Abstract
The toxigenic Inaba serotype of Vibrio cholerae O1 biotype El Tor reappeared in India in 1998 and 1999, almost 10 years after its last dominance in Calcutta in 1989. Extensive molecular characterization by ribotyping, restriction fragment length polymorphism, and pulsed-field gel electrophoresis indicated that recent Inaba strains are remarkably different from the earlier Inaba strains but are very similar to the prevailing V. cholerae O1 Ogawa El Tor biotype strains. The antibiograms of the Inaba strains were also similar to those of the recent V. cholerae Ogawa strains. These V. cholerae O1 Inaba strains appear to have evolved from the currently prevailing Ogawa strains and are likely to dominate in the coming years.
The disease cholera, caused by toxigenic strains of Vibrio cholerae belonging to the O1 or O139 serogroup, is characterized by the passing of voluminous watery stools, which rapidly leads to dehydration and, if left untreated, to death. V. cholerae O1 is further classified into two biotypes, classical and El Tor, and into two major serotypes, Inaba and Ogawa. The genes responsible for O1 antigen biosynthesis have been designated wbe (previously known as rfb) (18) and are localized on a 21.6-kb SacI fragment of DNA (7, 24). This region is highly conserved; the only changes observed between the Ogawa and Inaba serotypes are related to a mutation in the wbeT region (24, 27). V. cholerae O1 strains can undergo serotype conversion or switching between the Inaba and Ogawa serotypes (5, 8, 24). Observations of the epidemic that broke out in Latin America in 1991 have supported this notion. Extensive biochemical analyses and rRNA restriction fragment length polymorphism (RFLP) analysis have shown that the El Tor Inaba epidemic strains were unique to Latin America (13, 21). However, Ogawa isolates that were identical to the epidemic strain in all other respects began to appear in about the seventh month of the epidemic, suggesting that the epidemic strains had undergone a serotype conversion, possibly because of immune pressure in the population (13).
With the advent of the O139 serogroup in 1992 (17), the Inaba serotype of V. cholerae O1 was displaced in Calcutta and other parts of India (15) by the O139 serogroup, and the last Inaba predominance in Calcutta was observed in 1989 (17). The isolation of V. cholerae O1 belonging to the Inaba serotype became rare; the only isolation reported was from a cholera outbreak in Warangal, which was due to nontoxigenic V. cholerae O1 Inaba El Tor biotype strains (20). We began receiving some representative strains belonging to the Inaba serotype from Delhi in December 1998 and from Sewagram in November 1999. This study is an extensive molecular characterization of the recently isolated Inaba strains to determine their clonality and to evaluate their similarity to Inaba strains that were isolated in Calcutta in 1989.
The present study is part of the continuing nationwide surveillance program on cholera of the National Institute of Cholera and Enteric Diseases (NICED), Calcutta, India. In December 1998, we received a representative set of strains from Delhi, two of which were identified as V. cholerae O1 Inaba. In November 1999 we received a set of seven strains from Sewagram, six of which were identified as V. cholerae O1 Inaba and one of which was V. cholerae O1 Ogawa. The purity and identity of the strains were then confirmed by previously published procedures (15).
The strains were examined for resistance to ampicillin (10 μg), chloramphenicol (30 μg), cotrimoxazole (25 μg), ciprofloxacin (5 μg), furazolidone (100 μg), gentamicin (10 μg), neomycin (30 μg), nalidixic acid (30 μg), norfloxacin (10 μg), streptomycin (10 μg), and tetracycline (30 μg) with commercial disks (HiMedia, Mumbai, India). Antimicrobial susceptibility analysis was carried out as described previously (15).
The 7.5-kb BamHI fragment of plasmid pKK 3535 containing the 16S and 23S rRNA genes of Escherichia coli was used as the rRNA probe (4). The ctxA probe consisted of a 540-bp XbaI-ClaI fragment of ctxA cloned in pKTN901 using EcoRI linkers (11). The DNA probe for the RS element was a 2.7-kb NotI fragment from plasmid pCT5A11 (10). Genomic DNA for ribotyping and for studying the structure and organization of the CTX prophage was extracted as described previously (1). The transfer of digested DNA from a gel to a Hybond N+ membrane (Amersham Pharmacia Biotech, Little Chalfont, Buckinghamshire, England) and hybridizations with the rRNA probe for ribotyping and with the ctxA and RS1 probes for CTX genotyping were performed as described previously (1) using the ECL Nucleic Acid Detection System (Amersham Pharmacia Biotech). The membranes were then washed, exposed to Kodak Biomax film (Eastman Kodak Co., Rochester, N.Y.), and developed according to the manufacturer's instructions. Autoradiograms were digitally processed for documentation using the Gel Doc 2000 gel documentation system (Bio-Rad, Richmond, Calif.).
