Abstract
Watercress (Nasturtium officinale R. Br.) is an important aquatic herb species belonging to the Brassicaceae family. It has various medicinal properties and has been utilized for the treatment of cancer and other diseases; however, currently available genomic information regarding this species is limited. Here, we performed the first comprehensive analysis of the carotenoid biosynthesis pathway (CBP) genes of N. officinale, which were identified from next-generation sequencing data. We identified and characterized 11 putative carotenoid pathway genes; among these, nine full and two partial open reading frames were determined. These genes were closely related to CBP genes of the other higher plants in the phylogenetic tree. Three-dimensional structure analysis and multiple alignments revealed several distinct conserved motifs, including aspartate or glutamate residues, carotene-binding motifs, and dinucleotide-binding motifs. Quantitative reverse transcription-polymerase chain reaction results showed that the CBP was expressed in a tissue-specific manner: expression levels of NoPSY, NoPDS, NoZDS-p, NoCrtISO, NoLCYE, NoCHXE-p, and NoCCD were highest in the flower, whereas NoLCYB, NoCHXB, NoZEP, and NoNCED were highest in the leaves. Stems, roots, and seeds did not show a significant change in the expression compared to the leaves and flowers. High-performance liquid chromatography analysis of the same organs showed the presence of seven distinct carotenoid compounds. The total carotenoid content was highest in the leaves followed by flowers, seeds, stems, and roots. Among the seven individual carotenoids, the levels of six carotenoids (i.e., 13-Z-β-carotene, 9-Z-β-carotene, E-β-carotene, lutein, violaxanthin, and β-cryptoxanthin) were highest in the leaves. The highest content was observed for lutein, followed by E-β-carotene, and 9-Z-β-carotene; these carotenoids were much higher in the leaves compared to the other organs. The results will be useful references for further molecular genetics and functional studies involving this species and other closely related species.
1. Introduction
Watercress (Nasturtium officinale R. Br.) belongs to the Brassicaceae family, and it is cultivated worldwide in regions of Australia, Europe, India, North America, southern Africa, and sub-Saharan Africa, as a perennial herb or as an edible aquatic plant because of its nutraceutical properties.1 This raw leafy vegetable can be eaten in the form of salads, or it can be cooked and consumed in the same manner as other vegetables and used as ingredients in soups as well as a garnish in other dishes.2N. officinale is rich in vitamins A, B, C, and E; in addition, it is also rich in carotenoids, flavonoids, folic acid, glucosinolates, phenolics, protein, and minerals such as iodine, iron, calcium, and sulfur compounds.3,4 Recently, several studies have reported its antidiabetic, anticancer, anti-inflammatory, and antioxidant properties.4,5
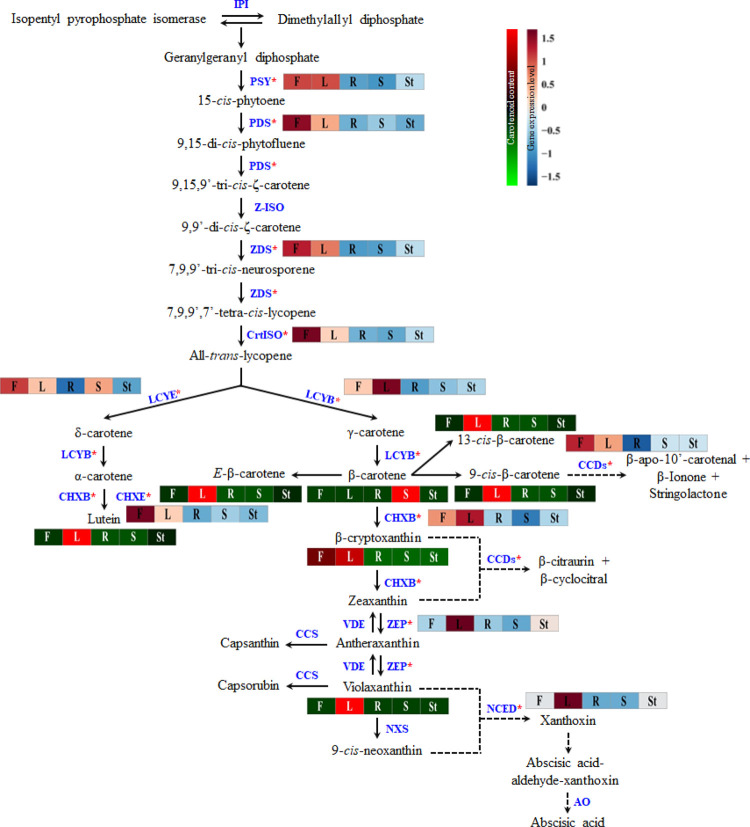
Among these components, carotenoids are a natural pigment that plays an important role in the photosynthetic organelles of algae, mosses, ferns, and other higher plants.6 In addition, they are also found in the membranes of photosynthetic bacteria such as phototropic bacteria and cyanobacteria.7 Carotenoids have received great attention because of their various health benefits and their ability to protect the skin against damaging free radicals, age-related macular degeneration, cancer, and cardiovascular diseases, as well as boosting immune system function.8,9 Recently, several reviews have summarized genes involved in the transcriptional regulation of the carotenoid biosynthesis pathway (CBP) in plants.7,10 The regulation of the CBP genes at the transcriptional level is critically important for the syntheses of photosynthetic pigments and plant hormones. The primary steps in the CBP are the conversion of two geranylgeranyl pyrophosphate (GGPP) molecules by the enzyme phytoene synthase (PSY) to generate phytoene by the condensation process. This step is the most important rate-limiting step in the CBP.8 Following these steps, the enzymes of phytoene desaturase (PDS), ζ-carotene isomerase (Z-ISO), ζ-carotene desaturase (ZDS), and carotenoid isomerase (CrtISO) carry out consecutive desaturations and result in the production of all-trans-lycopene. Furthermore, lycopene ε-cyclase and lycopene β-cyclase carry out cyclization, which results in the production of α-carotene and β-carotene; this step is a critical branch-point in the CBP.8 Both α-carotene and β-carotene undergo sequential hydroxylation by β-ring carotene hydroxylase (CHXB) and ε-ring carotene hydroxylase (CHXE), which results in the synthesis of lutein and zeaxanthin, respectively. β-cryptoxanthin was produced as an intermediate product during the hydroxylation of zeaxanthin. After this step, zeaxanthin is converted to antheraxanthin and violaxanthin by two successive epoxidations with the same enzyme zeaxanthin epoxidase (ZEP), and violaxanthin can be converted back to antheraxanthin and zeaxanthin by deepoxidations with the enzyme violaxanthin de-epoxidase. The last step in the CBP is the conversion of violaxanthin into 9-cis-neoxanthin via the enzyme neoxanthin synthase (NXS).7 In one branch, conversion of antheraxanthin to capsanthin occurred, and in the next step, the conversion of violaxanthin to capsorubin was transformed with the help of enzyme capsanthin-capsorubin synthase.7 In the other branch of the pathway, β-carotene, β-cryptoxanthin zeaxanthin, violaxanthin, and 9-cis-neoxanthin are catabolized into volatile and nonvolatile apocarotenoids by substrate cleavage enzymes, such as carotenoid cleavage dioxygenases (CCDs) and 9-cis-epoxycarotenoid dioxygenase (NCED) (Figure 1).
Figure 1.
