Summary
Overlapping clinical phenotypes and an expanding breadth and complexity of genomic associations are a growing challenge in the diagnosis and clinical management of Mendelian disorders. The functional consequences and clinical impacts of genomic variation may involve unique, disorder-specific, genomic DNA methylation episignatures. In this study, we describe 19 novel episignature disorders and compare the findings alongside 38 previously established episignatures for a total of 57 episignatures associated with 65 genetic syndromes. We demonstrate increasing resolution and specificity ranging from protein complex, gene, sub-gene, protein domain, and even single nucleotide-level Mendelian episignatures. We show the power of multiclass modeling to develop highly accurate and disease-specific diagnostic classifiers. This study significantly expands the number and spectrum of disorders with detectable DNA methylation episignatures, improves the clinical diagnostic capabilities through the resolution of unsolved cases and the reclassification of variants of unknown clinical significance, and provides further insight into the molecular etiology of Mendelian conditions.
Keywords: Episignatures, Neurodevelopmental disorders, DNA methylation, Epigenetics, Clinical diagnostics
Introduction
The diagnosis of Mendelian genetic disorders remains a challenge despite advancements in genomic sequencing. While the term “rare disorder” primarily reflects the population frequency of any specific condition, most of which have monogenetic (Mendelian) causation,1 it is estimated that 8% of the population are affected by a rare disorder.2,3 Diagnosis of Mendelian disorders is often complicated by non-specific clinical features, including the spectrum of neurodevelopmental delays and dysmorphic features,3 therefore a specific genetic finding is often required to establish a specific clinical diagnosis. The expanded use of gene panels and exome and genome sequencing has significantly improved diagnostic yield in Mendelian disorders.4 However, this technological advancement has increased the gap between our capacity to read and our ability to interpret the DNA sequence, as shown by the high prevalence of variants of unknown clinical significance (VUS).5 Rare-disease patients spend on average over 5 years on their diagnostic odyssey, and approximately half of patients presenting to medical genetics specialists are undiagnosed using traditional genetic diagnostics techniques.6 Whole-exome and whole-genome sequencing can help identify variants; however, the difficulty in predicting the impact of a VUS on protein-coding DNA and the lack of ability to predict their impact on non-coding DNA can still leave patients without a conclusive molecular diagnosis. Familial variant segregation studies, in silico prediction algorithms, and gene-specific functional studies may help resolve some VUS, but in the majority of cases, these analyses are not available, feasible, or conclusive.
One possible functional consequence of pathogenic variants in patients with genetic neurodevelopmental disorders is the alteration of genomic DNA methylation. DNA methylation is an epigenetic modification that changes the structural and chemical properties of DNA, impacting molecular mechanisms including chromatin assembly and gene transcription.7, 8, 9 Genomic DNA methylation patterns can be influenced by a variation in DNA sequence.10 These changes in DNA methylation, referred to as episignatures, are a functional consequence of disease-associated genetic variants and are emerging as highly accurate and stable biomarkers in a growing number of Mendelian disorders.11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28 Previous work by our group and others has demonstrated evidence of DNA methylation episignatures in a growing number of neurodevelopmental genetic disorders, which have previously been clinically validated as part of a diagnostic test called EpiSign.15,29,30 These episignatures are particularly evident in disorders involving chromatin remodeling genes. In addition to gene-specific episignatures, common DNA methylation profiles have been described for disorders resulting from pathogenic variants in genes encoding members of the same protein complexes17 and for multiple genes related to a specific syndrome gene,31 as well as for specific genic regions encoding particular protein domains.19
Germline inheritance of variants in Mendelian disorders implies an early developmental etiology of gene-specific episignatures that can be readily detectable in peripheral blood.7 The accessibility of peripheral blood provides the opportunity for a simple and cost-effective clinical implementation of the episignature analysis using genome-wide DNA methylation arrays.18,29 The clinical utility of DNA methylation episignatures has recently been demonstrated, with 57 out of 207 clinical samples testing positive for an episignature, giving an overall diagnostic yield of 27.6%.30 The main indications for episignature analysis included reclassification of genetic VUS as well as the screening of patients with no definitive genetic diagnosis but with a clinical presentation consistent with one of the mapped episignature disorders. However, the key limitation to the clinical application of a genome-wide DNA methylation assessment is the need to develop unique analytical methylation profiles for each Mendelian disorder, requiring expansion of reference databases and the development of sophisticated, machine-learning-based bioinformatic algorithms.32 Similarly, the ongoing national-scale study EpiSign-CAN, involving episignature analysis in thousands of patients with rare disorders, aims to provide a more comprehensive assessment of the clinical utility and the impact on the health system and to accelerate the rate of episignature discovery internationally (https://www.genomecanada.ca/en/beyond-genomics-assessing-improvement-diagnosis-rare-diseases-using-clinical-epigenomics-canada).
We previously reported a classification system that assessed 38 episignatures29 and now describe the addition of 19 episignatures to this classifier. The addition of these 19 episignatures expands the total number of clinically validated episignatures to 57, associated with 65 syndromes. We describe the improvements and refinements to the previously published multiclass episignature classifier29 and demonstrate its effectiveness in episignature analysis. By increasing the number of reference samples and disorder types in the EpiSign Knowledge Database (EKD), we can define further data complexity including novel gene sub-signatures and clinical associations. We also demonstrate the ability to sub-stratify some of the previously reported, closely related sub-signatures and highlight the analytical approach used to solve some of the more complex clinical cases.
Materials and methods
Patient samples
The discovery cohort included 235 peripheral blood samples from patients clinically diagnosed with or suspected of having 1 of 19 neurodevelopmental disorders and with a pathogenic variant in the corresponding gene, for which episignatures had not yet been identified or had not been previously included in the EpiSign multiclass classifier (Table 1 and S1). Unaffected controls were peripheral blood samples from individuals with no specific neurodevelopmental phenotype and no known pathogenic or suspected pathogenic variant in any of the episignature-related genes. These controls included a mix of samples from publicly available databases indicated to be “control,” “wild type,” or similar, and new samples from patients clinically assessed as not having a neurodevelopmental phenotype. Each unaffected control sample was assessed to ensure its DNA methylation was similar to previous healthy controls. The study was approved by the Western University Research Ethics Board (REB 106302 and REB 116108), and informed consent documents were reviewed and approved by the institutional review board (IRB) of Self Regional Healthcare. Some of the datasets used in this study are available publicly, as previously described.29 Sixteen of the 17 Chr16p11.2del samples are from GEO: GSE113967.33 Anonymized data for each subject is described in the study. The raw DNA methylation data for other samples are not available due to institutional and ethics restrictions.
Table 1.
List of episignatures and their corresponding syndromes and genes or genomic regions
| Syndrome | Signature abbreviation | Underlying gene or region | OMIM | Samples | In EpiSign V2 classifier |
|---|---|---|---|---|---|
| X-linked alpha-thalassemia/mental retardation syndrome (ATRX) | ATRX | ATRX | 301040 | 22 | yes |
| Arboleda-Tham syndrome (ARTHS) | ARTHS | KAT6A | 616268 | 18 | no |
| Autism, susceptibility to, 18 (AUTS18) | AUTS18 | CHD8 | 615032 | 28 | yes |
| Beck-Fahrner syndrome (BEFAHRS) | BEFAHRS | TET3 | 618798 | 16 | no |
| Blepharophimosis Intellectual disability SMARCA2 syndrome | BISS | SMARCA2 | 619293 | 5 | yes |
| Börjeson-Forssman-Lehmann syndrome (BFLS) | BFLS | PHF6 | 301900 | 16 | yes |
| Cerebellar ataxia, deafness, and narcolepsy, autosomal dominant (ADCADN) | ADCADN | DNMT1 | 604121 | 5 | yes |
| CHARGE syndrome | CHARGE | CHD7 | 214800 | 65 | yes |
| Chr16p11.2 deletion syndrome, 593-KB | Chr16p11.2del | Chr16p11.2 deletion | 611913 | 18 | no |
| Coffin-Siris syndrome-1,2 (CSS1,2) | CSS_c.6200a | ARID1B; ARID1A | 135900; 614607 | 4 | no |
| Coffin-Siris syndrome-1,2,3,4 (CSS1,2,3,4); Nicolaides-Baraitser syndrome (NCBRS) | BAFopathy | ARID1B; ARID1A; SMARCB1; SMARCA4; SMARCA2 | 135900; 614607; 614608; 614609; 601358 | 97 | yes |
| Coffin-Siris syndrome-4 (CSS4) | CSS4_c.2656a | SMARCA4 | 614609 | 3 | no |
| Coffin-Siris syndrome-9 (CSS9) | CSS9 | SOX11 | 615866 | 10 | no |
| Cohen-Gibson syndrome (COGIS); Weaver syndrome (WVS) | PRC2 | EED; EZH2 | 617561; 277590 | 7 | yes |
| Cornelia de Lange syndromes 1,2,3,4 (CDLS1,2,3,4) | CdLS | NIPBL; SMC1A; SMC3; RAD21 | 122470; 300590; 610759; 614701 | 57 | yes |
| Down syndrome | Down | Chr21 trisomy | 190685 | 40 | yes |
| Dystonia 28, childhood-onset (DYT28) | DYT28 | KMT2B | 617284 | 11 | no |
| Epileptic encephalopathy, childhood-onset (EEOC) | EEOC | CHD2 | 615369 | 8 | yes |
| Floating Harbor syndrome (FLHS) | FLHS | SRCAP | 136140 | 20 | yes |
| Gabriele-de Vries syndrome (GADEVS) | GADEVS | YY1 | 617557 | 10 | no |
| Genitopatellar syndrome (see also Ohdo syndrome, SBBYSS variant) (KAT6B) | GTPTS | KAT6B | 606170 | 4 | yes |
| Helsmoortel-van der Aa syndrome (HVDAS) | HVDAS_Ca | ADNP | 615873 | 13 | yes |
| Helsmoortel-van der Aa syndrome (HVDAS) | HVDAS_Ta | ADNP | 615873 | 23 | yes |
| Hunter McAlpine craniosynostosis syndrome | HMA | Chr5q35-qter duplication | 601379 | 4 | yes |
| Immunodeficiency-centromeric instability-facial anomalies syndrome 1 (ICF1) | ICF_1 | DNMT3B | 242860 | 8 | yes |