Pulsed-field gel electrophoresis (PFGE) was performed with the genomic DNA of the V. cholerae strains by preparing agarose plugs as described previously (12, 28). PFGE of the NotI (Takara, Shuzo Co., Ltd., Shiga, Japan)-digested inserts was performed by using the contour-clamped homogenous electric field (CHEF) method on the CHEF Mapper system (Bio-Rad) with 1% PFGE grade agarose in 0.5× TBE (44.5 mM Tris-HCl, 44.5 mM boric acid, 1.0 mM EDTA [pH 8.0]) for 40 h, 24 min. Run conditions were generated by the autoalgorithm mode of the CHEF Mapper PFGE system using a size range of 20 to 300 kb. The gels were stained for 30 min in Elix MilliQ water (Millipore Corporation, Bedford, Mass.) containing 1.0 μg of ethidium bromide per ml, then destained in Elix water for 15 min and photographed under UV light using the Gel Doc 2000 gel documentation system (Bio-Rad).
For molecular characterization we included two Inaba strains, V2 and V13, isolated in 1989 in Calcutta, two Inaba strains from Delhi isolated in 1998, six Inaba strains and one Ogawa strain isolated in 1999 in Sewagram, and four Ogawa strains from Calcutta isolated in 1998. Our rationale for examining these strains was to determine whether the Inaba strains that reappeared recently in Delhi and Sewagram bore any resemblance to the Inaba strains last isolated in Calcutta in 1989 or whether they belonged to a new clone.
The antibiotic susceptibility patterns of these strains revealed that V2 and V13, representing the Inaba strains isolated in 1989, were sensitive to nalidixic acid and streptomycin (16, 17), while the Inaba strains recently isolated in Sewagram and Delhi were resistant to these antibiotics (Table 1). Overall, the antibiograms of the recent Inaba strains matched those of the prevailing O1, Ogawa strains (9, 22). Resistance to cotrimoxazole, furazolidone, and streptomycin suggests the possibility of the presence of the SXT element in recently isolated Inaba strains. The SXT element is an approximately 62.0-kb self-transmissible, chromosomally integrating genetic element which carries resistances to the antibiotics sulfamethoxazole, trimethoprim, streptomycin, and furazolidone (26). The SXT element integrates into the chromosome by a site-specific mechanism independent of recA. The properties of the SXT element are very similar to those of the conjugative transposons. The widespread use of cotrimoxazole, a very popular and useful antimicrobial drug combination, may have provided an additional selective pressure for the sporadic emergence of the V. cholerae O1 Inaba strains in Delhi and Sewagram.
TABLE 1.
Antibiograms, ribotype patterns, and sizes of restriction enzyme-digested DNAs of V. cholera O1 strains isolated in India
| Strain no. (serotype) | Isolation yr, place | Antibiograma | RPb | Fragment size(s) (kb) with the following probe and restriction enzyme:
|
||||
|---|---|---|---|---|---|---|---|---|
|
ctxA
|
RS1
|
|||||||
| BglI | HindIII | PstI | BglII | PstI | ||||
| V2, (Inaba) | 1989, Calcutta | A Co Fz N S | PD | 18.5 | 20.0 | 10.0 | 20.0, 7.0, 3.6 | 30.0, 10.0 |