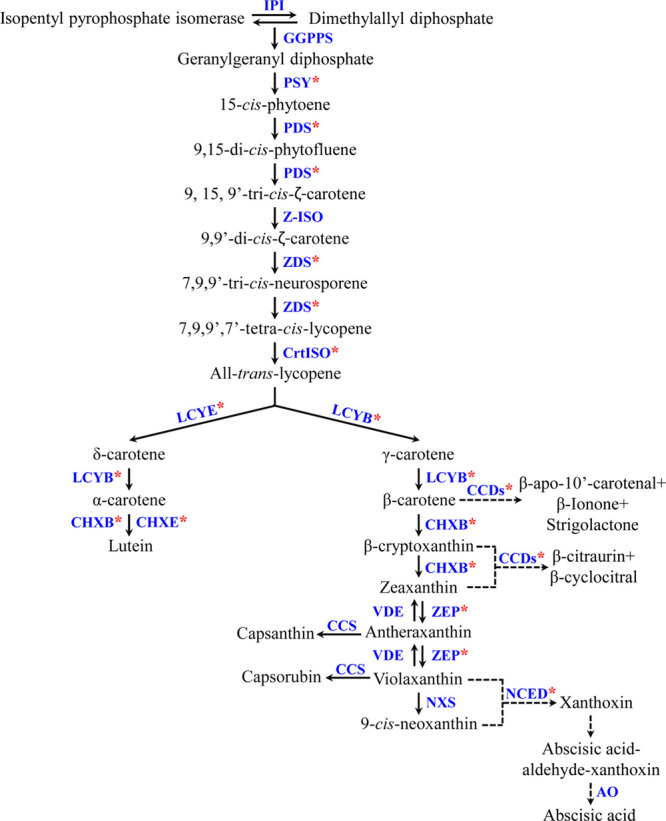
Schematic view of the CBP in Nasturtium officinale. Enzymes are shown in blue, and the pink asterisk represents the gene identified and characterized in this study. Solid black arrows denote biosynthesis, and dotted black arrows denote degradation of carotenoids. IPI-Isopentenyl pyrophosphate isomerase. The pathway scheme is adapted and modified from the study by Sathasivam et al.7
These CBP genes have been identified and characterized in several plants such as Arabidopsis, Chinese cabbage, citrus, Ixeris dentate, papaya, Scutellaria baicalensis, strawberry, and wolfberry.11−19 To date, there have only been a few studies regarding the molecular biology of N. officinale,1,20,21 and there are no published reports regarding the characterization and gene expression of CBP genes in N. officinale. In our laboratory, a cDNA library was constructed from the N. officinale seedling, and the transcript sequences were assembled using the trinity package (http://trinityrnaseq.github.io) and evaluated by Transrate S/W (http://trinityrnaseq.github.io). The raw read transcriptome sequences were submitted to the National Center for Biotechnology Information (NCBI), sequence read archive (SRA) database under the accession number SRR3490957.20 This study aimed to use these transcriptomic data to identify CBP genes in N. officinale.
This is the first report to identify and characterize the CBP genes (NoPSY, NoPDS, NoZDS-p, NoCrtISO, NoLCYB, NoLCYE, NoCHXB, NoCHXE-p, NoZEP, NoCCD, and NoNCED) in N. officinale. To validate the spatial distribution of the transcripts of CBP genes, we examined gene expression in various organs of N. officinale using quantitative reverse transcription-polymerase chain reaction (qRT-PCR). In addition, we analyzed the distribution of seven carotenoid compounds in the various organs of N. officinale using high-performance liquid chromatography (HPLC). The present study would improve our understanding of the differential carotenoid accumulation and biosynthetic pathway genes in various organs of N. officinale that are beneficial for human health. Our results can then increase our understanding of CBP genes and allow us to explore strategies that could improve the anticarcinogenic properties of N. officinale.
2. Results and Discussion
2.1. In Silico Identification, Protein Nomenclature, and Sequence Analysis of CBP Genes
CBP genes were identified from the transcriptomic data of N. officinale. Conserved regions of the sequences from previously identified and classified CBP genes from higher plants were used as queries to search against N. officinale transcript databases using the NCBI BLASTN (Basic Local Alignment Search Tool Nucleotide) program. The specific genes were identified and were subjected to NCBI’s open reading frame (ORF) finder program to identify whether the gene possesses the full ORF with a maximum nucleotide length. The full ORF with the highest number of nucleotides was then retrieved and subjected to structural and functional characterization by searching for conserved regions; moreover, the CBP gene that did not possess the full ORF was also taken for the characterization studies. In total, nine full ORF (NoPSY, NoPDS, NoCrtISO, NoLCYB, NoLCYE, NoCHXB, NoZEP, NoCCD, and NoNCED) CBP genes were determined, whereas NoZDS-p and NoCHXE-p showed partial ORFs in the cDNA library used in this study. All were submitted to GenBank (Figure S1 and Table 1). The GGPPS and Z-ISO genes were identified from the transcriptomic data; however, they possess a short ORF so these two genes were not taken for further analysis. The expression levels of these genes in the transcriptomic data are shown in Table S1. The predicted molecular weights and their estimated isoelectric points are shown in Table 1. The predicted molecular weights of some N. officinale CBP proteins are in accordance with those previously reported from other higher plant species such as Brassica napus,22Chelidonium majus,23Lycium chinenses,14S. baicalensis,18,24 and I. dentate.15 Signal IP analyses showed that the maximum values of the original shearing site (C score) were seen in NoCCD followed by NoZDS-p, NoCHXB, NoPDS, NoLCYE, NoPSY, NoNCED, NoZEP, NoCrtISO, NoLCYB, and then NoCHXE-p (P450 type), whereas maximum values of the synthesized shearing site (Y score) were highest in NoCCD followed by NoZDS-p, NoPSY, NoLCYE, NoCHXB, NoNCED, NoPDS, NoCrtISO, NoLCYB, NoZEP, and NoCHXE-p. The maximum values of the signal peptide (S score) were found in NoZDS-p followed by NoPSY, NoLCYE, NoCCDs, NoNCED, NoCHXB, NoCrtISO, NoPDS, NoLCYB, NoZEP, and NoCHXE-p (Table S2). No transmembrane region was detected in identified CBP genes of N. officinale. A similar result indicating that the CBP genes do not possess any transmembrane region was found in the green algae Tetraselmis suecica(25) and higher plants C. majus.23 Similarly, some CBP genes in the higher plants such as Banana (MaPSY1 and MaPSY3), wheat (TaPSY3), and Brassica napus (BnCCD) also do not possess any transmembrane region.26−28 Homology analysis using CDD showed that the CBP amino acid sequences had high similarity with other higher plant species, including the sequences of amino acids 66–407 for NoPSY, 4–564 for NoPDS, 1–137 for NoCrtISO, 56–501 for NoLCYB, 1–526 for NoLCYE, 1–303 for NoCHXB, 1–249 for NoZEP, 1–170 for NoCCD, 5–527 for NoNCED, 34–225 for NoCHXE-p, and 11–318 for NoZDS-p. In green algae, Dunaliella salina homology analysis of CBP genes by CCD showed high homology with other microalgae and higher plants.27,29,30 From these results, it can be inferred that N. officinale CBP genes are highly conserved when compared to genes of other higher plants and green algae.
Table 1. Molecular Characterization of CBP Genes in Nasturtium officinalea.
| gene names | NCBI accession no. | ORF (bp) | length (aa) | ORF type | MW (kDa) | pI |
|---|---|---|---|---|---|---|
| NoPSY | MT547989 | 1230 | 409 | full | 46 | 9.16 |
| NoPDS | MT547988 | 1695 | 564 | full | 62.8 | 7.10 |
| NoZDS-p | MT547991 | 955 | 318 | partial | ||
| NoCrtISO | MT547983 | 465 | 154 | full | 17.9 | 6.90 |
| NoLCYB | MT547985 | 1506 | 501 | full | 56.3 | 6.81 |
| NoLCYE | MT547986 | 1581 | 526 | full | 58.4 | 5.62 |
| NoCHXB | MT547981 | 921 | 306 | full | 34 | 8.45 |
| NoCHXE-p | MT547982 | 678 | 226 | partial | ||
| NoZEP | MT547990 | 750 | 249 | full | 26.8 | 6.63 |
| NoCCD | MT547984 | 519 | 172 | full | 19.6 | 4.89 |
| NoNCED | MT547987 | 1587 | 528 | full | 58 | 5.86 |
ORF, open reading frame; bp, base pair; aa, amino acid; MW, molecular weight; pI, isoelectric point; p, partial (not complete ORFs).
2.2. Phylogenetic and Homology Analysis
To investigate molecular evolutionary relationships between N. officinale CBP proteins and other higher CBP sequences, a neighbor-joining phylogenetic tree was constructed using all 11 CBP sequences and a set consisting of each specific CBP sequence. Phylogenetic analysis of each sequence showed that the N. officinale CBP proteins formed a cluster with other higher plants, whereas bacteria, chlorophyte, dinoflagellates, and heterokonts formed separate clades (Figure S2). Similar results were obtained from several studies; the phylogenetic analysis of plant CBP sequences with other species showed that they formed a separate cluster with higher plants.24,31,32 In addition, we constructed a phylogenetic tree with 11 CBP protein sequences shared among the eight species that show a high bootstrap value which indicated that the CBP genes are highly conserved. Hence, these identified 11 CBP proteins in N. officinale have a similar putative conserved function to that of the other higher plants, especially Arabidopsis thaliana (Figure 2). This supports the previous study result that the N. officinale transcriptomic data showed the highest similarity and annotation ratio to A. thaliana.20 Pairwise identity matrix of all the CBP sequences shared sequence identities with the A. thaliana amino acid sequence (Figure 2 and Table S3). In addition, bacteria, chlorophyta, dinoflagellates, and heterokonts showed less sequence identity when compared to N. officinale CBP amino acid sequences (Table S3). Previous studies reported that the CBP amino acid sequences of C. majus, S. baicalensis, and I. dentata showed high similarity with those in higher plant species.15,16,23,24 These results clearly showed that CBP genes may share higher sequence identities with higher plants, indicating that in higher plants, these identified CBP proteins are highly conserved in sequences and have a similar function to that of the other higher plants especially A. thaliana.
