Abstract
Live-cell fluorescence microscopy is broadly applied to study the dynamics of receptor-mediated cell signaling, but the availability of intracellular biosensors is limited. A biosensor based on the tandem SH2 domains from phospholipase C γ1 (PLCγ1), tSH2-WT, has been used to measure phosphorylation of the epidermal growth factor receptor (EGFR). Here, we found that tSH2-WT lacked specificity for phosphorylated EGFR, consistent with the known promiscuity of SH2 domains. Further, EGF-stimulated membrane recruitment of tSH2-WT differed qualitatively from the expected kinetics of EGFR phosphorylation. Analysis of a mathematical model suggested, and experiments confirmed, that the high avidity of tSH2-WT resulted in saturation of its target and interference with EGFR endocytosis. To overcome the apparent target specificity and saturation issues, we implemented two protein engineering strategies. In the first, we screened a combinatorial library generated by random mutagenesis of the C-terminal SH2 domain (cSH2) of PLCγ1 and isolated a mutant form (mSH2) with enhanced specificity for phosphorylated Tyr992 (pTyr992) of EGFR. A biosensor based on mSH2 faithfully reported the kinetics of EGFR phosphorylation but retained cross-reactivity similar to tSH2-WT. In the second approach, we isolated a pTyr992-binding protein (SPY992) from a combinatorial library generated by mutagenesis of the Sso7d protein scaffold. Compared to tSH2-WT and mSH2, SPY992 exhibited superior performance as a specific, moderate-affinity biosensor. We extended this approach to isolate a biosensor for EGFR pTyr1148 (SPY1148). This approach of integrating theoretical considerations with protein engineering strategies can be generalized to design and evaluate suitable biosensors for various phospho-specific targets.
Introduction
Receptor tyrosine kinases (RTKs) are cell surface receptors that mediate many key cellular processes including proliferation, differentiation, and cell migration (1, 2). Aberrant activation of RTKs and their downstream signaling pathways is a hallmark of most human cancers. Typically, ligand binding to the extracellular domain of the receptor results in activation of tyrosine kinase function in the cytoplasmic domain, which in turn results in autophosphorylation of specific residues in the receptor and phosphorylation of receptor-associated proteins. Thus, ligand binding triggers RTK activation and initiation of downstream signaling pathways. The ability to obtain spatially resolved measurements of receptor activation using live-cell imaging can uniquely enable a mechanistic understanding of the relationship between dynamic changes in signaling and resulting cell behavior. Yet, implementation of live-cell imaging using intracellular biosensors remains a considerable challenge.
Intracellular biosensors contain a molecular recognition element that should specifically recognize the target(s) of interest. However, there are only a small number of highly specific protein biosensors currently available. A second and arguably greater limitation is that biosensors may interfere with or otherwise modulate the signaling process they are meant to detect. In this latter context, we have previously outlined a theoretical framework for understanding the relationship between fidelity of biosensor readout and binding affinity of the biosensor for its target (3). Notably, an ideal biosensor should have an intermediate affinity for its target. Whereas low affinity results in very little binding, very high affinity may result in saturation of readout if the target species is present at high concentration, or perturb the target or its ability to stimulate endogenous downstream signaling molecules if the biosensor is present in excess. Conversely, biosensors with low affinities for their targets may not accurately reflect the activity of the target, resulting in a misleadingly low measure of signaling. Thus, both specificity and binding affinity determines the fidelity of the biosensor readout to dynamic changes in the concentration of the target species being detected. Here, we applied protein engineering tools in conjunction with the aforementioned theoretical principles to design protein biosensors targeting a specific phosphotyrosine (pTyr) site in the epidermal growth factor receptor (EGFR).
Ligand binding to the extracellular domain of EGFR results in receptor dimerization and subsequent autophosphorylation of multiple tyrosine residues in the cytoplasmic domain, leading to the recruitment of downstream signaling components that contain pTyr-binding SH2 domains. We evaluated the tandem SH2 domains from wild-type phospholipase C γ1 (PLCγ1) fused to enhanced green fluorescent protein (EGFP), hereafter denoted as tSH2-WT, as a biosensor for studying the dynamics of EGFR autophosphorylation at Tyr992. Fluorescent protein fusions of SH2 domains have been previously used to study EGFR signaling (4, 5). Consistent with previous reports of promiscuous binding of SH2 domains (6–8), we show that tSH2-WT lacked specificity for EGFR pTyr992. Furthermore, the juxtaposition of two SH2 domains in tSH2-WT resulted in intramolecular avidity; the two SH2 domains could simultaneously bind to two pTyr-containing epitopes. This resulted in high effective affinity of the interaction between tSH2-WT and EGFR, which in turn distorted the readout corresponding to the abundance of phosphorylated EGFR in the plasma membrane. Therefore, we investigated the use of two distinct strategies for generating more accurate intracellular biosensors for EGFR pTyr992. By screening a combinatorial library generated by random mutagenesis of the C-terminal SH2 domain of tSH2-WT, we isolated a mutant protein (mSH2) with enhanced specificity for pTyr992 EGFR. An mSH2-based biosensor faithfully reported the expected kinetics of Tyr992 phosphorylation. However, like tSH2-WT, mSH2 retained cross-reactivity with the Tyr1021 phosphorylation site in platelet-derived growth factor receptor (PDGFR).
To overcome the limitations imposed by promiscuous binding of SH2 domain proteins, we investigated the use of an “inert” protein scaffold with no known native binding interactions with intracellular proteins in mammalian cells. Specifically, we isolated an EGFR pTyr992-binding protein (SPY992) from a combinatorial library constructed by mutagenesis of the Sso7d protein from the hyperthermophilic archaeon Sulfolobus solfataricus (9). In previous studies, binding proteins for a wide spectrum of targets have been obtained by mutagenesis of the Sso7d protein (9–13). Notably, SPY992 not only exhibited a readout that was consistent with the expected phosphorylation dynamics of EGFR, but also no cross-reactivity with PDGFR. Moreover, unlike mSH2 and tSH2-WT, SPY992 did not affect receptor internalization. Finally, generation of an Sso7d-based intracellular biosensor for pTyr1148 EGFR confirmed the generalizability of our strategy to other pTyr targets.
Results
The tSH2-WT biosensor lacks specificity and does not faithfully track kinetics of EGFR phosphorylation
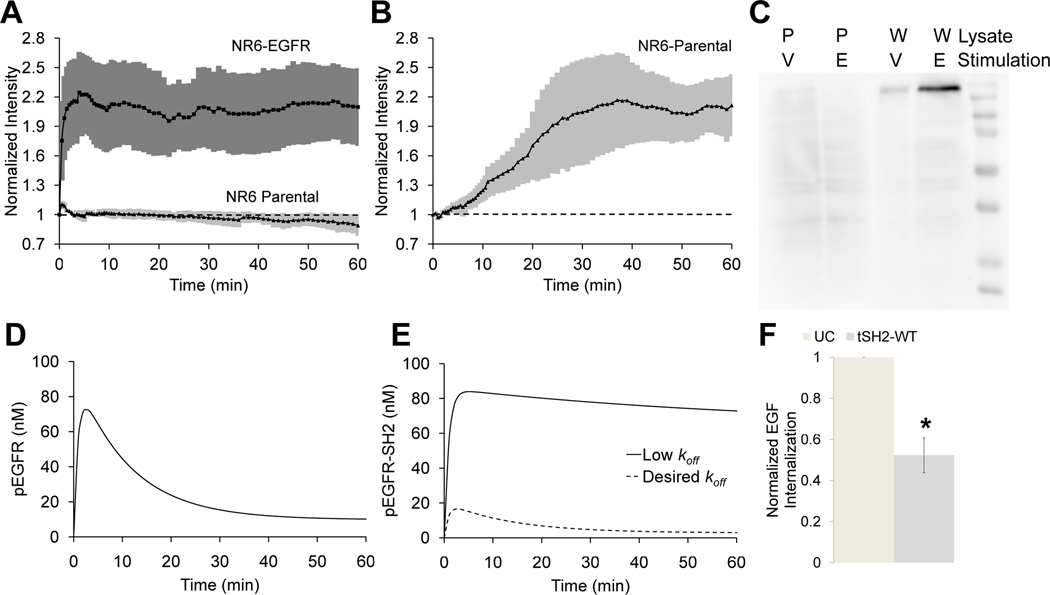
The paired PLCγ1 SH2 domain tSH2-WT has been shown to bind phosphorylated EGFR (pEGFR) and prevent tyrosine dephosphorylation of EGFR in vitro, and pTyr992 has been identified as its major binding site (14). Further, tSH2-WT tagged with a fluorescent protein has been used in live-cell imaging studies (5, 15). Therefore, we evaluated tSH2-WT tagged with EGFP as a biosensor to monitor the dynamics of EGFR phosphorylation at pTyr992. We expressed an EGFP-tSH2-WT fusion protein by transient transfection in parental NR6 mouse fibroblast cells (control) or NR6 cells expressing human EGFR, treated the cells with EGF, and observed membrane localization of tSH2-WT using total internal reflection fluorescence (TIRF) microscopy. The fluorescence signal increased substantially in response to EGF stimulation in EGFR-expressing cells, but not in parental cells (Fig. 1A), demonstrating that tSH2-WT may be useful for monitoring the kinetics of EGFR phosphorylation. The next step was to characterize the critical attributes of specificity and fidelity for this biosensor.