| V13 (Inaba) | 1989, Calcutta | A Co Fz N | PD | 19.0 | 23.0 | 10.0 | 16.0, 7.0, 3.6, 2.7 | 30.0, 10.0 |
| DO182 (Inaba) | 1998, Delhi | A Co Fz Na S | RIII | 14.0 | 21.0 | 6.2 | 16.0, 8.4, 2.7 | 30.0 |
| DO183 (Inaba) | 1998, Delhi | A Co Fz N Na S | RIII | 23.0 | 23.0 | 16.0 | 16.0, 7.0, 3.6, 2.7 | 30.0, 16.0 |
| SO85 (Inaba) | 1999, Sewagram | A Co Fz N Na S | RIII | 12.0 | 19.5 | 6.2 | 16.0, 8.4, 2.7 | 30.0 |
| SO86 (Inaba) | 1999, Sewagram | A Co Fz N Na S T | RIII | 13.0 | 19.5 | 6.3 | 16.0, 8.4, 2.7 | 30.0 |
| SO87 (Inaba) | 1999, Sewagram | A Cf Co Fz N Na S | RIII | 13.0 | 20.0 | 6.3 | 16.0, 8.4, 2.7 | 30.0 |
| SO88 (Ogawa) | 1999, Sewagram | A Co Fz N Na S | RIII | 14.0 | 20.5 | 6.2 | 16.0, 8.4, 2.7 | 30.0 |
| SO89 (Inaba) | 1999, Sewagram | A Co Fz N S T | RIII | 13.0 | 13.0 | 6.2 | 16.0, 8.4, 2.7 | 30.0 |
| SO90 (Inaba) | 1999, Sewagram | A Cf Co Fz Na S | RIII | 14.0 | 21.0 | 6.3 | 16.0, 8.4, 2.7 | 30.0 |
| SO84 (Inaba) | 1999, Sewagram | A Co Fz N Na S T | ND | ND | ND | ND | ND | ND |
| PG81 (Ogawa) | 1998, Calcutta | A Co Fz N Na | RIII | ND | 26.0 | 5.6 | 18.5, 8.6, 3.1 | 31.0 |
| PG117 (Ogawa) | 1998, Calcutta | A C Co Fz Na S | RIII | ND | 26.0 | 5.6 | 17.0, 8.6, 3.1 | 22.0 |
| CO840 (Ogawa) | 1995, Calcutta | A Co Fz N Na S | RIII | ND | 23.0 | 6.0 | ND | 23.0 |
A, ampicillin; Cf, ciprofloxacin; Co, cotrimoxazole; Fz, furazolidone; N, neomycin; Na, nalidixic acid; S, streptomycin; T, tetracycline.
RP, ribotype pattern; PD, previously described by Dalsgaard et al. (6); ND, not done.
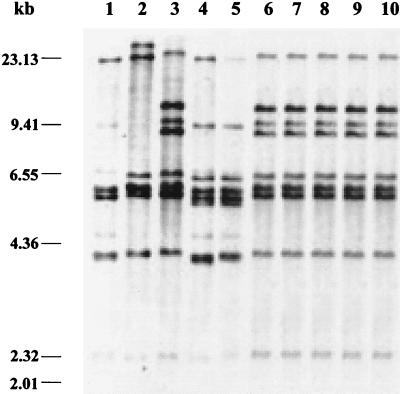
BglI ribotyping showed that the Inaba strains recently isolated in Delhi and Sewagram had a ribotype different from those of the Inaba strains isolated in 1989. The ribotype patterns of the Inaba strains isolated in 1989, V2 and V13 (Fig. 1, lanes 4 and 5), were different from the ribotype patterns shown by the V. cholerae reference strains SG24 (O139) (Fig. 1, lane 1), 569B (O1 classical Inaba) (Fig. 1, lane 2), and CO840 (O1 Ogawa El Tor) (Fig. 1, lane 3). On the other hand, the recent Inaba strains DO182 and DO183 isolated in Delhi in 1998 (Fig. 1, lanes 6 and 7) and the Inaba strains isolated in Sewagram in 1999, SO86 and SO90 (Fig. 1, lanes 8 and 9), showed ribotypes similar to that of the prevailing V. cholerae O1 Ogawa strain CO840 (the slight difference in the mobility of the bands is due to a gel anomaly), which is the RIII type, first described by Sharma et al. (22). The ribotypes of the recent V. cholerae Inaba strains indicate that these strains have molecular traits identical to those of the prevailing V. cholerae O1 Ogawa strains. This result also points out that the recent Inaba strains are quite different from the Inaba strains isolated in 1989, when Inaba was the dominant serotype (16, 17).
FIG. 1.
Ribotypes of representative V. cholerae strains. Lanes: 1, SG24 (O139); 2, 569B (O1, classical Inaba); 3, CO840 (O1, El Tor Ogawa); 4, V2 (O1, Inaba); 5, V13 (O1, Inaba); 6, DO182 (O1, Inaba); 7, DO183 (O1, Inaba); 8, SO86 (O1, Inaba); 9, SO90 (O1, Inaba); 10, SO88 (O1, Ogawa). The positions of λ HindIII molecular size markers are indicated on the left.
