Abstract
Humans diverge from other primates in numerous ways including their neuroanatomy and cognitive capacities. Human-specific features are particularly prominent in the cerebral cortex, which has undergone an expansion in size and acquired unique cellular composition and circuitry. Human-specific gene expression is postulated to explain neocortical anatomical differences across evolution. In particular, non-coding regulatory loci are strongly linked to human traits including progenitor proliferation and cortical size. In this review, we highlight emerging non-coding elements implicated in human cortical evolution, including roles for regulatory DNA and RNA. Further, we discuss the association of human-specific genetic changes with neurodevelopmental diseases.
Introduction
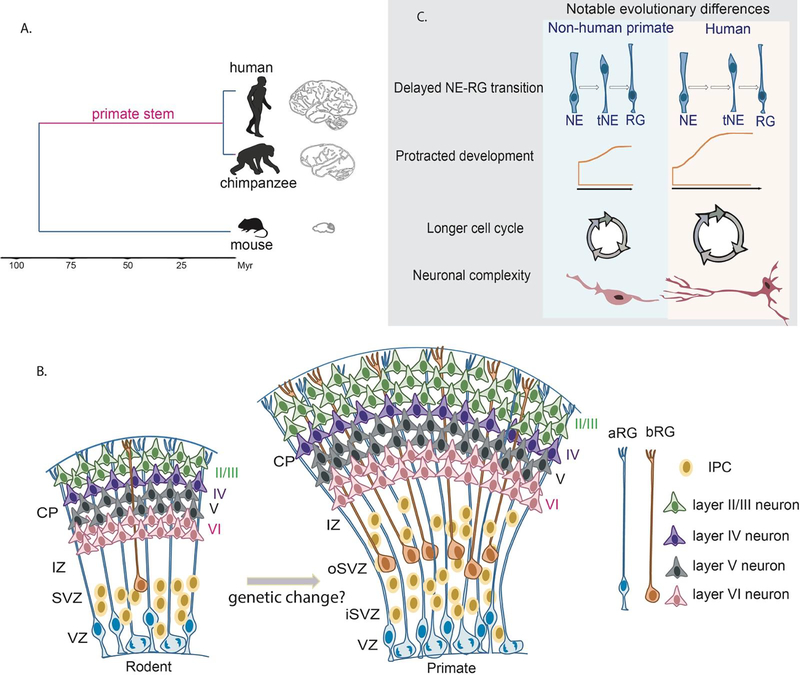
The evolution of the human brain with its cerebral cortex occurred over 300 million years [1–3]. The neocortex is a highly-organized and specialized structure which controls higher-order functions, such as cognition, language, reasoning and emotion. Compared to other primates, the neocortex has expanded tremendously in size and accounts for almost 80% of the total brain mass [3]. Additionally, human cortices contain more neurons and glia, which are themselves posited to be more complex. The cellular mechanism of evolutionary cortical expansion was first postulated in the radial unit hypothesis, which attributed size differences to the proliferative capacity of neural precursors [4]. Indeed, an evolutionary increase in progenitor number, including outer radial glia (oRG, also termed basal radial glia), along with an expansion of germinal zones, including the outer subventricular zone (OSVZ), is thought to underlie increased neuron number [5–7]. Humans also undergo longer gestation resulting in prolonged neurogenesis relative to other species--another factor posited to influence human brain development. Taken together, these anatomical and cellular changes collectively contribute to unique human cognitive capacities (Figure 1).
Figure 1. Evolutionary development of the human cortex.
A. Graph showing a phylogenetic tree indicating the brain morphology of chimpanzee and rodents compared to humans. Primate stem has been highlighted. B. Cartoon representing comparisons of rodent and primate neocortical development. Progenitors are designated by their locations in the cortex. Apical radial glia (aRGs) which localize to the ventricular zone (VZ) can undergo symmetric and asymmetric divisions to self-renew or produce neurons directly or indirectly by producing intermediate progenitors (IPC) in the subventricular zone (SVZ). Newborn neurons will migrate through the intermediate zone (IZ) and arrive to the cortical plate (CP) with first born neurons located deeper and late born neurons located superficially. In the developing primate brain, SVZ expansion leads to a new germinal zone, the outer subventricular zone (OSVZ), which includes proliferative IPCs and basal radial glia cells (bRGs). The expansion of basal progenitors in turn, leads to more upper layer (mainly layer II/III) neurons in the primate neocortex. C. Comparisons of notable cellular differences between human and non-human primate cortical development.
State-of-the art genomic and genetic analyses of humans and other species, including comparisons between modern and archaic humans, have implicated human-specific gene expression in cortical evolution [8]. Changes in non-coding regions have long been proposed to have a crucial role in human brain evolution [9]. This is increasingly supported by empirical data suggesting that gene regulatory regions, such as enhancers and noncoding RNAs play an instrumental role in brain evolution [10]. Indeed, broader and fine-tuned regulation of many genes may be achieved by modifications in gene regulatory mechanisms.