Fig. 1. The tSH2-WT biosensor lacks specificity and does not faithfully track EGFR phosphorylation kinetics.
(A) Normalized membrane recruitment of tSH2-WT in response to stimulation of NR6 parental cells and EGFR-expressing NR6 cells with EGF, as measured by TIRF microscopy (n = 5 cells from 5 independent experiments). (B) Normalized membrane recruitment of tSH2-WT in NR6 parental cells treated with sodium orthovanadate (n = 7 cells from 7 independent experiments). For (A) and (B), error bars indicating 95% confidence intervals are shaded. (C) Far-Western blotting analysis of NR6 parental cells (P) and NR6 cells expressing wild-type EGFR (W) stimulated with sodium orthovanadate for 60 min. (V) or EGF for 10 min. (E). Cell lysates were blotted and probed with tSH2-WT. Blot is representative of n = 3 independent experiments. (D) Kinetics of pEGFR predicted by the mathematical model. (E) Predicted effect of binding affinity on the interaction between an idealized biosensor and the target on biosensor readout. The desired koff is higher (lower affinity), and therefore biosensor binding does not dramatically perturb receptor dynamics. (F) Internalization of labeled EGF measured by flow cytometry in EGFR-expressing NR6 cells transfected with tSH2-WT, compared to untransfected cells (UC). *p <0.05 for comparison with UC. (n = 3 independent experiments).
To assess the specificity of tSH2-WT, we monitored the fluorescence readout upon treatment of NR6 parental cells with sodium orthovanadate, an inhibitor of protein tyrosine phosphatases. Because NR6 parental cells do not produce EGFR, any recruitment of tSH2-WT to the cell membrane must be due to recognition of other pTyr motifs. tSH2-WT produces a significant TIRF readout in orthovanadate-treated parental cells, indicating poor specificity (Fig. 1B). To further assess specificity of tSH2-WT, we conducted far-Western blotting analysis. Lysates of NR6 parental cells and EGFR-expressing NR6 cells treated with sodium orthovanadate or EGF were probed with GST-tagged tSH2-WT (Fig. 1C). Although a high-intensity band corresponding to the molecular mass of EGFR was observed in lysates of EGF-stimulated, EGFR-expressing cells as expected, the presence of other bands both cell lines confirms the promiscuity of tSH2-WT binding.
To investigate the fidelity of tSH2-WT as a biosensor for live-cell imaging, it is instructive to review the expected kinetics of EGFR phosphorylation, which is well-established in the literature (16–18). In response to a saturating dose of EGF well above its apparent binding KD (~1 nM), receptor activation at the cell surface peaks rapidly and then decreases because of enhanced endocytosis of the active receptor complex (19). These kinetics are typified by a simple mathematical model of EGFR dynamics (20–22) (see Materials and Methods) that calculates the characteristic, transient kinetics of pEGFR at the cell surface (Fig. 1D). In contrast to the kinetics of EGFR phosphorylation, the experimental readout from the tSH2-WT sensor showed little adaptation corresponding to internalization of the receptor (Fig. 1B). The cooperative binding of the two SH2 domains in tSH2-WT yields high binding avidity between the tSH2-WT biosensor and pEGFR (23). Kinetic model calculations further suggested that the apparent lack of adaptation seen in the experiments was consistent with high overall binding affinity (low dissociation rate constant) of tSH2-WT for pEGFR (Fig. 1E). Conversely, the model also predicted that a substantial increase in the dissociation rate constant (thus reducing the affinity) of biosensor binding to pEGFR would result in a readout that is consistent with the true time course of pEGFR (compare Fig. 1D and 1E). Consistent with the model assumptions, we found that the presence of tSH2-WT affected the kinetics of receptor internalization. We measured the internalization of fluorescently labeled EGF in EGFR-expressing NR6 cells, in the absence (control) or presence of tSH2-WT. A significant decrease in internalization of EGF in the cells expressing tSH2-WT, relative to control cells, was observed (Fig. 1F).
Taken together, our experimental data and modeling analysis lead to two insights. First, tSH2-WT is not a suitable biosensor, because it lacks specificity and does not faithfully track the expected kinetics of pEGFR. Second, the lack of fidelity of the tSH2-WT biosensor can be attributed to its high binding affinity for pEGFR; therefore the use of a biosensor with lower affinity for pEGFR would be predicted to alleviate this discrepancy.
The engineered protein mSH2 tracks the expected kinetics of EGFR phosphorylation and shows specificity for EGFR pTyr992 in live-cell imaging experiments
Use of a single SH2 domain instead of the tandem SH2 domains in tSH2-WT should eliminate the high affinity for pEGFR arising from the avidity effect. However, when a GST-tagged single SH2 domain – the C-terminal SH2 domain of the pair (cSH2) – was used to probe cell lysates in far-Western blotting assays, no specific band corresponding to the molecular mass of EGFR was detected (Fig 2A). We also assessed the use of cSH2 as a biosensor in TIRF microscopy. The kinetics of EGFR phosphorylation reported by cSH2 upon addition to EGF to EGFR-expressing NR6 cells were more consistent than tSH2-WT with the readout expected from the EGFR activation kinetics model (Fig. 2B). Nevertheless, the fluorescence readout upon treatment of NR6 parental cells with sodium orthovanadate indicated poor specificity of the cSH2 biosensor (Fig. 2B). This result was not surprising given that SH2 domains are inherently promiscuous; the consensus pYXXP motif bound by SH2 domains is present in many proteins (24).
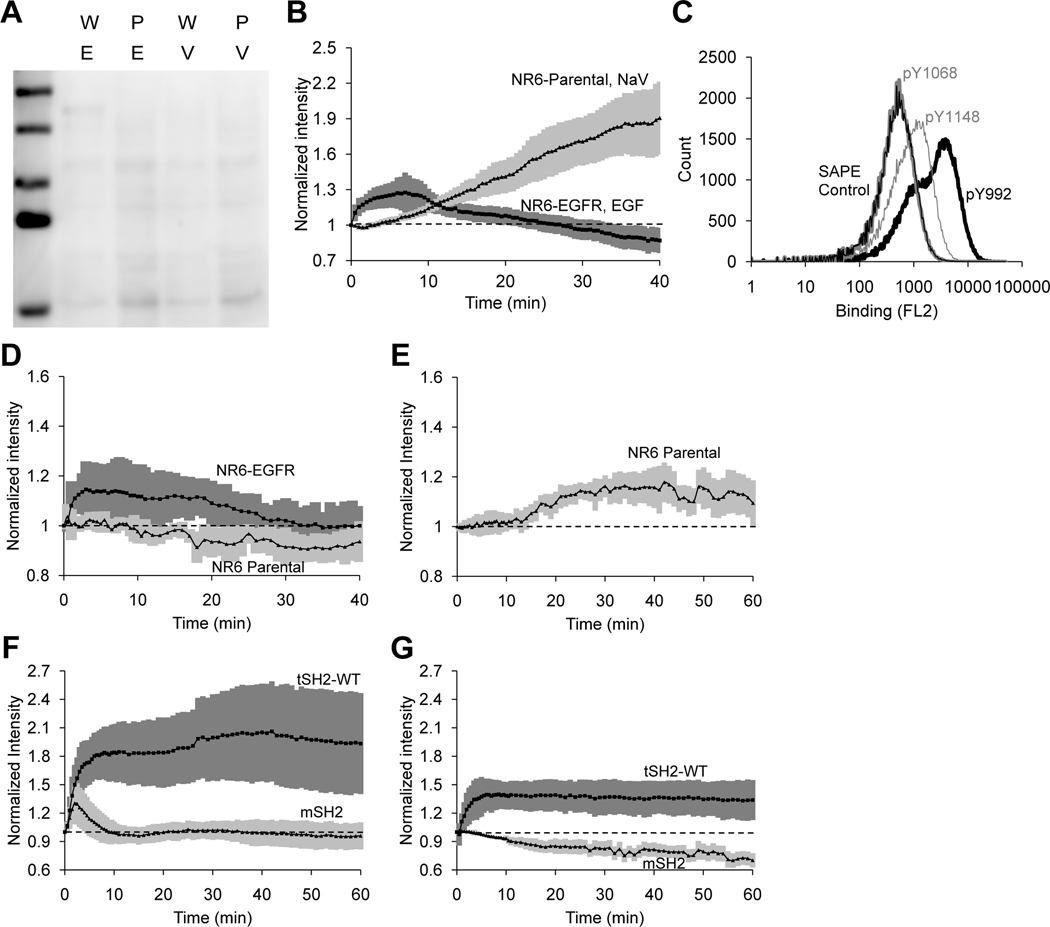
Fig 2. A mutant SH2 domain (mSH2) biosensor closely tracks the expected kinetics of EGFR phosphorylation and is more specific the cSH2 for the EGFR pTyr992.