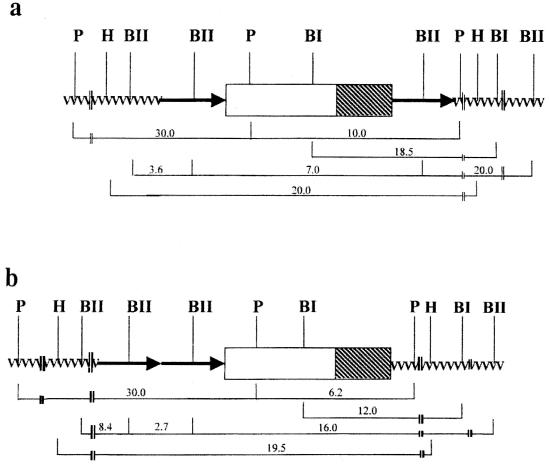
Southern hybridization with the ctxA probe using HindIII-digested genomic DNA detected only one band, but of varying size, in the Inaba strains. This result indicates that the CTX prophage is located at a single site in the chromosome, as HindIII does not have any recognition site within the CTX prophage (14). The arrangement and number of copies of the CTX prophage were determined by analysis of the Southern hybridization pattern generated separately with ctxA and RS1 probes using other restriction enzymes which do cut within the CTX prophage but not in ctxA. ctxA RFLP patterns generated with BglI- and PstI-digested genomic DNA showed a single band in all the strains (Table 1), suggesting the presence of a single copy of the CTX prophage. The CTX prophage genome has two regions: a 4.6-kb “core region” that includes ctxAB and a 2.4-kb region termed RS2. The integrated CTX prophage is frequently flanked by an element known as RS1, which is related to RS2 (25). These related elements contain three nearly identical open reading frames (ORFs), while RS1 contains an additional ORF (25). Southern hybridization with the RS1 probe was carried out to determine the organization of the RS sequences upstream and downstream of the core region. The restriction endonuclease PstI, which cuts within the core region (14), was used to digest genomic DNA, which was then hybridized with the RS1 probe. RS1 RFLP patterns generated with PstI-digested genomic DNA exhibited the presence of two bands of 30.0 and 10.0 kb in strains V2 and V13 and of 30.0 and 16.0 kb in strain DO183, while a single band of 30.0 kb appeared in the remaining strains (Table 1). Therefore, strains V2, V13, and DO183 each contain at least one copy of RS1 at both sides of the core region. When the results of hybridization of PstI-digested genomic DNA with the ctxA and RS1 probes were compared, a band common to both was observed for strains V2, V13, and DO183. The size of the common band was 10.0 kb in strains V2 and V13 and 16.0 kb in strain DO183 (Table 1). This result confirms the presence of an RS element downstream of the CTX prophage. The BglII restriction enzyme has a site in the RS region (25). As expected, when BglII-digested genomic DNA was hybridized with the RS1 probe, three bands were observed for V2 and four bands were observed for V13 and DO183; one of these, a 7.0-kb band, is actually the size of the CTX prophage (25). The presence of tandemly arranged RS sequences is not uncommon in toxigenic V. cholerae, and the size of RS1 is reported to be 2.7 kb (25). Since hybridization with the RS1 probe never showed the presence of a 2.7-kb band in V2, the possibility of tandemly arranged RS1 on either side of the core region can be excluded. The presence of a 2.7-kb band in V13 and DO183 indicates the possibility of tandemly arranged RS1 on either side of the core region. When the Inaba strains isolated in Sewagram and strain DO182 were digested with PstI, the RFLP data showed a single band upon hybridization with both the ctxA and RS1 probes (Table 1), indicating the presence of a single copy of the CTX prophage, while BglII digestion and hybridization with the RS1 probe showed a band of 2.7 kb, indicating the presence of two tandemly arranged copies of the RS1 element in these strains. Thus, strain DO183 and the Sewagram strains have a single copy of CTX prophage with one RS1 element upstream of the prophage. This organization is very similar to that of the prevailing Ogawa strains (22). Figure 2 shows schematic representations, based on RFLP analysis, of the CTX prophage of the Inaba strain V2, isolated in 1989 (Fig. 2a), and of strain SO85, which is typical of the Inaba strains recently isolated in Delhi and Sewagram (Fig. 2b).
FIG. 2.
