Abstract
Misdirected immunity gives rise to the autoimmune tissue inflammation of rheumatoid arthritis (RA), in which excess production of the cytokine TNF is an important pathogenic event. Mechanisms underlying the breakdown of self-tolerance are unclear, but T cells in the arthritic joint have a distinctive metabolic signature of ATPlo acetyl-CoAhi pro-inflammatory effector cells. Here, we identified a deficiency of mitochondrial aspartate production as a key abnormality in these autoimmune T cells. Shortage of mitochondrial aspartate disrupted the regeneration of the metabolic cofactor nicotinamide adenine dinucleotide (NAD), causing ADP-deribosylation of the endoplasmic reticulum (ER) sensor GRP78/BiP. As a result, ribosome-rich ER membranes expanded, promoting co-translational translocation and enhanced biogenesis of the transmembrane cytokine TNF. ERrich T cells were the predominant TNF producers in the arthritic joint. Transfer of intact mitochondria into T cells as well as supplementation of exogenous aspartate rescued the mitochondria-instructed expansion of ER membranes and suppressed TNF release and rheumatoid tissue inflammation.
Introduction
Rheumatoid arthritis (RA) is a classical autoimmune disease with a decade-long preclinical phase during which patients lose self-tolerance and begin to produce autoantibodies against post-translationally modified proteins1. During clinically apparent RA, innate and adaptive immune cells infiltrate into the synovial membrane and together with joint-endogenous cells form a tissue-destructive pannus. Tumor necrosis factor-α (TNF) functions as a key cytokine in RA pathogenesis and TNF inhibitors have fundamentally reshaped the therapeutic landscape of RA2,3. TNF is mostly considered an innate cytokine but recent single-cell transcriptomics of synovial cells have assigned the highest TNF transcripts to T cells4. Pathomechanism leading to aberrant TNF production are unknown, necessitating current anti-TNF therapies to rely exclusively on blocking the secreted cytokine. Lack and loss of efficacy and side effects have raised the demand for more precise targeting of inappropriate TNF secretion5. Also, the promise of early immune modulation in at-risk individuals prior to clinical disease requires molecular definition of underlying processes.
Cell type-specific TNF knockout mouse models have demonstrated distinct and nonredundant functions of T-cell and macrophage-derived TNF, both in anti-bacterial immunity and in induction of autoimmunity6. TNF-producing Th1 cell expansion in both blood and liver are typical for patients with autoimmune hepatitis7. Activated T cells, via the release of TNF, instruct myeloid cells to produce inflammasome-independent IL-1β to cause autoimmunity8. A distinct tissue-resident population of TNF-producing CD4+ T cells promotes mucosal development and mediates inflammation in necrotizing enterocolitis9. In RA, T cells are key pathogenic drivers required for very early, intermediate, and late stages of the disease process10,11. Required for autoantibody production, they also sustain synoviocyte proliferation, tissue inflammation, neoangiogenesis, and cartilage and bone erosion10,12. In mature synovitis, T cells are one of the most abundant cell types, comprising 30–50% of synovial tissue cells13. T-cell-directed TNF overexpression is sufficient to promote arthritis, wasting syndrome, and organ necrosis14. In RA patients, naïve CD4+ T cells are imprinted with a metabolic signature that biases their differentiation into short-lived effector T cells (SLEC) instead of long-lived memory precursor15. Key elements of the metabolic program are a slowdown in glycolysis and glucose shunting to the biosynthetic pentose phosphate pathway, facilitated by a shift in the PFKFB3/G6PD ratio16,17. A major abnormality is the transcriptional repression of SUCLG2, a key enzyme in the mitochondrial TCA cycle18. Unable to convert α-ketoglutarate into succinate and impaired in oxidative phosphorylation, RA T cells revert the TCA cycle and accumulate acetyl-CoA. Together with surplus NADPH production and a reductive environment, acetyl-CoA oversupply expedites lipogenesis and membrane formation, supporting the differentiation of tissue-invasive T cells16,18,19. Defects in mitochondrial DNA repair20 as well as misrouting of AMPK away from the lysosomal surface21 sustain the defect in mitochondrial fitness and enable persistent mTOR activation, promoting differentiation into SLEC despite the lack of ATP.
Current data show that the T cells’ disposition for membrane expansion extends to the endomembrane system and supports growth of the endoplasmic reticulum (ER), where it affects the biogenesis of the transmembrane molecule TNF. Mitochondrial insufficiency in RA T cells was coupled with enlargement of ER membranes. Specifically, lack of mitochondrial aspartate production prevented cytosolic NAD+ regeneration, causing deribosylation of the ER stress sensor GRP78/BiP and dissociation from the kinase/ endoribonuclease inositol-requiring enzyme 1 α (IRE1α). The resulting expansion of ribosome-rich ER membranes enhanced post-translational translocation of the type II transmembrane protein TNF, turning RA T cells into TNF superproducers. Transfer of intact mitochondria into T cells or aspartate supplementation successfully suppressed TNF production and treated synovial tissue inflammation. These data connect autoimmune tissue inflammation to dysfunctional inter-organelle communication and define mitochondrial intactness in T cells as a key component of tissue tolerance.
Results
Mitochondrial insufficiency promotes ER expansion.
Naïve CD4+ T cells from RA patients have a mitochondrial defect, manifesting with low oxygen consumption and low mitochondrial membrane potential (MMP) (Fig. 1a–c), while mitochondrial mass is similar between healthy and RA T cells (Extended Data Fig. 1). To understand how the lack of mitochondrial fitness impacts other organelles, we quantified endoplasmic reticulum (ER) mass. Flow cytometry for ER-positioned ATP-sensitive K+ channels (marked by ER Tracker) and confocal imaging for the ER-resident enzyme protein disulfide isomerase (PDI) revealed an inverse relationship between the mitochondrial membrane potential and the biomass of the ER (Fig. 1d,e). Cell size-restricted gating (Extended Data Fig. 2) confirmed the negative correlation between MMP and ER-Tracker intensity in all subpopulations (Fig. 1f). Compared to healthy T cells, CD4+ T cells from RA patients had a higher ER Tracker signal (Fig. 1g) and expressed more of the ER chaperone Calnexin (Fig. 1h, i). In contrast, ER Tracker signals in activated CD4+ T cells from patients with psoriatic arthritis (PsA) were indistinguishable from age-matched controls (Fig. 1j), indicating that the expansion of ER size is not simply a consequence of systemic inflammation. RA T cells synthesized significantly more phosphatidylcholine, one of the major phospholipids needed for biomembrane generation (Fig. 1k). Transmission electron microscopy confirmed highly abundant and elongated ER structures in activated CD4+ T cells from RA patients (Fig. 1l).
Fig. 1. Mitochondrial insufficiency and ER expansion in rheumatoid arthritis.

Naïve CD4+CD45RA+ T cells from patients with RA or psoriatic arthritis (PsA) and age-matched healthy individuals were stimulated for 72 h. (a-c) Reduced mitochondrial fitness in RA T cells. (a) Basal mitochondrial oxygen consumption rates (OCR) measured by Seahorse Analyzer. n=6 each. (b, c) Mitochondrial membrane potential measured by flow cytometry (MitoTracker Red). n=7 each. (d) Correlation of ER size (ER Tracker MFI) and mitochondrial membrane potential in healthy T cells by Linear regression (n=10). (e) Representative confocal microscopy imaging of the ER enzyme protein disulfide isomerase (PDI) and mitochondrial membrane potential (MitoTracker Red) from 3 independent experiments. Green arrow marks a cell with abundant ER, red arrow marks a cell with high mitochondrial activity. Scale bar, 10 μm. (f) Correlation of ER size (ER Tracker intensity) and mitochondrial membrane potential in cells gated based on cell size. Linear regression with confidence interval shown in gray around the regression line. (g-l) Expanded ER size in RA T cells. (g) Flow cytometric quantification of ER size (ER Tracker MFI) in RA and healthy control T cells; n = 8 each. (h, i) Confocal microscopy imaging of the ER chaperon protein calnexin in RA and healthy T cells. Scale bar, 10 μm. Single cell calnexin intensity quantification (n=50 T cells from 5 healthy and 5 RA samples). (j) Flow cytometric quantification of ER size in T cells from PsA patients and healthy donors; n = 8 each. (k) Intracellular phosphatidylcholine concentrations in healthy and RA T cells (Healthy: n=6; RA: n =8). (l) Representative transmission electron microscope image of healthy and RA T cells from 30 cells from 3 healthy and 3 RA samples. White arrows indicate ER. Scale bar,1 μm. All data are mean ± SEM. Two-tailed unpaired Mann-Whitney-Wilcoxon rank test (a, c, g-k). *P < 0.05, **P < 0.01, ***P < 0.001.
ER biogenesis and expansion are partially controlled by ER stress-related signals22,23. We therefore explored whether ER stress contributes to the ERrich phenotype of RA T cells. When activated, naive CD4+ T cells from RA patients disproportionally upregulated the ER stress gene signature (Fig. 2a). We verified that ER stress promotes expansion of the ER membrane system by overexpressing the active form of XBP1 (Xbp-1s) in healthy T cells and quantifying ER biomass (Extended Data Fig. 3a–e). To address whether immuno-modulatory treatment could affect ER biogenesis, we compared expression level of XBP1S in patients with or w/o Methotrexate (MTX) treatment; no significant difference was observed in these two patient cohorts (Extended Data Fig. 3f). To define whether mitochondrial activity regulates ER stress signaling, T cells were sorted into MMPlo and MMPhi subpopulations (Fig. 2b). ER stress gene expression was a feature of T cells with low mitochondrial activity (Fig. 2c). Distinct to RA T cells, PsA patient-derived T cells did not have abnormal ER stress signals (Fig. 2d).
Fig. 2. Mitochondrial insufficiency promotes ER expansion.
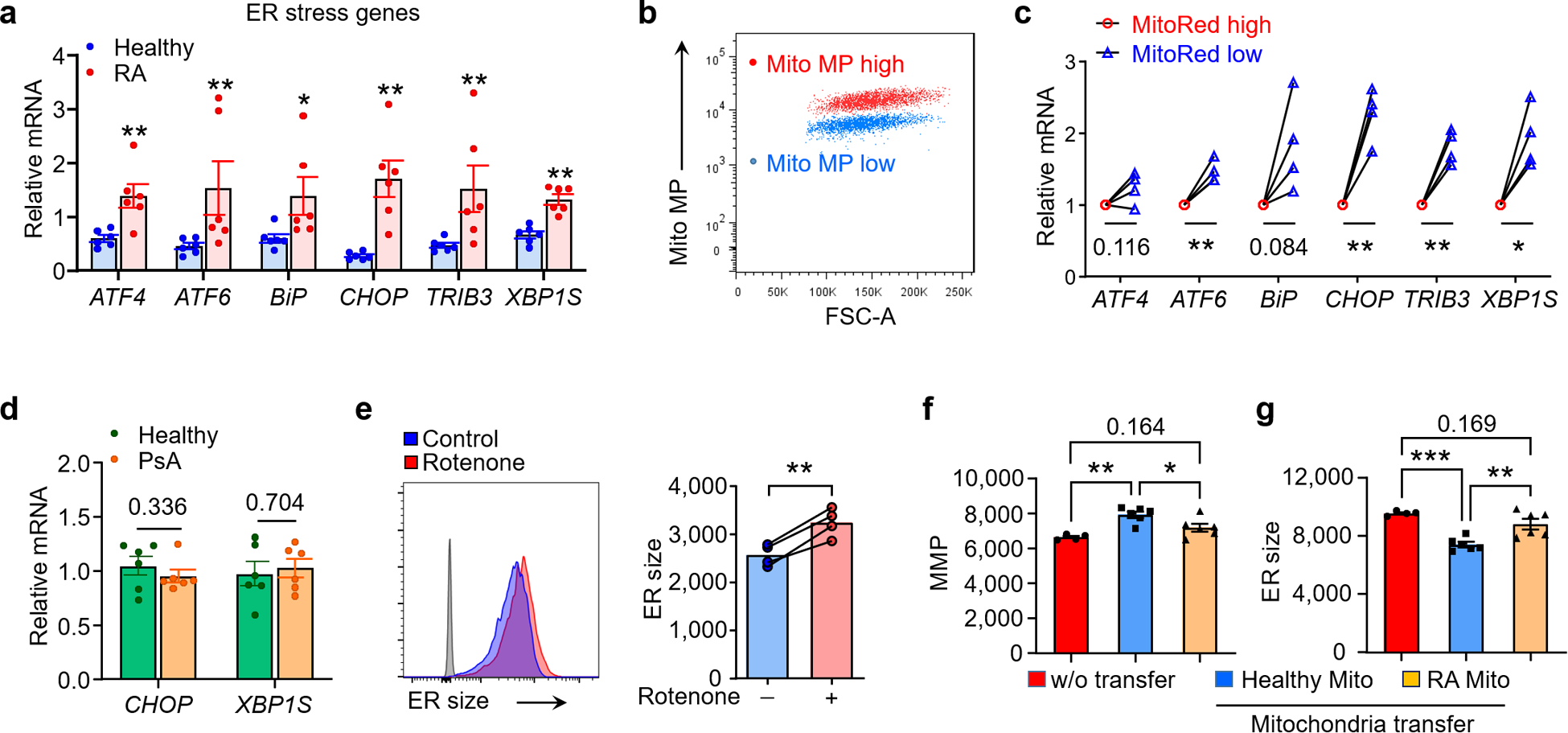
(a) ER stress gene expression (qPCR) in healthy and RA T cells (Healthy: n=6; RA: n =6). (b, c) Separation of two CD4+ T cell subpopulations based on mitochondrial membrane potential and qPCR analysis of ER stress genes in Mito MPhi and Mito MPlo T cells, n=4. (d) Comparison of CHOP, XBP1S mRNA level in healthy and PsA T cells, n=6. (e) Mitochondrial stress expands ER size. Healthy naïve T cells were activated for 72 h with or w/o the complex I inhibitor Rotenone (10nM). Histogram of ER Tracker staining and collective MFI, n=4. (f, g) Mitochondrial transfer corrects ER size. CD4+CD45RA+ T cells from RA patients were stimulated with anti-CD3/CD28 for 48 h. mitochondria isolated from healthy or RA CD4+ T cells were transferred into RA T cells (donor cell number/recipient cell number=10:1). Mitochondrial membrane potential (f) and ER size (g) in RA T cells after mitochondria transfer, n = 6. All data are mean ± SEM. Two-tailed unpaired Mann-Whitney-Wilcoxon rank test (a, d). Two-tailed paired t test (c, e). One-way ANOVA and post-ANOVA pair-wise two-group comparisons conducted with Tukey’s method (f, g). *P < 0.05, **P < 0.01, ***P < 0.001.