Figure 2.
Phylogenetic analysis based on the concatenated amino acid sequences of 11 CBP genes. The NJ phylogenetic tree was drawn with the Poisson-correction distance. The number at each node denotes the percentage in the bootstrap analysis (1000 replicates), whereas the numbers below the branch points represent bootstrap values. The outgroup is Eutrema salsugineum.
2.3. Multiple Alignments, Tertiary Structure, and Protein Localization Analysis
Multiple alignments and predicted three-dimensional (3D) structures of N. officinale CBP proteins showed highly conserved domains compared to the other higher plants15,19,33 and microalgae34,35 (Figures 3, 4, and S3). It is well known that protein function mainly depends on its 3D structure and its stability.36 The results showed that the conformations of α and β secondary structural elements and substrate-binding pockets were similar to those of A. thaliana, C. reinhardtii, and D. salina (data not shown). However, we found slight structural differences in the CBP proteins in the variable loop regions of the models; this might be due to their sequence identities being relatively low.37 This supports the multiple alignment and percent identity results of this study (Figure S3 and Table S3).
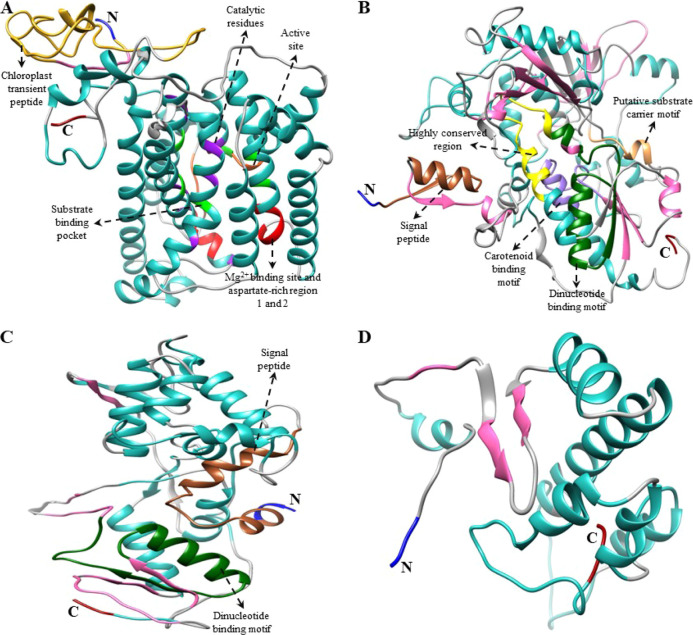
Figure 3.
Predicted 3D structure of upstream CBP genes of Nasturtium officinale. (A) NoPSY, (B) NoPDS, (C) NoZDS-p (partial ORF), and (D) NoCrtISO structures were generated using Chimera 1.14 software.63 The amino (NH2) and carboxyl (COOH) terminals are presented in blue and dark red, respectively. In these 3D structures, α-helices and β-strands are shown in light sea green and hot pink, respectively. For the sequence alignment of each gene, see Φιγυρε Σ3.
Figure 4.
Predicted 3D structure of downstream CBP genes of Nasturtium officinale. (A) NoLCYB, (B) NoLCYE,(C) NoCHXB-p, (D) NoZEP, (E) NoCCD, and (F) NoNCED structures were generated using Chimera 1.14 software.63 The amino (NH2) and carboxyl (COOH) terminals are shown in blue and dark red, respectively. In these 3D structures, α-helices and β-strands are shown in light sea green and hot pink, respectively. For sequence alignment of each gene, see Φιγυρε Σ3.
The predicted 3D structure of N. officinale CBP genes possesses a central hydrophobic substrate-binding pocket which was folded by α-helices and β-sheet strands; notably, the binding pocket was almost covered within the core of the α-helices. In addition, other domains including the aspartate-rich domain (ARD), carotene-binding domain (CBD), and dinucleotide-binding domain (DBD) signature motifs were found near the cavity, which may be required for enzyme activity.29 In detail, the key upstream pathway enzyme NoPSY possesses a conserved trans-isoprenyl diphosphate synthase domain and ARD in its structure. This result agreed with previous studies that showed the presence of these conserved domains in higher plants such as I. dentate and S. baicalensis.15,18 The second important gene in the CBP is NoPDS, which possesses both the CBD and DBD in its structure, whereas NoZDS-p shares similar identical features to NoPDS, which consists of a CBD in the C-terminal region and a DBD in the N-terminal region. This result was consistent with a previous study which showed that higher plants (Carica papaya, C. majus, I. dentate, and S. baicalensis) and marine green algae (D. salina) had PDS and ZDS consisting of these domains in their structures.15,18,23,35
Moving on to the downstream pathway genes, both NoLCYB and NoLCYE contain a DBD that is found in all lycopene cyclases and helps to bind flavin adenine dinucleotide (FAD). Moreover, a plant β-conserved region was also found in plant-type cyclases (CrtL) but not in bacterial CrtYm, and this may play a crucial role in the specific interaction between the cyclase and components of the membrane-associated enzymes.38 In addition, three well-conserved regions, namely, cyclase motif 1, 2, and charged regions, were also found, and these are potentially involved in substrate binding and catalysis.11 Similar LCYB and LCYE conserved domains were also found in some higher plant species (Arabidopsis, Capsicum annuum, and C. majus) and green algae (Haematococcus pluvialis).11,23,38,39 The common gene responsible for both the upstream and downstream branches of the CBP is NoCHXB, which consists of four histidine domains that may be involved in Fe2+ adhesion while hydroxylation takes place.15 Similarly, NoCCD and NoNCED consist of four highly conserved histidine residues (Figures 3, 4, and S3); this result was similar to the structural result obtained from Citrus CCD4a, CCD4b1, and CCD4c.40 Previous studies reported that these four histidine residues are involved in coordinating the Fe2+ cofactor required for activity and the aspartate or glutamate moieties that fix the positions of the histidines.41,42 From the multiple alignments and 3D structure analysis results, it can be inferred that most of the N. officinale CBP genes are highly conserved and that the genes are generally closely related to higher plants and algae. However, further detailed studies are required to understand the functions of the N. officinale CBP proteins identified in this study.
CBP sequences of N. officinale were analyzed using CELLO, ChloroP 1.1, TargetP 1.1, and WoLF PSORT web-based programs to predict the subcellular location of these proteins. Most of the N. officinale CBP proteins were, through consensus, predicted to be targeted to the chloroplast, whereas some of the CBP proteins were targeted to the cytoplasm or to the mitochondrion (Table 2). In A. thaliana, transgenic sweet potato, and in some other plants, most of the CBP genes were localized within the chloroplast, which show results similar to the results of this study.23,30,43 From these, we found that all the N. officinale CBP proteins share a highly conserved region with higher plants, as such subcellular location prediction also showed similar results to those obtained from the higher plants.
Table 2. Subcellular-Localization Predictions of N. officinale CBP Genesa.
| gene names | CELLO | chlorop 1.1 | Targetp | wolf psort | consensus prediction |
|---|---|---|---|---|---|
| NoPSY | CP | CP | CP | CP | CP |
| NoPDS | CP | CP | CP | CP | CP |
| NoZDS-p | CP | CP | CP | CP | CP |
| NoCrtISO | CYT/MT | other | other | CP | CP/CYT/MT/other |
| NoLCYB | CP | CP | other | CYT | CP/CYT/other |
| NoLCYE | CP/PM | CP | CP | CP | CP/PM |
| NoCHXB | CP/PM | CP | CP | CP | CP/PM |
| NoCHXE-p | CYT | CP | other | CP | CP/CYT/other |
| NoZEP | CP | CP | CP | CP | CP |
| NoCCD | CYT | other | other | CYT | CYT/other |
| NoNCED | CP/CYT | other | other | PER | CP/CYT/PER/other |
CP, chloroplast; CYT, cytoplasmic; MT, mitochondrion; PER, peroxisome; PM, plasma membrane.