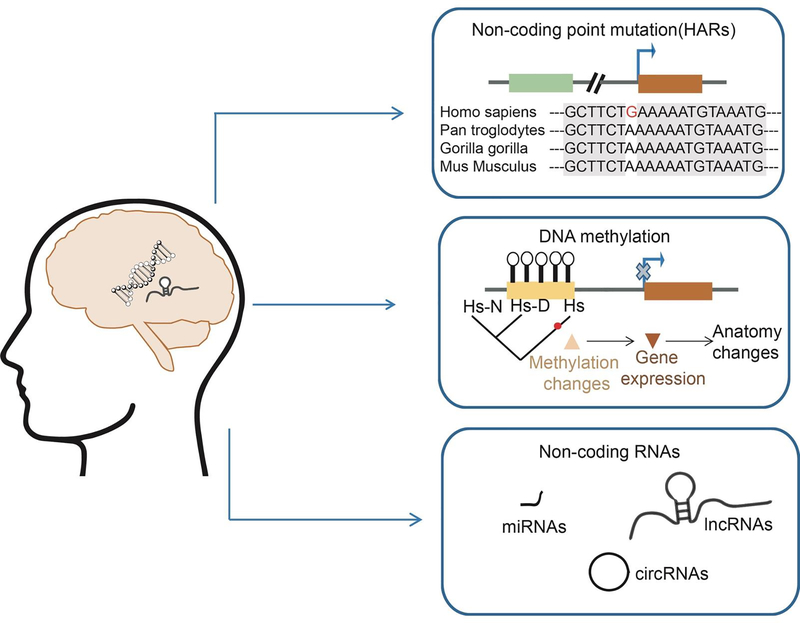
In this review, we describe emerging evidence that modern human brain divergence is impacted by noncoding regulatory elements, including regulatory DNA and RNA (Figure 2). We focus on recent evidence implicating changes in promoters, enhancers, and non-coding RNAs in cortical evolution [11,12]. We further highlight current methods for experimental investigation of evolutionary differences. Finally, we discuss known mutations in human-specific loci linked to neurodevelopmental disorders, including schizophrenia, autism spectrum disorder (ASD), and neurodegenerative diseases.
Figure 2. Non-coding regulatory elements in human brain evolution.
Cartoon showing genomic modifications during human brain evolution. Representation of epigenetic changes, non-coding DNA and RNA alterations which have been implicated in human brain evolution, including the HARs, genomic methylation, and diversity of non-coding RNAs.
REGULATORY DNA
Epigenetic differences and evolution
DNA methylation is a stable epigenomic modification implicated in cell identity across human cortical development [13]. Recent studies comparing DNA methylomes of neurons and oligodendrocytes across primates showed that human-specific differences depend upon both specific cell-type and the cytosine-context [14]. Compare to other primates, humans have significantly higher CH methylation (hypermethylation where H is equivalent to A,C or T) but reduced CG methylation (hypomethylation). CH hypermethylation is especially associated with human neurons and CG methylation discriminates neurons from oligodendrocytes. Further, human neuron-specific CG methylation is associated with neurological disease, and schizophrenia risk. Methylation studies have also detected differences discriminating modern humans, Neanderthals, and Denisovans, including within facial and speech anatomy-associated genes [15,16]. These epigenetic differences have been used to reconstruct the anatomy of Denisovans [17]. By analyzing these epigenetic marks, we can further understand how gene expression may have been controlled in non-extant populations.
These studies implicate methylation in the human genome as a force for modern human divergence and raise fascinating questions. How might epigenetic marks parlay into functional changes and how do methylation changes shape modern human brains? The ability to perform gene editing and evaluate brain development using organoids (see box) offers new opportunities to address these questions [18,19].
Human-accelerated regions
Human Accelerated Regions (HARs) are genomic regions, ranging in size, which are conserved in non-human primates and other euarchontoglires, and present an unexpectedly high rate of mutations in humans [12,20]. These fast-evolving regions were identified through comparative genomics, with some studies taking into consideration human SNPs [21–23]. Various groups have estimated the exact number of HARs in our genome, but differences in datasets and computational definitions have led to only partial consensus on these regions’ identification. At least 2700 HARs are estimated to be present in the human genome [20]. Further, based on epigenetic marks and functional assays, around 30–40% of HARs are predicted to function as regulatory enhancers [21,24]. Predicted functions for non-enhancer HARs are less defined.