(A) Far-Western blotting analysis of NR6 parental cells (P) and NR6 cells expressing wild-type EGFR (W) stimulated with sodium orthovanadate for 60 min. (V) or EGF for 10 min. (E). Cell lysates were blotted and probed with cSH2. Blot is representative of n = 3 independent experiments. (B) Normalized membrane recruitment of cSH2, measured by TIRF microscopy, in response to stimulation of EGFR-expressing NR6 cells with EGF (n = 21 cell analyzed from 5 independent experiments) or NR6 parental cells with sodium orthovanadate (n = 21 cells analyzed from 3 biological experiments). (C) Yeast cells displaying mSH2 on the cell surface were labeled with biotinylated synthetic peptides corresponding to the pTyr992 (pY992), pTyr1068 (pY1068), or pTyr1148 (pY1148) sites in EGFR, followed by secondary labeling with streptavidin-phycoerythrin (SA-PE). A control sample of yeast cells labeled only with SA-PE is also shown. n = 3 independent experiments. (D) Normalized membrane recruitment of mSH2, measured by TIRF microscopy, in response to EGF stimulation of NR6 parental cells (n = 7 cells analyzed from 7 independent experiments) or EGFR-expressing NR6 cells (n = 18 cells analyzed from 5 independent experiments). (E) Normalized membrane recruitment of mSH2, measured by TIRF microscopy, in response to sodium orthovanadate treatment of NR6 parental cells. (n = 7 cells from 7 independent experiments). (F–G) Normalized the expressing either the c’1000 EGFR truncation mutant (F, n = 5 cells from 5 independent experiments) or the c’1000F EGFR mutant with the Y992F mutation (G, n = 6 cells from 6 independent experiments). 95% confidence intervals for normalized recruitment are shaded in each figure.
To identify an improved biosensor, we generated a combinatorial library by random mutagenesis of cSH2 and screened for mutants that showed specific and detectable binding to pTyr992 of EGFR. A library of ~ 109 mutants was generated using yeast surface display and screened using magnetic selection and fluorescence-activated cell sorting (FACS) (25) with a synthetic peptide corresponding to the pTyr992 site as the target. Negative selection steps were used to eliminate proteins that bound to peptides corresponding to the nonphosphorylated Tyr992 site, other phosphorylation sites in EGFR, and putative tyrosine phosphorylation motifs in other proteins that have sequence homology to the pTyr motifs in EGFR. The population of cells obtained after magnetic selection and two rounds of FACS was plated, and single clones were analyzed (fig. S1A-D). DNA sequencing (table S1) identified a mutant form (mSH2) with a single amino acid substitution (C65G) at a residue that has been implicated in the determination of binding specificity of SH2 domains (26).
We used yeast surface-displayed mSH2 and synthetic peptides to assess the specificity of binding to the pTyr992 site in EGFR relative to other phosphorylation sites. mSH2 exhibited > 10-fold greater signal for binding to the pTyr992 peptide relative to the pTyr1148 peptide and no binding to the pTyr1068 peptide (Fig. 2C). To evaluate mSH2 as a biosensor, an mSH2-tdTomato or mSH2-EGFP fusion was transiently transfected into NR6 parental cells or into NR6 cells expressing either EGFR or variants thereof. The cells were treated with EGF or sodium orthovanadate, and membrane localization of mSH2 was observed using TIRF microscopy. Compared with the EGF-stimulated translocation of tSH2-WT in EGFR-expressing cells (Fig. 1A), the corresponding time course of mSH2 recruitment showed a significantly lower fold change (Fig. 2D, fig. S2A). A similar comparison held true for sodium orthovanadate treatment of NR6 parental cells that lack EGFR (Fig. 2E fig. S2B). More importantly, unlike tSH2-WT, mSH2 exhibited the transient, EGF-stimulated response that is characteristic of pEGFR on the cell surface. Finally, the intracellular concentration of the biosensor also affected readout in our model (fig. S3A–B). Nevertheless, it is important to note that higher affinity of interaction between the biosensor and pEGFR resulted in significantly greater distortion of the readout relative to the true pEGFR concentration compared with changes in biosensor concentration (fig. S3A–B).
To further evaluate the specificity of mSH2 for EGFR pTyr992, we compared the readout from mSH2 and tSH2-WT in NR6 cells expressing truncation mutants of EGFR, c’1000 or c’1000F. The cytoplasmic domain of EGFR is truncated at residue 1000 in the c’1000 mutant, leaving Tyr992 as the only major phosphorylation site. In the c’1000F variant, Tyr992 is mutated to phenylalanine and therefore cannot be phosphorylated at that site; however, it is established that c’1000F mediates intracellular signaling in response to EGF stimulation through heterodimerization with ErbB2 (27). Similar to the responses in cells expressing wild-type EGFR, there was a substantial difference between the EGF-stimulated responses of mSH2 and tSH2-WT in cells expressing the c’1000 mutant, in terms of both their magnitudes and shapes (Fig. 2F). The most compelling contrast was observed in the c’1000F-expressing cells: whereas tSH2-WT was recruited in response to EGF, albeit to a lesser extent than in cells expressing wild-type EGFR, mSH2 showed no discernable response to the activated EGFR lacking pTyr992 (Fig. 2G).
Taken together, these results demonstrate the suitability of the engineered mutant, mSH2, for live-cell imaging experiments. Unlike tSH2-WT, the mSH2 biosensor better tracks the kinetics of EGFR phosphorylation at the cell surface and exhibits greater specificity for its intended target, pTyr992. The findings also demonstrate that biosensors with improved specificity can be obtained through random mutagenesis of native protein domains and combinatorial screening of variants with prescribed properties.
The mSH2 biosensor exhibits cross-reactivity with PDGFR pTyr1021
To further test the binding specificity of mSH2, we performed far-Western blotting analysis of NR6 parental and EGFR-expressing cells treated with EG or sodium orthovanadate. Cell lysates were blotted and probed with GST-tagged mSH2 (Fig. 3A). A comparison with the corresponding analysis with tSH2-WT (Fig. 1C) indicated that mSH2 has greater specificity for EGFR. Notably, non-specific bands are absent in parental NR6 cells stimulated with EGF. However, a single high molecular weight band was seen in orthovanadate-stimulated parental NR6 cells. We hypothesized that this band could be the platelet derived growth factor β-receptor (PDGFR), because tSH2-WT is known to bind pTyr1021 of PDGFR (28–30). Consistent with this hypothesis, both mSH2 and tSH2-WT displayed on yeast cells bound to a synthetic peptide corresponding to the pTyr1021 site in PDGFR (Fig. 3B). We further evaluated the readout of mSH2 and tSH2-WT in NR6 parental cells, which do not produce EGFR, treated with platelet-derived growth factor (PDGF). Both tSH2-WT and mSH2 exhibited responsiveness to PDGF stimulation (Fig. 3C). The significantly higher readout for tSH2-WT relative to mSH2 may be attributed to the higher avidity of the tSH2-WT biosensor.
Fig. 3. mSH2 displays affinity for pTyr1021 of PDGFR in addition to pTyr992 of EGFR.
(A) Far-Western blotting analysis of NR6 parental cells (P) and NR6 cells expressing wild-type EGFR (W) stimulated with sodium orthovanadate (V) or EGF (E). Cell lysates were blotted and probed with mSH2. Blot is representative of n = 3 independent experiments. (B) Yeast cells displaying mSH2 or tSH2-WT on the cell surface were labeled with a biotinylated synthetic peptide corresponding to the pTyr1021 site in PDGFR, followed by secondary labeling with SA-PE. A control sample of yeast cells labeled with only SA-PE is also shown. n = 3 independent experiments. (C) Normalized membrane recruitment of mSH2 (n = 27 cells from 7 independent experiments) and tSH2-WT (n = 4 cells from 4 independent experiments) in response to PDGF stimulation of NR6 parental cells. 95% confidence intervals are shaded.
The non-specific binding of mSH2 to PDGFR and, consequently the responsiveness of the mSH2 biosensor to PDGF stimulation, is consistent with our experimental protocol for library screening, which did not incorporate a negative selection step to eliminate proteins that bound to phosphorylation motifs on PDGFR. These results highlight the importance of a carefully designed negative selection strategy for engineering intracellular biosensors with high specificity. These results also reveal the disadvantages of using protein domains with promiscuous binding as the starting point for engineering biosensors.
