and all DMD isoforms represented, except for Dp140 (Figure 6C, right panel). A clear delay of the exon 54–55 junction was also well visible, possibly suggesting the occurrence of a genomic event hampering the regular splicing of these two exons. In addition, a global significant reduction of muscular DMD transcript (M) is clearly visible in muscle biopsy-derived RNA (Figure 6C, right panel). WGS analysis identified a large inversion of about 15Mb encompassing the region of chrX: g.16147177_31662545inv (Figure 6D). The two breakpoints were localized in Xp22.2, position g.16147177_178, corresponding to intron 1 of the GRPR gene and in Xp21.1, position g.31662545_546, corresponding to intron 54 of the DMD gene, respectively. RepeatMasker analysis showed a 71 bp Short interspersed nuclear element (SINE) localized in intron 54 of the DMD gene and a 50 bp Long interspersed nuclear element (LINE) in intron 1 of the GRPR gene, both adjacent to the inversion events. This genomic inversion encompasses DMD exons 55–79 and includes the promoter regions of Dp116 and Dp71 (Figure 6E). Breakpoint junctions were amplified and sequenced, revealing the 5′–3′ unique junctions (Figure 6E, genomic locus). A microhomology (AG/TC) at the two breakpoints might have been possible, facilitating the inversion. The mother is heterozygous for the inversion, being a confirmed carrier (data not shown). Interestingly, this inversion impacts on transcription, since we identified two fusion transcripts: one is driven by the GRPR promoter, leading to exon 1 being adjacent to the inverted DMD exon 55; the second is driven by the M DMD promoter till exon 54, which becomes adjacent to the inverted GRPR ex2 (Figure 6E, fusion transcript panels). These two fusion transcripts account for the muscle biopsy FluiDMD profile, which detects all DMD exons. In native USCs, only the DMD transcript driven by the GRPR promoter is amplified starting from exon 55 (Figure 6E).
Figure 6.
Clinical features, transcriptional studies, and identification of mutation in DMD SE
(A) Pedigree of DMD patient SE’s family. The index individual (IV2, arrow) has a DMD diagnosis and carries the inversion of about 15 Mb encompassing the chrX region: g.16147177_31662545inv; the individual’s mother (III2) is a carrier of the DMD mutation with a high serum CK level and mosaic dystrophin pattern at muscle biopsy (panel B).
(B) Right panel (upper part) shows the immunostaining with Dys-2 dystrophin antibody of muscle sections from the normal control, SE, and his carrier mother. Only a cluster of a few revertant, dystrophin-positive fibers is seen in patient SE, while a mosaic dystrophin staining is observed in the mother. Histological analysis in DMD patient SE with H&E (lower part) shows diffuse dystrophic changes (variation in fiber size, fibrosis, inflammatory infiltrate, and pale necrotic fibers). WB (left panel) shows the complete absence of the dystrophin protein in DMD SE’s muscle (Dys2, C-terminal antibody). In the carrier mother, a fragment of normal molecular weight and reduced intensity is detectable.
(C) Expression of dystrophin transcript in native USCs from patient SE. Left panel: FluiDMD on native USCs shows the absence of amplification of region spanning exon 1–55 with only the Dp71 isoform represented. Central panel: FluiDMD on skeletal muscle shows the amplification of both full-length and shorter DMD isoforms, except for Dp140, which is usually undetected. Right panel: 2−ΔΔCt method was used to obtain the relative quantification of the DMD transcript between the DMD individual and the control. A global significant reduction of muscular dystrophin transcript (M) and the upregulation of P and Dp260 isoforms are observed. Ct, cycle threshold.
(D) Inversion of about 15 Mb encompassing the chrX region: g.16147177_31662545inv, revealed by WGS analysis. The breakpoints are localized in Xp22.2, position g.16147177_178, corresponding to intron 1 of the GRPR gene and in Xp21.1, position g.31662545_546, corresponding to intron 54 of the DMD gene.
(E) Genomic locus: the unique junctions A and B generated by the inversion were amplified and sequenced, and the inversion configuration was confirmed. The figure highlights the genomic coordinates of the novel junctions generated by the inversion; 2 bp homology (AG/TC) was observed at the breakpoints, possibly causing the genomic inversion by a microhomology-mediated recombination (MMR). Fusion transcript: two fusion transcripts were found in RNA from SKM, the first one between GRPR exon 1 and DMD exon 55 induced by the GRPR promoter (left) and the second one between DMD ex 54 and GRPR ex 2 derived from the DMD promoter (right).
Discussion
We analyzed USCs to define their transcriptional landscape also in relation to AON treatment and to identify subpopulations that might be disease related. We also used USCs as a source of DMD transcripts to address atypical mutations in undiagnosed DMD individuals, followed by WGS.