Many regulatory HARs reside in proximity to genes involved in brain development [25]. This led to the prediction that HARs influence neuronal development, including proliferation, differentiation, and axogenesis [10,24,26,27]. For example, 14 HARs are found within non-coding elements of NPAS3, which is linked to neural development [27,28]. A recent study using Hi-C, a technique to identify long range genomic interactions, in human fetal brain confirmed the physical interactions of several HARs with neurodevelopmental genes [10]. The authors identified three HARs predicted to drive expression of GLI2, GLI3 or TBR1, which are implicated in diverse aspects of cortical development. Inactivation of their enhancer activity in human neural progenitors using dCas9-VP64 (see box) led to decreased expression of the three target genes. These chromatin architecture analyses and epigenetic manipulations in developing human brain cells suggest HARs can influence diverse developmental stages and cellular processes.
HAR activity is also associated with layer specification and cortical size. This is particularly interesting given that over the course of evolution the human neocortex has undergone exceptional enlargement accompanied by increased neuronal generation [8]. The relationship between HARs and cortical size was evidenced by Boyd et al., who used mouse transgenic models to demonstrate differential enhancer activity by the human and the chimpanzee orthologs of HARE5 (HAR Enhancer 5), which promotes expression of Fzd8 [29]. Expression of human HARE5 in mice accelerated progenitor cell cycle and increased brain size. It remains to be seen whether experimental manipulation of HARE5 has a similar impact in primates.
Beyond cortical size, HARs are also implicated in higher human capacities such as language. Complex spoken language, a uniquely human characteristic, is linked to the FOXP2 locus that includes 12 HARs [30]. When introduced into mouse or zebrafish, these human and chimpanzee HAR sequences exhibit strikingly different spatial and expression patterns during brain development. Notably, these orthologous sequences contain distinct predicted binding sites for transcription factors, suggesting possible mechanisms by which FOXP2 expression may be differentially controlled across species. The direct interaction between these HARs and the FOXP2 promoter has not yet been demonstrated and the actual role of these specific HARs in human language is yet uncharacterized.
HARs have also been associated with characteristics of modern humans. For example, changes in loci discriminating modern humans have been linked to cortical surface area, including HARs which control expression of the transcription factor HEY2 [31]. Further, accelerated regions control the differential expression of 212 neurodevelopmental genes which distinguish modern and ancient humans and are postulated to affect cortical size and chromatin regulation [32]. These new data collectively implicate HARs both in macroscopic phenotypes discriminating human and other primates, as well as in subtle changes in the human lineage.
The literature to date suggests enhancer HARs are central for human brain evolution, yet many fundamental questions remain. In particular, the efforts to characterize and index the ~2700 HARs are not yet comprehensive and there is no common repository of datasets. Additionally, to date, high throughput investigation of HARs has been limited to cell culture models. While cell culture can provide broad insights into functions, these assays measure HARs outside their normal genomic context, and transient interrogation may provide just a snapshot of HAR function. Indeed, some HARs show temporal differences across developmental stages, reinforcing the value of in vivo investigation using stable expression in model systems [24,29]. Mouse models have been invaluable for assaying HARs across different tissues and developmental or adult stages, but their use raises concerns with the trans-environment. Remarkably, a vast majority of HARs apparently preserve differential activity regardless of the species tested, showing that regulatory function in cis is more relevant than the trans environment [33]. This provides rationale that HARs can be characterized in model organisms. Ideally, multi-pronged approaches using model organisms and species-specific iPSCs (see box) can provide complementary information.
NON-CODING RNAS
microRNAs
MicroRNAs (miRNAs) are short endogenous single-stranded RNAs (20–24 nts) which are especially relevant in the nervous system, where about 70% of the 2500 mature human miRNAs are expressed [34,35]. Functionally, miRNAs regulate gene expression by repressing translation or degrading their target mRNAs.
Comparison of human, chimpanzee and macaque neocortical transcriptomes led to the discovery of differential miRNA expression including human-specific changes [7]. For example, in silico analysis across 11 mammalian species identified one miR941, which is implicated in cell differentiation by controlling target genes in the hedgehog- and insulin-signaling pathways [36]. A recent study discovered corticospinal motor neuron (CSMN)-enriched miRNAs in the mammalian brain, and showed that misexpressing miR-409–3p in vivo and in vitro increased CSMN neurons at the expense of deep-layer neuron development [37].
miRNAs have also been implicated in multiple aspects of primate divergence including expansion of the germinal zones and increased neural progenitor complexity [34,38–40]. One of the first studies to investigate this question used deep miRseq of laser dissected macaque visual cortex to discover novel primate-specific miRNAs [41]. These miRNAs were specifically expressed in primate germinal zones, with target genes related to cell cycle and neurogenesis. More recent discoveries have used single cell sequencing to identify cell-specific miRNAs and their targets in human fetal brains across development [42]. Their discovery of some miRNAs which are great ape-specific lays the groundwork for future functional studies of miRNAs in brain evolution. Another recent study discovered two miRNAs enriched in ferret proliferative zones that play roles in regulating basal progenitor expansion and neural differentiation [43]. Notably, the validated targets of these ferret miRNAs are related to cell proliferation and neurogenesis. Likewise, miR934 has been shown to influence expression of genes associated with neurogenesis and early-born subplate neurons [44]. Collectively, these studies implicate miRNAs in the evolutionary expansion and function of the OSVZ and basal progenitors in primates.
