Abstract

Cancer is a leading cause of death worldwide and its incidence is unfortunately anticipated to rise in the next years. On the other hand, vascular endothelial growth factor receptor 2 (VEGFR-2) is highly expressed in tumor-associated endothelial cells, where it affects tumor-promoting angiogenesis. Therefore, VEGFR-2 is considered one of the most promising therapeutic targets for cancer treatment. Furthermore, some FDA-approved benzimidazole anthelmintics have already shown potential anticancer activities. Therefore, repurposing them against VEGFR-2 can provide a rapid and effective alternative that can be implicated safely for cancer treatment. Hence, 13 benzimidazole anthelmintic drugs were subjected to molecular docking against the VEGFR-2 receptor. Among the tested compounds, fenbendazole (FBZ, 1), mebendazole (MBZ, 2), and albendazole (ABZ, 3) were proposed as potential VEGFR-2 antagonists. Furthermore, molecular dynamics simulations were carried out at 200 ns, giving more information on their thermodynamic and dynamic properties. Besides, the anticancer activity of the aforementioned drugs was tested in vitro against three different cancer cell lines, including liver cancer (HUH7), lung cancer (A549), and breast cancer (MCF7) cell lines. The results depicted potential cytotoxic activity especially against both HUH7 and A549 cell lines. Furthermore, to improve the aqueous solubility of MBZ, it was formulated in the form of mixed micelles (MMs) which showed an enhanced drug release with better promising cytotoxicity results compared to the crude MBZ. Finally, an in vitro quantification for VEGFR-2 concentration in treated HUH7 cells has been conducted based on the enzyme-linked immunosorbent assay. The results disclosed that FBZ, MBZ, and ABZ significantly (p < 0.001) reduced the concentration of VEGFR-2, while the lowest inhibition was achieved in MBZ-loaded MMs, which was even much better than the reference drug sorafenib. Collectively, the investigated benzimidazole anthelmintics could be encountered as lead compounds for further structural modifications and thus better anticancer activity, and that was accomplished through studying their structure–activity relationships.
1. Introduction
Cancer is the second leading cause of mortality in the world, killing about 8 million people each year. Furthermore, it is a pity that cancer incidence is anticipated to rise by more than 50% in the next years.1−4 In comparison to healthy cells, cancer cells frequently require more oxygen and nutrients, but it is far from cancer’s only requirement. Therefore, researchers are paying efforts and attempts to discover and develop new therapies for many cancer types.5−9
Furthermore, in adults, angiogenesis is essential during tissue growth, repair, and pregnancy. In addition, angiogenesis is a basic underlying process in the pathogenesis of several human diseases, including cancer.10,11 Angiogenesis is a fundamental step in the turning of a benign tumor into a malignant one, where tumor masses are infiltrated by new blood vessels furnishing them with oxygen and nutrients to promote tumor growth and metastasis.12 Since its discovery in 1983, vascular endothelial growth factor (VEGF) has been considered the most important regulator of blood vessel formation and is a key mediator of neovascularization in cancer.10 Besides, VEGF can promote endothelial cell proliferation and motility. Hence, endothelial cells are abundantly represented in the malignant tissue.5,13,14 Cancer cells are not capable of growing nor metastasizing when they cannot express VEGF. Notably, VEGFs are overexpressed in several types of cancers including hepatocellular carcinoma, colorectal cancer, and breast cancer.12
The biological duties of the VEGFs are mediated by a family of protein tyrosine kinase receptors called VEGF receptor tyrosine kinases (VEGFR-TKs) including VEGFR-1, VEGFR-2, and VEGFR-3.15 VEGFR-1 and VEGFR-2 both enhance angiogenesis, whereas VEGFR-3 stimulation can induce lymphangiogenesis. Although VEGFR-1 has been shown to influence the function of VEGFR-2, VEGFR-2 has been revealed to mediate nearly all known VEGF cellular responses.12
When VEGF binds to VEGFR, it causes a conformational change in the receptor, which is followed by dimerization and phosphorylation of tyrosine residues. VEGF signaling through VEGFR-2 has been shown to play a key role in tumor angiogenesis regulation.16,17 As a result, VEGF/VEGFR-2 signaling represents a promising therapeutic target in cancer treatment.18−20 Therefore, inhibiting VEGFR-2 or downregulating its signaling is a pivotal strategy for developing novel drugs for a variety of human angiogenesis-dependent cancers.21
Additionally, concerning their common pharmacophoric features shared, VEGFR-2 antagonists showed four main structural features.18,22,23 These features are as follows: (i) a flat heteroaromatic ring system containing at least one H-bond acceptor, (ii) a central aryl ring (spacer), (iii) a pharmacophore moiety contains two groups of an H-bond acceptor and an H-bond donor (e.g., amide or urea), and (iv) a terminal hydrophobic group.12
On the other hand, drugs that have been clinically approved or experimentally evaluated for diseases other than cancer but have been revealed to have unexpected cytotoxicity against malignant cells could be treated as good repurposed anticancer candidates.24 Furthermore, several approved benzimidazole anthelmintic drugs were repurposed as anticancer agents.25−29 It is worth mentioning that fenbendazole possesses an efficient antiproliferative activity. It was introduced as a novel microtubule interfering drug with antineoplastic action that could be investigated as a promising anticancer agent.24 Besides, mebendazole was chosen for cancer cell suppression studies based on its pharmacokinetic properties.30 In addition, the phosphorylation of Bcl-2, which results in dosage and time-dependent intrinsic apoptotic action in melanoma cells driven by microtubule depolymerization, is one of the mebendazole’s anticancer mechanisms.30 Moreover, albendazole is a well-known FDA-approved anthelmintic medicine that is also cytotoxic to healthy cells and has been found as an anticancer agent. It was proved to have an extensive effect against human paclitaxel-resistant ovarian cancer cells.31
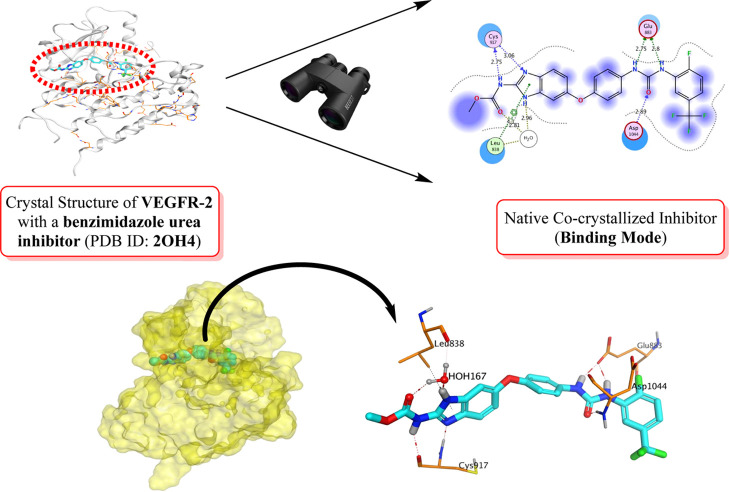
Accordingly, concerning the pivotal role of VEGFR-2 antagonism for cancer treatment and applying the repurposing approach (Figure 1), our perspective in this article was targeting VEGFR-2 through virtual screening on a small library of some FDA-approved benzimidazole anthelmintics (1–13) which were found to have a great structural similarity to the co-crystallized benzimidazole urea inhibitor (14) of VEGFR-2 (PDB ID: 2OH4(32)), as depicted in Figures 2 and 3. Also, we managed to enhance the solubility, dissolution, and the in vitro anticancer activity of one of the promising benzimidazole anthelmintics (MBZ) by preparing it in the form of mixed micelles (MMs) incorporating vitamin E.
Figure 1.
Graphical representation for the repurposing of benzimidazole anthelmintic drugs as VEGFR-2 antagonists.
Figure 2.

Chemical structures of FDA-approved benzimidazole-based anthelmintic agents; fenbendazole 1, mebendazole 2, albendazole 3, ricobendazole 4, cyclobendazole 5, oxibendazole 6, oxfendazole 7, dribendazole 8, parbendazole 9, bendazole 10, tiabendazole 11, triclabendazole 12, flubendazole 13, as well as the crystalline benzimidazole-urea inhibitor (14) as potent VEGFR-2 inhibitor.
Figure 3.
Crystal structure of VEGFR-2 (PDB: 2OH4) with a benzimidazole urea inhibitor shows its essential binding interactions.
Accordingly, we carried out molecular docking33 of the selected ligands on the 3D crystal structure of the mentioned VEGFR-2. Furthermore, molecular dynamics (MD) simulations were performed for the most promising members of the screened anthelmintics to confirm the obtained docking results and obtain more insights regarding the drug’s thermodynamic properties at the target receptor.34,35
Then, our proposed in silico studies were complemented through different in vitro studies on the best-screened candidates of benzimidazole anthelmintic drugs for their cytotoxicity and VEGFR-2 inhibiting potentials as well.
On the other hand, MBZ suffers from poor aqueous solubility which has been a major negative impact on its oral bioavailability. Therefore, a high dose of MBZ is administered to achieve a proper therapeutic effect, causing many adverse effects. High doses of MBZ cause anemia and liver damage.36 Moreover, studies have shown evidence of teratogenic effects of MBZ in rats and mice. To overcome poor aqueous solubility and enhance the bioavailability of MBZ at a lower dose, MMs were prepared to contain a hydrophobic block that enables the poorly soluble drugs to be incorporated as an internal core and a hydrophilic block as a surrounding shell improving its activity.37
1.1. Rationale of the Work
Compounds with the benzimidazole carbamide scaffold have been introduced as potent inhibitors of the VEGFR-2 and endothelium-specific receptor tyrosine kinase (TIE-2).32 The respective prototypical compound of such chemical class was albendazole being identified through initial high-throughput screening efforts and originally targeted as an anthelmintic agent.38 The X-ray crystallographic structure of the top active benzimidazole-urea compound in complex with VEGFR-2 showed favored ligand anchoring at the ATP-specific site mediating polar contacts with Cys919 of the hinge region (Figure 3). While the methoxycarbamido functional group of the crystalline ligand extended toward the solvent front region, the rest of the skeleton showed deep anchoring toward the back pocket region of the receptor with its urea moiety being sandwiched between Asp1046 and Glu885 at the Asp-Phe-Gly (DFG) motif of the VEGFR-2 activation loop. The compound exhibited potent in vitro activity against both VEGFR-2 and TIE-2 with IC50 values down to single-digit nanomolar concentrations.32
Owing to the great conserved structure of the ATP binding site of several human kinases’ receptors39 as well as sharing the 2-methoxycarbamido-1H-benzimidazole core skeleton as with different anthelmintic agents, it was highly rationalized to explore the VEGFR-2 inhibition activity of market-released benzimidazole-based anthelmintic drugs acting as promising anticancer agents. The latter has driven us to determine the efficiency of FDA-approved benzimidazole-based anthelmintic drugs (Figure 3) against VEGFR-2 using in silico and in vitro approaches, while the preliminary data obtained from this study would guide further optimization within the future based on structure–activity relationships (SARs) studied being attaining. Adopting this drug repurposing approach possesses the advent of assuring medical safety because these FDA-approved benzimidazole-based anthelmintic drugs have already been tested in animal models, undergone all the essential clinical trials, and are already infrastructured toward manufacture at large scales.40,41
2. Results and Discussion
2.1. Docking Studies
Molecular docking stimulation of fenbendazole 1, mebendazole 2, albendazole 3, ricobendazole 4, cyclobendazole 5, oxibendazole 6, oxfendazole 7, dribendazole 8, parbendazole 9, bendazole 10, tiabendazole 11, triclabendazole 12, flubendazole 13, and the benzimidazole urea inhibitor 14 into the active site of VEGFR-2 receptor was done. They got stabilized at the VEGFR-2 binding site by variable several electrostatic bonds. The order of strength of binding was as follows: the benzimidazole urea inhibitor (14, docked) > fenbendazole (1) > mebendazole (2) > albendazole (3) > ricobendazole 4 > cyclobendazole 5 > oxibendazole 6 > oxfendazole 7 > dribendazole 8 > parbendazole 9 > bendazole 10 > tiabendazole 11 > triclabendazole 12 > flubendazole 13.
Most drugs showed nearly binding modes similar to the benzimidazole urea co-crystallized inhibitor. Many poses were obtained with better binding modes and interactions inside the receptor pocket. The poses with the most acceptable scores (related to the stability of the pose) and rmsd_refine values (related to the closeness of the selected pose to the original ligand position inside the receptor pocket) were selected. Results of scores and different interactions with the amino acids of protein pocket are depicted in Table 1.
Table 1. Receptor Interactions and Binding Energies of the Tested Anthelmintic Drugs (1–13) and the Docked Benzimidazole–Urea Inhibitor (14) at the Binding Site of the VEGFR-2 Receptor.
| no. | anthelmintic drug | Sa | rmsdb | amino acid bonds | distance (A) |
|---|---|---|---|---|---|
| 1 | fenbendazole | –8.18 | 1.08 | Leu838/H2O bridged H bond | 2.74 |
| Leu838/H2O bridged H bond | 2.89 | ||||
| Cys917/H-donor | 2.96 | ||||
| Cys917/H-acceptor | 3.12 | ||||
| Leu838/pi-H | 3.98 | ||||
| Val846/pi-H | 4.69 | ||||
| 2 | mebendazole | –8.12 | 0.93 | Leu838/H2O bridged H bond | 2.68 |
| Leu838/H2O bridged H bond | 2.93 | ||||
| Cys917/H-donor | 2.97 | ||||
| Cys917/H-acceptor | 3.08 | ||||
| Leu838/pi-H | 4.03 | ||||
| 3 | albendazole | –7.91 | 1.15 | Leu838/H2O bridged H bond | 2.74 |
| Leu838/H2O bridged H bond | 2.90 | ||||
| Cys917/H-donor | 3.01 | ||||
| Cys917/H-acceptor | 3.12 | ||||
| Leu838/pi-H | 4.01 | ||||
| 4 | ricobendazole | –7.85 | 1.31 | Leu838/H2O bridged H bond | 2.77 |
| Cys917/H-donor | 2.89 | ||||
| Leu838/H2O bridged H bond | 2.99 | ||||
| Cys917/H-acceptor | 3.03 | ||||
| Leu838/pi-H | 4.09 | ||||
| 5 | cyclobendazole | –7.60 | 1.66 | Leu838/H2O bridged H bond | 2.83 |
| Cys917/H-donor | 2.86 | ||||
| Leu838/H2O bridged H bond | 2.97 | ||||
| Cys917/H-acceptor | 3.02 | ||||
| Leu838/pi-H | 4.06 | ||||
| 6 | oxibendazole | –7.58 | 1.05 | Cys917/H-donor | 2.80 |
| Leu838/H2O bridged H bond | 2.92 | ||||
| Cys917/H-acceptor | 3.09 | ||||
| Leu838/pi-H | 4.03 | ||||
| 7 | oxfendazole | –7.54 | 1.69 | Leu838/H2O bridged H bond | 2.75 |
| Cys917/H-acceptor | 3.63 | ||||
| Leu838/pi-H | 4.04 | ||||
| 8 | dribendazole | –7.53 | 1.31 | Cys917/H-donor | 2.85 |
| Leu838/H2O bridged H bond | 2.91 | ||||
| Cys917/H-acceptor | 3.06 | ||||
| Leu838/pi-H | 4.03 | ||||
| Val846/pi-H | 4.82 | ||||
| 9 | parbendazole | –7.46 | 1.06 | Cys917/H-donor | 2.80 |
| Leu838/H2O bridged H bond | 2.92 | ||||
| Cys917/H-acceptor | 3.05 | ||||
| Leu838/pi-H | 4.06 | ||||
| Val846/pi-H | 4.72 | ||||
| 10 | bendazole | –6.84 | 0.70 | ||
| 11 | tiabendazole | –6.62 | 0.59 | Leu838/H2O bridged H bond | 2.82 |
| Leu838/H2O bridged H bond | 2.92 | ||||
| Cys917/H-acceptor | 3.30 | ||||
| Leu838/pi-H | 3.97 | ||||
| 12 | triclabendazole | –6.40 | 1.18 | Leu838/H2O bridged H bond | 2.83 |
| Cys917/H-acceptor | 3.22 | ||||
| Leu838/pi-H | 3.89 | ||||
| 13 | flubendazole | –5.94 | 1.05 | Leu838/H2O bridged H bond | 2.72 |
| Leu838/H2O bridged H bond | 2.88 | ||||
| Cys917/H-donor | 2.96 | ||||
| Cys917/H-acceptor | 3.09 | ||||
| Leu838/pi-H | 4.06 | ||||
| Val846/pi-H | 4.78 | ||||
| 14 | benzimidazole urea inhibitor (GIG) | –11.15 | 1.39 | Cys917/H-donor | 2.82 |
| Glu883/H-donor | 2.86 | ||||
| Glu883/H-donor | 2.89 | ||||
| Leu838/H2O bridged H bond | 2.92 | ||||
| Asp1044/H-acceptor | 2.94 | ||||
| Cys917/H-acceptor | 3.10 | ||||
| Leu838/pi-H | 4.02 |
S: score of a compound inside the binding pocket of protein (kcal/mol).
rmsd/refine: root-mean-square deviations among heavy atoms of crystallized structure (prerefinement) and those of the obtained binding mode (postrefinement).