RNA-seq data defined transcriptomic signatures of USCs and support their use as a DMD cell model
Our RNA-seq study demonstrates that during USC myogenesis, a set of MyoD-regulated genes are consistently deregulated (both in DMD and in control individuals) as observed in other MyoD-induced cell types.19 In addition, specific markers related to myogenesis and muscular functions are upregulated. As we previously reported,9 direct exogenous expression of MyoD induces trans-differentiation of USCs into myoblast-like cells. According to this, functional enrichment analysis showed that native USCs partially recapitulate the DMD muscle phenotype and that only a few genes associated with DMD muscle signatures, like inflammation, are deregulated. Following USC myogenic transformation, the vast majority of known DMD deregulated gene sets are markedly deregulated, especially those involved in muscle contraction mechanisms, as expected, thus highlighting dystrophin deficiency and DMD-related RNA features.
In DMD myogenic USCs, we observed an evident deregulation of transcripts associated with muscle structural genes, development, and differentiation pathways, indicating an aberrant myogenesis, as also suggested by the activation of myogenesis genes. Indeed, dystrophin interacts with other anchoring proteins and plays a key role in maintaining the stability of the sarcolemma, therefore regulating the expression and localization of several myogenesis-related signaling molecules such as kinases.11 Dystrophin is also produced in satellite cells, where it is involved in controlling the satellite cell polarity and asymmetric division. Therefore, lack of dystrophin may influence their symmetrical division and may decrease the generation of new myogenic progenitors.33
Among the deregulated gene sets, we found that the mitochondrial gene pathway is invariably altered, not surprisingly since impaired mitochondrial respiratory function with Complex I dysfunction and elevated mH2O2 levels is known to occur in DMD muscle.25,34,35 Notably, deregulation of mitochondrial pathways was reverted by AON exon-44-skipping treatment, suggesting mitochondria might be an appropriate target to be studied in USCs.
A MYC-signaling-related path is also upregulated in DMD myogenic USCs. MYC targets are known to predict tumor aggressiveness in breast tumors.36 Very interestingly, DMD is a known oncosuppressor, a role that is now more explored in cancers, especially brain tumors.37, 38, 39, 40 It is appealing to consider USCs as pertinent cells to study cancer, like in DMD-related tumors such as sarcoma and glioblastoma, but also in other cancer types.
AON transfection inducing the skipping of exon 44 and the restoration of the dystrophin protein expression9 reverted most of the DMD deregulated genes, especially those belonging to myogenesis, inflammation, and muscle contraction gene sets. In a previous study performed in human control primary myotubes treated by U7snRNA-E53, this was not observed.41 The authors, however, used control cells and not DMD-derived cells. Further data are needed to confirm that USCs can be a valuable cell model in evaluating exon-skipping effects.9,42
Obtaining meaningful omics data from large, rare disease patient cohorts is challenging. DMD is a rare neuromuscular disorder with an estimated incidence of 1 in 5,000 male births, and large patient cohort studies are possible only by international collaborations.43
Further complicating the issue of having consistent replicates, many DMD mutations are private, ultrarare, and occur in a small group of individuals.44 It is therefore unlikely to find DMD muscle biopsy from individuals carrying the same genomic mutation to be considered as replicates. Nevertheless, the large dataset of RNA-seq output included in this paper represents a proof of concept supporting USCs as mirroring DMD muscle transcriptome and suggesting that they can be useful to study DMD disease pathomechanisms. This is corroborated further by our genomic data, where USCs were successfully used to address DMD atypical mutation identification.
Celector technology for USC sub-population studies
Deconvolution analysis of RNA-seq data clearly suggests different cell-type compositions of USCs. It is known that mesenchymal cells, such as USCs, are a heterogeneous cell population displaying distinct morphologic and expression characteristics in cultures.9,42,45 Unfortunately, the lack of already-known specific and unique cell surface markers for the identification and isolation of each cell subpopulation makes cell sorting difficult and imprecise. To gather information about USC cellular composition, we used Celector technology. This novel method revealed for the first time the presence of at least three distinct cell fractions that were eluted at different velocities and collected in separated tubes for subsequent analysis, without any additional manipulation and use of antigen-specific antibodies.
The profile of these fractions was, however, similar in the control samples but quite different in DMD cells with possible disease-related characteristics.
In DMD cells, CD markers were uneven, and DMD isoforms were differentially detected in each fraction and among DMD individuals carrying different mutations. The Dp140 isoform, which is known to be a developmentally regulated isoform, uniquely expressed in some adult brain areas, was found expressed in the F2 fraction of a DMD individual carrying an exon 46–47 deletion. This surprising finding suggests that the F2 subpopulation might be used to study DMD brain-related functions.46 Furthermore, based on published data reporting the possibility to convert the urinary cells in neurons,47,48 we may speculate that specific USC fractions might be most suitable to be induced as neuronal cells, with a more general utility for exploring other brain circuits. In addition, we suggest that cell morphology in adherent cells might not be related to expression characteristics, since both rounded and elongated USCs, after sorting, are still present in cultures.