The engineered SPY992 biosensor has specificity for EGFR pTyr992 and shows no cross-reactivity with PDGFR
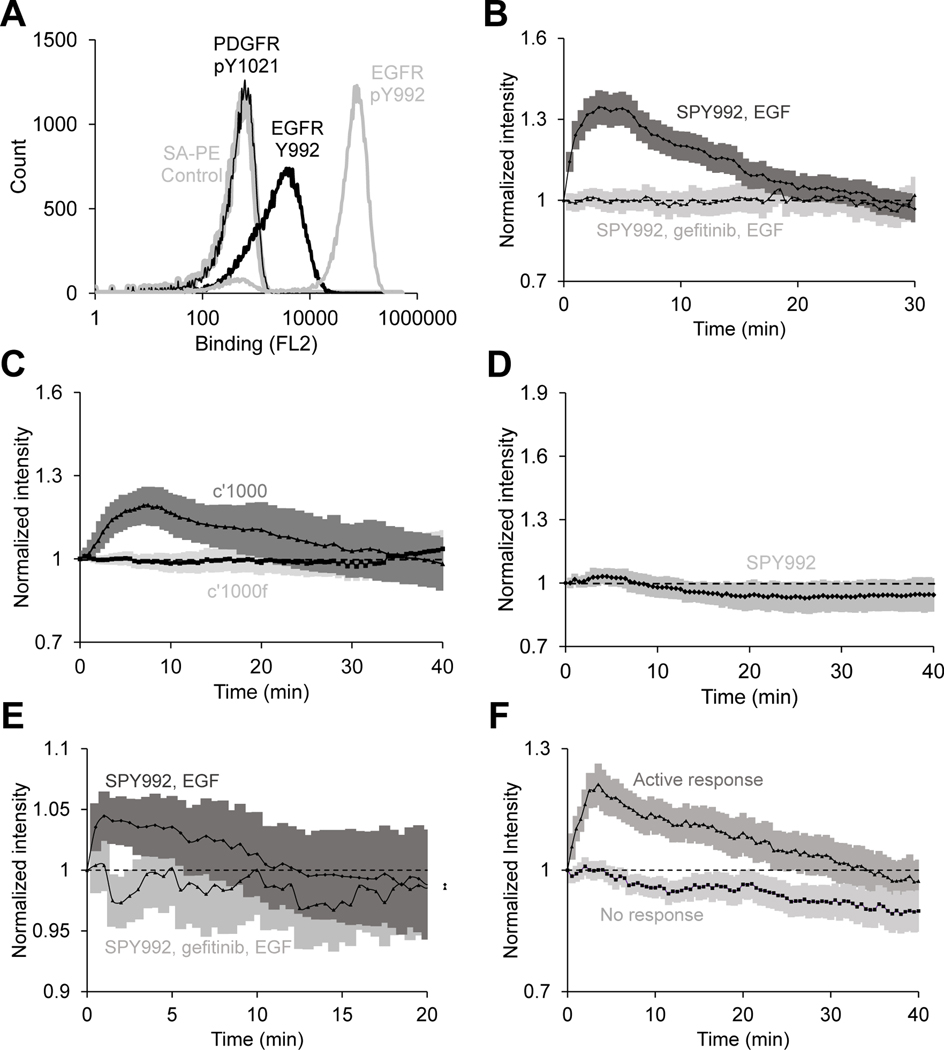
The cross-reactivity of mSH2 with PDGFR may be attributed to the promiscuous binding of SH2 domains. Therefore, we hypothesized that an “inert” protein, one with no known intracellular interactions in mammalian cells, is likely to be better suited for use as a de novo scaffold for generation of intracellular biosensors with high specificity. Accordingly, to obtain an improved pTyr992 biosensor with no cross-reactivity to PDGFR, we screened a combinatorial library constructed by mutagenesis of the Sso7d protein scaffold as previously described (9). A library of ~ 108 Sso7d mutants in yeast surface display format was screened by magnetic selection and FACS, using a synthetic peptide corresponding to the pTyr992 site as the target. Negative selection steps were used to eliminate proteins that bound to peptides corresponding to the nonphosphorylated EGFR Tyr992, other phosphorylation sites in EGFR, putative tyrosine phosphorylation motifs in other proteins that have sequence homology to the pTyr motifs in EGFR, and PDGFR pTyr1021. SPY992 (table S1) was identified after magnetic selection and three rounds of FACS. Analysis of SPY992 binding using yeast surface–displayed SPY992 and peptides corresponding to the phosphorylated and nonphosphorylated Tyr992 site in EGFR and the pTyr1021 site of PDGFR confirmed the specificity of SPY992 for the pTyr992 site in EGFR (Fig. 4A).
Fig 4. The SPY992 biosensor closely tracks the expected kinetics of EGFR phosphorylation and displays increased specificity for EGFR pTyr992 site of EGFR in live-cell imaging.
(A) Yeast cells displaying SPY992 on the cell surface were labeled with biotinylated synthetic peptides corresponding to the pTyr992 or Tyr992 sites in EGFR and the pTyr1021 site in PDGFR, followed by secondary labeling with SA-PE. A control sample of yeast cells labeled with only SA-PE is also shown. n = 3 independent experiments. (B) Normalized membrane recruitment of SPY992 in response to EGF stimulation of EGFR-expressing NR6 cells pretreated with gefitinib (n = 27 cells from 2 independent experiments) or not pretreated (n = 110 cells from 5 independent experiments), as measured by TIRF microscopy. p <0.0001 by two-way ANOVA. (C) Normalized membrane recruitment of SPY992 in response to EGF stimulation of NR6 cells expressing the c’1000 EGFR truncation mutant (n = 20 cells from 4 independent experiments) and NR6 cells expressing the c’1000F EGFR mutant with the Y992F mutation (n = 23 cells from 5 independent experiments). p <0.0001 by two-way ANOVA. (D) Normalized membrane recruitment of SPY992 in response to PDGF stimulation of NR6 parental cells (n = 21 cells from 4 independent experiments). (E) Normalized membrane recruitment of SPY992 in response to EGF stimulation of MDA-MB-231 cells pretreated with gefitinib for 5 min. (n = 97 cells from 4 independent experiments) or not pretreated (n = 52 cells from 6 independent experiments). p <0.0001 by two-way ANOVA. (F) Normalized membrane recruitment of SPY992 in EGFR-expressing NR6 cells in response to stimulation with a sub-threshold concentration of EGF (300 pM). Data from cells with an active response (n = 105 cells from 5 independent experiments) and no response (n = 95 cells from 5 independent experiments) are shown. p <0.0001 by two-way ANOVA. 95% confidence intervals are shaded (B–F).
We next evaluated the use of SPY992 as a biosensor in EGFR-expressing NR6 cells. The readout from SPY992 in EGF-stimulated cells was consistent with the expected readout (Fig. 4B). In contrast, no response was observed in EGF-stimulated cells treated with the EGFR inhibitor gefitinib. Additionally, no SPY992 response was observed in NR6 parental cells stimulated with EGF (fig. S4A). The SPY992 response was similar to that of mSH2 in NR6 parental cells treated with sodium orthovanadate (fig. S4B). Further, similar to mSH2, SPY992 was recruited to the membrane upon EGF stimulation in NR6 cells expressing the c’1000 EGFR mutant, but not in cells expressing the c’1000F mutant that lacks the Tyr992 phosphorylation site (Fig. 4C). However, strikingly different from tSH2-WT and mSH2, SPY992 did not respond to PDGF stimulation in NR6 parental cells lacking EGFR (Fig. 4D, fig. S4C). Thus, unlike tSH2-WT and mSH2, SPY992 does not exhibit cross-reactivity with PDGFR. We further confirmed that SPY992 reported the kinetics of EGFR phosphorylation in MDA-MB-231 breast cancer epithelial cells (Fig. 4E). SPY992 showed an expected readout in EGF-stimulated cells, although the signal intensity was lower than that observed in NR6 cells (Fig. 4E). Finally, we also observed that SPY992 did not significantly impact internalization of EGF in NR6 cells, unlike tSH2-WT or mSH2, both of which cause a reduction in EGF internalization, as measured by flow cytometry analysis using fluorescently labeled EGF (fig. S4D).
Having validated SPY992 as an effective biosensor for pTry992 EGFR, we investigated the kinetics of Tyr992 phosphorylation in EGFR-expressing NR6 cells upon stimulation with EGF at a concentration (300 pM) lower than the apparent KD of EGF binding to EGFR (~ 1 nM). The cells exhibited a bimodal SPY992 readout under these conditions. No SPY992 readout was observed in a fraction of cells; however, the remaining cells showed a response consistent with the expected readout, similar to cells stimulated with a saturating concentration of EGF (Fig. 4F).
Taken together, these results demonstrate the superiority of SPY992 over mSH2 and tSH2-WT as a specific biosensor for live-cell imaging of EGFR phosphorylation at Tyr992. Our results also show that generation of intracellular biosensors by combinatorial screening of libraries derived from non-native, inert protein scaffolds is a powerful approach.
SPY1148 reports kinetics of Tyr1148 EGFR phosphorylation
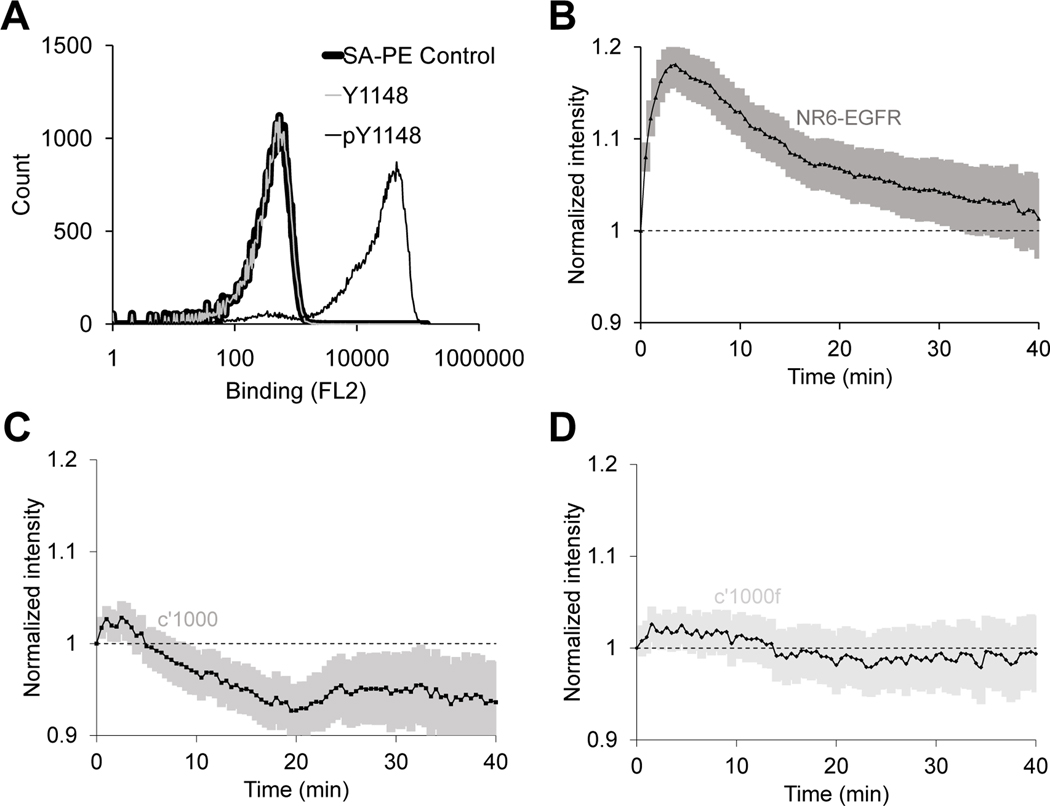
We screened the library of Sso7d variants to isolate a mutant protein, SPY1148 (table S1), that bound to a phosphopeptide corresponding to the Tyr1148 phosphorylation site (pTyr1148) in EGFR. SPY1148 bound to the pTyr1148 phosphopeptide, but not a peptide corresponding to the nonphosphorylated Tyr1148 site in EGFR in vitro (Fig. 5A). Subsequently, we evaluated SPY1148 as a biosensor in EGFR-expressing NR6 cells. SPY1148 showed an expected readout in response to EGF stimulation, similar to SPY992 (Fig. 5B). We further evaluated the specificity of SPY1148 by observing the readout upon EGF stimulation in cells expressing the c’1000 or c’1000F EGFR mutants (Fig. 5, C and D). SPY1148 showed no response in cells expressing these mutants that lack the Tyr1148 phosphorylation site. These results confirm the broader applicability of our approach for generating biosensors specific to other phosphorylation sites in EGFR.