| Immunodeficiency-centromeric instability-facial anomalies syndromes 2,3,4 (ICF2,3,4) | ICF_2_3_4 | ZBTB24; CDCA7; HELLS | 614069; 616910; 616911 | 7 | yes |
| Intellectual developmental disorder with seizures and language delay (IDDSELD) | IDDSELD | SETD1B | 619000 | 10 | yes |
| Kabuki syndromes 1,2 (KABUK1,2) | Kabuki | KMT2D; KDM6A | 147920; 300867 | 149 | yes |
| KDM2B-related syndrome | KDM2B | KDM2B | unofficial | 9 | no |
| Autosomal dominant intellectual developmental disorder-65 (MRD65) | KDM4B | KDM4B | 619320 | 6 | no |
| Kleefstra syndrome 1 (KLEFS1) | Kleefstra | EHMT1 | 610253 | 32 | yes |
| Koolen de Vreis syndrome (KDVS) | KDVS | KANSL1 | 610443 | 11 | yes |
| Luscan-Lumish syndrome (LLS) | LLS | SETD2 | 616831 | 4 | no |
| Menke-Hennekam syndromes 1,2 (MKHK1,2) | MKHK_ID4a | CREBBP; EP300 | 618332; 618333 | 13 | no |
| Intellectual developmental disorder, X-linked, syndromic, Armfield type (MRXSA) | MRXSA | FAM50A | 300261 | 6 | no |
| Mental retardation, autosomal dominant 23 (MRD23) | MRD23 | SETD5 | 615761 | 25 | yes |
| Mental retardation, autosomal dominant 51 (MRD51) | MRD51 | KMT5B | 617788 | 7 | yes |
| Intellectual developmental disorder, X-linked 93 (MRX93) | MRX93 | BRWD3 | 300659 | 11 | yes |
| Intellectual developmental disorder, X-linked 97 (MRX97) | MRX97 | ZNF711 | 300803 | 15 | yes |
| Intellectual developmental disorder, X-linked syndromic, Nascimento-type (MRXSN) | MRXSN | UBE2A | 300860 | 4 | yes |
| Intellectual developmental disorder, X-linked, Snyder-Robinson type (MRXSSR) | MRXSSR | SMS | 309583 | 17 | yes |
| Intellectual developmental disorder, X-linked, syndromic, Claes-Jensen type (MRXSCJ) | MRXSCJ | KDM5C | 300534 | 49 | yes |
| Myopathy, lactic acidosis, and sideroblastic anemia 2 (MLASA2) | MLASA2 | YARS2 | 613561 | 11 | no |
| Ohdo syndrome, SBBYSS variant (SBBYSS) | SBBYSS | KAT6B | 603736 | 10 | yes |
| Phelan-McDermid syndrome (PHMDS) | PHMDS | Chr22q13.3 deletion | 606232 | 11 | no |
| Rahman syndrome (RMNS) | RMNS | HIST1H1E | 617537 | 8 | yes |
| Renpenning syndrome (RENS1) | RENS1 | PQBP1 | 309500 | 8 | no |
| Rubinstein-Taybi syndrome 1 (RSTS1) | RSTS1 | CREBBP | 180849 | 37 | no |
| Rubinstein-Taybi syndromes 1,2 (RSTS1,2) | RSTS | CREBBP; EP300 | 180849; 613684 | 39 | yes |
| Rubinstein-Taybi syndrome 2 (RSTS2) | RSTS2 | EP300 | 613684 | 29 | no |
| Sotos syndrome 1 (SOTOS1) | Sotos | NSD1 | 117550 | 69 | yes |
| Tatton-Brown-Rahman syndrome (TBRS) | TBRS | DNMT3A | 615879 | 27 | yes |
| Velocardiofacial syndrome (VCFS) | VCFS | Chr22q11.2 deletion | 192430 | 11 | no |
| Wiedemann-Steiner syndrome (WDSTS) | WDSTS | KMT2A | 605130 | 42 | yes |
| Williams-Beuren deletion syndrome (WBS) | Williams | Chr7q11.23 deletion | 194050 | 22 | yes |
| Williams-Beuren duplication syndrome (Chr7q11.23 duplication syndrome) | Dup7 | Chr7q11.23 duplication | 609757 | 13 | yes |
| Wolf-Hirschhorn syndrome (WHS) | WHS | Chr4p16.13 deletion | 194190 | 12 | yes |
Episignatures that encompass a specific region or variant within a gene.
Sample processing
Peripheral blood DNA was extracted using standard techniques. Bisulfite conversion was performed with 500 ng of genomic DNA using the Zymo EZ-96 DNA Methylation Kit (D5004), and bisulfite-converted DNA was used as input to the Illumina Infinium HumanMethylation450 (450K array) or MethylationEPIC BeadChip array (EPIC array). Array data were generated according to the manufacturer’s protocol. Sample quality control was performed using the R minfi package version 1.35.2.34
Methylation data analysis
The data analysis pipeline was adapted from previously described methods,29 as summarized in Figure S1. IDAT files containing methylated and unmethylated signal intensities were imported into R 4.0.3 for analysis. Normalization was performed using the Illumina normalization method with background correction using the minfi package. Probes with a detection p value > 0.01, probes located on the X and Y chromosomes, probes that contained SNPs at the CpG interrogation or single-nucleotide extension sites, and probes that are known to cross-react with other genomic locations were removed.35,36
For each cohort (set of case samples for a particular syndrome/episignature), a set of controls was chosen using the R package matchit version 3.0.2,37 matched for age, sex, and array type. To increase signal specificity, controls consisted of samples from healthy/unaffected individuals and other episignature samples and included batch controls. For each case sample, two to ten controls were used (case:control ratio of 1:2 to 1:10), resulting in matched control cohorts with a mean size of 53 samples (range 30–74) (Table S2). Additional controls from other episignature syndromes were included in some analyses to differentiate between closely related signatures: Arboleda-Tham syndrome (ARTHS)/Ohdo syndrome; SBBYSS variant (SBBYSS)/Genitopatellar syndrome (GTPTS); and Rubinstein-Taybi syndromes 1 and 2 (RSTS1/RSTS2), as described in detail in the Results. Principal-component analysis (PCA) was performed prior to episignature analysis to identify and remove control outliers. Probes with beta values of 0 and the top 1% most variable (variance) probes within the case or control samples were removed. Combined filtering yielded on average approximately 650,000 probes for subsequent analysis.
Methylation levels (beta values) were logit-transformed to M-values and the transformed values used for linear regression modeling using the limma package version 3.45.19.38 Estimated blood cell proportions39 were added to the model matrix as confounding variables. The generated p values were moderated using the eBayes function. Probes that had a mean methylation difference of less than 5% between the case and control samples were removed.
Probe selection parameters were optimized depending on the cohort size and signal differences to enhance separation between the case and control samples as evaluated using hierarchical clustering and multidimensional scaling (MDS) plots. The parameters used were as follows: a probe “score,” the area under the receiver’s operating curve (AUC), and a probe-to-probe methylation correlation. First, a probe score was generated as previously described29 by multiplying the absolute value of the mean methylation difference by the negative value of the log-transformed Benjamini-Hochberg-adjusted p value. For some cohorts (typically small cohorts), non-adjusted p values were used. The 800–1,000 probes with the highest scores were selected, and receiver-operating characteristic (ROC) curve analysis was applied, yielding 160–500 probes. Lastly, we calculated the Pearson’s correlation coefficients for the selected probes and removed highly correlated probes. Using the final set of selected probes, we performed hierarchical clustering using the R package gplots version 3.1.0 using the heatmap.2 function with Ward’s method, and MDS was performed by scaling of the pairwise Euclidean distances between samples. Hierarchical clustering was assessed to ensure the case and control samples were properly clustered, and MDS plots were assessed to identify the set of probes that generated the greatest distance between the case and control samples. Leave-one-out sample cross-validation was performed for each sample in each episignature cohort and evaluated using hierarchical clustering, MDS, and methylation variant pathogenicity (MVP) plots (MVP plots described below).
The e1071 R package version 1.7–4 was used to train a support vector machine (SVM) and for the construction of a multiclass prediction model as previously described.29 Each cohort of case samples was trained against the control samples present in the EKD. Controls consisted of samples from unaffected individuals and other episignature samples (Table 1). Seventy-five percent of control samples were used for training and 25% were used for testing. This was repeated four times so that each control sample was used at least once for testing (4-fold training/testing cross-validation). A final classifier for each cohort was made by training case samples against all control samples to generate the EpiSign V3 clinical classifier.30 SVM decision values were converted to probability scores according to Platt’s scaling method,40 which were then used to create the MVP plots. The MVP score predicts the probability that a sample’s methylation pattern matches a given episignature, with scores closest to one indicating the highest probability.
Results
Identification of disorder-specific episignatures
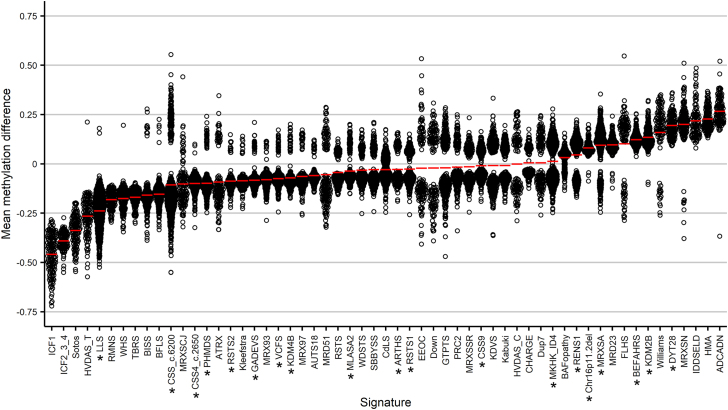
We have previously described the EpiSign (EpiSign V2) classifier, which included 38 episignatures, which encompassed 60 genes or genomic regions, related to 49 Mendelian neurodevelopmental disorders present in the EKD.29,30 We applied our analysis pipeline to 16 additional cohorts involving pathogenic variants in 14 genes or genomic regions, enabling the identification of 19 novel DNA methylation episignatures (Table 1): ARTHS; Beck-Fahrner syndrome (BEFAHRS); Chr16p11.2 deletion syndrome, 593-KB; Coffin-Siris syndrome-1,2 (CSS1,2; genes ARID1B, ARID1A); CSS4; CSS9; Dystonia 28, childhood-onset (DYT28); Gabriele-de Vries syndrome (GADEVS); KDM2B-related syndrome; autosomal dominant intellectual developmental disorder-65 (MRD65); Luscan-Lumish syndrome (LLS); Menke-Hennekam syndromes 1,2 (MKHK1,2); intellectual developmental disorder, X-linked, syndromic, Armfield type (MRXSA); myopathy, lactic acidosis, and sideroblastic anemia 2 (MLASA2); Phelan-McDermid syndrome (PHMDS); Renpenning syndrome (RENS1); RSTS1; RSTS2; and Velocardiofacial syndrome (VCFS). To identify probes with more robust changes in methylation, for each episignature, we first removed probes that had a mean methylation difference of less than 5% between the case and control samples. After filtering, there was a median across the 19 episignatures of 11,709 probes remaining. The final set of selected probes for each episignature consisted of 100–500 differentially methylated probes that best separated the case samples from controls (Figure S2, Table S3). The probes for the 19 new episignatures were then added to and compared with the probes from the previously reported episignatures. Mean methylation levels of these classifier probes showed hypomethylation in 40 (70%) and hypermethylation in 17 (30%) of the episignatures. Thirty-six (63%) of episignatures showed moderate methylation differences (between −10% and +10%), 12 (21%) had a larger decrease in methylation, and 9 (16%) had a larger increase in methylation (Figure 1). While trends in episignature methylation changes generally reflect global methylation changes, ongoing work focused on the detailed analysis of the broader genomic methylation patterns will provide further insights into the molecular and functional aspects of these epigenomic changes.