By analyzing docking results of the selected anthelmintics, it was found that most of the selected compounds manifested very close binding scores and modes compared to the co-crystallized inhibitor (GIG) at the VEGFR-2 target receptor. FBZ (1), MBZ (2), and ABZ (3) were found to have the best binding affinities and modes against VEGFR-2 with binding scores of −8.18, −8.12, and −7.91 kcal/mol, respectively (Table 1). These energy values were close to that of the docked GIG inhibitor (binding energy = −11.15 kcal/mol). When we put eyes on the binding interactions of FBZ (1) with the VEGFR-2 target receptor, we could reveal that it showed two water bridged hydrogen bonds with Leu838, interacted with Cys917 through both hydrogen bond donor and acceptor, and bound to Leu838 and Val846 via pi–H bonds. However, regarding the binding interactions of MBZ (2) and ABZ (3), both revealed that they are bound to Leu838 through two water bridged hydrogen bonds, interacted with Cys917 through hydrogen bond donor and acceptor, and bound to Leu838 via a pi–H bond as well.
The detailed binding modes of the docked benzimidazole urea inhibitor (GIG, 14) and all of the tested anthelmintics (1–13) are presented in Table 1. Nevertheless, the 3D-protein positioning, as well as 3D-binding interactions of the best selected three anthelmintics (1, 2, and 3), is described in Table 2.
Table 2. 3D View of Binding Interactions and the 3D Positioning of the Best-Docked Benzimidazole Anthelmintic Drugs (1, 2, and 3) within the VEGFR-2 Receptor Pocket (PDB: 2OH4) Compared to the Benzimidazole Urea Inhibitor (Docked, 14).
2.2. MD Simulations
For gaining an in-depth understanding of the relative stability and thermodynamic behavior of the assigned ligand–protein complexes across a specific time, a MD investigation was conducted. The latter computational tool is considered particularly beneficial for investigating the conformational spaces of ligand–protein complexes which are considered more advantageous over other different computational techniques including mechanics-energy minimization and molecular docking approaches concerning the sole analysis of static images.42 Exhibiting relevant ligand–protein docking interactions, the top-docked binding modes of the benzimidazole-based anthelmintic compounds (FBZ, MBZ, and ABZ) in addition to the crystalline reference inhibitor (GIG), at the VEGFR-2 active site, were enrolled within a 200 ns explicit MD simulation for comparative dynamic investigation.
2.2.1. Analyzing the Stability of Protein–Ligand Complexes
Under 200 ns MD simulation, all investigated anthelmintic compounds showed significant thermodynamic stability at the hVEGFR-2 active site which was confirmed via monitoring the root-mean standard-deviation (rmsd) trajectories. Typically, the rmsd determines the deviation of the molecules in relation to a designated reference/original structure. This analytical parameter would indicate ligand–protein stability and confirm the validity of the MD simulation protocol being adopted. High rmsd values are indicative of significant conformational changes and target instability.43 However, high complex rmsds are correlative for minimal ligand–protein affinity having the ligand unable to be maintained at the protein’s pocket throughout the MD simulation interval.44
In reference to the backbone, the obtained VEGFR-2 protein’s rmsds showed overall typical MD simulation behaviors (Figure 4A). At the beginning of MD runs and over the initial 20 ns frames, the protein backbone rmsds elevated due to the release of constraining applied during the prior minimization and equilibration stages. After the initial 20 ns, steady protein rmsds were depicted over more than half the MD run (>150 ns), except for limited fluctuations being illustrated at FBZ-associated protein across the 50–60 ns time-frames. Nearly all simulated proteins leveled off at almost similar rmsds throughout the equilibration plateau and until the MD simulation ends. Comparable rmsd trajectories were depicted for ABZ-, MBZ-, and GIG-bounded VEGFR-2 protein, following their respective equilibration with average values of 3.34 ± 0.18, 3.59 ± 0.37, and 3.96 ± 0.23 Å, respectively. However, slightly higher values were assigned for FBZ-bound protein (5.12 ± 0.31 Å), being correlated to its depicted limited fluctuations as well as late equilibration following the 50 ns of the MD simulation timeframes. The ABZ-bound VEGFR-2 protein managed to achieve the steadiest rmsd trajectories, illustrating minimal standard deviation (SD) following the attained equilibrium. All described VEGFR-2 protein thermodynamic behaviors emphasize the successful target protein convergence across the designated MD simulation timeframe. Additionally, the above-described protein rmsds inferred the adequacy of the prior system minimization/relaxation and thermal equilibration as well as the adopted 200 ns MD runs that required no further extensions.
Figure 4.

rmsd trajectory analysis of the examined anthelmintic compounds and reference inhibitor in bound with VEGFR-2 target across the 200 ns explicit MD runs. (A) Protein’s backbone-rmsds; (B) ligand–protein complex backbone-rmsds; and (C) only ligand backbone-rmsds (Å), along MD timeframe (ns).
To insure the maintenance of the simulated ligand at the VEGFR-2 ATP-binding site, the combined ligand–protein complex rmsd deviations were monitored having the protein backbones as their reference frame (Figure 4B). All simulated VEGFR-2/ligand complexes successfully attained their respective thermodynamic stability state showing backbone rmsd plateau despite limited fluctuations. Although differential backbone rmsds were shown across the initial MD frames, each of the ABZ, MBZ, and GIG models was capable to converge across the second 100 ns timeline, achieving the final rmsd value at 3.80 ± 0.33 Å. Nonetheless, the latter dynamic behaviors were nontypical for the FBZ–protein complex because the depicted model showed two equilibration plateau; an initial equilibration around 20–40 ns and a latter one beyond 50 ns until the end of the MD simulation run. Such a dynamic behavior suggested significant FBZ conformation change beyond 50 ns. Nevertheless, the FBZ ligand itself was suggested to be confined within the target’s binding site because a slight complex rmsd shift was depicted between both equilibria (3.87 ± 0.23 → 4.49 ± 0.18 Å) and such attained values were not higher than those depicted for the protein’s rmsds. On the other hand, compounds ABZ, MBZ, and crystalline reference GIG achieved earlier equilibrations of steadier complex rmsd trajectories and lower comparative average values (3.30 ± 0.29, 3.62 ± 0.29, and 3.97 ± 0.30 Å, respectively). All the above findings highlight the significant ligand’s retainment of the investigated anthelmintic agents within the protein active site, which was highly comparable to the crystalline GIG potent VEGFR-2 inhibitor.
Further analysis was proceeded through monitoring the only ligand’s rmsd tones in relation to the reference backbone frame of the protein which was considered as a relevant stability indicator of ligand’s confinement within the pocket as well as simulated protein convergence (Figure 4C). Interestingly, both MBZ and GIG showed the steadiest and lowest average ligand rmsd trajectories across the whole MD simulation runs (1.52 ± 0.29 and 1.68 ± 0.22 Å, respectively) inferring limited ligand conformational changes within the VEGFR-2 binding site. Concerning the ABZ compound, limited fluctuations were depicted at the initial MD simulation frames where subsequently the rmsds reached equilibration maintaining the initial ligand’s conformation/orientation across the same timeframes where the respective VEGFR-2 protein showed successful convergence (beyond the 20 ns). Notably, the average ABZ ligand rmsd value following equilibration (1.29 ± 0.29 Å) was nearly comparable to those of both MBZ and GIG suggesting further stability of these ligand-docked poses within the target site. In concordance with the above FBZ–protein rmsd trajectories, the ligand showed stable trajectories until the 50 ns time frames where after that the rmsds were elevated and attained stable trajectories until the end of the MD simulation. Such a dynamic behavior ensures the significant change of the FBZ compound attaining a second conformation/orientation following the 50 ns simulation run. Notably, the rmsd tones of each VEGFR-2 protein did not exceed the 1.5-fold of those of their corresponding ligand in complex, which further confirms the successful convergence of the ligand–protein complexes and infers the suitability of 200 ns MD simulation runs requiring no further extension.
The ligand–protein global stability was further investigated through monitoring both the radii of gyration (Rg) and solvent-accessible surface area (SASA) trajectories of the complex entities along with the whole MD timeframe. Typically, the estimated radii of gyration of the investigated complexes permitted the exploration of the complex rigidity and compactness where this stability parameter accounts for the complex’s mass-weighted root-mean-square distance relative to its common mass center. In these regards, minimal Rgs with achieved plateau around the average value would be correlated to the sustained stability/compactness of an investigated complex.45 On similar bases, decreased SASA trajectories confer ligand–protein structural shrinkage due to the influence of solvent-surface charges, which would lead to more conformational compactness and stability. The latter has been correlated to the SASA calculation which estimates the molecular surface area being assessable to solvent molecules, providing a quantitative measurement of the complex-solvent interaction.46
Herein, the steadiest Rg trajectories were assigned for the crystalline ligand, GIG, in complex with VEGFR-2, showing an average value of 20.65 ± 0.13 Å (Figure 5A). Concerning both FBZ and MBZ, the complexes seemed to be expanding at the initial MD simulation frames, with the MBZ–protein complex being at higher Rg tones (max values 21.08 and 21.59 Å, respectively). However, both systems achieved compactness and significant contraction following the 80 ns and until the MD ends. The ABZ–protein complex, on the other hand, showed initial compactness for more than half the MD runs (20.68 ± 0.13 Å), while after that the complex seemed to be expanding until the next 40 ns where then it finally attained significant compactness with the lowest Rg trajectories among all investigated molecules (20.16 Å). Interestingly, the four investigated ligand–protein complexes converged around similar Rg values (20.68 ± 0.19 Å) at 200 ns, confirming the comparable compactness and stability profiles of the three anthelmintic models as well as the crystalline potent inhibitor. Significant intra-/intermolecular interactions among the investigated complexes were further confirmed through the calculated complex SASA tones (Figure 5B). Following the 60 ns MD timeframe, comparable complex SASA trajectories were assigned for the ABZ– and MBZ–protein complexes as well as the crystalline inhibitor (172.82 ± 3.46, 173.07 ± 2.77, and 172.19 ± 3.20 nm2, respectively), conferring preferential ligand confinement within the VEGFR-2 binding site. Notably, the FBZ–protein complex illustrated significantly reduced SASA tones (165.78 ± 3.23 nm2) following 50 ns and until the end of the simulation run, suggesting better ligand–protein interactions, particularly for the second ligand’s conformation, because binding is a solvent-substitution process.
Figure 5.

Global stability profiles of obtained Rg and SASA tones of the examined anthelmintic compounds and reference inhibitor complexed with VEGFR-2 target across the 200 ns explicit MD run. (A) Complex Rg (Å); (B) complex SASA tones (nm2), along MD timeframe (ns).
Because the presented rmsd analysis highlights the significant ligand-target stability for the examined ligands, it was beneficial to further investigate the local protein flexibility and how this could be contributed to the ligand–protein binding. The fluctuation of VEGFR-2’s residues was monitored by estimating the RMS-fluctuation (RMSF) stability validation parameter being able to highlight the residue-wise contribution within the target protein stability. Typically, RMSF provides a valuable evaluation of the target’s residue dynamic behavior represented as both fluctuation and flexibility, through estimating the average deviation of each protein’s amino acid in relation to its respective reference position across time.47 Within the presented article, the difference RMSF (ΔRMSF) was a better estimation of the protein local flexibility being the RMSF difference for each ligand-bound protein relative to the VEGFR-2 apo state (ΔRMSF = apo RMSF – holo RMSF). Adopting a ΔRMSF cutoff value of 0.30 Å was relevant for estimating the significant alterations within the protein’s structural movements, meaning that amino acids with ΔRMSF above 0.30 were considered of limited mobility.48 Investigating the RMSF trajectories essentially execute for a trajectory region considered stable. Based on the above protein’s rmsd analysis (Figure 4A), the VEGFR-2 proteins targets were of significant conformational stability along the 200 ns MD simulations for all systems despite the limited fluctuations for the FBZ system. Therefore, the backbone RMSF calculations were reasoned to be estimated across the whole MD simulation trajectories.
Throughout the ΔRMSF analysis, the free terminal residues showed a typical fluctuation pattern with the highest negative ΔRMSF values in comparison to the core residues (Figure 6). The latter could be reasoned for the lower extent of intramolecular interactions among the terminal residues as compared to those of core amino acids which have been considered ideal behavior throughout MD simulation runs. Notably, patterns of high fluctuation were illustrated for all ligand-bound VEGFR-2 amino acids near the N-terminal relative to those settled near the carboxy end (average −3.90 ± 0.51 vs 0.12 ± 1.43 Å). Almost all residue ranges within the C-lobe and N-lobe of the kinase domain illustrated significant immobility depicting positive ΔRMSF values. Interestingly, the residue ranges Ser875–Leu880 of the N-lobe and Lys1053–Arg1059 at C-lobe showed the highest immobility profiles with ΔRMSF up to 4.13 and 4.74 Å, respectively. Such dynamic behavior confers significant influence of ligand’s binding upon the stability of these residue ranges (particularly at C-lobe) or in other terms the pivotal role of these residues for the ligand stability at the VEGFR-2’s active site. This came in great agreement with several reported studies investigating the potential antiangiogenic VEGFR-2 inhibition activity of naturally occurring metabolites and chemical library deposits.49−54 However, it is worth noting that both residue ranges were proven to possess relatively conserved hydrogen bond interactions among the constituting residues as well as with the binding inhibitors.55 On the contrary, the residue range Ala943–Glu993 was of the most flexible pattern (ΔRMSF down to the highest negative values −9.92 Å) inferring the negligible contribution of such residues within the ligand–protein interactions. This was also consistent with the reported data by where the residue range of Thr940–Glu989 possessed an insignificant impact upon the catalytic activity of the VEGFR-2 protein.56
Figure 6.