Fig 5. The SPY1148 biosensor responds to EGFR phosphorylation and displays increased specificity for the pTyr1148 site of EGFR in live-cell imaging experiments.
(A) Yeast cells displaying SPY1148 on the cell surface were labeled with biotinylated synthetic peptides corresponding to the pTyr1148 or Tyr1148 sites in EGFR, followed by secondary labeling with streptavidin-PE (SAPE). A control sample of yeast cells labeled with only SAPE is also shown. (B–D) Normalized membrane recruitment of SPY1148 in response to EGF stimulation of NR6 cells expressing wild-type EGFR (B, n = 213 cells from 8 independent experiments), the c’1000 EGFR truncation mutant (C, n = 144 cells from 5 independent experiments), or the c’1000F EGFR mutant with the Y992F mutation (D, n = 144 cells from 5 independent experiments). EGFR compared to c’1000 and c’1000f are significantly different, p <0.0001 by two-way ANOVA) 95% confidence intervals are shaded in (B–D).
Discussion
Live-cell imaging studies of intracellular dynamics are limited by the availability of suitable biosensors. Here, we show that a previously described biosensor using the tandem SH2 domains of PLCγ1 (tSH2-WT) suffers from two major limitations in the context of studying EGFR phosphorylation dynamics. First, tSH2-WT lacks specificity, consistent with the inherent promiscuity of SH2 domain recognition (6–8). Second, EGF-stimulated translocation of tSH2-WT, although robust in magnitude, exhibits kinetics that qualitatively deviate from the expected transient changes in abundance of pEGFR at the cell surface.
Analysis of a basic kinetic model shows that the inability of tSH2-WT to faithfully track cell-surface pEGFR can be explained by its high binding affinity and occupancy of more than one site in the dimerized pEGFR complex. The avidity of the two linked SH2 domains in tSH2-WT results in high overall binding affinity with pEGFR in cells. As discussed earlier, high affinity of the biosensor-target interaction can result in saturation of the biosensor readout or perturbation of the target’s dynamics (3). Indeed, mutant SH2 domains evolved for high-affinity pTyr binding have been shown to antagonize pTyr-mediated signaling (31), and binding of tSH2-WT to pTyr992 from EGFR prevents tyrosine dephosphorylation in vitro (14). Additionally, binding of SH2 domain biosensors may alter the kinetics of EGFR phosphorylation by protecting SH2-bound phosphosites from phosphatase-mediated dephosphorylation (32). Our modeling results suggest that tSH2-WT perturbs EGFR dynamics by interfering with endocytosis of the complex. Importantly, our experiments confirm that tSH2-WT does indeed affect internalization of receptor-bound EGF.
We applied protein engineering tools to overcome limitations associated with high target binding affinity and lack of specificity of the tSH2-WT biosensor. Live-cell imaging studies confirmed that the mSH2 biosensor indeed exhibited specificity for EGFR pTyr992. Most importantly, the readout from the mSH2 biosensor was qualitatively consistent with the kinetics of EGFR phosphorylation in response to EGF treatment. Phosphorylation of cell surface EGFR peaks and then shows adaptation, in large part because of ligand-induced endocytosis. The plasma membrane translocation of mSH2 shows a nearly complete recovery during the course of chronic EGF stimulation, a much greater extent of adaptation than previously inferred from population measurements for the same cell line (33). In that study, a general phosphotyrosine-recognizing antibody was used to quantify pEGFR, and a mild acid strip was used to distinguish cell-surface and internalized pEGFR. The quantitative differences between these measurements might reflect distinct, site-specific kinetics of EGFR phosphorylation or differential endocytosis of the various phosphorylated forms of EGFR. Development of other phospho-specific biosensors will allow such hypotheses to be addressed.
Although mSH2 exhibited greater specificity for pTyr992 of EGFR than did tSH2-WT, it also retained affinity for the pTyr1021 site of PDGFR (28–30). We employed extensive negative selection steps using synthetic peptides corresponding to proteins containing phosphorylation motifs homologous to EGFR. However, negative selection against the PDGFR site, which lacks homology with the sequence surrounding Tyr992 of EGFR, was not carried out; SH2 domain binding is based on the consensus pYXXP motif present in both the pTyr992 site in EGFR and the pTyr1021 site in PDGFR (24). This result underscores the importance of the negative selection strategy in our approach.
To overcome reduced specificity resulting from promiscuous interaction of SH2 domains, and to assess the use of “inert” protein scaffolds, we screened a combinatorial library derived from the Sso7d protein scaffold to isolate a mutant protein – SPY992 – that selectively binds the Tyr992 phosphorylation site in EGFR and exhibits no cross-reactivity with PDGFR. Evaluation of SPY992 in TIRF microscopy studies confirmed the superiority of SPY992 over mSH2 and tSH2-WT. Notably, the SPY992 readout was specific to phosphorylation of Tyr992 in EGFR, and unlike mSH2 and tSH2-WT, SPY992 was not responsive to PDGFR phosphorylation. In addition, SPY992 also did not perturb receptor endocytosis. Validation of SPY1148, a protein similarly isolated as an effective biosensor for pTyr1148 EGFR, suggests generalizability of this approach.
Taken together, the isolation of mSH2, SPY992, and SPY1148 illustrates two distinct strategies for the generation of intracellular biosensors. In the case of mSH2, a native protein with promiscuous binding for a phosphotyrosine-containing motif was mutagenized to select for mutants that were more specific than the original protein for binding to EGFR pTyr992. In contrast, SPY992 and SPY1148 were constructed by creating binding affinity and specificity for target phosphopeptides through mutagenesis of a protein that has no known phosphotyrosine binding. Negative selection steps are critical in both strategies to eliminate proteins that bind to non-target species. From a practical standpoint, it is reasonable to assume that the negative selection steps are more likely to require greater stringency when a native protein with promiscuous binding is used as the starting point for biosensor development. Thus, it is conceivable that using protein scaffolds with no native binding for intracellular proteins (9, 34) may be better suited for engineering intracellular biosensors.
Studies on EGFR Tyr992 phosphorylation in cells stimulated with a sub-saturating concentration of EGF revealed a bimodal SPY992 response. Notably, a substantial fraction of the cells analyzed showed no detectable SPY992 readout under these conditions. This result may be explained by heterogeneity in receptor expression or reduced receptor occupancy with sub-saturating EGF concentrations. In cells with lower EGFR copy number, it is conceivable that protein tyrosine phosphatase activity could set a threshold receptor occupancy, below which there is negligible net phosphorylation of EGFR at Tyr992 (35). Alternatively, the bimodal response might be a consequence of competition between the biosensor and endogenous PLCγ1 and other proteins that bind pTyr992 EGFR. Either case illustrates that the SPY992 biosensor does not interfere with endogenous cellular processes due to its modest binding affinity. A biosensor with low binding affinity for its target phosphorylation site is particularly relevant in the context of EGFR. The amino acid composition of residues surrounding the phosphosites in EGFR have likely evolved to avoid off-pathway phosphorylation by intracellular kinases, a consequence of which is weaker binding to adapter proteins (36).
Specificity and binding affinity are critical properties that affect biosensor performance. Our results show that specificity of binding can be engineered by judicious choice of a native protein or protein scaffold and careful negative selections during combinatorial library screening. However, the optimal binding affinity is context-specific; it depends on factors such as the abundance of the target and the presence of adapter proteins competing for the target as well as their binding affinities for the target. The optimal binding affinity of the biosensor represents a trade-off: high affinity affects cellular processes associated with the target, whereas low affinity reduces the signal-to-noise ratio. From the practical standpoint of biosensor design by protein engineering, a useful heuristic may be to select a binding protein with the lowest affinity that gives a workable readout in the cell type of interest. Generally, combinatorial library screens can identify several binding proteins with a range of binding affinities (13, 37).