Figure 1.
Methylation differences of probes used for episignatures
Methylation differences between cases and controls for the microarray probes that make up each episignature for the newly identified and previously reported episignatures. Red lines indicate mean methylation for each episignature. Asterisk indicates new episignatures and/or those that have not previously been included in the multiclass classifier.
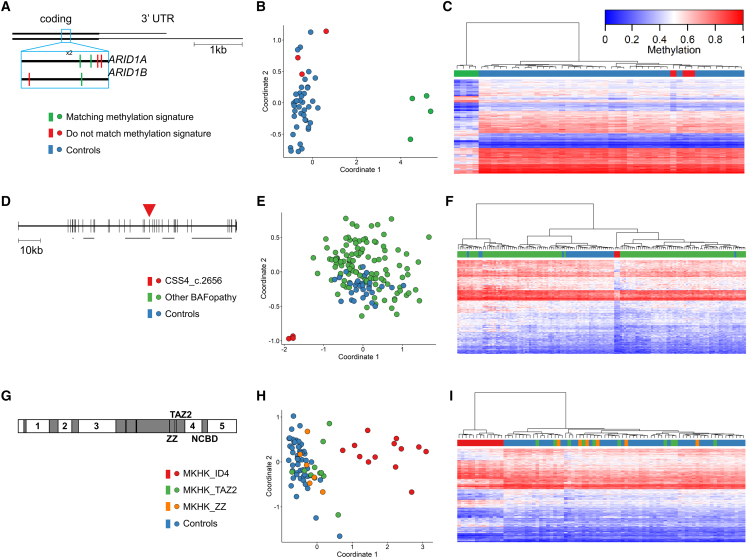
In addition to the common gene-level episignatures, we identified novel distinct sub-gene level signatures associated with specific gene regions and domains. Six cases with variants near position c.6200 in the last exon of ARID1A or ARID1B were shown not to match the BAFopathy signature. These included cases with missense mutations in ARID1A: c.6232G>A,p.(Glu2078Lys) (x2), c.6254T>G,p.(Leu2085Arg), and c.6275C>A,p.(Ala2092Glu) and ARID1B: c.6032A>T,p.(Glu2011Val) and c.6133T>C,p.(Cys2045Arg). In addition, the nearby BAFopathy-positive sample ARID1A:c.6269A>G,p.(His2090Arg) was included for comparison. By iterative assessment, 4 of the 7 samples were determined to share a common DNA methylation profile, outlining the boundary for this sub-gene episignature (Figures 2A–2C).
Figure 2.
Gene region- or variant-specific sub-signatures
(A) The last exon of ARID1A and ARID1B shown with the location of seven variants in the c.6200 region colored by whether they match the c.6200 episignature or not. (B and C) MDS (B) and hierarchical clustering (C) plots of the seven samples showing that the four central samples have a matching episignature, while the outer three cluster with controls. For hierarchical clustering plots, each row represents one microarray probe, and each column represents one sample. (D) Gene diagram of SMARCA4 (NM_001128849.1) showing the location of the three c.2656A>G variants in exon 19 (red arrowhead). The five horizontal gray bars indicate the locations of protein domains: QLQ, HSA, helicase ATP-binding, helicase C-terminal, and bromodomain. (E and F) MDS (E) and hierarchical clustering (F) showing that the three CSS4 samples with the above variant cluster separately from controls and from other BAFopathy samples. (G) Protein diagram of CREBBP/EP300 showing the location of protein domains (gray boxes) and intrinsically disordered (ID) domains (numbered). (H and I) MDS (H) and hierarchical clustering (I) showing the MKHK_ID4 samples clustering separately from controls and from other MKHK samples.
Three separate patients with CSS4 caused by the same variant SMARCA4:c.2656A>G,p.(Met886Val) did not match the general BAFopathy episignature but also clustered separately from controls, indicating the presence of a separate, distinct episignature (Figures 2D–2F and S3). Additional cases in the EKD with nearby variants in SMARCA4 were also tested: an unresolved case with variant c.2620C>T,p.(Arg874Cys) and two samples that matched the BAFopathy episignature and had variants c.2932C>G,p.(Arg978Gly) and c.2933G>A,p.(Arg978Gln). However, a consistent episignature could not be found when any of these additional samples were included, providing further support for the distinct episignature related to the SMARCA4:c.2656A>G,p.(Met886Val) variant specifically.
MKHK1 and MKHK2 are caused by pathogenic variants in exons 30/31 of CREBBP and EP300, respectively. Variants in these exons that affect additional downstream regions of the protein, such as frameshift variants, are shown to cause RSTS. Exons 30 and 31 include a ZZ domain, a TAZ2 domain, and an intrinsically disordered linker (ID4).41 We evaluated 31 samples with variants in these domains but were not able to identify an episignature common to all 31 samples. We therefore examined each domain separately and were able to identify a distinct episignature for the 13 samples in the ID4 domain (episignature MKHK_ID4) but not for the ZZ or TAZ2 samples (Figures 2G–2I).
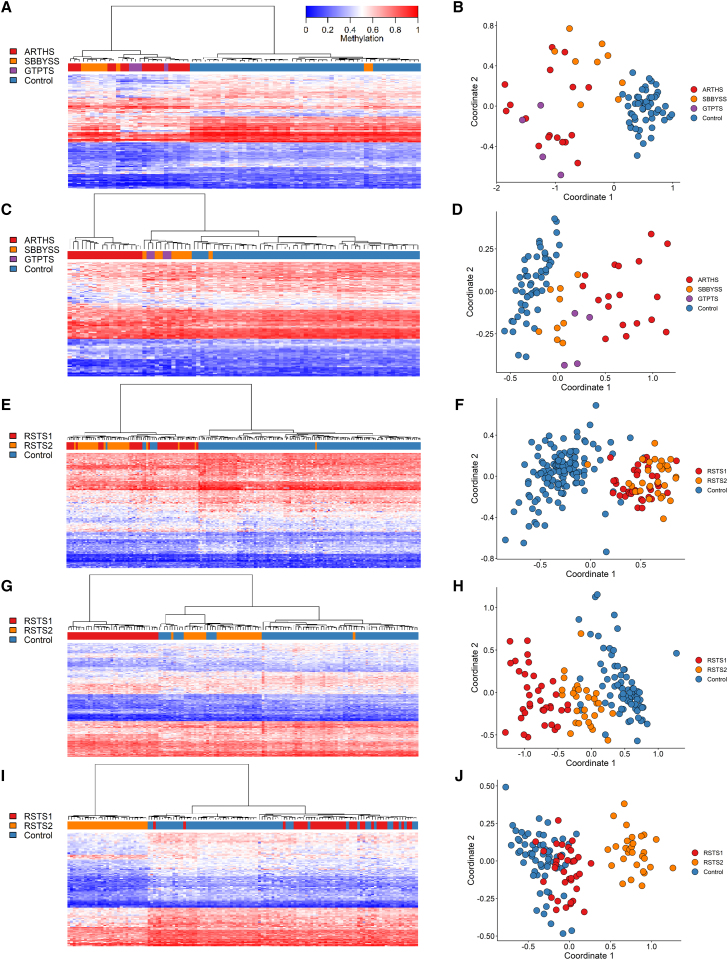
Syndromes caused by the same or by functionally related genes (similar function or part of the same protein complex) can be difficult to distinguish using episignatures. We previously reported separate episignatures for GTPTS and SBBYSS, which are both caused by pathogenic variants in KAT6B.29 We have now identified an episignature for ARTHS, which is caused by pathogenic variants in KAT6A. We first used our standard pipeline for identifying differentially methylated probes between ARTHS and control samples. The identified probe set showed sensitivity for ARTHS, as all ARTHS samples could be distinguished from controls based on supervised clustering. However, it lacked specificity in relation to GTPTS and SBBYSS, as ARTHS samples were interspersed with GTPTS and SBBYSS samples (Figures 3A and 3B). By performing probe selection with GTPTS and SBBYSS samples in the control cohort, we were able to identify probes that were both highly sensitive and specific for ARTHS in relation to GTPTS, SBBYSS, and controls. Using this updated probe set, ARTHS samples clustered separately from controls and from GTPTS and SBBYSS samples (Figures 3C, 3D, and 4).
Figure 3.
Identifying episignatures to distinguish between closely related syndromes
Hierarchical clustering and MDS plots are shown for each episignature. For hierarchical clustering plots, each row represents one microarray probe, and each column represents one sample. (A and B) ARTHS probe selection using only ARTHS and control samples. (C and D) ARTHS probe selection when GTPTS and SBBYSS samples are included as controls. (E and F) The previously reported RSTS (RSTS1/RSTS2 combined) episignature. (G and H) The RSTS1 episignature generated by including RSTS2 samples as control. (I and J) the RSTS2 episignature generated by including RSTS1 samples as control.
