Abstract
CD133 has been recognized as a prominent biomarker for cancer stem cells (CSCs), which promote tumor relapse and metastasis. Here, we developed a clinically relevant, stable, and peptide-based positron emission tomography (PET) tracer, [64Cu]CM-2, for mapping CD133 protein in several kinds of cancers. Through the incorporation of a 6-aminohexanoic acid (Ahx) into the N terminus of a CM peptide, we constructed a stable peptide tracer [64Cu]CM-2, which exhibited specific binding to CD133-positive CSCs in multiple preclinical tumor models. Both PET imaging and ex vivo biodistribution verified the superb performance of [64Cu]CM-2. Furthermore, the matched physical and biological half-life of [64Cu]CM-2 makes it a state-of-the-art PET tracer for CD133. Therefore, [64Cu]CM-2 PET may not only enable the longitudinal tracking of CD133 dynamics in the cancer stem cell niche but also provide a powerful and noninvasive imaging tool to track down CSCs in refractory cancers.
Introduction
Cancer recurrence and metastasis are the major causes of tumor-associated deaths. A subset of pluripotent tumor progenitor cells has been identified as cancer stem cells (CSCs).1 Increasing evidence has shown that CSCs are capable of self-renewal, long-term propagation, and long-distance dissemination and can drive tumor initiation and relapse. Therefore, therapeutic strategies designed to specifically eliminate CSCs are critical for preventing cancer relapse and for improving the overall survival of patients.2 Unfortunately, CSCs maintain a high level of resistance to conventional chemo- and radiotherapies, which makes targeting CSCs challenging.3
Furthermore, CSCs usually represent a very minor subpopulation of cancer cells in bulky tumors, and their abundance is highly heterogeneous among different types of tumors; this makes the detection of CSCs in tumors particularly challenging.4 Conventional diagnostic tools, such as magnetic resonance (MR) and immunohistochemistry (IHC), are insufficient to accurately delineate the CSCs in tumors.5,6 Hence, the development of noninvasive imaging tools that can quantitatively map the location and abundance of CSCs in lesions is exceptionally important and may not only facilitate the exploration of the fundamental biological functions of CSCs but also foster the development of CSC-targeting therapeutics. Among all current clinically relevant diagnostic techniques, positron emission tomography (PET) with high sensitivity and quantitative attributes has been well recognized as a superb imaging tool to annotate the biological status of tumors by using metabolic or molecular targeting radiotracers.7−14 Therefore, the development of CSC-specific PET tracers will be a critical step on the way to tackling CSCs.
CD133, also known as prominin-1, is a five-transmembrane glycoprotein originally detected in hematopoietic progenitor cells and neuroepithelial stem cells.15 Recently, CD133 was further discovered to be expressed in a variety of cancers including brain, colon, pancreatic, lung, liver, prostate, and ovarian cancers as well as melanomas, sarcomas, and several kinds of leukemia.16−23 Moreover, a multitude of studies have suggested that CD133 is an essential biomarker and therapeutic target for CSCs.24,25 To enable noninvasive detection of CSCs in tumors, several studies have reported the development of optical or radioprobes for CD133.26,27 Nevertheless, PET tracers that are ultrasensitive, suitable for long-term in vivo monitoring, and can be cleared quickly by the kidneys have not yet been developed. Here, we report the development of a stable peptide-based PET tracer for CD133 (Scheme 1).
Scheme 1. Schematic Illustration of Our Approach.
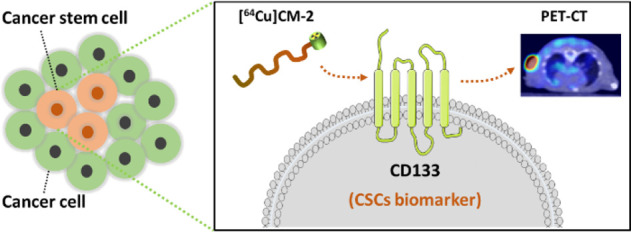
[64Cu]CM-2 was designed as a PET radiotracer for CD133 to detect the cancer stem cell populations in tumors.
Results and Discussion
CM-1 is a CD133 binding peptide, which was originally identified in a screen and shown to have high binding affinity for CD133 (KD = 7.37 nM).28 A CM-conjugated second near-infrared (NIR-II) probe was constructed and demonstrated as a superior imaging probe for CD133 detection in a tumor model. Due to the high binding affinity and quick renal excretion of the CM peptide, we envisioned that the CM peptide could be an ideal targeting moiety for more clinically relevant PET probes. With this hypothesis in mind, we synthesized a peptide called CM-1, which presents an N-terminal 1,4,7,10-tetraazacyclododecane-1,4,7,10-tetraacetic acid (DOTA) as a chelator for 64Cu labeling (Figure 1A). The radiotracer [64Cu]CM-1 was then produced with satisfactory radiocharacteristics (Table 1). However, it showed fast degradation in mouse serum, as approximately 22.4 and 20.1% of intact [64Cu]CM-1 remained after 1 and 2 h incubation, respectively (Figure 1B). The poor stability of [64Cu]CM-1 limits its clinical usefulness.
Figure 1.
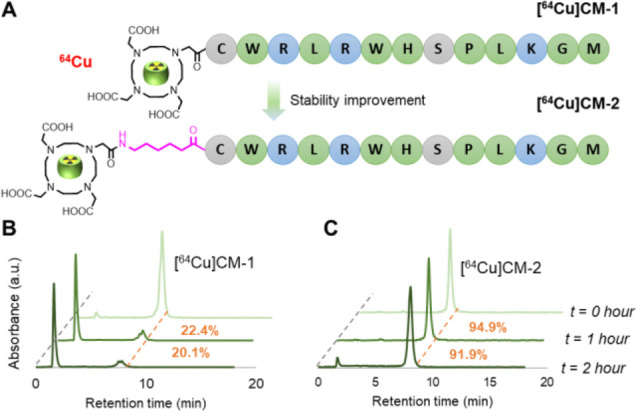
(A) Chemical structures of radiotracers [64Cu]CM-1 and [64Cu]CM-2. In the latter, an Ahx was positioned at the N terminus to space the peptide and DOTA. (B, C) In vitro stability of [64Cu]CM-1 (B) and [64Cu]CM-2 (C). Both radiotracers were incubated in mouse serum for 1 and 2 h at 37 °C and then analyzed by radio HPLC. Orange dashed lines indicate the peaks of the intact radiotracers. The numbers in orange indicate the fraction of intact radiotracers.