In summary, we demonstrate the integration of theoretical considerations with two distinct protein engineering strategies to design intracellular biosensors for site-specific phosphorylation of EGFR. Our approach serves as a blueprint for the design and evaluation of intracellular biosensors, towards the development of a broad palette of highly specific live-cell imaging reagents on par with phospho-specific antibodies used for immunoblotting.
Materials and Methods
Base model of EGF receptor dynamics
We adopt a highly simplified description of receptor dynamics similar to the classic model of Wiley and Cunningham (22). This model does not consider complexities such as, e.g., receptor dimerization. In the absence of the SH2 biosensor, the dynamic equations and initial conditions are as follows.
The model assumes that free, surface receptor (R) is produced via de novo synthesis at a constant rate, Vsyn. It is consumed via basal turnover of the membrane, with first-order rate constant kt. These two parameters determine the initial density of R. At time zero, the ligand EGF is added to the external medium with concentration [L], and this concentration is held constant in the model (i.e., its depletion is considered negligible). Formation of the ligand-receptor complex on the cell surface (Cs) occurs by mass-action kinetics (forward rate constant kf, reverse rate constant kr). Internalization of Cs is characterized by the first-order endocytic rate constant, ke, which is greater than kt; thus, ligand binding induces overall receptor downregulation. Finally, the internalized ligand-receptor complex (Ci) is degraded with first-order rate constant kdeg.
The values of the kinetic parameters described above were assigned according to order-of-magnitude estimates: Vsyn = 1 nM/min (cytosolic volume basis); kt = 0.01 min−1; kf = 0.1 nM−1min−1; kr = ke = 0.1 min−1; kdeg = 0.05 min−1. The initial receptor expression on the surface is Vsyn/kt = 100 nM, or roughly ~105 molecules/cell.
Introduction of the SH2 biosensor in model
The SH2 biosensor binds to the phosphorylated EGF receptor. Receptor phosphorylation is presumably rapid upon ligand binding, and therefore we assume that all ligated receptors are phosphorylated (or, rapidly come to an equilibrium of fast phosphorylation and dephosphorylation rates). Conversely, we assume that dissociation of ligand from the receptor, or of the biosensor from an unligated receptor, is followed by rapid dephosphorylation. After internalization, the EGF-EGFR complex remains phosphorylated, with its phosphorylation sites exposed to the cytoplasm (38–40). Phosphorylated receptors, whether on the surface or internalized, bind the SH2 biosensor with mass-action kinetics (association rate constant kon, dissociation rate constant koff). With these assumptions in place, the full set of equations and initial conditions are as follows.
In this model, the new species are the receptor species with asterisk superscripts, signifying that the SH2 biosensor is bound, and [B], the concentration of biosensor in the cytosol. Besides the aforementioned kon and koff, the other new rate constant is keSH2, the endocytic rate constant for the ligated receptor with SH2 bound. We fixed kon = 0.1 nM−1min−1 and varied koff = KDkon. The high-affinity variant was koff = 1 min−1 (KD = 10 nM), whereas the low-affinity variant was koff = 100 min−1 (KD = 1 μM). Note that, in both cases, koff >> kr; therefore, the abundance of R* is negligible. As for the parameter keSH2, we investigated the hypothesis that SH2 binding interferes with endocytosis and chose keSH2 = 0.1ke (i.e., keSH2 = kt). Finally, one must specify the initial value of [B]. This parameter was set in the range of 100 nM – 1 μM as is typical for expressed fluorescent protein fusions, as indicated in the corresponding figure caption.
Numerical implementation of model
All calculations were performed in the freely available Virtual Cell software environment (21), using the Combined Stiff Solver (IDA/CVODE) solver option. The Biomodel ‘SH2 biosensor’ is publicly available via www.vcell.org (note that the default units of VCell are μM and s, so we have externally converted those to units of nM and min).
Library generation
The C-terminal SH2 domain from PLCγ1 (cSH2) was amplified using primers FP1 and RP1 from a plasmid containing the Rat PLCG1 gene (5) by error prone PCR as described (11). All primers were obtained from Integrated DNA Technologies (IDT), and sequences are listed in table S2. Nucleotide analogues 8-oxo-dGTP and dPTP (Trilink Biotechnologies) were added at a final concentration of 10μM to Taq 2X Master Mix (New England Biolabs, NEB) along with 0.2μM of each primer and 10 ng of template DNA. The PCR product after 30 cycles was visualized on a 1% agarose gel and then used as a template for amplification by Phusion DNA polymerase (NEB) using primers PCTF1 and PCTR1 to generate at least 50μg of final PCR product containing the sequences necessary for homologous recombination. The PCR product was concentrated by using Pellet Paint™ (Novagen). A yeast display library was generated using the PCR product through homologous recombination as described (25). Briefly, the yeast display vector pCTCON was linearized by restriction digestion with NheI and BamHI (NEB) and concentrated using Pellet Paint. 4μg of linearized vector and 12μg of PCR product was added to 400μl of electrocompetent yeast cells and electroporation was performed using a 2 mm electrode gap cuvette in a Bio-Rad Gene Pulser system (Bio-Rad) at 2.5kV and 25μF. Four such transformations were performed, and following a one hour outgrowth in 32 ml of 1:1 mix of 1M Sorbitol and YPD (10 g/l yeast extract, 20 g/l peptone, 20 g/l dextrose), the electroporated cells were expanded in 1L of SDCAA (20 g/l dextrose, 5 g/l Casamino acids, 6.7 g/l yeast nitrogen base, 5.40 g/l Na2HPO4, 7.45 g/l NaH2PO4). Simultaneously, an aliquot was grown on SDCAA agar plates to estimate library size. A library of ~ 108 Sso7d mutants that was previously constructed and characterized was used in this study (9).
Isolation of mutant proteins binding the pTyr992 and pTyr1148 sites in EGFR
Peptides listed in table S3 were obtained from Genscript. All peptides were at least 90% pure and contained an N-Terminal biotin moiety. The yeast display library of cSH2 mutants generated, or the Sso7d library, was grown in SGCAA (20 g/l galactose, 5 g/l Casamino acids, 6.7 g/l yeast nitrogen base, 5.40 g/l Na2HPO4, 7.45 g/l NaH2PO4) to induce surface protein expression. Binders to non-target species were eliminated by negative selection using magnetic sorting as described (25). Biotinylated peptides were immobilized on biotin binder beads (Life Technologies). An equimolar mix of all bead-immobilized peptides other than the target peptide was prepared. This mix was incubated with ~1010 cells of the induced cSH2 library, or ~ 109 cells of the Sso7d library, for an hour and the beads were removed using a magnet. The remaining cells were incubated with beads containing the target peptide for 30 minutes. Following three washes in 1 ml PBS-BSA (PBS + 0.1% BSA), the beads were collected and grown in SDCAA. These expanded cells were again grown in SGCAA to induce surface protein expression and FACS was performed using a MoFlo cell sorter (Beckman Coulter Inc.). For cytometry approximately 5×107 induced cells were labeled simultaneously with an anti-c-myc chicken antibody (Life technologies) to quantify surface protein expression and a biotinylated peptide to quantify binding to the peptide. The secondary antibodies used were Streptavidin-PE and goat-anti-chicken conjugated to Alexa-633 (Life technologies). Cells obtained after the magnetic selection step were sorted by successive rounds of FACS and the final pool of cells was grown on SDCAA plates. Yeast colonies were sequenced by Genewiz. Further analysis of the sorted population and individual clones were performed using Accuri C6 analyzer (BD Biosciences).
Recombinant expression and purification of SH2 domain mutants
mSH2, cSH2 and tSH2-WT were cloned from the yeast display vector into the pET42 vector using the restriction sites SpeI and XhoI to generate GST-tagged fusions. Primers FP2 and RP2 were used to introduce the restriction sites by PCR to facilitate cloning of mSH2 and cSH2 whereas primers FP3 and RP2 were used for cloning of tSH2-WT. Sequences were verified by colony sequencing using a T7 reverse primer by Genewiz. Subsequently, plasmids were introduced into Rosetta Cells™ (EMD Biosciences). Protein expression was induced by addition of 1mM IPTG followed by incubation at 20 °C for 12 hours with shaking at 250 rpm. After induction, cells were harvested by centrifugation (4800g, 10min), resuspended in PBS with 0.2mM PMSF (Sigma Aldrich) and lysed by sonication (2 sec ON, 5 sec OFF) for 7 minutes. The lysate was cleared by centrifugation and filtration through a 0.22 μm filter and purified using a GSTrap FF column (GE Healthcare Bio-Sciences) on a Bio-Rad Biologic LP system using a linear glutathione gradient. The eluted fractions were analyzed by SDS PAGE and pure fractions were combined and dialyzed into 4L of PBS using a 3.5kDa MWCO filter (Thermo Fisher). Protein concentrations were determined by bicinchoninic acid (BCA) assay (Thermo scientific).