Difference RMSF analysis across VEGFR-2 residues bound to the investigated anthelmintic compounds and reference ligand across the 200 ns MD runs. The protein’s backbone-ΔRMSFs were determined considering independent 200 ns MD run for holo VEGFR-2 states (in complex with investigated ligands or crystalline reference inhibitor, GIG) against the unliganded/apo state (PDB ID: 1VR2). Trajectories of ΔRMSF are illustrated as functional residue numbers (residues 814-up to-1169).
Concerning the comparative local stability of ligand-bound VEGFR-2 proteins, trends of high negative ΔRMSF values were depicted for FBZ-bound amino acids as compared to those of other investigated anthelmintic agents as well as reference inhibitors. These trends were recognized along with several VEGFR-2 residue ranges while being most noted for the flexible Ala943–Glu993 range as well as Asp1062–Lys1068 residue range at the C-lobe domain. On the other hand, the highest residue-wise stability and rigidity profiles were assigned for the ABZ being comparable to that of the crystalline potent inhibitor, which further confirms the relevant stability of such systems as being previously discussed within the rmsd findings.
Further investigation of ligand-VEGFR-2 interactions was proceeded through comparative studying the furnished ΔRMSF values and specific flexibility of the pocket’s canonical lining amino acids. Interestingly, several key pocket residues, as well as vicinal residues, showed significant inflexibility having ΔRMSF values beyond the threshold index 0.30 Å (Table 3). The hinge region residues depicted moderate immobility profiles with both Cys917 and Lys918 represented as the most rigid ones among such residue range, exhibiting comparable ΔRMSF values (0.76 ± 0.10 Å) across the four investigated ligands. The latter infers the important role of these two hinge residues for anchoring the polar carbamoyl of anthelmintic agents as well as the urea scaffold of the crystalline ligand through hydrogen bond interactions. Both the hydrophobic back-pocket and ATP-association cleft showed moderate rigidity profiles for all ligands. Such a significant inflexibility pattern highlights the preferential and deep anchoring of all anthelmintic compounds within the binding site of the kinase domain. Despite being short-length ligands, the anthelmintic agents managed to achieve a relevant residue-wise stability profile for the deep hydrophobic back pocket being comparable to that of the crystalline potent inhibitor. Such important observation further confirms the potential inhibition activity of these anthelmintic agents.
Table 3. Estimated ΔRMSFa Values for Ligand-VEGFR-2 Proteins across the 200 ns MD Runs.
| canonical subsites | comprising residues | FBZ | MBZ | ABZ | GIG |
|---|---|---|---|---|---|
| hinge region | Val914 | 0.35 | 0.33 | 0.34 | 0.438 |
| Glu915 | 0.46 | 0.45 | 0.45 | 0.50 | |
| Phe916 | 0.47 | 0.45 | 0.45 | 0.44 | |
| Cys917 | 0.72 | 0.65 | 0.66 | 0.66 | |
| Lys918 | 0.91 | 0.81 | 0.84 | 0.80 | |
| ATP-binding cleft | Leu838 | 1.34 | 1.19 | 1.32 | 1.27 |
| Ala864 | 1.15 | 0.98 | 1.07 | 1.05 | |
| Val865 | 0.78 | 0.63 | 0.76 | 0.68 | |
| Lys866 | 0.70 | 0.60 | 0.67 | 0.61 | |
| Glu883 | 0.78 | 0.85 | 0.92 | 1.13 | |
| Gly920 | 1.15 | 1.01 | 1.15 | 1.16 | |
| Leu1033 | 0.64 | 0.58 | 0.71 | 0.67 | |
| hydrophobic deep back pocket | Ile886 | 0.39 | 0.52 | 0.54 | 0.77 |
| Leu887 | 1.04 | 0.99 | 1.01 | 1.16 | |
| Ile890 | 0.96 | 0.87 | 0.82 | 0.99 | |
| Val896 | 0.62 | 0.53 | 0.66 | 0.68 | |
| Val897 | 0.45 | 0.28 | 0.47 | 0.46 | |
| Val912 | 0.34 | 0.28 | 0.16 | 0.50 | |
| Leu1017 | 0.36 | 0.31 | 0.47 | 0.41 | |
| Cys1043 | 0.59 | 0.54 | 0.68 | 0.66 | |
| Ile1042 | 0.82 | 0.19 | 0.77 | 0.82 | |
| glycine-rich region | Gly841 | 1.28 | 1.11 | 1.23 | 1.26 |
| Ala842 | 1.31 | 1.19 | 1.26 | 1.31 | |
| Phe843 | 1.49 | 1.42 | 1.44 | 1.49 | |
| Gly844 | 1.14 | 1.01 | 1.09 | 1.11 | |
| Gln845 | 1.39 | 1.04 | 1.41 | 1.23 | |
| Val846 | 2.11 | 1.88 | 2.11 | 2.03 | |
| catalytic loop | Cys1022 | 0.48 | 0.46 | 0.65 | 0.69 |
| Ile1023 | 0.50 | 0.38 | 0.61 | 0.70 | |
| His1024 | 0.33 | 0.28 | 0.38 | 0.41 | |
| Arg1025 | 0.33 | 0.43 | 0.55 | 0.48 | |
| Asp1026 | 0.30 | 0.31 | 0.34 | 0.31 | |
| activation loop DFG motif (Asp–Phe–Gly) and vicinal residues | Cys1043 | 0.42 | 0.09 | 0.45 | 0.43 |
| Asp1044 | 0.27 | 0.06 | 0.26 | 0.22 | |
| Phe1045 | 0.45 | 0.23 | 0.42 | 0.49 | |
| Gly1046 | 0.29 | –0.01 | 0.29 | 0.29 | |
| Leu1047 | 0.46 | 0.08 | 0.44 | 0.54 | |
| Ala1048 | 0.99 | 0.13 | 0.90 | 1.07 | |
| Arg1049 | 0.69 | –0.39 | 0.66 | 0.74 |
ΔRMSF values were determined for each ligand-associated VEGFR-2 protein in relation to unliganded/apo state (PDB ID: 1VR2). ΔRMSF > 0.30 Å immobility threshold was set, where amino acids with significant inflexibility/immobility profiles showed values above this designated cutoff.
Interestingly, the glycine-rich region exhibited the highest immobility profiles with ΔRMSF values up to 2.03 ± 0.11 Å being assigned for the Val846 hydrophobic residue. The latter could be reasoned for the conserved hydrogen bonding among the region comprising residues as well as the close proximity of Val846 toward the ATP-binding site inferring its pivotal role for the stability of the investigated ligand and crystalline inhibitor.39 Moving toward the significant loops near the canonical substrate binding site, higher flexibility trends were assigned for residues of the DFG motif of the activation segment rather than those of the catalytic loop. Notably, the ΔRMSF values for the activation loop DFG motif were lower than those of any of the binding site subsites. Residues including Asp1044 and Gly1046 as well as Arg1049 only for the FBZ–protein model were below the 0.3 Å threshold or even at negative values inferring their insignificant role in stabilizing the binding ligands. Interestingly, significantly higher flexibility was assigned for activation loop residues in complex with FBZ which may be due to the fact that the ligand exhibited conformational shift throughout the MD simulation. Based on the ΔRMSF furnished stability results across different pocket subsites, comparable dynamic behaviors were depicted for the three anthelmintic agents in relation to the crystalline inhibitor. Despite adopting altered conformation/orientation, the FBZ-bound protein residues exhibited almost comparable inflexibility and stability profiles to those of ABZ, MBZ, and the crystalline inhibitor (GIG). All of which are in good agreement with the above-described MD behaviors illustrated through the analysis of Rg, SASA, and rmsd findings. However, examining the differential conformations of investigated ligand-VEGFR-2 complexes as well as time-evolution of ligand-VEGFR-2 binding interactions at selected frames would provide valuable information regarding the nature and conformationally directed ligands’ affinity toward the VEGFR-2 binding site.
2.2.2. Conformational and Intermolecular Binding Analysis
Analysis of key conformational alterations across the MD simulation timeframe was performed through examining the ligand–protein models at trajectories of the start and final timeframes. For each ligand-VEGFR-2 model, frames at 0 and 200 ns were extracted and minimized to a gradient of 1 × 10–3 kcal mol–1 A–2 using the MOE-package. Stable ligand-target binding profiles were depicted for almost all investigated anthelmintic agents and reference inhibitors. Interestingly, both ABZ and MBZ showed deeper anchoring into the VEGFR-2 pocket at the MD simulation end (Figure 7A,B). However, the initial polar interaction between both ligand’s benzimidazole carbamoyl scaffold and the main chain of Cys917 (CO and NH) were maintained at overall frequencies of 75.51 and 77.34% for ABZ and MBZ, respectively, across the MD simulation. Notably, the occupancy of the Cys917 (C=O) mainchain hydrogen bond pair is always around twofold higher than that of the Cyst917 (NH) mainchain functionality. Several hydrophobic pocket residues, including Leu838, Val846, Val897, Phe916, Leu1033, and Phe1045, maintained the sandwich-like conformation around the linear hydrophobic skeleton of both ABZ and MBZ until 200 ns MD runs. This was observed because these residues kept short distances from both ligand’s structure along the 200 ns simulation run as being presented within the given heatmap (Figure 7A,B lower panels). On the other hand, few ionizable residues such as Lys866, Glu883, and Asp1044 might have imposed unfavored repulsion forces with the ligand’s hydrophobic terminal. This was particularly obvious with MBZ where the terminal benzoyl of MBZ showed 45° rotation from its initial docked position to minimize steric hindrances as well as unfavored contacts with Lys866 and Glu883 (Figure 7B).
Figure 7.
Conformational and intermolecular distance analysis of simulated ligand–protein complexes. (A) FBZ; (B) MBZ; (C) ABZ; and (D) GIG. Upper panels are overlaid snapshots of the ligand-VEGFR-2 complexes at 0 and 200 ns of MD runs. The VEGFR-2 proteins are illustrated in red and green 3D representation (cartoon) relative to the last and initially extracted frames, respectively. Both ligands (represented as sticks) and polar hydrogen bond interactions (dashed lines) are colored in correspondence to their respective extracted frames. Lower panels represent the heatmap representation of the time evolution of intermolecular distances between binding ligand and protein residues functionalities during the whole MD simulation. Polar interaction (hydrogen bonding) distances were measured between the designated interacting donor-H···acceptor, while the distances of hydrophobic interactions were measured from the nearest interacting atom of the ligand to the Cα of a particular residue. The values of the intermolecular distances were conditionally formatted through a color scale from 0 Å (green) and up to 10 Å (white) using the Microsoft EXCEL spreadsheets.
The third anthelmintic compound, FBZ, showed a significant conformational shift for its terminal aromatic moiety, yet kept the rest of the compound without relevant conformational/positional changes (Figure 7C). The latter could explain the second equilibration plateau beyond the 50 ns at the previously described rmsd tones (Figure 4). Interestingly, the initial close proximity of the phenyl substitution at Lys866 might have caused such depicted orientation flip, allowing a more favored position where the aromatic group was at an ideal T-shaped π–π stacking with Phe1045. However, such a new orientation allowed the phenyl substitution to face another ionizable residue, Arg1049, yet stabilization imposed by Phe1045 might overcompensate any depicted ring-residue repulsions. Finally, the great superimposition of the FBZ’s benzimidazole carbamoyl scaffold at initial and 200 ns allowed stronger hydrogen bond pairing with Cys917 main chain with slightly higher frequencies (80.17%) as compared to other anthelmintic compounds. Moving toward the crystalline inhibitor, minimal conformational changes were depicted for the ligand’s core skeleton, while the terminal aromatic moiety was the most being altered (Figure 7D). The hydrogen pair with Cys917 was also maintained, yet at a much higher frequency (91.61%) with the preferentiality for the carbonyl mainchain functionality of this hinge residue. Another polar hydrogen bond pairing was deduced through analyzing the running trajectories including urea carbonyl: Asp1044 mainchain (53.20%), urea NH: Glu883 sidechain (3.35%), and fluoro substitutions: His1024 NH sidechain (0.25%) hydrogen bond pairs. The heat map for ligand-residue intermolecular distance showed the much more close-range hydrogen-donor acceptor distances for Asp1044 polar interactions over those of Glu883 and His1024 across the simulation where both weak bonds were lost by the end of the MD simulation. Finally, the crystalline ligand showed close range hydrophobic distances with more pocket lining nonpolar residues which further stabilize the ligand at the target pocket.
2.2.3. Free Binding Energy Calculations
The calculation of the binding free energy was performed to understand the ligand-VEGFR-2 interaction nature, investigate comparative ligand-directed pocket affinities, and obtain more insights regarding individual residue-wise energy contributions.57 In these regards, the molecular mechanics Poisson/Boltzmann’s surface area (MM/PBSA) calculations were used to estimate the binding-free energies, where lower negative values confer less ligand’s affinity toward the target binding site.58 Accuracy of the MM/PBSA calculation is considered comparable to the free-energy perturbation methods, yet with the advantage of lower computational expenditures.58 Both single trajectory approach and only SASA model (ΔGTotal = ΔGMolecular Mechanics + ΔGPolar + ΔGAPolar) were adopted. Extracted and saved representative frames along the entire 200 ns MD runs were used for estimating each term of the binding energy as well as their average values. The latter was reasoned because ligand–protein complexes rmsds rapidly achieved the convergence/equilibration plateau after few initial MD trajectories (Figure 4B).
To our delight, all simulated anthelmintic agents illustrated significant binding-free energies and in turn affinities toward the VEGFR-2’s binding site (Table 4). Comparable binding-free energies at significant negative values were assigned for the three anthelmintic agents; −105.28 ± 11.82, −99.52 ± 23.18, and −96.28 ± 39.37 kJ/mol for FBZ, MBZ, and ABZ, respectively. Interestingly, the latter binding free energy pattern came in great concordance with the preliminary docking investigation showing preferential higher docking scores for FBZ, followed by that of MBZ and then ABZ. Similarly, higher binding-free energy was depicted for the crystalline reference inhibitor being at almost 1.5-fold those of the investigated ligands. This preferential binding affinity highlights the significant impact of the fluorinated aromatic carbamide tail substitution in guiding the ligand-VEGFR-2 binding and mediating preferential ligand anchoring at the deep target pocket. Considering the cited potent inhibitory bioactivity of the crystalline compound against VEGFR-2 and serving as a relevant antiangiogenic agent,32 the nearly matchable binding affinity data for these anthelmintic agents imply their promising activities toward this same protein target.