2.4. Comparison of N. officinale CBP Gene Sequences with Other Plant CBP Sequences and Its Impact on Carotenoid Accumulation
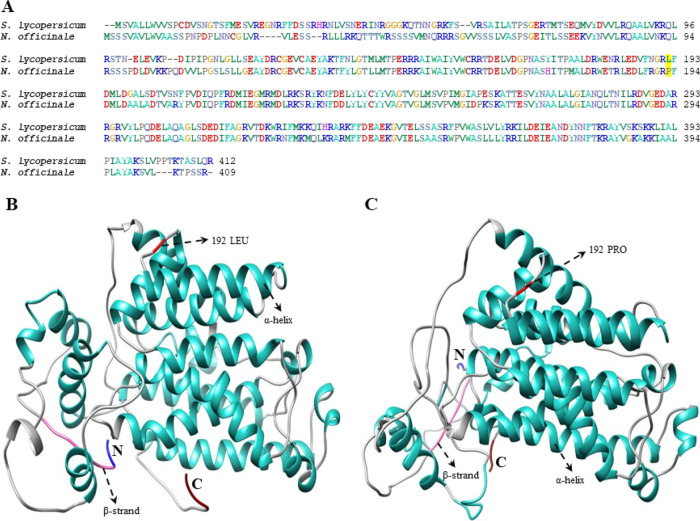
In several crop species, PSY is one of the most important and rate-limiting enzymes in the CBP. NoPSY genes consist of five distinctive PSY motifs (Figure S3), and they possess a putative PSY active site (Figure 3). When compared to A. thaliana the highest level of sequence difference was found at the chloroplast transit peptide (TP) in the N-terminal region (Figure S3). This result was similar to the previous study result observed in Brassica and other PSY sequences.22 Because of high-sequence variability at the N-termini, TPs are well recognized and bound by protein import complexes (translocons) in the Toc (translocon at the outer envelope membrane of chloroplasts) and Tic (translocon at the inner envelope membrane of chloroplasts),22 which efficiently targets the nuclear-encoded proteins to plastids.44 Various types of translocons are drawn together in plastids of diverse tissue types (e.g., photosynthetic vs nonphotosynthetic) and developmental stages.45 However, no functional prediction of TP was performed still, but, it is now possible merely based on TP sequence data. In addition, a previous study reported that in tomatoes, a mutation in the 192 position of the PSY1 gene (P192L) causes a change in the proline to leucine substitution which leads to delayed accumulation of carotenoids in the fruit; this might be due to a decrease of PSY enzymatic activity. In addition, they found that in the P192L mutant the accumulation of phytoene, lycopene, and β-carotene was much lower when compared to the nonmutated line.46 Comparison of NoPSY with other PSY genes showed that there was no mutation that occurs at the 192 position of the NoPSY genes (Figure 5), which leads to accumulation of carotenoids in N. officinale (Figure 6). The second most important enzyme in the CBP is PDS. NoPDS contained GXGX2GX3AX2LX3GX6EX5GG, a secondary structure consisting of a β sheet-α helix-β sheet configuration which is called DBD fold, which shares high similarity with A. thaliana (Figures S2 and S3). However, the structure-functional association of PDS remains unclear, and in the future, it needs to be characterized.47LCYB and LCYE are the two major types of cyclase in plants. A catalytically active domain (involved in the FAD-binding site, along with the substrate-binding site) and a transmembrane domain were discovered through the structure analysis of NoLCYB. In addition, the NoLCYB protein also possesses the FAD domain at 77–467 amino acid residues (Figure S3). Previous studies reported that lycopene cyclase uses FAD as a cofactor.48 Moise et al.49 reported that most of the enzymes involved in the transformation of lycopene and carotene are membrane-bound. Similarly, in NoLCYB the binding site is close to the membrane helix and the FAD-binding domain is present near the enzyme (data not shown). Functional analysis of LCY-B2 in the yellow-fleshed Kapoho variety reaveled an A/C sequence polymorphism at 607 positions that could result in the amino acid change and lead to a decrease in the enzymatic function in red-fleshed papaya.12 However, the sequence analysis of NoLCYB showed the presence of nucleotide ‘A’ at the 607 positions (Figure S4), indicating that NoLCYB gene encodes a fully functional enzyme in N. officinale, leading to the highest accumulation of carotenoid content (Figure 6). The CCD belongs to a family of oxygenases, which particularly cleaves the carotenoids into apocarotenoids (ABA and strigolactones). The NoCCDs contain hydrophobic patches in their structures (Figure S3), which might allow them to interact with the nonpolar lipids of plastoglobule, where the CCD is localized.50 In addition, several amino acids were identified in the hydrophobic patches, and these might represent interacting structural elements.33 The comparison of N. officinale CBP gene sequences with other CBP gene sequences showed high sequence similarity with higher plants. However, the structure-functional relationship in most of the CBP genes remains unclear. Subsequent functional analysis will improve our understanding of particular gene functions and interactions.
Figure 5.
Comparison of tomato and watercress PSY nucleotide sequences. The tomato PSY sequence was retrieved from the previous manuscript published by Gady et al.46 (A) Multiple alignments of the SlPSY and NoPSY protein sequence were performed with the BioEdit program. The yellow highlighted represents the change in the amino acid sequence. Predicted 3D structures, (B) SlPSY, and (C) NoPSY. The amino (NH2) and carboxyl (COOH) termini are presented in blue and dark red, respectively. In these structures, α-helices and β-strands are shown in light sea green and hot pink, respectively. The changes in the amino acid sequence in the structural region are indicated.
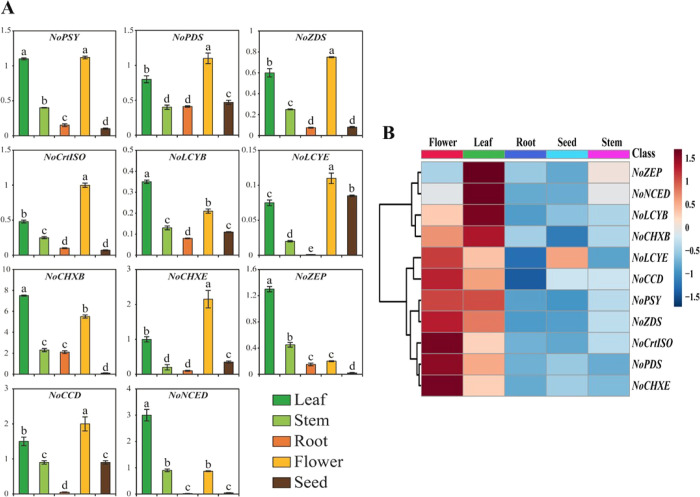
Figure 6.
Relative gene expression profiles of 11 CBP genes of Nasturtium officinale. (A) Transcriptional levels of CBP genes were analyzed in different tissues such as leaf, stem, root, flower, and seed using qRT-PCR analysis. The relative gene expression was calculated using ubiquitin-conjugating enzyme 9 (UBC9). Results are given as the means of triplicates ± SD. Letters a–e denote significant differences (p < 0.05). (B) Heat map showing the expression profiles of CBP genes in five different tissues namely leaf, stem, root, flower, and seed. The heat map was generated using fold change values obtained from qRT-PCR. The tree view of hierarchical clustering was used to show the organ-specific expression of CBP genes. A gradient color bar at the top is used to illustrate whether the CBP genes are upregulated (red) or downregulated (green).
2.5. Expression Levels of CBP Genes in Different Organs of N. officinale
qRT-PCR analysis was used to determine the expression patterns of CBP genes in the different plant organs such as leaves, stems, roots, flowers, and seeds of N. officinale (Figure 6). The result showed that the CBP genes were constitutively expressed in N. officinale. Among the identified CBP genes, NoCHXB showed the highest expression levels. The key enzyme NoPSY was highly expressed in leaves and flowers, whereas the lowest levels of expression were observed in the stems, roots, and seeds. The upstream pathway genes, NoPDS and NoZDS-p, exhibited similar expression patterns to that of NoPSY, which is strongly high in the leaves and flowers, whereas it was relatively low in the other organs. Among the downstream CBP genes, NoCrtISO, NoLCYE, NoCHXE-p, and NoCCD were highly expressed in the flowers, whereas lower levels of expression were observed in the leaves, stems, roots, and seeds. Expression levels of NoLCYB, NoCHXB, NoZEP, and NoNCED were highest in the leaves, whereas the lowest expression levels were observed in stems, roots, flowers, and seeds. This result was consistent with a previous study that showed that in Brassica rapa, most of the CBP genes are highly expressed in the flower and leaves.51 From the gene expression analysis results, most of the N. officinale CBP genes played a similar role to their orthologs in other species. For instance, in A. thaliana, it was reported that AtPSY, AtPDS, AtZDS, and AtZEP genes play important roles in the CBP.52 In these studies, most of the CBP genes were significantly expressed in flowers (NoPSY, NoPDS, NoZDS-p, NoCrtISO, NoLCYE, NoCHXE-p, and NoCCD) and leaves (NoLCYB, NoCHXB, NoZEP, and NoNCED). Expression profiles of CBP genes accrued in this study will help contribute to future genetic research in N. officinale and enhance the carotenoid content through metabolic engineering.