The different expression profiles between control and DMD cells found in both deconvolution and Celector analysis indeed indicate different development stages of cells underlining its cell-specific sorting performance. Indeed, Celector is capable of sorting cells with higher stemness and plasticity, as observed in amniotic fluid stem cells, which share similar characteristics with USCs because of their renal origin,49, 50, 51 therefore being an attractive tool to finely separate USC populations to be used for different aims. These results lead the basis for new studies, in which the deep characterization of each cell fraction will elucidate the possible biological applications of the USC sub-populations.
USC RNA profile and WGS successfully identify “undetected”, rare, atypical DMD mutations
Bridging USCs with diagnostic applications, we demonstrated that these cells can be used to consistently profile DMD RNA, addressing the genomic definition of rare, atypical mutations. Although about 98% of DMD mutations can be identified by standard methods, a very small percentage (likely less than 1%) of DMD individuals remain undiagnosed and orphan of a genetic diagnosis.44 These individuals are often defined as “undiagnosed”, a very popular term currently. More appropriately, they could be defined as “undetected”, since the clinical diagnosis was made, but the mutation not identified, due to insufficient detection rates of routine diagnostic methods. FluiDMD gene transcriptional profiling followed by WGS analysis successfully allowed the identification of all causative mutations in the 3 undiagnosed or undetected DMD individuals. In all cases, FluiDMD alone was able to precisely identify the exonic critical region involved in the mutation, where absent exons or retarded exon-exon junctions were seen. In patient PF, a pathogenic intron 10 small variation creates a very strong (scored 0.44) novel donor splice site, which induced the recognition of two intron 10 downstream cryptic acceptor splice sites. This small pathogenic variation causes the production of two out-of-framing transcripts, each of them incorporating one of the two alternative pseudoexons. The out-of-frame transcripts hamper DYS translation and cause a severe DMD phenotype. This is a novel, rare, and deep intronic DMD mutation. It is not unusual, however; a few similar ones have been previously described.52
In patients DI and SE, we found two large and never-before-reported inversions in the DMD gene, further supporting the value of WGS in detecting novel complex rearrangements.53, 54, 55
Indeed, though in a few cases, complex CNVs, such as large insertions, deletions, translocations, and inversions, were recently identified in the DMD gene by long-read WGS, long-read gene-specific DMD sequencing, and nanochannel-based NGS technology.56, 57, 58, 59 We successfully identified two inversions using short-read WGS. Short-read WGS preserves high read quality and sequencing coverage depth, obviously very useful for accurate variation identification, reconstructing complex variations, and finely recognizing the breakpoints.
Patient DI’s inversion is a classic inversion whose 3′ breakpoint localizes in an Xp22.12 non-coding, intergenic region, and its 5′ breakpoint localizes inside DMD intron 2. A small deletion/insertion occurs at the 3′ breakpoint due to a 2 bp deletion (delAA) and to an insertion of 3 bp (insTTC). Adjacent to the 3′ breakpoint, a GAATA-like motif is observed. Not surprisingly, intron 2 is a huge intron (170,318 bp), extremely rich in mobile elements. RepeatMasker analysis indeed showed a 201 bp LINE1 element, which is involved in the inversion. This element might have primed a non-homologous recombination event. Functionally, and as expected, this inversion causes the complete lack of all full-length DMD isoforms. Patient DI may thus only express the Dp71 isoform. Nevertheless, his muscle biopsy shows a complete lack of DYS protein. The Dp71 isoform is known to play essential functions in the brain;60 therefore, protein production in this individual might be preserved in brain areas only via a tissue-specific expression. Indeed, patient DI is a 36-year-old man with classic DMD motor milestones but with fully preserved cognitive functions. He has two university degrees and excellent intellectual skills. This extremely peculiar mutation, which knocks out all DMD full-length isoforms in terms of either protein translation or complete transcription knockout, might therefore be very useful in studying the effects of pure Dp71-related functions. Patient SE shows a large inversion of about 15 Mb, whose breakpoints, however, are different, being localized at the 3′ of DMD intron 54 and at the 5′ of intron 1 of the GRPR gene. Gastrin-releasing polypeptide (GRPR or Bombesin, MIM: 137260) is a receptor, networking within the autocrine loop and involved in cell growth, being highly expressed in neuroendocrine cells, and was found overexpressed in a variety of cancers such as small-cell lung carcinoma. GRPR is highly expressed in GABAergic interneurons of the lateral nucleus of the amygdala and exerts an excitatory function in these interneurons with a cascade inhibitory effect. Some animal studies supported a causal relationship between GRPR gene expression and amygdala-dependent memory for fear.61 Apparently, no relationship exists between GRPR and DMD gene function, although the fear-related aspects are of interest, since they are invariably present in DMD individuals.46,62 As pure speculation, the inversion breakpoint within the GRPR gene, causing its knockout, could have contributed to worsening the fear-related phenotype.
