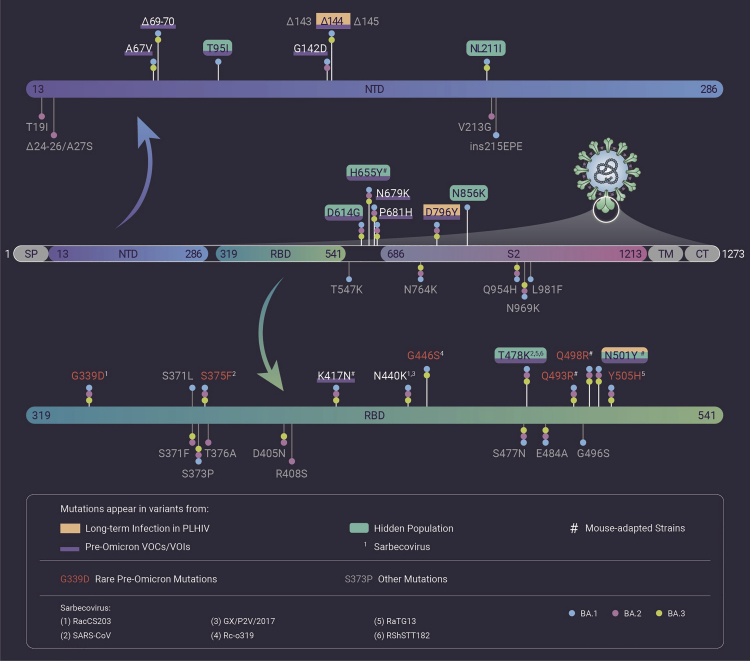
The coronavirus disease 2019 (COVID-19) pandemic has ravaged the world for nearly 2 years, with continuous emergence of new variants of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Omicron, the latest Variant of Concern (VOC), has spread to all six continents, leading to a rapid increase in confirmed COVID-19 cases and surpassing Delta as the dominant variant in many countries. So far, Omicron includes three sublineages, BA.1, BA.2, and BA.3, with BA.1 as the most prevalent one. The most unsettling feature of Omicron is the large number of mutations (50/51/43 mutations, hereafter ordered as BA.1/BA.2/BA.3) in its genome, a number far higher than that in the earlier VOCs. What is more concerning, 33/28/27 of these mutations are located on the spike (S) protein, with 15/16/15 concentrated in the receptor-binding domain (RBD) (Figure 1). Many mutations present in the Omicron variant have been detected in the Alpha, Beta, Gamma, Delta, and other SARS-CoV-2 variants as well and have been associated with enhanced severity, transmissibility, or immune escape. Such mutations in RBD quickly raised questions about the efficacy of current neutralizing antibodies and vaccines against Omicron, as RBD is the target of most SARS-CoV-2-neutralizing antibodies and the immunogen of many vaccines. Recently, scientists worldwide have been racing for the answers to this question. However, another astonishing fact about Omicron is that it contains many mutations rarely seen in other VOCs or Variants of Interest (VOIs) before the emergence of Omicron (rare pre-Omicron mutations), which signifies the mystery of Omicron and the evolutionary distance with the current VOCs, especially Delta. This raises another question with regard to the origins of Omicron: where did Omicron, with so many mutations, originate from? More specifically, how could Omicron suddenly appear with so many previously rare mutations, particularly in the S protein? Currently, there are three theories to this question. In this commentary, we provide analysis on the possibility of each theory with current evidence.
Figure 1.
Mutations in Omicron S protein that are present in other SARS-CoV-2 variants or sarbecoviruses
Mutations present in lineages associated with pre-Omicron VOCs and VOIs are drawn based on 75% prevalence in at least one lineage (outbreak.info, 2021-12-31 accessed). Mutations present in sarbecoviruses are drawn based on the following sequences: SARS-CoV (NC_004718), GX/P2V/2017 (EPI_ISL_410542), RaTG13 (EPI_ISL_402131), RacCS203 (MW251308), RShSTT182 (EPI_ISL_852604), and Rc-o319 (LC556375). Rare mutations are defined as existing in less than 0.02% of the 6,664,979 records on Gisaid.org (2021-12-31 accessed). Numbering of all mutation sites is based on the SARS-CoV-2 prototype strain (EPI_ISL_402119).