Table 4. Total Free Binding Energies (ΔGTotal binding ± SD) and Dissected Energy Terms Regarding Investigated Anthelmintic Agents and Reference Inhibitor at the VEGFR-2 Active Site.
| ligand–protein complex |
||||
|---|---|---|---|---|
| energy (kJ/mol ± SD) | FBZ | MBZ | ABZ | GIG |
| ΔGvan der Waal | –158.88 ± 10.57 | –164.78 ± 35.98 | –155.90 ± 16.17 | –247.06 ± 49.17 |
| ΔGElectrostatic | –23.68 ± 35.00 | –29.06 ± 44.78 | –23.72 ± 21.09 | –36.04 ± 52.00 |
| ΔGSolvation; Polar | 93.36 ± 47.82 | 110.30 ± 38.65 | 98.82 ± 38.73 | 160.76 ± 49.30 |
| ΔGSolvation; SASA | –16.08 ± 1.91 | –15.95 ± 2.46 | –15.48 ± 1.63 | –25.07 ± 4.78 |
| ΔGTotal binding | –105.28 ± 11.82 | –99.52 ± 23.18 | –96.28 ± 39.37 | –147.41 ± 19.77 |
The furnished free binding energies illustrated the dominance of van der Waal potentials within the binding energy calculations of each simulated ligand. Higher electrostatic and lipophilic energy contributions were depicted for the crystalline reference over those of the anthelmintic agents due to possessing profound aromatic characteristics and hydrogen bond features (high numbers of hydrogen bong acceptors) at its terminal substitutions. This was highly reasoned because the crystallized ligand depicted a higher hydrogen bond number/frequency with VEGFR-2 pocket residues than any of the investigated anthelmintic agents. It is worth mentioning that the total nonpolar interactions (ΔGvan der Waal plus ΔGSASA) were higher for the crystalline inhibitor as compared to the anthelmintic compounds, which could be correlated to the large surface area of the VEGFR-2 pocket. Interestingly, several reported data have considered the pocket of VEGFR-2 as deep and more hydrophobic than other comparable targets possessing conserved nonpolar residues lining the target’s pocket.32,39,56 Being with a large surface area and hydrophobic, the VEGFR-2 pocket would favor higher hydrophobic interactions with GIG as this ligand can attain more extended structural conformations at the target’s binding site. On the other hand, deep anchoring of anthelmintic agents at the target pocket while attaining relevant could be achieved through furnishing favored strong hydrophilic binding interactions with VEGFR-2 pocket key amino acids. This was clearly presented throughout the above illustrated intermolecular bonding as well as conformational analyses.
All furnished data confirm the important role of hydrophobic/polar combined functional groups at the terminal fluorinated aromatic carbamide tail substitutions for anchoring the reference ligand at the VEGFR-2 pocket. However, these particular polar functional moieties might serve as double-bladed influencers upon ligand-target binding. This could be reasoned that such functional groups impose higher ΔGsolvation for the GIG–protein system, compromising the ligand’s anchoring as the binding process is considered a solvent-substitution one. Therefore, these anthelmintic compounds could be optimized through future developmental approaches via introducing ionizable groups with relevant lipophilic characters (e.g., tetrazole ring) has been considered significant for ΔGsolvation reduction, ligand-VEGFR-2 binding extension, and in turn potential target inhibition.
Free binding energy decomposition via g_mmpbsa scripts for identifying the key amino acids enrolled in the furnished free binding energies.58 Interestingly, almost comparable residue-wise energy contributions were depicted for three anthelmintic agents except for limited differences regarding the magnitude of energy contribution (Figure 8A–C). This was expected as these benzimidazole agents showed close values of the dissected energy terms and total free binding energies. The significant residues showing favored contribution (high negative values) within the ligand–protein binding energy include the following: Val914, Phe916, and Cys917 (hinge region); Leu838, Ala864, Gly920, and Leu1033 (ATP-binding cleft); Val897 and Cys1043 (hydrophobic back pocket); Val846 of the glycine-rich region; and finally Cys1043, Phe1045, and Leu1047 at the activation loop. On the other hand, the crystalline ligand GIG shared many of the residue-wise energy contributions being comparable to those of the anthelmintic compounds, however, of significantly higher magnitudes (Figure 8D). Moreover, additional residues such as Ile886, Leu887, Ile890, Leu1017, and His1024 were depicted as significant for the binding of the crystalline ligand at the protein pocket.
Figure 8.

Binding-free energy/residue decomposition illustrating the protein residue contribution at ligand–protein complex ΔGTotalbinding calculation. (A) FBZ; (B) MBZ; (C) ABZ; and (D) GIG.
The nature of the top energy-contributing residues is mostly hydrophobic which further emphasizes the predominance of the van der Waals potentials for binding the investigated ligands deep into the target binding site. However, the strong polar contacts with the hinge Cys917 provided a significant role in Coulomb’s electrostatic potentials for enforced ligand–protein biding. On the contrary, several other VEGFR-2 pocket-lining amino acids depicted positive energy contribution values with all ligands, which inferred repulsion forces and unfavored ligand-VEGFR-2 affinity. Among these energy-positive contributing residues are the ionizable Lys866, Glu883, Glu915, Asp1044, and Arg1049 amino acids. The furnished residue-wise energy contributions were consistent with ΔRMSF results, showing significant immobility and rigidity for all high energy-negative contributing residues while depicting higher fluctuations for the energy-positive contributors inferring significant repulsion and instability. Notably, the number of the energy-positive contributing residues was less in the case of GIG as compared to any of the anthelmintic agents where Lys866 and Arg1049 lacked unfavored energy contributions. Based on the previous conformational analysis, three of the repulsive residues (Lys866, Glu883, and Asp1044) showed close proximity toward the terminal aromatic substitutions of both FBZ and MBZ, the terminal propyl in ABZ, or the central aromatic ring of GIG ligands. That is why the substitution of the anthelmintics’ terminal structure with polar functionality (e.g., carbamide or guanidine moiety as close analogue) was suggested to overcompensate the repulsive-derived binding energy contributions through providing preferential polar binding interactions with Glu883 and/or Asp1044. Concerning the other side of the simulated ligands, the hinge Glu915 residues impose repulsive force against the aromatic phenyl ring within the core benzimidazole scaffold of all simulated compounds.
Certain residue-wise energy contributions were characteristic for the FBZ and GIG simulated systems. Unlike the other investigated anthelmintic agents, FBZ lacked the energy-positive contribution by Lys866, while expressed nonprecedential repulsive forces by Arg1049 as well as attractive favored binding interactions with Val865. The latter was reasoned because FBZ adopted a second different conformation following 50 ns simulation where its terminal aromatic tail became at close proximity toward Arg1049 rather than Lys866. Moving toward GIG, the ATP-binding specific residue, Glu883, imposed much higher positive energy contributions against the ligand-binding as compared to anthelmintic agents. Based on the conformational analysis, the Glu883 negatively charged residues were at additional closeness toward the fluorinated ring substitution. On the contrary, the unfavored positive-energy contribution by Asp1044 was much reduced in the case of GIG because being best oriented toward the polar carbamide rather than the hydrophobic parts of the ligand. Based on the above differential residue-wise contribution, it became highly reasoned that the fluorinated aromatic carbamide terminal substitution could impose a double-bladed impact upon the preferential anchoring of GIG within the VEGFR-2 pocket, which redeems further structure-based modification and optimization.
In brief, the MD findings were in great concordance with docking ordering values for the examined anthelmintic compounds and co-crystallized ligand. The free binding energy furnished through MM/PBSA was GIG > FBZ > MBZ > ABZ being at comparable ordering as obtained by the docking simulation findings. This may be reasoned for the strong polar interactions mediated by the ligand’s benzimidazole carbamoyl scaffold at the VEGFR-2 hinge region, while only the ligand’s terminal aromatic functionality would provide the differential ligand-target binding interactions. The same findings were consistent through protein ΔRMSF and ligand-target hydrogen bond analysis of the MD simulation trajectories. Nevertheless, the MD simulation provided one of the most interesting findings that have been highlighted, where FBZ managed to attain a second orientation/conformation beyond the 50 ns MD run. FBZ showed a significant conformational shift for its terminal aromatic moiety, yet kept the rest of the compound without relevant conformational/position changes (Figure 7A). Despite such conformational shift, FBZ managed to attain stabilized orientation within the VEGFR-2 binding site and top calculated free binding energy values across the investigated anthelmintic agents. This was suggested for a more favored position attained by the aromatic group where it formed an ideal T-shaped π–π stacking with Phe1045 overcompensating any depicted ring-residue unfavored electrostatic interactions. The latter observation further confirms the fact that the ligand’s terminal aromatic functionality would provide the differential ligand-target binding interactions. This was obvious with the superior free-energy of binding for co-crystallized ligand (GIG) which was correlated to the significant role of the polar/hydrophobic combined functionalities of its terminal fluorinated aromatic carbamide tail substitution for anchoring within the VEGFR-2 binding site.
2.3. Characterization of the Prepared MBZ-Loaded MMs
Obviously, the solubilization of a hydrophobic drug can be enhanced by its structure in biological media, and thus, the drug bioavailability and stability could be improved. The nanoscopic size of micelles (<100 nm) lets them be readily carried to the systemic circulation. Besides, MM possesses drug-loading efficiency and higher stability compared to micelles with single components.37 Incorporation of vitamin-E in nanocarriers has been proved to improve the hydrophobicity of the drug delivery system, enhance the solubility of the loaded poorly soluble drugs, increase the biocompatibility of the polymeric drug carriers, and improve the anticancer potential of the anticancer agents by reversing the cellular drug resistance via simultaneous administration. In addition to being a powerful antioxidant, vitamin E demonstrated its anticancer potential by inducing apoptosis in various cancer cell lines.59
2.3.1. Particle Size, PDI, and Zeta Potential
The particle size of MBZ-loaded MMs was found to be 110.8 ± 1.1 nm, as shown in Figure 9. The polydispersity index (PDI) is a measure of the distribution homogeneity of molecular size in a given sample. Low PDI indicates that the BBV dispersion is homogeneous in nature.60 The PDI was 0.17 ± 0.014, indicating good size homogeneity of the micellar solution.
Figure 9.
(A) Size and size distribution of the micelles, (B) zeta potential of the MBZ-loaded MMs.
MMs with ZP > 30 mV were considered electrophoretically stable. MBZ-loaded MMs showed a negative ZP value of −22.8 ± 1.3 mV, and the negative charge is due to the presence of sodium deoxycholate (SDC) in the micelles.61
2.3.2. Transmission Electron Microscopy
To ensure the morphological structure of the prepared MBZ-loaded MMs, they were inspected using negative stain transmission electron microscopy (TEM), as shown in Figure 10. The shape of the prepared MMs was almost spherical and uniform in shape.
Figure 10.
TEM micrograph of MBZ-loaded MMs. Scale bar in 200 (A) and 100 nm (B).
2.3.3. Entrapment Efficiency and Drug Loading
The entrapment efficiency of MBZ in the prepared MMs was 25 ± 2%. The drug loading capacity expressed as % of MBZ from the weight of the micelles forming materials was 1%.
2.3.4. In Vitro Dissolution of MBZ and MBZ-Loaded MMs
The cumulative percent of MBZ released, as a function of time, from prepared lyophilized MMs in comparison to plain drug powder is illustrated in Figure 11. Dissolution testing was performed in 0.1 N HCl (USP dissolution media of MBZ). Dissolution is well known as an important indicator of absorption and eventual bioavailability.62
Figure 11.
Dissolution profiles of MBZ drug and MBZ-loaded MMs in 0.1 N HCl medium.
Interestingly, lyophilized MBZ-loaded MMs showed almost 100% (97 ± 0.02) release of MBZ after 2 h, while MBZ plain powder showed only 34 ± 1.3%. The preparation of MBZ in the form of MMs considerably improved its dissolution ability; consequently, this will increase the absorption of MBZ from GIT and provide a greater benefit in the biological system.
2.4. In Vitro Cytotoxic Activity
Based on the previously discussed in silico studies, three anthelmintic drugs, namely, FBZ (1), MBZ (2), and ABZ (3), together with MBZ-loaded MMs, and plain MMs were tested for their cytotoxic activity against three cancer cell lines, including liver cancer (HUH7) cell line, lung cancer (A549) cell line, and breast cancer (MCF7) cell lines. Different concentrations of drugs were prepared in dimethyl sulfoxide (DMSO) ranging from (1, 10, 100, and 1000 μg/mL), and incubated with the cancer cells for 48 h, the cell viability was tested using 3-[4,5-methylthiazol-2-yl]-2,5-diphenyl-tetrazolium bromide (MTT). Tested drugs showed potent cytotoxic activity against the liver cancer cell line (Figure 12), while MBZ-loaded MMs was the best cytotoxic agent, with IC50 (6.54 μg/mL), which was even better than both plain MMs and MBZ free form (IC50 75.19 and 32.42 μg/mL, respectively).
Figure 12.

Cytotoxic effect of the tested compounds against different cancer cell lines. (A) HUH7% cell viability upon treatment with a series of tested compounds, (B) A549% cell viability upon treatment with a series of tested compounds. (C) MCF7% cell viability upon treatment with a series of tested compounds. Concentrations were used starting from 1 to 1000 μM for 48 h, and the cytotoxicity effect was detected by the MTT assay (n = 3).
Testing anticancer activity against lung cancer showed that FBZ, MBZ, and ABZ had promising antilung cancer activity with IC50 (5.74, 22.45, and 32.98 μg/mL, respectively). Free MBZ showed slightly higher cytotoxic activity compared to MBZ-loaded MMs (31.93 μg/mL).
From another point, free FBZ, MBZ, and ABZ showed low cytotoxic activity against breast cancer cell line (MCF7), with IC50 values (216.38, 948.46, and 854.33 μg/mL, for FBZ, MBZ, and ABZ respectively). These results prove the high affinity of chosen drugs for VEGFR-2, as VEGFR-2 was noted to be poorly expressed in the MCF-7 cells both in vitro and in vivo.63 Interestingly, MBZ-loaded MMs showed higher anticancer activity against MCF7 with IC50 (26.09 mg/mL), compared to plain MMs (240.99 μg/mL) and free MBZ (948.46 μg/mL) (Table 5).