RepeatMasker analysis showed a 71 bp SINE element localized in intron 54 of the DMD gene and a 50 bp LINE element in intron 1 of the GRPR. The presence of these two mobile elements does not suggest a specific mutation mechanism, although their proximity may have facilitated non-homologous recombination and inversion. This inversion is also extremely peculiar for its effect on transcription. It generates two fusion transcripts, one driven by the GRPR gene with composition GRPR exon 1 and DMD exon 55–exon 79, and the other driven by the DMD M promoter with composition of DMD exon 1 m–exon 54 and GRPR exon 2.
The different transcriptional profile in the USCs and skeletal muscle we have observed, characterized by the lack of the DMD promoter-driven transcript in USCs, which, according to our USC transcriptional data, would be expected since the M isoform should be transcribed, is surprising. Epigenetic chromatin reshaping, due to the very large genomic rearrangement, may have occurred, causing the silencing of the M promoter, as already described for other genomic regions.63
Patient SE’s inversion was addressed by RNA analysis, since the delayed 54–55 exon-exon junction likely reflects the occurrence of the intron 54 breakpoint, and its low-level representation in muscle might be related to trans-splicing events between the two fusion transcripts, which may form a chimeric RNA.64,65 The WGS reads seem to exclude a highly represented mosaicism, which, however, cannot be completely ruled out.66
Conclusions
We showed that USCs are a robust tool for studying DMD, with a large landscape of utilities, from transcriptomic profiling to diagnostics. The intriguing possibility of sorting out cell subpopulations using Celector may further allow for the isolation of specific cells that may exhibit different functional properties and give rise to several cell lineages, with many implications in research and therapeutic investigations. We believe that these autologous cells will have a variety of applications for both DMD and other rare genetic disease research and diagnostics. As a final consideration, the genomic (RNA profile followed by WGS) approach we have adopted might be the gold standard to identify undetected DMD mutations, very likely applicable to many other similar, undetected, pathogenic gene variations.
Acknowledgments
This study was financially supported by the EU SOLVE-RD project (grant no. 779257 to A.F.) and by Duchenne Parent Project Italy (Urinary Stem Cell biobank, grant no. 11/2018 to A.F.). Acknowledgments are also due to Sarepta Therapeutics for the research grant “Improving time to diagnosis in Dystrophinopathies and other Limb Girdle Muscular Dystrophies” (2018, to A.F.) and to the Telethon Genetic BioBank (GTB12001D) and the Eurobiobank network for providing individuals’ samples. A.F., V.A.S., E.P., and T.M. are HCP representatives of the European Reference Network (ERN) Euro-NMD (ern-euro-nmd.eu).
Declaration of interests
A.F. is the Principal Investigator of Sarepta Therapeutics Essence and MIS51ON clinical trial for DMD. A.F. is PI of ongoing grants on DMD diagnosis funded by PTC Therapeutics and Sarepta Therapeutics. All other authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xhgg.2021.100054.
Data and code availability
RNA-seq data from this study have been deposited in the GEO database under accession number GEO: GSE162108.
Web resources
Babraham Bioinformatics, https://www.bioinformatics.babraham.ac.uk/projects/fastqc/
Integrative Genomics Viewer, http://software.broadinstitute.org/software/igv/
RepeatMasker, http://www.repeatmasker.org/
Supplemental information
References
- 1.Falzarano M.S., Ferlini A. Urinary stem cells as tools to study genetic disease: overview of the literature. J. Clin. Med. 2019;8:E627. doi: 10.3390/jcm8050627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meola G., Bugiardini E., Cardani R. Muscle biopsy. J. Neurol. 2012;259:601–610. doi: 10.1007/s00415-011-6193-8. [DOI] [PubMed] [Google Scholar]
- 3.Elston D.M., Stratman E.J., Miller S.J. Skin biopsy: Biopsy issues in specific diseases. J. Am. Acad. Dermatol. 2016;74:1–16, quiz 17–18. doi: 10.1016/j.jaad.2015.06.033. [DOI] [PubMed] [Google Scholar]
- 4.Ji X., Wang M., Chen F., Zhou J. Urine-derived stem cells: the present and the future. Stem Cells Int. 2017;2017:4378947. doi: 10.1155/2017/4378947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou T., Benda C., Dunzinger S., Huang Y., Ho J.C., Yang J., Wang Y., Zhang Y., Zhuang Q., Li Y., et al. Generation of human induced pluripotent stem cells from urine samples. Nat. Protoc. 2012;7:2080–2089. doi: 10.1038/nprot.2012.115. [DOI] [PubMed] [Google Scholar]