2.6. Analysis of Carotenoid Content in Different Organs of N. officinale
Carotenoids were analyzed and identified through coelution with 20 authentic standards. Among these, only seven different carotenoids were detected from the different N. officinale organs (Figure 7). The total carotenoid content in the different organs of N. officinale varied significantly, ranging from 3.44 to 2383.27 μg/g of dry weight. The highest total carotenoid level was noted in the leaves followed by stems, flowers, seeds, and roots. Total carotenoid contents in N. officinale leaves were found to be 7.57, 10.83, 360.01, and 692.81 times higher than those found in roots, stems, seeds, and flowers, respectively. Among the seven carotenoids, levels of six carotenoids, namely, 13-Z-β-carotene, 9-Z-β-carotene, E-β-carotene, lutein, violaxanthin, and β-cryptoxanthin, were highest in the leaves, whereas in leaves the α-carotene was not detected. Notably, 9-Z-β-carotene, E-β-carotene, and lutein levels were much higher compared to the other individual carotenoids.
Figure 7.

Carotenoid content in the different tissues of Nasturtium officinale. For HPLC analysis, samples were harvested from 2-month-old plants. Results are given as the means of triplicates ± SD. Letters a–e denote significant differences (p < 0.05).
Considering all the watercress organs, lutein levels in the leaves were 425.95, 252.97, 12.13, and 6.87 times higher than those in the root, seed, flower, and stem, respectively, while E-β-carotene content was 1578.74, 74.70, 11.0, and 8.14 times higher than that in the root, seed, flower, and stem, respectively. Likewise, 9-Z-β-carotene contents in the leaves were 1272.2, 454.36, 9.31, and 7.87 times higher than those in the root, seed, flower, and stem, respectively. In the root, only 9-Z-β-carotene, E-β-carotene, and lutein were detected. 13-Z-β-carotene levels were mainly accumulated in the leaf, with its content reaching 8.88 and 7.69 times higher than the levels in the stem and flower, respectively, whereas it was not detected in root and seed. Interestingly, the violaxanthin and α-carotene were detected only in leaves and seed, respectively. The levels of β-cryptoxanthin were slightly higher in the leaves compared to the flower, whereas it was not detected in stem, roots, and seed. In terms of overall individual carotenoid content, α-carotene levels showed the lowest accumulation when compared to other carotenoids detected in this study (Figure 7). This finding is consistent with previous studies conducted in Allium sativum,53B. rapa,17C. majus,23 and M. charantia,54 wherein it was found that the contents of carotenoids were significantly high in the leaves compared to the other plant organs.
2.7. Relationship between Carotenoid Content and Gene Expression
Except for α-carotene, most of the other individual carotenoids were significantly accumulated in leaves (Figures 7 and 8); however, the enhanced transcription of few CBP genes (NoPSY, NoPDS, NoZDS-p, NoCrtISO, NoLCYE, NoCHXE-p, and NoCCD) was observed in N. officinale flowers (Figures 6 and 8). This showed that most of the CBP gene expression and carotenoid accumulation patterns were not correlated and that the highest gene expression does not always lead to a significant accumulation of carotenoids.24,29 From another point of view, it can be explained that the CBP is regulated at multiple levels, not only at the transcriptional level but also at the translational level.55 In addition, the CBP gene expression and carotenoid content are controlled by the combination of cis-regulatory elements in the upstream promoter region and untranslated regions.56 Moreover, protein modifications may be one of the main reasons behind the mismatched accumulation patterns of carotenoid accumulation and CBP gene expression.57 Previously, several studies reported that soluble carbohydrates play a crucial role in carotenoid metabolism.58 There was strong coordination between the photosynthetic machinery and carotenoids; this might be due to the hypothesis that carotenoid pathway gene expression was repressed in leaves in response to high glucose levels.58 To support this hypothesis, Mortain-Bertrand et al.59 conducted a study in tomatoes and found that the genes related to the carotenoid and MEP pathway were repressed because of high glucose levels. In contrast, few studies have reported that accumulation of sugar in leaves inhibits photoinhibition which will trigger carotenoid accumulation in leaves.60,61 This contradictory result was obtained in leaves of N. officinale, which might be due to the high soluble sugar level present in leaves. These are all the possible reasons for the mismatch accumulation of carotenoid content and gene expression in N. officinale.
Figure 8.
Overview of carotenoid pathway gene expression and carotenoid accumulation changes in different plant organs of N. officinale. Each colored box (left to right) under each gene and compound represents F-Flower; L-Leaf; R-Root; S-Seed; St-Stem. The scale bar indicates the transformed average value of gene expression level and metabolites, and the colored square boxes (gene expression level (green to red) and carotenoid content (dark blue to dark red)) represent the relative gene expression level and metabolite abundance in different plant organs.
3. Conclusions
In conclusion, carotenoid metabolism has been widely studied in plants because of its importance to plants and humans. Here, we present for the first time comprehensive molecular characterization and analysis of CBP genes in N. officinale. This study will therefore improve our understanding of the molecular mechanisms regulating carotenoid accumulation in N. officinale, and this can subsequently serve as a valuable resource for genetic manipulation. Interestingly, by in silico analysis, we predict that most CBP genes were localized in the chloroplast. However, in the future, further studies are necessary to perform an in vivo localization study to confirm using an organelle (e.g., chloroplast) specific marker to validate it. In addition, how each CBP gene contributes to the dynamic assembly and association of the multifaceted complex carotenoid metabolons that must form in the suborganellar location also needs to be studied. The knowledge may facilitate fine modification of the carotenoids intended for specific organelles and increase the nutritional content of edible tissues for human benefits.
4. Experimental Section
4.1. Plant Material
Seeds of N. officinale (Lot No. 3056631) were acquired from Asia Seed Co, Ltd., Seoul, Republic of Korea. Seeds were sowed in a plastic pot (size: 11 × 11 cm) filled with commercial perlite. The pots were kept in the greenhouse of Chungnam National University (Daejeon, Korea), and the seeds were allowed to grow for 2 months. Plants were subjected to irrigation following a 2-day interval. Three biological replicates of the samples were collected from different plant organs, namely, the leaves, stems, roots, flowers, and seeds. Harvested samples were flash-frozen in liquid nitrogen and then stored at −80 °C until RNA extraction and HPLC analysis.
4.2. In Silico Identification and Sequence Analysis of CBP Genes
CBP gene sequences were retrieved from the N. officinale transcriptomic data obtained in our laboratory. An Illumina NextSeq500 platform was used to analyze the cDNA using the commercial service of Seeders, Inc. (Daejeon, South Korea). Raw reads of the transcriptome sequence are available on the NCBI SRA database under the accession number SRR3490957. Obtained sequences were then subjected to in silico BLAST on the NCBI database. Meanwhile, sequences were also analyzed using online servers and public databases, including the PFAM (http://pfam.xfam.org/search) and NCBI Conserved Domain Database (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) databases, to predict putative protein signature motifs. Secondary structure and signal peptide analyses were performed using the SOPMA program (https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_sopma.html) and the SignalP 4.0 Server (http://www.cbs.dtu.dk/services/SignalP-4.0/), respectively. Predicted subcellular locations of the CBP proteins were identified using the CELLO (http://cello.life.nctu.edu.tw/), ChloroP 1.1 (http://www.cbs.dtu.dk/services/ChloroP/), TargetP 1.1 (http://www.cbs.dtu.dk/services/TargetP-1.1/index.php), and WoLF PSORT (https://wolfpsort.hgc.jp/) tools. The theoretical pI (isoelectric point)/molecular weight was then calculated using the compute pI/molecular weight tool on the ExPASy platform (https://web.expasy.org/compute_pi/).
4.3. Structural Analysis of CBP Gene
Multiple sequence alignment was performed using BioEdit 7.2.5 (Therapeutics, Carlsbad, CA, USA).62 CBP protein sequences were submitted to the Phyre2 online web server (www.sbg.bio.ic.ac.uk/phyre2/html/page.cgi) for homology modeling and for 3D structure analysis. 3D structures were predicted using Chimera 1.14 software (https://www.cgl.ucsf.edu/chimera/).63 Conserved signature motifs among the CBP genes were found using the Multiple Expectation maximizations for Motif Elicitation tool (http://meme.nbcr.net/).