First, Omicron could have circulated and evolved in a hidden population. In this theory, the mystery lies in the question of how a variant could secretly evolve for over a year without being detected? A possible explanation is that Omicron, or its ancestral variant, evolved in a remote area of the world where nucleic acid tests are rarely conducted. In fact, nucleic acid tests in South Africa might have already caught a glimpse of Omicron's ancestor. For example, a variant discovered on July 28, 2021 (EPI_ISL_4652284) carries 20 mutations in the S protein and shares seven of them with Omicron (T95I, NL211I, T478K, N501Y, D614G, H655Y, and N856K). Notably, N856K is extremely rare in pre-Omicron mutations (507 out of 6,664,979 records; Gisaid.org, 2021-12-31 accessed), which suggests this variant as an ancestor of Omicron (Figure 1).
Second, Omicron may have evolved from a cat-and-mouse game between the virus and host in some immunosuppressed patients; for example, AIDS patients infected with SARS-CoV-2. Some scientists, such as Dr. Andrew Rambaut from the University of Edinburgh, found the hidden population theory hard to believe, because the long period of time the virus would need to stay hidden in a group of people seems improbable. Instead, they proposed that Omicron evolved from people living with HIV (PLHIV).1 A recent study showed that depletion of CD4+ T cells leads to significantly slower clearance of SARS-CoV-2 in mouse models,2 suggesting PLHIV, after being infected, would carry SARS-CoV-2 for a longer time than HIV-negative COVID-19 patients owing to lack of CD4+ cells. This persistent infection allows continuous mutation of SARS-CoV-2 through intra-host evolution within PLHIV (Figure 1).3 Furthermore, the immune system of PLHIV could generate neutralizing antibodies targeting the newly evolved viral strains but be unable to clear the virus owing to HIV infection. Thus, like an endless cat-and-mouse game, antibodies serve as a mild selective pressure that drives the adaptation of SARS-CoV-2 to the changing antibody environment, leading to the continuous addition of new mutations. Notably, as the most targeted region by neutralizing antibodies, RBD is probably under the heaviest selective pressure, which also explains the disproportionately greater mutations in RBD. Therefore, given a sufficient number of PLHIV and a long period of time, it is entirely possible for a variant with mutations as many as Omicron to evolve and become robust enough to spread in the real world.
Third, Omicron could have originated from adaptation in animal reservoirs and been transmitted back to human, exemplified by the SARS-CoV-2 variants with mutations from adapting to mink (e.g., F486L) that were transmitted back to human in the Netherlands and Denmark in 2020. For example, Omicron could be the result of SARS-CoV-2 transmitting back from mouse after adaptation. Several studies have demonstrated Omicron mutations generated in mouse-adapted SARS-CoV-2 variants by serial passaging in mouse.4 Specifically, both Q493R and Q498R emerged in two different variants (EPI_ISL_1666328 and MACo3), while K417N, N501Y, and H655Y have also emerged in various mouse-adapted variants (Figure 1). Notably, Q493R and Q498R are rare in pre-Omicron mutations (569 and 432 out of 6,664,979 records, respectively; Gisaid.org, 2021-12-31 accessed). These mutations suggest the potential mouse origins of Omicron, as their emergence in laboratories could also happen in nature. Furthermore, the suspect animals of SARS-CoV-2 transmitting back are not limited to mouse. Besides mink, many wild animals have been reported to be infected by SARS-CoV-2, including the ones infected in zoos (e.g., lions, tigers) and in the wild (e.g., snow leopards, pumas, gorilla, and white-tailed deer). Surveillance for SARS-CoV-2 infection can be easily conducted for animals living in zoos, but becomes much more difficult for the ones living in the wild, which raises the question: could SARS-CoV-2 transmit back to human from wild animals, especially the ones not under surveillance? In a recent survey, the SARS-CoV-2 infection rate of the white-tailed deer in North America has reached up to 70%, with three different lineages detected (B.1.2, B.1.582, and B.1.596) in multiple locations.5 Although no Omicron S mutation besides D614G was found from the variants detected in white-tailed deer (108 records from Odocoileus virginianus; Gisaid.org, 2021-12-31 accessed), the risk of Omicron, or future variants, originating from adaptation in other wild animals and transmitting back to human should be considered. In addition, several rare pre-Omicron mutations in Omicron were found in sarbecoviruses closely related to SARS-CoV-2, although only one or two of them exist in the same sarbecovirus, which suggests the examined sarbecoviruses are not likely to be the origins of Omicron (Figure 1). However, considering the close evolutionary links of sarbecoviruses with SARS-CoV-2 and our limited knowledge on sarbecoviruses, another question should be considered: could unknown sarbecoviruses be associated with the origins of Omicron? More research should be conducted to collect evidence regarding these questions.