- 6.Bento G., Shafigullina A.K., Rizvanov A.A., Sardão V.A., Macedo M.P., Oliveira P.J. Urine-derived stem cells: applications in regenerative and predictive medicine. Cells. 2020;9:573. doi: 10.3390/cells9030573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guan J., Zhang J., Li H., Zhu Z., Guo S., Niu X., Wang Y., Zhang C. Human Urine Derived Stem Cells in Combination with β-TCP Can Be Applied for Bone Regeneration. PLoS ONE. 2015;10:e0125253. doi: 10.1371/journal.pone.0125253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schosserer M., Reynoso R., Wally V., Jug B., Kantner V., Weilner S., Buric I., Grillari J., Bauer J.W., Grillari-Voglauer R. Urine is a novel source of autologous mesenchymal stem cells for patients with epidermolysis bullosa. BMC Res. Notes. 2015;8:767. doi: 10.1186/s13104-015-1686-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Falzarano M.S., D’Amario D., Siracusano A., Massetti M., Amodeo A., La Neve F., Maroni C.R., Mercuri E., Osman H., Scotton C., et al. Duchenne muscular dystrophy myogenic cells from urine-derived stem cells recapitulate the dystrophin genotype and phenotype. Hum. Gene Ther. 2016;27:772–783. doi: 10.1089/hum.2016.079. [DOI] [PubMed] [Google Scholar]
- 10.Neri M., Rossi R., Trabanelli C., Mauro A., Selvatici R., Falzarano M.S., Spedicato N., Margutti A., Rimessi P., Fortunato F., et al. The genetic landscape of dystrophin mutations in Italy: a nationwide study. Front. Genet. 2020;11:131. doi: 10.3389/fgene.2020.00131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ghahramani Seno M.M., Trollet C., Athanasopoulos T., Graham I.R., Hu P., Dickson G. Transcriptomic analysis of dystrophin RNAi knockdown reveals a central role for dystrophin in muscle differentiation and contractile apparatus organization. BMC Genomics. 2010;11:345. doi: 10.1186/1471-2164-11-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Falzarano M.S., Scotton C., Passarelli C., Ferlini A. Duchenne muscular dystrophy: from diagnosis to therapy. Molecules. 2015;20:18168–18184. doi: 10.3390/molecules201018168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spitali P., Rimessi P., Fabris M., Perrone D., Falzarano S., Bovolenta M., Trabanelli C., Mari L., Bassi E., Tuffery S., et al. Exon skipping-mediated dystrophin reading frame restoration for small mutations. Hum. Mutat. 2009;30:1527–1534. doi: 10.1002/humu.21092. [DOI] [PubMed] [Google Scholar]
- 14.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liao Y., Smyth G.K., Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 16.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aran D., Hu Z., Butte A.J. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017;18:220. doi: 10.1186/s13059-017-1349-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roda B., Reschiglian P., Alviano F., Lanzoni G., Bagnara G.P., Ricci F., Buzzi M., Tazzari P.L., Pagliaro P., Michelini E., Roda A. Gravitational field-flow fractionation of human hemopoietic stem cells. J. Chromatogr. A. 2009;1216:9081–9087. doi: 10.1016/j.chroma.2009.07.024. [DOI] [PubMed] [Google Scholar]
- 19.Fratter C., Dalgleish R., Allen S.K., Santos R., Abbs S., Tuffery-Giraud S., Ferlini A. EMQN best practice guidelines for genetic testing in dystrophinopathies. Eur. J. Hum. Genet. 2020;28:1141–1159. doi: 10.1038/s41431-020-0643-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bovolenta M., Scotton C., Falzarano M.S., Gualandi F., Ferlini A. Rapid, comprehensive analysis of the dystrophin transcript by a custom micro-fluidic exome array. Hum. Mutat. 2012;33:572–581. doi: 10.1002/humu.22017. [DOI] [PubMed] [Google Scholar]
- 21.Robinson J.T., Thorvaldsdóttir H., Wenger A.M., Zehir A., Mesirov J.P. Variant review with the integrative genomics viewer. Cancer Res. 2017;77:e31–e34. doi: 10.1158/0008-5472.CAN-17-0337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bergstrom D.A., Penn B.H., Strand A., Perry R.L., Rudnicki M.A., Tapscott S.J. Promoter-specific regulation of MyoD binding and signal transduction cooperate to pattern gene expression. Mol. Cell. 2002;9:587–600. doi: 10.1016/s1097-2765(02)00481-1. [DOI] [PubMed] [Google Scholar]