Far Western blotting
Far-Western blotting was performed following the protocol of (8). Cells were lysed in lysis buffer containing protease and phosphatase inhibitors (50mM HEPES, 100mM NaCl, 10% Glycerol, 1% Triton X-100, 1mM sodium orthovanadate, 50mM β-glycerophosphate, 5mM NaF, 1mM EGTA, 10mM sodium pyrophosphate, 10μg/ml each of Aprotinin, Leupeptin, Pepstatin A and Chymostatin). Lysates from NR6 parental cells and EGFR-overexpressing NR6 cells, stimulated with EGF (Peprotech) or sodium orthovanadate (Thermo Fisher), were electrophoresed on a 7.5% SDS-PAGE gel and transferred to PVDF membranes using standard protocols. The membranes were blocked overnight at 4°C in 5% nonfat dry milk in TBST with 1 mM sodium orthovanadate and 5 mM EDTA. Primary incubation was performed using 20 nM mSH2-GST, 20 nM tSH2-WT-GST and 100nM cSH2-GST and the membranes were then washed with TBST and blocked for 1 hour at room temperature in 5% nonfat dry milk in TBST. Anti-GST HRP (GE Healthcare Bio-Sciences) was used to probe for the GST tag and SuperSignal® West Femto substrate (Thermo Fisher Scientific) was used to obtain a chemiluminescent readout.
Cell Culture
Parental mouse NR6 cells (41) were cultured at 37°C, 5% CO2 in Dulbecco’s MEMα medium supplemented with 7.5% fetal bovine serum, 1% sodium pyruvate, 1% non-essential amino acids and 1% PSG (L-glutamine and antibiotics penicillin and streptomycin). Three variants of this cell line with full length EGFR (wild-type), C-terminal truncation mutants (c’1000) and Y to F mutant of c’1000 (c’1000F) (5, 42) were cultured in same media, additionally supplemented with 350 μg/ml G418 antibiotic. MDA-MB-231 cells were cultured at 37°C, 5% CO2 in Dulbecco’s MEM medium supplemented with 10% fetal bovine serum and 1% PSG. All tissue culture reagents were purchased from Thermo Fisher Scientific. EGF and PDGF were obtained from R&D Systems and Peprotech, respectively. EGF concentration was 10 nM unless otherwise specified. PDGF concentration used was 15 ng/mL. Sodium orthovanadate (Sigma-Aldrich) and gefitinib (R&D Systems) were used at concentrations of 1 mM and 500 nM respectively. NR6 and MDA-MB-231 cell lines were transiently transfected with tSH2-WT-EGFP, mSH2-EGFP, cSH2-EGFP, SPY992-EGFP, SPY1148-EGFP, or mSH2-tdTomato using Lipofectamine 3000 with Plus reagent (Thermo Fisher Scientific) per the manufacturer’s protocol and a plasmid amount of 700ng.
TIRF imaging and image analysis
Using PCR amplification with primers tdEF1 and tdXR1 tdTomato from tdTomato-Lifeact was cloned into EcoRI and XhoI digested pET28 vector. Then mSH2 was subcloned into this construct from the yeast display vector using NheI and BamHI to generate a tdTomato fusion. These fusions were then cloned into pBM-IRES-puro through the restriction sites NotI and HindIII using primers FP4 and tdHR2. Each of the biosensors, cSH2, mSH2, SPY992 and SPY1148 were subcloned into the vector containing EGFP-tSH2-WT (pEGFP-C1 plasmid with fusion of tSH2-WT to C-terminus of EGFP (5)) vector through BglII and EcoRI replacing tSH2-WT. Cells were detached with a brief trypsin-EDTA treatment and suspended in growth medium. After centrifugation at 300 × g for 5 min, the cells were resuspended in growth medium and counted using a Beckman Coulter Counter. Adhesive surfaces were prepared on clean, sterile 3.5 cm glass plates (MatTek), which were coated with 25μg/ml Fibronectin (R&D systems) for 1 hour at room temperature, washed with culture medium. NR6 and MDA-MB-231 cells were plated at 50,000 and 125,000 cells, respectively, and allowed to spread for 24 h. The cells were transiently transfected with fluorescent protein-tagged biosensors (tSH2-WT-EGFP, mSH2-EGFP, cSH2-EGFP, SPY992-EGFP, SPY1148-EGFP, or mSH2-tdTomato) for 24 h, then were subsequently serum-starved in imaging buffer (20 mM HEPES pH 7.4, 125 mM NaCl, 5 mM KCl, 1.5 mM MgCl2, 1.5 mM CaCl2, 10 mM glucose and 2 mg/ml fatty-acid-free BSA) for 4 h prior to imaging. Mineral oil was layered on top of the buffer to prevent evaporation and cellular oxidation during the experiment. Growth factors and inhibitors were diluted in the same buffer before adding to the cells. Images were acquired using TIRF microscopy with excitation of EGFP (488nm) and tdTomato (561 nm) achieved with incident beam energy of roughly 20–50 mJ per image. A 40× water immersion objective (Zeiss Achroplan, 0.8 NA) and 0.63× camera mount was used. Digital images were acquired using a cooled charge-coupled device (ORCA ER; Hamamatsu Photonics) and MetaMorph software (Universal Imaging). In each image, the intensity of the acellular region was defined as background and subtracted from the intensity of each pixel prior to further analysis. Images were analyzed using MATLAB and ImageJ. For each cell, the background-subtracted average intensity was calculated for each frame and then normalized by the average intensity measured prior to stimulation.
Analysis of EGFR internalization
NR6 cells expressing EGFR were transfected with tSH2-WT-EGFP, mSH2-EGFP, or SPY992-EGFP using Lipofectamine with Plus reagent as per the manufacturer’s protocol. The cells were removed from plate with 0.05% trypsin, inactivated with culture medium, and washed with Dulbecco’s Phosphate Buffered Saline (DPBS) at room temperature. The cells were resuspended DPBS at 37°C supplemented with biotinylated EGF complexed to Alexa Fluor 647 Streptavidin (Thermo Fisher). The cells were incubated away from light for 15 minutes at 37°C, washed with ice-cold DPBS and resuspended gently in ice-cold acid strip buffer (50 mM glycine-HCl, 100 mM NaCl, 2 M urea). The cells were then immediately washed with ice-cold DPBS, resuspended in ice-cold DPBS and analyzed by flow cytometry using Accuri C6 analyzer (BD Biosciences). EGF uptake was measured in cells transfected with biosensor (gated on EGFP fluorescence).
Statistical analysis
For comparison of the time courses for different biosensors, ANOVA (two-factor with replication) analysis of normalized intensity was performed and a p value was obtained for the sample being the source of variation. For analysis of EGF internalization, a two-tailed t-test was used to compare the median fluorescence of cells expressing the biosensor, with that from untransfected cells, using data from three independent experiments.
Supplementary Material
Fig. S1: Analysis of intermediate cell populations during library screening
Fig. S2: Normalized recruitment of tSH2-WT and mSH2 biosensors.
Fig. S3: Effect of changing binding affinity or biosensor concentration on readout from the biosensor.
Fig. S4: Comparison of biosensor responses.
Table S1. Protein sequences of mSH2, SPY992, and SPY1148.
Table S2. Primer sequences.
Table S3. Peptides used for library screening.
Acknowledgments:
The authors thank Mr. Shah Md. Toufiqur Rahman for assistance with image analysis.
Funding: This work was supported by NIH grant EB015712.
Footnotes
Competing interests: The authors declare no competing interests.
Data and Materials Availability: All the data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials.
