Abstract
A rapid protocol for subtyping vancomycin-resistant enterococci by pulsed-field gel electrophoresis is reported. The procedure is simple and potentially cost-effective and allows reproducible subtyping of the strains in approximately 1 day.
Vancomycin-resistant enterococci (VRE) have been isolated with increased frequency in all major medical centers in the United States, Canada, and Western Europe (10). Several typing methods, such as phage typing (13), serotyping (15), biotyping (4), biochemical fingerprinting (12) and, more recently, DNA restriction fragment analysis (8), total plasmid profile analysis (14), random amplified PCR (9), pulsed-field gel electrophoresis (PFGE) (21), and ribotyping (6), have been used for epidemiological investigations of enterococcal outbreaks and for subtyping of enterococcal strains. These methods vary in their reproducibility and discriminatory ability, with PFGE reported (2, 3, 6, 16) to be superior to the others. Therefore, PFGE is currently considered to be the “gold standard” for subtyping enterococci and has been used extensively for molecular epidemiological characterization of VRE outbreaks. However, despite this fact, a standardized, optimal procedure for PFGE typing of VRE is not available at the present time. In 1990, Murray et al. (21) developed one of the first PFGE protocols for typing enterococci, and their procedure has been used, in its original form (2, 7, 12, 16, 18) or with minor modifications (5, 11, 20, 22), by numerous investigators. However, the studies used PFGE typing in order to address specific clinical or research issues, and they were not designed to simplify or improve the PFGE typing protocol. Therefore, the typing procedures were not necessarily optimal, since they involved the use of numerous reagents and buffers, were laborious, and required several days before the results could be evaluated.
The goal of the studies described in this paper was to develop a simple, rapid, and reproducible procedure for PFGE typing of vancomycin-resistant Enterococcus faecium and Enterococcus faecalis (the two species that account for virtually all human VRE infections [10]), which would also be suitable for subtyping other enterococci, including vancomycin-susceptible strains and, potentially, other gram-positive bacteria.
Bacterial strains.
Our enterococcal strain collection contained 70 VRE isolates (4 E. faecalis strains, including ATCC strain 51299, and 66 E. faecium strains) and 10 vancomycin-sensitive enterococci (all E. faecalis strains, including ATCC strain 29212). The strains were kept in 30% glycerol–70% L broth at −80°C, and they were plated on L-agar medium before plugs were prepared. The nonenterococcal isolates analyzed were three strains of Staphylococcus aureus (including ATCC strains 25923 and 29213), two strains of Listeria monocytogenes (including the CDC standard strain H2446), two strains of Streptococcus pyogenes (ATCC strains 14919 and 12353), and one strain of Lactobacillus casei.
Standard procedure.
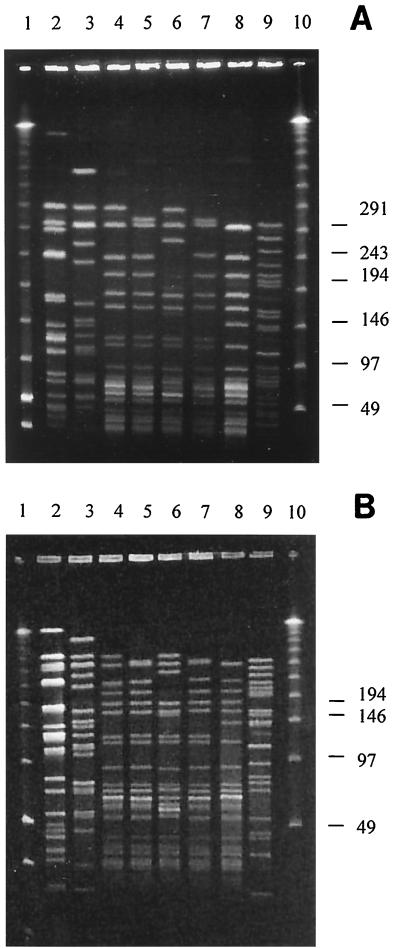
We used the procedure of Murray et al. (21), which has been used by many investigators (2, 7, 12, 16, 18) for PFGE typing of enterococci (including VRE), as the standard protocol from which we developed (and against which we evaluated) our procedure. PFGE patterns obtained from eight enterococcal strains analyzed by the standard procedure are shown in Fig. 1A.
FIG. 1.