Table 1. Quality Control Results for [64Cu]CM-1, [64Cu]CM-2, and [64Cu]CM-3a.
| tracer | [64Cu]CM-1 | [64Cu]CM-2 | [64Cu]CM-3c |
|---|---|---|---|
| retention time | 8.18 | 8.36 | 8.28 |
| cLogP | –2.84 | –1.93 | –2.15 |
| radiochemical yield (%) | >99 | >99 | >99 |
| molar activity (GBq μmol–1) | >74 | >74 | >74 |
| radiochemical purity (%)b | >98 | >98 | >98 |
| chelator | DOTA | DOTA | NODAGA |
Radiochemical yield (RCY), molar activity, and radiochemical purity of the as-prepared tracer. Data are expressed as mean values (n = 7).
The radiochemical purity was determined by HPLC with the conditions as follows: column, YMC-Triat-C18 column (4.6 mm i.d. × 150 mm, 5 μm); solvent gradient, 10–90% acetonitrile (0.1% trifluoroacetic acid (TFA)), 20 min; flow rate, 1 mL/min.
[64Cu]CM-3 is an analog of [64Cu]CM-2 while differing by the chelator type. NODAGA: 1,4,7-triazacyclononane,1-glutaric acid-4,7-acetic acid.
To improve the stability of CM-1, peptide stabilization strategies, such as macrocyclization, stapling, d-amino acid replacement, and others, can be undertaken.11,29−33 Among them, N-terminal modification is an effective and direct way to ameliorate peptide stability.8,14,34 Moreover, it generally preserves the specific binding mode of the original peptide. We therefore synthesized a peptide called CM-2, in which a 6-aminohexanoic acid (Ahx) linker is anchored between the N terminus of the peptide and the DOTA (Figure 1A). [64Cu]CM-2 had a similar radiocharacteristic profile to [64Cu]CM-1 (Table 1). Moreover, Ahx modification led to an increase in the lipophilicity of the peptide, as the partition coefficient (cLogP) of [64Cu]CM-2 became larger than that of [64Cu]CM-1 (−1.93 vs −2.84). Expectedly, the stability of [64Cu]CM-2 was significantly improved: approximately 94.9 and 91.9% of intact tracers remained after 1 h and 2 h of incubation, respectively (Figure 1C). This result establishes [64Cu]CM-2 as a promising PET tracer for preclinical animal study.
Next, we measured CD133 protein levels in five cancer cell lines, including murine melanoma B16F10, human hepatocellular carcinoma Huh-7, human glioma U87MG, human melanoma Bowes, and human breast cancer MDA-MB231. Flow cytometry analysis showed that Huh-7 cells have the highest CD133 levels, as approximately 74.5% of the cells were CD133-positive (Figure 2A). However, for B16F10, U87MG, and Bowes cell lines, the CD133-positive populations were 1–3% of the total (Figure 2A and Figure S1). Moreover, we detected no CD133 expression in MDA-MB231 cells (Figure S1). These results are consistent with those reported previously.23,35
Figure 2.
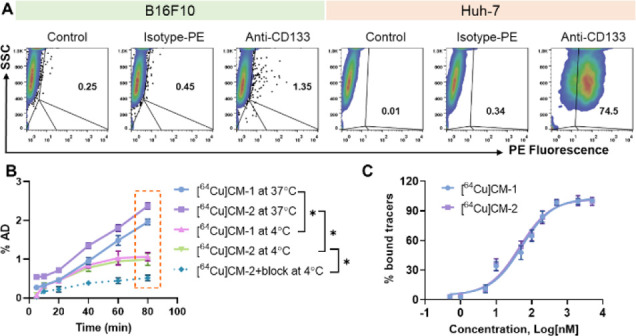
(A) Flow cytometry analysis of the levels of CD133 protein in different cancer cell lines. PE: phycoerythrin; SSC: side-scatter; the cells gated in the upper right panels were assigned as CD133-positive cells, and their proportions are indicated in the panels (number in black %). (B) Cellular uptake of [64Cu]CM-2 in Huh-7 cells at 4 or 37 °C. The block study was performed using antiCD133 (0.1 mg/mL) as a blocking agent. Statistical analysis was performed for 80 min uptakes using two-way ANOVA, followed by Bonferroni’s multiple comparisons test, n = 3, *P < 0.05. (C) Saturation binding curves of [64Cu]CM-1 and [64Cu]CM-2 determined using a Huh-7 cell-based assay.
To examine whether Ahx incorporation alters the binding affinity of the CM peptide, we compared the uptake of [64Cu]CM-1 and [64Cu]CM-2 by Huh-7 cells (Figure 2B). CM peptides are positively charged in cell buffered conditions, and they might cross the cell membrane via a non-CD133-mediated endocytosis pathway.36 Moreover, Ahx is an aliphatic linker, which may further enhance the nonspecific uptake of [64Cu]CM-2. Indeed, [64Cu]CM-2 showed a higher uptake than [64Cu]CM-1 at all measured time points at 37 °C (Figure 2B). To inhibit energetically promoted nonspecific uptake, we then measured cellular binding of the tracers at 4 °C. [64Cu]CM-2 displayed an almost identical uptake profile to that of [64Cu]CM-1 (Figure 2B), indicating that [64Cu]CM-2 has a similar binding affinity to [64Cu]CM-1. In addition, we directly measured the binding affinity of the tracers via a cell-based assay. As shown in Figure 2C, [64Cu]CM-2 showed a binding affinity (IC50) of 44.95 nM, which is similar to that of [64Cu]CM-1 (IC50 = 52.55 nM).
To investigate whether [64Cu]CM-2 can be used to image CD133-positive CSCs populations in mice, we first performed a PET imaging study with [64Cu]CM-2 in a Huh-7 tumor-bearing BALB/C nude mouse model. [64Cu]CM-2 showed apparent accumulation and retention in Huh-7 tumors (Figure 3A, Figure S2, and Table S1). In addition, the tracer showed a quick uptake and slow clearance by the liver, which is due partially to the relatively high hydrophobicity of CM-2, which might undergo oxidative degradation in the liver before excretion. We also noticed that the degraded products were efficiently excreted via the renal clearance pathway (Figure S2 and Table S1).
Figure 3.