4.4. Phylogenetic Analysis and Percent Identity Matrix
The phylogenetic tree was constructed using the MEGA7 software.64 Neighbor-joining (NJ) phylogenetic trees65 were constructed using the Poisson model. Robustness of the trees was estimated by performing 1000 bootstrap replicates.66 The percent identity matrix between the CBP amino acid sequences was calculated using Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/), and identities were calculated from the pairwise multiple sequence alignment.67
4.5. RNA Extraction and cDNA Synthesis
Total RNA was extracted from the leaves, stem, root, flower, and seed of the plant. Each sample was ground into a fine powder using liquid nitrogen. Then, 100 mg of each sample was transferred to a fresh 1.5-mL microcentrifuge tube. Total RNA was extracted using the Plant Total RNA Mini Kit (Geneaid, Taiwan), according to the manufacturer’s protocols. RNA concentration and quality were determined using a NanoVue Plus spectrophotometer (GE Health Care Life Sciences, USA) and through 1% agarose gel electrophoresis, respectively. Extracted total RNAs were reverse transcribed to cDNA using the ReverTra Ace-α-kit (Toyobo Co. Ltd., Osaka, Japan), according to the manufacturer’s protocols; afterward, the cDNAs templates were diluted 20-fold with nuclease-free water for downstream experiments.
4.6. CBP Gene Expression
For qRT-PCR, the ubiquitin-conjugating enzyme 9 (UBC9) gene was used as an internal control. Specific primers for the N. officinale CBP and UBC9 genes were designed using the online Primer3 software.68 Primers used in this study are shown in Table S4. Relative gene expression was calculated using UBC9. qRT-PCR conditions used in our study followed the protocol described by Tuan et al.18 For calculating the gene expression, the ΔCt method was used.69 The visualization and expression of CBP genes in the heatmap and hierarchical clustering were analyzed using heatmapper software.70 Three biological replicates were used for all the PCR reactions.
4.7. Extraction of Carotenoids and HPLC Analysis
Carotenoids were extracted and analyzed using HPLC following the protocol reported by Ha et al.71 For HPLC analysis, 300 mg of fine powder samples were mixed with 3 mL of ethanol containing 0.1% ascorbic acid (w/v), and the mixture was vortexed and incubated for 10 min at 85 °C in a water bath. For saponification, potassium hydroxide (120 μL, 80% w/v) was added, and then the samples were immediately placed on ice for 5 min to terminate the reaction. Then, 1.5 mL of ice-cold deionized water and 0.05 mL of internal standard β-apo-8′-carotenal (1.25 μg) were added to this mixture. Carotenoids were extracted thrice using hexane (1.5 mL) and centrifuged at 140 × g for 5 min at 4 °C. The combined extracts were dried under nitrogen gas and were redissolved in 0.25 mL of 50:50 (v/v) dichloromethane/methanol. These mixtures were filtered through a 0.50 μm PTFE filter (Advantec, Tokyo, Japan) into brown screw cap vials (Thermo Fisher Scientific, USA). The carotenoids were separated using a HPLC Agilent 1100 system (Massy, France) equipped with a photodiode array detector using a C30 YMC column (250 × 4.6 mm, 3 μm, Water Corporation, MA, USA), and the chromatogram is obtained at 450 nm. HPLC conditions and gradient programs used followed the protocol described by Ha et al.71 The concentrations of individual carotenoids were quantified using the retention time and their co-elution with β-apo-8′-carotenal, an internal standard, and were quantitated with reference to the corresponding calibration curves of standards. All carotenoid standards were purchased from CaroteNature (Lupsingen, Switzerland).
4.8. Statistical Analysis
In this study, all results are expressed as the mean ± standard deviation (SD) of three independent biological replicates. Data were all analyzed by analysis of variance with Duncan’s multiple range tests to compare means with a significant level of p < 0.05 using the Statistical Analysis System version 9.2 (SAS Institute Inc., Cary, NC, USA, 2009).
Acknowledgments
This work was supported by the Incheon National University Research Grant in 2021, Republic of Korea, and a grant from the Next-generation BioGreen 21 Program (PJ015665), Korea.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.1c04802.
The nucleotide sequence and deduced amino acid sequences of CBP genes; phylogeny of deduced CBP amino acid sequences along with other CBP sequences; amino acid alignment of CBP genes with other CBP genes; comparison of Carica papaya (ACR61334) and N. officinale LCYB nucleotide sequences; the expression levels of CBP genes in the transcriptomic data; analysis of CBP gene sequences using the SignalP program; GenBank accession numbers of CBP genes used for identity matrix; and list of primers used in qRT-PCR analysis to determine mRNA expression levels of N. officinale CBP genes (PDF)
Author Contributions
∥ R.S. and S.J.B. contributed equally to this work.
The authors declare no competing financial interest.
Supplementary Material
References
- Bong S. J.; Jeon J.; Park Y. J.; Kim J. K.; Park S. U. Identification and analysis of phenylpropanoid biosynthetic genes and phenylpropanoid accumulation in watercress (Nasturtium officinale R. Br.). 3. BioTechniques 2020, 10, 1–8. 10.1007/s13205-020-02244-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz R. M. S.; Vieira M. C.; Silva C. L. M. Effect of heat and thermosonication treatments on watercress (Nasturtium officinale) vitamin C degradation kinetics. Innov. Food Sci. Emerg. 2008, 9, 483–488. 10.1016/j.ifset.2007.10.005. [DOI] [Google Scholar]
- Pourhassan-Moghaddam M.; Zarghami N.; Mohsenifar A.; Rahmati-Yamchi M.; Gholizadeh D.; Akbarzadeh A.; de la Guardia M.; Nejati-Koshki K. Watercress-based gold nanoparticles: biosynthesis, mechanism of formation and study of their biocompatibility in vitro. Micro. Nano Lett. 2014, 9, 345–350. 10.1049/mnl.2014.0063. [DOI] [Google Scholar]
- Gonçalves E. M.; Cruz R. M. S.; Abreu M.; Brandão T. R. S.; Silva C. L. M. Biochemical and colour changes of watercress (Nasturtium officinale R. Br.) during freezing and frozen storage. J. Food Eng. 2009, 93, 32–39. 10.1016/j.jfoodeng.2008.12.027. [DOI] [Google Scholar]
- Sadeghi H.; Mostafazadeh M.; Sadeghi H.; Naderian M.; Barmak M. J.; Talebianpoor M. S.; Mehraban F. In vivo anti-inflammatory properties of aerial parts of Nasturtium officinale. Pharm. Biol. 2014, 52, 169–174. 10.3109/13880209.2013.821138. [DOI] [PubMed] [Google Scholar]
- Park C. H.; Park Y. E.; Yeo H. J.; Yoon J. S.; Park S.-Y.; Kim J. K.; Park S. U. Comparative analysis of secondary metabolites and metabolic profiling between diploid and tetraploid Morus alba L. J. Agric. Food Chem. 2021, 69, 1300–1307. 10.1021/acs.jafc.0c06863. [DOI] [PubMed] [Google Scholar]
- Sathasivam R.; Radhakrishnan R.; Kim J. K.; Park S. U. An update on biosynthesis and regulation of carotenoids in plants. S. Afr. J. Bot. 2021, 140, 290–302. 10.1016/j.sajb.2020.05.015. [DOI] [Google Scholar]