Schematic representations of the organization of the CTX prophage (not to scale) as deduced by Southern hybridization data for V. cholerae O1 Inaba strains represented by V2 (a) and V. cholerae O1 Inaba strains represented by SO85 (b). Arrows and boxes represent the RS elements and the core region of the CTX prophage, respectively, with the hatched portion of the box representing the ctxAB gene. Restriction site abbreviations: BI, BglI; BII, BglII; P, PstI; H, HindIII.
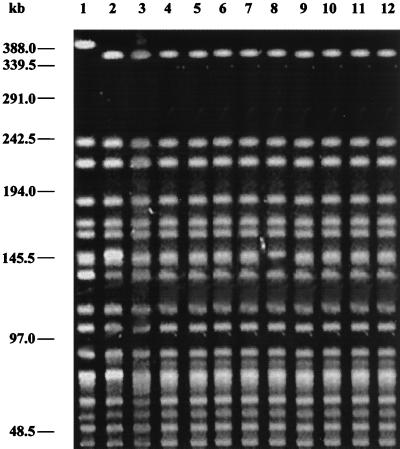
Analysis of PFGE patterns showed that the Inaba strains isolated recently (Fig. 3, lanes 3 to 7) had a profile different from those of strains V2 (Fig. 3, lane 1) and V13 (Fig. 3, lane 2), representing the Inaba strains isolated in 1989. The recent Inaba strains (Fig. 3, lanes 3 to 7) differed from strain CO840 (Fig. 3, lane 8) by the presence of more than one band in the 145.5-kb region. Interestingly, O1 Ogawa strains isolated during 1998 (Fig. 3, lanes 9 to 12) had a PFGE profile identical to that of the recently isolated Inaba strains, indicating that the Inaba strains that have emerged recently are similar to the prevailing O1 Ogawa strains.
FIG. 3.
PFGE profiles of V. cholerae O1 El Tor biotype strains. Lanes: 1, V2 (Inaba); 2, V13 (Inaba); 3, DO182 (Inaba); 4, DO183 (Inaba); 5, SO85 (Inaba); 6, SO88 (Ogawa); 7, SO90 (Inaba); 8, CO840 (Ogawa); 9, PG11 (Ogawa); 10, PG81 (Ogawa); 11, PG117 (Ogawa); 12, PG299 (Ogawa). The positions of bacteriophage λ ladder molecular size markers are indicated on the left.
V. cholerae O1 strains are known to interconvert between the Ogawa and Inaba forms (5, 8, 24). The frequency of conversion of Ogawa to Inaba is approximately 10−5 (3), whereas conversion from Inaba to Ogawa is rare and may be strain dependent (13). In vivo seroconversion correlates well with the host immune response, and this is supported by observations with germ-free mice (19) and a clinical study carried out by Sheehy et al. (23). The wbeT gene of the highly conserved wb region is responsible for the serotype conversion, as there is a single-nucleotide change within the gene, resulting in a TGA stop codon and a truncated 32-kDa wbeT protein (24). The product of the wbeT gene of V. cholerae O1 is not essential for O antigen biosynthesis but is required for determining the Ogawa serotype specificity (24). A variety of changes in wbeT could produce an Inaba strain by leading to truncated wbeT proteins of various sizes due to reading frameshifts. Thus, Inaba strains are effectively wbeT mutants and presumably have arisen as a result of selection due to the immune response during cholera infection (24). This study demonstrates that V. cholerae O1 Inaba, after predominating until 1989 (17), suddenly disappeared and that the Inaba strains that have emerged in different parts of the country in 1998 to 1999 could have evolved from the prevailing V. cholerae O1 Ogawa El Tor biotype.
Acknowledgments
This work was supported, in part, by the Japan International Cooperation Agency (JICA/NICED project O54-1061-E-O).