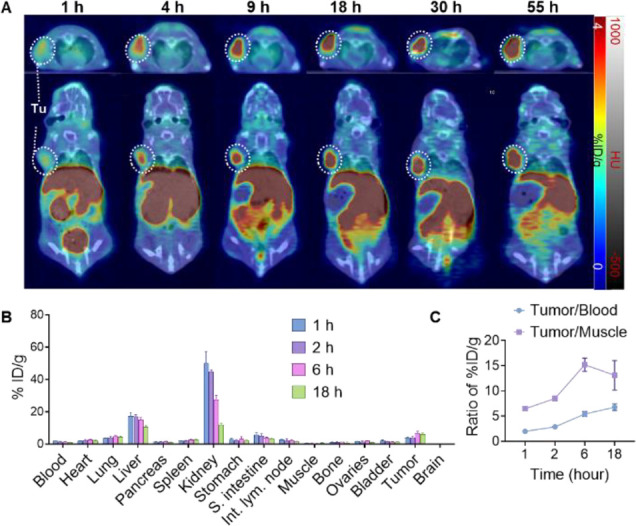
(A) Series of coregistered sectional PET-CT images of Huh-7 tumor-bearing BALB/C nude mice at different time points p.i. of [64Cu]CM-2 (17 MBq/mouse, 0.2 mL, ∼0.5 nmol of CM-2). Upper panels: axial view; lower panels: coronal view. White dashed circles indicate Huh-7 tumors. (B) Ex vivo biodistribution of [64Cu]CM-2 in major organs at different time points after i.v. injection (3.7 MBq per mouse, 0.1 mL, ∼0.1 nmol of CM-2). S. intestine: small intestine; Int. lym. node: intestine lymph node. Data represent means ± SD, n = 3 for each point. (C) Uptake ratios of tumor to blood (T/B) and tumor to muscle (T/M) calculated from the ex vivo biodistribution data. Data represent means ± SD, n = 3.
To verify our observations made using PET imaging, we further conducted an ex vivo biodistribution study. Major organs were excised from the Huh-7 tumor-bearing mice, and the tracer uptake was expressed in % injection dose per gram of tissue (% ID/g). The mean uptakes in the Huh-7 tumors were 3.77, 4.23, 6.89, and 6.19% ID/g at 1, 2, 6, and 18 h after intravenous (i.v.) injection, respectively (Figure 3B and Table S2). An ex vivo autoradiography was performed to dissect the tracer’s spatial distribution in the tumor. As shown in Figure S3, intense activity was shown in the central area of the tumor, indicating a superb tumor penetrating ability of the tracer. The highest uptake was shown in the kidney, while the intensity manifested a fast drop from 1 to 18 h postinjection (p.i.), indicating quick kidney–bladder clearance of [64Cu]CM-2. The uptake ratios of tumor to organs are usually used to depict the imaging contrast of PET images. We then calculated the tumor to blood (T/B) and tumor to muscle (T/M) ratios. For the four time points studied, the highest T/B ratio occurred at 18 h p.i. In contrast, the highest T/M ratio was achieved at 6 h p.i. (Figure 3C). These results suggest that 2–18 h p.i. would be a suitable time window for [64Cu]CM-2 PET imaging. To confirm the CD133-specific uptake of the tracer, blocking PET studies by coinjection of an excess of the unlabeled peptide or antiCD133 monoclonal antibody (1 mg/kg) was performed (Figure S4 and Figure 4, respectively). As a result, we observed a significant decrease of the tracer uptake in the Huh-7 tumor of the blocking group, suggesting a CD133-specific uptake in the tumors.
Figure 4.
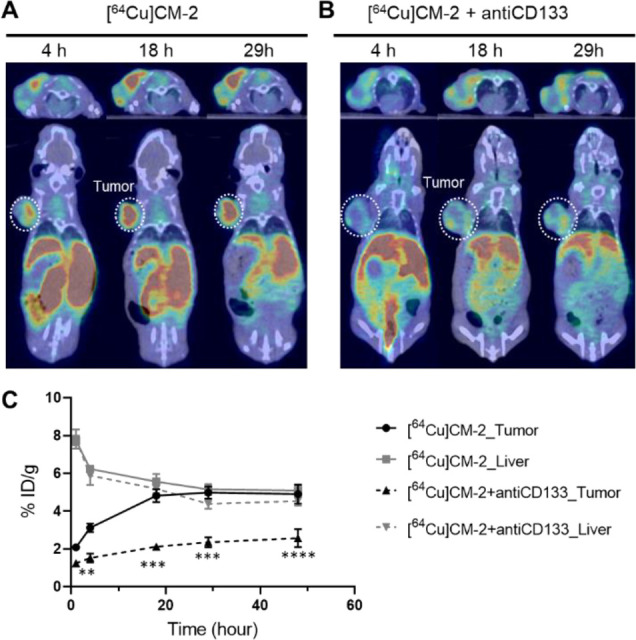
A blocking PET study to verify the specificity of the tracer. Series of coregistered sectional PET-CT images of Huh-7 tumor-bearing BALB/C nude mice at different time points p.i. of (A) [64Cu]CM-2 (18.5 MBq/mouse, 0.2 mL, ∼0.5 nmol of CM-2) or (B) [64Cu]CM-2 (18.5 MBq/mouse, 0.2 mL, ∼0.5 nmol of CM-2) + antiCD133 (1 mg/kg, administered 2 min prior to [64Cu]CM-2 injection). Upper panels: axial view; lower panels: coronal view. White dashed circles indicate Huh-7 tumors. (C) Quantification of the % ID/g in the Huh-7 tumor and the liver from panels A and B. Statistical analysis was performed using two-way ANOVA, followed by Bonferroni’s multiple comparisons test, n = 3, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Having demonstrated the capability of [64Cu]CM-2 PET to visualize CSCs in CD133-overexpressing Huh-7 tumors, we then investigated the general applicability of this imaging modality in several other kinds of tumor models with low CD133 expression. First, we performed a PET-CT study in a B16F10 allograft model, as [64Cu]CM-2 PET is anticipated to provide a more realistic reflection of the tracer’s behavior in the human body. Similarly to the observation in Huh-7 xenograft, the tracer showed apparent uptake and retention in the B16F10 tumors (Figure 5A, Figure S5, and Table S3). The ex vivo biodistribution study validated the PET results (Figure 5B and Table S4). Notably, we observed a high tracer uptake in the intestine and stomach at 0.5 and 2 h p.i., while the radioactivity in the two organs was significantly decreased at 18 and 30 h p.i. This suggests that a waiting time of several hours after injection is recommended to achieve the best PET imaging quality in clinical settings. Finally, the T/B and T/M ratios suggested that higher imaging contrast can be achieved even in animals bearing low CD133-expressing tumors (Figure 5C).
Figure 5.