- Sathasivam R.; Ki J. S. A review of the biological activities of microalgal carotenoids and their potential use in healthcare and cosmetic industries. Mar. Drugs 2018, 16, 26. 10.3390/md16010026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathasivam R.; Radhakrishnan R.; Hashem A.; Abd_Allah E. F. Microalgae metabolites: A rich source for food and medicine. Saudi J. Biol. Sci. 2019, 26, 709–722. 10.1016/j.sjbs.2017.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley L.; Yuan Y. W. Transcriptional regulation of carotenoid biosynthesis in plants: So many regulators, so little consensus. Front. Plant Sci. 2019, 10, 1017. 10.3389/fpls.2019.01017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham F. X.; Pogson B.; Sun Z. R.; McDonald K. A.; DellaPenna D.; Gantt E. Functional analysis of the beta and epsilon lycopene cyclase enzymes of Arabidopsis reveals a mechanism for control of cyclic carotenoid formation. Plant Cell 1996, 8, 1613–1626. 10.1105/tpc.8.9.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devitt L. C.; Fanning K.; Dietzgen R. G.; Holton T. A. Isolation and functional characterization of a lycopene beta-cyclase gene that controls fruit colour of papaya (Carica papaya L.). J. Exp. Bot. 2010, 61, 33–39. 10.1093/jxb/erp284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M.; Ikoma Y.; Matsumoto H.; Sugiura M.; Hyodo H.; Yano M. Accumulation of carotenoids and expression of carotenoid biosynthetic genes during maturation in citrus fruit. Plant Physiol. 2004, 134, 824–837. 10.1104/pp.103.031104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C.; Ji J.; Wang G.; Li Z. D.; Wang Y. R.; Fan Y. J. Over-expression of LcPDS, LcZDS, and LcCRTISO, genes from wolfberry for carotenoid biosynthesis, enhanced carotenoid accumulation, and salt tolerance in tobacco. Front. Plant Sci. 2020, 11, 119. 10.3389/fpls.2020.00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy C. S.; Lee S. H.; Yoon J. S.; Kim J. K.; Lee S. W.; Hur M.; Koo S. C.; Meilan J.; Lee W. M.; Jang J. K.; Hur Y.; Park S. U.; Kim Y. B. Molecular cloning and characterization of carotenoid pathway genes and carotenoid content in Ixeris dentata var. albiflora. Molecules 2017, 22, 1449. 10.3390/molecules22091449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan B. C.; Joseph L. M.; Deng W. T.; Liu L. J.; Li Q. B.; Cline K.; McCarty D. R. Molecular characterization of the Arabidopsis 9-cis epoxycarotenoid dioxygenase gene family. Plant J. 2003, 35, 44–56. 10.1046/j.1365-313X.2003.01786.x. [DOI] [PubMed] [Google Scholar]
- Tuan P. A.; Kim J. K.; Lee J.; Park W. T.; Kwon do Y.; Kim Y. B.; Kim H. H.; Kim H. R.; Park S. U. Analysis of carotenoid accumulation and expression of carotenoid biosynthesis genes in different organs of Chinese cabbage (Brassica rapa Subsp pekinensis). EXCLI J. 2012, 11, 508–516. [PMC free article] [PubMed] [Google Scholar]
- Tuan P. A.; Kim Y. B.; Kim J. K.; Arasu M. V.; Al-Dhabi N. A.; Park S. U. Molecular characterization of carotenoid biosynthetic genes and carotenoid accumulation in Scutellaria baicalensis Georgi. EXCLI J. 2015, 14, 146–157. 10.17179/excli2014-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H. S.; Chen M. D.; Wen Q. F.; Li Y. P. Isolation and characterization of the carotenoid biosynthetic genes LCYB, LCYE and CHXB from strawberry and their relation to carotenoid accumulation. Sci. Hortic. 2015, 182, 134–144. 10.1016/j.scienta.2014.12.007. [DOI] [Google Scholar]
- Jeon J.; Bong S. J.; Park J. S.; Park Y. K.; Arasu M. V.; Al-Dhabi N. A.; Park S. U. De novo transcriptome analysis and glucosinolate profiling in watercress (Nasturtium officinale R. Br.). BMC Genom. 2017, 18, 401. 10.1186/s12864-017-3792-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voutsina N.; Payne A. C.; Hancock R. D.; Clarkson G. J.; Rothwell S. D.; Chapman M. A.; Taylor G. Characterization of the watercress (Nasturtium officinale R. Br.; Brassicaceae) transcriptome using RNASeq and identification of candidate genes for important phytonutrient traits linked to human health. BMC Genom. 2016, 17, 378. 10.1186/s12864-016-2704-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Emparán A.; Quezada-Martinez D.; Zúñiga-Bustos M.; Cifuentes V.; Iñiguez-Luy F.; Federico M. L. Functional analysis of the Brassica napus L. phytoene synthase (PSY) gene family. PLoS One 2014, 9, e114878 10.1371/journal.pone.0114878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathasivam R.; Yeo H. J.; Park C. H.; Choi M.; Kwon H.; Sim J. E.; Park S. U.; Kim J. K. Molecular characterization, expression analysis of carotenoid, xanthophyll, apocarotenoid pathway genes, and carotenoid and xanthophyll accumulation in Chelidonium majus L. Plants 2021, 10, 1753. 10.3390/plants10081753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuan P. A.; Kim J. K.; Lee S.; Chae S. C.; Park S. U. Molecular characterization of carotenoid cleavage dioxygenases and the effect of gibberellin, abscisic acid, and sodium chloride on the expression of genes involved in the carotenoid biosynthetic aathway and carotenoid accumulation in the callus of Scutellaria baicalensis Georgi. J. Agric. Food Chem. 2013, 61, 5565–5572. 10.1021/jf401401w. [DOI] [PubMed] [Google Scholar]
- Sathasivam R.; Ki J. S. Differential transcriptional responses of carotenoid biosynthesis genes in the marine green alga Tetraselmis suecica exposed to redox and non-redox active metals. Mol. Biol. Rep. 2019, 46, 1167–1179. 10.1007/s11033-018-04583-9. [DOI] [PubMed] [Google Scholar]
- Flowerika; Alok A.; Kumar J.; Thakur N.; Pandey A.; Pandey A. K.; Upadhyay S. K.; Tiwari S. Characterization and expression analysis of phytoene synthase from bread wheat (Triticum aestivum L.). PLoS One 2016, 11, e0162443 10.1371/journal.pone.0162443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur N.; Pandey A.; Shivani; Kumar P.; Pandey P.; Kesarwani A. K.; Mantri S. S.; Awasthi P.; Tiwari S. Regulation of banana phytoene synthase (MaPSY) expression, characterization and their modulation under various abiotic stress conditions. Front. Plant Sci. 2017, 8, 462. 10.3389/fpls.2017.00462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X.-T.; Jia L.-D.; Duan M.-Z.; Chen X.; Qiao C.-L.; Ma J.-Q.; Zhang C.; Jing F.-Y.; Zhang S.-S.; Yang B.; Zhang L. Y.; Li J. N. Genome-wide identification and expression profiling of the carotenoid cleavage dioxygenase (CCD) gene family in Brassica napus L. PLoS One 2020, 15, e0238179 10.1371/journal.pone.0238179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q. L.; Zheng J. L.; Liu J. H. Transcription activation of beta-carotene biosynthetic genes at the initial stage of stresses as an indicator of the increased beta-carotene accumulation in isolated Dunaliella salina strain GY-H13. Aquat. Toxicol. 2020, 222, 105472 10.1016/j.aquatox.2020.105472. [DOI] [PubMed] [Google Scholar]
- Han Y.; Zheng Q. S.; Wei Y. P.; Chen J.; Liu R.; Wan H. J. In silico identification and analysis of phytoene synthase genes in plants. Genet. Mol. Res. 2015, 14, 9412–9422. 10.4238/2015.August.14.5. [DOI] [PubMed] [Google Scholar]
- Chen Y.; Li F. Q.; Wurtzel E. T. Isolation and characterization of the Z-ISO gene encoding a missing component of carotenoid biosynthesis in plants. Plant Physiol. 2010, 153, 66–79. 10.1104/pp.110.153916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z. D.; Wu G. X.; Ji J.; Wang G.; Tian X. W.; Gao H. L. Cloning and expression of a zeta-carotene desaturase gene from Lycium chinense. J. Genet. 2015c, 94, 287–294. 10.1007/s12041-015-0519-8. [DOI] [PubMed] [Google Scholar]
- Ahrazem O.; Rubio-Moraga A.; Berman J.; Capell T.; Christou P.; Zhu C. F.; Gómez-Gómez L. The carotenoid cleavage dioxygenase CCD2 catalysing the synthesis of crocetin in spring crocuses and saffron is a plastidial enzyme. New Phytol. 2016, 209, 650–663. 10.1111/nph.13609. [DOI] [PubMed] [Google Scholar]
- Cui H. L.; Wang Y. C.; Qin S. Molecular evolution of lycopene cyclases involved in the formation of carotenoids in eukaryotic algae. Plant Mol. Biol. Rep. 2011, 29, 1013–1020. 10.1007/s11105-011-0297-2. [DOI] [Google Scholar]
- Zhu Y. H.; Jiang J. G.; Yan Y.; Chen X. W. Isolation and characterization of phytoene desaturase cDNA involved in the beta-carotene biosynthetic pathway in Dunaliella salina. J. Agric. Food Chem. 2005, 53, 5593–5597. 10.1021/jf0506838. [DOI] [PubMed] [Google Scholar]
- DePristo M. A.; Weinreich D. M.; Hartl D. L. Missense meanderings in sequence space: A biophysical view of protein evolution. Nat. Rev. Genet. 2005, 6, 678–687. 10.1038/nrg1672. [DOI] [PubMed] [Google Scholar]
- Garg R.; Jhanwar S.; Tyagi A. K.; Jain M. Genome-wide survey and expression analysis suggest diverse roles of glutaredoxin gene family members during development and response to various stimuli in rice. DNA Res. 2010, 17, 353–367. 10.1093/dnares/dsq023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugueney P.; Badillo A.; Chen H. C.; Klein A.; Hirschberg J.; Camara B.; Kuntz M. Metabolism of cyclic carotenoids - a model for the alteration of this biosynthetic pathway in Capsicum annuum chromoplasts. Plant J. 1995, 8, 417–424. 10.1046/j.1365-313X.1995.08030417.x. [DOI] [PubMed] [Google Scholar]