- 23.Rosenberg A.S., Puig M., Nagaraju K., Hoffman E.P., Villalta S.A., Rao V.A., Wakefield L.M., Woodcock J. Immune-mediated pathology in Duchenne muscular dystrophy. Sci. Transl. Med. 2015;7:299rv4. doi: 10.1126/scitranslmed.aaa7322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pellegrini C., Zulian A., Gualandi F., Manzati E., Merlini L., Michelini M.E., Benassi L., Marmiroli S., Ferlini A., Sabatelli P., et al. Melanocytes--a novel tool to study mitochondrial dysfunction in Duchenne muscular dystrophy. J. Cell. Physiol. 2013;228:1323–1331. doi: 10.1002/jcp.24290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hughes M.C., Ramos S.V., Turnbull P.C., Rebalka I.A., Cao A., Monaco C., Varah N.E., Edgett B.A., Huber J.S., Tadi P., et al. Early myopathy in Duchenne muscular dystrophy is associated with elevated mitochondrial H2O2 emission during impaired oxidative phosphorylation. J. Cachexia Sarcopenia Muscle. 2019;10:643–661. doi: 10.1002/jcsm.12405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang D., Wei G., Li P., Zhou X., Zhang Y. Urine-derived stem cells: A novel and versatile progenitor source for cell-based therapy and regenerative medicine. Genes Dis. 2014;1:8–17. doi: 10.1016/j.gendis.2014.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Benda C., Zhou T., Wang X., Tian W., Grillari J., Tse H.F., Grillari-Voglauer R., Pei D., Esteban M.A. Urine as a source of stem cells. Adv. Biochem. Eng. Biotechnol. 2013;129:19–32. doi: 10.1007/10_2012_157. [DOI] [PubMed] [Google Scholar]
- 28.He W., Zhu W., Cao Q., Shen Y., Zhou Q., Yu P., Liu X., Ma J., Li Y., Hong K. Generation of mesenchymal-like stem cells from urine in pediatric patients. Transplant. Proc. 2016;48:2181–2185. doi: 10.1016/j.transproceed.2016.02.078. [DOI] [PubMed] [Google Scholar]
- 29.Jouni M., Si-Tayeb K., Es-Salah-Lamoureux Z., Latypova X., Champon B., Caillaud A., Rungoat A., Charpentier F., Loussouarn G., Baró I., et al. Toward personalized medicine: using cardiomyocytes differentiated from urine-derived pluripotent stem cells to recapitulate electrophysiological characteristics of type 2 Long QT syndrome. J. Am. Heart Assoc. 2015;4:e002159. doi: 10.1161/JAHA.115.002159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kogelman B., Khmelinskii A., Verhaart I., Vliet L.V., Bink D.I., Aartsma-Rus A., Putten M.V., Weerd L.V. Influence of full-length dystrophin on brain volumes in mouse models of Duchenne muscular dystrophy. PLoS ONE. 2018;13:e0194636. doi: 10.1371/journal.pone.0194636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Doorenweerd N., Mahfouz A., van Putten M., Kaliyaperumal R.T.’, Hoen P.A.C., Hendriksen J.G.M., Aartsma-Rus A.M., Verschuuren J.J.G.M., Niks E.H., Reinders M., et al. Author correction: Timing and localization of human dystrophin isoform expression provide insights into the cognitive phenotype of Duchenne muscular dystrophy. Sci. Rep. 2018;8:4058. doi: 10.1038/s41598-018-22154-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gazzoli I., Pulyakhina I., Verwey N.E., Ariyurek Y., Laros J.F., ’t Hoen P.A., Aartsma-Rus A. Non-sequential and multi-step splicing of the dystrophin transcript. RNA Biol. 2016;13:290–305. doi: 10.1080/15476286.2015.1125074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dumont N.A., Wang Y.X., von Maltzahn J., Pasut A., Bentzinger C.F., Brun C.E., Rudnicki M.A. Dystrophin expression in muscle stem cells regulates their polarity and asymmetric division. Nat. Med. 2015;21:1455–1463. doi: 10.1038/nm.3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuznetsov A.V., Winkler K., Wiedemann F.R., von Bossanyi P., Dietzmann K., Kunz W.S. Impaired mitochondrial oxidative phosphorylation in skeletal muscle of the dystrophin-deficient mdx mouse. Mol. Cell. Biochem. 1998;183:87–96. doi: 10.1023/a:1006868130002. [DOI] [PubMed] [Google Scholar]
- 35.Schuh R.A., Jackson K.C., Khairallah R.J., Ward C.W., Spangenburg E.E. Measuring mitochondrial respiration in intact single muscle fibers. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2012;302:R712–R719. doi: 10.1152/ajpregu.00229.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schulze A., Oshi M., Endo I., Takabe K. MYC targets scores are associated with cancer aggressiveness and poor survival in ER-positive primary and metastatic breast cancer. Int. J. Mol. Sci. 2020;21:8127. doi: 10.3390/ijms21218127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Y., Marino-Enriquez A., Bennett R.R., Zhu M., Shen Y., Eilers G., Lee J.C., Henze J., Fletcher B.S., Gu Z., et al. Dystrophin is a tumor suppressor in human cancers with myogenic programs. Nat. Genet. 2014;46:601–606. doi: 10.1038/ng.2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luce L.N., Abbate M., Cotignola J., Giliberto F. Non-myogenic tumors display altered expression of dystrophin (DMD) and a high frequency of genetic alterations. Oncotarget. 