To examine whether mitochondria-derived signals determine ER size and function, we quantified ER-associated membranes after inhibiting the electron transport chain complex I with Rotenone (10 nM). Impaired electron transport prompted an increase in ER mass (Fig. 2e). Other mitochondrial respiration inhibitors (Piericidin A, Antimycin A, Oligomycin) had similar effects (Extended Data Fig. 4). To repair defective mitochondria in RA T cells, exogenous mitochondria were harvested and transferred into RA recipient cells (Extended Data Fig. 5a). Mitochondrial transfer at a donor/recipient cell ratio of 10:1 yielded excellent improvement in MMP (Extended Data Fig. 5b,c). Delivery of exogenous mitochondria from healthy T cells into RA T cells restored the MMP to normal levels (Fig. 2f) and suppressed ER biomass by 30% (Fig. 2g). Mitochondria harvested from RA T cells failed to restore the ER size.
These data identified mitochondrial metabolism as a key regulator of ER biogenesis and classified CD4+ T cells from RA patients as MMPlo ERrich.
Mitochondria-derived aspartate controls ER size.
The fact that supplying healthy mitochondria corrected the inappropriate expansion of ER membranes in RA T cells directed attention to mitochondria-ER inter-organelle communication. Metabolites of mitochondrial origin have been recognized as signal carriers controlling different cellular functions24,25. In previous work, we have described a shift in the α-ketoglutarate (α-KG)/succinate ratio as a distinguishing feature of RA T cells, imposed by disruption of the TCA cycle18. To address which mitochondrial signals restrain ER expansion and clarify how the ER senses impaired mitochondrial fitness, we screened key mitochondrial intermediates. Exogenous α-KG left ER mass unaffected (Fig. 3a), while succinate, malate, and more so, oxaloacetate and aspartate rapidly suppressed formation of ER membranes in RA T cells (Fig. 3a). Aspartate is synthesized in the mitochondrial matrix from malate-oxaloacetate by the transaminase GOT2 and then transported into the cytosol. Under conditions of NADH availability, cytosolic aspartate is converted back into malate to reenter the mitochondria through the malate-aspartate shuttle (Fig. 3b). Activation of healthy T cells resulted in accumulation of aspartate (4 nmol/106 cells), while RA T cells generated less than 2.5 nmol/106 cells (Fig. 3c). Similarly, RA T cells lacked behind in producing adequate concentrations of oxaloacetate, the precursor of aspartate in the TCA cycle (Fig. 3d). The aspartatelo phenotype was not due to insufficient glutamic-oxaloacetic transaminases; GOT1 and GOT2 transcript concentrations were indistinguishable in healthy and RA T cells (Extended Data Fig. 6). Exogenous aspartate corrected the ER stress gene signature (Fig. 2e) and inhibited the inappropriate biosynthesis of phosphatidylcholine, a membrane-building precursor required for ER biomass generation (Fig. 3f). Maintenance of cellular aspartate concentrations required intactness of mitochondrial metabolism. Inhibiting mitochondrial respiration through the complex I inhibitor Rotenone dramatically decreased aspartate levels in healthy T cells (Fig. 3g), while the transfer of healthy mitochondria into RA T cells restored aspartate generation (Fig. 3h). Knockdown of GOT2, the gate keeper enzyme in aspartate synthesis, successfully mimicked the aspartatelo phenotype of RA T cells (Fig. 3i), and healthy T cells responded to GOT2 loss-of-function with increased formation of ER membranes and upregulation of the ER stress gene signature (Fig. 3j,k). Exogenous aspartate rescued the ER expansion induced by GOT2 knockdown (Fig. 3j).
Fig. 3. Mitochondria-derived aspartate controls ER size.

Naïve CD4+CD45RA+ T cells from patients with rheumatoid arthritis (RA), and age-matched healthy individuals were stimulated for 72 h. (a) Mitochondrial intermediates determine ER size. RA T cells were supplemented with the indicated mitochondrial intermediates (all 1 mM) and ER size was quantified flow cytometrically, n=6. (b) Scheme for the malate/aspartate shuttle. (c, d) RA T cells are aspartate/oxaloacetate deficient. Intracellular aspartate (Healthy: n=8; RA: n =8) (c) and oxaloacetate (Healthy: n=6; RA: n=6) (d) concentrations in T cells. (e, f) Aspartate inhibits ER stress signals and phosphatidylcholine synthesis. RA T cells were treated with aspartate for 3 days. ER stress gene expression (n=5) (e) and Phosphatidylcholine content (n=4) (f) were measured. (g, h) Aspartate concentrations depend on intact mitochondrial function. Intracellular aspartate concentrations were measured after treatment of healthy T cells with the complex I inhibitor Rotenone (10nM; n=3) or after the transfer of healthy mitochondria into RA T cells (n=3). (i-k) Glutamic-Oxaloacetic Transaminase 2 (GOT2) regulates ER size. GOT2 was knocked down by si-RNA in healthy T cells. (i) Intracellular aspartate levels, n=5; (j) ER size quantified by flow cytometric analysis (ER Tracker MFI), n=4; (k) ER stress genes quantified by qPCR, n=4. All data are mean ± SEM. One-way ANOVA and post-ANOVA pair-wise two-group comparisons conducted with Tukey’s method (a, j), P-values from comparison with control group. Two-tailed paired t test (e-i, k). Two-tailed unpaired Mann-Whitney-Wilcoxon rank test (c, d). *P < 0.05, **P < 0.01, ***P < 0.001.
These data identified the amino acid aspartate as a sensitive biomarker of mitochondrial respiration and as a signal transducer between mitochondria and the ER membrane system.
Aspartate supplementation suppresses tissue inflammation.
To test whether the aspartatelo phenotype in RA T cells is disease-relevant and if supplementation of aspartate can protect tissue from inflammatory damage, we induced synovitis in human synovium-engrafted NSG mice and treated the chimeras with aspartate. RA PBMC induced robust synovitis, which was successfully suppressed by aspartate treatment (Fig. 4a). Transcriptomic analysis of the tissue lesions revealed a marked beneficial effect of aspartate supplementation lowering T cell receptor (TRB) expression and curtailing TBX21 and RORG mRNA accrual (Fig. 4b). Immunohistochemical analysis confirmed that the frequency of tissue-residing CD3+ IFN-γ+ T cells was highly dependent on aspartate availability, declining by 65% with aspartate supplementation (Fig. 4c). mRNA levels for the inflammatory mediators IFN-γ, IL-17, IL-6, TNF, IL-1β all decreased (Fig. 4d), indicating successful containment of tissue inflammation. Conversely, the anti-inflammatory genes TGFB and IL10 were not affected.
Fig. 4. Aspartate is anti-inflammatory.
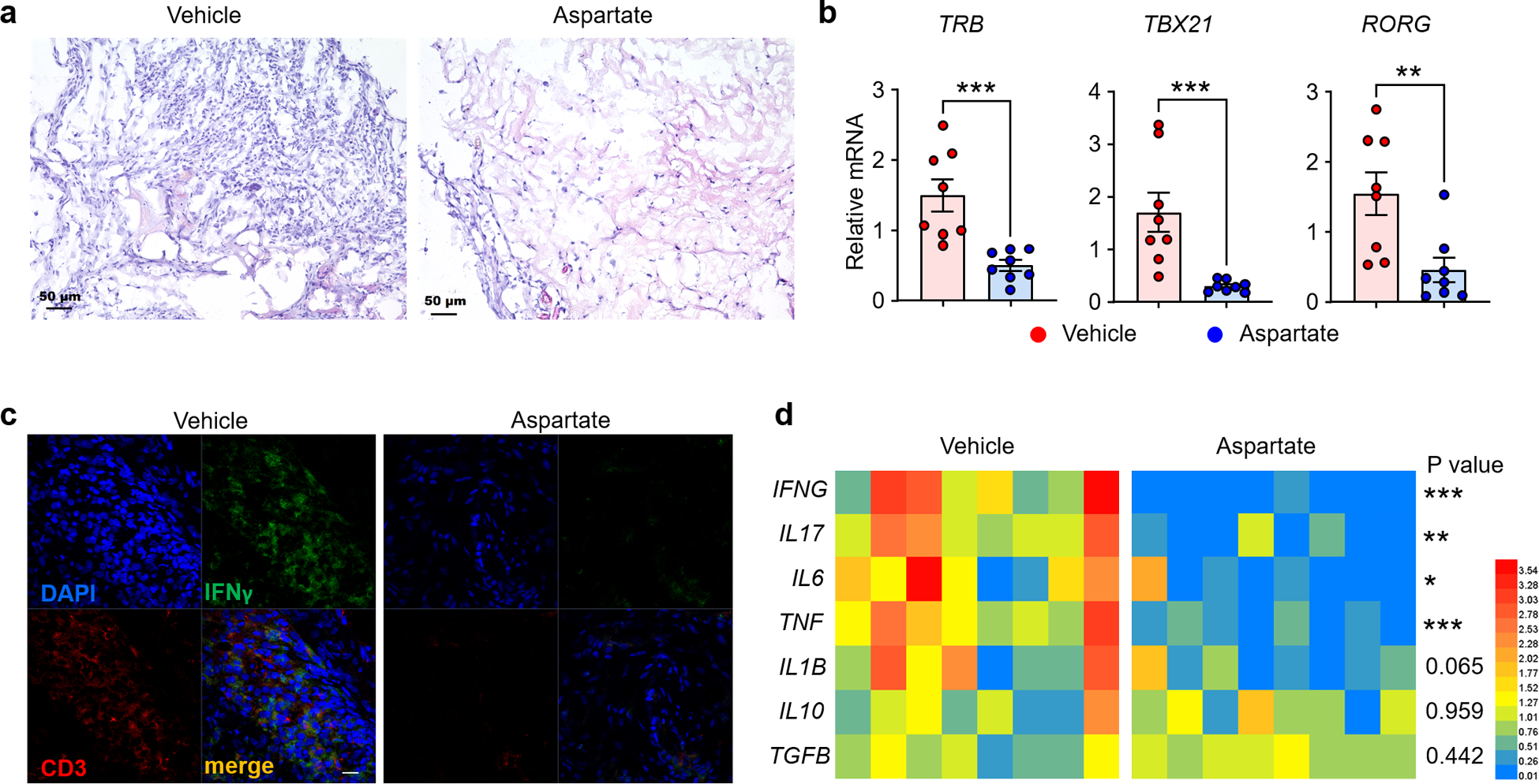
Synovitis was induced in chimeric mice engrafted with human synovial tissue and immuno-reconstituted with RA PBMC. Chimeras were treated with vehicle or aspartate i.p. 5 mg/kg. (a) Representative H&E staining of explanted synovial tissue, n=8 (b) Tissue transcriptomic analysis (qPCR) of synovial explants. Shown are data for T cell receptor (TRB) transcripts and the lineage-determining transcription factors TBX21 and RORC (n=8). (c) Representative image of co-immunofluorescence staining for IFN-γ-producing CD3+ T cells in the synovial tissue, n=8; scale bar,10 μm. (d) Tissue transcriptomic analysis of key inflammatory cytokines. 8 tissues in each study arm. All data are mean±SEM. Two-tailed unpaired Mann-Whitney-Wilcoxon rank test. *P < 0.05, **P < 0.01, ***P < 0.001.
Together, these data identified the mitochondrial intermediate aspartate as a regulator of ER mass and as an anti-inflammatory mediator.
Aspartate regulates BiP ADP-ribosylation.
The malate-aspartate shuttle functions primarily to translocate electrons produced during glycolysis into the mitochondria for oxidative phosphorylation26. Essentially, aspartate is dispatched from mitochondria as an electron acceptor, facilitating the cytoplasmic regeneration of NAD+ from NADH, and then malate reenters the mitochondria as an electron carrier (Fig. 3b). To test whether NAD+ and NADH concentrations are affected by aspartate shortage, we measured intracellular NAD+/NADH in healthy and patient-derived CD4+ T cells. Healthy T cells reached NAD+ levels of 550 pmol/106 cells and NADH concentrations of 70 pmol/106 cells, resulting in a NAD+/NADH ratio exceeding 7. With reduced NAD+ and elevated NADH, RA T cells achieved only an NAD+/NADH ratio of 3 (Fig. 5a–c). Treatment with aspartate or transfer of exogenous mitochondria significantly improved NAD+ regeneration (Fig. 5d, e). Inhibiting mitochondrial respiration with the complex I inhibitor Rotenone essentially prevented conversion of NADH into NAD+ (Fig. 5f), implicating aspartate and mitochondrial intactness in maintaining the NAD+/NADH balance. To examine whether NAD+ functions similarly to aspartate in regulating ER mass/function, RA T cells were activated with or without NAD+ supplement. Like aspartate, exogenous NAD+ lowered the ER biomass (Fig. 5g, h) and reduced the ER stress signature (Fig. 5i).
Fig. 5. Aspartate is required for NAD regeneration and ADP-ribosylation of BiP.