- Lao Y. M.; Jin H.; Zhou J.; Zhang H. J.; Cai Z. H. Functional characterization of a missing branch component in Haematococcus pluvialis for control of algal carotenoid biosynthesis. Front. Plant Sci. 2017, 8, 1341. 10.3389/fpls.2017.01341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigo M. J.; Alquézar B.; Alós E.; Medina V.; Carmona L.; Bruno M.; al-Babili S.; Zacarías L. A novel carotenoid cleavage activity involved in the biosynthesis of citrus fruit-specific apocarotenoid pigments. J. Exp. Bot. 2013, 64, 4461–4478. 10.1093/jxb/ert260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang F. C.; Molnár P.; Schwab W. Cloning and functional characterization of carotenoid cleavage dioxygenase 4 genes. J. Exp. Bot. 2009, 60, 3011–3022. 10.1093/jxb/erp137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing S. A. J.; Gabelli S. B.; Echeverria I.; Vogel J. T.; Guan J. C.; Tan B. C.; Klee H. J.; McCarty D. R.; Amzel L. M. Structural insights into maize viviparous14, a key enzyme in the biosynthesis of the phytohormone abscisic acid. Plant Cell 2010, 22, 2970–2980. 10.1105/tpc.110.074815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang C.; Zhai H.; Xue L. Y.; Zhao N.; He S. Z.; Liu Q. C. A lycopene beta-cyclase gene, IbLCYB2, enhances carotenoid contents and abiotic stress tolerance in transgenic sweetpotato. Plant Sci. 2018, 272, 243–254. 10.1016/j.plantsci.2018.05.005. [DOI] [PubMed] [Google Scholar]
- Li H. M.; Chiu C. C. Protein transport into chloroplasts. Annu. Rev. Plant Biol. 2010, 61, 157–180. 10.1146/annurev-arplant-042809-112222. [DOI] [PubMed] [Google Scholar]
- Yan J. M.; Campbell J. H.; Glick B. R.; Smith M. D.; Liang Y. Molecular characterization and expression analysis of chloroplast protein import components in tomato (Solanum lycopersicum). PLoS One 2014, 9, e95088 10.1371/journal.pone.0095088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gady A. L. F.; Vriezen W. H.; van de Wal M. H. B. J.; Huang P. P.; Bovy A. G.; Visser R. G. F.; Bachem C. W. B. Induced point mutations in the phytoene synthase 1 gene cause differences in carotenoid content during tomato fruit ripening. Mol. Breeding 2012, 29, 801–812. 10.1007/s11032-011-9591-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M. Y.; Gan Z. B.; Cui Y.; Shi C. L.; Shi X. M. Structure and function characterization of the phytoene desaturase related to the lutein biosynthesis in Chlorella protothecoides CS-41. Mol. Biol. Rep. 2013, 40, 3351–3361. 10.1007/s11033-012-2410-5. [DOI] [PubMed] [Google Scholar]
- Elleuch F.; Hlima H. B.; Barkallah M.; Baril P.; Abdelkafi S.; Pichon C.; Fendri I. Carotenoids overproduction in Dunaliella sp.: Transcriptional changes and new insights through lycopene beta cyclase regulation. Appl. Sci. 2019, 9, 5389. 10.3390/app9245389. [DOI] [Google Scholar]
- Moise A. R.; Al-Babili S.; Wurtzel E. T. Mechanistic aspects of carotenoid biosynthesis. Chem. Rev. 2014, 114, 164–193. 10.1021/cr400106y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio A.; Rambla J. L.; Santaella M.; Gómez M. D.; Orzaez D.; Granell A.; Gómez-Gómez L. Cytosolic and plastoglobule-targeted carotenoid dioxygenases from Crocus sativus are both involved in beta-ionone release. J. Biol. Chem. 2008, 283, 24816–24825. 10.1074/jbc.M804000200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P. R.; Zhang S. J.; Zhang S. F.; Li F.; Zhang H.; Cheng F.; Wu J.; Wang X. W.; Sun R. F. Carotenoid biosynthetic genes in Brassica rapa: comparative genomic analysis, phylogenetic analysis, and expression profiling. BMC Genom. 2015, 16, 492. 10.1186/s12864-015-1655-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Sola M. A.; Rodríguez-Concepción M. Carotenoid biosynthesis in Arabidopsis: a colorful pathway. Arabidopsis Book 2012, 10, e0158 10.1199/tab.0158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuan P. A.; Kim J. K.; Kim H. H.; Lee S. Y.; Park N. I.; Park S. U. Carotenoid accumulation and characterization of cDNAs encoding phytoene synthase and phytoene desaturase in garlic (Allium sativum). J. Agric. Food Chem. 2011, 59, 5412–5417. 10.1021/jf2009827. [DOI] [PubMed] [Google Scholar]
- Cuong D. M.; Arasu M. V.; Jeon J.; Park Y. J.; Kwon S. J.; Al-Dhabi N. A.; Park S. U. Medically important carotenoids from Momordica charantia and their gene expressions in different organs. Saudi J. Biol. Sci. 2017, 24, 1913–1919. 10.1016/j.sjbs.2017.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai S.; Behra R.; Nestler H.; Suter M. J. F.; Sigg L.; Schirmer K. Linking toxicity and adaptive responses across the transcriptome, proteome, and phenotype of Chlamydomonas reinhardtii exposed to silver. Proc. Natl. Acad. Sci. USA 2014, 111, 3490–3495. 10.1073/pnas.1319388111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koul A.; Sharma D.; Kaul S.; Dhar M. K. Identification and in silico characterization of cis-acting elements of genes involved in carotenoid biosynthesis in tomato. 3. BioTechniques 2019, 9, 287. 10.1007/s13205-019-1798-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q. L.; Guo S. N.; Wen F.; Zhang X. L.; Wang C. C.; Si L. F.; Zheng J. L.; Liu J. H. Transcriptional and physiological responses of Dunaliella salina to cadmium reveals time-dependent turnover of ribosome, photosystem, and ROS-scavenging pathways. Aquat. Toxicol. 2019, 207, 153–162. 10.1016/j.aquatox.2018.12.007. [DOI] [PubMed] [Google Scholar]
- Fanciullino A.-L.; Bidel L.; Urban L. Carotenoid responses to environmental stimuli: integrating redox and carbon controls into a fruit model. Plant Cell Environ. 2014, 37, 273–289. 10.1111/pce.12153. [DOI] [PubMed] [Google Scholar]
- Mortain-Bertrand A.; Stammitti L.; Telef N.; Colardelle P.; Brouquisse R.; Rolin D.; Gallusci P. Effects of exogenous glucose on carotenoid accumulation in tomato leaves. Physiol. Plant. 2008, 134, 246–256. 10.1111/j.1399-3054.2008.01130.x. [DOI] [PubMed] [Google Scholar]
- Apel K.; Hirt H. Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 2004, 55, 373–399. 10.1146/annurev.arplant.55.031903.141701. [DOI] [PubMed] [Google Scholar]
- Urban L.; Alphonsout L. Girdling decreases photosynthetic electron fluxes and induces sustained photoprotection in mango leaves. Tree Physiol. 2007, 27, 345–352. 10.1093/treephys/27.3.345. [DOI] [PubMed] [Google Scholar]
- Hall T. A. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Pettersen E. F.; Goddard T. D.; Huang C. C.; Couch G. S.; Greenblatt D. M.; Meng E. C.; Ferrin T. E. UCSF chimera - A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Kumar S.; Stecher G.; Tamura K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N.; Nei M. The neighbor-joining method - a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Confidence Limits on Phylogenies: An Approach Using the Bootstrap. Evolution 1985, 39, 783–791. 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Madeira F.; Park Y. M.; Lee J.; Buso N.; Gur T.; Madhusoodanan N.; Basutkar P.; Tivey A. R. N.; Potter S. C.; Finn R. D.; Lopez R. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019, 47, W636–W641. 10.1093/nar/gkz268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S.; Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 2000, 132, 365–386. [DOI] [PubMed] [Google Scholar]
- Silver N.; Best S.; Jiang J.; Thein S. L. Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol. Biol. 2006, 7, 33. 10.1186/1471-2199-7-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babicki S.; Arndt D.; Marcu A.; Liang Y.; Grant J. R.; Maciejewski A.; Wishart D. S. Heatmapper: web-enabled heat mapping for all. Nucleic Acids Res. 2016, 44, W147–W153. 10.1093/nar/gkw419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha S.-H.; Kim J. K.; Jeong Y. S.; You M.-K.; Lim S.-H.; Kim J.-K. Stepwise pathway engineering to the biosynthesis of zeaxanthin, astaxanthin and capsanthin in rice endosperm. Metab. Eng. 2019, 52, 178–189. 10.1016/j.ymben.2018.11.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.