Table 5. IC50 of the Tested Compounds on Different Cell Lines.
| IC50 (μg/mL) |
|||
|---|---|---|---|
| compound | HUH7 | A549 | MCF7 |
| FBZ (1) | 28.23729 | 5.74467 | 216.38202 |
| MBZ (2) | 32.42383 | 22.45513 | 948.46979 |
| ABZ (3) | 21.1508 | 32.98596 | 854.33276 |
| MBZ-loaded MMs | 6.54465 | 31.9354 | 26.09982 |
| plain MMs | 75.19966 | 472.62847 | 240.99073 |
| 5FU | 14.8919 | 19.6689 | ND |
| Taxol | 6.68 | 37.72706 | 25.61472 |
| Dox. | 4.95317 | 6.35791 | 11.40258 |
However, the enhanced cytotoxic activity of plain MMs incorporating vitamin E only or MBZ-loaded MMs is attributed to higher endocytosis by the cells and lower efflux of MBZ by the P-group pumps, allowing the MBZ to remain inside the cells for a longer duration and resulting in higher cytotoxicity (26.1 μg/mL).59,64,65 Furthermore, the enhanced cytotoxicity of MMs may be due to the presence of F127 and vitamin E, which are known to be inhibitors of the P-group and reduce the efflux of drugs.59,66 Vitamin E also shows both in vitro and in vivo anticancer activities against various cancer cells due to its apoptosis-inducing properties.67
2.5. Cell-Based VEGFR-2 Assay
To further validate the previously investigated in silico study, in vitro quantification for VEGFR-2 concentration in treated HUH7 cells with the drug under investigation has been conducted based on enzyme-linked immunosorbent assay. The results disclosed that all tested drugs significantly (p < 0.001) reduce the concentration of VEGFR2 from (3255 ± 312 pg/mL) for control to (1787 ± 37.3 pg/mL) for FBZ, (2041 ± 30.1 pg/mL) for MBZ, and (1213 ± 21.5 pg/mL) for ABZ, while the lowest inhibition for VEGFR-2 was achieved in MBZ-loaded MMs (860.8 ± 312 pg/mL), which was even much better than the reference drug, sorafenib (1073 ± 41.1) (Figure 13) (Table 6).
Figure 13.
Schematic diagram illustrating the concentration of VEGFR-2 in HUH7 cells treated with the IC50 of tested drugs after 48 h using ELISA technique (n = 3), *p < 0.05, ***p < 0.001 compared to control.
Table 6. Concentration of VEGFR-2 in HUH7 Cells Treated with the IC50 of Tested Drugs after 48 h.
| compound | VEGFR-2 (pg/mL) |
|---|---|
| control | 3255 ± 181 |
| FBZ (1) | 1787 ± 64.6 |
| MBZ (2) | 2041 ± 52.1 |
| ABZ (3) | 1213 ± 37.2 |
| MBZ-loaded MMs | 860.8 ± 57.1 |
| sorafenib | 1073 ± 41.1 |
3. SAR Studies
Therefore, based on the stabilities and binding scores of benzimidazole anthelmintic drugs to VEGFR-2, we could put eyes on the SAR that showed very promising and interesting activities against VEGFR-2, and hence the best expected antineoplastic activity. Moreover, concerning the anticancer activity of benzimidazole derivatives, the best affinity of anthelmintic drugs toward VEGFR-2 was attained when benzimidazole scaffold substituted at position 6 with phenyl sulfanyl group, FBZ (1), with benzoyl group, MBZ (2), or with propylthio group, ABZ (3). Furthermore, the studied SAR revealed that better activity could be accomplished when benzimidazole scaffold is substituted at position 2 with carbamate than any other group. Besides, it was noticed that substitution of benzimidazole scaffold at position 6 showed better activity against VEGFR-2 than unsubstituted ones (Figure 14).
Figure 14.

SAR study of the tested benzimidazole anthelmintics as VEGFR-2 antagonists.
4. Conclusions
Thirteen benzimidazole anthelmintic drugs were subjected to molecular docking studies as promising anticancer agents targeting VEGFR-2. The tested drugs showed comparable binding mode toward the ATP binding pocket of the VEGFR-2 receptor as compared to the potent co-crystallized benzimidazole urea inhibitor especially for FBZ (1), MBZ (2), and ABZ (3). Moreover, MD simulations revealed the great stability and binding affinity of the aforementioned investigated anthelmintic agents toward the VEGFR-2 active site. Achieving comparable free-binding energy and thermodynamic behavior along the 200 ns simulation time further confirms the suitability of these benzimidazole-based anthelmintic agents for repurposing approach targeting cancer’s angiogenesis pathway. Also, the enhanced release of MBZ from the MMs in USP dissolution media of MBZ ensured better oral availability after administration. Thus, allowing the administration of lower doses of MBZ consequently decreasing its side effects. Hence, these three promising drugs besides the MBZ-loaded MMs were subjected to further in vitro screening using three different cancer cell lines, including HUH7, A549, and MCF7 cell lines to confirm their anticancer activities. Accordingly, IC50 values for FBZ were 28.23, 5.74, and 216.38, for MBZ were 32.42, 22.45, and 948.46, for ABZ were 21.15, 32.98, and 854.33, and for MBZ-loaded MMs showed better promising in vitro cytotoxicity values of 6.54, 31.93, and 26.09 concerning HUH7, A549, and MCF7 cell lines, respectively. The MBZ-loaded MMs showed increased in vitro cytotoxicity in MCF7, HUH7, and neutral effect on A549 compared to free FBZ, MBZ, and ABZ. In conclusion, MMs could be used for enhancing the bioavailability and anticancer activity of MBZ. Finally, an in vitro quantification for VEGFR-2 concentration in treated HUH7 cells has been conducted based on an enzyme-linked immunosorbent assay. All tested drugs significantly (p < 0.001) reduced the concentration of VEGFR-2 from (3255 ± 312 pg/mL) for control to (1787 ± 37.3 pg/mL), (2041 ± 30.1 pg/mL), and (1213 ± 21.5 pg/mL) for FBZ, MBZ, and ABZ, respectively, while the lowest inhibition was achieved in MBZ-loaded MMs (860.8 ± 312 pg/mL), which was even much better than the reference drug sorafenib (1073 ± 41.1). Hence, MM nanoformulations could be used for the delivery of MBZ in cancer treatment. Additionally, the investigated benzimidazole anthelmintics can be treated as lead compounds for further structural modifications. That was accomplished after shedding light on the SARs improving their anticancer activity.
5. Materials and Methods
5.1. Materials
FBZ, MBZ, ABZ, and vitamin E were kindly gifted from Adwia, Pharaonic, and EVA pharmaceutical companies, Egypt. Pluronic F127 was procured from Sigma-Aldrich. SDC was purchased from BASF Co. (Florham Park, New Jersey, USA). Acetonitrile HPLC grade and ethanol 96% HPLC grade were purchased from CARLO ERBA Reagents (France). Ammonium acetate and hydrochloric acid were obtained from Merck (Darmstadt, Germany). Deionized (DI) water from the Ultrapure (type 1) water system (Direct-Q3 UV) was used for the preparation of all buffer and water-based solutions.
5.2. Docking Studies
Molecular docking studies of 13 benzimidazole anthelmintic drugs (1–13) at the VEGFR-2 receptor (PDB: 2OH4)32 using MOE 2019.0102 drug design software68,69 were performed to evaluate their predicted affinity as potent VEGFR-2 antagonists compared to the co-crystallized benzimidazole urea inhibitor (GIG, 14).
5.2.1. Preparation of the Tested Benzimidazole Anthelmintic Drugs
The chemical structures of the tested benzimidazole anthelmintic drugs were downloaded from the PubChem database (https://pubchem.ncbi.nlm.nih.gov/). They were converted into their 3D forms and checked for their chemical structures and the formal charges on atoms to be prepared for docking.70−74 Then, energy minimization and automatic calculation of the partial charges were also done as previously described.75−79 Finally, they were imported with the isolated co-crystallized benzimidazole urea inhibitor in the same database and saved as an MDB file before docking calculations with the target VEGFR-2 receptor.
5.2.2. Target VEGFR-2 Receptor Optimization
The X-ray structure of the target VEGFR-2 receptor complex was downloaded from the Protein Data Bank (http://www.rcsb.org/, PDB: 2OH4, resolution of 2.05 Å).32 It was prepared for docking studies as follows: adding hydrogen atoms with their standard 3D geometry, and checking for any errors in the atom’s connection and the type through automatic correction as described earlier.80−83 The applied force field was Amber10: EHT, and the co-crystallized ligand site was selected to be the docking site to show the different interactions with it in the complex structure.84−86
5.2.3. Docking of the Tested Drugs to the Target VEGFR-2 Receptor Active Site
The prepared database containing both the 13 benzimidazole anthelmintics (1–13) together with the benzimidazole urea co-crystallized inhibitor (GIG, 14) was docked using the MOE 2019 suite.68,87 The applied methodology was as follows:71,88−90 the docking was initiated as a general process after loading the file of the target protein active site. The docking site was selected to be the ligand site, triangle matcher was chosen as the placement methodology, and London dG was selected as the scoring methodology.91 The refinement methodology was selected as a rigid receptor and the scoring methodology was GBVI/WSA dG for the selection of the best poses.11,86 The MDB file was generally docked automatically. After the end of the docking process, the obtained poses were carefully studied, and the best ones for each having the best interactions and scores with the protein pocket were selected.
5.3. MD Simulations
Best docked models of promising anthelmintic agent leads, as well as reference inhibitor, in complex with VEGFR-2, represented the starting coordinates for 200 ns explicit MD runs via GROMACS-2019 package.92−94 The automatic CHARMM-General ForceField program (https://cgenff.umaryland.edu/) was used for ligand parameterization and topology file generation.95 The CHARMM36m forcefield was considered suitable for simulated proteins.33,89,96 Models were solely solvated in TIP3P cubic 3D-box at periodic boundary conditions with 10 Å marginal distances between protein and 3D-box sides.97 The VEGFR-2 amino acids were set at their respective standard ionization within physiological pH 7.0. The entire system net charge was neutralized via sufficient chloride and sodium ions introduced using the Monte-Carlo ion-placement method.98
Following construction, the systems were minimized through 5 ps under the steepest descent algorithm99 for optimizing the system geometry.80 Under a constant number of particles, volume, and temperature (NVT) ensembles (303.15 K), the minimized system was equilibrated for 100 ps under the Berendsen-temperature coupling approach.100 A second equilibration was done at a constant number of particles, pressure, and temperature (NPT) ensembles (303.15 K/1 atm pressure) under the regulation of the Parrinello–Rahman barostat approach.101 A 1000 KJ/mol.nm2 force constant was used for all heavy atoms restrainment and original protein folding maintenance throughout the minimization/equilibration stages. Finally, 200 ns MD runs were produced under NPT ensembles using particle mesh Ewald algorithm for long-range electrostatic interaction estimation.102 Linear constraint LINCS method allowed modeling of all covalent bond lengths (including hydrogen atoms) at 2 fs integration time step.103 The van der Waals and Coulomb nonbonded potentials were truncated at 10 Å via Verlet cutoff schemes.104
Analysis tools, such as rmsd, RMSF, Rg, and SASA were estimated using GROMACS analysis scripts. The ΔRMSF was determined for each simulated liganded/holo protein relative to VEGFR-2 unliganded/apo state (PDB ID: 1VR2; ΔRMSF is RMSFapo–holo). The apo VEGFR-2 was subjected to similar preparation, minimization, equilibration, and 200 ns production stages, yet without ligand preparations. Visual Molecular Dynamics V-1.9.3 (VMD; University of Illinois, Urbana-Champaign, USA) was used for ligand–protein intermolecular hydrogen-bond across the whole MD timeframes. Hydrogen bond (acceptor···H-donor) cutoff values for angles and distances were established at 20° and 3.0 Å, respectively.48,105 Using GROMACS “g_mmpbsa” module, the ligand–protein free binding energies were estimated through MM/PBSA energy calculation applied on representative frames of whole MD run (200 ns). This calculation approach provided more insights concerning ligand–protein magnitude affinity, nature of the ligand–protein binding, and protein residue-associated contributions at the free binding energy calculations.58 Schrödinger-Pymol V.2.0.6 was used for representing the ligand-VEGFR-2 within the depicted conformational analysis at specific frames.106,107
5.4. MMs Loaded Mebendazole Preparation
MBZ-loaded MMs were prepared by using thin-film hydration followed by the sonication method.62,108 Briefly, 10 mM concentration of F127 and SDC at molar ratio (7:3) with vitamin E (10% w/w) and MBZ (5% w/w) were dissolved in ethanol. The solvent was removed under low pressure, in a rotary vacuum evaporator set at a temperature of 40 °C. Then, 10 mL of DI water was added to the thin film and sonicated in a water bath sonicator for 10 min at room temperature. The resultant micellar solution was centrifuged at 10,000 rpm for 10 min (Sigma 3K30, Germany) to remove the unentrapped MBZ. The mixed micellar solution was kept at −80 °C for 2 h and then lyophilized (Alpha 2–4, CHRIST, Osterode am Harz, Germany) for 18 h. Similarly, plain MMs (without MBZ) were also prepared.
5.4.1. Characterization of the Prepared MBZ-Loaded MMs
5.4.1.1. Determination of Particle Size and Zeta Potential
The average size of the micelles and their PDI (size distribution) were determined by the dynamic light scattering method (Malvern Instruments, Malvern, UK). Cell temperature was kept at 25 °C with a detection angle of 90°. All measurements were performed in triplicate after appropriate reconstitution and dilution using DI water. The zeta potential of the prepared MMs was measured using the same instrument. All measurements were carried out in triplicate and the data are represented as mean ± SD.
5.4.1.2. Transmission Electron Microscopy
The morphology of the lyophilized MBZ-loaded MMs was observed by TEM (JEM-2100, JEOL, Tokyo, Japan). The lyophilized powder was reconstituted with DI water and shook for the formation of MMs. A drop (10 μL) of the prepared micellar solution was submitted on a carbon-coated copper grid and left to dry until a thin film is formed. This film was negatively stained with phosphotungstic acid (2% w/v) aqueous solution for 5 min and then air-dried at room temperature for 10 min before viewing and photography by TEM.
5.4.1.3. Drug Loading and Encapsulation Efficiency
10 mg of lyophilized MBZ-loaded MMs was accurately weighed and 5 mL of ethanol was added and sonicated for 2–3 min, and the samples were measured by HPLC using an Agilent 1100 HPLC (Agilent Technologies, Santa Clara, CA) instrument. Chromatographic separation of the samples was attained by a μBondapak, C18, 150 × 4.6 mm, 5 μm column (Waters, USA). The mobile phase was composed of acetonitrile 25% and buffer 75% (7.5 g of ammonium acetate in 1000 mL of DI water). The mobile phase was set at a flow rate of 1.2 mL/min with UV detection at 250 nm. The injection volume was 20 μL.109 A calibration curve in ethanol was constructed in the concentration range of 1.6–240 μg/mL (linearity R2 = 0.9988). Encapsulation efficiency (EE) and drug loading (DL) was calculated according to the following equations
5.4.1.4. In Vitro Drug Release Study
The dissolution test was performed by adding 5 mg of MBZ drug and an amount of lyophilized MMs powder equivalent to 5 mg of MBZ was introduced into the USP II dissolution apparatus (Hanson Research, SR8PLUS, USA) filled with 100 mL of 0.1 N HCl in minivessels, maintained at 37 ± 0.5 °C, and agitated with a paddle rotating at a speed of 75 rpm.109 Five-milliliter samples were withdrawn periodically at predetermined time intervals of 5, 15, 30, 60, and 120 min and replaced instantly by equal amounts of dissolution medium to maintain the initial volume. The dissolved amounts of MBZ at each interval were determined by HPLC as previously mentioned. A calibration curve in 0.1 N HCl was constructed for the concentration range of 1.6–20 μg/mL (linearity R2 = 0.9906).109
5.5. In Vitro Cytotoxic Assay
5.5.1. Cell Culture and Maintenance
Human hepatoma (HUH7), lung small cell adenocarcinoma (A549), and breast cancer (MCF7) cell lines were propagated in Dulbecco’s modified Eagle’s medium. All mediums were supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin antibiotics (Seralab, UK). For growth maintenance, the cells were incubated at 37 °C in 5% humidified CO2.