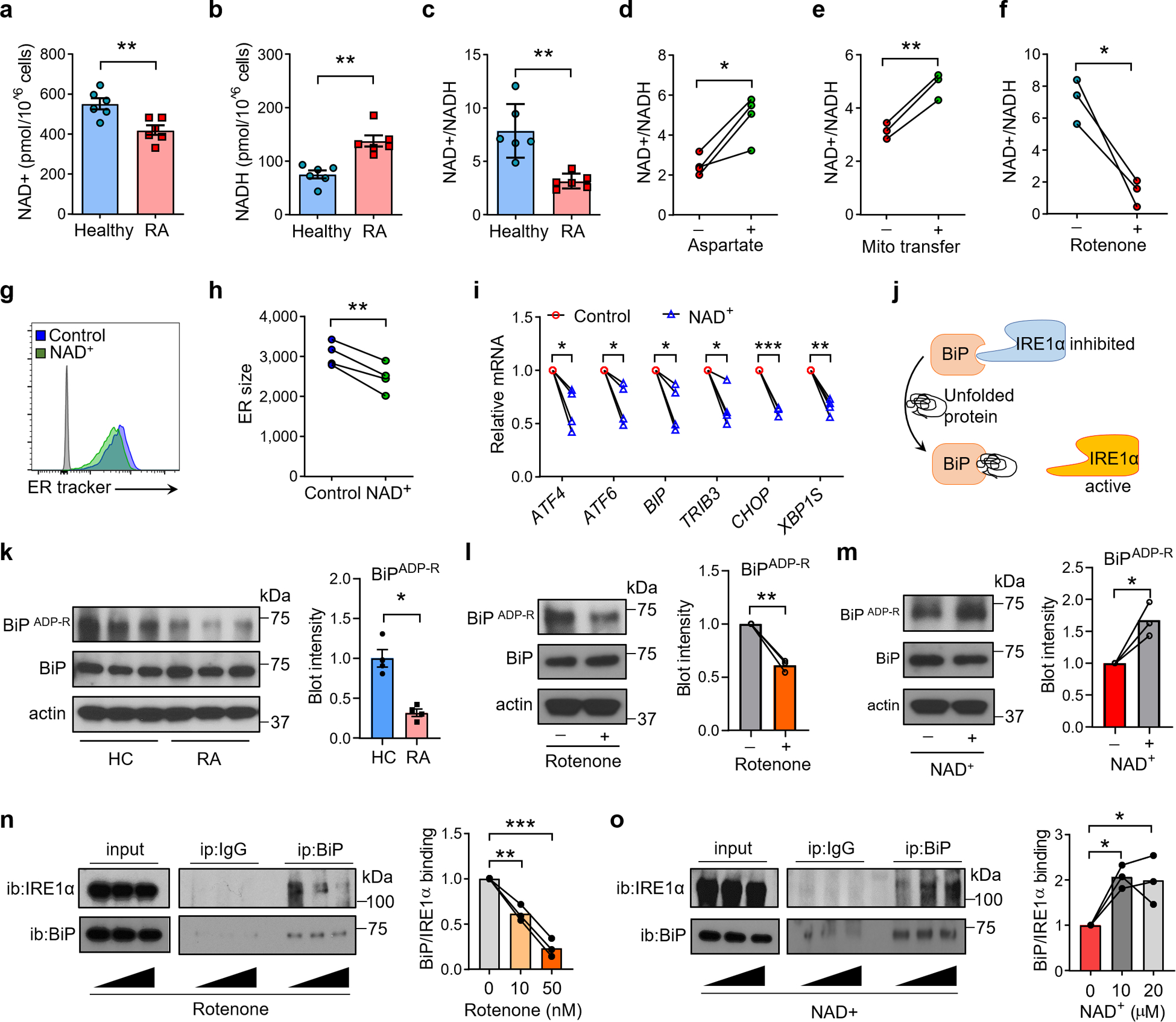
Naïve CD4+CD45RA+ T cells from patients with rheumatoid arthritis (RA), and age-matched healthy individuals were stimulated for 72 h. (a-c) NAD+ deficiency in RA T cells. Quantification of intracellular NAD+, NADH and the NAD/NADH ratio in healthy and RA T cells, n=6. (d-f) Aspartate and intact mitochondria regenerate NAD+. (d) NAD+/NADH ratios in RA T cells treated with or w/o aspartate, n=4. (e) NAD+/NADH ratios in RA T cells with or w/o mitochondrial transfer, n=3. (f) NAD+/NADH ratios in healthy T cells treated with or w/o the complex I inhibitor Rotenone (10 nM), n=3. (g-i) NAD+ controls ER size and ER stress. RA T cells were treated with or w/o NAD. ER size was determined flow cytometrically (ER Tracker MFI). (g, h) Representative histograms. ER size measurements from 4 experiments. (i) ER stress gene expression profiling (qPCR) in RA T cells treated with or w/o NAD+, n=4. (j-o) NAD-dependent ribosylation of BiP prevents ER expansion and stabilizes Ire-1a binding. (j) Scheme of IRE1-α activity controlled by BiP in the ER lumen. (k) ADP-ribosylation of BiP in healthy and RA T cells, n=3. (l) ADP-ribosylation of BiP in healthy CD4+ T cells treated with or w/o Rotenone (10 nM) for 24 h. n=3. (m) ADP-ribosylation of BiP in RA CD4+ T cells treated with or w/o NAD (10 μM) for 24 h. n=3. (n) BiP- IRE-1α binding in healthy CD4+ T cells treated with 0, 10, 50 nM rotenone for 24 h. n=3. (o) BiP- IRE-1α binding in activated RA CD4+ T cells treated with 0, 10, 20 μM NAD for 24 h. n=3. All data are mean ± SEM. Two-tailed unpaired Mann-Whitney-Wilcoxon rank test (a-c, k). Two-tailed paired t test (d-i). One-way ANOVA and post-ANOVA pair-wise two-group comparisons conducted with Tukey’s method (n, o). *P < 0.05, **P < 0.01, ***P < 0.001.
Collectively, these data delineated a mechanistic connection between insufficient mitochondrial aspartate production, failed regeneration of the electron acceptor NAD+ and expansion of the ER membrane system.
GRP78/BiP is a master regulator for the ER stress response27,28, capable to bind to all three major ER stress mediators (PERK, ATF6 and IRE1α). Under ER stress conditions, BiP senses and binds to unfolded proteins, releasing PERK, ATF6 and IRE and triggering their respective ER stress pathways (Fig. 5j). Given the low NAD+ availability in RA T cells, we tested whether NAD-dependent ADP-ribosylation is relevant for BiP function. When compared to healthy T cells, a much lower fraction of BiP molecules in RA T cells were ADP-ribosylated (Fig. 5k). The BiP-ADP-Rlo phenotype could be reproduced by blocking the electron transport chain with Rotenone (Fig. 3l) or Piercidin A (Extended Data Fig. 7). In contrast, surplus exogenous NAD+ restored ADP-ribosylation of BiP in RA T cells (Fig. 5m).
To link ADP-ribosylation to BiP function, we analyzed the binding between BiP and its target protein IRE1α by immunoprecipitation. Increasing doses of Rotenone disrupted the binding of BiP to IRE1α (Fig. 5n), whereas NAD+ supplementation doubled BiP-IRE1α complex formation (Fig. 5o).
Together, these data define NAD+-dependent ADP-ribosylation of BiP as an on-off switch of ER stress signals, placing mitochondrial fitness upstream of ER size and function, and specifying aspartate as a mitochondria-to-ER messenger.
Expansion of the rough ER increases co-translational translocation in RA T cells.
To explore the functional consequence of ER expansion in RA T cells, we focused on cytokine production/secretion, which relies heavily on the endomembrane system. We first quantified ribosome-rich rough ER in healthy and patient-derived T cells (Fig. 6a–d). ER biomass was expanded in RA T cells, as indicated by the increased load of the ER chaperone calnexin while global ribosomal protein levels were indistinguishable in healthy and RA T cells (Fig. 6a, b). To relate the expansion of the ER in RA T cells to their functional behavior, we focused on the ribosome-bonded rough ER, responsible for co-translational translocation. Immunoblotting showed high purity of rough ER when isolated out of the T cell cytosol (Extended Data Fig. 8a). T cells responded to activation with a dramatic increase in the ribosome-occupied rough ER membrane system (Extended Data Fig. 8b). RA T cells had a >2fold enlargement of ribosome-occupied ER membrane sheets (Fig. 6c, d).
Fig. 6. Expansion of the rough ER increases co-translational translocation in RA T cells.

Naïve CD4+CD45RA+ T cells from RA patients and age-matched controls were stimulated for 72 h. (a-e) Enrichment of rough ER in RA T cells. (a, b) Immunoblot analysis of the ER chaperon protein calnexin and the ribosomal proteins L-17 and S-7 in CD4+ T cells from 5 healthy individuals and 6 RA patients. (c, d) Rough ER was isolated from healthy and RA T cells 2 h after restimulation. (c) Calnexin, ribosomal S7 and β-actin were quantified by immunoblotting. (d) Quantification of blot intensity for rough ER in each group, n=5 healthy and 4 RA. (e) TNF is the predominant cytokine expressed by activated naïve CD4+ T cells. Transcripts for T cell effector cytokines in activated naïve CD4+ T cells quantified by qPCR, n=3. (f) Healthy and RA T cells express similar level of TNF mRNA. TNF mRNA concentrations in healthy and RA T cells before (n=8 healthy and 8 RA) and after (n=5 healthy and 7 RA) PMA/ION stimulation (qPCR). (g) T cell stimulation induces enrichment of ER-bound mRNA for secretory proteins. Fold change of ER-bound mRNA for secretory proteins and intracellular proteins after PMA/ION stimulation, n=3. (h) Enrichment for ER-bound TNF mRNA in RA T cells. Rough ER was isolated from healthy and RA T cells 2 h after stimulation. mRNA associated with rough ER was quantified by qPCR. n=6. All data are mean ± SEM. Two-tailed unpaired Mann-Whitney-Wilcoxon rank test (b, d, f, h). *P < 0.05, **P < 0.01.
Membrane-integrated and secretory protein synthesis involves localization of ribosomes to the cytosolic surface of the ER and co-translational docking of the mRNA/ribosome complex on the ER membrane29,30. To understand the functional implications of the enlarged rough ER in RA T cells, we analyzed mRNAs contained in the ER-bonded ribosomes. Considering that the rough ER sheets serve as the main site of synthesis for secreted and membrane-integrated proteins, we concentrated on T cell effector cytokines. In naïve CD4+ T cells, transcripts for the lineage-determining effector cytokines IFN-γ, IL-4 and IL-17 were barely detectable, but IL2, and even more so, TNF mRNA were abundant (Fig. 6e). Considering the critical role of TNF in the RA disease process, we focused on the transcription and translation of this cytokine. The overall pool of TNF mRNA in resting and stimulated T cells from healthy individuals and patients were indistinguishable (Fig. 6f). To explore whether the expansion of ER membrane sheets in RA T cells impacts TNF transcription, ER-bound mRNAs were measured upon PMA/Ionomycin activation. As expected, non-secretory cytoplasmic proteins, including the glycolytic enzyme GAPDH and the cytoskeletal protein ACTIN, were underrepresented amongst ER-bound mRNAs (Fig. 6g). mRNA for the 4 cytokines IFN-γ, IL-2, IL-17 and TNF were all highly enriched, with IL2 and TNF mRNA being >100fold more abundant at the ER membrane (Fig. 6g). With their expanded ER morphology, RA CD4+ T cells recruited a higher proportion of TNF mRNA to the organelle’s surface (Fig. 6h), where selected mRNAs become subject to co-translational translocation31.
T cells rich in rough ER are TNF superproducers.
The redistribution of TNF mRNA resulted in highly efficient TNF biogenesis (Fig. 7a–c). In patient-derived T cells, confocal imaging revealed a strong signal for TNF co-localizing with calnexin as well as embedded into the plasma membrane (Fig. 7a). Intracellular staining yielded higher concentrations of TNF protein in RA compared to healthy T cells (Fig. 7b). Quantification of secreted TNF confirmed the increased cytokine release in patient-derived cells (Fig. 7c). We tested whether the unfolded protein response (UPR), a stress response program triggered by the accumulation of unfolded proteins in the ER lumen, is sufficient to enhance TNF biogenesis and can mimic the ERrich phenotype of RA T cells. Treatment of healthy CD4+ T cells with tunicamycin failed to upregulate TNF synthesis (Extended Data Fig. 9a,b), suggesting that the mitochondria-induced ER expansion in RA T cells represents a state of ER stress without ER dysfunction.
Fig. 7. ERrich RA T cells are TNF-superproducers.

Naïve CD4+CD45RA+ T cells from RA patients and age-matched controls were stimulated for 72 h. (a) Representative confocal image of the ER chaperone protein Calnexin and TNF in healthy and RA T cells, n=3 independent experiments; scale bar,10 μm. (b) Flow cytometric measurement of intracellular TNF in CD4+ T cells from RA patients and healthy individuals before and after PMA/ION stimulation (Healthy: n=5; RA: n =5). (c) TNF secreted into the extracellular space by unstimulated (Healthy: n=4; RA: n =6) and stimulated (Healthy: n=4; RA: n =5) RA and control CD4+ T cells. (d-i) Mitochondrial function and aspartate control TNF production. (d, e) Electron transfer was inhibited in healthy T cells with Rotenone (10 nM) or Piercidin A (10 pM). TNF was measured by flow cytometry, n=4 in each series. (f) GOT2 knockdown in healthy T cells, combined with or without aspartate rescue. TNF was measured by flow cytometry, n=4. (g) TNF production in RA T cells treated with or w/o aspartate, n=3. (h) TNF production in RA T cells reconstituted with or w/o healthy mitochondria, n=3. (i) TNF production in RA T cells treated with or w/o NAD+, n=4. (j) Scheme showing the aspartate-NAD-BiP pathway controlling TNF secretion. All data are mean ± SEM. Two-tailed unpaired Mann-Whitney-Wilcoxon rank test (b, c). Two-tailed paired t test (d-i). *P < 0.05, **P < 0.01, ***P < 0.001.
The mechanistic link between ER expansion and enhanced recruitment of TNF mRNA to the ER surface resulting in a TNF superproducer phenotype raised the question whether correcting the growth of the endomembrane system could repair the uncontrolled cytokine release. Disrupting mitochondrial function with the respiration inhibitors Rotenone and Piercidin A or by knocking down GOT2 mRNA transformed healthy T cells into TNFhi producers (Fig. 7d–f). GOT2 lose-of-function could be rescued by supplementation of aspartate (Fig. 7f). Three interventions repaired the excessive TNF secretion of RA T cells: the supplementation of exogenous aspartate or NAD+ and the transfer of intact mitochondria into the patients’ T cells (Fig. 7g–i). Interestingly, asparagine, the amino acid structurally similar to aspartate, failed to affect ER size and TNF production (Extended Data Fig. 9c,d), indicating a unique role of aspartate in the NAD+/NADH metabolism. Finally, we tested the metabolic intermediates pyruvate and α-ketobutyrate, which can regenerate NAD+ through a distinct mechanism32. Both metabolites inhibited ER expansion in RA T cells and suppressed TNF production (Extended Data Fig. 9e,f).
Collectively, these data indicate that the enlargement of ribosome-occupied ER sheets in RA T cells have profound implications for TNF biogenesis by enhancing the process of co-translational translocation (Fig. 7j).
Enhanced TNF production by CD4+ T cells is arthritogenic.