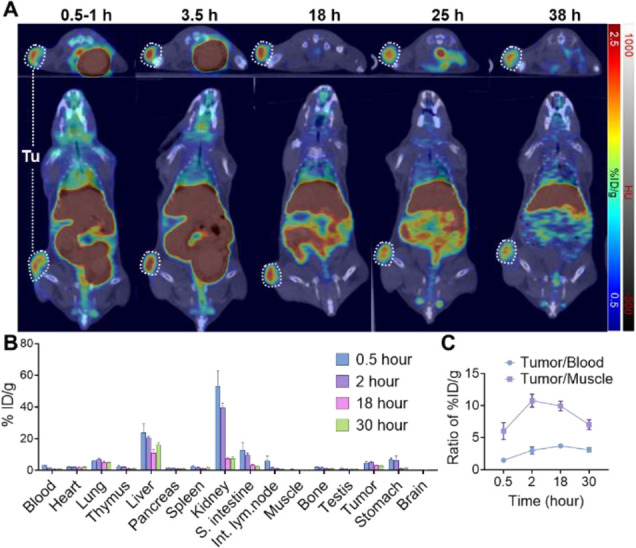
(A) Series of coregistered sectional PET-CT images of C57BL/6J mice at different time points p.i. of [64Cu]CM-2 (17 MBq/mouse, 0.2 mL, ∼0.5 nmol of CM-2). Upper panels: axial view; lower panels: coronal view. White dashed circles indicate B16F10 tumors. (B) Ex vivo biodistribution of [64Cu]CM-2 in major organs at different time points after i.v. injection (3.7 MBq per mouse, 0.1 mL, ∼0.1 nmol of CM-2). S. intestine: small intestine; Int. lym. node: intestine lymph node. Data represent means ± SD, n = 3 for each point. (C) Uptake ratios of tumor to blood (T/B) and tumor to muscle (T/M) calculated from the ex vivo biodistribution data. Data represent means ± SD, n = 3.
[64Cu]CM-2 PET was also performed in Bowes, U87MG, and MDA-MB231 xenografts (Figure S6A). We observed notable tracer uptake at the former two xenografts. However, we only observed background levels of uptake of the tracer in the MDA-MB231 xenograft tumor, which is consistent with the flow cytometry analysis (Figure S1). Furthermore, the uptake values quantified from the PET images correlated well with the expression levels of CD133 in the tumor cells (Figure S6B).
To this end, we have demonstrated that [64Cu]CM-2 is a robust tracer for PET imaging of CD133-specific CSCs. However, this tracer shows high liver uptake, impeding its clinical utility in imaging tumors located at the liver. Moreover, it may cause potential liver toxicities. NODAGA has been reported to be a superior chelator than DOTA to form a more stable complex with 64Cu and thus decrease the liver uptake of the radioactivity.37 We then synthesized a new tracer, [64Cu]CM-3, with a NODAGA as a chelator (Table 1). Unfortunately, [64Cu]CM-3 showed almost identical liver uptake with [64Cu]CM-2, as evidenced by PET scan in normal mice (Figure S7). Moreover, PET imaging in tumor mice suggested that [64Cu]CM-3 exhibited a lower tumor uptake than [64Cu]CM-2 (Figure S8A–F). The in vitro cellular internalization assay further indicated that [64Cu]CM-3 exhibited a weaker binding to Huh-7 and B16F10 cells compared to [64Cu]CM-2 (Figure S8G,H). Collectively, our results suggested that NODAGA is not a suitable chelator for the CM peptide. Further optimization of the linker structure or replacement of DOTA by other types of chelators should be considered to decrease the liver uptake of the radioactivity.
Based on the CSC hypothesis, the CSCs comprise only a small subpopulation of cells in bulky tumors. This makes the development of CSC-specific imaging tools for naturally occurring tumors a formidable challenge. Moreover, CD133 levels are sensitive to the tumor microenvironment. Studies have shown that CD133 is overexpressed in hypoxic tumors,38 which further confers malignant properties to CSCs. Therefore, the development of CD133-targeting imaging probes for CSC detection is much more difficult than the development of tracers targeting biomarkers of normal cancer cells.
Here, we developed a peptide-based PET tracer for CD133 by harnessing stable peptide technology. Through the incorporation of an Ahx into the N terminus of a CM peptide, we constructed a stable peptide tracer [64Cu]CM-2, which exhibited specific binding to CD133-positive CSCs in multiple preclinical tumor models. Compared to previously reported CD133-targeting radiotracers (Table S5), our [64Cu]CM-2 shows apparent advantages. Most of the reported radiotracers are antibody-based immunotracers, which usually show unfavorable pharmacokinetics and poor tissue penetration.39−42 In contrast, [64Cu]CM-2 showed fast accumulation in the tumors and exhibited excellent tissue penetration ability. Recently, a peptide-based PET tracer 68Ga-DOTA-LS7 has been reported for CD133. However, this tracer showed relatively low tumor uptake (the highest uptake is ∼2.24% ID/g in HCT116 tumors).43 In comparison, [64Cu]CM-2 showed significantly higher uptake in tumors (6.89%ID/g in Huh-7 and 4.99% ID/g in B16F10). Moreover, it showed durable tumor retention and fast renal clearance, enabling its high potential for long-term monitoring of CD133 in tumors.
Conclusions
To our knowledge, the [64Cu]CM-2 is the first 64Cu-labeled peptide PET tracer for CD133. The matched physical and biological half-life of [64Cu]CM-2 makes it a state-of-the-art PET tracer for CD133. Therefore, [64Cu]CM-2 PET may not only enable the longitudinal tracking of CD133 dynamics in the cancer stem cell niche but also provide a powerful and noninvasive imaging tool to track down CSCs in refractory cancers.
Materials and Methods
Reagents and Instruments
All chemicals for peptide synthesis and radiolabeling were purchased from Wako Pure Chemical Industries (Osaka, Japan), Macrocyclics (Dallas, Texas, USA), or Sigma-Aldrich (St. Louis, MO, USA). 64Cu was produced in-house at the National Institute of Radiological Science (Chiba, Japan) with 98% radionuclidic purity. 68Ga was obtained from a 68Ge/68Ga generator (ITM, Munich, Germany).