5.5.2. Evaluation of Cell Proliferation by MTT Assay
After treatment with variable concentrations of the compounds, the HUH7, A549, and MCF7 viable cell percentages were evaluated by the MTT assay as reported previously,102,110−113 with a small modification. In brief, the cell count and viability were evaluated by the trypan blue dye-based method, and then, cancer cells (1 × 104 cells/well) were seeded using a 96-well plate. The cancer cells were then kept overnight for attachment. On the next day, the medium was exchanged completely with a fresh one, and then different concentrations of the synthesized compounds (0, 0.1, 1, 10, and 100 μM) were examined on each cell line. Thereafter, cells were permitted to grow for 24 h, and then, 10 μL of the MTT (5 mg/mL) was added to each well for 4 h before completion of the incubation period. After the incubation accomplishment, the formazan crystals were dissolved by adding 100 μL of DMSO to each well and left for 20 min, and then, the 96 well plates were waved for 5 min to assure the dye homogeneity in the solution. After the reaction occurs, a BioTek microplate reader was used to measure the color development at 490 nm. Based on the MTT results attained, we have chosen the most outstanding cytotoxic compound to investigate its mode of action.
5.6. VEGFR-2 Cell-Based Quantification
HUH7 cells were treated with the IC50 values of the investigated drugs. After 48 h, cell extraction buffer was used to lyse treated and nontreated cells, and then, the standard diluent buffer was used to dilute the lysate formed over the range of the assay and then measured for vascular endothelial growth factor (ab213476—Human VEGF Receptor 2 SimpleStep ELISA Kit). All procedures for standard ELISA techniques were done according to the methodology described earlier.114
Acknowledgments
The authors would like to thank the Deanship of Scientific Research at Majmaah University, Saudi Arabia, for supporting this work under the Project R-2021-287.
Author Contributions
A.A.E. and K.M.D. equally contributed to this work. Conceptualization: A.C. and A.A.A.; data curation: A.A.E., K.M.D., and A.A.A.; formal analysis: K.M.D. and A.A.A.; funding acquisition: A.C. and A.A.A.; investigation: A.A.E. and A.A.A.; methodology: K.M.D., A.A.B., M.A.T., S.S.E., A.B.S., M.M., and A.A.A.; project administration: A.A.E. and A.A.A.; resources: A.C. and A.A.A.; software: A.A.E., K.M.D., and A.A.A.; supervision: A.A.A.; validation: K.M.D., A.A.B., M.A.T., and A.A.A.; visualization: A.A.E., K.M.D., A.A.B., M.A.T., and A.A.A.; writing—original draft: A.A.E., K.M.D., A.A.B., M.A.T., and A.A.A.; and writing—review and editing: A.A.E., K.M.D., A.A.B., M.A.T., A.C., A.B.S., M.M., and A.A.A. All authors approved the final version of the manuscript.
The authors declare no competing financial interest.
References
- Martínez-Jiménez F.; Muiños F.; Sentís I.; Deu-Pons J.; Reyes-Salazar I.; Arnedo-Pac C.; Mularoni L.; Pich O.; Bonet J.; Kranas H.; Gonzalez-Perez A.; Lopez-Bigas N. A compendium of mutational cancer driver genes. Nat. Rev. Cancer 2020, 20, 555–572. 10.1038/s41568-020-0290-x. [DOI] [PubMed] [Google Scholar]
- Samra R. M.; Soliman A. F.; Zaki A. A.; Ashour A.; Al-Karmalawy A. A.; Hassan M. A.; Zaghloul A. M. Bioassay-guided isolation of a new cytotoxic ceramide from Cyperus rotundus L. South Afr. J. Bot. 2021, 139, 210–216. 10.1016/j.sajb.2021.02.007. [DOI] [Google Scholar]
- Ghanem A.; Emara H. A.; Muawia S.; Abd El Maksoud A. I.; Al-Karmalawy A. A.; Elshal M. F. Tanshinone IIA synergistically enhances the antitumor activity of doxorubicin by interfering with the PI3K/AKT/mTOR pathway and inhibition of topoisomerase II: in vitro and molecular docking studies. New J. Chem. 2020, 44, 17374–17381. 10.1039/d0nj04088f. [DOI] [Google Scholar]
- Ma C.; Taghour M. S.; Al-Karmalawy A.; Belal A.; Mehany A.; Mostafa N.; Nabeeh A.; Eissa I. Design and Synthesis of New Quinoxaline Derivatives as Potential Histone Deacetylase Inhibitors Targeting Hepatocellular Carcinoma: In Silico, In Vitro, and SAR Studies. Front. Chem. 2021, 648, 725135. 10.3389/fchem.2021.725135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobbi P. G.; Ferreri A. J. M.; Ponzoni M.; Levis A. Hodgkin lymphoma. Crit. Rev. Oncol. Hematol. 2013, 85, 216–237. 10.1016/j.critrevonc.2012.07.002. [DOI] [PubMed] [Google Scholar]
- Alesawy M. S.; Al-Karmalawy A. A.; Elkaeed E. B.; Alswah M.; Belal A.; Taghour M. S.; Eissa I. H. Design and discovery of new 1, 2, 4-triazolo [4, 3-c] quinazolines as potential DNA intercalators and topoisomerase II inhibitors. Arch. Pharmazie 2021, 354, e2000237 10.1002/ardp.202000237. [DOI] [PubMed] [Google Scholar]
- El-Shershaby M. H.; Ghiaty A.; Bayoumi A. H.; Al-Karmalawy A. A.; Husseiny E. M.; El-Zoghbi M. S.; Abulkhair H. S. From triazolophthalazines to triazoloquinazolines: A bioisosterism-guided approach toward the identification of novel PCAF inhibitors with potential anticancer activity. Bioorg. Med. Chem. 2021, 42, 116266. 10.1016/j.bmc.2021.116266. [DOI] [PubMed] [Google Scholar]
- Eliaa S. G.; Al-Karmalawy A. A.; Saleh R. M.; Elshal M. F. Empagliflozin and Doxorubicin Synergistically Inhibit the Survival of Triple-Negative Breast Cancer Cells via Interfering with the mTOR Pathway and Inhibition of Calmodulin: In Vitro and Molecular Docking Studies. ACS Pharmacol. Transl. Sci. 2020, 3, 1330–1338. 10.1021/acsptsci.0c00144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elshal M. F.; Eid N.; El-Sayed I.; El-Sayed W.; Ali Al-Karmalawy A. Concanavalin-A shows synergistic cytotoxicity with tamoxifen via inducing apoptosis in estrogen receptor-positive breast cancer: In vitro and molecular docking studies. Pharm. Sci. 2021, 10.34172/ps.2021.22. [DOI] [Google Scholar]
- Holmes D. I.; Zachary I. The vascular endothelial growth factor (VEGF) family: angiogenic factors in health and disease. Genome Biol. 2005, 6, 209. 10.1186/gb-2005-6-2-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taher R. F.; Al-Karmalawy A. A.; Abd El Maksoud A. I.; Khalil H.; Hassan A.; El-Khrisy E.-D. A.; El-Kashak W. A. Two new flavonoids and anticancer activity of Hymenosporum flavum: in vitro and molecular docking studies. J. HerbMed Pharmacol. 2021, 10, 443. 10.34172/jhp.2021.52. [DOI] [Google Scholar]
- Mahdy H. A.; Ibrahim M. K.; Metwaly A. M.; Belal A.; Mehany A. B. M.; El-Gamal K. M. A.; El-Sharkawy A.; Elhendawy M. A.; Radwan M. M.; Elsohly M. A.; Eissa I. H. Design, synthesis, molecular modeling, in vivo studies and anticancer evaluation of quinazolin-4 (3H)-one derivatives as potential VEGFR-2 inhibitors and apoptosis inducers. Bioorg. Chem. 2020, 94, 103422. 10.1016/j.bioorg.2019.103422. [DOI] [PubMed] [Google Scholar]
- Sun S.; Schiller J. H.; Gazdar A. F. Lung cancer in never smokers--a different disease. Nat. Rev. Cancer 2007, 7, 778–790. 10.1038/nrc2190. [DOI] [PubMed] [Google Scholar]
- Gaber A. A.; El-Morsy A. M.; Sherbiny F. F.; Bayoumi A. H.; El-Gamal K. M.; El-Adl K.; Al-Karmalawy A. A.; Ezz Eldin R. R.; Saleh M. A.; Abulkhair H. S. Pharmacophore-linked pyrazolo[3,4-d]pyrimidines as EGFR-TK inhibitors: Synthesis, anticancer evaluation, pharmacokinetics, and in silico mechanistic studies. Arch. Pharmazie 2021, e2100258 10.1002/ardp.202100258. [DOI] [PubMed] [Google Scholar]
- Holmes K.; Roberts O. L.; Thomas A. M.; Cross M. J. Vascular endothelial growth factor receptor-2: structure, function, intracellular signalling and therapeutic inhibition. Cell. Signal. 2007, 19, 2003–2012. 10.1016/j.cellsig.2007.05.013. [DOI] [PubMed] [Google Scholar]
- Shibuya M.; Claessonwelsh L. Signal transduction by VEGF receptors in regulation of angiogenesis and lymphangiogenesis. Exp. Cell Res. 2006, 312, 549–560. 10.1016/j.yexcr.2005.11.012. [DOI] [PubMed] [Google Scholar]
- Strawn L. M.; Shawver L. K. Tyrosine kinases in disease: overview of kinase inhibitors as therapeutic agents and current drugs in clinical trials. Expet Opin. Invest. Drugs 1998, 7, 553–573. 10.1517/13543784.7.4.553. [DOI] [PubMed] [Google Scholar]
- Lee K.; Jeong K.-W.; Lee Y.; Song J. Y.; Kim M. S.; Lee G. S.; Kim Y. Pharmacophore modeling and virtual screening studies for new VEGFR-2 kinase inhibitors. Eur. J. Med. Chem. 2010, 45, 5420–5427. 10.1016/j.ejmech.2010.09.002. [DOI] [PubMed] [Google Scholar]
- El-Helby A.-G. A.; Sakr H.; Eissa I. H.; Al-Karmalawy A. A.; El-Adl K. Benzoxazole/benzothiazole-derived VEGFR-2 inhibitors: Design, synthesis, molecular docking, and anticancer evaluations. Arch. Pharmazie 2019, 352, 1900178. 10.1002/ardp.201900178. [DOI] [PubMed] [Google Scholar]
- El-Helby A.-G. A.; Sakr H.; Eissa I. H.; Abulkhair H.; Al-Karmalawy A. A.; El-Adl K. J. A. d. P. Design, synthesis, molecular docking, and anticancer activity of benzoxazole derivatives as VEGFR-2 inhibitors. Arch. Pharmazie 2019, 352, 1900113. 10.1002/ardp.201900113. [DOI] [PubMed] [Google Scholar]
- Abdullaziz M. A.; Abdel-Mohsen H. T.; El Kerdawy A. M.; Ragab F. A. F.; Ali M. M.; Abu-Bakr S. M.; Girgis A. S.; El Diwani H. I. Design, synthesis, molecular docking and cytotoxic evaluation of novel 2-furybenzimidazoles as VEGFR-2 inhibitors. Eur. J. Med. Chem. 2017, 136, 315–329. 10.1016/j.ejmech.2017.04.068. [DOI] [PubMed] [Google Scholar]
- Xie Q.-Q.; Xie H.-Z.; Ren J.-X.; Li L.-L.; Yang S.-Y. Pharmacophore modeling studies of type I and type II kinase inhibitors of Tie2. J. Mol. Graph. Model. 2009, 27, 751–758. 10.1016/j.jmgm.2008.11.008. [DOI] [PubMed] [Google Scholar]
- Eskander R. N.; Tewari K. S. Incorporation of anti-angiogenesis therapy in the management of advanced ovarian carcinoma—mechanistics, review of phase III randomized clinical trials, and regulatory implications. Gynecol. Oncol. 2014, 132, 496–505. 10.1016/j.ygyno.2013.11.029. [DOI] [PubMed] [Google Scholar]
- Dogra N.; Kumar A.; Mukhopadhyay T. Fenbendazole acts as a moderate microtubule destabilizing agent and causes cancer cell death by modulating multiple cellular pathways. Sci. Rep. 2018, 8, 11926. 10.1038/s41598-018-30158-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khattab M.; Al-Karmalawy A. A. Computational repurposing of benzimidazole anthelmintic drugs as potential colchicine binding site inhibitors. Future Med. Chem. 2021, 13, 1623. 10.4155/fmc-2020-0273. [DOI] [PubMed] [Google Scholar]
- Al-Karmalawy A. A.; Khattab M. Molecular modelling of mebendazole polymorphs as a potential colchicine binding site inhibitor. New J. Chem. 2020, 44, 13990–13996. 10.1039/d0nj02844d. [DOI] [Google Scholar]
- Son D.-S.; Lee E.-S.; Adunyah S. E. The antitumor potentials of benzimidazole anthelmintics as repurposing drugs. Immune Network 2020, 20, e29 10.4110/in.2020.20.e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav S.; Narasimhan B.; kaur H. Perspectives of benzimidazole derivatives as anticancer agents in the new era. Anti Cancer Agents Med. Chem. 2016, 16, 1403–1425. 10.2174/1871520616666151103113412. [DOI] [PubMed] [Google Scholar]
- Králová V.; Hanušová V.; Stanková P.; Knoppová K.; Cánová K.; Skálová L. Antiproliferative effect of benzimidazole anthelmintics albendazole, ricobendazole, and flubendazole in intestinal cancer cell lines. Anti Cancer Drugs 2013, 24, 911–919. 10.1097/cad.0b013e3283648c69. [DOI] [PubMed] [Google Scholar]