By controlling gene expression, miRNAs have the potential to significantly influence species-specific brain modifications. Recent studies catalog miRNAs and raise intriguing questions including how human-specific miRNAs regulate function of specific cell subtypes and what are their downstream targets? It is also interesting to consider exactly how miRNAs functionally diverge, as it was recently shown that most primate-specific miRNAs share highly conserved RNA structures [45]. Finally, it is fascinating to consider how miRNAs may have influenced the modern human divergence from our closest relative archaic humans including Denisovans and Neanderthals.
IncRNAs and circRNAs
In contrast to short miRNAs, long-non coding (lncRNA) transcripts are more than 200 nt, with an average length of ~3000 nt [46]. The structure of IncRNAs is similar to that of mRNAs, with 5’ and 3’ modifications, introns and exons, as well as cytoplasmic localization [11]. To date, the human genome is reported to contain more than 16,000 lncRNA genes, about 40% percent of which are expressed in the brain [47]. Further, about one-third of lncRNAs are primate-specific, with a large fraction found in the brain [48]. LncRNAs are implicated in cortical development, including neural stem cell maintenance, differentiation, and neural maturation [49].
Collectively, these data argue that lncRNA may play important roles in brain evolution. In 2006, Pollard et al. discovered the first HAR, called HAR1, which falls within the lncRNA HAR1F [50]. HAR1F is specifically expressed in human Cajal–Retzius neurons during early stages of cortical development although its function is unknown. More recent discoveries implicate a primate lncRNA, LncND, in cortical expansion, as it is expressed in neural progenitors and regulates Notch signaling [51]. Thus, the specific expression patterns of lncRNAs may provide potent spatial and temporal control of traits relevant for evolution. As many lncRNAs act as enhancers, it will be interesting to evaluate if this is the case for human-specific lncRNAs [47].
Recent studies also highlight an emerging function of circular RNAs (circRNAs) in species evolution. circRNAs are single-strand RNAs molecules composed of 1–5 exons formed into a circle as a result of non-canonical back-splicing events [52]. By comparing circRNA expression systematically across humans, non-human primates, and mice, a majority of circRNAs were found to be human-specific [53,54]. While functions for these in evolution are unknown, circRNAs themselves have been implicated in control of translation and are abundant in the brain [55]. These remarkable observations indicate that the future study of circRNAs can give valuable insights into cortical evolution.
NON-CODING REGULATION AND DISEASE
Given their role in evolutionary features which rely upon development, it is not surprising that mutations in non-coding regions, in particular within HARs, are implicated in neurodevelopmental diseases [56]. Doan et al. found a rare de novo duplication of a HAR located upstream of NR2F2, a gene implicated in ASD risk, with higher frequency in ASD probands than in control siblings [26]. Moreover, they identified HARs predicted to be active in the brain and which interact with genes associated with ASD, brain malformations and cortical development. Notably, rare biallelic mutations in these HARs were estimated to contribute to approximately 5% of consanguineous ASD cases. GWAS studies of schizophrenic individuals also detected an enrichment for HAR-regulated genes involved in signaling related to GABAergic neurons, which is dysregulated in affected individuals [57]. In comparison, Primate Accelerated Regions (PARs) were less enriched, suggesting that changes in human fast-evolving regions may have facilitated functional changes. These data point to the influence of mutations in noncoding HARs upon developmental disorders and for human traits such as social behavior and cognitive capabilities.
While developmental diseases and cognitive dysfunction are the most common phenotypes deriving from disruption of HAR activity, recent studies show HAR mutations are also implicated in older-age onset diseases. Specifically, 93 enhancer HARs active in the nervous system have been studied for their association with cancer and Alzheimer’s [58]. Numerous miRNAs and lncRNAs have also been associated with neurodegenerative disorders, but human-specific changes have not yet been linked to these diseases [59]. Collectively, these studies highlight that the continued discovery of human-specific non-coding regulatory elements can give valuable insights into the etiology and treatment of neurological disease.
Summary
Comparative genomic and chromosome architecture studies have shown that a powerful mediator of human brain evolution resides in the non-coding regulatory elements of our genome. Here, we described some of these known changes and their implicated functional roles in cortical evolution. Given the new tools of single-cell (sc) RNA sequencing, genomic editing, and species-specific cerebral organoids, we will continue to gain a richer resolution of how these changes influence function in specific cell subpopulations and across developmental stages. By interrogating non-coding regulatory loci, together with protein-coding changes, we will move closer to understanding what makes our brains different from our closest living and extinct relatives. As mutations within human-specific non-coding loci continue to be associated with various disease contexts, future studies can provide molecular insights into the basis of neurological diseases.