PFGE patterns of SmaI-digested DNAs of enterococcal strains. (A) Results obtained using the standard procedure of Murray et al. (21). (B) Results obtained using our procedure. Lane 1, low-range PFGE markers; lanes 2 and 3, E. faecalis strains, including the ATCC strains 29212 (lane 2) and 51299 (lane 3); lanes 4 through 9, E. faecium strains; lane 10, λ ladder. Molecular sizes are shown in kilobases on the right.
Optimization of plug preparation.
In order to improve the typing procedure, we examined the value of, and eliminated, several reagents (including deoxycholic acid, which is toxic) used by Murray et al. (21) during plug preparation (Table 1). In addition, we examined the efficacies of various concentrations of three lytic enzymes (lysozyme, mutanolysin, and lysostaphin) instead of or in addition to lysozyme (which is used in the standard procedure) for lysing the VRE strains and the effect of shortening the lysis and proteolysis steps (Table 1). Furthermore, we determined that eliminating the proteinase K digestion step (1) did not have a negative effect on the quality of the DNA in the plugs.
TABLE 1.
Optimization of PFGE protocol for typing of enterococci
| Parameter examineda | Details | Optimal procedure or solution |
|---|---|---|
| Reagents eliminated (during plug preparation) | RNase, Brij 58, deoxycholate, NaCl (used in reference 21), and PMSFb (used in reference 11) | 50 mM Tris-HCl (pH 8), 50 mM EDTA, mutanolysin (1,250 U/ml), lysozyme (2.5 mg/ml), proteinase K (1.5 mg/ml) |
| Lytic enzymes | Lysozyme (2.5 and 5 mg/ml), lysostaphin (25 and 50 μg/ml), mutanolysin (1,250 and 2,500 U/ml) | Combination of lysozyme and mutanolysin (2.5 mg/ml and 1,250 U/ml, respectively) |
| Lysis duration | Four different time periods (10 min and 1, 2, and 6 h) were examined | 10 min at 37°C |
| Proteolysis duration | Three different time periods (2, 4, and 8 h) were examined | 2 h at 55°C |
| Restriction digestion | Preincubation with restriction enzyme buffer for 0.1, 1.5, 4, and 6 h, before introducing the restriction enzyme; incubation (2, 4, 6, 16, and 20 h) with various concentrations (10 to 50 units per plug, in 10-unit increments) of the restriction enzyme | Brief (10 min) preincubation with restriction enzyme buffer, followed by incubation (2 h) with restriction enzyme (30 units per plug) |
| Agarose concentration | 0.8 to 1.2%, in 0.1% increments | 0.8% |
| Electrophoresis conditions | Block 1, two initial switching times (3.5 and 5 s) and two final switching times (20 and 25 s); block 2, two initial switching times (1 and 3.5 s) and two final switching times (5 and 12.5 s); duration of electrophoresis,c 16, 20, and 24 h | Block 1, voltage, 200 V, initial time, 3.5 s, final time, 25 s, duration, 12 h; block 2, voltage, 200 V, initial time, 1 s, final time, 5 s, duration, 8 h |
| Staining and destaining | Staining for 0.1, 0.5, and 2 h; destaining for 0.1, 2, and 10 h | Staining, 10 min; destaining, 15 min |
One value at a time was changed.
MSE, phenylmethylsulfonyl fluoride.
After the initial and switching times were optimized.
Optimization of restriction digestion and electrophoresis parameters.
Previous studies (21) established that digestion with SmaI is best for PFGE analysis of enterococci; thus, we used that enzyme in all of our PFGE typing experiments. However, because other investigators (19) reported that digestion with ApaI may be useful in PFGE typing of VRE strains, we examined whether the DNA in plugs prepared by our procedure was suitable for digestion with that enzyme. DNA of 20 randomly selected strains was incubated (25°C; 2 h) with ApaI, and PFGE typing revealed complete DNA digestion (data not shown). Thus, the DNA obtained by our procedure is suitable for analysis by both enzymes routinely used for PFGE typing of enterococci.
We also determined the conditions required for the most rapid, optimal digestion of the enterococcal DNA and the value of using various agarose concentrations and electrophoresis conditions for optimal resolution of the SmaI-generated DNA fragments (Table 1).
Optimization of staining and destaining.
Primarily because of safety and environmental concerns, we chose not to consider the approach (17) of including ethidium bromide in the agarose-electrophoresis buffer in order to eliminate the time required to stain gels after electrophoresis. However, we determined the shortest staining and destaining times yielding optimal results (Table 1).
Modified PFGE protocol.