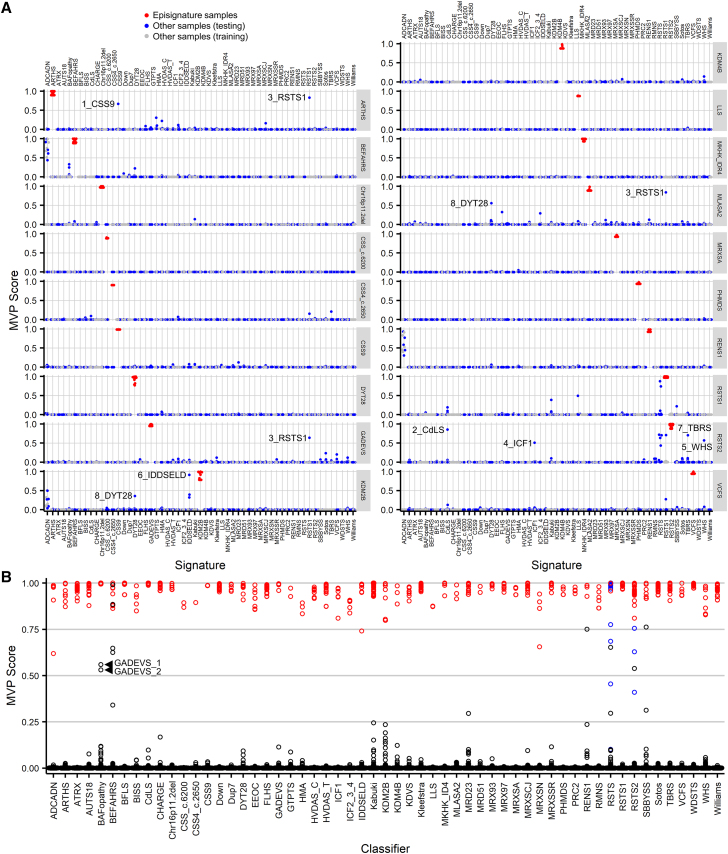
Figure 4.
Support vector machine-based classifiers for concurrent episignature detection
(A) Each of the 19 new episignatures were evaluated using 4-fold cross-validation. For each fold, a different 25% of samples were used for testing (blue), and the remaining 75% of samples were used for training (gray). The case samples for each episignature are shown in red. The eight testing samples referenced in the text are labeled. (B) The final classifiers for all 57 episignatures. The case samples for each episignature are shown in red, and all other samples are in black. Non-case RSTS samples (for example, RSTS1 samples in the RSTS2 column) are in blue. Arrowheads indicate the two GADEVS samples with high BAFopathy scores.
We previously reported a signature for RSTS that included both RSTS1 (CREBBP) and RSTS2 (EP300) samples.29 Using this episignature, we were able to differentiate RSTS1 and RSTS2 samples from controls but not from each other (Figures 3E and 3F). By expanding the size of the reference cohorts from 39 to 66 samples and applying the strategy of including the alternate disorder samples in the control cohort, we were able to identify RSTS1- and RSTS2-specific episignatures (Figures 3G–3J).
EpiSign V3 classifier enables concurrent screening of 57 episignatures
We used an SVM-based approach to develop a multiclass classifier enabling a sensitive and specific DNA methylation screening for all 57 distinct episignatures using a previously described strategy.29 We used a training/testing experimental design to validate the episignatures. For each episignature, all case samples plus 75% of all other syndrome/episignature samples and unaffected controls were used for training, and the remaining 25% were used for testing. This was repeated four times so that each sample was used once for testing (and three times for training). Testing MVP scores and mean training MVP scores are shown in Figure 4A. Overall, MVP data showed a high level of accuracy. The classifiers were highly sensitive, with all cohort cases receiving a high score above 0.5 for their episignature. They were also highly specific, with only eight samples (3.4%) scoring above 0.5 for an alternate cohort episignature. In addition, of the approximately 1,200 unaffected controls used as testing samples for each of the 19 episignatures (22,718 individual MVP scores), only five had an MVP score above 0.1 and none had over 0.25. Unsupervised clustering showed that five of the eight samples clearly did not match the secondary episignatures despite their unexpectedly high MVP scores. Sample 1_CSS9 with variant SOX11:c.250G>A,p.(Gly84Ser) had a high score for the ARTHS classifier (Figure 4A), but when compared to other ARTHS samples, it clustered with controls (Figure S4). Sample 2_CdLS with variant RAD21:c.218del, p.(Tyr73Serfs∗13) had a high score for the RSTS2 classifier (Figure 4A) but clustered separately from both controls and RSTS2 samples (Figures S5A and S5B). Sample 3_RSTS1 with variant CREBBP:c.4507T>C, p.(Tyr1503His) had elevated scores for the ARTHS, GADEVS, and MLASA classifiers (Figure 4A) but clustered separately from controls and from the three secondary cohorts (Figure S6). Sample 4_ICF1, with DNMT3B variants c.310C>T,p.(Arg104∗) and 2162T>C,p.(Ile721Thr), had a high score for the RSTS2 classifier (Figure 4A) but clustered separately from controls and RSTS2 samples (Figures S5C and S5D). Sample 5_WHS with variant 4p16.3p15.2(68,345–24,136,683)x1 had a high score for the RSTS2 classifier but clustered with controls (Figures S5E and S5F).
Sample 6_IDDSELD with a deletion in 12q24.31 had a high score for the KDM2B classifier (Figure 4A). While this sample clustered distinctly from controls and KDM2B samples, its separation from KDM2B was not as clear as with the five previously described samples (Figure S7). Sample 7_TBRS with variant DNMT3A:c.2525A>G,p.(Gln842Arg) had a high score for the RSTS2 classifier (Figure 4A). MDS showed overlap with RSTS2 samples; however, the hierarchical clustering heatmap methylation pattern differed from RSTS2 samples (Figures 4G and 4H). Sample 8_DYT28 with variant KMT2B:c.4844C>T,p.(Ser1615Leu) had a high score for the MLASA2 classifier (Figure 4A) and clustered with other MLASA2 samples (Figures S8A and S8B). While this sample also clustered well with other DYT28 samples (Figures S8E–S8G), the heatmap results showed hypermethylation compared to others in the DYT28 cohort (Figure S8E). Sample 8_DYT28 also scored higher than expected, although below 0.5 at 0.36, for episignature KDM2B (Figure 4A); however, unsupervised clustering using the KDM2B episignature probes showed that the sample did not cluster well with either KDM2B samples or controls (Figures S8C and S8D).
Besides these eight samples, all ADCADN samples had elevated MVP scores for the BEFAHRS classifier (Figure 4A). ADCADN is caused by activating mutations in the DNA methyltransferase DNMT1, whereas BEFAHRS is caused by deactivating mutations in the DNA demethylase TET3, with both resulting in overall hypermethylation. However, unsupervised clustering is able to clearly distinguish between the two methylation profiles (Figures S9A and S9B). A similar observation is seen in relation to ADCADN samples and the RENS1 classifier (Figures 4A, S9C, and S9D).
To increase specificity of the final classifiers, for each cohort, the case samples were trained against all other episignature samples and controls in the EKD. These final classifiers were added to our previously reported classifiers to create the 57 episignature multiclass system for sample classification (Figure 4B). This reduced the non-specific MVP scores for most of the previously discussed samples; however, two GADEVS samples (GADEVS_1 and GADEVS_2) had MVP scores over 0.5 for the previously reported BAFopathy episignature (Figure 4B). Examination of the full GADEVS cohort found that five others had elevated BAFopathy episignature MVP scores, from 0.01 to 0.11, with the remaining three being less than 0.01. Leave-one-out cross-validation of the two GADEVS samples, which scored highest for BAFopathy, showed that they specifically matched other GADEVS samples (Figures S10A–S10F). Unsupervised clustering of GADEVS and BAFopathy samples using the BAFopathy episignature probes showed all GADEVS samples clustered with controls except for the one GADEVS sample that scored highest for BAFopathy (GADEVS_1), which clustered near other BAFopathy samples (Figures S10G and S10H), suggesting that this sample at least partially matches both the GADEVS and BAFopathy episignatures.
Screening unresolved cases
The 19 new classifiers were used to assess a cohort of samples from the EKD, which were previously assessed using EpiSign V2 but were unsolved. Nineteen samples had an MVP score greater than 0.5 for one of the new classifiers. Hierarchical clustering and MDS analysis ruled out the majority of these cases, leaving three that clustered with their target cohorts.
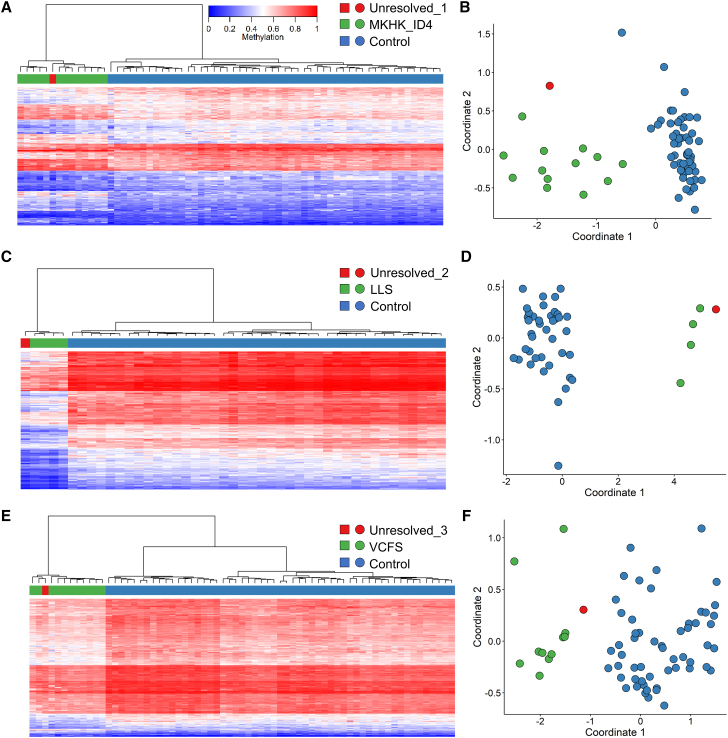
Sample 1 (Unresolved_1) had an MVP score for MKHK_ID4 of 0.98, and unsupervised clustering showed that this sample clustered with other MKHK_ID4 samples (Figures 5A and 5B). Follow-up with the submitting clinician confirmed that the patient carried a subsequently identified CREBBP exon 31 pathogenic variant and had a clinical diagnosis of MKHK, confirming the EpiSign findings. Sample 2 (Unresolved_2) had an MVP score for LLS of 0.93, and unsupervised clustering showed that this sample clustered with other LLS samples (Figures 5C and 5D), but further clinical information was not available for follow up. Sample 3 (Unresolved_3) is from a 5-year-old male with the variant UBE2A:c.283C>T,p(.Arg95Cys) and phenotype of the Nascimento form of syndromic X-linked mental retardation (MRXSN); however, previous EpiSign analysis ruled out the MRXSN episignature. This sample had an MVP score for VCFS of 0.64, and unsupervised clustering showed that this sample clustered near other VCFS samples (Figures 5E and 5F). Array comparative genomic hybridization showed that the patient did not have the VCFS-associated Chr22q11.2 deletion. Clinical follow-up confirmed this subject has an intellectual disability, a congenital heart defect, and dysmorphism consistent with MRXSN. This subject is described in greater detail by Cordeddu et al. as patient #7.42
Figure 5.