2017;8:145–155. doi: 10.18632/oncotarget.10426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mauduit O., Delcroix V., Lesluyes T., Pérot G., Lagarde P., Lartigue L., Blay J.Y., Chibon F. Recurrent DMD deletions highlight specific role of Dp71 isoform in soft-tissue sarcomas. Cancers (Basel) 2019;11:922. doi: 10.3390/cancers11070922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ruggieri S., De Giorgis M., Annese T., Tamma R., Notarangelo A., Marzullo A., Senetta R., Cassoni P., Notarangelo M., Ribatti D., Nico B. Dp71 expression in human glioblastoma. Int. J. Mol. Sci. 2019;20:5429. doi: 10.3390/ijms20215429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Domenger C., Allais M., François V., Léger A., Lecomte E., Montus M., Servais L., Voit T., Moullier P., Audic Y., Le Guiner C. RNA-Seq analysis of an antisense sequence optimized for exon skipping in Duchenne patients reveals no off-target effect. Mol. Ther. Nucleic Acids. 2018;10:277–291. doi: 10.1016/j.omtn.2017.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takizawa H., Hara Y., Mizobe Y., Ohno T., Suzuki S., Inoue K., Takeshita E., Shimizu-Motohashi Y., Ishiyama A., Hoshino M., et al. Modelling Duchenne muscular dystrophy in MYOD1-converted urine-derived cells treated with 3-deazaneplanocin A hydrochloride. Sci. Rep. 2019;9:3807. doi: 10.1038/s41598-019-40421-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Szigyarto C.A., Spitali P. Biomarkers of Duchenne muscular dystrophy: current findings. Degener. Neurol. Neuromuscul. Dis. 2018;8:1–13. doi: 10.2147/DNND.S121099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nallamilli B.R.R., Chaubey A., Valencia C.A., Stansberry L., Behlmann A.M., Zeqiang M., Mathur A., Shnenoy S., Ganapathy V., Jagannathan L., et al. A single NGS-based assay covering the entire genomic sequence of the DMD gene facilitates diagnostic and newborn screening confirmatory testing. Hum. Mutat. 2021;42:626–638. doi: 10.1002/humu.24191. [DOI] [PubMed] [Google Scholar]
- 45.Mo M., Wang S., Zhou Y., Li H., Wu Y. Mesenchymal stem cell subpopulations: phenotype, property and therapeutic potential. Cell. Mol. Life Sci. 2016;73:3311–3321. doi: 10.1007/s00018-016-2229-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ricotti V., Mandy W.P., Scoto M., Pane M., Deconinck N., Messina S., Mercuri E., Skuse D.H., Muntoni F. Neurodevelopmental, emotional, and behavioural problems in Duchenne muscular dystrophy in relation to underlying dystrophin gene mutations. Dev. Med. Child Neurol. 2016;58:77–84. doi: 10.1111/dmcn.12922. [DOI] [PubMed] [Google Scholar]
- 47.Liu D., Rychkov G., Al-Hawwas M., Manaph N.P.A., Zhou F., Bobrovskaya L., Liao H., Zhou X.F. Conversion of human urine-derived cells into neuron-like cells by small molecules. Mol. Biol. Rep. 2020;47:2713–2722. doi: 10.1007/s11033-020-05370-1. [DOI] [PubMed] [Google Scholar]
- 48.Xu G., Wu F., Gu X., Zhang J., You K., Chen Y., Getachew A., Zhuang Y., Zhong X., Lin Z., et al. Direct Conversion of Human Urine Cells to Neurons by Small Molecules. Sci. Rep. 2019;9:16707. doi: 10.1038/s41598-019-53007-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Casciaro F., Zia S., Forcato M., Zavatti M., Beretti F., Bertucci E., Zattoni A., Reschiglian P., Alviano F., Bonsi L., et al. Unravelling Heterogeneity of Amplified Human Amniotic Fluid Stem Cells Sub-Populations. Cells. 2021;10:158. doi: 10.3390/cells10010158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Monteiro Carvalho Mori da Cunha M.G., Zia S., Oliveira Arcolino F., Carlon M.S., Beckmann D.V., Pippi N.L., Luhers Graça D., Levtchenko E., Deprest J., Toelen J. Amniotic Fluid Derived Stem Cells with a Renal Progenitor Phenotype Inhibit Interstitial Fibrosis in Renal Ischemia and Reperfusion Injury in Rats. PLoS ONE. 2015;10:e0136145. doi: 10.1371/journal.pone.0136145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Takizawa H., Sato M., Aoki Y. Exon skipping in directly reprogrammed myotubes obtained from human urine-derived cells. J. Vis. Exp. 2020;159 doi: 10.3791/60840. [DOI] [PubMed] [Google Scholar]
- 52.Tuffery-Giraud S., Miro J., Koenig M., Claustres M. Normal and altered pre-mRNA processing in the DMD gene. Hum. Genet. 2017;136:1155–1172. doi: 10.1007/s00439-017-1820-9. [DOI] [PubMed] [Google Scholar]