The results of the above-described studies optimized the PFGE protocol for typing VRE. Briefly, bacteria from overnight L-agar cultures were harvested and washed twice with cell suspension buffer (100 mM Tris-HCl [pH 8] and 100 mM EDTA), and the suspensions were diluted with cell suspension buffer to a final optical density at 610 nm (1-cm light path) of 3.7 to 4.0 (ca. 2.5 × 109 CFU/ml). Aliquots (0.2 ml each) of the suspensions were lysed (Table 2), an equal volume of 1.2% molten SeaKem Gold agarose (FMC BioProducts, Rockland, Maine) containing 1% sodium dodecyl sulfate was added, and the mixtures were poured into 2-cm by 1-cm by 1.5-mm reusable plug molds (Bio-Rad Laboratories, Hercules, Calif.) and allowed to solidify at 4°C for 10 min.
TABLE 2.
Comparison between the standard and modified protocols for PFGE typing of enterococci
| Procedure | Standard protocol (21) | Modified protocol |
|---|---|---|
| Lysis | 6 mM Tris-HCl (pH 7.6), 1 M NaCl, 100 mM EDTA, 0.5% Brij 58, 0.2% deoxycholate, 0.5% sarcosyl, RNase (20 μg/ml), lysozyme (1 mg/ml); overnight at 37°C | 50 mM Tris-HCl (pH 8), 50 mM EDTA, mutanolysin (1,250 U/ml), lysozyme (2.5 mg/ml), proteinase K (1.5 mg/ml); 10 min at 37°C |
| Proteolysis | 0.5 M EDTA, 1% sarcosyl, 50 μg of proteinase K/ml; overnight at 50°C | 0.5 M EDTA, 1% sarcosyl, and 400 μg of proteinase K/ml; 2 h at 55°C |
| Washing | 3 times in TEa; 30 min each | 3 times in H2O, 50°C, 10 min each; 3 times in TE, 50°, 10 min each |
| Restriction digestion | Preincubation, none; digestion, SmaI (20 U; 6 h at 25°C) | Preincubation, 10 min at 30°C; digestion, SmaI (30 U; 2 h at 30°C) |
| Washing | TE (1 h at 37°C) | None |
| Electrophoresis | 200 V, pulse time increased from 5 to 35 s over 30 h; total time, 30 h | Block 1, 200 V, initial time, 3.5 s, final time, 25 s, 12 h; block 2, 200 V, initial time, 1 s, final time, 5 s; 8 h; total time, 20 h |
| Staining and destaining | 1.5 to 10 h | 25 min |
| Total duration | 3 to 7 days | ca. 28 h |
TE, Tris-EDTA buffer.
The proteolysis, washing, electrophoresis (performed using the CHEF Mapper or CHEF DR III apparatus [Bio-Rad]), and staining and destaining steps were performed as described in Table 2. This modified PFGE protocol, which includes a significantly faster method for making plugs (ca. 4 h instead of the usual 3 to 4 days) and a shorter electrophoresis time (20 h instead of 30 to 40 h) than does the standard protocol, allowed VRE typing to be performed in ca. 28 h instead of the 3 to 7 days required for the standard PFGE protocol (the major differences between the standard protocol and our modified protocol are summarized in Table 2).
Discriminatory ability and reproducibility of our procedure.
PFGE patterns obtained from eight enterococcal strains analyzed by our modified procedure are shown in Fig. 1B. The bands in Fig. 1A are better separated than the bands in Fig. 1B, and the improved resolution probably is due to the significantly longer electrophoresis time used in the standard procedure (30 h, versus 20 h in our modified protocol). However, the resolution achieved by our modified procedure provides the same level of discrimination among the VRE strains as does the standard procedure (i.e., the strains identified as separate clones by the standard procedure were also differentiated by our protocol [Fig. 1]). Thus, we found that our modified electrophoresis parameters are sufficient for generating easily interpretable PFGE banding patterns of VRE strains. The electrophoresis time can be shortened further if the results are urgently required (e.g., 16 h of electrophoresis provides good separation; data not shown) or can be prolonged if improved resolution of the bands is desired.
Analysis of the entire enterococcal strain collection by our modified procedure yielded clear banding patterns in all cases (data not shown). In addition, repeated typing (at least three times per strain) of 10 randomly chosen VRE isolates yielded identical patterns.
Applicability for typing of other gram-positive bacteria.