- Doudican N.; Rodriguez A.; Osman I.; Orlow S. J. Mebendazole induces apoptosis via Bcl-2 inactivation in chemoresistant melanoma cells. Mol. Cancer Res. 2008, 6, 1308–1315. 10.1158/1541-7786.mcr-07-2159. [DOI] [PubMed] [Google Scholar]
- Chu S. W.; Badar S.; Morris D. L.; Pourgholami M. H. Potent inhibition of tubulin polymerisation and proliferation of paclitaxel-resistant 1A9PTX22 human ovarian cancer cells by albendazole. Anticancer Res. 2009, 29, 3791. [PubMed] [Google Scholar]
- Hasegawa M.; Nishigaki N.; Washio Y.; Kano K.; Harris P. A.; Sato H.; Mori I.; West R. I.; Shibahara M.; Toyoda H.; Wang L.; Nolte R. T.; Veal J. M.; Cheung M. Discovery of novel benzimidazoles as potent inhibitors of TIE-2 and VEGFR-2 tyrosine kinase receptors. J. Med. Chem. 2007, 50, 4453–4470. 10.1021/jm0611051. [DOI] [PubMed] [Google Scholar]
- Elmaaty A. A.; Darwish K. M.; Khattab M.; Elhady S. S.; Salah M.; Hamed M. I. A.; Al-Karmalawy A. A.; Saleh M. M. In a search for potential drug candidates for combating COVID-19: computational study revealed salvianolic acid B as a potential therapeutic targeting 3CLpro and spike proteins. J. Biomol. Struct. Dyn. 2021, 1–28. 10.1080/07391102.2021.1918256. [DOI] [PubMed] [Google Scholar]
- Soltan M. A.; Elbassiouny N.; Gamal H.; Elkaeed E. B.; Eid R. A.; Eldeen M. A.; Al-Karmalawy A. A. In Silico Prediction of a Multitope Vaccine against Moraxella catarrhalis: Reverse Vaccinology and Immunoinformatics. Vaccines 2021, 9, 669. 10.3390/vaccines9060669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltan M. A.; Eldeen M. A.; Elbassiouny N.; Mohamed I.; El-damasy D. A.; Fayad E.; Abu Ali O. A.; Raafat N.; Eid R. A.; Al-Karmalawy A. A. Proteome Based Approach Defines Candidates for Designing a Multitope Vaccine against the Nipah Virus. Int. J. Mol. Sci. 2021, 22, 9330. 10.3390/ijms22179330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Torre-Iglesias P. M.; García-Rodriguez J. J.; Torrado G.; Torrado S.; Torrado-Santiago S.; Bolás-Fernández F. Enhanced bioavailability and anthelmintic efficacy of mebendazole in redispersible microparticles with low-substituted hydroxypropylcellulose. Drug Des. Dev. Ther. 2014, 8, 1467. 10.2147/dddt.s65561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin W.; Tan X.; Wen J.; Meng Y.; Zhang Y.; Li H.; Jiang D.; Song H.; Zheng W. A Novel Dantrolene Sodium-Loaded Mixed Micelle Containing a Small Amount of Cremophor EL: Characterization, Stability, Safety and Pharmacokinetics. Molecules 2019, 24, 728. 10.3390/molecules24040728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton J. Albendazole: a broad spectrum anthelminthic for treatment of individuals and populations. Curr. Opin. Infect. Dis. 2002, 15, 599–608. 10.1097/00001432-200212000-00008. [DOI] [PubMed] [Google Scholar]
- Shaik F.; Cuthbert G. A.; Homer-Vanniasinkam S.; Muench S. P.; Ponnambalam S.; Harrison M. A. Structural Basis for Vascular Endothelial Growth Factor Receptor Activation and Implications for Disease Therapy. Biomolecules 2020, 10, 1673. 10.3390/biom10121673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pushpakom S.; Iorio F.; Eyers P. A.; Escott K. J.; Hopper S.; Wells A.; Doig A.; Guilliams T.; Latimer J.; McNamee C.; Norris A.; Sanseau P.; Cavalla D.; Pirmohamed M. Drug repurposing: progress, challenges and recommendations. Nat. Rev. Drug Discov. 2019, 18, 41–58. 10.1038/nrd.2018.168. [DOI] [PubMed] [Google Scholar]
- Cha Y.; Erez T.; Reynolds I. J.; Kumar D.; Ross J.; Koytiger G.; Kusko R.; Zeskind B.; Risso S.; Kagan E.; Papapetropoulos S.; Grossman I.; Laifenfeld D. Drug repurposing from the perspective of pharmaceutical companies. Br. J. Pharmacol. 2018, 175, 168–180. 10.1111/bph.13798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karplus M.; Petsko G. A. Molecular dynamics simulations in biology. Nature 1990, 347, 631–639. 10.1038/347631a0. [DOI] [PubMed] [Google Scholar]
- Arnittali M.; Rissanou A. N.; Harmandaris V. Structure Of Biomolecules Through Molecular Dynamics Simulations. Procedia Comput. Sci. 2019, 156, 69–78. 10.1016/j.procs.2019.08.181. [DOI] [Google Scholar]
- Liu K.; Watanabe E.; Kokubo H. Exploring the stability of ligand binding modes to proteins by molecular dynamics simulations. J. Comput. Aided Mol. Des. 2017, 31, 201–211. 10.1007/s10822-016-0005-2. [DOI] [PubMed] [Google Scholar]
- Likić V. A.; Gooley P. R.; Speed T. P.; Strehler E. E. A statistical approach to the interpretation of molecular dynamics simulations of calmodulin equilibrium dynamics. Protein Sci. 2005, 14, 2955–2963. 10.1110/ps.051681605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirolli D.; Sciandra F.; Bozzi M.; Giardina B.; Brancaccio A.; De Rosa M. C. Insights from molecular dynamics simulations: structural basis for the V567D mutation-induced instability of zebrafish alpha-dystroglycan and comparison with the murine model. PLoS One 2014, 9, e103866 10.1371/journal.pone.0103866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson N. C.; Daggett V. A comparison of multiscale methods for the analysis of molecular dynamics simulations. J. Phys. Chem. B 2012, 116, 8722–8731. 10.1021/jp302103t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Souza A. S.; Pacheco B. D. C.; Pinheiro S.; Muri E. M. F.; Dias L. R. S.; Lima C. H. S.; Garrett R.; de Moraes M. B. M.; Puzer L.; Puzer L. 3-Acyltetramic acids as a novel class of inhibitors for human kallikreins 5 and 7. Bioorg. Med. Chem. Lett 2019, 29, 1094–1098. 10.1016/j.bmcl.2019.02.031. [DOI] [PubMed] [Google Scholar]
- Li J.; Zhou N.; Luo K.; Zhang W.; Li X.; Wu C.; Bao J. In silico discovery of potential VEGFR-2 inhibitors from natural derivatives for anti-angiogenesis therapy. Int. J. Mol. Sci. 2014, 15, 15994–16011. 10.3390/ijms150915994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitranshi N.; Gupta V.; Kumar S.; Graham S. Exploring the Molecular Interactions of 7,8-Dihydroxyflavone and Its Derivatives with TrkB and VEGFR2 Proteins. Int. J. Mol. Sci. 2015, 16, 21087–21108. 10.3390/ijms160921087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L.; Wang X.; Feng J.; Jia Y.; Xu F.; Xu W. Discovery of novel vascular endothelial growth factor receptor 2 inhibitors: a virtual screening approach. Chem. Biol. Drug Des. 2012, 80, 893–901. 10.1111/cbdd.12036. [DOI] [PubMed] [Google Scholar]
- Sharma N.; Sharma M.; Rahman Q. I.; Akhtar S.; Muddassir M. Quantitative structure activity relationship and molecular simulations for the exploration of natural potent VEGFR-2 inhibitors: an in silico anti-angiogenic study. J. Biomol. Struct. Dynam. 2021, 39, 2806–2823. 10.1080/07391102.2020.1754916. [DOI] [PubMed] [Google Scholar]
- Dash R.; Junaid M.; Mitra S.; Arifuzzaman M.; Hosen S. M. Z. Structure-based identification of potent VEGFR-2 inhibitors from in vivo metabolites of a herbal ingredient. J. Mol. Model. 2019, 25, 98. 10.1007/s00894-019-3979-6. [DOI] [PubMed] [Google Scholar]
- Rampogu S.; Baek A.; Zeb A.; Lee K. W. Exploration for novel inhibitors showing back-to-front approach against VEGFR-2 kinase domain (4AG8) employing molecular docking mechanism and molecular dynamics simulations. BMC Cancer 2018, 18, 264. 10.1186/s12885-018-4050-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.; Yang S.; Jiao Y.; Liu H.; Yuan H.; Lu S.; Ran T.; Yao S.; Ke Z.; Xu J.; Xiong X.; Chen Y.; Lu T. An integrated virtual screening approach for VEGFR-2 inhibitors. J. Chem. Inf. Model. 2013, 53, 3163–3177. 10.1021/ci400429g. [DOI] [PubMed] [Google Scholar]
- McTigue M. A.; Wickersham J. A.; Pinko C.; Showalter R. E.; Parast C. V.; Tempczyk-Russell A.; Gehring M. R.; Mroczkowski B.; Kan C. C.; Villafranca J. E.; Appelt K. Crystal structure of the kinase domain of human vascular endothelial growth factor receptor 2: a key enzyme in angiogenesis. Structure 1999, 7, 319–330. 10.1016/s0969-2126(99)80042-2. [DOI] [PubMed] [Google Scholar]
- Cavasotto C. N. Binding Free Energy Calculation Using Quantum Mechanics Aimed for Drug Lead Optimization. Methods Mol. Biol. 2020, 2114, 257–268. 10.1007/978-1-0716-0282-9_16. [DOI] [PubMed] [Google Scholar]
- Kumari R.; Kumar R.; Lynn A. g_mmpbsa—A GROMACS Tool for High-Throughput MM-PBSA Calculations. J. Chem. Inf. Model. 2014, 54, 1951–1962. 10.1021/ci500020m. [DOI] [PubMed] [Google Scholar]
- Muddineti O. S.; Ghosh B.; Biswas S. Current trends in the use of vitamin E-based micellar nanocarriers for anticancer drug delivery. Expet Opin. Drug Deliv. 2017, 14, 715–726. 10.1080/17425247.2016.1229300. [DOI] [PubMed] [Google Scholar]
- Boseila A. A.; Rashed H. M.; Sakr T. M.; Abdel-Reheem A. Y.; Basalious E. B. Superiority of DEAE-Dx-Stabilized Cationic Bile-Based Vesicles over Conventional Vesicles for Enhanced Hepatic Delivery of Daclatasvir. Mol. Pharm. 2019, 16, 4190–4199. 10.1021/acs.molpharmaceut.9b00517. [DOI] [PubMed] [Google Scholar]
- Boseila A. A.; Abdel-Reheem A. Y.; Basalious E. B. Design of bile-based vesicles (BBVs) for hepatocytes specific delivery of Daclatasvir: Comparison of ex-vivo transenterocytic transport, in-vitro protein adsorption resistance and HepG2 cellular uptake of charged and β-sitosterol decorated vesicles. PLoS One 2019, 14, e0219752 10.1371/journal.pone.0219752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanitwatthanasak S.; Sagis L. M. C.; Chitprasert P. Pluronic F127/Pluronic P123/vitamin E TPGS mixed micelles for oral delivery of mangiferin and quercetin: Mixture-design optimization, micellization, and solubilization behavior. J. Mol. Liq. 2019, 274, 223–238. 10.1016/j.molliq.2018.10.089. [DOI] [Google Scholar]
- Comşa Ş.; Cîmpean A. M.; Raica M. The story of MCF-7 breast cancer cell line: 40 years of experience in research. Anticancer Res. 2015, 35, 3147. [PubMed] [Google Scholar]
- Biswas S.; Kumari P.; Lakhani P. M.; Ghosh B. Recent advances in polymeric micelles for anti-cancer drug delivery. Eur. J. Pharmaceut. Sci. 2016, 83, 184–202. 10.1016/j.ejps.2015.12.031. [DOI] [PubMed] [Google Scholar]
- Yu W.; Liao Q. Y.; Hantash F. M.; Sanders B. G.; Kline K. Activation of extracellular signal-regulated kinase and c-Jun-NH2-terminal kinase but not p38 mitogen-activated protein kinases is required for RRR-α-tocopheryl succinate-induced apoptosis of human breast cancer cells. Cancer Res. 2001, 61, 6569. [PubMed] [Google Scholar]
- Cheng X.; Yan H.; Jia X.; Zhang Z. Preparation and in vivo/in vitro evaluation of formononetin phospholipid/vitamin E TPGS micelles. J. Drug Target. 2016, 24, 161–168. 10.3109/1061186x.2015.1064435. [DOI] [PubMed] [Google Scholar]
- de Melo-Diogo D.; Gaspar V. M.; Costa E. C.; Moreira A. F.; Oppolzer D.; Gallardo E.; Correia I. J. Combinatorial delivery of Crizotinib–Palbociclib–Sildenafil using TPGS-PLA micelles for improved cancer treatment. Eur. J. Pharm. Biopharm. 2014, 88, 718–729. 10.1016/j.ejpb.2014.09.013. [DOI] [PubMed] [Google Scholar]
- Chemical Computing Group Inc. Molecular operating environment (MOE); Chemical Computing Group Inc, 1010 Sherbooke St. West, Suite# 910, Montreal, 2016.