Screening unresolved cases
Samples with MVP scores greater than 0.5 were further assessed by unsupervised clustering plots. Hierarchical clustering and MDS plots are shown for each case. For hierarchical clustering plots, each row represents one microarray probe, and each column represents one sample. (A) Sample Unresolved_1, a previously unresolved case that matches the MKHK_ID4 episignature. (B) Sample Unresolved_2, a previously unresolved case that matches the LLS episignature. (C) Sample Unresolved_3, a previously unresolved case that matches the VCFS episignature.
Discussion
Expanding the EpiSign classifier by 19 episignatures
Peripheral blood DNA methylation episignatures have emerged as highly specific biomarkers in a growing number of Mendelian disorders.30,43,44 This study significantly expands on our previous work29 by describing 19 new episignatures, bringing the total to 57. The expanding landscape of Mendelian episignatures includes genes and disorders beyond those with direct involvement of chromatin regulatory mechanisms. Twenty seven (71%) of our 38 previously reported episignatures represent chromatinopathies, while in the present study, chromatinopathies accounted for only 10 (53%) of the episignatures reported. Five of the new episignatures detect syndromes caused by pathogenic variants in histone remodeling genes: ARTHS (KAT6A), DYT28 (KMT2B), LLS (SETD2), MRD65 (KDM4B), and the as-yet-unnamed syndrome related to KDM2B. Three are associated with syndromes caused by transcription factors: GADEVS (YY1), CSS9 (SOX11), and RENS1 (PQBP1). Another three episignatures define syndromes caused by copy-number variation: Chr16p11.2 deletion syndrome, PHMDS caused by Chr22q13.3del, and VCFS caused by Chr22q11.2del. The previously reported RSTS episignature has been refined into two distinct episignatures that can now differentiate between RSTS1 (CREBBP) and RSTS2 (EP300). The sensitivity of BAFopathy detection has been improved with the identification of two sub-signatures for specific regions or variants in ARID1A, ARID1B, and SMARCA4, which cause CSS1, CSS2, and CSS4. Another region/domain-level signature, the MKHK_ID4 episignature defines the subset of MKHK1 and MKHK2 caused by pathogenic variants in the CREBBP/EP300 ID4 domain. The final three episignatures are for BEFAHRS, caused by the DNA demethylase TET3; MLASA2, caused by the mitochondrial gene YARS2; and MRXSA, caused by FAM50A, which has a role in mRNA splicing.45
Each episignature consists of the 100–500 CpGs that best distinguish the samples of the given cohort from all other samples and which therefore have applications for clinical diagnostic testing.30 The initial identification of differentially methylated probes based only on methylation difference and p value, without additional filtering, identified a median of 11,709 probes per cohort. Combined, these changes represent over 100,000 individual differentially methylated CpGs. Future studies will be needed to investigate the biological significance of these changes. For example, to examine the genomic location of differentially methylated CpGs. It will also be necessary to identify the functions of genes that overlap changes in DNA methylation and explore in more detail the relationships between episignatures. Identifying such functional consequences may help explain why certain CpGs or regions exhibit changes in DNA methylation and may provide insight into the mechanisms behind syndrome-specific phenotypes.
Approximately 5%–10% of pathogenic variants may be mosaic,46 which presents a challenge for the clinical use of episignatures and genetic diagnostic tests in general. If such mutations occur early in development and affect multiple tissues it is likely that DNA methylation differences will be exhibited in peripheral blood, albeit at lower levels reflective of the degree of mosaicism. Episignatures with more robust methylation differences will likely enable lower levels of mosaicism detection than ones for less pronounced episignatures. Mutations that occur later in development and affect specific tissues, such as a mosaicism that only affects neural tissue, would not be detected using an episignature test, which relies on peripheral blood samples. Further analysis of representative patient cohorts with mosaicism will be needed to determine thresholds of detection independently for each episignature.
Studies have used the 450K or EPIC arrays, which assess approximately 450,000 and 850,000 CpGs, respectively, to identify differences in DNA methylation between ethnic/racial groups. While one study found 26,262 differentially methylated CpGs between two populations,47 several others found changes limited to a few hundred to a few thousand CpGs.48, 49, 50, 51, 52 Additional studies will be needed to determine whether these differences affect the accuracy of episignatures. Excluding ethnicity-associated CpGs from episignature analysis, similar to how SNP-associated and other potentially confounding CpGs are currently excluded (see Materials and methods), could help account for potential ethnic diversity.
Variant-, region-, and domain-specific episignatures
Previous work showed that pathogenic variants in one gene can sometimes lead to more than one episignature depending on where in the gene the variant occurs.19 Furthering this concept, we have identified three episignatures specific to a sub-section of a gene. The CSS1 and CSS2 genetic region-specific sub-signature was observed in cases with missense variants surrounding the c.6200 region within the ARID1A (CSS1) and ARID1B (CSS2) genes (Figures 2A–2C). The paralogs ARID1A and ARID1B are exchangeable core components of the BAF chromatin remodeling complex. Pathogenic variants in either gene lead to recognizable clinical features of CSS.53 Previous studies suggested a broad distribution of pathogenic variants across ARID1A and ARID1B,53 while a recent study proposed a model for ARID1A-mediated DNA and protein complex interactions,54 with two key domains identified: the N-terminal ARID domain responsible for DNA binding and the C-terminal domain of unknown function, recently annotated as BAF250_C.54,55 This new CSS_c.6200 episignature consists of variants within the BAF250_C domain. The nearest variants assessed that do not match this signature also lie within the BAF250_C domain (Figures 2A–2C), suggesting further specificity within the domain.
An extreme example of sub-signature specificity is evident in another BAFopathy gene, SMARCA4 (involved in CSS4), and was observed in multiple cases with the same pathogenic variant c.2656A>G (Figures 2D and 2E). SMARCA4 is an ATPase subunit of the BAF complex with a critical role in regulating chromatin structure and transcription,56 with previously described variability in clinical presentation.57 The SMARCA4:c.2656A>G,p.(Met886Val) variant is in the helicase ATP-binding domain, which lies between the mutational hotspot HAS domain and the helicase C-terminal domain.58 One other sample with a variant in the helicase domain did not match this sub-signature, indicating that this is a variant-specific and not domain-specific episignature.
A domain-specific episignature associated with the ID4 domain was seen in the MKHK cohort. MKHK is caused by pathogenic variants in CREBBP (MKHK 1) and EP300 (MKHK 2).59 CREBBP and EP300 are both transcriptional coactivators and histone acetyltransferases60 that, when mutated, result in a common pathogenic mechanism involving aberrant chromatin regulation. Variants in both genes were assessed for a potential episignature. Though an overarching common episignature for MKHK types 1 and 2 was not identified, samples with pathogenic variants within ID4 of both genes clustered together and separately from all other MKHK and control samples (Figures 2F and 2G). This provides a unique instance where episignature discovery resulted in a domain-specific sub-signature across two paralogs emerging without a disorder-specific syndrome episignature defined first. Additional case samples along with detailed clinical descriptions will be necessary to determine if these sub-signatures could be associated with a specific clinical presentation within the associated syndrome.
Achieving episignature specificity in closely related disorders
Specificity of episignature classifiers can be ensured by training each classifier against samples from all other episignatures. However, as more episignatures are defined, particularly when they represent similar syndromes or genes, an additional step may be needed. The inclusion of samples from cohorts with similar episignatures in the control sample sets during the initial probe selection allows for additional specificity by deprioritizing probes with concurrent methylation changes between the two overlapping episignatures. Using this strategy, we were able to separate the previously reported combined RSTS1/RSTS2 episignature into almost fully distinct RSTS1 and RSTS2 episignatures. While some RSTS2 samples cluster near RSTS1 samples (Figure 3H) and a few RSTS1 and RSTS2 samples have high MVP scores for the reciprocal episignature (Figures 4A and 4B), these cases can be resolved by the combination of clustering analysis and MVP scores.
A similar challenge was encountered when assessing the ARTHS cohort, caused by mutations in KAT6A.61 ARTHS has some clinical overlap with two other syndromes that have defined episignatures: SBBYSS and GTPTS.62 SBBYSS and GTPTS are both caused by pathogenic variants in KAT6B. KAT6A and KAT6B are paralogous lysine acetyltransferases within the conserved MYST family and form a complex with other proteins to modulate gene expression via histone acetylation.62 Therefore, the disruption of this protein complex due to pathogenic variants or the loss-of-function of either KAT6A or KAT6B likely impacts the same downstream pathways and leads to similar and overlapping DNA methylation changes across the genome during development. Despite the significant overlap between ARTHS and the SBBYSS and GTPTS episignatures (Figures 3A and 3B), we were able to define an ARTHS episignature by implementing the same method used to differentiate RSTS1 and RSTS2 episignatures (Figures 3C and 3D). This approach will be important going forward as more similar syndromes, both in phenotypic presentation or molecular mechanism, are assessed for episignatures, and differences in DNA methylation patterns between such syndromes are more difficult to ascertain.