TNF is a prime therapeutic target in RA and specific blockade of the cytokine is now considered standard therapy33. The propensity of the patients’ CD4+ T cells to release high amounts of TNF therefore raised the question whether such T cells have disease relevance. Rheumatoid synovial lesions consist of a mixture of cell types, most prominently, T cells, B cells, macrophages, and synovial fibroblasts. To implicate some or all cell populations in TNF production, we first analyzed recently published single cell RNAseq data4. Tissue-residing T cells contained abundant amounts of TNF mRNA, while macrophages and B cells were low positive, and fibroblasts were negative. In a second step, we allocated TNF production to the different cellular subsets by determining intracellular TNF by flow cytometry. Synovial tissues from RA patients were disaggregated and stimulated with LPS/PMA/ionomycin in the presence of the Golgi blocker Brefeldin A (BFA). Intracellular TNF was measured cytometrically in T cells (CD45+CD3+), B cells (CD45+CD19+) and macrophages (CD45+CD68+). In leukocyte-rich synovial tissues (Fig. 8a), 80% of T cells and B cells were TNF+, while only 40% of macrophages produced TNF (Fig. 8b, c). On a per cell basis, TNF was highly abundant in T cells (Fig. 8d), multifold higher than in B cells and macrophages. In leukocyte-poor RA tissues, B cells were barely detected (Fig. 8e), but most T cells were able to produce high amounts of TNF (Fig. 8f–h).
Fig. 8. TNF-producing CD4+ T cells function as key arthritogenic effector cells.

(a, e) Cellular composition of leucocyte-rich and leukocyte-poor tissues collected from patients with rheumatoid synovitis. (b-d, f-h) TNF is a product of tissue T cells. Flow cytometric analysis of intracellular TNF in T cells, B cells and macrophages after stimulation with LPS/PMA/ION/BFA for 4h. (b, f) Histogram of TNF staining. (c, g) Frequencies of TNF-producing cell populations. (d, h) MFI of TNF staining in different cell populations. (i-k) Spontaneous TNF production in T cells and macrophages residing in the synovium. Freshly harvested synovial tissue from RA patients was incubated with or w/o the secretion inhibitor BFA for 4 h, before cells were dissociated from the tissue and intracellular TNF was detected by flow cytometry. TNF+ CD45+ CD68+ macrophages (i) and TNF+ CD45+ CD3+ T cells (j) in synovial tissue before and after BFA treatment. (k) Fold change in the frequency of TNF+ macrophage and TNF+ T cells after BFA treatment. n=3 tissues. (l-n) TNF-producing CD4+ T cells are an absolute requirement for rheumatoid synovitis. Rheumatoid synovitis was induced in human synovial tissues engrafted into NSG mice. CD4+ T cells from RA patients were transfected with control or TNF siRNA and adoptively transferred into the chimeric mice. Synovial grafts were explanted two weeks later. (l) H&E staining of explanted synovial tissues. Scale bar; 50 μm. (m) Immuno-fluorescence staining of CD3+ T cells in synovial infiltrates. Scale bar; 10 μm. (n) Synovial tissue transcriptome for TRB, TBET, RORG and other key inflammatory markers (n=8). All data are mean ± SEM. Two-tailed paired t test (k). Two-tailed unpaired Mann-Whitney-Wilcoxon rank test (n). *P < 0.05, **P < 0.01, ***P < 0.001.
To validate that TNF production is a feature of tissue-embedded cells and not just detectable after LPS/PMA/ION stimulation of disaggregated cells, we established a method to analyze cytokine production in freshly harvested synovial tissues from RA patients. Cytokine secretion from the cells was blocked by treating intact tissue slices with BFA for 4 h. Subsequently, cells were isolated from the tissue and intracellular TNF was detected by flow cytometry (Fig. 8i–k). In fresh synovial tissues, about 40% of synovial CD68+ macrophages produced TNF, with or without BFA treatment, indicating intracellular retention of TNF in tissue-residing macrophages. In contrast, tissue-residing T cells appeared to immediately release TNF into the tissue microenvironment. The frequencies of TNF+ T cells increased 10–20-fold upon BFA-induced secretion blockade (Fig. 8k). These data identified synovial T cells as a dominant cellular source of TNF, in both leukocyte-poor and leukocyte-rich tissue lesions.
To investigate whether T cell-derived TNF is relevant for synovial inflammation, we relied on a humanized mouse model in which human synovial tissue is engrafted into NSG mice and the chimeric host is immuno-reconstituted with peripheral blood mononuclear cells (PBMC) from RA patients19,34. Prior to the PBMC transfer, FACS-sorted CD4+ T cells were transfected with control siRNA or siRNA targeting TNF. The knockdown lowered TNF transcripts to about 50% of controls, mimicking a physiologic situation. Histological evaluation of explanted synovial tissues demonstrated that suppressing T cell-derived TNF was strongly anti-inflammatory. Control chimeras, injected with control siRNA transfected CD4+ T cells, developed robust synovitis. Tissues harvested from the control mice were densely infiltrated with CD3+ T cells (Fig. 8l, m). Tissue transcriptomic analysis revealed abundance of TCR, TBX21 and RORC transcripts (Fig. 8n). CD4+ T cells with intact TNF production infiltrated into the synovial tissue space and triggered induction of IFNG, IL17, IL21, TNF, IL6 and IL1B transcription (Fig. 8n). Synovial explants harvested from mice reconstituted with CD4+TNFlo T cells had few tissue-infiltrating cells (Fig. 8I), density of tissue-residing CD3+ T cells was low (Fig. 8m), and all inflammatory genes were expressed at low abundance (Fig. 8n). In a parallel approach, we tested whether reconstitution of intact mitochondria affected synovitis. Analysis of the synovial explants documented obvious anti-inflammatory potency of mitochondria transfer. Synovial grafts harvested from mice reconstituted with RA T cells transferred with intact mitochondria had few tissue-infiltrating cells (Extended Data Fig. 10a) and low-density CD3+ T cell infiltrates (Extended Data Fig. 10b). Transcripts for key inflammatory genes were consistently low (Extended Data Fig. 10c). Collectively, these data established that TNF-producing T cells preferentially home to the synovial tissue environment and are indispensable for the induction and maintenance of synovitis.
Discussion
Recognition of autoantigen has been considered the major mechanism of autoimmunity, but recent studies have emphasized the importance of the intracellular environment in regulating T cell tolerance. Specifically, metabolic conditions have emerged as a guide to pathogenic T cell functions, with a failure of mitochondrial function and misdirected protein trafficking promoting differentiation of RA CD4+ T cells into short-lived effector cells, prone to execute tissue damage15,18,21. A key effector pathway in RA is the unrestrained production of TNF, a defect exploited by anti-TNF therapeutics. Data presented here attribute TNF release to tissue-residing T cells that have dysfunctional mitochondria, a bloated, ribosome-bonded ER, and highly efficient co-translational translocation. The molecular components of this pathogenic pathway begin with inadequate mitochondrial aspartate synthesis causing deficient regeneration of cytoplasmic NAD+ and sensing this defect by the ER through failed ADP-ribosylation of the ER chaperone BiP. Un-ribosylated BiP releases IRE1α, prompting expansion of ribosome-occupied ER sheets. Implicating the protein synthesis function of the ER in RA pathogenesis adds a new dimension to the conceptual understanding of autoimmunity and provides novel strategies to treat tissue inflammation by replenishing aspartate, restoring BiP ADP-ribosylation or by interfering with co-translational translocation.
While organelle function is recognized as a critical determinant of T cell antigen responsiveness, a role for ER morphology in autoimmunity has not been proposed. Current data implicate ER size, function, and co-translational translocation in TNF biogenesis, and thus in pro-inflammatory T cell effector functions. As the major site of protein synthesis, folding and transport, ER function ultimately governs the fate of secretory cytokines. Functional analysis of T cells mapped the underlying defect in unconstrained TNF secretion to the expansion of ER membranes that provide surface docking sites for the recruitment of mRNA:ribosome complexes. The ER is considered a highly dynamic organelle, its structure rapidly adapting to the cellular environment. Consisting of sheets and tubules, the peripheral ER is believed to shift the ratio of these structures to best serve the cell’s needs. Cells synthesizing large amounts of proteins favor formation of sheets, whereas cells involved in lipogenesis preferentially build tubules32. While the ER’s role in handling cellular stress is well established, signals determining ER size and shape are less well defined. The transcription factor XBP-1 appears essential in ER biogenesis, possibly by controlling secretory pathway genes and membrane phospholipid synthesis23,35. Data presented here predict that it is the morphology of the ER that deviates protective immunity to tissue-damaging immunity and exposes the host to uncontrolled TNF release.
Mechanistic experiments localized the signal upstream of the ER defect to the mitochondria. As the hub for protein, lipid and steroid synthesis, the ER must closely communicate with mitochondria to assess bioenergetic state and needs. ER-mitochondria communicate through membrane contacts36, but could also involve soluble messengers. Disruption of ER-mitochondria interactions triggers ER stress responses37–39, while ER-stressed cells tend to increase mitochondria activity and ATP production to restore energy reservoirs40,41. However, prolonged ER stress induces mitochondrial swelling and functional collapse and, if unresolved, results in mitochondria-dependent cell death. Information flux from the ER to the mitochondria relies largely on calcium transduced from ER stores to the mitochondrial matrix. The opposite direction of information flux, mitochondria affecting ER homeostasis, has remained elusive. Since direct signaling between the ER and the mitochondria is mostly facilitated by tubular structures, we suspected secreted mitochondrial intermediates to drive the expansion of ER sheets. Screening key mitochondrial metabolites implicated succinate and its oxidation products in communicating the metabolic state of mitochondria, whereas α-KG was ineffective in inducing ER shape changes. Oxaloacetate and its downstream product, the amino acid aspartate, were most effective in controlling ER expansion. Aspartate functions as a critical component of the malate-aspartate shuttle, responsible for transporting electrons across the mitochondrial membrane through NADH to NAD conversion. Pinpointing aspartate as the mitochondrial messenger governing ER morphology placed the sensing mechanism into the cytoplasm.
ERrich RA T cells were poor in aspartate, had accumulated NADH and lacked the hydrogen/electron acceptor NAD. The shortage of aspartate resulted from a combination of two mitochondrial defects; one in the TCA cycle providing insufficient carbon backbone and one in the electron transport chain failing to deliver NAD. We have defined two molecular defects in the mitochondria of RA T cells18,20. Faulty repair of mitochondrial DNA results in a non-functional respiration chain20 and transcriptional repression of succinate-CoA ligase GDP-forming subunit β (SUCLG2) leads to reversal of the TCA by intersecting the production of succinate18. Thus, the aspartatelo state of RA T cells is a consequence of mitochondrial malfunction.
Supporting aspartate biosynthesis is an essential role of mitochondrial respiration32,42, designating the amino acid as an ideal messenger of mitochondrial intactness. Aspartate exits mitochondria to function as an electron acceptor in the cytosol, facilitating the regeneration of NAD+ from NADH. The mitochondrial inner membrane is impermeable to oxaloacetate43, relying on aspartate to shuttle electrons across the membrane. Replenishing aspartate was highly effective in re-educating RA T cells. Aspartate supplementation was sufficient to reverse the inflated ER membrane system and had potent anti-inflammatory effects in vivo, validating the relevance of mitochondria-ER communication in autoimmune tissue inflammation.
Ultimately, aspartate deficiency reflected the failure of the electron transport chain and at the same time aggravated cytoplasmic NAD+ shortage. The ER senses NAD deficiency by relying on NAD’s function as a co-substrate for post-translational modification, specifically, the ADP-ribosylation of an ER-positioned protein. The ER chaperone and signaling regulator GRP78/BiP monitors ER stress signals by controlling the activation of the transmembrane ER stress sensors (IRE1, PERK, and ATF6) through a binding-release mechanism44,45. Upon sensing and binding of unfolded proteins, BiP releases IRE1, PERK and ATF6, activating parallel pathways in the ER stress program. The substrate-binding ability of BiP is fine-tuned by NAD-dependent ADP-ribosylation. Increasing burden of unfolded proteins in the ER results in decreased BiP ribosylation46, while lowering of the unfolded protein flux stabilizes BiP-client binding through enhanced ADP ribosylation. Two potential ADP ribosylation sites (Arg470, Arg492) have been identified within BiP’s substrate binding domain (SBD), with ADP-ribosylation destabilizing the binding of peptide substrates and inactivating the chaperone. Data presented here directly linked intracellular NAD+ concentrations to BiP ribosylation. In aspartatelo/NADlo RA T cells, BiP was de-ribosylated and released the ER stress sensor IRE1, communicating mitochondrial malfunction to the ER membrane system. Both, exogenous aspartate, and NAD+ reestablished ER homeostasis and corrected the pro-inflammatory phenotype of RA T cells. Conversely, targeting mitochondrial respiration in healthy T cells was sufficient to deplete intracellular aspartate and NAD+ generation, phenocopied the RA-associated ER expansion and induced TNF-superproducing effector cells. Also, knockdown of the aminotransferase GOT2 transformed healthy T cells into TNF-releasing pro-inflammatory effector cells, clearly identifying aspartate generation as the pinnacle defect.
Understood as a key pathogenic element in RA, T cells have adopted fundamentally different bioenergetic pathways that directly underpin their disease-inducing properties. Characteristic traits, such as clonal expansion, cytokine release, migratory behavior and tissue invasiveness are all dependent on alterations in fuel selection and handling of energy carriers19–21. While extracellular signals, such as nutrient availability have not been excluded as determining factors in the metabolic reprogramming, cell-intrinsic abnormalities seem to be causative47. Cellular organelles, including lysosomes, mitochondria, and the endomembrane system, are now recognized to contribute to RA by deviating T cell differentiation towards short-lived effector T cells (SLECs)20,21,47. Current data suggest a hierarchical order, with mitochondrial malfunction giving rise to ER reshaping and stress responses. Consequently, approaches to re-engineer auto-aggressive RA T cells into host-protective memory T cells should focus on repairing the mitochondria. In an effort to explore new therapeutic strategies, we validated several therapeutic interventions in an in vivo system of synovial inflammation. Mitochondria-ER miscommunication, and the resulting excessive TNF production could be repaired by transferring healthy mitochondria into RA T cells. Technically less demanding was the provision of exogenous NAD and aspartate, which suppressed TNF production and successfully restrained synovial tissue inflammation. Other metabolites capable of regenerating cytoplasmic NAD may be equally suitable to treat RA.
Direct targeting of TNF with biologic therapies has revolutionized the management of multiple autoimmune diseases33,48. However, low rates of disease remission, the development of adverse effects and the generation of antibodies against biologic TNF inhibitors33 all curb the efficacy of anti-TNF therapy. Defining TNF superproducing T cells as the major perpetrator in synovial inflammation and rectifying the defect by replenishing missing mitochondrial metabolites may give rise to new classes of immunomodulatory therapies that interfere with upstream pathologies instead of inhibiting the final product of pro-inflammatory effector cells. Targeting the mitochondria-ER crosstalk may facilitate highly effective, pathogenesis-inspired therapeutic interventions.