The origins of Omicron remain mysterious after analyzing the S protein mutations, especially the rare pre-Omicron mutations. It appears all three theories may contribute to the generation of Omicron, making the origins of Omicron a complicated question involving three different origins. As a possible scenario, a SARS-CoV-2 variant could be transferred to a remote area where travel was not so convenient and started circulating in a hidden population, until being carried out and spread as the new Omicron variant. During this period, it may gain mutations from other SARS-CoV-2 lineages (e.g., B.1.1), from adaptation to animals living in close proximity to human (e.g., mouse), or from intra-host evolution in immunosuppressed patients.
With regard to the origins of Omicron, it is important to keep in mind that any seemingly unlikely opinions may turn out to be true when new information emerges, as long as these opinions are evidence based. This is a lesson learned from the simultaneous outbreaks of two different Ebola viruses, Ebola Zaire and Ebola Sudan, in 1976, which resulted in two vastly different death rates (88% and 53%, respectively). We may never pinpoint the origins of Omicron during the COVID-19 pandemic, as much evidence may have already disappeared in time. However, we must be vigilant to these possible ways of generating variants such as Omicron. If any of these possibilities are true, there will be more variants following Omicron until actions are taken to address them. As new variants continue to emerge and there exists no way to precisely predict the direction of mutations, the best strategy to control the spread of SARS-CoV-2 is still blocking the transmission immediately after new cases appear, as demonstrated by the zero-COVID policy in a few countries, and to ensure the herd immunity has been built through vaccine sharing in the whole world and vaccination being implemented worldwide.
Acknowledgments
Declaration of interests
The authors declare no competing interests.
Published Online: January 13, 2022
References
- 1.Kupferschmidt K. Where did 'weird' Omicron come from? Science. 2021;374:1179. doi: 10.1126/science.acx9738. [DOI] [PubMed] [Google Scholar]
- 2.Israelow B., Mao T., Klein J., et al. Adaptive immune determinants of viral clearance and protection in mouse models of SARS-CoV-2. Sci. Immunol. 2021;6:eabl4509. doi: 10.1126/sciimmunol.abl4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Karim F., Moosa M., Gosnell B., et al. Persistent SARS-CoV-2 infection and intra-host evolution in association with advanced HIV infection. medRxiv. 2021 doi: 10.1101/2021.06.03.21258228. [DOI] [Google Scholar]
- 4.Wei C., Shan K.J., Wang W., et al. Evidence for a mouse origin of the SARS-CoV-2 Omicron variant. J. Genet. Genomics. 2021 doi: 10.1016/j.jgg.2021.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hale V.L., Dennis P.M., McBride D.S., et al. SARS-CoV-2 infection in free-ranging white-tailed deer. Nature. 2021 doi: 10.1038/s41586-021-04353-x. [DOI] [PMC free article] [PubMed] [Google Scholar]