References and Notes
- 1.Cadena DL, Gill GN, Receptor tyrosine kinases. FASEB J. 6, 2332–2337 (1992). [DOI] [PubMed] [Google Scholar]
- 2.Ullrich A, Schlessinger J, Signal transduction by receptors with tyrosine kinase activity. Cell. 61, 203–212 (1990). [DOI] [PubMed] [Google Scholar]
- 3.Haugh JM, Live-cell fluorescence microscopy with molecular biosensors: what are we really measuring? Biophys J. 102, 2003–2011 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Antczak C, Bermingham A, Calder P, Malkov D, Song K, Fetter J, Djaballah H, Domain-based biosensor assay to screen for epidermal growth factor receptor modulators in live cells. Assay Drug Dev Technol. 10, 24–36 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haugh JM, Meyer T, Active EGF receptors have limited access to PtdIns(4,5)P(2) in endosomes: implications for phospholipase C and PI 3-kinase signaling. J Cell Sci. 115, 303–310 (2002). [DOI] [PubMed] [Google Scholar]
- 6.Machida K, Mayer BJ, Nollau P, Profiling the global tyrosine phosphorylation state. Mol. Cell. Proteomics 2, 215–233 (2003). [DOI] [PubMed] [Google Scholar]
- 7.Machida K, Thompson CM, Dierck K, Jablonowski K, Kärkkäinen S, Liu B, Zhang H, Nash PD, Newman DK, Nollau P, Pawson T, Renkema GH, Saksela K, Schiller MR, Shin D-G, Mayer BJ, High-throughput phosphotyrosine profiling using SH2 domains. Mol. Cell 26, 899–915 (2007). [DOI] [PubMed] [Google Scholar]
- 8.Nollau P, Mayer BJ, Profiling the global tyrosine phosphorylation state by Src homology 2 domain binding. Proc. Natl. Acad. Sci. U. S. A 98, 13531–13536 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gera N, Hussain M, Wright RC, Rao BM, Highly Stable Binding Proteins Derived from the Hyperthermophilic Sso7d Scaffold. J. Mol. Biol 409, 601–616 (2011). [DOI] [PubMed] [Google Scholar]
- 10.Hussain M, Lockney D, Wang R, Gera N, Rao BM, Avidity-mediated virus separation using a hyperthermophilic affinity ligand. Biotechnol. Prog 29, 237–246 (2013). [DOI] [PubMed] [Google Scholar]
- 11.Gera N, Hill AB, White DP, Carbonell RG, Rao BM, Design of pH sensitive binding proteins from the hyperthermophilic Sso7d scaffold. PLoS One. 7, e48928 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cruz-Teran CA, Carlin KB, Efimenko K, Genzer J, Rao BM, Targeted Mutagenesis and Combinatorial Library Screening Enables Control of Protein Orientation on Surfaces and Increased Activity of Adsorbed Proteins. Langmuir. 32, 8660–8667 (2016). [DOI] [PubMed] [Google Scholar]
- 13.Carlin KB, Cruz-Teran CA, Kumar JP, Gomes C, Rao BM, Combinatorial Pairwise Assembly Efficiently Generates High Affinity Binders and Enables a “Mix-and-Read” Detection Scheme. ACS Synth. Biol 5, 1348–1354 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Rotin D, Margolis B, Mohammadi M, Daly RJ, Daum G, Li N, Fischer EH, Burgess WH, Ullrich A, Schlessinger J, SH2 domains prevent tyrosine dephosphorylation of the EGF receptor: identification of Tyr992 as the high-affinity binding site for SH2 domains of phospholipase C gamma. EMBO J. 11, 559–567 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stauffer TP, Meyer T, Compartmentalized IgE receptor-mediated signal transduction in living cells. J Cell Biol. 139, 1447–1454 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Salazar G, González A, Novel mechanism for regulation of epidermal growth factor receptor endocytosis revealed by protein kinase A inhibition. Mol. Biol. Cell 13, 1677–1693 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schmidt-Glenewinkel H, Reinz E, Eils R, Brady NR, Systems biological analysis of epidermal growth factor receptor internalization dynamics for altered receptor levels. J. Biol. Chem 284, 17243–17252 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wiley HS, Herbst JJ, Walsh BJ, a Lauffenburger D, Rosenfeld MG, Gill GN, The role of tyrosine kinase activity in endocytosis, compartmentation, and down-regulation of the epidermal growth factor receptor. J. Biol. Chem 266, 11083–11094 (1991). [PubMed] [Google Scholar]
- 19.Sorkin A, Goh LK, Endocytosis and intracellular trafficking of ErbBs. Exp Cell Res. 315, 683–696 (2009). [DOI] [PubMed] [Google Scholar]
- 20.Baass PC, Di Guglielmo GM, Authier F, Posner BI, Bergeron JJ, Compartmentalized signal transduction by receptor tyrosine kinases. Trends Cell Biol. 5, 465–470 (1995). [DOI] [PubMed] [Google Scholar]
- 21.Resasco DC, Gao F, Morgan F, Novak IL, Schaff JC, Slepchenko BM, Virtual Cell: Computational tools for modeling in cell biology. Wiley Interdiscip. Rev. Syst. Biol. Med 4, 129–140 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wiley HS, Cunningham DD, A steady state model for analyzing the cellular binding, internalization and degradation of polypeptide ligands. Cell. 25, 433–440 (1981). [DOI] [PubMed] [Google Scholar]
- 23.Ottinger EA, Botfield MC, Shoelson SE, Tandem SH2 domains confer high specificity in tyrosine kinase signaling. J Biol Chem. 273, 729–735 (1998). [DOI] [PubMed] [Google Scholar]
- 24.Songyang Z, Shoelson SE, Chaudhuri M, Gish G, Pawson T, Haser WG, King F, Roberts T, Ratnofsky S, Lechleider RJ, et al. , SH2 domains recognize specific phosphopeptide sequences. Cell. 72, 767–778 (1993). [DOI] [PubMed] [Google Scholar]
- 25.Gera N, Hussain M, Rao BM, Protein Selection using Yeast Cell Surface Display. Methods (2012), doi: 10.1016/j.ymeth.2012.03.014. [DOI] [PubMed] [Google Scholar]
- 26.Songyang Z, Gish G, Mbamalu G, Pawson T, Cantley LC, A single point mutation switches the specificity of group III Src homology (SH) 2 domains to that of group I SH2 domains. J. Biol. Chem 270, 26029–32 (1995). [DOI] [PubMed] [Google Scholar]
- 27.Sasaoka T, Langlois WJ, Bai F, Rose DW, Leitner JW, Decker SJ, Saltiel A, Gill GN, Kobayashi M, Draznin B, Olefsky JM, Involvement of ErbB2 in the signaling pathway leading to cell cycle progression from a truncated epidermal growth factor receptor lacking the C-terminal autophosphorylation sites. J Biol Chem. 271, 8338–8344 (1996). [DOI] [PubMed] [Google Scholar]
- 28.Ji QS, Chattopadhyay A, Vecchi M, Carpenter G, Physiological requirement for both SH2 domains for phospholipase C-gamma1 function and interaction with platelet-derived growth factor receptors. Mol Cell Biol. 19, 4961–4970 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klinghoffer RA, Duckworth B, Valius M, Cantley L, Kazlauskas A, Platelet-derived growth factor-dependent activation of phosphatidylinositol 3-kinase is regulated by receptor binding of SH2-domain-containing proteins which influence Ras activity. Mol Cell Biol. 16, 5905–5914 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pascal SM, Singer AU, Gish G, Yamazaki T, Shoelson SE, Pawson T, Kay LE, Forman-Kay JD, Nuclear magnetic resonance structure of an SH2 domain of phospholipase C-gamma 1 complexed with a high affinity binding peptide. Cell. 77, 461–472 (1994). [DOI] [PubMed] [Google Scholar]
- 31.Kaneko T, Huang H, Cao X, Li X, Li C, Voss C, Sidhu SS, Li SSC, Superbinder SH2 Domains Act as Antagonists of Cell Signaling. Sci. Signal 5, ra68–ra68 (2012). [DOI] [PubMed] [Google Scholar]
- 32.Jadwin JA, Curran TG, Lafontaine AT, White FM, Mayer BJ, Src homology 2 domains enhance tyrosine phosphorylation in vivo by protecting binding sites in their target proteins from dephosphorylation. J. Biol. Chem 293, 623–637 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haugh JM, Schooler K, Wells A, Wiley HS, Lauffenburger DA, Effect of epidermal growth factor receptor internalization on regulation of the phospholipase C-gamma1 signaling pathway. J Biol Chem. 274, 8958–8965 (1999). [DOI] [PubMed] [Google Scholar]
- 34.Hussain M, Gera N, Hill AB, Rao BM, Scaffold Diversification Enhances Effectiveness of a Superlibrary of Hyperthermophilic Proteins. ACS Synth. Biol 2, 6–13 (2013). [DOI] [PubMed] [Google Scholar]
- 35.Haugh JM, Schneider IC, Lewis JM, On the cross-regulation of protein tyrosine phosphatases and receptor tyrosine kinases in intracellular signaling. J. Theor. Biol 230, 119–132 (2004). [DOI] [PubMed] [Google Scholar]
- 36.Cantor AJ, Shah NH, Kuriyan J, Deep mutational analysis reveals functional trade-offs in the sequences of EGFR autophosphorylation sites. Proc. Natl. Acad. Sci. U. S. A 115, E7303–E7312 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jaggi M, Rao PS, Smith DJ, Wheelock MJ, Johnson KR, Hemstreet GP, Balaji KC, E-cadherin phosphorylation by protein kinase D1/protein kinase C{mu} is associated with altered cellular aggregation and motility in prostate cancer. Cancer Res. 65, 483–92 (2005). [PubMed] [Google Scholar]
- 38.Baass PC, Di Guglielmo GM, Authier F, Posner BI, Bergeron JJ, Compartmentalized signal transduction by receptor tyrosine kinases. Trends Cell Biol. 5, 465–70 (1995). [DOI] [PubMed] [Google Scholar]
- 39.Haugh JM, Schooler K, Wells A, Wiley HS, Lauffenburger DA, Effect of epidermal growth factor receptor internalization on regulation of the phospholipase C-gamma1 signaling pathway. J. Biol. Chem 274, 8958–65 (1999). [DOI] [PubMed] [Google Scholar]
- 40.Haugh JM, Localization of Receptor-Mediated Signal Transduction Pathways: The Inside Story. Mol. Interv 2, 292–307 (2002). [DOI] [PubMed] [Google Scholar]
- 41.Pruss RM, Herschman HR, Variants of 3T3 cells lacking mitogenic response to epidermal growth factor. Proc Natl Acad Sci U S A. 74, 3918–3921 (1977). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen P, Xie H, Sekar MC, Gupta K, Wells A, Epidermal growth factor receptor-mediated cell motility: phospholipase C activity is required, but mitogen-activated protein kinase activity is not sufficient for induced cell movement. J Cell Biol. 127, 847–857 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1: Analysis of intermediate cell populations during library screening
Fig. S2: Normalized recruitment of tSH2-WT and mSH2 biosensors.
Fig. S3: Effect of changing binding affinity or biosensor concentration on readout from the biosensor.
Fig. S4: Comparison of biosensor responses.
Table S1. Protein sequences of mSH2, SPY992, and SPY1148.
Table S2. Primer sequences.
Table S3. Peptides used for library screening.