All radio–high-performance liquid chromatography (HPLC) analyses for the radiotracers were performed using a JASCO HPLC system (JASCO, Tokyo, Japan) coupled with a YMC-Triat-C18 column (4.6 mm i.d. × 150 mm, 5 μm, Waters, Milford, MA, USA). A flow rate of 1 mL/min was used. The gradient started with 90% solvent A (0.1% trifluoroacetic acid [TFA] in water) and 10% solvent B (0.1% TFA in acetonitrile [MeCN]), and, 20 min later, ended with 0% solvent A and 100% solvent B. Effluent radioactivity was measured using a NaI (TI) scintillation detector system (Ohyo Koken Kogyo, Tokyo, Japan). A 1480 Wizard autogamma counter (PerkinElmer, Waltham, MA, USA) was used to measure radioactivity as expressed in counts of radioactivity per minute (CPM) accumulating in cells and animal tissues. A dose calibrator (IGC-7 Curiemeter; Aloka, Tokyo, Japan) was used for the other radioactivity measurements. PE anti-human CD133 (Prominin-1) Monoclonal Antibody (EMK08, PE, eBioscience, USA), PE anti-mouse CD133 Antibody (Rat IgG2a, λ, cat. 141204, CA, USA), PE Mouse IgG2b (κ Isotype Ctrl Antibody, cat. 400314, BioLegend, CA, USA), and PE anti-human IgG Fc Antibody (Rat IgG2a, κ, cat. 410708) were used for flow cytometry analysis.
Cell Line and Animals
The murine melanoma B16F10, human glioma U87MG, human melanoma Bowes, and human breast cancer MDA-231 were directly obtained from the ATCC (Virginia, USA). Human hepatocellular carcinoma Huh-7 was obtained from the Cell Resource Center for Biomedical Research at Tohoku University. Cells were cultured according to the manufacturer’s instructions. All cell lines were maintained and passaged in a humidified CO2 incubator (37 °C/5% CO2); B16F10, U87MG, and Huh-7 cells were cultured in Dulbecco’s modified Eagle medium (DMEM) with 10% fetal bovine serum and 1% of penicillin/streptomycin. Bowes cells were cultured in Eagle’s minimum essential medium (EMEM) medium with 10% fetal bovine serum and 1% HEPES. MDA-MB231 cells were cultured in ATCC-formulated Leibovitz’s L-15 medium with 10% fetal bovine serum.
Female BALB/c nude–/– mice (7 weeks old) and C57BL/6J Jms mice (7 weeks old) were purchased from Japan SLC (Shizuoka, Japan). All animals received humane care, and the Animal Ethics Committee of the National Institute of Radiological Sciences approved all experiments. All experiments were conducted according to the recommendations of the Committee for the Care and Use of Laboratory Animals, National Institute of Radiological Sciences, and all animal studies were approved by the Animal Ethics Committee of the National Institute for Quantum and Radiological Science and Technology. A B16F10 tumor-bearing mouse model using C57BL/6J Jms was prepared via a left flank subcutaneous injection of 100 μL of cells (1 × 106 cells/mouse). Mice were used for PET imaging when tumor volumes reached approximately 100. All other tumor-bearing mouse models using BALB/c nude–/– mice were prepared via a left flank subcutaneous injection or hindleg subcutaneous injection of 100 μL of cells (5 × 106 cells/mouse). Mice were used for PET imaging or radionuclide therapy when tumor volume reached approximately 50–100 mm3.
Synthesis of CM-1/2/3 Peptides
Peptides CM-1 and CM-2 were synthesized manually using a Fmoc-based solid-phase peptide synthesis strategy as illustrated in Schemes S1 and S2, respectively. 1,4,7,10-Tetraazacyclododecane-1,4,7,10-tetraacetic acid (DOTA) was used as a chelator for 64Cu labeling. For DOTA/NOTA/NOTAGA conjugation, DOTA/NOTA/NOTAGA was preactivated using N-(3-dimethylaminopropyl)-N-ethylcarbodiimide hydrochloride (EDC·HCl, 2 mg) at a molar ratio for DOTA/EDC·HCl/N-hydroxysuccinimide (NHS) of 10:5:4 in dimethyl sulfoxide (DMSO, 2 mL) for 3 h. Then the DPA-conjugated resin (100 mg; 0.37 mmol/g) was suspended in 1 mL of N-methyl-2-pyrrolidone (NMP). Afterward, the activated DOTA/NOTA/NOTAGA and 100 μL of DIEA were coadded to the resin. The reaction mixture was gently bubbled by N2 gas for 2 h. Finally, the peptides were cleaved from the resin and purified by HPLC. The collected product eluents were lyophilized and redissolved in sodium acetate buffer (100 mM, pH 5.0) at a concentration of 1 mg/mL for use in radiolabeling reactions.
Radionuclide Labeling
Twenty micrograms of peptide in 10 μL of 0.1 M sodium acetate buffer (pH 5.0) was reacted with approximately 370 MBq (10 mCi) of the 64CuCl2 solution (0.1 M NaOAc, pH 5) at room temperature for 10 min. After incubation, the reaction mixture was analyzed by radio-HPLC.
Peptide Stability Assays
For the stability test in saline, 10 μL of radiotracers (∼100 μCi, in NaOAc) was added to 990 μL of saline buffer and incubated at 37 °C with slight agitation for 1, 2, and 4 h. Aliquots of the solution were removed at each time point and then injected into the radio-HPLC for analysis. For the stability in mouse serum, 10 μL of radiotracers (∼100 μCi) were added to 90 μL of mouse serum (freshly prepared) and incubated at 37 °C with slight agitation for 1, 2, and 4 h. Aliquots (20 μL) were removed at each time point, and 100 μL of MeCN and water (1:1, v/v) were added. Then, the mixture was centrifuged for 10 min at a speed of 10,000 rpm. The supernatant was then analyzed using radio-HPLC. For the stability in saline, the radiotracers were incubated in saline (>95%, v), and then aliquots were removed at each time point for subsequent radio-HPLC analysis.
Partition Coefficient Test
The partition coefficients of [64Cu]CM-1 and [64Cu]CM-2 were determined by measuring the distribution of the radioactivity associated with the tracer in equal volumes of 1-octanol and ultrapure water. Briefly, 0.5 MBq of the radiolabeled compound was loaded into an Eppendorf tube containing 500 μL of ultrapure water and 500 μL of 1-octanol (Merck). After vigorous mixing for 2 h at room temperature, samples were centrifuged at 15,000 rpm for 1 min to ensure complete separation of the solvents. Then, 100 μL aliquots of each layer were withdrawn and pipetted into separate test tubes. The samples were then counted in a gamma counter, and the cLogD was calculated as counts in octanol/counts in aqueous solution. Three independent experiments were performed in duplicate.
Cellular Uptake and Inhibition Experiment
The Huh7 cells were plated in 12-well plates with a seeding density of 1 × 105 cells per well and were maintained in DMEM medium supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin. Radiolabeled tracers in medium (740 KBq/1 mL) were added to each well, and the cells were incubated at 37 or 4 °C for 5, 10, 20, 40, 60, and 80 min. After incubation, the medium was removed, and the cells were washed with cold PBS three times. Then, 300 μL of 0.5 M NaOH was added, and the cell lysate was collected in 1 mL tubes. Radioactivity in each tube was measured with an autogamma counter.