Box - Methods
High-throughput approaches have been used to identify and evaluate the activity of regulatory DNA elements. While scRNA-seq produces a map of differential gene expression across cell types, scATAC-seq generates a list of the open genomic regions within precise stages and cell types. A synergy of these two methods now enables a clearer understanding of changing DNA regulation and gene expression across time and space [60]. New technological advances offer possibilities for functionally screening non-coding elements in mammalian cells. CRISPR interference (CRISPRi) uses sgRNAs complementary to specific DNA regions to drive nuclease-dead Cas9 (dCas9) and interfere with the gene expression. For example, dCas9-KRAB is widely used to repress enhancers and promoters [61,62]. Furthermore, massively parallel reporter assays (MPRAs) can interrogate enhancer activity and single-base variants in a library of regulatory elements using RNA reporters [21]. These approaches permit analyses of libraries of regulatory regions in different conditions allowing an exhaustive view of their activity. A challenge for studying human-specific traits is the need to evaluate functions in vivo, within a tissue architecture and at the level of circuitry and behavior. However, there are ethical and practical issues with obtaining and using human and non-human primate tissues. Brain organoids generated from various species have helped obviate these limitations by providing in vitro models of brain development [19,63,64]. Recent studies have also used hybrid human-chimpanzee brain organoids derived from tetraploid iPSCs [65,66]. This model enables a direct cross-species comparison and resolves the issues of cis- or trans- regulation of non-coding elements, thus providing a new alternative to existing methods.
Highlights.
Differences in DNA methylomes discriminate modern and ancestral humans
Human-accelerated regions transcriptionally control genes involved in cortical development
Post-transcriptional mechanisms including miRNAs, IncRNAs, and circRNAs influence human brain evolution
Mutations in human-specific loci are associated with neurological diseases
Acknowledgements
We thank Craig Lowe and members of the Silver lab for careful reading of this manuscript. This work was supported by the NIH (R01NS083897, R01NS120667, R21MH119813, and R01NS110388 to D.L.S), and the Ruth K. Broad Foundation Fellowship to J.L.
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
*paper of special interest
**paper of outstanding interest
- 1.Neubauer S, Hublin JJ, Gunz P: The evolution of modern human brain shape. Sci Adv 2018, 4:eaao5961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Debra L Silver PR, Elizabeth A Grove,, Tarik F H Haydar TK, Wieland B Huttner,, Zoltán Molnár J L R, Nenad Sestan,, Michael P Stryker MS, Maria Antonietta Tosches aCAW: Evolution and Ontogenetic Development of Cortical Structures. In The Neocortex. Edited by Singer W TJSaPR: MIT Press; 2019. vol 27.] [Google Scholar]
- 3.Sousa AMM, Meyer KA, Santpere G, Gulden FO, Sestan N: Evolution of the Human Nervous System Function, Structure, and Development. Cell 2017, 170:226–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rakic P: Specification of cerebral cortical areas. Science (New York, NY) 1988, 241:170–176. [DOI] [PubMed] [Google Scholar]
- 5.Ostrem B, Di Lullo E, Kriegstein A: oRGs and mitotic somal translocation - a role in development and disease. Curr Opin Neurobiol 2017, 42:61–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dehay C, Kennedy H, Kosik KS: The outer subventricular zone and primate-specific cortical complexification. Neuron 2015, 85:683–694. [DOI] [PubMed] [Google Scholar]
- 7.Somel M, Liu X, Tang L, Yan Z, Hu H, Guo S, Jiang X, Zhang X, Xu G, Xie G, et al. : MicroRNA-driven developmental remodeling in the brain distinguishes humans from other primates. PLoS Biol 2011, 9:e1001214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heide M, Huttner WB: Human-Specific Genes, Cortical Progenitor Cells, and Microcephaly. Cells 2021, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.King M-C, Wilson AC: Evolution at two levels in humans and chimpanzees. Science (New York, NY) 1975, 188:107–116. [DOI] [PubMed] [Google Scholar]
- 10. Won H, Huang J, Opland CK, Hartl CL, Geschwind DH: Human evolved regulatory elements modulate genes involved in cortical expansion and neurodevelopmental disease susceptibility. Nat Commun 2019, 10:2396.31160561 **This study examines genomic interactions between HARs and target genes, and assesses functions of several HARs using epigenomic techniques.
- 11.Zimmer-Bensch G: Emerging Roles of Long Non-Coding RNAs as Drivers of Brain Evolution. Cells 2019, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Franchini LF, Pollard KS: Human evolution: the non-coding revolution. BMC Biology 2017, 15:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Price AJ, Collado-Torres L, Ivanov NA, Xia W, Burke EE, Shin JH, Tao R, Ma L, Jia Y, Hyde TM, et al. : Divergent neuronal DNA methylation patterns across human cortical development reveal critical periods and a unique role of CpH methylation. Genome Biol 2019, 20:196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jeong H, Mendizabal I, Berto S, Chatterjee P, Layman T, Usui N, Toriumi K, Douglas C, Singh D, Huh I, et al. : Evolution of DNA methylation in the human brain. Nat Commun 2021, 12:2021.33795684 *This study identifies human-specific methylation differences in a cell-type specific fashion.