REFERENCES
- 1.Bag P K, Maiti S, Sharma C, Ghosh A, Basu A, Mitra R, Bhattacharya S K, Nakamura S, Yamasaki S, Takeda Y, Nair G B. Rapid spread of the new clone of Vibrio cholerae O1 biotype El Tor in cholera endemic areas in India. Epidemiol Infect. 1998;121:245–251. doi: 10.1017/s0950268898001423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhaskaran K. Recombination of characters between mutant stock of Vibrio cholerae strain 162. J Gen Microbiol. 1960;23:47–54. doi: 10.1099/00221287-23-1-47. [DOI] [PubMed] [Google Scholar]
- 3.Bhaskaran K, Gorrill R H. A study of antigenic variation in Vibrio cholerae. J Gen Microbiol. 1957;16:721–729. doi: 10.1099/00221287-16-3-721. [DOI] [PubMed] [Google Scholar]
- 4.Brosius J, Ullrich A, Raber M A, Garg A, Dull J J, Gutell R R, Noler H F. Construction and fine mapping of recombinant plasmids containing the rrnB ribosomal RNA operon of Escherichia coli. Plasmid. 1981;6:112–118. doi: 10.1016/0147-619x(81)90058-5. [DOI] [PubMed] [Google Scholar]
- 5.Colwell R R, Huq A, Chowdhury M A R, Brayton P R, Xu B. Serogroup conversion of Vibrio cholerae. Can J Microbiol. 1995;41:946–950. doi: 10.1139/m95-131. [DOI] [PubMed] [Google Scholar]
- 6.Dalsgaard A, Skov M N, Serichantalergs O, Echeverria P, Meza R, Taylor D N. Molecular evolution of Vibrio cholerae O1 strains isolated in Lima, Peru, from 1991 to 1995. J Clin Microbiol. 1997;35:1151–1156. doi: 10.1128/jcm.35.5.1151-1156.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fallarino A, Mavrangelos C, Stroeher U H, Manning P A. Identification of additional genes required for O-antigen biosynthesis in Vibrio cholerae O1. J Bacteriol. 1997;179:2147–2153. doi: 10.1128/jb.179.7.2147-2153.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gangarosa E J, Sonati A, Saghari H, Feeley J C. Multiple serotypes of Vibrio cholerae from a case of cholera. Lancet. 1967;i:646–648. doi: 10.1016/s0140-6736(67)92542-1. [DOI] [PubMed] [Google Scholar]
- 9.Garg P, Chakraborty S, Basu I, Datta S, Rajendran K, Bhattacharya T, Yamasaki S, Bhattacharya S K, Takeda Y, Nair G B, Ramamurthy T. Expanding multiple antibiotic resistance among clinical strains of Vibrio cholerae isolated from 1992 to 1997 in Calcutta, India. Epidemiol Infect. 2000;124:393–399. doi: 10.1017/s0950268899003957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gennaro M L, Greenaway P J, Broadbent D A. The expression of biologically active cholera toxin in Escherichia coli. Nucleic Acids Res. 1982;10:4883–4890. doi: 10.1093/nar/10.16.4883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaper J B, Jr, Morris J G, Nishibuchi M. DNA probes for pathogenic Vibrio species. In: Tenover F C, editor. DNA probes for infectious disease. Boca Raton, Fla: CRC Press, Inc.; 1992. pp. 65–77. [Google Scholar]
- 12.Kurazono H, Okuda J, Takeda Y, Nair G B, Albert M J, Sack R B, Chongsanguan M, Chaichumpa W. Vibrio cholerae O139 Bengal isolated from India, Bangladesh and Thailand are clonal as determined by pulsed-field gel electrophoresis. J Infect. 1994;29:109–110. doi: 10.1016/s0163-4453(94)95357-0. [DOI] [PubMed] [Google Scholar]
- 13.Manning P A, Stroeher U H, Morona R. Molecular basis for O-antigen biosynthesis in Vibrio cholerae O1: Ogawa-Inaba switching. In: Wachsmuth K, Blake P A, Olsvik O, editors. Vibrio cholerae and cholera: molecular to global perspectives. Washington, D.C.: ASM Press; 1994. pp. 77–94. [Google Scholar]
- 14.Mekalanos J J. Duplication and amplification of toxin genes in Vibrio cholerae. Cell. 1983;35:253–263. doi: 10.1016/0092-8674(83)90228-3. [DOI] [PubMed] [Google Scholar]