Flow Cytometry Analysis of CD133 Expression
To assess the CD133 expression in the five kinds of cancer cell lines, cells were harvested and suspended in phosphate-buffered saline, followed by separate incubation with a matched antibody at 4 °C for 1 h. Then, cells were washed in cold phosphate-buffered saline and analyzed on the BD LSRII flow cytometer (Becton Dickinson, New Jersey, USA). The results were analyzed using FlowJo version 10 (Becton Dickinson, New Jersey, USA).
Ex Vivo Biodistribution
Formulated tracer (1.85 MBq/100 μL) was injected into B16F10-bearing C57BL/6J mice via the tail vein. Three mice were sacrificed by cervical dislocation at 0.5, 2, 18, and 30 h after injection. Major organs, including the total blood, heart, liver, lung, thymus, spleen, pancreas, kidneys, stomach, muscle, small intestine, intestinal lymph node, testis, muscle, bone, brain, blood, and the tumor were quickly harvested and weighed. The radioactivity in these organs was measured using the autogamma counter. Results are expressed as the percentage of injected dose per gram of wet tissue (% ID/g). All radioactivity measurements were decay-corrected.
Small-Animal PET Study
PET scans were conducted using an Inveon PET scanner (Siemens Medical Solutions, Knoxville, TN, USA), which provides 159 transaxial slices with 0.796 mm (center-to-center) spacing, a 10 cm transaxial field of view, and a 12.7 cm axial field of view. All list-mode acquisition data were sorted into three-dimensional (3D) sinograms, which were then Fourier-rebinned into two-dimensional (2D) sinograms (frames × min: 4 × 1, 8 × 2, and 8 × 5). Experimental mice were kept in the prone position under anesthesia with 1–2% (v/v) isoflurane during the scan. The tracers (10–17 MBq/100–200 μL) were injected via a preinstalled tail vein catheter. Immediately after the injection, a dynamic scan in 3D list mode was acquired for 60 min (n = 3). Maximum-intensity projection (MIP) images were obtained for all mice. PET dynamic images were reconstructed by filtered back-projection using Hanning’s filter with a Nyquist cutoff of 0.5 cycle/pixel, which was summed using analysis software (ASIPro VM, Siemens Medical Solutions, Siemens, Munich, Germany). Volumes of interest, including the heart, liver, kidney, and tumor, were placed using the ASIPro software. The radioactivity was decay-corrected for the injection time and expressed as the percent of the total injection dose/per gram tissue (% ID/g).
Statistical Analysis
Data were analyzed using Prism version 8.0 software (GraphPad Software, La Jolla, CA). Comparisons among groups were performed using two-way ANOVA with Bonferroni’s multiple comparisons posttest or unpaired two-tailed Student’s t test. Data are presented as the mean ± SEM or the mean ± SD. The threshold for statistical significance was set as P < 0.05.
Acknowledgments
We would like to thank the staff of the National Institutes for Quantum and Radiological Sciences and Technology (QST) for their support with cyclotron operation, radioisotope production, radiosynthesis, and animal experiments. We sincerely thank the financial support from the JSPS KAKENHI grant nos. 21H02873, 21K07659, and 21H03635; the AMED Moonshot Research and Development Program (grant no. 21zf0127003h001), JSPS International Joint Research Program (JPJSBP120207203) and the Beijing Jishuitan Hospital Elite Young Scholar Programme XKGG2021.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.1c04711.
Peptide synthesis and characterization; HPLC chromatograms for stability analysis; whole-body PET images; immunofluorescence data; ex vivo biodistribution data (PDF)
Author Contributions
⊥ K.H. and X.M. contributed equally to this work.
Author Contributions
K.H. and M.-R.Z conceived the project. K.H., X.M., L.X., Y.Z., M.H., H.O., L.Z., K.N., and H.S. performed the experiment. K.H., R.S., W.W, and M.-R.Z. wrote the manuscript, and all authors discussed the data.
The authors declare no competing financial interest.
Supplementary Material
References
- Clevers H. The cancer stem cell: premises, promises and challenges. Nat. Med. 2011, 17, 313–319. 10.1038/nm.2304. [DOI] [PubMed] [Google Scholar]
- Beck B.; Blanpain C. Unravelling cancer stem cell potential. Nat. Rev. Cancer 2013, 13, 727–738. 10.1038/nrc3597. [DOI] [PubMed] [Google Scholar]
- Frank N. Y.; Schatton T.; Frank M. H. The therapeutic promise of the cancer stem cell concept. J. Clin. Invest. 2010, 120, 41–50. 10.1172/JCI41004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eramo A.; Lotti F.; Sette G.; Pilozzi E.; Biffoni M.; Di Virgilio A.; Conticello C.; Ruco L.; Peschle C.; De Maria R. Identification and expansion of the tumorigenic lung cancer stem cell population. Cell Death Differ. 2008, 15, 504–514. 10.1038/sj.cdd.4402283. [DOI] [PubMed] [Google Scholar]
- Hart L. S.; El-Deiry W. S. Invincible, but not invisible: imaging approaches toward in vivo detection of cancer stem cells. J. Clin. Oncol. 2008, 26, 2901–2910. 10.1200/JCO.2008.16.9573. [DOI] [PubMed] [Google Scholar]
- Han J.; Won M.; Kim J. H.; Jung E.; Min K.; Jangili P.; Kim J. S. Cancer stem cell-targeted bio-imaging and chemotherapeutic perspective. Chem. Soc. Rev. 2020, 49, 7856–7878. 10.1039/D0CS00379D. [DOI] [PubMed] [Google Scholar]
- Rudin M.; Weissleder R. Molecular imaging in drug discovery and development. Nat. Rev. Drug Discov. 2003, 2, 123–131. 10.1038/nrd1007. [DOI] [PubMed] [Google Scholar]