Assessing complex cases with more than one potential positive result
All samples used to define the 57 episignatures were tested at least once against each new episignature classifier to generate the MVP probability scores (Figure 4A) to ensure specificity. ADCADN samples scored high (MVP over 0.5) for both BEFAHRS and RENS (Figure 4A), but unsupervised clustering demonstrated clear grouping for each cohort (Figure S9). Some samples, however, showed less distinct unsupervised clustering and required further assessment. Sample 8_DYT28 had a variant in KMT2B, which is associated with DYT28, which is characterized by childhood-onset dystonia. This sample had high MVP scores and distinct unsupervised clustering for two episignatures: DYT28 and MLASA2. MLASA2 is caused by mutations in YARS2 and is a mitochondrial respiratory chain disorder63 characterized by skeletal myopathy, lactic acidosis, and sideroblastic anemia. The MLASA2 episignature is overall hypomethylated (Figure 1) but contains a block of hypermethylated probes, as shown on the heatmap, where strongly hypomethylated probes in controls are less hypomethylated in MLASA2 samples (Figure S2M). Sample 8_DYT28 exhibits more hypermethylation than other samples present in the DYT28 cohort (Figure S8C), with the DYT28 episignature also presenting with mean hypermethylation overall (Figure 1). In addition to the two already noted MVP scores over 0.5, sample 8_DYT28 had a moderate score for the hypermethylated KDM2B episignature at 0.36, although unsupervised clustering was less conclusive than either the DYT28 or MLASA2 results (Figure S8). Therefore, the unexpectedly high MVP scores for sample 8_DYT28 may be due to non-specific overlap of hypermethylated probes. However, the possibility of a pathogenic variant in YARS2 or KDM2B has not been ruled out, and sequencing of these genes for this subject should be considered.
Another example of such overlap was observed in previously established, strongly hypomethylated episignature samples (ICF1, TBRS, and Wolf-Hirschhorn syndrome [WHS]; Figure 1) that demonstrated moderate MVP scores for RSTS2 (Figure 4A), which also exhibits overall hypomethylation. The ICF1 (4_ICF1) and WHS (5_WHS) samples showed unsupervised clustering that ruled out RSTS2 (Figures S5C–S5F, respectively), but the TBRS sample was more ambiguous, clustering closer to the RSTS2 samples than to the controls (Figures S5G and S5H). It is important to note that while the TBRS sample clustered with the controls in the MDS plot (Figure S5H), the hierarchical clustering heatmap showed differences from RSTS2 samples with the sample clustering in a branch separate from the other RSTS2 samples (Figure S5G). In this instance, a review of overlapping probes and their relative methylation could also be used to determine if the observed MVP score for this TBRS sample represents non-specific hypomethylation overlap with RSTS2 rather than a true episignature match when this sample is used for testing; however, the possibility that this sample contains variants in genes associated with both TBRS and RSTS2 has not been ruled out.
Two samples with documented pathogenic variants in YY1 associated with GADEVS presented with elevated MVP scores for the BAFopathy episignature (Figure 4B). Both samples exhibited an MVP score greater than 0.5; however, one sample (GADEVS_1) clustered closer to BAFopathy samples than to controls (Figures S10A and S10B). YY1 is a DNA-binding factor that can activate or repress gene expression via cofactor recruitment, the disruption of binding sites, or conformational DNA changes and plays an important role in embryogenesis, differentiation, DNA replication, and cellular proliferation.64 The BAF complex is also important in embryonic development,65,66 and a recent study has demonstrated the interaction between YY1 and seven of the BAF complex subunits in mouse embryonic stem cells, which promotes proliferation.67 Further comparison of YY1 and BAF complex subunit SMARCA4-binding sites showed a significant overlap, suggesting that YY1 may work with the BAF complex to maintain pluripotency.67 Therefore, it is possible that the elevated MVP scores observed in the GADEVS samples could represent a functional overlap between YY1 and the BAF complex that results in similar differentially methylated regions. Further investigation into the overlap of the probes within these two episignatures is required.
Finally, a subject sample (6_IDDSELD) with a 1.5 Mb deletion on chromosome 12 including SETD1B, assessed clinically and by episignature analysis as positive for IDDSELD, demonstrated a high MVP score for KDM2B (Figure 4A). The deletion starts approximately 215 kb upstream of the KDM2B start site. While unsupervised MDS clustering showed that this sample clustered away from controls (Figure S7B), the hierarchical clustering indicated that the sample was more like the KDM2B cases than the controls (Figure S7A). This high MVP score and unsupervised clustering indicate a partial match to the KDM2B episignature, which could potentially be a result of KDM2B upstream regulatory elements that may be impacted by the deletion observed in this sample, potentially resulting in decreased KDM2B expression and possible changes to the methylome. While the vast majority of cases present with highly specific episignatures, these examples demonstrate an approach to assessing more complex cases and the importance of reviewing supervised and unsupervised algorithm outputs, as well as gene function and mean episignature methylation patterns. We have previously discussed in more detail the clinical implementation and use of episignatures, including the use of clustering analysis with MVP scores for clinical diagnose.30
Conclusions
This study expands the number of defined episignatures in Mendelian disorders to 57. In addition to seven imprinting disorders and two trinucleotide repeat expansion disorders,18 EpiSign V3 now screens for a total of 74 syndromes, which further broadens the clinical utility of DNA methylation analysis as a screening tool, an additional approach to unresolved cases, and a method for VUS reclassification. Further clinical adoption will benefit from the development of specific guidelines for episignature assessment within the scope of general guidelines for the interpretation of functional evidence in genetic testing.68 The continued refinement of existing episignatures, including the characterization of sub-signatures and the addition of disorders assessed by new episignatures, will be required as the number of disorders and complexity of the data and clinical associations continue to expand. The epigenetic landscape during development, depicted by Conrad Waddington as a ball atop a hill with multiple intersecting paths to follow, represents developmental “choices” that cells must make that are influenced by epigenetic changes that alter the possible paths to choose from.69 DNA methylation has emerged as a reliable marker for these changes within the epigenetic landscape. Ongoing large-scale studies, such as EpiSign-CAN, are expected to provide insight into the real-world applications and the health-system impact of DNA methylation episignature assessment in the diagnosis of genetic disorders. International advisories such as the currently ongoing International Rare Disorders Research Consortium70 “Working Group on Integrating New Technologies for the Diagnosis of Rare Diseases” are focused on developing guidelines for the establishment of diagnostic standards for new molecular technologies including diagnostic DNA methylation analysis in Mendelian disorders. Finally, the broadening clinical utility of DNA methylation testing for the diagnosis of Mendelian disorders highlights the need for the expansion of the current ACMG recommendations68 for the application of the functional evidence in genetic variant interpretation.
Data and code availability
Some of the datasets used in this study are available publicly as previously described.29 Sixteen of the 17 Chr16p11.2del samples are from GEO: GSE113967.33 Anonymized data for each subject is described in the study. The raw DNA methylation data for other samples are not available due to institutional and ethics restrictions. The software used in this study is publicly available with software packages and versions described in the Materials and methods.
Acknowledgments
Funding for this study is provided in part by the London Health Sciences Molecular Diagnostics Development Fund and the Genome Canada Genomic Applications Partnership Program. The research conducted at the Murdoch Children's Research Institute was supported by the Victorian Government's Operational Infrastructure Support Program. The Chair in Genomic Medicine awarded to J.C. is generously supported by The Royal Children's Hospital Foundation. Funding was provided by the Italian Ministry of Health (Ricerca Corrente to A.C.; 5x1000, CCR-2017-23669081, and RCR-2020-23670068_001 to M.T.) and the Italian Ministry of Research (FOE 2019 to M.T.). The authors wish to acknowledge Care4Rare for providing some of the patient samples. Support for this study is provided in part by the MKHK Association.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xhgg.2021.100075.