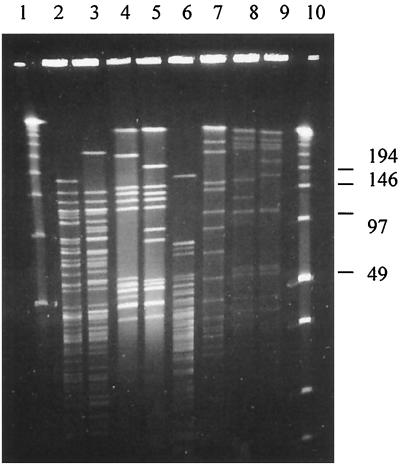
We analyzed a small collection of gram-positive, nonenterococcal bacteria by our modified procedure, without custom selecting the restriction enzyme or optimizing the PFGE parameters for each species. In all instances, a complete digest of plug-embedded DNA was obtained (Fig. 2), which suggests that the modified protocol also can be used (with some species-specific modifications in bacterial cell density, restriction enzymes, and fine-tuning of electrophoresis conditions) for molecular epidemiological analysis of gram-positive bacteria other than enterococci.
FIG. 2.
PFGE patterns of SmaI-digested DNAs of various gram-positive, nonenterococcal bacteria analyzed by our protocol. Lane 1, λ ladder; lanes 2 and 3, L. monocytogenes strains; lanes 4 and 5, S. pyogenes strains; lane 6, L. casei strain; lanes 7, 8, and 9, S. aureus strains; lane 10, low-range PFGE marker. Molecular sizes are shown in kilobases on the right.
In conclusion, the protocol described in this article allows rapid (ca. 28 h) and reproducible PFGE typing of enterococci and, potentially, other gram-positive bacteria. Moreover, although we have not calculated the actual cost of performing PFGE typing by our simplified procedure, it is likely, because of the reduction in reagents and personnel time, that the procedure is a cost-saving alternative to previously published PFGE typing protocols. We expect that the procedure will be of value during epidemiological investigations of VRE outbreaks and for comparative characterization of VRE strains isolated in different geographic loci.
Acknowledgments
We gratefully acknowledge Judith Johnson, Connie Mackinson, Zemphira Alavidze, and Ekaterine Chighladze for their help in subculturing and cataloguing the bacterial strains used in this study. M.K. was supported by an International Training and Research in Emerging Infectious Diseases grant from the Fogarty International Center, National Institutes of Health.
REFERENCES
- 1.Bannerman T L, Hancock G A, Tenover F C, Miller J M. Pulsed-field gel electrophoresis as a replacement for bacteriophage typing of Staphylococcus aureus. J Clin Microbiol. 1995;3:551–555. doi: 10.1128/jcm.33.3.551-555.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barbier N, Saulnier P, Chachaty E, Dumontier S, Andremont A. Random amplified polymorphic DNA typing versus pulsed-field gel electrophoresis for epidemiological typing of vancomycin-resistant enterococci. J Clin Microbiol. 1996;34:1096–1099. doi: 10.1128/jcm.34.5.1096-1099.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chiew Y F, Hall L M. Comparison of three methods for the molecular typing of Singapore isolates of enterococci with high-level aminoglycoside resistances. J Hosp Infect. 1998;3:223–230. doi: 10.1016/s0195-6701(98)90278-x. [DOI] [PubMed] [Google Scholar]
- 4.Facklam R R, Collins M D. Identification of Enterococcus species isolated from human infections by a conventional test scheme. J Clin Microbiol. 1989;27:731–734. doi: 10.1128/jcm.27.4.731-734.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fridkin S C, Yokoe D S, Whitney C G, Onderdonk A, Hooper D C. Epidemiology of a dominant clonal strain of vancomycin-resistant Enterococcus faecium at separate hospitals in Boston, Massachusetts. J Clin Microbiol. 1998;36:965–970. doi: 10.1128/jcm.36.4.965-970.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gordillo M E, Singh K V, Murray B E. Comparison of ribotyping and pulsed-field gel electrophoresis for subspecies differentiation of strains of Enterococcus faecalis. J Clin Microbiol. 1993;31:1570–1574. doi: 10.1128/jcm.31.6.1570-1574.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gottberg A, Nierop W, Duse A, Kassel M, McCarthy K, Brink A, Meyers M, Smego R, Koornhof H. Epidemiology of glycopeptide-resistant enterococci colonizing high-risk patients in hospitals in Johannesburg, Republic of South Africa. J Clin Microbiol. 2000;2:905–909. doi: 10.1128/jcm.38.2.905-909.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hall L M, Duke B, Guiney M, Williams R. Typing of Enterococcus species by DNA restriction fragment analysis. J Clin Microbiol. 1992;30:915–919. doi: 10.1128/jcm.30.4.915-919.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hsueh P R, Teng L J, Pan H J, Chen Y C, Wang L H, Chang S C, Ho S W, Luh K T. Emergence of vancomycin-resistant enterococci at a university hospital in Taiwan: persistence of multiple species and multiple clones. Infect Control Hosp Epidemiol. 1999;12:828–833. doi: 10.1086/501592. [DOI] [PubMed] [Google Scholar]