- 15. Gokhman D, Nissim-Rafinia M, Agranat-Tamir L, Housman G, Garcia-Perez R, Lizano E, Cheronet O, Mallick S, Nieves-Colon MA, Li H, et al. : Differential DNA methylation of vocal and facial anatomy genes in modern humans. Nat Commun 2020, 11:1189.32132541 *This study compiles methylation differences between modern and ancient humans affecting genes relevant for human evolution.
- 16.Gokhman D, Lavi E, Prüfer K, Fraga MF, Riancho JA, Kelso J, Pääbo S, Meshorer E, Carmel L: Reconstructing the DNA methylation maps of the Neandertal and the Denisovan. Science (New York, NY) 2014, 344:523–527. [DOI] [PubMed] [Google Scholar]
- 17.Gokhman D, Mishol N, de Manuel M, de Juan D, Shuqrun J, Meshorer E, Marques-Bonet T, Rak Y, Carmel L: Reconstructing Denisovan Anatomy Using DNA Methylation Maps. Cell 2019, 179:180–192 e110. [DOI] [PubMed] [Google Scholar]
- 18.Trujillo CA, Rice ES, Schaefer NK, Chaim IA, Wheeler EC, Madrigal AA, Buchanan J, Preissl S, Wang A, Negraes PD, et al. : Reintroduction of the archaic variant of NOVA1 in cortical organoids alters neurodevelopment. Science 2021, 371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Benito-Kwiecinski S, Giandomenico SL, Sutcliffe M, Riis ES, Freire-Pritchett P, Kelava I, Wunderlich S, Martin U, Wray GA, McDole K, et al. : An early cell shape transition drives evolutionary expansion of the human forebrain. Cell 2021, 184:2084–2102 e2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hubisz MJ, Pollard KS: Exploring the genesis and functions of Human Accelerated Regions sheds light on their role in human evolution. Curr Opin Genet Dev 2014, 29:15–21. [DOI] [PubMed] [Google Scholar]
- 21. Uebbing S, Gockley J, Reilly SK, Kocher AA, Geller E, Gandotra N, Scharfe C, Cotney J, Noonan JP: Massively parallel discovery of human-specific substitutions that alter enhancer activity. Proc Natl Acad Sci U S A 2021, 118. **This study uses MPRA approaches to comprehensively test enhancer activity across HARs.
- 22.Pollard KS, Salama SR, King B, Kern AD, Dreszer T, Katzman S, Siepel A, Pedersen JS, Bejerano G, Baertsch R, et al. : Forces shaping the fastest evolving regions in the human genome. PLoS genetics 2006, 2:e168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Prabhakar S, Noonan JP, Pääbo S, Rubin EM: Accelerated evolution of conserved noncoding sequences in humans. Science (New York, NY) 2006, 314:786–786. [DOI] [PubMed] [Google Scholar]
- 24.Capra JA, Erwin GD, McKinsey G, Rubenstein JLR, Pollard KS: Many human accelerated regions are developmental enhancers. Philosophical Transactions of the Royal Society B: Biological Sciences 2013, 368:20130025–20130025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haygood R, Fedrigo O, Hanson B, Yokoyama K-D, Wray GA: Promoter regions of many neural- and nutrition-related genes have experienced positive selection during human evolution. Nature Genetics 2007, 39:1140–1144. [DOI] [PubMed] [Google Scholar]
- 26. Doan RN, Bae B-I, Cubelos B, Chang C, Hossain AA, Al-Saad S, Mukaddes NM, Oner O, Al Saffar M, Balkhy S, et al. : Mutations in Human Accelerated Regions Disrupt Cognition and Social Behavior. Cell 2016. **This paper discovers and investigates mutations in HARs associated with neurodevelopmental diseases including Autism.
- 27.Kamm GB, Pisciottano F, Kliger R, Franchini LF: The developmental brain gene NPAS3 contains the largest number of accelerated regulatory sequences in the human genome. Molecular biology and evolution 2013, 30:1088–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kamm GB, López-Leal R, Lorenzo JR, Franchini LF: A fast-evolving human NPAS3 enhancer gained reporter expression in the developing forebrain of transgenic mice. Philosophical Transactions of the Royal Society B: Biological Sciences 2013, 368:20130019–20130019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Boyd JL, Skove SL, Rouanet JP, Pilaz L-J, Bepler T, Gordân R, Wray GA, Silver DL: Human-chimpanzee differences in a FZD8 enhancer alter cell-cycle dynamics in the developing neocortex. Current biology: CB 2015, 25:772–779.25702574 *This paper is amongst the first to test the functional impact of HARs upon cortical size in mouse models.