Supplemental information
References
- 1.Nguengang Wakap S., Lambert D.M., Olry A., Rodwell C., Gueydan C., Lanneau V., Murphy D., Le Cam Y., Rath A. Estimating cumulative point prevalence of rare diseases: analysis of the Orphanet database. Eur. J. Hum. Genet. 2020;28:165–173. doi: 10.1038/s41431-019-0508-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baird P.A., Anderson T.W., Newcombe H.B., Lowry R.B. Genetic disorders in children and young adults: a population study. Am. J. Hum. Genet. 1988;42:677–693. [PMC free article] [PubMed] [Google Scholar]
- 3.Kvarnung M., Nordgren A. Intellectual disability & rare disorders: A diagnostic challenge. Adv. Exp. Med. Biol. 2017;1031:39–54. doi: 10.1007/978-3-319-67144-4_3. [DOI] [PubMed] [Google Scholar]
- 4.Schwarze K., Buchanan J., Taylor J.C., Wordsworth S. Are whole-exome and whole-genome sequencing approaches cost-effective? A systematic review of the literature. Gen. Med. 2018;20:1122–1130. doi: 10.1038/gim.2017.247. [DOI] [PubMed] [Google Scholar]
- 5.Eisenberger T., Neuhaus C., Khan A.O., Decker C., Preising M.N., Friedburg C., Bieg A., Gliem M., Charbel Issa P., Holz F.G., et al. Increasing the yield in targeted next-generation sequencing by implicating CNV analysis, non-coding exons and the overall variant load: the example of retinal dystrophies. PLoS One. 2013;8:e78496. doi: 10.1371/journal.pone.0078496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wise A.L., Manolio T.A., Mensah G.A., Peterson J.F., Roden D.M., Tamburro C., Williams M.S., Green E.D. Genomic medicine for undiagnosed diseases. Lancet. 2019;394:533–540. doi: 10.1016/S0140-6736(19)31274-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schubeler D. Function and information content of DNA methylation. Nature. 2015;517:321–326. doi: 10.1038/nature14192. [DOI] [PubMed] [Google Scholar]
- 8.Gopalakrishnan S., Van Emburgh B.O., Robertson K.D. DNA methylation in development and human disease. Mutat. Res. 2008;647:30–38. doi: 10.1016/j.mrfmmm.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jin Z., Liu Y. DNA methylation in human diseases. Genes Dis. 2018;5:1–8. doi: 10.1016/j.gendis.2018.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Velasco G., Francastel C. Genetics meets DNA methylation in rare diseases. Clin. Genet. 2019;95:210–220. doi: 10.1111/cge.13480. [DOI] [PubMed] [Google Scholar]
- 11.Choufani S., Cytrynbaum C., Chung B.H., Turinsky A.L., Grafodatskaya D., Chen Y.A., Cohen A.S., Dupuis L., Butcher D.T., Siu M.T., et al. NSD1 mutations generate a genome-wide DNA methylation signature. Nat. Commun. 2015;6:10207. doi: 10.1038/ncomms10207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kernohan K.D., Cigana Schenkel L., Huang L., Smith A., Pare G., Ainsworth P., Care4Rare Canada C., Boycott K.M., Warman-Chardon J., Sadikovic B. Identification of a methylation profile for DNMT1-associated autosomal dominant cerebellar ataxia, deafness, and narcolepsy. Clin. Epigenet. 2016;8:91. doi: 10.1186/s13148-016-0254-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hood R.L., Schenkel L.C., Nikkel S.M., Ainsworth P.J., Pare G., Boycott K.M., Bulman D.E., Sadikovic B. The defining DNA methylation signature of Floating-Harbor Syndrome. Sci. Rep. 2016;6:38803. doi: 10.1038/srep38803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Butcher D.T., Cytrynbaum C., Turinsky A.L., Siu M.T., Inbar-Feigenberg M., Mendoza-Londono R., Chitayat D., Walker S., Machado J., Caluseriu O., et al. CHARGE and kabuki syndromes: gene-specific DNA methylation signatures identify epigenetic mechanisms linking these clinically overlapping conditions. Am. J. Hum. Genet. 2017;100:773–788. doi: 10.1016/j.ajhg.2017.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aref-Eshghi E., Rodenhiser D.I., Schenkel L.C., Lin H., Skinner C., Ainsworth P., Pare G., Hood R.L., Bulman D.E., Kernohan K.D., et al. Genomic DNA methylation signatures enable concurrent diagnosis and clinical genetic variant classification in neurodevelopmental syndromes. Am. J. Hum. Genet. 2018;102:156–174. doi: 10.1016/j.ajhg.2017.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schenkel L.C., Aref-Eshghi E., Skinner C., Ainsworth P., Lin H., Pare G., Rodenhiser D.I., Schwartz C., Sadikovic B. Peripheral blood epi-signature of Claes-Jensen syndrome enables sensitive and specific identification of patients and healthy carriers with pathogenic mutations in KDM5C. Clin. Epigenet. 2018;10:21. doi: 10.1186/s13148-018-0453-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aref-Eshghi E., Bend E.G., Hood R.L., Schenkel L.C., Carere D.A., Chakrabarti R., Nagamani S.C.S., Cheung S.W., Campeau P.M., Prasad C., et al. BAFopathies' DNA methylation epi-signatures demonstrate diagnostic utility and functional continuum of Coffin-Siris and Nicolaides-Baraitser syndromes. Nat. Commun. 2018;9:4885. doi: 10.1038/s41467-018-07193-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aref-Eshghi E., Bend E.G., Colaiacovo S., Caudle M., Chakrabarti R., Napier M., Brick L., Brady L., Carere D.A., Levy M.A., et al. Diagnostic utility of genome-wide DNA methylation testing in genetically unsolved individuals with suspected hereditary conditions. Am. J. Hum. Genet. 2019;104:685–700. doi: 10.1016/j.ajhg.2019.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bend E.G., Aref-Eshghi E., Everman D.B., Rogers R.C., Cathey S.S., Prijoles E.J., Lyons M.J., Davis H., Clarkson K., Gripp K.W., et al. Gene domain-specific DNA methylation episignatures highlight distinct molecular entities of ADNP syndrome. Clin. Epigenet. 2019;11:64. doi: 10.1186/s13148-019-0658-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aref-Eshghi E., Bourque D.K., Kerkhof J., Carere D.A., Ainsworth P., Sadikovic B., Armour C.M., Lin H. Genome-wide DNA methylation and RNA analyses enable reclassification of two variants of uncertain significance in a patient with clinical Kabuki syndrome. Hum. Mutat. 2019;40:1684–1689. doi: 10.1002/humu.23833. [DOI] [PubMed] [Google Scholar]
- 21.Krzyzewska I.M., Maas S.M., Henneman P., Lip K.V.D., Venema A., Baranano K., Chassevent A., Aref-Eshghi E., van Essen A.J., Fukuda T., et al. A genome-wide DNA methylation signature for SETD1B-related syndrome. Clin. Epigenet. 2019;11:156. doi: 10.1186/s13148-019-0749-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ciolfi A., Aref-Eshghi E., Pizzi S., Pedace L., Miele E., Kerkhof J., Flex E., Martinelli S., Radio F.C., Ruivenkamp C.A.L., et al. Frameshift mutations at the C-terminus of HIST1H1E result in a specific DNA hypomethylation signature. Clin. Epigenet. 2020;12:7. doi: 10.1186/s13148-019-0804-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Choufani S., Gibson W.T., Turinsky A.L., Chung B.H.Y., Wang T., Garg K., Vitriolo A., Cohen A.S.A., Cyrus S., Goodman S., et al. DNA methylation signature for EZH2 functionally classifies sequence variants in three PRC2 complex genes. Am. J. Hum. Genet. 2020;106:596–610. doi: 10.1016/j.ajhg.2020.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cappuccio G., Sayou C., Tanno P.L., Tisserant E., Bruel A.L., Kennani S.E., Sa J., Low K.J., Dias C., Havlovicova M., et al. De novo SMARCA2 variants clustered outside the helicase domain cause a new recognizable syndrome with intellectual disability and blepharophimosis distinct from Nicolaides-Baraitser syndrome. Gen. Med. 2020;22:1838–1850. doi: 10.1038/s41436-020-0898-y. [DOI] [PubMed] [Google Scholar]
- 25.Schenkel L.C., Aref-Eshghi E., Rooney K., Kerkhof J., Levy M.A., McConkey H., Rogers R.C., Phelan K., Sarasua S.M., Jain L., et al. DNA methylation epi-signature is associated with two molecularly and phenotypically distinct clinical subtypes of Phelan-McDermid syndrome. Clin. Epigenet. 2021;13:2. doi: 10.1186/s13148-020-00990-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haghshenas S., Levy M.A., Kerkhof J., Aref-Eshghi E., McConkey H., Balci T., et al. Detection of a DNA methylation signature for the intellectual developmental disorder, X-linked, syndromic, armfield type. Int. J. Mol. Sci. 2021;22 doi: 10.3390/ijms22031111. https://www.mdpi.com/1422-0067/22/3/1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Radio F.C., Pang K., Ciolfi A., Levy M.A., Hernandez-Garcia A., Pedace L., Pantaleoni F., Liu Z., de Boer E., Jackson A., et al. SPEN haploinsufficiency causes a neurodevelopmental disorder overlapping proximal 1p36 deletion syndrome with an episignature of X chromosomes in females. Am. J. Hum. Genet. 2021;108:502–516. doi: 10.1016/j.ajhg.2021.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aref-Eshghi E., Kerkhof J., Pedro V.P., France G.D., Barat-Houari M., Ruiz-Pallares N., Andrau J.C., Lacombe D., Van-Gils J., Fergelot P., et al. Evaluation of DNA methylation episignatures for diagnosis and phenotype correlations in 42 mendelian neurodevelopmental disorders. Am. J. Hum. Genet. 2021;108:1161–1163. doi: 10.1016/j.ajhg.2021.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aref-Eshghi E., Kerkhof J., Pedro V.P., Groupe D.I.F., Barat-Houari M., Ruiz-Pallares N., Andrau J.C., Lacombe D., Van-Gils J., Fergelot P., et al. Evaluation of DNA methylation episignatures for diagnosis and phenotype correlations in 42 mendelian neurodevelopmental disorders. Am. J. Hum. Genet. 2020;106:356–370. doi: 10.1016/j.ajhg.2020.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sadikovic B., Levy M.A., Kerkhof J., Aref-Eshghi E., Schenkel L., Stuart A., McConkey H., Henneman P., Venema A., Schwartz C.E., et al. Clinical epigenomics: genome-wide DNA methylation analysis for the diagnosis of Mendelian disorders. Gen. Med. 2021;23:1065–1074. doi: 10.1038/s41436-020-01096-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aref-Eshghi E., Schenkel L.C., Lin H., Skinner C., Ainsworth P., Pare G., Rodenhiser D., Schwartz C., Sadikovic B. The defining DNA methylation signature of Kabuki syndrome enables functional assessment of genetic variants of unknown clinical significance. Epigenetics. 2017;12:923–933. doi: 10.1080/15592294.2017.1381807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sadikovic B., Levy M.A., Aref-Eshghi E. Functional annotation of genomic variation: DNA methylation episignatures in neurodevelopmental Mendelian disorders. Hum. Mol. Genet. 2020;29:R27–R32. doi: 10.1093/hmg/ddaa144. [DOI] [PubMed] [Google Scholar]