- 53.Pettersson M., Grochowski C.M., Wincent J., Eisfeldt J., Breman A.M., Cheung S.W., Krepischi A.C.V., Rosenberg C., Lupski J.R., Ottosson J., et al. Cytogenetically visible inversions are formed by multiple molecular mechanisms. Hum. Mutat. 2020;41:1979–1998. doi: 10.1002/humu.24106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Plesser Duvdevani M., Pettersson M., Eisfeldt J., Avraham O., Dagan J., Frumkin A., Lupski J.R., Lindstrand A., Harel T. Whole-genome sequencing reveals complex chromosome rearrangement disrupting NIPBL in infant with Cornelia de Lange syndrome. Am. J. Med. Genet. A. 2020;182:1143–1151. doi: 10.1002/ajmg.a.61539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pons L., Bouvagnet P., Bakloul M., Di Filippo S., Buisson A., Chatron N., Labalme A., Metton O., Mitchell J., Diguet F., et al. Supravalvular Aortic Stenosis Caused by a Familial Chromosome 7 Inversion Disrupting the ELN Gene Uncovered by Whole-Genome Sequencing. Mol. Syndromol. 2019;10:209–213. doi: 10.1159/000500215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sanchis-Juan A., Stephens J., French C.E., Gleadall N., Mégy K., Penkett C., Shamardina O., Stirrups K., Delon I., Dewhurst E., et al. Complex structural variants in Mendelian disorders: identification and breakpoint resolution using short- and long-read genome sequencing. Genome Med. 2018;10:95. doi: 10.1186/s13073-018-0606-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xie Z., Sun C., Zhang S., Liu Y., Yu M., Zheng Y., Meng L., Acharya A., Cornejo-Sanchez D.M., Wang G., et al. Long-read whole-genome sequencing for the genetic diagnosis of dystrophinopathies. Ann. Clin. Transl. Neurol. 2020;7:2041–2046. doi: 10.1002/acn3.51201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xie Z., Sun C., Liu Y., Yu M., Zheng Y., Meng L., Wang G., Cornejo-Sanchez D.M., Bharadwaj T., Yan J., et al. Practical approach to the genetic diagnosis of unsolved dystrophinopathies: a stepwise strategy in the genomic era. J. Med. Genet. 2020;25 doi: 10.1136/jmedgenet-2020-107113. jmedgenet-2020-107113. [DOI] [PubMed] [Google Scholar]
- 59.Barseghyan H., Tang W., Wang R.T., Almalvez M., Segura E., Bramble M.S., Lipson A., Douine E.D., Lee H., Délot E.C., et al. Next-generation mapping: a novel approach for detection of pathogenic structural variants with a potential utility in clinical diagnosis. Genome Med. 2017;9:90. doi: 10.1186/s13073-017-0479-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Daoud F., Candelario-Martínez A., Billard J.M., Avital A., Khelfaoui M., Rozenvald Y., Guegan M., Mornet D., Jaillard D., Nudel U., et al. Role of mental retardation-associated dystrophin-gene product Dp71 in excitatory synapse organization, synaptic plasticity and behavioral functions. PLoS ONE. 2008;4:e6574. doi: 10.1371/journal.pone.0006574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shumyatsky G.P., Tsvetkov E., Malleret G., Vronskaya S., Hatton M., Hampton L., Battey J.F., Dulac C., Kandel E.R., Bolshakov V.Y. Identification of a signaling network in lateral nucleus of amygdala important for inhibiting memory specifically related to learned fear. Cell. 2002;111:905–918. doi: 10.1016/s0092-8674(02)01116-9. [DOI] [PubMed] [Google Scholar]
- 62.Lee A.J., Buckingham E.T., Kauer A.J., Mathews K.D. Descriptive Phenotype of Obsessive Compulsive Symptoms in Males With Duchenne Muscular Dystrophy. J. Child Neurol. 2018;33:572–579. doi: 10.1177/0883073818774439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jamil M.A., Sharma A., Nuesgen N., Pezeshkpoor B., Heimbach A., Pavlova A., Oldenburg J., El-Maarri O. F8 Inversions at Xq28 Causing Hemophilia A Are Associated With Specific Methylation Changes: Implication for Molecular Epigenetic Diagnosis. Front. Genet. 2019;10:508. doi: 10.3389/fgene.2019.00508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lei Q., Li C., Zuo Z., Huang C., Cheng H., Zhou R. Evolutionary Insights into RNA trans-Splicing in Vertebrates. Genome Biol. Evol. 2016;8:562–577. doi: 10.1093/gbe/evw025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lu Z.X., Jiang P., Xing Y. Genetic variation of pre-mRNA alternative splicing in human populations. Wiley Interdiscip. Rev. RNA. 2012;3:581–592. doi: 10.1002/wrna.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kesari A., Neel R., Wagoner L., Harmon B., Spurney C., Hoffman E.P. Somatic mosaicism for Duchenne dystrophy: evidence for genetic normalization mitigating muscle symptoms. Am. J. Med. Genet. A. 2009;149A:1499–1503. doi: 10.1002/ajmg.a.32891. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data from this study have been deposited in the GEO database under accession number GEO: GSE162108.