Materials and Methods
Patients and samples
Patients enrolled into the study fulfilled the diagnostic criteria for RA and tested positive for rheumatoid factor and/or anti-CCP antibodies. All patients (n=120) recruited had active disease and clinical characteristics are presented in Supplementary Table 1. The following criteria excluded individuals from enrollment: current or previous diagnosis of cancer, uncontrolled medical disease, chronic inflammatory syndrome. Age-matched healthy donors without a personal history of cancer or autoimmune disease served as controls. All patients and controls provided informed consent and they did not receive compensation. All studies were approved by Institutional Review Board (IRB) Stanford and Institutional Review Board (IRB) Mayo Clinic.
Cell preparation and culture
Peripheral blood mononuclear cells (PBMCs) were isolated by gradient centrifugation with Lymphocyte Separation Medium (Lonza). CD4+CD45RA+ naive T cells were isolated from PBMCs with the EasySep Human Naïve CD4+ T Cell Enrichment Kit (STEMCELL Technologies). Purity of cell populations was consistently > 95%. Anti-CD3/anti-CD28-coated Dynabeads (Gibco) were used to activate naïve CD4+ T cells at a ratio of 2 cells: 1 bead for 72 h. Cell Activation Cocktail (PMA/Ionomycin) (Biolegend) was used to stimulate cytokine production in T cells with or without Brefeldin A (eBioscience) for 2 h.
Reagents
The pCMV5-Flag-XBP1s plasmid was purchased from Addgene. Human TNF siRNA, GOT2 siRNA and control siRNA were obtained from Thermo Fisher Scientific. The mitochondrial respiration inhibitors Antimycin A, Rotenone and Oligomycin were purchased from Agilent Technologies, Piericidin A was from Cayman Chemical. NAD+ was obtained from Cayman Chemical. α-ketoglutaric acid, succinic acid, L-aspartic acid, α-ketobutyric acid, L-asparagine were from Sigma-Aldrich. Malic acid was from Santa Cruz Biotechnology. MitoTracker Red and MitoTracker Green were purchased from Thermo Fisher Scientific. Assay kits for aspartate, oxaloacetate, NAD+/NADH and phosphatidylcholine were obtained from Sigma-Aldrich. Elisa kit for human TNF was from Thermo Fisher Scientific.
Intracellular TNF measurement
To measure intracellular TNF production, CD4+CD45RA+ naive T cells were activated for 72 h and stimulated with PMA/ION/BFA for 2 h. 4% PFA was used for fixation and 0.1% saponin to permeabilize the cells. Cells were stained with PE anti-human TNF (1:100, BD, Cat# 554513) antibody for 1 h. Flow cytometry was performed on an LSR II flow cytometer (BD Biosciences). Data were analyzed with FlowJo software (Tree Star Inc).
Labelling of ER membranes
ER tracker green (Thermo Fisher Scientific, Cat# E34251) was used to stain the ER-localized ATP-sensitive K+ channels following the manufacturer’s instruction. Flow cytometry was performed to detect ER tracker signal on an LSR II flow cytometer (BD Biosciences). Intensity of ER tracker staining was taken as a correlate of the size of ER membranes. Data were analyzed with FlowJo software (Tree Star Inc).
Transmission electron microscopy
Cells were resuspended in McDowell’s and Trump’s fixative for 1 hr at room temperature and then pelleted with a microcentrifuge. Cells were washed with 0.1M phosphate buffer for 5 minutes (twice). Liquid agar was added to the cell pellet and the cells were resuspended followed immediately by centrifugation. Once the sample had cooled, the agar was removed, and the sample pellet removed with a razor blade and placed into 0.1M phosphate buffer. Following 2 rinses in 0.1M phosphate buffer (pH 7.2), the sample was placed in 1% osmium tetroxide in the same buffer for 1 hr at room temperature. The sample was rinsed 2 times in distilled water and dehydrated in an ethanolic series culminating in two changes of 100% acetone. The cell pellet was then placed in a mixture of Spurr resin and acetone (1:1) for 30 min, followed by 2 hr in 100% resin with 2 changes. The cell pellet was placed into 100% Spurr resin in an embedding mold and polymerized at 65° C for 12 h or longer. Ultrathin (70–90 nm) sections were cut on an ultramicrotome with a diamond knife, stained with lead citrate and examined with a JEOL 1400 transmission electron microscope.
Rough ER isolation
Endoplasmic Reticulum Isolation Kit (Sigma-Aldrich, Catalog Number: ER0100) was used to isolate rough ER.
Measurement of mitochondrial membrane potential
MitoTracker Red (Thermo Fisher Scientific, Cat# M7512) is a fluorescent dye that stains mitochondria in live cells and its accumulation is dependent upon mitochondrial membrane potential (MMP). Staining intensity was analyzed with an LSR II flow cytometer (BD Biosciences) and data were analyzed with FlowJo software (Tree Star Inc).
Immunoblotting
Cellular proteins or rough ER proteins were extracted with RIPA buffer (Sigma-Aldrich). Protein expression levels were examined by western blotting as previously described1. Monoclonal antibodies specific for ribosomal protein L17, ribosomal protein S7 and BiP were purchased from Santa Cruz Biotechnology. Antibodies specific for calnexin and IRE1α were from Cell Signaling Technology. β-actin expression detected with anti-β-actin antibody (Cell Signaling Technology, 8H10D10) served as the internal control.
Real time PCR
Total RNA or Rough ER bound RNA were extracted with Trizol (Thermo Fisher Scientific) and Direct-zol RNA MiniPrep Kit (ZYMO Research). cDNA was synthesized using Maxima First Strand cDNA Synthesis Kits (Thermo Fisher Scientific). Quantitative PCR analyses were performed using SYBR Green qPCR Master Mix (Bimake) and gene expression was normalized to β-actin. Primers are listed in Supplementary Table 2.
Mitochondria transfer
On day 0, naïve CD4+ T cells from healthy donor were isolated and activated with anti-CD3/CD28 beads. On day 3, naïve CD4+ T cells from RA patient were purified and activated. On day 5, 4 million healthy CD4+ T cells were harvested, and mitochondria were isolated using Mitochondria Isolation Kit for Cultured Cells (Thermo Fisher Scientific, Catalog number: 89874). In parallel, 0.2 million activated RA CD4+ T cells were collected and spun at 300g for 5 minutes. Resuspend cells were mixed with the mitochondria suspension and carefully resuspended. The cell/mitochondria mixture and the control cell/PBS mixture were centrifuged at 1,500g for 5 minutes. After washing, the cells were stimulated with anti-CD3/CD28 beads for one more day.
Human synovial tissue–NSG mouse chimeras
As previously reported2,3, NSG mice from the Jackson Laboratory were maintained and bred under specific pathogen-free conditions on a 12/12 h light/dark cycle at 20–22°C with free access to water and food. Animal housing facilities were monitored for infection with specific pathogens every 6 weeks and the health status of all animals was checked daily. Both male and female mice were randomly used at 8–12 weeks of age. After 7 days, the mice were infused with 10 million PBMC collected from RA patients with active disease. In some experiments, CD4+ T cells in PBMC were FACS sorted and transfected with siRNA targeting TNF or control siRNA before the cells were injected into the mice. Alternatively, healthy mitochondria were transferred into RA CD4+ T cells prior to the immune reconstitution. For these experiments, CD4+ T cells were FACS sorted and mitochondria from healthy T cells were transferred into RA CD4+ T cells as described above. On day 14, synovial tissues were explanted from the chimeric mice, OCT-embedded (4583; Sakura Finetek USA) or shock-frozen for further experiments (tissue staining or RNA extraction). All in vivo experiments were carried out in accordance with the guidelines of the Institutional Animal Care and Use Committee.
Immunohistochemistry
Frozen sections of synovial tissues were stained with mouse anti-human CD3 (1:100; DAKO, Clone F7.2.38) and rabbit anti-human IFN-γ (1:100, Abcam, ab25101). Alexa Fluor 594 anti-mouse IgG (1:200, Thermo Fisher Scientific, A-11032) and Alexa Fluor 488 anti-rabbit IgG (1:200, Thermo Fisher Scientific, A-11034) were used as secondary antibodies. Images of CD3/IFN-γ staining were obtained using a LSM710 confocal microscope (Carl Zeiss) with a Plan-Neofluar × 40/1.3-NA oil objective lens.
Immunofluorescence
To visualize intracellular proteins, cells were collected, fixed with 4% PFA for 10 min and permeabilized using 0.5% saporin. The following primary antibodies were used: ant- calnexin rabbit mAb (1:100, Cell signaling technology), anti-protein disulfide isomerase (PDI) mouse mAb (1:100, Thermo Fisher Scientific). The following secondary antibodies were used: Alexa Fluor 594 goat anti-mouse IgG (1:200, Thermo Fisher Scientific, A-11032), Alexa Fluor 488 goat anti-rabbit IgG (1:200, Thermo Fisher Scientific, A-11008). Nuclei were stained with DAPI. The LSM710 system (Carl Zeiss) with a Plan Apochromat 63×/1.40-NA oil DICIII objective lens (Carl Zeiss) was used to acquire images.
BiP ADP-ribosylation
5×106 activated naïve CD4+ T cells were lysed with IP Lysis Buffer (Thermo Fisher Scientific, Catalog#: 87788) containing protease and phosphatase inhibitors. BiP protein in whole cell lysates was pulled down by incubation with agarose-conjugated anti-BiP antibody (Santa Cruz Biotechnology, Catalog #: sc-13539) for 4 h at 4°C. Agarose was washed 5 times with IP lysis buffer and boiled for 5 min in loading buffer. Eluted protein was separated by SDS-page and ADP-ribosylated BiP was detected using antibody against ADP-ribose (Cell Signaling Technology, Catalog#: 83732).
BiP/ IRE1α binding
Cells were lysed with IP Lysis Buffer (Thermo Fisher Scientific, Catalog#: 87788) containing protease and phosphatase inhibitors. Whole cell lysates were incubated with 2μg anti-BiP antibody (Santa Cruz Biotechnology, Catalog#: sc-13539) and Protein A/G PLUS-Agarose (Santa Cruz Biotechnology, Catalog#: sc-2003) for 4 h at 4°C. Normal rat IgG (Santa Cruz Biotechnology, Catalog#: sc-2026) was used as IgG control. The immunocomplexes were washed with IP lysis buffer 5 times, then eluted with loading buffer and separated by SDS-PAGE. Immunoblotting for IRE1α was performed following standard procedures for Western blotting.
Quantification and Statistical Analysis
Statistical analyses were performed using GraphPad Prism software (GraphPad Software). To compare data within two groups, the paired Wilcoxon test or the Mann-Whitney test were used when the sample size per group was >5. Parametric t test was only used if the sample size per group was <=5. To adjust for multiple testing, we used Hochberg’s step-down method to control for a family-wise-error rate at the 0.05 levels. One-way ANOVA was used and pair-wise comparison using Tukey’s method was applied for comparisons between 3 or more groups. All data points were included in the analysis and no outliers were detected using Grubbs’ test. All data are presented as mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001 and statistical parameters are presented in each figure legend.
Extended Data
Extended Figure 1.
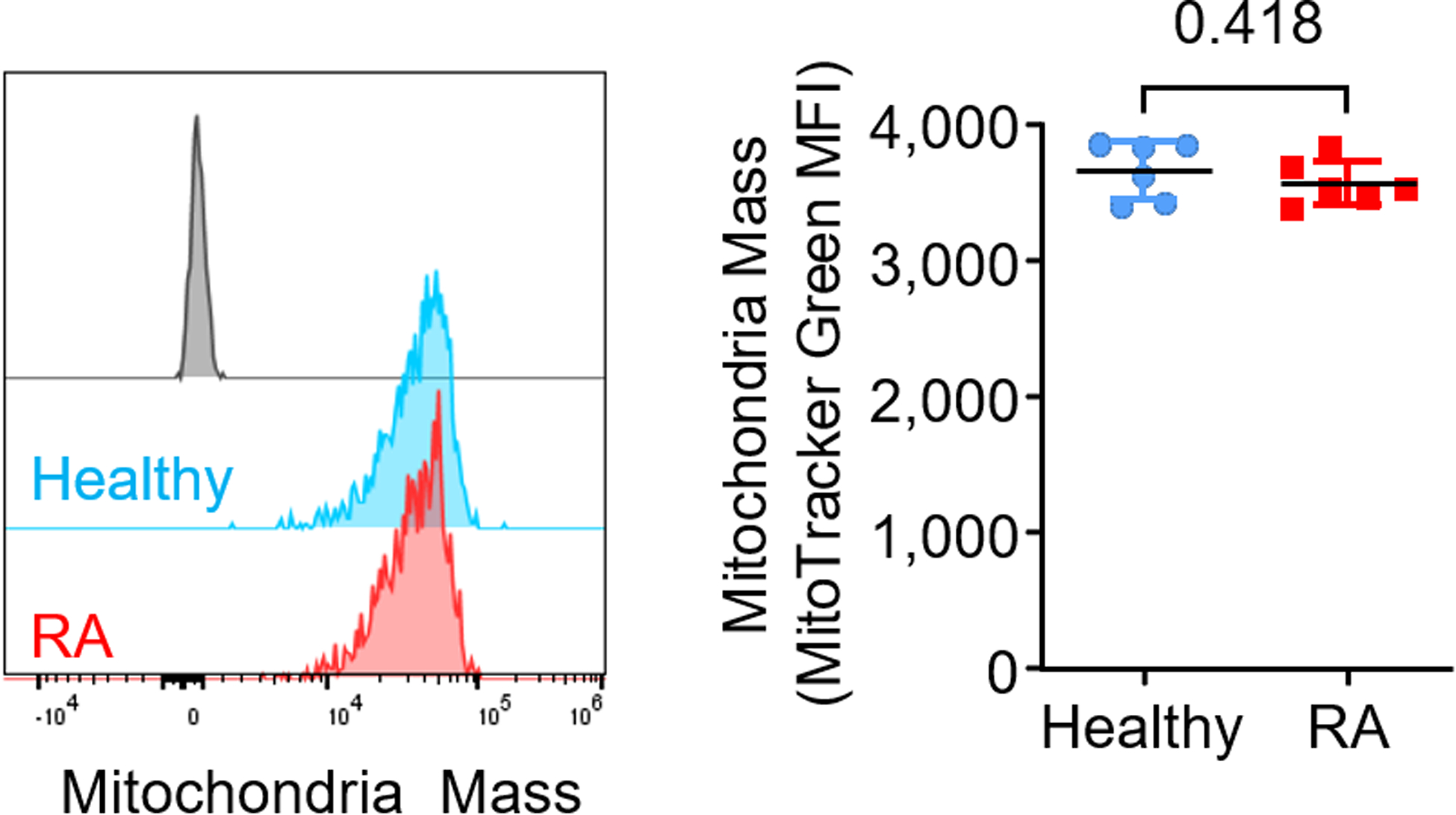
CD4+CD45RA+ T cells were stimulated for 72 h. Flow cytometric quantification of mitochondria mass (MitoTrack Green MFI); n = 6 healthy and 6 RA. Data are mean ± SEM. Two-tailed unpaired Mann-Whitney-Wilcoxon rank test.