- 30. Caporale AL, Gonda CM, Franchini LF: Transcriptional Enhancers in the FOXP2 Locus Underwent Accelerated Evolution in the Human Lineage. Mol Biol Evol 2019. *This study uses zebrafish and mouse models to assess enhancer activity of HARs associated with FOXP2.
- 31.Tilot AK, Khramtsova EA, Liang D, Grasby KL, Jahanshad N, Painter J, Colodro-Conde L, Bralten J, Hibar DP, Lind PA, et al. : The Evolutionary History of Common Genetic Variants Influencing Human Cortical Surface Area. Cereb Cortex 2021, 31:1873–1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Moriano J, Boeckx C: Modern human changes in regulatory regions implicated in cortical development. BMC Genomics 2020, 21:304.32299352 *This study evaluates activity of HARs discriminating modern and ancient humans.
- 33.Ryu H, Inoue F, Whalen S, Williams A, Kircher M, Martin B, Alvarado B, Samee MAH, Keough K, Thomas S, et al. : Massively parallel dissection of human accelerated regions in human and chimpanzee neural progenitors. bioRxiv 2018:256313. [Google Scholar]
- 34.Prodromidou K, Matsas R: Species-Specific miRNAs in Human Brain Development and Disease. Front Cell Neurosci 2019, 13:559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rajman M, Schratt G: MicroRNAs in neural development: from master regulators to fine-tuners. Development 2017, 144:2310–2322. [DOI] [PubMed] [Google Scholar]
- 36.Hu HY, He L, Fominykh K, Yan Z, Guo S, Zhang X, Taylor MS, Tang L, Li J, Liu J, et al. : Evolution of the human-specific microRNA miR-941. Nat Commun 2012, 3:1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Diaz JL, Siththanandan VB, Lu V, Gonzalez-Nava N, Pasquina L, MacDonald JL, Woodworth MB, Ozkan A, Nair R, He Z, et al. : An evolutionarily acquired microRNA shapes development of mammalian Cortical projections. Proc Natl Acad Sci U S A 2020, 117:29113–2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fietz SA, Lachmann R, Brandi H, Kircher M, Samusik N, Schröoder R, Lakshmanaperumal N, Henry I, Vogt J, Riehn A, et al. : Transcriptomes of germinal zones of human and mouse fetal neocortex suggest a role of extracellular matrix in progenitor self-renewal. Proceedings of the National Academy of Sciences 2012, 109:11836–11841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kosik KS, Nowakowski T: Evolution of New miRNAs and Cerebro-Cortical Development. Annu Rev Neurosci 2018, 41:119–137. [DOI] [PubMed] [Google Scholar]
- 40.Chinnappa K, Máarquez-Galera Á, Prieto-Colomina A, Nomura Y, Cárdenas A, López-Atalaya JP, Borrell V: <em>MIR3607</em> regulates cerebral cortex development via activation of Wnt/βCat signaling. bioRxiv 2019:729939. [Google Scholar]
- 41.Arcila ML, Betizeau M, Cambronne XA, Guzman E, Doerflinger N, Bouhallier F, Zhou H, Wu B, Rani N, Bassett DS, et al. : Novel Primate miRNAs Coevolved with Ancient Target Genes in Germinal Zone-Specific Expression Patterns. Neuron 2014, 81:1255–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nowakowski TJ, Rani N, Golkaram M, Zhou HR, Alvarado B, Huch K, West JA, Leyrat A, Pollen AA, Kriegstein AR, et al. : Regulation of cell-type-specific transcriptomes by microRNA networks during human brain development. Nat Neurosci 2018, 21:1784–179230455455 *This study uses single-cell sequencing of human fetal brains to identify miRs, including ape-specific miRs in neural progenitors.
- 43.Tomasello U, Klingler E, Niquille M, Mule N, de Vevey L, Prados J, Santinha AJ, Platt R, Borrell V, Jabaudon D, et al. : MiR-137 and miR-122, two outer subventricular zone-enriched non-coding RNAs, regulate basal progenitor expansion and neuronal differentiation. bioRxiv 2021:2021.2004.2001.438039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Prodromidou K, Vlachos IS, Gaitanou M, Kouroupi G, Hatzigeorgiou AG, Matsas R: MicroRNA-934 is a novel primate-specific small non-coding RNA with neurogenic function during early development. Elife 2020, 9 *This study demonstrates a miR linked to control of genes involved in early corticogenesis including subplate neurons.