- 33.Siu M.T., Butcher D.T., Turinsky A.L., Cytrynbaum C., Stavropoulos D.J., Walker S., Caluseriu O., Carter M., Lou Y., Nicolson R., et al. Functional DNA methylation signatures for autism spectrum disorder genomic risk loci: 16p11.2 deletions and CHD8 variants. Clin. Epigenet. 2019;11:103. doi: 10.1186/s13148-019-0684-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aryee M.J., Jaffe A.E., Corrada-Bravo H., Ladd-Acosta C., Feinberg A.P., Hansen K.D., Irizarry R.A. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics. 2014;30:1363–1369. doi: 10.1093/bioinformatics/btu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen Y.A., Lemire M., Choufani S., Butcher D.T., Grafodatskaya D., Zanke B.W., Gallinger S., Hudson T.J., Weksberg R. Discovery of cross-reactive probes and polymorphic CpGs in the Illumina Infinium HumanMethylation450 microarray. Epigenetics. 2013;8:203–209. doi: 10.4161/epi.23470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pidsley R., Zotenko E., Peters T.J., Lawrence M.G., Risbridger G.P., Molloy P., Van Djik S., Muhlhausler B., Stirzaker C., Clark S.J. Critical evaluation of the Illumina MethylationEPIC BeadChip microarray for whole-genome DNA methylation profiling. Genome Biol. 2016;17:208. doi: 10.1186/s13059-016-1066-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ho D., Imai K., King G., Stuart E.A. MatchIt: Nonparametric preprocessing for parametric causal inference. J. Stat. Softw. 2011;42:28. [Google Scholar]
- 38.Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Houseman E.A., Accomando W.P., Koestler D.C., Christensen B.C., Marsit C.J., Nelson H.H., Wiencke J.K., Kelsey K.T. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinform. 2012;13:86. doi: 10.1186/1471-2105-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Platt J.C. MIT Press; 1999. Probabilistic Outputs for Support Vector Machines and Comparisons to Regularized Likelihood Methods. [Google Scholar]
- 41.Contreras-Martos S., Piai A., Kosol S., Varadi M., Bekesi A., Lebrun P., Volkov A.N., Gevaert K., Pierattelli R., Felli I.C., et al. Linking functions: an additional role for an intrinsically disordered linker domain in the transcriptional coactivator CBP. Sci. Rep. 2017;7:4676. doi: 10.1038/s41598-017-04611-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cordeddu V., Macke E.L., Radio F.C., Lo Cicero S., Pantaleoni F., Tatti M., Bellacchio E., Ciolfi A., Agolini E., Bruselles A., et al. Refinement of the clinical and mutational spectrum of UBE2A deficiency syndrome. Clin. Genet. 2020;98:172–178. doi: 10.1111/cge.13775. [DOI] [PubMed] [Google Scholar]
- 43.Sadikovic B., Aref-Eshghi E., Levy M.A., Rodenhiser D. DNA methylation signatures in mendelian developmental disorders as a diagnostic bridge between genotype and phenotype. Epigenomics. 2019;11:563–575. doi: 10.2217/epi-2018-0192. [DOI] [PubMed] [Google Scholar]
- 44.Haghshenas S., Bhai P., Aref-Eshghi E., Sadikovic B. Diagnostic utility of genome-wide DNA methylation analysis in mendelian neurodevelopmental disorders. Int. J. Mol. Sci. 2020;21 doi: 10.3390/ijms21239303. https://www.mdpi.com/1422-0067/21/23/9303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee Y.R., Khan K., Armfield-Uhas K., Srikanth S., Thompson N.A., Pardo M., Yu L., Norris J.W., Peng Y., Gripp K.W., et al. Mutations in FAM50A suggest that Armfield XLID syndrome is a spliceosomopathy. Nat. Commun. 2020;11:3698. doi: 10.1038/s41467-020-17452-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.D'Gama A.M., Walsh C.A. Somatic mosaicism and neurodevelopmental disease. Nat. Neurosci. 2018;21:1504–1514. doi: 10.1038/s41593-018-0257-3. [DOI] [PubMed] [Google Scholar]
- 47.Natri H.M., Bobowik K.S., Kusuma P., Crenna Darusallam C., Jacobs G.S., Hudjashov G., Lansing J.S., Sudoyo H., Banovich N.E., Cox M.P., et al. Genome-wide DNA methylation and gene expression patterns reflect genetic ancestry and environmental differences across the Indonesian archipelago. PLoS Genet. 2020;16:e1008749. doi: 10.1371/journal.pgen.1008749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Carja O., MacIsaac J.L., Mah S.M., Henn B.M., Kobor M.S., Feldman M.W., Fraser H.B. Worldwide patterns of human epigenetic variation. Nat. Ecol. Evol. 2017;1:1577–1583. doi: 10.1038/s41559-017-0299-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Galanter J.M., Gignoux C.R., Oh S.S., Torgerson D., Pino-Yanes M., Thakur N., et al. Differential methylation between ethnic sub-groups reflects the effect of genetic ancestry and environmental exposures. eLife. 2017;6:e20532. doi: 10.7554/eLife.20532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Giri A.K., Bharadwaj S., Banerjee P., Chakraborty S., Parekatt V., Rajashekar D., Tomar A., Ravindran A., Basu A., Tandon N., et al. DNA methylation profiling reveals the presence of population-specific signatures correlating with phenotypic characteristics. Mol. Genet. Genom. 2017;292:655–662. doi: 10.1007/s00438-017-1298-0. [DOI] [PubMed] [Google Scholar]
- 51.McKennan C., Naughton K., Stanhope C., Kattan M., O'Connor G.T., Sandel M.T., Visness C.M., Wood R.A., Bacharier L.B., Beigelman A., et al. Longitudinal data reveal strong genetic and weak non-genetic components of ethnicity-dependent blood DNA methylation levels. Epigenetics. 2021;16:662–676. doi: 10.1080/15592294.2020.1817290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Song M.A., Seffernick A.E., Archer K.J., Mori K.M., Park S.Y., Chang L., Ernst T., Tiirikainen M., Peplowska K., Wilkens L.R., et al. Race/ethnicity-associated blood DNA methylation differences between Japanese and European American women: an exploratory study. Clin. Epigenet. 2021;13:188. doi: 10.1186/s13148-021-01171-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bogershausen N., Wollnik B. Mutational landscapes and phenotypic spectrum of SWI/SNF-related intellectual disability disorders. Front. Mol. Neurosci. 2018;11:252. doi: 10.3389/fnmol.2018.00252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sandhya S., Maulik A., Giri M., Singh M. Domain architecture of BAF250a reveals the ARID and ARM-repeat domains with implication in function and assembly of the BAF remodeling complex. PLoS One. 2018;13:e0205267. doi: 10.1371/journal.pone.0205267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Finn R.D., Coggill P., Eberhardt R.Y., Eddy S.R., Mistry J., Mitchell A.L., Potter S.C., Punta M., Qureshi M., Sangrador-Vegas A., et al. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 2016;44:D279–D285. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schuettengruber B., Martinez A.M., Iovino N., Cavalli G. Trithorax group proteins: switching genes on and keeping them active. Nat. Rev. Mol. Cell Biol. 2011;12:799–814. doi: 10.1038/nrm3230. [DOI] [PubMed] [Google Scholar]
- 57.Kosho T., Okamoto N., Coffin-Siris Syndrome International C. Genotype-phenotype correlation of Coffin-Siris syndrome caused by mutations in SMARCB1, SMARCA4, SMARCE1, and ARID1A. Am. J. Med. Genet. C Sem. Med. Genet. 2014;166C:262–275. doi: 10.1002/ajmg.c.31407. [DOI] [PubMed] [Google Scholar]
- 58.Li D., Ahrens-Nicklas R.C., Baker J., Bhambhani V., Calhoun A., Cohen J.S., Deardorff M.A., Fernandez-Jaen A., Kamien B., Jain M., et al. The variability of SMARCA4-related Coffin-Siris syndrome: do nonsense candidate variants add to milder phenotypes? Am. J. Med. Genet. A. 2020;182:2058–2067. doi: 10.1002/ajmg.a.61732. [DOI] [PubMed] [Google Scholar]
- 59.Menke L.A., study D.D.D., Gardeitchik T., Hammond P., Heimdal K.R., Houge G., Hufnagel S.B., Ji J., Johansson S., Kant S.G., et al. Further delineation of an entity caused by CREBBP and EP300 mutations but not resembling Rubinstein-Taybi syndrome. Am. J. Med. Genet. A. 2018;176:862–876. doi: 10.1002/ajmg.a.38626. [DOI] [PubMed] [Google Scholar]
- 60.Bedford D.C., Kasper L.H., Fukuyama T., Brindle P.K. Target gene context influences the transcriptional requirement for the KAT3 family of CBP and p300 histone acetyltransferases. Epigenetics. 2010;5:9–15. doi: 10.4161/epi.5.1.10449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Arboleda V.A., Lee H., Dorrani N., Zadeh N., Willis M., Macmurdo C.F., Manning M.A., Kwan A., Hudgins L., Barthelemy F., et al. De novo nonsense mutations in KAT6A, a lysine acetyl-transferase gene, cause a syndrome including microcephaly and global developmental delay. Am. J. Hum. Genet. 2015;96:498–506. doi: 10.1016/j.ajhg.2015.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wiesel-Motiuk N., Assaraf Y.G. The key roles of the lysine acetyltransferases KAT6A and KAT6B in physiology and pathology. Drug resistance updates : reviews and commentaries in antimicrobial and anticancer chemotherapy. 2020;53:100729. doi: 10.1016/j.drup.2020.100729. [DOI] [PubMed] [Google Scholar]
- 63.Riley L.G., Cooper S., Hickey P., Rudinger-Thirion J., McKenzie M., Compton A., Lim S.C., Thorburn D., Ryan M.T., Giege R., et al. Mutation of the mitochondrial tyrosyl-tRNA synthetase gene, YARS2, causes myopathy, lactic acidosis, and sideroblastic anemia--MLASA syndrome. Am. J. Med. Genet. 2010;87:52–59. doi: 10.1016/j.ajhg.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gordon S., Akopyan G., Garban H., Bonavida B. Transcription factor YY1: structure, function, and therapeutic implications in cancer biology. Oncogene. 2006;25:1125–1142. doi: 10.1038/sj.onc.1209080. [DOI] [PubMed] [Google Scholar]
- 65.Panamarova M., Cox A., Wicher K.B., Butler R., Bulgakova N., Jeon S., Rosen B., Seong R.H., Skarnes W., Crabtree G., et al. The BAF chromatin remodelling complex is an epigenetic regulator of lineage specification in the early mouse embryo. Development. 2016;143:1271–1283. doi: 10.1242/dev.131961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang H., Wang X., Li J., Shi R., Ye Y. BAF complex in embryonic stem cells and early embryonic development. Stem Cells Int. 2021;2021:6668866. doi: 10.1155/2021/6668866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang J., Wu X., Wei C., Huang X., Ma Q., Huang X., Faiola F., Guallar D., Fidalgo M., Huang T., et al. YY1 positively regulates transcription by targeting promoters and super-enhancers through the BAF complex in embryonic stem cells. Stem Cell Rep. 2018;10:1324–1339. doi: 10.1016/j.stemcr.2018.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Brnich S.E., Abou Tayoun A.N., Couch F.J., Cutting G.R., Greenblatt M.S., Heinen C.D., Kanavy D.M., Luo X., McNulty S.M., Starita L.M., et al. Recommendations for application of the functional evidence PS3/BS3 criterion using the ACMG/AMP sequence variant interpretation framework. Genome Med. 2019;12:3. doi: 10.1186/s13073-019-0690-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Waddington C.H. Allen & Unwin; 1957. The Strategy of the Genes; a Discussion of Some Aspects of Theoretical Biology. [Google Scholar]
- 70.Cutillo C.M., Austin C.P., Groft S.C. A global approach to rare diseases research and orphan products development: the International Rare Diseases Research Consortium (IRDiRC) Adv. Exp. Med. Biol. 2017;1031:349–369. doi: 10.1007/978-3-319-67144-4_20. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Some of the datasets used in this study are available publicly as previously described.29 Sixteen of the 17 Chr16p11.2del samples are from GEO: GSE113967.33 Anonymized data for each subject is described in the study. The raw DNA methylation data for other samples are not available due to institutional and ethics restrictions. The software used in this study is publicly available with software packages and versions described in the Materials and methods.