Extended Figure 2.
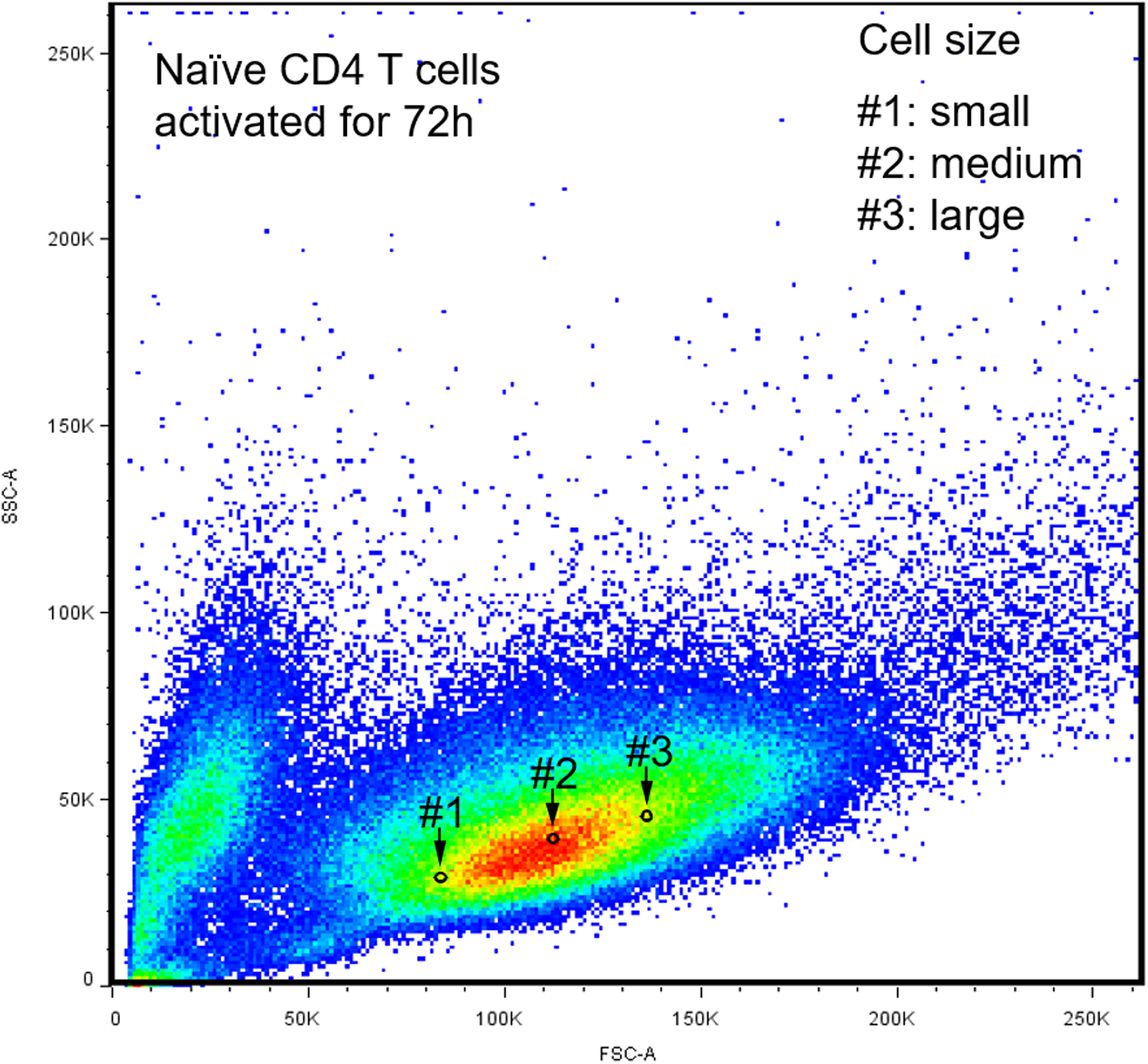
CD4+CD45RA+ T cells were stimulated for 72 h. ER biomass was determined with ER tracker and mitochondrial function was assessed with the mitochondrial membrane potential. Gate #1: small cellular size. Gate #2: medium cellular size. Gate #3: large cellular size.
Extended Figure 3.
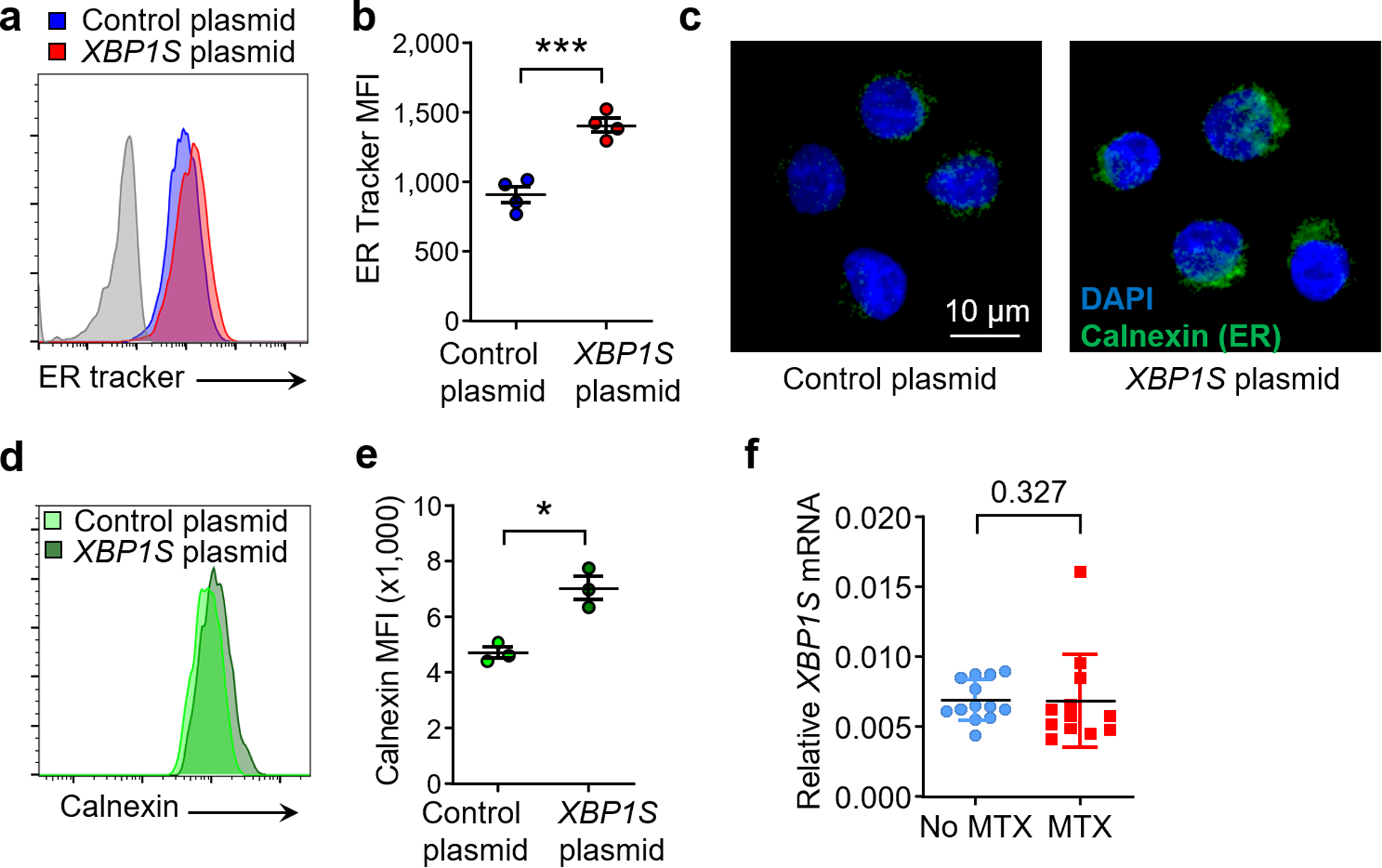
Naive CD4+ T cells were stimulated and transfected with control or XBP1S overexpression plasmid before the ER size was determined. (a, b) Flow cytometry for ER Tracker staining; n=4. (c) Confocal microscopy imaging of the ER protein calnexin. Scale bar, 10 μm, n=3 independent experiments. (d, e) Flow cytometry for calnexin expression; n=3. (f) XBP1S expression in T cells from patients treated with or w/o Methotrexate (MTX) (MTX: n=13; w/o MTX: n=12). All data are mean ± SEM. Two-tailed paired t test (b, e). Unpaired Mann-Whitney-Wilcoxon rank test (f). *P < 0.05, ***P < 0.001.
Extended Figure 4.

Healthy naive CD4+ T cells were stimulated for 72 h in the presence of the mitochondrial respiration inhibitors Piericidin A (10 pM), Antimycin A (10 nM) or Oligomycin (1 nM). ER size was determined by flow cytometry measuring ER tracker (n=4). Data are mean ± SEM. One-way ANOVA and post-ANOVA pair-wise two-group comparisons conducted with Tukey’s method. **P < 0.01.
Extended Figure 5.
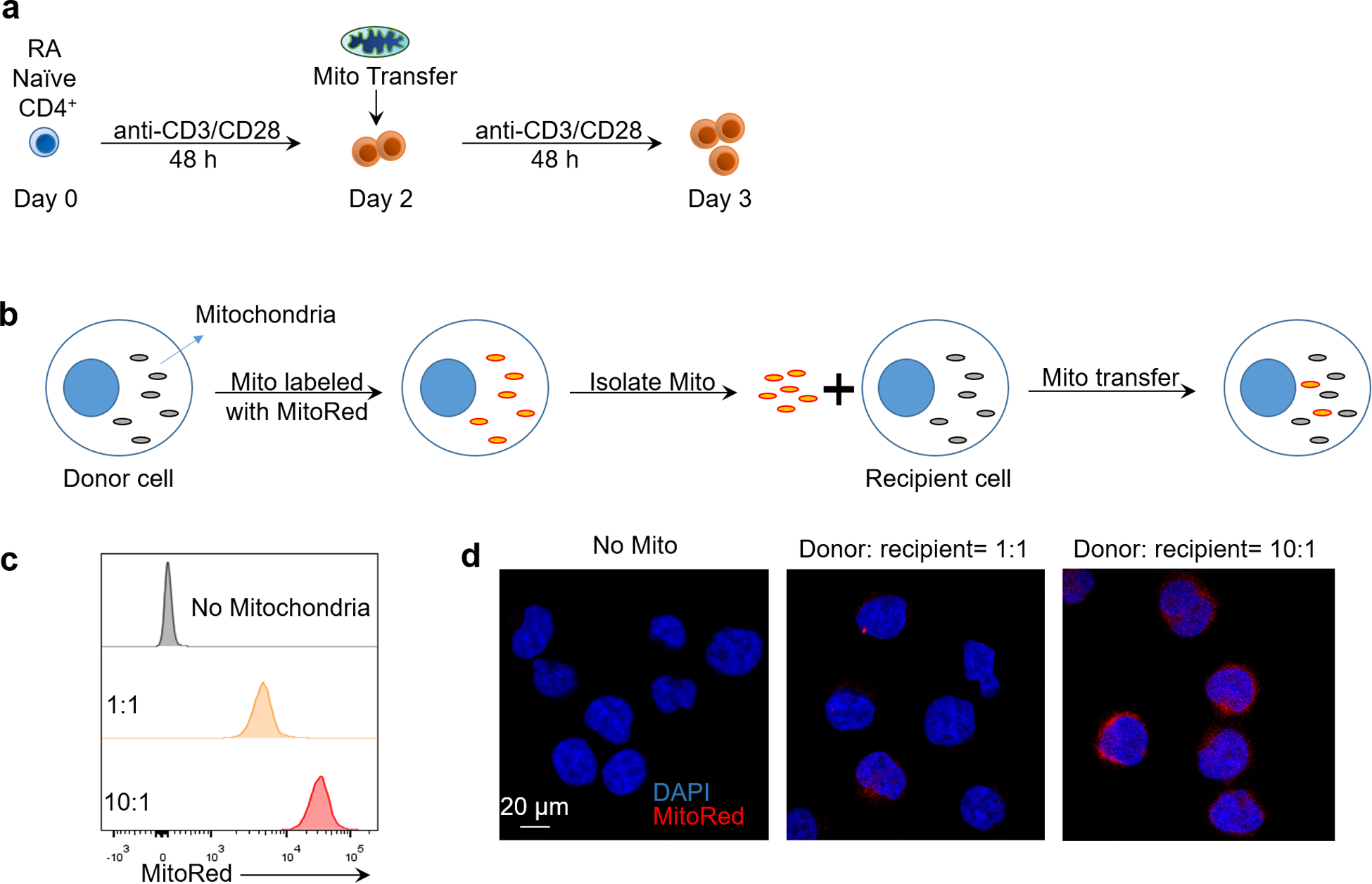
(a) Experimental scheme for mitochondria transfer into RA T cells. (b) Experimental scheme for mitochondria transfer in Jurkat T cells. Mitochondria were labeled with MitoTrackerRed and isolated, then transferred into recipient cells. (c) Flow cytometric analysis of MitoTracker Red intensity after mitochondria transfer. Ratio indicates donor cell number/recipient cell number. (d) Representative confocal imaging of exogenous mitochondria transferred into Jurkat T cells, n=3 independent experiments, scale bar, 20 μm.
Extended Figure 6.