- 45.McCreight JC, Schneider SE, Wilburn DB, Swanson WJ: Evolution of microRNA in primates. PLoS One 2017, 12:e0176596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gao Y, Li S, Lai Z, Zhou Z, Wu F, Huang Y, Lan X, Lei C, Chen H, Dang R: Analysis of Long NonCoding RNA and mRNA Expression Profiling in Immature and Mature Bovine (Bos taurus) Testes. Front Genet 2019, 10:646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Briggs JA, Wolvetang EJ, Mattick JS, Rinn JL, Barry G: Mechanisms of Long Non-coding RNAs in Mammalian Nervous System Development, Plasticity, Disease, and Evolution. Neuron 2015, 88:861–877. [DOI] [PubMed] [Google Scholar]
- 48.Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, et al. : The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res 2012, 22:1775–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wei CW, Luo T, Zou SS, Wu AS: The Role of Long Noncoding RNAs in Central Nervous System and Neurodegenerative Diseases. Front Behav Neurosci 2018, 12:175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pollard KS, Salama SR, Lambert N, Lambot M-A, Coppens S, Pedersen JS, Katzman S, King B, Onodera C, Siepel A, et al. : An RNA gene expressed during cortical development evolved rapidly in humans. Nature 2006, 443:167–172. [DOI] [PubMed] [Google Scholar]
- 51. Rani N, Nowakowski TJ, Zhou H, Godshalk SE, Lisi V, Kriegstein AR, Kosik KS: A Primate IncRNA Mediates Notch Signaling during Neuronal Development by Sequestering miRNA. Neuron 2016, 90:1174–1188.27263970 **This study discovers a primate-specific IncRNA implicated in cortical neurogenesis.
- 52.Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J: The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet 2019, 20:675–691. [DOI] [PubMed] [Google Scholar]
- 53.Dong R, Ma XK, Chen LL, Yang L: Increased complexity of circRNA expression during species evolution. RNA Biol 2017, 14:1064–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Santos-Rodriguez G, Voineagu I, Weatheritt RJ: Evolutionary dynamics of circular RNAs in primates. bioRxiv 2021:2021.2005.2001.442284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hoye ML, Silver DL: Decoding mixed messages in the developing cortex: translational regulation of neural progenitor fate. Curr Opin Neurobiol 2021, 66:93–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Baresic A, Nash AJ, Dahoun T, Howes O, Lenhard B: Understanding the genetics of neuropsychiatric disorders: the potential role of genomic regulatory blocks. Mol Psychiatry 2020, 25:6–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xu K, Schadt EE, Pollard KS, Roussos P, Dudley JT: Genomic and Network Patterns of Schizophrenia Genetic Variation in Human Evolutionary Accelerated Regions. Molecular biology and evolution 2015, 32:1148–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen H, Li C, Zhou Z, Liang H: Fast-Evolving Human-Specific Neural Enhancers Are Associated with Aging-Related Diseases. Cell Syst 2018, 6:604–611 e604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Godlewski J, Lenart J, Salinska E: MicroRNA in Brain pathology: Neurodegeneration the Other Side of the Brain Cancer. Noncoding RNA 2019, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Przytycki PF, Pollard KS: CellWalker integrates single-cell and bulk data to resolve regulatory elements across cell types in complex tissues. Genome Biol 2021, 22:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kampmann M: CRISPRi and CRISPRa Screens in Mammalian Cells for Precision Biology and Medicine. ACS Chem Biol 2018, 13:406–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pickar-Oliver A, Black JB, Lewis MM, Mutchnick KJ, Klann TS, Gilcrest KA, Sitton MJ, Nelson CE, Barrera A, Bartelt LC, et al. : Targeted transcriptional modulation with type I CRISPR-Cas systems in human cells. Nat Biotechnol 2019, 37:1493–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Camp JG, Badsha F, Florio M, Kanton S, Gerber T, Wilsch-Bräuninger M, Lewitus E, Sykes A, Hevers W, Lancaster M, et al. : Human cerebral organoids recapitulate gene expression programs of fetal neocortex development. Proceedings of the National Academy of Sciences 2015:201520760–201520766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Otani T, Marchetto MC, Gage FH, Simons BD, Livesey FJ: 2D and 3D Stem Cell Models of Primate Cortical Development Identify Species-Specific Differences in Progenitor Behavior Contributing to Brain Size. Cell stem cell 2016, 18:467–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Agoglia RM, Sun D, Birey F, Yoon SJ, Miura Y, Sabatini K, Paşca SP, Fraser HB: Primate cell fusion disentangles gene regulatory divergence in neurodevelopment. Nature 2021, 592:421–427.* [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Gokhman D, Agoglia RM, Kinnebrew M, Gordon W, Sun D, Bajpai VK, Naqvi S, Chen C, Chan A, Chen C, et al. : Human-chimpanzee fused cells reveal cis-regulatory divergence underlying skeletal evolution. Nat Genet 2021, 53:467–476.33731941 *These studies establish new models for studying evolution using cell fusions from human and chimpanzee.