- Diab R. T.; Abdelsamii Z.; Abd-Elaal E. H.; Al-Karmalawy A. A.; AboDya N. E. Design and Synthesis of a New Series of 3,5-Disubstituted-1,2,4-Oxadiazoles as Potential Colchicine Binding Site Inhibitors: Antiproliferative activity, Molecular docking, and SAR Studies. New J. Chem. 2021, 45, 21657. 10.1039/d1nj02885e. [DOI] [Google Scholar]
- El-Shershaby M. H.; El-Gamal K. M.; Bayoumi A. H.; El-Adl K.; Alswah M.; Ahmed H. E. A.; Al-Karmalamy A. A.; Abulkhair H. S. The antimicrobial potential and pharmacokinetic profiles of novel quinoline-based scaffolds: synthesis and in silico mechanistic studies as dual DNA gyrase and DHFR inhibitors. New J. Chem. 2021, 45, 13986. 10.1039/d1nj02838c. [DOI] [Google Scholar]
- Mahmoud A.; Mostafa A.; Al-Karmalawy A. A.; Zidan A.; Abulkhair H. S.; Mahmoud S. H.; Shehata M.; Elhefnawi M. M.; Ali M. A. Telaprevir is a potential drug for repurposing against SARS-CoV-2: computational and in vitro studies. Heliyon 2021, 7, e07962 10.1016/j.heliyon.2021.e07962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz M. A.; Shehab W. S.; Al-Karmalawy A. A.; EL-Farargy A. F.; Abdellattif M. H. Design, Synthesis, Biological Evaluation, 2D-QSAR Modeling, and Molecular Docking Studies of Novel 1H-3-Indolyl Derivatives as Significant Antioxidants. Int. J. Mol. Sci. 2021, 22, 10396. 10.3390/ijms221910396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Gizawy H. A.; Boshra S. A.; Mostafa A.; Mahmoud S. H.; Ismail M. I.; Alsfouk A. A.; Taher A. T.; Al-Karmalawy A. A. Pimenta dioica (L.) Merr. Bioactive Constituents Exert Anti-SARS-CoV-2 and Anti-Inflammatory Activities: Molecular Docking and Dynamics, In Vitro, and In Vivo Studies. Molecules 2021, 26, 5844. 10.3390/molecules26195844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamed M. I. A.; Darwish K. M.; Soltane R.; Chrouda A.; Mostafa A.; Abo Shama N. M.; Elhady S. S.; Abulkhair H. S.; Khodir A. E.; Elmaaty A. A.; Al-karmalawy A. A. β-Blockers bearing hydroxyethylamine and hydroxyethylene as potential SARS-CoV-2 Mpro inhibitors: rational based design, in silico, in vitro, and SAR studies for lead optimization. RSC Adv. 2021, 11, 35536–35558. 10.1039/d1ra04820a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abo Elmaaty A.; Hamed M. I. A.; Ismail M. I.; Elkaeed E.; Abulkhair H.; Khattab M.; Al-Karmalawy A. A. Computational Insights on the Potential of Some NSAIDs for Treating COVID-19: Priority Set and Lead Optimization. Molecules 2021, 26, 3772. 10.3390/molecules26123772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Karmalawy A. A.; Eissa I. H. J. P. S. Molecular docking and dynamics simulations reveal the potential of anti-HCV drugs to inhibit COVID-19 main protease. Pharm. Sci. 2021, 27, S109. 10.34172/ps.2021.3. [DOI] [Google Scholar]
- Kandeil A.; Mostafa A.; Kutkat O.; Moatasim Y.; Al-Karmalawy A. A.; Rashad A. A.; Kayed A. E.; Kayed A. E.; El-Shesheny R.; Kayali G.; Ali M. A. Bioactive Polyphenolic Compounds Showing Strong Antiviral Activities against Severe Acute Respiratory Syndrome Coronavirus 2. Pathogens 2021, 10, 758. 10.3390/pathogens10060758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Karmalawy A. A.; Farid M. M.; Mostafa A.; Ragheb A. Y.; Mahmoud S.; Shama N. M. A.; GabAllah M.; Mostafa-Hedeab G.; Marzouk M. M.; Marzouk M. M. Naturally Available Flavonoid Aglycones as Potential Antiviral Drug Candidates against SARS-CoV-2. Molecules 2021, 26, 6559. 10.3390/molecules26216559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raslan M. A.; Taher R. F.; Al-Karmalawy A. A.; El-Ebeedy D.; Metwaly A. G.; Elkateeb N. M.; Ghanem A.; Elghaish R. A.; Abd El Maksoud A. I. Cordyline fruticosa (L.) A. Chev. leaves: isolation, HPLC/MS profiling and evaluation of nephroprotective and hepatoprotective activities supported by molecular docking. New J. Chem. 2021, 45, 22216. 10.1039/d1nj02663a. [DOI] [Google Scholar]
- Zaki A. A.; Ashour A.; Elhady S. S.; Darwish K. M.; Al-Karmalawy A. A. Calendulaglycoside A Showing Potential Activity Against SARS-CoV-2 Main Protease: Molecular Docking, Molecular Dynamics, and SAR Studies. J. Tradit., Complementary Med. 2021, 10.1016/j.jtcme.2021.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltane R.; Chrouda A.; Mostafa A.; Al-Karmalawy A. A.; Chouaïb K.; dhahri A.; Pashameah R. A.; Alasiri A.; Kutkat O.; Shehata M.; Jannet H. B.; Gharbi J.; Ali M. A. Strong Inhibitory Activity and Action Modes of Synthetic Maslinic Acid Derivative on Highly Pathogenic Coronaviruses: COVID-19 Drug Candidate. Pathogens 2021, 10, 623. 10.3390/pathogens10050623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdallah A. E.; Alesawy M. S.; Eissa S. I.; El-Fakharany E. M.; Kalaba M. H.; Sharaf M. H.; Abo Shama N. M.; Mahmoud S. H.; Mostafa A.; Al-Karmalawy A. A.; Elkady H. Design and synthesis of new 4-(2-nitrophenoxy)benzamide derivatives as potential antiviral agents: molecular modeling and in vitro antiviral screening. New J. Chem. 2021, 45, 16557. 10.1039/d1nj02710g. [DOI] [Google Scholar]
- Mahmoud A.; Kotb E.; Alqosaibi A. I.; Al-Karmalawy A. A.; Al-Dhuayan I. S.; Alabkari H. In vitro and in silico characterization of alkaline serine protease from Bacillus subtilis D9 recovered from Saudi Arabia. Heliyon 2021, 7, e08148 10.1016/j.heliyon.2021.e08148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki A. A.; Al-Karmalawy A. A.; El-Amier Y. A.; Ashour A. Molecular docking reveals the potential of Cleome amblyocarpa isolated compounds to inhibit COVID-19 virus main protease. New J. Chem. 2020, 44, 16752–16758. 10.1039/d0nj03611k. [DOI] [Google Scholar]
- Mahmoud D. B.; Ismail W. M.; Moatasim Y.; Kutkat O.; ElMeshad A. N.; Ezzat S. M.; El Deeb K. S.; El-Fishawy A. M.; Gomaa M. R.; Kandeil A.; Al-karmalawy A. A.; Ali M. A.; Mostafa A. Delineating a potent antiviral activity of Cuphea ignea extract loaded nano-formulation against SARS-CoV-2: In silico and in vitro studies. J. Drug Deliv. Sci. Technol. 2021, 66, 102845. 10.1016/j.jddst.2021.102845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elebeedy D.; Elkhatib W. F.; Kandeil A.; Ghanem A.; Kutkat O.; Alnajjar R.; Saleh M. A.; Abd El Maksoud A. I.; Badawy I.; Al-Karmalawy A. A. Anti-SARS-CoV-2 activities of tanshinone IIA, carnosic acid, rosmarinic acid, salvianolic acid, baicalein, and glycyrrhetinic acid between computational and in vitro insights. RSC Adv. 2021, 11, 29267–29286. 10.1039/d1ra05268c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raslan M. A.; Fikry R.; Al-Karmalawy A. A.; El-Ebeedy D.; Metwaly A. G.; Elkateeb N. M.; Ghanem A. A.; Elghaish R. A.; Abd El Maksoud A. Cordyline fruticose (L.) A. Chev. Leaves: Isolation, HPLC/MS profiling and evaluation of nephroprotective and hepatoprotective activities supported by molecular docking. New J. Chem. 2021, 45, 22216. 10.1039/d1nj02663a. [DOI] [Google Scholar]
- Khattab M.; Al-Karmalawy A. A. Revisiting Activity of Some Nocodazole Analogues as a Potential Anticancer Drugs Using Molecular Docking and DFT Calculations. Front. Chem. 2021, 9, 628398. 10.3389/fchem.2021.628398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Karmalawy A. A.; Dahab M. A.; Metwaly A. M.; Elhady S. S.; Elkaeed E. B.; Eissa I. H.; Darwish K. M. Molecular Docking and Dynamics Simulation Revealed the Potential Inhibitory Activity of ACEIs Against SARS-CoV-2 Targeting the hACE2 Receptor. Front. Chem. 2021, 9, 661230. 10.3389/fchem.2021.661230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoala T.; Al-Karmalawy A. A.; Germoush M. O.; ALshamrani S. M.; Abdein M. A.; Awad N. S. Nanobiotechnological Approaches to Enhance Potato Resistance against Potato Leafroll Virus (PLRV) Using Glycyrrhizic Acid Ammonium Salt and Salicylic Acid Nanoparticles. Horticulturae 2021, 7, 402. 10.3390/horticulturae7100402. [DOI] [Google Scholar]
- Elia S. G.; Al-Karmalawy A. A.; Nasr M. Y.; Elshal M. F. Loperamide potentiates doxorubicin sensitivity in triple-negative breast cancer cells by targeting MDR1 and JNK and suppressing mTOR and Bcl-2: In vitro and molecular docking study. J. Biochem. Mol. Toxicol. 2021, e22938 10.1002/jbt.22938. [DOI] [PubMed] [Google Scholar]
- Elhady S. S.; Abdelhameed R. F. A.; Malatani R. T.; Alahdal A. M.; Bogari H. A.; Almalki A. J.; Mohammad K. A.; Ahmed S. A.; Khedr A. I. M.; Darwish K. M. Molecular Docking and Dynamics Simulation Study of Hyrtios erectus Isolated Scalarane Sesterterpenes as Potential SARS-CoV-2 Dual Target Inhibitors. Biology 2021, 10, 389. 10.3390/biology10050389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Páll S.; Abraham M. J.; Kutzner C.; Hess B.; Lindahl E. In Tackling Exascale Software Challenges in Molecular Dynamics Simulations with GROMACS, Solving Software Challenges for Exascale; Markidis S., Laure E., Eds.; Springer International Publishing: Cham, 2015; pp 3–27. [Google Scholar]
- El-Demerdash A.; Al-Karmalawy A. A.; Abdel-Aziz T. M.; Elhady S. S.; Darwish K. M.; Hassan A. H. E. Investigating the structure–activity relationship of marine natural polyketides as promising SARS-CoV-2 main protease inhibitors. RSC Adv. 2021, 11, 31339–31363. 10.1039/d1ra05817g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanommeslaeghe K.; Hatcher E.; Acharya C.; Kundu S.; Zhong S.; Shim J.; Darian E.; Guvench O.; Lopes P.; Vorobyov I.; Mackerell A. D. Jr. CHARMM general force field: A force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J. Comput. Chem. 2010, 31, 671. 10.1002/jcc.21367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J.; Rauscher S.; Nawrocki G.; Ran T.; Feig M.; de Groot B. L.; Grubmüller H.; MacKerell A. D. Jr. CHARMM36m: an improved force field for folded and intrinsically disordered proteins. Nat. Methods 2017, 14, 71–73. 10.1038/nmeth.4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh A. H.; Abdelwaly A.; Darwish K. M.; Eissa A. A. H. M.; Chittiboyina A.; Helal M. A. Deciphering the molecular basis of the kappa opioid receptor selectivity: A Molecular Dynamics study. J. Mol. Graph. Model. 2021, 106, 107940. 10.1016/j.jmgm.2021.107940. [DOI] [PubMed] [Google Scholar]
- Ross G. A.; Rustenburg A. S.; Grinaway P. B.; Fass J.; Chodera J. D. Biomolecular Simulations under Realistic Macroscopic Salt Conditions. J. Phys. Chem. B 2018, 122, 5466–5486. 10.1021/acs.jpcb.7b11734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alnajjar R.; Mostafa A.; Kandeil A.; Al-Karmalawy A. A. Molecular docking, molecular dynamics, and in vitro studies reveal the potential of angiotensin II receptor blockers to inhibit the COVID-19 main protease. Heliyon 2020, 6, e05641 10.1016/j.heliyon.2020.e05641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golo V. L.; Shaĭtan K. V. Dynamic attractor for the Berendsen thermostat an the slow dynamics of biomacromolecules. Biofizika 2002, 47, 611. [PubMed] [Google Scholar]
- Tuble S. C.; Anwar J.; Gale J. D. An Approach to Developing a Force Field for Molecular Simulation of Martensitic Phase Transitions between Phases with Subtle Differences in Energy and Structure. J. Am. Chem. Soc. 2004, 126, 396–405. 10.1021/ja0356131. [DOI] [PubMed] [Google Scholar]
- Darden T.; York D.; Pedersen L. Particle mesh Ewald: An N·log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993, 98, 10089–10092. 10.1063/1.464397. [DOI] [Google Scholar]
- Hess B.; Bekker H.; Berendsen H. J. C.; Fraaije J. G. E. M. LINCS: A linear constraint solver for molecular simulations. J. Comput. Chem. 1997, 18, 1463–1472. . [DOI] [Google Scholar]
- Páll S.; Hess B. A flexible algorithm for calculating pair interactions on SIMD architectures. Comput. Phys. Commun. 2013, 184, 2641–2650. 10.1016/j.cpc.2013.06.003. [DOI] [Google Scholar]
- Albuquerque S. O.; Barros T. G.; Dias L. R. S.; Lima C. H. d. S.; Azevedo P. H. R. d. A.; Flores-Junior L. A. P.; dos Santos E. G.; Loponte H. F.; Pinheiro S.; Dias W. B.; Muri E. M. F.; Todeschini A. R. Biological evaluation and molecular modeling of peptidomimetic compounds as inhibitors for O-GlcNAc transferase (OGT). Eur. J. Pharmaceut. Sci. 2020, 154, 105510. 10.1016/j.ejps.2020.105510. [DOI] [PubMed] [Google Scholar]
- PyMOL Molecular Graphics System, 2.0.6; Schrödinger, LLC: New York, NY, 2016; Impact, Schrödinger, LLC, New York, NY, 2016; Prime, Schrödinger, LLC, New York, NY, 2020.
- Elmaaty A. A.; Alnajjar R.; Hamed M. I. A.; Khattab M.; Khalifa M. M.; Al-Karmalawy A. A. Revisiting activity of some glucocorticoids as a potential inhibitor of SARS-CoV-2 main protease: theoretical study. RSC Adv. 2021, 11, 10027–10042. 10.1039/d0ra10674g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patra A.; Satpathy S.; Shenoy A.; Bush J.; Kazi M.; Hussain M. D. Formulation and evaluation of mixed polymeric micelles of quercetin for treatment of breast, ovarian, and multidrug resistant cancers. Int. J. Nanomed. 2018, 13, 2869. 10.2147/ijn.s153094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consultation documents: the international pharmacopoeia. WHO Drug Inf. 2016, 30, 414–476. [Google Scholar]
- Abu Almaaty A. H.; Toson E. E. M.; El-Sayed E.-S. H.; Tantawy M. A. M.; Fayad E.; Abu Ali O. A.; Zaki I. 5-Aryl-1-Arylideneamino-1H-Imidazole-2 (3H)-Thiones: Synthesis and In Vitro Anticancer Evaluation. Molecules 2021, 26, 1706. 10.3390/molecules26061706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Far M.; Elmegeed G. A.; Eskander E. F.; Rady H. M.; Tantawy M. A. Novel modified steroid derivatives of androstanolone as chemotherapeutic anti-cancer agents. Eur. J. Med. Chem. 2009, 44, 3936–3946. 10.1016/j.ejmech.2009.04.020. [DOI] [PubMed] [Google Scholar]
- Tantawy M. A.; Shaheen S.; Kattan S. W.; Alelwani W.; Barnawi I. O.; Elmgeed G. A.; Nafie M. S. Cytotoxicity, in silico predictions and molecular studies for androstane heterocycle compounds revealed potential antitumor agent against lung cancer cells. J. Biomol. Struct. Dyn. 2020, 1–14. 10.1080/07391102.2020.1856188. [DOI] [PubMed] [Google Scholar]
- Tantawy M. A.; Sroor F. M.; Mohamed M. F.; El-Naggar M. E.; Saleh F. M.; Hassaneen H. M.; Abdelhamid I. A. Molecular docking study, cytotoxicity, cell cycle arrest and apoptotic induction of novel chalcones incorporating thiadiazolyl isoquinoline in cervical cancer. Anti Cancer Agents Med. Chem. 2020, 20, 70–83. 10.2174/1871520619666191024121116. [DOI] [PubMed] [Google Scholar]
- Tantawy M. A.; El-Sherbeeny N. A.; Helmi N.; Alazragi R.; Salem N.; Elaidy S. M. Synthetic antiprotozoal thiazolide drug induced apoptosis in colorectal cancer cells: implications of IL-6/JAK2/STAT3 and p53/caspases-dependent signaling pathways based on molecular docking and in vitro study. Mol. Cell. Biochem. 2020, 469, 143–157. 10.1007/s11010-020-03736-4. [DOI] [PubMed] [Google Scholar]