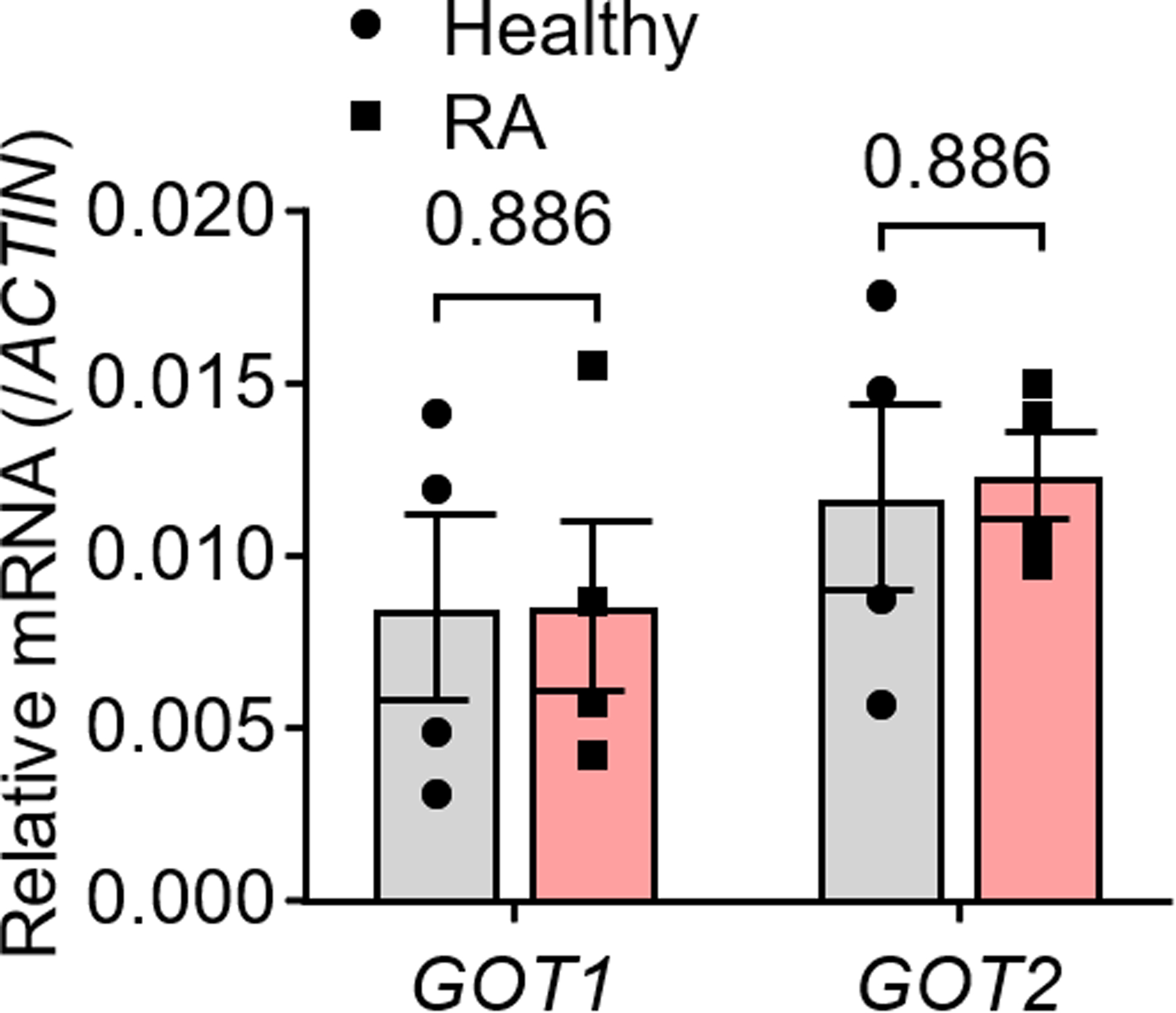
Peripheral blood CD4+CD45RA+ T cells from RA patients and age-matched healthy individuals were isolated and stimulated for 72 h. mRNA levels of GOT1 and GOT2 were determined by qPCR. n = 4 in each group. All data are mean ± SEM. Two-tailed unpaired Mann-Whitney-Wilcoxon rank test.
Extended Figure 7.

ADP-ribosylation of BiP in healthy CD4+ T cells treated with or w/o Piercidin A (10 pM) for 24 h. n = 2.
Extended Figure 8.
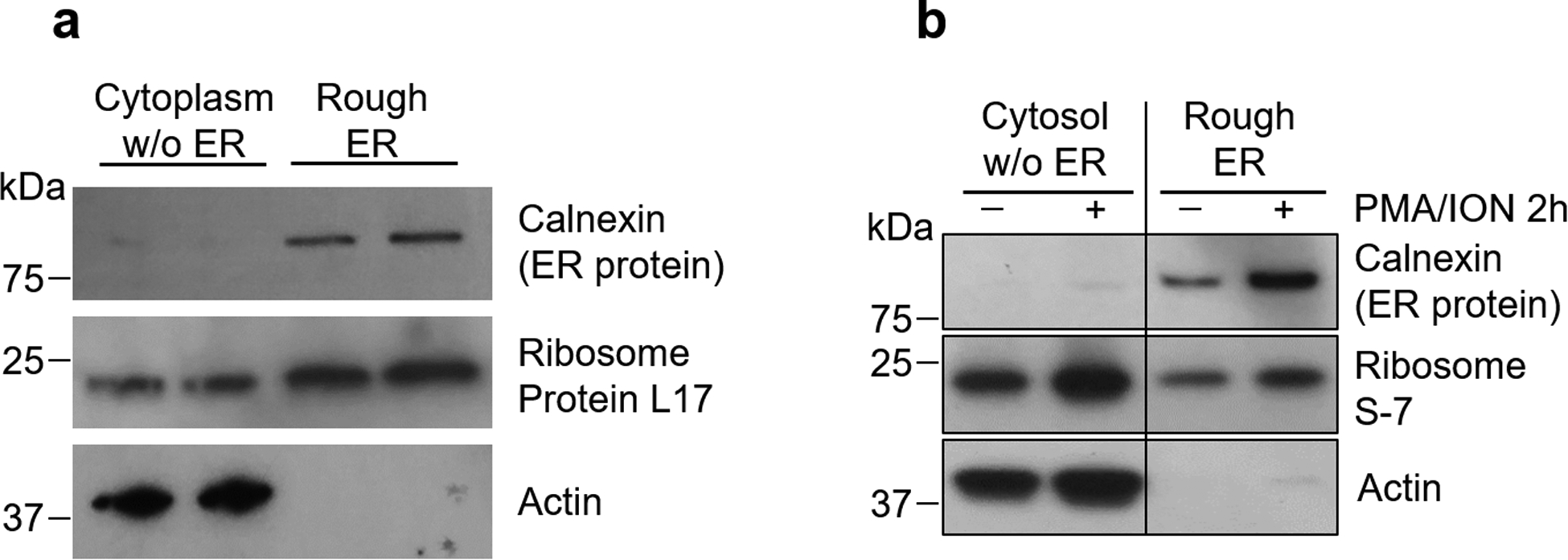
(a) Naïve CD4+ T cells were purified from peripheral blood mononuclear cells and stimulated with anti-CD3/CD28 for 72 h. The rough ER was isolated by calcium precipitation and the isolate was immunoblotted for the ER protein calnexin, the ribosomal protein L17 and the cytoplasmic protein a-actin. (b) Healthy CD4+ T cells were activated with PMA/Ionomycin for 2 h before isolation of the rough ER and immunoblotting of the ER protein calnexin, the ribosomal protein S7 and the cytosolic protein β-actin.
Extended Figure 9.
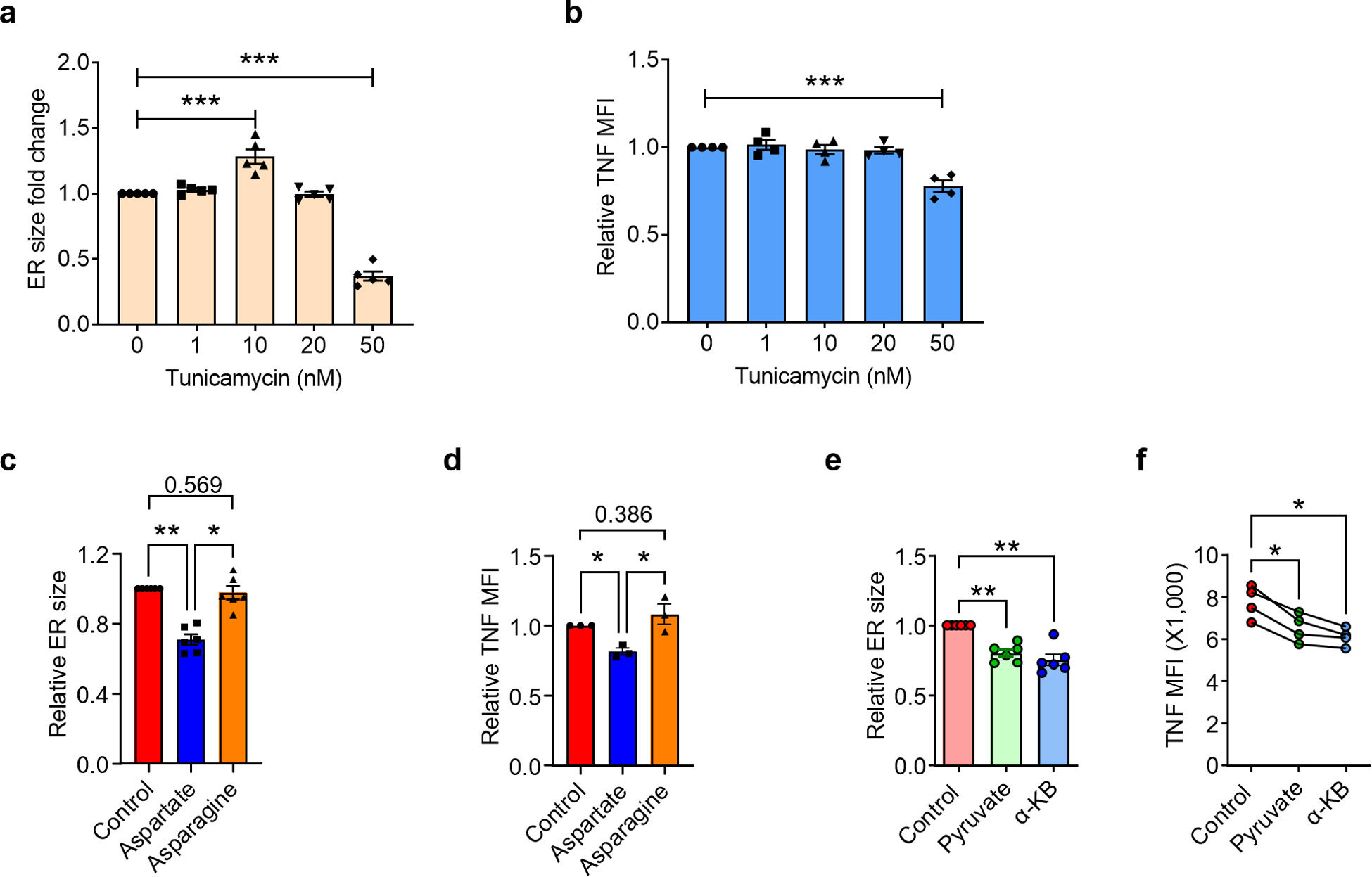
Naïve CD4+ T cells were purified from PBMCs and stimulated with anti-CD3/CD28 beads for 72 h in the presence of the indicated molecules. ER size quantified by MFI of ER tracker staining and TNF production measured by intracellular staining of TNF after PMA/ION stimulation for 2 h in the presence of the secretion inhibitor BFA. (a) Fold change of ER size after Tunicamycin treatment compared to the control group, n=6. (b) Fold change of TNF production after Tunicamycin treatment compared to the control group, n=4. (c) ER size change after Aspartate (1 mM) or Asparagine (1 mM) treatment, n=6. (d) TNF production after aspartate and asparagine treatment, n=3. (e) ER size change after pyruvate (1 mM) or α-KB (1 mM) treatment, n=6. (f) TNF production after pyruvate or α-KB treatment, n=4. All data are mean ± SEM, one-way ANOVA and post-ANOVA pair-wise two-group comparisons conducted with Tukey’s method. *P < 0.05, **P < 0.01, ***P < 0.001.
Extended Figure 10.
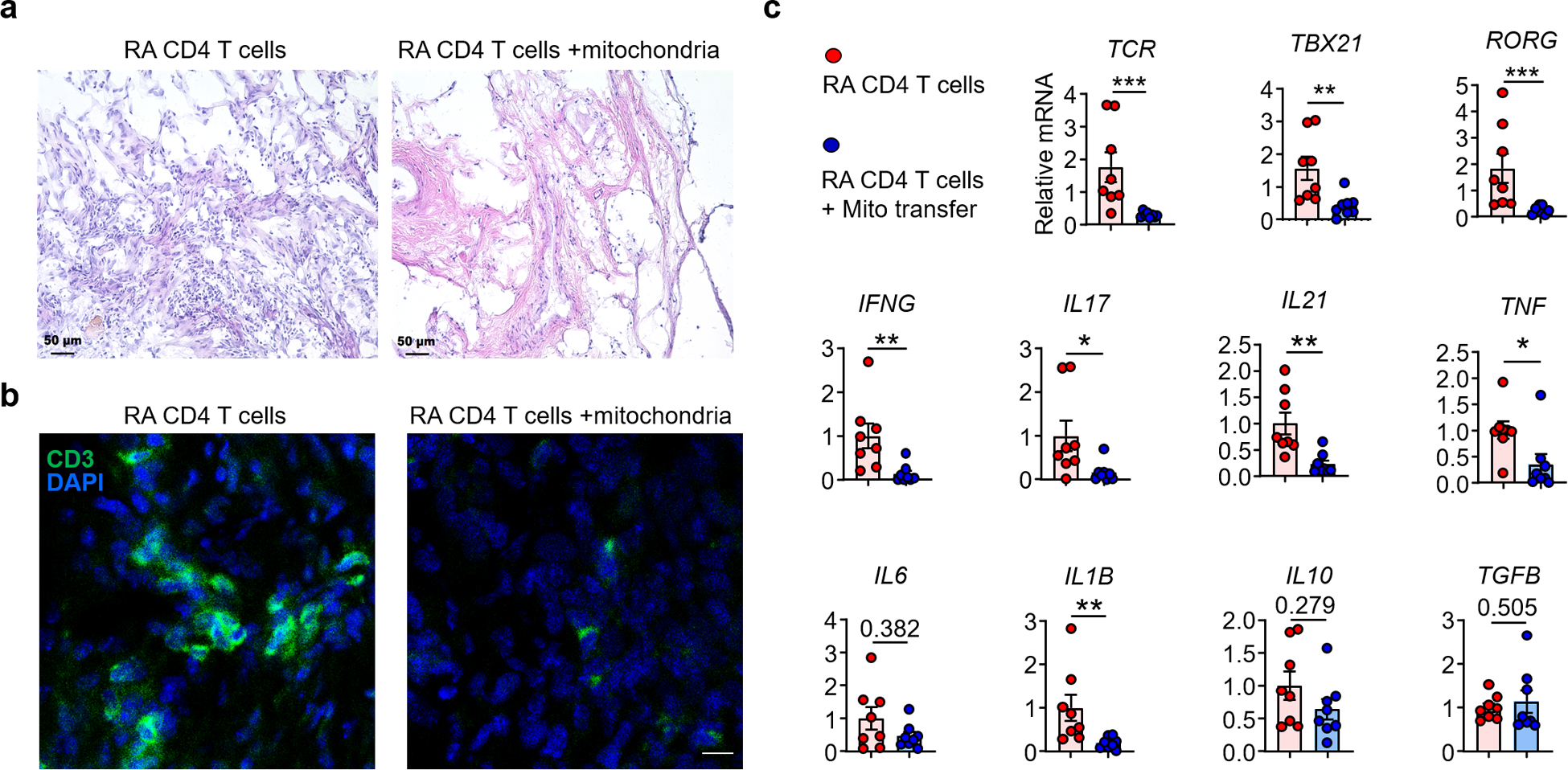
Mitochondria were isolated from healthy T cells and transferred into RA CD4+ T cells prior to their adoptive transfer into synovium-NSG chimeras. Explanted synovial grafts were analyzed by immunohistochemical staining and tissue transcriptomics (RT-PCR). 8 tissues in each group. (a) H&E staining of synovial tissue sections. Scale bar; 50 μm. (b) Immunofluorescence staining for CD3+ T cells in synovial infiltrates. Representative images. Scale bar; 10 μm. (c) Gene expression profiling (RT-PCR) of TRB, TBET, RORG and other key inflammatory markers (n=8). All data are mean ± SEM. Two-tailed unpaired Mann-Whitney-Wilcoxon rank test. *P < 0.05, **P < 0.01, ***P < 0.001.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health (R01AR042527, R01AI108906, R01HL142068, and P01HL129941 to CMW and R01AI108891, R01AG045779, U19AI057266, and R01AI129191 to JJG).