- 15.Mukhopadhyay A K, Garg S, Mitra R, Basu A, Dutta D, Bhattacharya S K, Shimada T, Takeda T, Takeda Y, Nair G B. Temporal shifts in traits of Vibrio cholerae strains isolated from hospitalized patients in Calcutta: a 3-year (1993–1995) analysis. J Clin Microbiol. 1996;34:2537–2543. doi: 10.1128/jcm.34.10.2537-2543.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nair G B, Ramamurthy T, Garg S, Takeda T, Takeda Y. Characteristics of Vibrio cholerae isolated from hospitalized patients with acute diarrhoea in Calcutta, India: a four year analysis. LabMedica Int. 1993;X:29–33. [Google Scholar]
- 17.Ramamurthy T, Pal A, Bhattacharya M K, Bhattacharya S K, Chowdhury A S, Takeda Y, Takeda T, Pal S C, Nair G B. Serovar, biotype, phage type, toxigenicity and antibiotic susceptibility patterns of Vibrio cholerae isolated during two consecutive cholera seasons (1989–1990) in Calcutta. Indian J Med Res. 1992;95:125–129. [PubMed] [Google Scholar]
- 18.Reeves P R, Hobbs M, Valvano M A, Skurnik M, Whitfield C, Coplin D, Kido N, Klena J, Maskell D, Raetz C R H, Rick P D. Bacterial polysaccharide synthesis and gene nomenclature. Trends Microbiol. 1996;4:495–503. doi: 10.1016/s0966-842x(97)82912-5. [DOI] [PubMed] [Google Scholar]
- 19.Sack R B, Miller C E. Progressive changes of Vibrio serotypes in germ-free mice inoculated with Vibrio cholerae. J Bacteriol. 1969;99:688–695. doi: 10.1128/jb.99.3.688-695.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saha P K, Koley H, Mukhopadhyay A K, Bhattacharya S K, Nair G B, Ramakrishna B S, Krishnan S, Takeda T, Takeda Y. Nontoxigenic Vibrio cholerae O1 serotype Inaba biotype El Tor associated with a cluster of cases of cholera in Southern India. J Clin Microbiol. 1996;34:1114–1117. doi: 10.1128/jcm.34.5.1114-1117.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Salazar-Lindo E, Seminario-Carrasco L, Carrillo-Paradi C, Gayoso-Villaflor A. The United States-Japan Cooperative Medical Science Program. Twenty-Seventh Joint Conference on Cholera and Related Diarrhoeal Diseases. Bethesda, Md: National Institutes of Health; 1991. The cholera epidemic in Peru; pp. 9–13. [Google Scholar]
- 22.Sharma C, Nair G B, Mukhopadhyay A K, Bhattacharya S K, Ghosh R K, Ghosh A. Molecular characterization of Vibrio cholerae O1 biotype El Tor strains isolated between 1992 and 1995 in Calcutta, India: evidence for the emergence of a new clone of the El Tor biotype. J Infect Dis. 1997;175:1134–1141. doi: 10.1086/516453. [DOI] [PubMed] [Google Scholar]
- 23.Sheehy T W, Sprinz H, Angerson W S, Formal S B. Laboratory Vibrio cholerae infection in the United States. JAMA. 1966;197:321–326. [PubMed] [Google Scholar]
- 24.Stroeher U H, Kanageorgos L E, Morona R, Manning P A. Serotype conversion in Vibrio cholerae O1. Proc Natl Acad Sci USA. 1992;89:2566–2570. doi: 10.1073/pnas.89.7.2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Waldor M K, Rubin E J, Pearson G D N, Kimsey H, Mekalanos J J. Regulation, replication and integration functions of the V. cholerae CTXφ are encoded by region RS2. Mol Microbiol. 1997;24:917–926. doi: 10.1046/j.1365-2958.1997.3911758.x. [DOI] [PubMed] [Google Scholar]
- 26.Waldor M K, Tschape H, Mekalanos J J. A new type of conjugative transposon encodes resistance to sulfamethoxazole, trimethoprim, and streptomycin in Vibrio cholerae O139. J Bacteriol. 1996;178:4157–4165. doi: 10.1128/jb.178.14.4157-4165.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yamasaki S, Garg S, Nair G B, Takeda Y. Distribution of Vibrio cholerae O1 antigen biosynthesis genes among O139 and other non-O1 serogroups of Vibrio cholerae. FEMS Microbiol Lett. 1999;179:115–121. doi: 10.1111/j.1574-6968.1999.tb08716.x. [DOI] [PubMed] [Google Scholar]
- 28.Yamasaki S, Nair G B, Bhattacharya S K, Yamamoto S, Kurazono H, Takeda Y. Cryptic appearance of a new clone of Vibrio cholerae O1 biotype El Tor in Calcutta, India. Microbiol Immunol. 1997;41:1–6. doi: 10.1111/j.1348-0421.1997.tb01165.x. [DOI] [PubMed] [Google Scholar]