- 10.Huycke M M, Sahm D F, Gilmore M S. Multiple-drug resistant enterococci: the nature of the problem and an agenda for the future. Emerg Infect Dis. 1998;2:239–249. doi: 10.3201/eid0402.980211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jensen L B, Ahrens P, Dons L, Jones R N, Hammerum A M, Aarestrup F M. Molecular analysis of Tn1546 in Enterococcus faecium isolated from animals and humans. J Clin Microbiol. 1998;36:437–442. doi: 10.1128/jcm.36.2.437-442.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuhn I, Burman L G, Haeggman S, Tullus K, Murray B E. Biochemical fingerprinting compared with ribotyping and pulsed-field gel electrophoresis of DNA for epidemiological typing of enterococci. J Clin Microbiol. 1995;33:2812–2817. doi: 10.1128/jcm.33.11.2812-2817.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuhnen E, Rommelsheim K, Andries L. Combined use of phage typing, enterococcinotyping and species differentiation of group D streptococci as an effective epidemiological tool. Zentbl Bakteriol Mikrobiol Hyg. 1987;266:586–595. doi: 10.1016/s0176-6724(87)80242-0. [DOI] [PubMed] [Google Scholar]
- 14.Luginbuhl L M, Rotbart H A, Facklam R R, Roe M H, Elliot J A. Neonatal enterococcal sepsis: case-control study and description of an outbreak. Pediatr Infect Dis J. 1987;11:1022–1026. [PubMed] [Google Scholar]
- 15.Maekava S, Yoshioka M, Kumamoto Y. Proposal of a new scheme for the serological typing of Enterococcus faecalis strains. Microbiol Immunol. 1992;36:671–681. doi: 10.1111/j.1348-0421.1992.tb02070.x. [DOI] [PubMed] [Google Scholar]
- 16.Malathum K, Singh K V, Weinstock G M, Murray B E. Repetitive sequence-based PCR versus pulsed-field gel electrophoresis for typing of Enterococcus faecalis at the subspecies level. J Clin Microbiol. 1998;36:211–215. doi: 10.1128/jcm.36.1.211-215.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matushek M G, Bonten M J, Hayden M K. Rapid preparation of bacterial DNA for pulsed-field gel electrophoresis. J Clin Microbiol. 1996;10:2598–2600. doi: 10.1128/jcm.34.10.2598-2600.1996. . (Erratum, 2:536, 1997.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miranda A G, Singh K V, Murray B E. DNA fingerprinting of Enterococcus faecium by pulsed-field gel electrophoresis may be a useful epidemiologic tool. J Clin Microbiol. 1991;12:2752–2757. doi: 10.1128/jcm.29.12.2752-2757.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morris J G, Jr, Shay D K, Hebden J N, McCarter R J, Jr, Perdue B E, Jarvis W, Johnson J A, Dowling T C, Polish L B, Schwalbe R S. Enterococci resistant to multiple antimicrobial agents, including vancomycin: establishment of endemicity in a university medical center. Ann Intern Med. 1995;4:250–259. doi: 10.7326/0003-4819-123-4-199508150-00002. [DOI] [PubMed] [Google Scholar]
- 20.Morrison D, Woodford N, Barrett S P, Sisson P, Cookson B D. DNA banding pattern polymorphism in vancomycin-resistant Enterococcus faecium and criteria for defining strains. J Clin Microbiol. 1999;4:1084–1091. doi: 10.1128/jcm.37.4.1084-1091.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murray B E, Singh K V, Heath J D, Sharma B R, Weinstock G M. Comparison of genomic DNAs of different enterococcal isolates using restriction endonucleases with infrequent recognition sites. J Clin Microbiol. 1990;9:2059–2063. doi: 10.1128/jcm.28.9.2059-2063.1990. . (Erratum, 2:418, 1991.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perlada D E, Smulian A G, Cushion M T. Molecular epidemiology and antibiotic susceptibility of enterococci in Cincinnati, Ohio: a prospective citywide survey. J Clin Microbiol. 1997;35:2342–2347. doi: 10.1128/jcm.35.9.2342-2347.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]