- Hu K.; Shang J.; Xie L.; Hanyu M.; Zhang Y.; Yang Z.; Xu H.; Wang L.; Zhang M.-R. PET Imaging of VEGFR with a Novel 64Cu-Labeled Peptide. ACS Omega 2020, 5, 8508–8514. 10.1021/acsomega.9b03953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu K.; Wu W.; Xie L.; Geng H.; Zhang Y.; Hanyu M.; Zhang L.; Liu Y.; Nagatsu K.; Suzuki H.; Guo J.; Wu Y.; Li Z.; Wang F.; Zhang M. Whole-body PET tracking of a D-dodecapeptide and its radiotheranostic potential for PD-L1 overexpressing tumors. Acta Pharm. Sin. B 2021, 10.1016/j.apsb.2021.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L.; Hu K.; Shao T.; Hou L.; Zhang S.; Ye W.; Josephson L.; Meyer J. H.; Zhang M.-R.; Vasdev N.; Wang J.; Xu H.; Wang L.; Liang S. H. Recent developments on PET radiotracers for TSPO and their applications in neuroimaging. Acta Pharm. Sin. B 2021, 11, 373–393. 10.1016/j.apsb.2020.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuan H.; Masayuki H.; Xie L.; Zhang Y.; Kotaro N.; Hisashi S.; Zhang M. R. Developing native peptide-based radiotracers for PD-L1 PET imaging and improving imaging contrast by pegylation. Chem. Commun. 2019, 55, 4162–4165. 10.1039/C9CC00445A. [DOI] [PubMed] [Google Scholar]
- Xie L.; Hu K.; Duo Y.; Shimokawa T.; Kumata K.; Zhang Y.; Jiang C.; Zhang L.; Nengaki N.; Wakizaka H.; Cao Y.; Zhang M.-R. Off-tumor IDO1 target engagements determine the cancer-immune set point and predict the immunotherapeutic efficacy. J. ImmunoTher. Cancer 2021, 9, e002616 10.1136/jitc-2021-002616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu K.; Xie L.; Zhang Y.; Hanyu M.; Yang Z.; Nagatsu K.; Suzuki H.; Ouyang J.; Ji X.; Wei J.; Xu H.; Farokhzad O. C.; Liang S. H.; Wang L.; Tao W.; Zhang M.-R. Marriage of black phosphorus and Cu2+ as effective photothermal agents for PET-guided combination cancer therapy. Nat. Commun. 2020, 11, 2778. 10.1038/s41467-020-16513-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu K.; Xie L.; Hanyu M.; Zhang Y.; Li L.; Ma X.; Nagatsu K.; Suzuki H.; Wang W.; Zhang M.-R. Harnessing the PD-L1 interface peptide for positron emission tomography imaging of the PD-1 immune checkpoint. RSC Chem. Biol. 2020, 1, 214–224. 10.1039/D0CB00070A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glumac P. M.; LeBeau A. M. The role of CD133 in cancer: a concise review. Clin. Transl. Med. 2018, 7, 18. 10.1186/s40169-018-0198-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S. K.; Clarke I. D.; Terasaki M.; Bonn V. E.; Hawkins C.; Squire J.; Dirks P. B. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003, 63, 5821–5828. [PubMed] [Google Scholar]
- Florek M.; Haase M.; Marzesco A. M.; Freund D.; Ehninger G.; Huttner W. B.; Corbeil D. Prominin-1/CD133, a neural and hematopoietic stem cell marker, is expressed in adult human differentiated cells and certain types of kidney cancer. Cell Tissue Res. 2005, 319, 15–26. 10.1007/s00441-004-1018-z. [DOI] [PubMed] [Google Scholar]
- Liu G.; Yuan X.; Zeng Z.; Tunici P.; Ng H.; Abdulkadir I. R.; Lu L.; Irvin D.; Black K. L.; Yu J. S. Analysis of gene expression and chemoresistance of CD133+ cancer stem cells in glioblastoma. Mol. Cancer 2006, 5, 67. 10.1186/1476-4598-5-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W.; Li H.; Tao K.; Li R.; Song Z.; Zhao Q.; Zhang F.; Dou K. Expression and clinical significance of the stem cell marker CD133 in hepatocellular carcinoma. Int. J. Clin. Pract. 2008, 62, 1212–1218. 10.1111/j.1742-1241.2008.01777.x. [DOI] [PubMed] [Google Scholar]
- Yoshii Y.; Furukawa T.; Kiyono Y.; Watanabe R.; Mori T.; Yoshii H.; Asai T.; Okazawa H.; Welch M. J.; Fujibayashi Y. Internal radiotherapy with copper-64-diacetyl-bis (N4-methylthiosemicarbazone) reduces CD133+ highly tumorigenic cells and metastatic ability of mouse colon carcinoma. Nucl. Med. Biol. 2011, 38, 151–157. 10.1016/j.nucmedbio.2010.08.009. [DOI] [PubMed] [Google Scholar]
- Wang C.; Xie J.; Guo J.; Manning H. C.; Gore J. C.; Guo N. Evaluation of CD44 and CD133 as cancer stem cell markers for colorectal cancer. Oncol. Rep. 2012, 28, 1301–1308. 10.3892/or.2012.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke C. C.; Liu R. S.; Yang A. H.; Liu C. S.; Chi C. W.; Tseng L. M.; Tsai Y. F.; Ho J. H.; Lee C. H.; Lee O. K. CD133-expressing thyroid cancer cells are undifferentiated, radioresistant and survive radioiodide therapy. Eur. J. Nucl. Med. Mol. Imaging 2013, 40, 61–71. 10.1007/s00259-012-2242-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng H.-L.; Liu Y.-Q.; Yang L.-J.; Bian X.-C.; Yang Z.-L.; Gu B.; Zhang H.; Wang C.-J.; Su X.-L.; Zhao X.-M. Expression of CD133 correlates with differentiation of human colon cancer cells. Cancer Biol. Ther. 2010, 9, 216–223. 10.4161/cbt.9.3.10664. [DOI] [PubMed] [Google Scholar]
- Rappa G.; Fodstad O.; Lorico A. The stem cell-associated antigen CD133 (Prominin-1) is a molecular therapeutic target for metastatic melanoma. Stem Cells 2008, 26, 3008–3017. 10.1634/stemcells.2008-0601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vora P.; Venugopal C.; Salim S. K.; Tatari N.; Bakhshinyan D.; Singh M.; Seyfrid M.; Upreti D.; Rentas S.; Wong N.; Williams R.; Qazi M. A.; Chokshi C.; Ding A.; Subapanditha M.; Savage N.; Mahendram S.; Ford E.; Adile A. A.; McKenna D.; McFarlane N.; Huynh V.; Wylie R. G.; Pan J.; Bramson J.; Hope K.; Moffat J.; Singh S. The rational development of CD133-targeting immunotherapies for glioblastoma. Cell Stem Cell 2020, 26, 832–844.e6. 10.1016/j.stem.2020.04.008. [DOI] [PubMed] [Google Scholar]