Footnotes
Declaration of Interests
The authors have declared that no conflict of interest exists.
Data availability
All data in this study are available within the paper, source data are provided with this paper. Any other relevant data are available from the corresponding author upon reasonable request.
References
- 1.Deane KD, Norris JM & Holers VM Preclinical rheumatoid arthritis: identification, evaluation, and future directions for investigation. Rheum Dis Clin North Am 36, 213–241, doi: 10.1016/j.rdc.2010.02.001 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Feldmann M, Brennan FM & Maini RN Role of cytokines in rheumatoid arthritis. Annu Rev Immunol 14, 397–440, doi: 10.1146/annurev.immunol.14.1.397 (1996). [DOI] [PubMed] [Google Scholar]
- 3.Maini RN & Taylor PC Anti-cytokine therapy for rheumatoid arthritis. Annu Rev Med 51, 207–229, doi: 10.1146/annurev.med.51.1.207 (2000). [DOI] [PubMed] [Google Scholar]
- 4.Zhang F et al. Defining inflammatory cell states in rheumatoid arthritis joint synovial tissues by integrating single-cell transcriptomics and mass cytometry. Nat Immunol 20, 928–942, doi: 10.1038/s41590-019-0378-1 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rubbert-Roth A et al. Failure of anti-TNF treatment in patients with rheumatoid arthritis: The pros and cons of the early use of alternative biological agents. Autoimmun Rev 18, 102398, doi: 10.1016/j.autrev.2019.102398 (2019). [DOI] [PubMed] [Google Scholar]
- 6.Grivennikov SI et al. Distinct and nonredundant in vivo functions of TNF produced by t cells and macrophages/neutrophils: protective and deleterious effects. Immunity 22, 93–104, doi: 10.1016/j.immuni.2004.11.016 (2005). [DOI] [PubMed] [Google Scholar]
- 7.Bovensiepen CS et al. TNF-Producing Th1 Cells Are Selectively Expanded in Liver Infiltrates of Patients with Autoimmune Hepatitis. J Immunol 203, 3148–3156, doi: 10.4049/jimmunol.1900124 (2019). [DOI] [PubMed] [Google Scholar]
- 8.Jain A et al. T cells instruct myeloid cells to produce inflammasome-independent IL-1beta and cause autoimmunity. Nat Immunol 21, 65–74, doi: 10.1038/s41590-019-0559-y (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schreurs R et al. Human Fetal TNF-alpha-Cytokine-Producing CD4(+) Effector Memory T Cells Promote Intestinal Development and Mediate Inflammation Early in Life. Immunity 50, 462–476 e468, doi: 10.1016/j.immuni.2018.12.010 (2019). [DOI] [PubMed] [Google Scholar]
- 10.Weyand CM & Goronzy JJ T-cell-targeted therapies in rheumatoid arthritis. Nat Clin Pract Rheumatol 2, 201–210, doi: 10.1038/ncprheum0142 (2006). [DOI] [PubMed] [Google Scholar]
- 11.Weyand CM & Goronzy JJ Immunometabolism in early and late stages of rheumatoid arthritis. Nat Rev Rheumatol 13, 291–301, doi: 10.1038/nrrheum.2017.49 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Slowikowski K, Wei K, Brenner MB & Raychaudhuri S Functional genomics of stromal cells in chronic inflammatory diseases. Curr Opin Rheumatol 30, 65–71, doi: 10.1097/BOR.0000000000000455 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Isaacs JD Therapeutic T-cell manipulation in rheumatoid arthritis: past, present and future. Rheumatology (Oxford) 47, 1461–1468, doi: 10.1093/rheumatology/ken163 (2008). [DOI] [PubMed] [Google Scholar]
- 14.Probert L et al. Wasting, ischemia, and lymphoid abnormalities in mice expressing T cell-targeted human tumor necrosis factor transgenes. J Immunol 151, 1894–1906 (1993). [PubMed] [Google Scholar]
- 15.Weyand CM & Goronzy JJ The immunology of rheumatoid arthritis. Nat Immunol 22, 10–18, doi: 10.1038/s41590-020-00816-x (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang Z et al. Restoring oxidant signaling suppresses proarthritogenic T cell effector functions in rheumatoid arthritis. Sci Transl Med 8, 331ra338, doi: 10.1126/scitranslmed.aad7151 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang Z, Fujii H, Mohan SV, Goronzy JJ & Weyand CM Phosphofructokinase deficiency impairs ATP generation, autophagy, and redox balance in rheumatoid arthritis T cells. J Exp Med 210, 2119–2134, doi: 10.1084/jem.20130252 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu B et al. Succinyl-CoA Ligase Deficiency in Pro-inflammatory and Tissue-Invasive T Cells. Cell Metab 32, 967–980 e965, doi: 10.1016/j.cmet.2020.10.025 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shen Y et al. Metabolic control of the scaffold protein TKS5 in tissue-invasive, proinflammatory T cells. Nat Immunol 18, 1025–1034, doi: 10.1038/ni.3808 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Y et al. The DNA Repair Nuclease MRE11A Functions as a Mitochondrial Protector and Prevents T Cell Pyroptosis and Tissue Inflammation. Cell Metab 30, 477–492 e476, doi: 10.1016/j.cmet.2019.06.016 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wen Z et al. N-myristoyltransferase deficiency impairs activation of kinase AMPK and promotes synovial tissue inflammation. Nat Immunol 20, 313–325, doi: 10.1038/s41590-018-0296-7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bommiasamy H et al. ATF6alpha induces XBP1-independent expansion of the endoplasmic reticulum. J Cell Sci 122, 1626–1636, doi: 10.1242/jcs.045625 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sriburi R, Jackowski S, Mori K & Brewer JW XBP1: a link between the unfolded protein response, lipid biosynthesis, and biogenesis of the endoplasmic reticulum. J Cell Biol 167, 35–41, doi: 10.1083/jcb.200406136 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martinez-Reyes I & Chandel NS Mitochondrial TCA cycle metabolites control physiology and disease. Nat Commun 11, 102, doi: 10.1038/s41467-019-13668-3 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zaslona Z & O’Neill LAJ Cytokine-like Roles for Metabolites in Immunity. Mol Cell 78, 814–823, doi: 10.1016/j.molcel.2020.04.002 (2020). [DOI] [PubMed] [Google Scholar]
- 26.Chinopoulos C Acute sources of mitochondrial NAD(+) during respiratory chain dysfunction. Exp Neurol 327, 113218, doi: 10.1016/j.expneurol.2020.113218 (2020). [DOI] [PubMed] [Google Scholar]
- 27.Korennykh A & Walter P Structural basis of the unfolded protein response. Annu Rev Cell Dev Biol 28, 251–277, doi: 10.1146/annurev-cellbio-101011-155826 (2012). [DOI] [PubMed] [Google Scholar]
- 28.Jager R, Bertrand MJ, Gorman AM, Vandenabeele P & Samali A The unfolded protein response at the crossroads of cellular life and death during endoplasmic reticulum stress. Biol Cell 104, 259–270, doi: 10.1111/boc.201100055 (2012). [DOI] [PubMed] [Google Scholar]
- 29.Voorhees RM & Hegde RS Toward a structural understanding of co-translational protein translocation. Curr Opin Cell Biol 41, 91–99, doi: 10.1016/j.ceb.2016.04.009 (2016). [DOI] [PubMed] [Google Scholar]
- 30.Elvekrog MM & Walter P Dynamics of co-translational protein targeting. Curr Opin Chem Biol 29, 79–86, doi: 10.1016/j.cbpa.2015.09.016 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schwarz DS & Blower MD The endoplasmic reticulum: structure, function and response to cellular signaling. Cell Mol Life Sci 73, 79–94, doi: 10.1007/s00018-015-2052-6 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sullivan LB et al. Supporting Aspartate Biosynthesis Is an Essential Function of Respiration in Proliferating Cells. Cell 162, 552–563, doi: 10.1016/j.cell.2015.07.017 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rubbert-Roth A et al. TNF inhibitors in rheumatoid arthritis and spondyloarthritis: Are they the same? Autoimmun Rev 17, 24–28, doi: 10.1016/j.autrev.2017.11.005 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Li Y et al. Deficient Activity of the Nuclease MRE11A Induces T Cell Aging and Promotes Arthritogenic Effector Functions in Patients with Rheumatoid Arthritis. Immunity 45, 903–916, doi: 10.1016/j.immuni.2016.09.013 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shaffer AL et al. XBP1, downstream of Blimp-1, expands the secretory apparatus and other organelles, and increases protein synthesis in plasma cell differentiation. Immunity 21, 81–93, doi: 10.1016/j.immuni.2004.06.010 (2004). [DOI] [PubMed] [Google Scholar]
- 36.Phillips MJ & Voeltz GK Structure and function of ER membrane contact sites with other organelles. Nat Rev Mol Cell Biol 17, 69–82, doi: 10.1038/nrm.2015.8 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hayashi T & Su TP Sigma-1 receptor chaperones at the ER-mitochondrion interface regulate Ca(2+) signaling and cell survival. Cell 131, 596–610, doi: 10.1016/j.cell.2007.08.036 (2007). [DOI] [PubMed] [Google Scholar]
- 38.Sebastian D et al. Mitofusin 2 (Mfn2) links mitochondrial and endoplasmic reticulum function with insulin signaling and is essential for normal glucose homeostasis. Proc Natl Acad Sci U S A 109, 5523–5528, doi: 10.1073/pnas.1108220109 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Simmen T et al. PACS-2 controls endoplasmic reticulum-mitochondria communication and Bid-mediated apoptosis. EMBO J 24, 717–729, doi: 10.1038/sj.emboj.7600559 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Knupp J, Arvan P & Chang A Increased mitochondrial respiration promotes survival from endoplasmic reticulum stress. Cell Death Differ 26, 487–501, doi: 10.1038/s41418-018-0133-4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bravo R et al. Increased ER-mitochondrial coupling promotes mitochondrial respiration and bioenergetics during early phases of ER stress. J Cell Sci 124, 2143–2152, doi: 10.1242/jcs.080762 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Birsoy K et al. An Essential Role of the Mitochondrial Electron Transport Chain in Cell Proliferation Is to Enable Aspartate Synthesis. Cell 162, 540–551, doi: 10.1016/j.cell.2015.07.016 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Davila A et al. Nicotinamide adenine dinucleotide is transported into mammalian mitochondria. Elife 7, doi: 10.7554/eLife.33246 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bettigole SE & Glimcher LH Endoplasmic reticulum stress in immunity. Annu Rev Immunol 33, 107–138, doi: 10.1146/annurev-immunol-032414-112116 (2015). [DOI] [PubMed] [Google Scholar]
- 45.Gardner BM, Pincus D, Gotthardt K, Gallagher CM & Walter P Endoplasmic reticulum stress sensing in the unfolded protein response. Cold Spring Harb Perspect Biol 5, a013169, doi: 10.1101/cshperspect.a013169 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chambers JE, Petrova K, Tomba G, Vendruscolo M & Ron D ADP ribosylation adapts an ER chaperone response to short-term fluctuations in unfolded protein load. J Cell Biol 198, 371–385, doi: 10.1083/jcb.201202005 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu B, Goronzy JJ & Weyand CM Metabolic Fitness of T Cells in Autoimmune Disease. Immunometabolism 2, doi: 10.20900/immunometab20200017 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kalliolias GD & Ivashkiv LB TNF biology, pathogenic mechanisms and emerging therapeutic strategies. Nat Rev Rheumatol 12, 49–62, doi: 10.1038/nrrheum.2015.169 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
References for Methods
- 1.Yang Z et al. Restoring oxidant signaling suppresses proarthritogenic T cell effector functions in rheumatoid arthritis. Sci Transl Med 8, 331ra338, doi: 10.1126/scitranslmed.aad7151 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wen Z et al. N-myristoyltransferase deficiency impairs activation of kinase AMPK and promotes synovial tissue inflammation. Nat Immunol 20, 313–325, doi: 10.1038/s41590-018-0296-7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li Y et al. Deficient Activity of the Nuclease MRE11A Induces T Cell Aging and Promotes Arthritogenic Effector Functions in Patients with Rheumatoid Arthritis. Immunity 45, 903–916, doi: 10.1016/j.immuni.2016.09.013 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data in this study are available within the paper, source data are provided with this paper. Any other relevant data are available from the corresponding author upon reasonable request.