- Gaedicke S.; Braun F.; Prasad S.; Machein M.; Firat E.; Hettich M.; Gudihal R.; Zhu X.; Klingner K.; Schüler J.; Herold-Mende C. C.; Grosu A.-L.; Behe M.; Weber W.; Mäcke H.; Niedermann G. Noninvasive positron emission tomography and fluorescence imaging of CD133<sup>+</sup> tumor stem cells. Proc. Natl. Acad. Sci. U. S. A. 2014, 111, E692–E701. 10.1073/pnas.1314189111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho J. H.; Kim A. R.; Kim S. H.; Lee S. J.; Chung H.; Yoon M. Y. Development of a novel imaging agent using peptide-coated gold nanoparticles toward brain glioma stem cell marker CD133. Acta Biomater. 2017, 47, 182–192. 10.1016/j.actbio.2016.10.009. [DOI] [PubMed] [Google Scholar]
- Wang W.; Ma Z.; Zhu S.; Wan H.; Yue J.; Ma H.; Ma R.; Yang Q.; Wang Z.; Li Q.; Qian Y.; Yue C.; Wang Y.; Fan L.; Zhong Y.; Zhou Y.; Gao H.; Ruan J.; Hu Z.; Liang Y.; Dai H. Molecular Cancer Imaging in the Second Near-Infrared Window Using a Renal-Excreted NIR-II Fluorophore-Peptide Probe. Adv. Mater. 2018, 30, 1800106. 10.1002/adma.201800106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu K.; Geng H.; Zhang Q.; Liu Q.; Xie M.; Sun C.; Li W.; Lin H.; Jiang F.; Wang T.; Wu Y.-D.; Li Z. An In-tether Chiral Center Modulates the Helicity, Cell Permeability, and Target Binding Affinity of a Peptide. Angew. Chem., Int. Edit. 2016, 55, 8013–8017. 10.1002/anie.201602806. [DOI] [PubMed] [Google Scholar]
- White C. J.; Yudin A. K. Contemporary strategies for peptide macrocyclization. Nat. Chem. 2011, 3, 509. 10.1038/nchem.1062. [DOI] [PubMed] [Google Scholar]
- Reguera L.; Rivera D. G. Multicomponent Reaction Toolbox for Peptide Macrocyclization and Stapling. Chem. Rev. 2019, 119, 9836–9860. 10.1021/acs.chemrev.8b00744. [DOI] [PubMed] [Google Scholar]
- Hu K.; Jiang Y.; Xiong W.; Li H.; Zhang P.-Y.; Yin F.; Zhang Q.; Geng H.; Jiang F.; Li Z.; Wang X.; Li Z. Tuning peptide self-assembly by an in-tether chiral center. Sci. Adv. 2018, 4, eaar5907 10.1126/sciadv.aar5907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu K.; Yin F.; Yu M.; Sun C.; Li J.; Liang Y.; Li W.; Xie M.; Lao Y.; Liang W.; Li Z.-G. In-Tether Chiral Center Induced Helical Peptide Modulators Target p53-MDM2/MDMX and Inhibit Tumor Growth in Stem-Like Cancer Cell. Theranostics 2017, 7, 4566–4576. 10.7150/thno.19840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H.; Yang J.; Gallazzi F.; Miao Y. Effects of the amino acid linkers on the melanoma-targeting and pharmacokinetic properties of 111In-labeled lactam bridge-cyclized alpha-MSH peptides. J. Nucl. Med. 2011, 52, 608–616. 10.2967/jnumed.110.086009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brugnoli F.; Grassilli S.; Piazzi M.; Palomba M.; Nika E.; Bavelloni A.; Capitani S.; Bertagnolo V. In triple negative breast tumor cells, PLC-β2 promotes the conversion of CD133high to CD133low phenotype and reduces the CD133-related invasiveness. Mol. Cancer 2013, 12, 165. 10.1186/1476-4598-12-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu K.; Li W.; Yu M.; Sun C.; Li Z. Investigation of Cellular Uptakes of the In-Tether Chiral-Center-Induced Helical Pentapeptides. Bioconjugate Chem. 2016, 27, 2824–2827. 10.1021/acs.bioconjchem.6b00492. [DOI] [PubMed] [Google Scholar]
- Ghosh S. C.; Pinkston K. L.; Robinson H.; Harvey B. R.; Wilganowski N.; Gore K.; Sevick-Muraca E. M.; Azhdarinia A. Comparison of DOTA and NODAGA as chelators for 64Cu-labeled immunoconjugates. Nucl. Med. Biol. 2015, 42, 177–183. 10.1016/j.nucmedbio.2014.09.009. [DOI] [PubMed] [Google Scholar]
- Yoshii Y.; Furukawa T.; Kiyono Y.; Watanabe R.; Waki A.; Mori T.; Yoshii H.; Oh M.; Asai T.; Okazawa H.; Welch M. J.; Fujibayashi Y. Copper-64-diacetyl-bis (N4-methylthiosemicarbazone) accumulates in rich regions of CD133+ highly tumorigenic cells in mouse colon carcinoma. Nucl. Med. Biol. 2010, 37, 395–404. 10.1016/j.nucmedbio.2009.12.011. [DOI] [PubMed] [Google Scholar]
- Lang J.; Lan X.; Liu Y.; Jin X.; Wu T.; Sun X.; Wen Q.; An R. Targeting cancer stem cells with an 131I-labeled anti-AC133 monoclonal antibody in human colorectal cancer xenografts. Nucl. Med. Biol. 2015, 42, 505–512. 10.1016/j.nucmedbio.2015.01.003. [DOI] [PubMed] [Google Scholar]
- Glumac P. M.; Gallant J. P.; Shapovalova M.; Li Y.; Murugan P.; Gupta S.; Coleman I. M.; Nelson P. S.; Dehm S. M.; LeBeau A. M. Exploitation of CD133 for the Targeted Imaging of Lethal Prostate Cancer. Clin. Cancer Res. 2020, 26, 1054–1064. 10.1158/1078-0432.CCR-19-1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- She X.; Qin S.; Jing B.; Jin X.; Sun X.; Lan X.; An R. Radiotheranostic Targeting Cancer Stem Cells in Human Colorectal Cancer Xenografts. Mol. Imaging Biol. 2020, 22, 1043–1053. 10.1007/s11307-019-01467-7. [DOI] [PubMed] [Google Scholar]
- Wyszatko K.; Valliant J.; Sadeghi S.; Singh S. PET imaging cancer stem cells using a novel zirconium-89 labelled fully human anti-CD133 antibody. J. Nucl. Med. 2021, 62, 154–154. [Google Scholar]
- Liu Y.; Yao X.; Wang C.; Wang M.; Wang Y.; Ye M.; Liu Y. Peptide-based 68Ga-PET radiotracer for imaging CD133 expression in colorectal cancer. Nucl. Med. Commun. 2021, 1144. 10.1097/MNM.0000000000001435. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


